ABSTRACT
The question of how many species of Fungi there are has occasioned much speculation, with figures mostly posited from around half a million to 10 million, and in one extreme case even a sizable portion of the spectacular number of 1 trillion. Here we examine new evidence from various sources to derive an updated estimate of global fungal diversity. The rates and patterns in the description of new species from the 1750s show no sign of approaching an asymptote and even accelerated in the 2010s after the advent of molecular approaches to species delimitation. Species recognition studies of (semi-)cryptic species hidden in morpho-species complexes suggest a weighted average ratio of about an order of magnitude for the number of species recognized after and before such studies. New evidence also comes from extrapolations of plant:fungus ratios, with information now being generated from environmental sequence studies, including comparisons of molecular and fieldwork data from the same sites. We further draw attention to undescribed species awaiting discovery in biodiversity hot spots in the tropics, little-explored habitats (such as lichen-inhabiting fungi), and material in collections awaiting study. We conclude that the commonly cited estimate of 1.5 million species is conservative and that the actual range is properly estimated at 2.2 to 3.8 million. With 120,000 currently accepted species, it appears that at best just 8%, and in the worst case scenario just 3%, are named so far. Improved estimates hinge particularly on reliable statistical and phylogenetic approaches to analyze the rapidly increasing amount of environmental sequence data.
BACKGROUND
In 1825, Elias Magnus Fries (1794–1878) predicted that the fungi would prove to be the largest group in the vegetable kingdom, analogous to the insects in the animal kingdom. Notwithstanding that fungi are not actually part of the plant kingdom, how right he has proved to be as the bicentenary of his prediction approaches. By the 1960s a few mycologists were speculating that there might be as many fungal as plant species, but almost no attempts to calculate estimates from the available data were made. As concern over the conservation of biodiversity in general grew in the subsequent decades, culminating in the signing of the Convention on Biological Diversity in 1992, more precise figures on species numbers of all kinds of organisms were required. A series of estimates of the number of fungi settled on figures ranging from 500,000 to almost 10 million species, with 1.5 to perhaps 5 million receiving most support among mycologists. A recent study even predicts up to a trillion species of microorganisms globally (1); how many of these are supposed to be fungi is not specified, but if this estimate holds true and only 1% of these were fungi, the global estimate of fungal diversity would be a thousand times higher than the current highest estimate of 10 million species.
Different extrapolation techniques have been used to arrive at global fungal species richness estimates, including publication rates of new taxa (2), plant:fungus ratios (3, 4) similar to plant:insect ratios first used in Erwin’s famous study (5), quantitative macroecological grid-based approaches (6–8), methods based on environmental sequence data including plant:fungus ratios (9, 10), and ecological scaling laws (1).
This article provides updated background information on the number of described species and explores new evidence obtained from rates of species discovery, extrapolations from fungus:plant ratios, and molecular sequence data from environmental samples. We conclude by indicating where undescribed species are likely to be found and recommend a working number suitable for general use in estimates of global and regional species richness.
We focus here on the numbers of species in the kingdom Fungi, that is, inclusive of the Cryptomycota and Microsporidia, and also including lichen-forming fungi. We do not, however, consider other organisms that mycologists study as fungi, such as the fungal analogues Mycetozoa (as an unranked supergroup or in the kingdom Protozoa, phylum Amoebozoa) and Oomycota and Hyphochytriomycota (in the kingdom Heterokonta = Straminipila). However, we caution that some data sets and papers discussed in the following sections are based on fungi in the broad sense and thus include information from the analogue phyla. While readers should bear this in mind, the numbers of known taxa in these groups are relatively modest, so their occasional unavoidable inclusion will not materially affect the interpretations presented here.
EVIDENCE FROM PUBLICATION RATES OF NEW TAXA
Numbers of Described Species
The number of new species described in each decade, as recorded in the Index Fungorum database (http://www.indexfungorum.org), shows three distinct phases (Fig. 1): an ascending phase in which progress was fast in the 1750s to 1860s, a steep phase as microscopy came into general use and there was intensive collecting in hitherto barely explored parts of the world in the 1870s to 1880s, and then a relatively constant phase from the 1890s to the present day. The partially higher figures in the first decades of the “constant phase” reflect continuing exploration and a number of prolific individual mycologists. The rate of description over the past 40 years has, however, remained fairly constant at around 1,300 per year (Fig. 2), with peaks generally relating to particular major monographic works.
FIGURE 1.

Numbers of newly introduced species names of fungi for each decade from 1750 to 2010. Based on data from the Index Fungorum database provided by P. M. Kirk.
FIGURE 2.

Numbers of newly introduced species names of fungi for each year from 1975 to 2015. Note that the data for 2015 were incomplete when this work went to press. Based on data from the Index Fungorum database provided by P. M. Kirk.
Of special note is the somewhat steep increase in the rate since 2010 to around 1,800 per year. This resurgence in species description is largely attributed to the increasing use of molecular techniques for species delimitation and is of particular note because this increase followed, and even accelerated after, the ending of the separate naming of morphs of the same species in 2011 (when the provision for a Latin diagnosis or description was also eliminated). Prior to that date, the totals for annually described “species” included separate names given to different morphs of the same species.
The number of newly named species does not, however, equal the number that have been truly described for the first time. It is much easier to name a fungus as new to science rather than to ascertain that it has previously been named. This is not simply a matter of comparing material to that of previously recognized species in the same genus but requires meticulous detective work, because not uncommonly, species were described in genera they do not belong to. Further, species concepts change, for example, in genera where morphologically identical fungi were regarded as different species if they occurred on different plant species. The number of accepted fungi can only be obtained by counting those recognized as “good” in each genus. This has been done since 1943 through the 10 editions of Ainsworth & Bisby’s Dictionary of the Fungi and now annually through Species Fungorum inputs to the Catalogue of Life (http://www.catalogueoflife.org/annual-checklist/). An analysis of these data shows that the total number of existing species names runs at around 2 to 3 times that of currently accepted species (Fig. 3), a figure consistent with the ratio of 2.6 derived from an analysis of names in monographic works (11). The number of described good fungal species currently stands at around 120,000.
FIGURE 3.
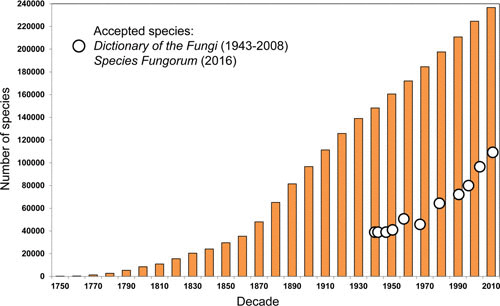
Growth in the total catalogued number of species names of fungi by decade from 1750 compared with the global number of accepted species. Based on figures adopted in the 10 editions of Ainsworth & Bisby’s Dictionary of the Fungi for 1943–2008 and data in the Index Fungorum and Species Fungorum (Catalogue of Life) databases provided by P. M. Kirk.
Unlike the situation in many other groups of organisms (12), the number of good fungal species continues to rise steeply decade by decade, with no indication of leveling off. The steepness of the curve would be even greater if there were more mycologists actively involved in species discovery, using either morphological or molecular approaches.
The number of species being recognized is necessarily impacted by the species concepts used. While in the premolecular era, species circumscriptions were based almost entirely on morphological and sometimes biochemical and cultural features, the incorporation of molecular information has led to a refining of species concepts. There is, however, a plethora of species concepts in biology (13, 14) and no single objective criterion that can be applied universally. Because the purpose of names is to provide a means of communication, a pragmatic species concept is essential, which may be defined as the following: “a species is what it is useful to give a species name to.” (15). There have been numerous attempts to apply mechanistic systems for species recognition in phylogenetic trees, but the consolidated species concept has much to commend it and ultimately provides a broad consensus on how to recognize fungal species. The consolidated species concept adopts a polyphasic approach combining morphological, ecological, and phylogenetic species concepts (16) and is being increasingly taken up by mycologists. While in some situations this may lead to more species being recognized than otherwise would have been the case, in others it results in formerly separately distinguished species being united. Therefore, over- and underestimations in particular cases are usually balanced out. A rarely considered notion is the time scale: a proportion of “species” is in active speciation, with incomplete lineage sorting, yielding it difficult to resolve good species (17); in such cases, determining the number of species is philosophical, since that number is in active flux. A large number of new fungal species continue to be described with no molecular data, either due to the age of the underlying material (e.g., unveiled in older collections) or by taxonomists who do not have access to molecular techniques; assessing the proportion of good species in such taxa is therefore difficult.
Patterns in Taxon Discovery
Some predictable patterns in the recognition of published taxa in the rank of genus and above have been recognized and applied to a broad set of organisms, including fungi (2). By combining the full classifications of around 1.2 million species of all groups of organisms, these authors found, as might have been expected, that accumulation curves of higher taxa over time showed that these were more completely described and in many cases approached an asymptote. The use of the higher taxon approach was then validated by comparing the actual number of known species with that predicted from the accumulation curves. The authors then fitted asymptotic regression models to the actual curves for different groups to provide estimates of undiscovered species numbers. In the case of fungi, this led to a total number of expected fungal species of 611,000 (standard error ± 297,000). This conclusion was, however, misleading, because the analysis was based on the assumption that there were only 43,271 catalogued fungal species, rather than the nearly 100,000 accepted at that time (18). Intriguingly, a recalculation using their method with the latter species number generates a figure of around 1,400,000—not far out of line with estimates derived from extrapolations based on plant:fungus ratios (see below).
EVIDENCE FROM SPECIES RECOGNITION STUDIES
Unknown fungi are now considered to come mostly from two sources: (i) newly discovered species by means of traditional inventory methods in little-studied areas and habitats and (ii) newly discovered lineages through environmental sequencing. A further, neglected but significant source of unrecognized fungal diversity is the restudy of known taxa, applying species recognition techniques to molecular data, using either the fungal barcoding locus ITS (19) or multilocus approaches. Exemplar taxa are the fly agaric, Amanita muscaria, and the golden chanterelle, Cantharellus cibarius, which contain numerous, often regionally distributed species (20, 21). The record is currently set by the basidiolichen fungus Cora glabrata, which has been shown to contain at least 189 species (6, 22). We surveyed 45 such studies published between 1998 and 2016, including studies of non-lichenized and lichenized forms, and computed the ratio of species recognized in the target group before and after the study (Table 1). Most studies started out with a single or few species, whereas two looked at complexes of up to 18 species—Protoparmelia (23) and Usnea (24)—and one revised a complex of 71 species accepted prior to the study, in the genus Coprinellus (25). The posterior:prior ratios in the number of species recognized ranged from 0.67 (from 18 to 12 species in a group of Usnea [24]) to 189 (from 1 to 189 species in the basidiolichen Cora glabrata) (6, 22). Other high ratios were found for Dictyonema with 59 (26), Cladosporium cladosporioides with 22 (27), Sticta weigelii with 15 (28), Microbotryum violaceum with 11 (29), and Fusarium graminearum with 7 (30). Environmental sequence and genomic data revealed high ratios for the genera Archaeorhizomyces, with 145 (31), and Cyphobasidium with 26 (32), as well as for the still barely explored Cryptomycota, with 138 (33, 34), in which the true figure could be much higher. The median ratio over all evaluated studies was 3.0, and the arithmetic mean was 14.5 (Table 1).
TABLE 1.
Selected species recognition studies in different groups of fungi
| Taxon | Genusa | Lifestyle | Coverageb | Rangec | Ratiod | Factore | Adjustedf | Weightg | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Acantholichen pannarioides | 1 | Lichenized | Neotropical | Neotropical | 6 | 1 | 6 | 8 | 100 |
| Amanita muscaria | 500 | Ectomycorrhizal | N. Hemisphere | Global | 6 | 2 | 12 | 6,000 | 20 |
| Archaeorhizomycetes | 0 | Soil fungus | N. Hemisphere | Global | 145 | 3 | 500 | 500 | 31 |
| Aspergillus flavus | 266 | Saprotrophic | Australia | Global | 2 | 6 | 12 | 3,192 | 101 |
| Aspergillus fumigatus | 266 | Human pathogenic | Global | Global | 2 | 1 | 2 | 532 | 102 |
| Auxarthron zuffianum | 15 | Saprotrophic | Global | Global | 2 | 1 | 2 | 30 | 103 |
| Beauveria | 9 | Entomopathogenic | Global | Global | 4 | 1 | 4 | 36 | 104 |
| Blastomyces dermatitidis | 1 | Human pathogenic | N. America | Global | 2 | 1 | 2 | 2 | 105 |
| Botrytis cinerea | 54 | Phytopathogenic | Europe | Global | 2 | 6 | 12 | 648 | 106 |
| Calonectria pauciramosa | 34 | Phytopathogenic | Global | Global | 3 | 2 | 6 | 204 | 107 |
| Cenococcum geophilum | 1 | Ectomycorrhizal | N. America | Global | 5 | 6 | 30 | 30 | 108 |
| Cladosporium cladosporioides | 150 | Saprotrophic | Global | Global | 22 | 1 | 22 | 3,300 | 27 |
| Coccidioides immitis | 3 | Human pathogenic | N. America | N. America | 2 | 1 | 2 | 6 | 103 |
| Coniophora puteana | 20 | Saprotrophic | Global | Global | 3 | 1 | 3 | 60 | 109 |
| Coprinellus | 100 | Saprotrophic | Global | Global | 1,2 | 1 | 1.2 | 120 | 25 |
| Cora | 1 | Lichenized | Subglobal | Global | 189 | 1 | 189 | 189 | 6, 22 |
| Corella | 0 | Lichenized | Central/S. America | Central/S. America | 11 | 1 | 11 | 11 | 26 |
| Cryptomycota | 0 | Aquatic | N. America | Global | 39 | 6 | 234 | 234 | 33 |
| Cyphellostereum | 1 | Lichenized | Central/S. America | Global | 15 | 3 | 45 | 45 | 26 |
| Cyphobasidium | 0 | Lichenicolous | N. America | Global | 26 | 4 | 100 | 100 | 32 |
| Dictyonema | 6 | Lichenized | Subglobal | Global | 59 | 3 | 177 | 1062 | 26 |
| Fusarium graminearum | 111 | Phytopathogenic | Global | Global | 7 | 1 | 7 | 777 | 30 |
| Grosmannia clavigera | 1 | Phytopathogenic | N. America | N. America | 2 | 1 | 2 | 2 | 110 |
| Hymenoscyphus albidus | 155 | Phytopathogenic | Europe | Europe | 2 | 1 | 2 | 310 | 111 |
| Lasiodiplodia theobromae | 9 | Phytopathogenic | Pantropical | Pantropical | 3 | 1 | 3 | 27 | 112 |
| Letharia | 2 | Lichenized | N. Hemisphere | N. Hemisphere | 3 | 1 | 3 | 6 | 113 |
| Lobariella | 5 | Lichenized | Neotropical | Neotropical | 3.8 | 1 | 3.8 | 19 | 114 |
| Metarhizium anisopliae | 9 | Entomopathogenic | N. America | Global | 2 | 6 | 12 | 108 | 115 |
| Microbotryum violaceum | 87 | Phytopathogenic | Europe/N. America | Global | 11 | 4 | 44 | 3,828 | 29 |
| Neofusicoccum parvum/ribis | 13 | Phytopathogenic | Regional/host | Global | 2 | 60 | 150 | 1,950 | 116 |
| Paracoccidioides brasiliensis | 1 | Human pathogenic | S. America | S. America | 4.0 | 1 | 4 | 4 | 117 |
| Parmelia | 95 | Lichenized | Europe/N. America | N. Hemisphere | 3 | 1.5 | 4.5 | 428 | 118 |
| Parmelia saxatilis | 1 | Lichenized | N. America | Global | 4 | 3 | 12 | 12 | 119 |
| Parmotrema reticulatum | 348 | Lichenized | Subglobal | Subglobal | 4 | 1 | 4 | 1,392 | 120 |
| Phialocephala fortinii | 1 | Endophytic | Europe | Global | 7 | 6 | 42 | 42 | 121 |
| Protoparmelia | 20 | Lichenized | Global | Global | 1.6 | 1 | 1.6 | 32 | 23 |
| Puccinia monoica | 4,000 | Phytopathogenic | N. America | Global | 1.8 | 6 | 10.5 | 42,000 | 35 |
| Rhizoplaca melanophthalma | 11 | Lichenized | N. America | Global | 2.5 | 6 | 15 | 165 | 122 |
| Stachybotrys chartarum | 44 | Saprotrophic | N. America | Global | 2 | 1 | 2 | 88 | 123 |
| Sticta fuliginosa | 114 | Lichenized | Global | Global | 15 | 1 | 45 | 5,130 | 28 |
| Sticta weigelii | 114 | Lichenized | Global | Global | 23 | 1 | 23 | 2,622 | 28 |
| Strobilomyces | 20 | Ectomycorrhizal | Asia | Global | 3.5 | 6 | 21 | 420 | 124, 125 |
| Tricholoma scalpturatum | 200 | Ectomycorrhizal | Europe | Global | 2 | 6 | 12 | 2,400 | 126 |
| Uncinocarpus reesii | 3 | Saprotrophic | Global | Global | 2 | 1 | 2 | 6 | 103 |
| Usnea | 338 | Lichenized | N. Hemisphere | N. Hemisphere | 0.7 | 1 | 1 | 338 | 24 |
Number of species recognized in the corresponding genus in reference 18.
Origin of material used in the study.
Global distribution of the group.
Ratio of the number of species recognized after versus before the study.
Geographic adjustment.
Adjusted ratio based on raw ratio and geographic adjustment.
Weight based on adjusted ratio and original number of species recognized in the corresponding genus in reference 18 (in some instances [Acantholichen, Sticta, Usnea] further adjusted based on unpublished data).
To obtain a more reliable picture, we adjusted these ratios by geographic area covered. If a species complex had a global distribution prior to the study, and if based on material from a single geographic area, it was found to represent several species with presumably restricted distribution, the detected ratio was adjusted by the factor 2 (expanding to both hemispheres in cases in which one hemisphere was covered), the factor 3 (for tropical fungi for which a single tropical region was covered), and up to the factor 6 (for cosmopolitan fungi for which a single region within a single continent was studied). These factors were derived from selected global studies, for example, in the genera Cladosporium (27), Cora (6, 22), Fusarium (30), and Sticta (28). In some cases, we added unpublished data to the adjusted values; thus, for Archaeorhizomyces we currently recognize at least 500 species (B. E. Smith and R. Lücking, unpublished data), and for Cyphobasidium, we recognize about 100 species from deposited GenBank sequences (32). The thus adjusted ratios ranged from 0.7 (Usnea) to 435 (Archaeorhizomyces), with a median value of 10.5 and an arithmetic mean of 39.3.
To predict the global species richness of Fungi using these raw and adjusted ratios, we employed two further adjustments. First, we weighted the ratios according to the number of species recognized in a genus in the 2008 Dictionary of the Fungi (18). For instance, a genus in which a single species had been recognized received a low weight (1.0), since even a high number of newly recognized species will only contribute minimally to global fungal species richness. This adjustment avoided a high impact of extraordinary ratios such as in the previously monotypic genus Cora. In contrast, for groups in which a large number of species had already been recognized, the ratios based on a study of one or a few species were upweighted, e.g., in the genus Puccinia (35), with 4,000 accepted species. In addition, to account for ecological bias in the evaluated studies, we applied a group weight according to the major lifestyle attributed to each genus; to that end, we divided the approximately 100,000 known fungi in 2008 as follows: 40,000 saprotrophic (weight 4.0), 20,000 phytopathogenic (weight 2.0), 20,000 lichenized (weight 2.0), 8,000 ectomycorrhizal (weight 0.8), 5,000 endophytic (weight 0.5), 3,000 entomopathogenic (weight 0.2), 2,000 aquatic (weight 0.2), 500 endomycorrhizal (weight 0.05), 500 marine (weight 0.05), 400 soil (weight 0.04), and 100 human pathogenic (weight 0.01). The categories aquatic, marine, and soil are used here for cases where the habitat is known but not the biology. These proportions denote estimates based on known fungi in 2008, not estimated proportions of extrapolated global species richness, in which, presumably, categories such as plant pathogens and soil fungi will have much higher values. Using these weights, the overall arithmetic mean of the posterior:prior ratio for all evaluated studies amounted to 11.3. This suggests that even without including important sources of new species discoveries, the already accepted Fungi may contain up to 10 times as many species as currently recognized, resulting in more than 1 million estimated species. Remarkably, this is about 4 times higher than the currently estimated synonymy ratio of 2.6, suggesting that a large proportion of these unrecognized species have no names available.
EVIDENCE FROM EXTRAPOLATIONS BASED ON PLANT:FUNGUS RATIOS
The widely cited and accepted plant:fungus ratio-derived global estimate of 1.5 million species of fungi was based on information from several independent sources (3): (i) the numbers of fungi reported from all habitats in the United Kingdom and sites within the United Kingdom compared with the number of plant species present, (ii) the numbers of host-specific fungi on particular plant species, (iii) percentages of new species being discovered, and (iv) extrapolations to the global level based on conservative estimates for the number of plant species, making allowances to discount separately named morphs of the same species. The 1.5 million figure was considered conservative because, while numbers of fungi from all sources, including organisms other than plants and soil, were included in the species totals for particular places, no allowances were made for undiscovered fungi in and on the millions of insects predicted to exist.
Some figures used in these early extrapolations have subsequently been revised upward. Ten years later, it was pointed out that the number of plant species and the fungus:plant ratio were too conservative and the vast number of insect fungi had not been considered. As a consequence, some revised estimates ranged between 2.7 and 9.9 million fungal species (4).
The total inventory of fungal species found in field surveys in a particular site continues to increase year by year, even with regular visits spanning several decades, while the number of plant species remains more or less unchanged. Esher Common (Surrey, United Kingdom) is by far the most investigated site by field mycologists in the world, and to date about 3,400 species have been found (B. M. Spooner, personal communication). Because the number of vascular plant species remains at 420, this now gives a fungus:plant ratio of 8:1.
Some preliminary results from next-generation sequencing of soil samples from a wood within the second most field-surveyed site in the world, the Slapton Ley National Nature Reserve in southwest England, are illuminating in relation to the fungus:plant ratios (G. W. Griffith, B. S. Dentinger, and D. L. Hawksworth, unpublished). The wood studied has 1,136 species of fungi and 88 vascular plant species recorded during field surveys, a ratio of 12.9:1. However, just 686 sequence-based species (accounting only for taxa represented by five or more sequences) were recovered from soil samples (and a further 153 taxa were represented by one to four sequences). What is intriguing is that of the 164 named genera represented in the next-generation sequencing data set, only 32 were among the 549 genera collected in the fieldwork, which included fungi on plant parts and plant litter, not just sporophores emerging from the soil. The actual ratio in a site may therefore be substantially higher than indicated by traditional inventory techniques.
The interpretation of some data sets needs to be considered in the light of such long-term investigations. Ratios comparing fungal sequences recovered from environmental samples with plants growing on the sample sites have given some surprising ratios. For example, data from a forest in North Carolina gave fungus:plant ratios of 19:1 (491 fungi and 26 plants) and 13:1 (616 fungi and 48 plants), leading to a suggestion that there may be 3.5 to 5.1 million fungal species worldwide (9). A more modest ratio of 7.5:1 (1,500 fungi and <200 plants) was obtained from boreal forest soils in Alaska (36). In contrast, Tedersoo et al.’s (37) impressive study of 365 globally distributed soil samples using metabarcoding molecular methods (see below) led the authors to conclude that fungal species richness had actually been overestimated by 1.5 to 2.5 times from that based on plant:fungus species ratios. Any such extrapolation from soil samples alone to the total fungal biota at a site is, however, unsound as evidenced by the above data from the Slapton reserve. Much of the actual fungal species diversity in a site will inevitably be missed in soil sampling, because most is above ground, on and in plants and animals of all kinds, in water, in lichens, on rocks, etc.
Collections made in brief smash-and-grab collecting trips or even extending over several years will only start to reveal what fungi may actually be present at a site. Studies such as that of Piepenbring and collaborators (38), in which 567 species of fungi and 311 species of plants were found along a 500-m pathway in Panama over 2 years, giving a ratio of just 1.8:1, have to be interpreted in this context. The ratio resulting from that study would be expected to rise significantly if the period over which it was conducted had been 2 decades rather than 2 years, and with increased attention to microscopic fungi.
When considering fungi of all biologies and in all ecological niches in a site, the ratio can be expected to vary depending on the geographical location, as recognized some 25 years ago (3). It is not surprising, therefore, that the studies of fungus:plant ratios in a site, obtained by field survey and molecular approaches, have generated ranges from 1.8 to 19.1:1. The mean of the figures cited in this section yields a ratio of 9.8:1, suggesting that the ratio of around 6:1 arrived at in 1991 may be conservative. With the currently accepted number of approximately 380,000 vascular plant species (39), this adjusted ratio would predict a number of nearly 3.8 million fungal species.
EVIDENCE FROM ENVIRONMENTAL SEQUENCING TECHNIQUES
The advent of obtaining taxon-specific, molecular sequence data from environmental samples, most commonly soils but also water and plant tissues, has generated a new source of data for estimating global species numbers. It is commonly found that numerous sequences thus obtained do not have any matches with those from named fungi in GenBank (see below), and this has led to speculations that previous estimates have been too low.
Generalizing data from environmental samples is problematic for several reasons, including that the broader geographic context of detected operational taxonomic units (OTUs) is usually unknown. For example, using 454-sequencing technology, two 0.25-g soil samples from an Alaskan forest collected just 1 m apart had only 14% of fungal species in common (36). In a pyrosequencing study in the Andes, 1,839 species-like taxa were found, of which 25% were most similar to other unidentified environmental samples, with significant differences between forests at different altitudes (40). The most comprehensive study of soil samples to date is that of Tedersoo et al. (37), who analyzed 2-g soil samples from pooled soil in 5-cm deep cores taken from 365 global sites. They obtained 80,486 fungal OTUs, used a 98% sequence similarity for species recognition, and did not consider almost half in further analyses because they were singletons and potentially artifacts; in that study, plant:fungus richness declined toward the poles. However, in concluding that extrapolations from plant:fungus ratios were unjustified (see above), the authors did not take into account fungi other than those in soil, such as those in and on aerial parts of plants, in decaying vegetation, lichen-fungi, and entomogenous fungi.
Especially instructive is a study of 928 swabs of dust samples across the continental United States by direct PCR and using high-throughput sequencing (HTS) to analyze them (41); not only were 38,473 fungal taxa detected, but there were clear geographic patterns with a predictive value of placing a sample within 230 km. Of the 38,473 taxa, however, sequences of about 40% could not be matched with any named species (R. R. Dunn, personal communication).
Just how many unidentified sequences generated from environmental samples represent undescribed species is uncertain. Compared with the 120,000 formally recognized species (see above), as of 25 November 2016, there were only 34,878 named fungal species in GenBank, but there were a further 94,059 species-level OTUs with no names (C. L. Schoch, personal communication); these figures have increased by 68% and a staggering 117%, respectively, in the past 5 years. However, only a minute fraction of environmental sequences are deposited individually in GenBank; the vast majority is placed in the Sequence Read Archive (SRA) (42). Currently, the SRA, on the query “(fungal OR Fungi) AND (internal transcribed spacer [or, ITS] OR ITS1 OR ITS2),” returns 179 studies, 1,822 biosamples (i.e., environmental samples), and 14,334 experiments or HTS runs, containing 928 million fungal ITS reads, with an average length of 353 bases (SRA: https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=search_obj; accessed 12 March 2017). In contrast, using the same query, GenBank (43) returns 993,987 sequences obtained predominantly through Sanger sequencing (GenBank: https://www.ncbi.nlm.nih.gov/genbank; accessed 12 March 2017). Thus, at present there are almost 1,000 times more HTS reads than Sanger sequences. Only 3 years ago, this ratio was 18:1, which means it has increased by a factor of more than 50 in this short period.
Obviously, a proportion of the unidentified sequences can be expected to represent known but as yet unsequenced species, especially considering that about 85,000 of the currently accepted fungal species have no sequence data available. In the case of Mortierella, a reference set of type and other strains was sequenced and then compared with unnamed sequences in GenBank (44); the authors modeled the effects of increasing type strain sequencing and found a linear relationship with the number of strains that could be identified, and they predicted a number close to that of the already described species.
One of the most significant problems with using environmental sequence data to extrapolate global fungal species richness is the approach to define OTUs. Due to the volume of data generated and the usual broad taxonomic range detected, alignment-based methods that would allow us to critically define species as phylogenetic lineages are impossible to use as routine techniques. Thus, environmental sequence data are analyzed using clustering techniques, and OTUs as equivalents to species are defined on thresholds, ranging between 95 and 99% depending on the study. This approach has several shortcomings. First, species do not exhibit fixed sequence thresholds; in contrast, sequence divergence and hence the barcoding gap depend on the evolutionary age of the species complex. Second, commonly used thresholds of 95 to 97% are unrealistic. Assuming that the ITS barcoding locus has a length of, e.g., 500 to 600 bases, 5% divergence would correspond to 25 to 30 bases. In most studied species, infraspecific ITS variation is much lower, with an average of about 1%. Theoretically, a more realistic threshold of 99% would greatly increase the number of OTUs detected in a data set. However, this effect is more than outweighed by the sensitivity of clustering methods to variation in the data, including the common CAFIE (carry-forward-incomplete-extension) sequencing error common to all base flow technologies (45) (see below).
This error and other problems with clustering techniques can lead to an overestimation of the number of OTUs at several orders of magnitude, which may result in serious bias when using such estimates for global extrapolations. We tested this effect with SRA data of the soil fungus Archaeorhizomyces (Smith and Lücking, unpublished). Through a blast script, we obtained 106,563 ITS reads from the SRA that were reasonably close to the type species, Archaeorhizomyces finlayi. Clustering analysis using USEARCH (46) resulted in the following numbers of OTUs depending on the set threshold: 2,658 at 95%, 5,793 at 97%, 10,630 at 98%, and 28,435 at 99%. Thus, the difference between the 95% and the 99% threshold is an order of magnitude. Alignment-based phylogenetic analysis suggests the presence of approximately 500 species-level lineages in this data set. Thus, depending on the threshold level used for species-level separations, clustering could overestimate the actual species richness in this case by a factor of between 5 and 50.
Lücking and colleagues (45) showed that severe problems with estimating OTUs through clustering emerge from even minor sequencing errors. The CAFIE error is common to all next-generation sequencing technologies, because it represents a statistical error of the distribution of bases among wells on a plate through a given base flow. Through careful adjustment, this error can be held to a minimum of less than one erroneously phased insertion per read, i.e., about 0.3% assuming an ITS1 or ITS2 read length of approximately 300 bases. This proportion is well within the expected infraspecific variation even at a high level of 99% similarity and thus theoretically should disappear as background signal. However, the random distribution of this error across the entire sequence, which cannot be recognized by quality filtering methods, especially not in environmental samples where no expected sequence patterns exist for comparison, causes clustering methods to interpret these differences as taxonomic, leading to serious overestimations by distribution sequences of the same taxon, but with different minor errors, into separate clusters. This is because clustering methods work with local alignments of small, highly similar sequences, whereas phylogenetic methods require alignment of sequences of a data set to template sequences obtained from Sanger sequencing. As a consequence, in a broad alignment including Sanger reference sequences, erroneous and phased indels will be located in mostly gapped columns, which have practically no influence on the resulting topology, whereas in clustering methods, they will frequently be aligned with non-homologous base calls and falsely interpreted as substitutions (45).
In the above (45), a proportion of less than 1% erroneously phased indels caused by CAFIE errors in the ITS of a single species caused overestimation of the number of taxa by a factor of 35 at a 95% threshold level, 137 at 97%, and 980 at 99%. Thus, instead of disappearing as background signal, the sequencing error had a tremendous effect on OTU evaluation when in that case it was known that all sequence data came from a single target species. Even after removing all erroneous indels, clustering continued to overestimate species richness by a factor of 4 at a 95% threshold level, 8 at 97%, and 116 at 99%, due to minor remaining infragenomic variation and other sequencing errors. In contrast, alignment-based phylogenetic analysis of a broad set of nearly 2,000 reads recovered a single species, even with all sequence errors included. Thus, while environmental sequencing continues to accumulate a vast amount of important data on unrecognized fungi, extrapolations from these data must be taken with great care when based on clustering methods. An extreme example is a recent study which predicts up to a trillion species of microorganisms globally based on OTUs derived from environmental sequence data (1), using ecological scaling laws which, put in simple terms, predict how the global, limited number of N individuals would divide into S species, taking into account resource competition based on body size and other factors. This number, while certainly intriguing, appears to be exaggerated, and it would be interesting to see what alternative prediction would be obtained when defining the underlying taxonomic entities in this analysis through alignment-based phylogenetic methods.
In spite of these shortcomings, it is clear that environmental sequencing is now the major source of discovering novel fungal taxa, at least in the same order of magnitude as taxa recovered in species recognition studies or perhaps much higher. It follows that to properly catalog these fungi, a naming system for voucherless sequences must be developed, and proposals on how to do that are currently under consideration (47–51).
In addition to discovering new species, environmental sequencing may reveal unknown higher fungal taxa up to the class level. The best-known example is the recently discovered class Archaeorhizomycetes (31, 52). In Tedersoo and colleagues’ study (37), ∼6% of all fungal OTUs below the phylum level could not be assigned to any known class. Further clustering of unidentified fungal sequences at 70% sequence similarity revealed 14 distinct taxonomic groups comprising >7 OTUs, suggesting that there are several deeply divergent class-level fungal lineages that have not yet been described or previously sequenced. A follow-up study by the same group (53) also revealed numerous new lineages at least at the order level.
WHERE ARE THE UNDESCRIBED FUNGI?
If there are at least 1.5 million or more fungi on Earth, and we know of just 120,000, where can the missing 1.38 million species be? As outlined, three major sources for unrecognized fungal diversity seem to emerge: (i) geographic areas and ecological habitats that are largely understudied, particularly in tropical regions and biodiversity hot spots, (ii) ecologically cryptic fungi that occur in the environment but usually do not manifest themselves via discernible structures other than microscopic hyphae and mycelia, and (iii) so-called cryptic species hidden under well-established names.
Biodiversity Hot Spots
It is generally recognized that missing species of all groups of organisms will be found in biodiversity hot spots (54). A novel quantitative method to identify hot spots of unrecognized species richness is the grid approach (6–8), which relies on linear interpolation and hence gives more reliable estimates than nonlinear extrapolation. The area in question is divided into grids, and observed richness per grid is linearly correlated with predictive macroenvironmental parameters and sampling effort, using at least one well-sampled grid for calibration. This method can be applied to both traditional inventories and data based on environmental sequencing, as long as species are reliably delimited and macroenvironmental predictors can be reasonably derived from the data. Thus far, the method has been employed for lichen-forming fungi in the families Graphidaceae and Trypetheliaceae and the genus Cora, resulting in estimates ranging between 1.5 and 4 times the number of known species.
South America is generally recognized as having major hot spots, and various studies point to high levels of novelty there. By combining data from basidiomes and sequence data from ectomycorrhizal roots in a Dicymbe corymbosa forest in Guyana, Henkel and collaborators (55) estimated that over 250 species were associated with this one tree species in this single site. López-Quintero and colleagues (56) found that of 632 macromycete species in Colombian Amazonian forests, 52% could not be identified to the species level, a significant proportion likely representing undescribed species. Truong and coworkers (57) collected 1,430 basidiomes during just four collecting expeditions in southern South American Nothofagaceae-dominated forests; they generated 439 OTUs out of 957 specimens, of which 308 (ca. 70%) did not match any in the UNITE dynamic database at 97 to 99% similarity and thus did not correspond to any “species hypothesis” currently in the database.
Little-Explored Habitats
There are an enormous number of potential sites for fungi in any locality, and in the case of the tropics at least 31 ecological niches meriting study can be recognized, many requiring different techniques and specialists to explore (58). While no site on Earth can yet be considered to have a complete inventory of the fungi present, some habitats stand out as particularly underexplored.
The situation can be exemplified by the fungi which obligately grow on lichens, the lichenicolous fungi. With a few notable exceptions, this niche was largely overlooked by lichenologists and other mycologists until the mid-1970s. In 1976, there were just 457 species known worldwide (59), but by 2016 that number exceeded 1,800 (http://www.lichenicolous.net/); that represents a 394% increase over 40 years, or 98.5% per decade. This growth is reflected at the more local level, where in Great Britain and Ireland the number known rose from just 218 species in 1983 to 403 in 2003, an increase of 85% over 20 years (60); the number has now passed 500 and continues to rise (D. L. Hawksworth, unpublished). That figure compares with around 1,900 lichenized species in the same region, implying by extrapolation to the global scale that there may be as many as 25% as many lichenicolous fungi as lichenized species, i.e., around 5,000 species. This hypothesis is supported by studies of these fungi in areas where they have not previously been investigated; for example, of 189 species reported from Isla Navarino in Chile on the basis primarily of collections made in just 1 year, 6 genera and 60 species (32%) were new to science (61). And these figures do not include the so-called endolichenic fungi that can be cultured from lichen thalli or are only known from sequence data (e.g., 62, 63). In addition, emerging studies suggest the presence of highly specific, yet morphologically cryptic, basidiomycete yeasts and other basidiomycetes in the cortex of lichen thalli, which add yet another dimension to unknown fungal diversity (32, 51).
The experience with lichenicolous fungi appears or can be expected to be paralleled in other little-explored habitats, including bryophytes (64–66), algae (67, 68), endophytic fungi inside vascular plants (69–71), tropical foliicolous and fungicolous fungi (72), mammal guts (73), insect guts (74–77) and exoskeletons (78, 79), on and in rocks (80, 81), and in deep sea and ocean sediments (82), to name just a few.
Cryptic Species
Biologically distinct species which are morphologically indistinguishable, so-called cryptic species, were recognized 2 decades ago as one place where some of the predicted “missing fungi” might be discovered, since physiological and other biological divergence evidently often precedes morphological divergence in the evolution of fungal species. Based on studies of selected complexes then available, it was suggested that the number of known fungi might rise by a factor of 5 or more for this single reason (83). Molecular studies reveal that cryptic speciation is widespread through diverse groups of fungi, including plant pathogens, clinically important fungi, lichen-forming fungi, and mushrooms. Recognition of such cryptic speciation is indeed playing an increasingly important role in species discovery, as outlined above, and the ratio derived from current studies, 11.3:1, is about twice as high as estimated by Hawksworth and Rossman (83).
Existing Reference Collections
It has been estimated that over half of the projected 70,000 undiscovered plant species have already been collected and await description (84). Similarly, most reference collections of fungi, whether of dried specimens (fungaria) or living cultures (culture or microbial resource collections), include material that is not named to the species level, and in some cases not even to the genus level. Further, some collections and isolates referred to named fungi not uncommonly prove, on critical examination, to be different species. It has been suggested that around 20,000 fungal species have been collected but are as yet undescribed (83), but that is likely to be an underestimate because many mycologists have large backlogs of material awaiting formal description.
One of the challenges in the linking of newly obtained sequence data with named material in collections, particularly those that are type material, is fixing the application of the species name. Currently, the 34,878 species with sequences in GenBank (see above) represent just 29% of the 120,000 known species. Obtaining sequences for the missing 71% of named accepted species is essential to determine the novelty of recovered sequences from the environment, specimens, or isolates. While some researchers have had considerable success in recovering DNA from dried specimens (85), even from a dried Hygrophorus basidiome collected as far back as 1794 (86), this is not always possible (87). Aged dried material generally exhibits high levels of DNA degradation, making access through conventional extraction and PCR methods difficult (88, 89). In view of the serendipity of obtaining satisfactory results, collection curators are often unwilling to allow parts of often scant, irreplaceable type specimens to be destroyed for DNA extraction. Several attempts are now being made at using high-throughput sequencing technologies to obtain sequence data from highly fragmented DNA (88). One of the main problems in this respect is contamination, since even for highly variable loci, it will be a challenge to piece small fragments together without the risk of generating artificial chimeras. Therefore, efforts should be made to identify highly diagnostic regions in the ITS or other loci that allow the identification of a taxon even when only short fragments are at hand (89), as exemplified by comparing the ITS1 and ITS2 subregions (90).
There is a particular problem regarding the type species of generic names. An impressive 5,317 generic names of fungi are represented in GenBank (C. L. Schoch, personal communication), but in many cases the sequences are not from the name-fixing type species. A concerted attempt has been initiated to sequence the type species of genera currently not represented in DNA databases. This involves recollection and/or isolation from the geographical area of the original material and from the same host or substrate when no DNA has been recovered from the type material (91). Sequenced material of species can then in some cases be designated as an interpretive type—an epitype (92). Such an effort involves global collaboration of sequencing laboratories with colleagues in countries where high unknown diversity is located.
TOWARD A WORKING NUMBER OF GLOBAL FUNGAL SPECIES RICHNESS
There is general acceptance among mycologists that the global number of fungal species is a seven-digit figure, in the range 1 to 5 million (10), with “at least” 1.5 million now predominating, though a possible order of magnitude higher has been hinted at (93) and a minimum figure of 611,000 to 712,000 was arrived at (2, 94), leaving aside the figure of potentially billions of fungi extrapolated by Locey and Lennon (1). A recalculation of the number computed via Mora and colleagues’ approach (2) gives a figure of about 1.4 million (see above), close to the 1.5 million estimate which has come to be widely accepted by other biologists (54, 95–97), notwithstanding a few estimates that fail to appreciate the difficulty of inventorying fungi (98) or that equate numbers detected in soil alone as indicative of the total number at a site (37).
Unfortunately, the different techniques used to estimate global species richness are in part additive and in part overlapping or redundant, so that a combination of estimates from these studies is difficult. Thus, the revised estimate of nearly 3.8 million fungal species based on an updated fungus:plant ratio of 9.8:1 and 380,000 vascular plant species should include fungal species of all sorts with all types of ecologies and detected by different methodologies, including environmental sequencing and species delimitation methods. This number should then be congruent with approaches such as publication rates or scaling methods, but these approaches result in widely disparate estimates. Alternatively, an additive approach would be based on the three major sources of unrecognized fungal species richness, which can be reasonably sorted in descending order into (i) environmental sequencing, (ii) cryptic speciation, and (iii) other novel discoveries through traditional field work. For cryptic speciation, the above survey of established literature results in a weighted ratio of about an order of magnitude, i.e., more than 1 million additional species if based on 120,000 previously known species. If novel species from environmental sequencing are at least as high in number, this would add at least another million.
We conservatively assume that other novel discoveries (excluding species delimitation methods and environmental sequencing) will occur at a proportion of at best 10% of the latter two approaches. This assumption is based on the rate of 1,300 newly described species per year until about 2010, before the onset of rigorous species delimitation methods and environmental sequencing (see above). At this rate, it would take nearly 100 years to double the number of known species. Thus, while environmental sequencing and species delimitation methods would contribute about 2 million new taxa, other novel discoveries would add at best another 120,000 taxa within a reasonable time frame, for a total of a little over 2.2 million. This would give a range of between 2.2 (additive approach) and 3.8 million (global ratio approach), very much in line with the previously updated estimates of between 2.3 and 3 million (4, 99) and precisely narrowing down the range of 1 to 5.1 million given by Blackwell (10) by 1.2 to 1.3 million on either side.
We therefore propose to replace previous estimates of global fungal species richness with this updated range of 2.2 to 3.8 million. This estimate is likely to be further improved on when reliable statistical approaches to analyze the huge amount of environmental sequence data become available. An interesting approach would be to combine the following techniques: (i) a geographically and ecologically broad sample of environmental sequence data, (ii) alignment-based species recognition methods to properly estimate OTU diversity, (iii) species-based niche modeling to establish macro- and microecological patterns, and (iv) a grid-based interpolation of global species richness, taking into account sampling effort. We predict that such an analysis will result in estimates that might lie well above the revised conservative estimate of 2.2 million species and likely even beyond 3.8 million species. But even if continuing to adopt the previous, now appearing highly conservative number of 1.5 million species, the discovery and formal description of the missing fungi remain a daunting task.
ACKNOWLEDGMENTS
We thank Robert R. Dunn, Gareth W. Griffiths, Conrad L. Schoch, and Brian M. Spooner for providing previously unpublished data. We are especially indebted to Paul M. Kirk for providing the raw data from Index Fungorum, on which Figs. 1 to 3 were based.
REFERENCES
- 1.Locey KJ, Lennon JT. 2016. Scaling laws predict global microbial diversity. Proc Natl Acad Sci USA 113:5970–5975 10.1073/pnas.1521291113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. 2011. How many species are there on Earth and in the ocean? PLoS Biol 9:e1001127 10.1371/journal.pbio.1001127. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawksworth DL. 1991. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol Res 95:641–655 10.1016/S0953-7562(09)80810-1. [DOI] [Google Scholar]
- 4.Hawksworth DL. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 105:1422–1432 10.1017/S0953756201004725. [DOI] [Google Scholar]
- 5.Erwin TL. 1982. Tropical forests: their richness in Coleoptera and other arthropod species. Coleopt Bull 36:74–75. [Google Scholar]
- 6.Lücking R, Dal-Forno M, Sikaroodi M, Gillevet PM, Bungartz F, Moncada B, Yánez-Ayabaca A, Chaves JL, Coca LF, Lawrey JD. 2014. A single macrolichen constitutes hundreds of unrecognized species. Proc Natl Acad Sci USA 111:11091–11096 10.1073/pnas.1403517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lücking R, Johnston MK, Aptroot A, Kraichak E, Lendemer JC, Boonpragob K, Cáceres MES, Ertz D, Ferraro LI, Jia ZF, Kalb K, Mangold A, Manoch L, Mercado-Díaz JA, Moncada B, Mongkolsuk P, Papong K, Parnmen S, Peláez RN, Poengsungnoen V, Rivas Plata E, Saipunkaew W, Sipman HJM, Sutjaritturakan J, Van den Broeck D, Von Konrat M, Weerakoon G, Lumbsch HT. 2014. One hundred and seventy five new species of Graphidaceae: closing the gap or a drop in the bucket? Phytotaxa 189:7–38 10.11646/phytotaxa.189.1.4. [DOI] [Google Scholar]
- 8.Aptroot A, Cáceres MES, Johnston MK, Lücking R. 2016. How diverse is the lichenized fungal family Trypetheliaceae (Ascomycota: Dothideomycetes): a quantitative prediction of global species richness. Lichenologist 48:983–1011 10.1017/S0024282916000463. [DOI] [Google Scholar]
- 9.O’Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol 71:5544–5550 10.1128/AEM.71.9.5544-5550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackwell M. 2011. The fungi: 1, 2, 3 ... 5.1 million species? Am J Bot 98:426–438 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 11.Hawksworth DL. 1992. The need for a more effective biological nomenclature for the 21st century. Bot J Linn Soc 109:543–567 10.1111/j.1095-8339.1992.tb01450.x. [DOI] [Google Scholar]
- 12.Costello MJ, Wilson SP. 2011. Predicting the number of known and unknown species in European seas using rates of description. Glob Ecol Biogeogr 20:319–330 10.1111/j.1466-8238.2010.00603.x. [DOI] [Google Scholar]
- 13.Richards RA. 2010. The Species Problem: a Philosophical Analysis. Cambridge University Press, Cambridge, United Kingdom. 10.1017/CBO9780511762222 [DOI] [Google Scholar]
- 14.Kunz W. 2012. Do Species Exist? Principles of Taxonomic Classification. Wiley-Blackwell, Weinheim, Germany. 10.1002/9783527664283 [DOI] [Google Scholar]
- 15.Hawksworth DL. 1996. Microbial collections as a tool in biodiversity and biosystematic research, p 26–35. In Samson RA, Stalpers JA, van de Mei D, Stouthamer AH (ed), Culture Collections to Improve the Quality of Life. Proceedings of the Eighth International Congress. Centraalbureau voor Schimmelcultures and World Federation for Culture Collections, Baarn, The Netherlands. [Google Scholar]
- 16.Quaedvlieg W, Binder M, Groenewald JZ, Summerell BA, Carnegie AJ, Burgess TI, Crous PW. 2014. Introducing the consolidated species concept to resolve species in the Teratosphaericaeae. Persoonia 33:1–40 10.3767/003158514X681981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavitt SD, Divakar PK, Crespo A, Lumbsch HT. 2016. A matter of time: understanding the limits of the power of molecular data for delimiting species boundaries. Herzogia 29:479–492 10.13158/heia.29.2.2016.479. [DOI] [Google Scholar]
- 18.Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008. Ainsworth & Bisby’s Dictionary of the Fungi, 10th ed. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 19.Schoch CL, et al, Fungal Barcoding Consortium, Fungal Barcoding Consortium Author List. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geml J, Laursen GA, O’Neill K, Nusbaum HC, Taylor DL. 2006. Beringian origins and cryptic speciation events in the fly agaric (Amanita muscaria). Mol Ecol 15:225–239 10.1111/j.1365-294X.2005.02799.x. [DOI] [PubMed] [Google Scholar]
- 21.Buyck B, Hofstetter V. 2011. The contribution of tef-1 sequences to species delimitation in the Cantharellus cibarius complex in the southeastern USA. Fung Div 49:35–46 10.1007/s13225-011-0095-z. [DOI] [Google Scholar]
- 22.Lücking R, et al. 2016. Turbo-taxonomy to assemble a megadiverse lichen genus: seventy new species of Cora (Basidiomycota: Agaricales: Hygrophoraceae), honouring David Leslie Hawksworth’s seventieth birthday. Fungal Diversity 81:1–69. 10.1007/s13225-016-0374-9. [DOI] [Google Scholar]
- 23.Singh G, Dal Grande F, Divakar PK, Otte J, Leavitt SD, Szczepanska K, Crespo A, Rico VJ, Aptroot A, Cáceres MES, Lumbsch HT, Schmitt I. 2015. Coalescent-based species delimitation approach uncovers high cryptic diversity in the cosmopolitan lichen-forming fungal genus Protoparmelia (Lecanorales, Ascomycota). PLoS One 10:e0124625 10.1371/journal.pone.0124625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mark K, Saag L, Leavitt SD, Will-Wolf S, Nelsen MP, Tõrra T, Saag A, Randlane T, Lumbsch HT. 2016. Evaluation of traditionally circumscribed species in the lichen-forming genus Usnea, section Usnea (Parmeliaceae, Ascomycota) using a six-locus dataset. Organ Div Evol 16:497–524. 10.1007/s13127-016-0273-7. (Erratum, doi:10.1007/s13127-016-0311-5.) [DOI] [Google Scholar]
- 25.Nagy LG, Házi J, Vágvölgyi C, Papp T. 2012. Phylogeny and species delimitation in the genus Coprinellus with special emphasis on the haired species. Mycologia 104:254–275 10.3852/11-149. [DOI] [PubMed] [Google Scholar]
- 26.Dal Forno M. 2015. Evolution and diversity of the Basidiolichen clade Dictyonema (Agaricales: Hygrophoraceae). PhD dissertation, College of Science, Environmental Science and Public Policy, George Mason University, Fairfax, VA. [Google Scholar]
- 27.Bensch K, Groenewald JZ, Dijksterhuis J, Starink-Willemse M, Andersen B, Summerell BA, Shin H-D, Dugan FM, Schroers H-J, Braun U, Crous PW. 2010. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud Mycol 67:1–94 10.3114/sim.2010.67.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moncada B, Lücking R, Suárez A. 2014. Molecular phylogeny of the genus Sticta (lichenized Ascomycota: Lobariaceae) in Colombia. Fung Div 64:205–231 10.1007/s13225-013-0230-0. [DOI] [PubMed] [Google Scholar]
- 29.Le Gac M, Hood ME, Fournier E, Giraud T. 2007. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution 61:15–26 10.1111/j.1558-5646.2007.00002.x. [DOI] [PubMed] [Google Scholar]
- 30.O’Donnell K, Kistler HC, Tacke BK, Casper HH. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci USA 97:7905–7910 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menkis A, Urbina H, James TY, Rosling A. 2014. Archaeorhizomyces borealis sp. nov. and a sequence-based classification of related soil fungal species. Fungal Biol 118:943–955 10.1016/j.funbio.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Spribille T, Tuovinen V, Resl P, Vanderpool D, Wolinski H, Aime MC, Schneider K, Stabentheiner E, Toome-Heller M, Thor G, Mayrhofer H, Johannesson H, McCutcheon JP. 2016. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science 353:488–492 10.1126/science.aaf8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarus KL, James TY. 2015. Surveying the biodiversity of the Cryptomycota using a targeted PCR approach. Fungal Ecol 14:62–70 10.1016/j.funeco.2014.11.004. [DOI] [Google Scholar]
- 34.Livermore JA, Mattes TE. 2013. Phylogenetic detection of novel Cryptomycota in an Iowa (United States) aquifer and from previously collected marine and freshwater targeted high-throughput sequencing sets. Environ Microbiol 15:2333–2341 10.1111/1462-2920.12106. [DOI] [PubMed] [Google Scholar]
- 35.Roy BA, Vogler DR, Bruns TD, Szaro TM. 1998. Cryptic species in the Puccinia monoica complex. Mycologia 90:846–853 10.2307/3761326. [DOI] [Google Scholar]
- 36.Taylor DL, Herriott IC, Stone KE, McFarland JW, Booth MG, Leigh MB. 2010. Structure and resilience of fungal communities in Alaskan boreal forest soils. Can J Res 40:1288–1301 10.1139/X10-081. [DOI] [Google Scholar]
- 37.Tedersoo L, et al. 2014. Global diversity and geography of soil fungi. Science 346:1256688 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 38.Piepenbring M, Hofmann TA, Unterseher M, Kost G. 2012. Species richness of plants and fungi in western Panama: towards a fungal inventory in the neotropics. Biodivers Conserv 21:2181–2193 10.1007/s10531-011-0213-y. [DOI] [Google Scholar]
- 39.Royal Botanic Gardens Kew. 2017. State of the World’s Plants Report 2017. Royal Botanic Gardens Kew, London, United Kingdom. [Google Scholar]
- 40.Geml J, Nouhra ER, Wicaksono CY, Pastor N, Fernandez L, Becerra AG. 2012. Mycota of the Andean Yungas forests: assessments of fungal diversity and habitat partitioning in a threatened ecosystem. Inoculum 63:18. [Google Scholar]
- 41.Grantham NS, Reich BJ, Pacifici K, Laber EB, Menninger HL, Henley JB, Barberán A, Leff JW, Fierer N, Dunn RR. 2015. Fungi identify the geographic origin of dust samples. PLoS One 10:e0122605 10.1371/journal.pone.0122605. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration. 2011. The sequence read archive. Nucleic Acids Res 39(Database):D19–D21 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res 41(D1):D36–D42 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy LG, Petkovits T, Kovács GM, Voigt K, Vágvölgyi C, Papp T. 2011. Where is the unseen fungal diversity hidden? A study of Mortierella reveals a large contribution of reference collections to the identification of fungal environmental sequences. New Phytol 191:789–794 10.1111/j.1469-8137.2011.03707.x. [DOI] [PubMed] [Google Scholar]
- 45.Lücking R, Lawrey JD, Gillevet PM, Sikaroodi M, Dal-Forno M, Berger SA. 2014. Multiple ITS haplotypes in the genome of the lichenized basidiomycete Cora inversa (Hygrophoraceae): fact or artifact? J Mol Evol 78:148–162 10.1007/s00239-013-9603-y. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 10.1093/bioinformatics/btq461. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH. 2011. Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biol Rev 25:38–47 10.1016/j.fbr.2011.01.001. [DOI] [Google Scholar]
- 48.Hibbett D. 2016. The invisible dimension of fungal diversity. Science 351:1150–1151 10.1126/science.aae0380. [DOI] [PubMed] [Google Scholar]
- 49.de Beer ZW, Marincowitz S, Duong TA, Kim JJ, Rodrigues A, Wingfield MJ. 2016. Hawksworthiomyces gen. nov. (Ophiostomatales), illustrates the urgency for a decision on how to name novel taxa known only from environmental nucleic acid sequences (ENAS). Fungal Biol 120:1323–1340 10.1016/j.funbio.2016.07.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Hawksworth DL, Hibbett DS, Kirk PM, Lücking R. 2016. (308–310) Proposals to permit DNA sequence data to serve as types of names of fungi. Taxon 65:899–900 10.12705/654.31. [DOI] [Google Scholar]
- 51.Lücking R, Moncada M. 2017. Dismantling Marchandiomphalina into Agonimia (Verrucariaceae) and Lawreymyces gen. nov. (Corticiaceae): setting a precedent to the formal recognition of thousands of voucherless fungi based on type sequences. Fung Div 84:119–138. 10.1007/s13225-017-0382-4 [DOI] [Google Scholar]
- 52.Rosling A, Cox F, Cruz-Martinez K, Ihrmark K, Grelet GA, Lindahl BD, Menkis A, James TY. 2011. Archaeorhizomycetes: unearthing an ancient class of ubiquitous soil fungi. Science 333:876–879 10.1126/science.1206958. [DOI] [PubMed] [Google Scholar]
- 53.Tedersoo L, Bahram M, Puusepp R, Nilsson RH, James TY. 2017. Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 5:42 10.1186/s40168-017-0259-5. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheffers BR, Joppa LN, Pimm SL, Laurance WF. 2012. What we know and don’t know about Earth’s missing biodiversity. Trends Ecol Evol 27:501–510 10.1016/j.tree.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 55.Henkel TW, Aime MC, Chin MML, Miller SL, Vilgalys R, Smith ME. 2012. Ectomycorrhizal fungal sporocarp diversity and discovery of new taxa in Dicymbe monodominant forests of the Guiana Shield. Biodivers Conserv 21:2195–2220 10.1007/s10531-011-0166-1. [DOI] [Google Scholar]
- 56.López-Quintero CA, Straatsma G, Franco-Molano AE, Boekhoet T. 2012. Macrofungal diversity in Colombian Amazon forests varies with regions and regimes of disturbance. Biodivers Conserv 21:2221–2243 10.1007/s10531-012-0280-8. [DOI] [Google Scholar]
- 57.Truong C, Mujic AB, Healy R, Kuhar F, Furci G, Torres D, Niskanen T, Sandoval-Leiva PA, Fernández N, Escobar JM, Moretto A, Palfner G, Pfister D, Nouhra E, Swenie R, Sánchez-García M, Matheny PB, Smith ME. 2017. How to know the fungi: combining field inventories and DNA-barcoding to document fungal diversity. New Phytol 214:913–919 10.1111/nph.14509. [DOI] [PubMed] [Google Scholar]
- 58.Hawksworth DL, Minter DW, Kinsey GC, Cannon PF. 1997. Inventorying a tropical fungal biota: intensive and extensive approaches, p 29–50. In Janardhanan KK, Rajendran C, Natarajan K, Hawksworth DL (ed), Tropical Mycology. Oxford & IBH Publishing, New Dehli, India. [Google Scholar]
- 59.Clauzade G, Roux C. 1976. Les Champignons Lichénicoles non Lichénisés. Univ. des Sciences et Techn. du Languedoc, Laboratoire de Systématique et Géobotanique Méditerranéenne, Inst. de Botanique, Montpellier, France. [Google Scholar]
- 60.Hawksworth DL. 2003. The lichenicolous fungi of Great Britain and Ireland: an overview and annotated checklist. Lichenologist 35:191–232 10.1016/S0024-2829(03)00027-6. [DOI] [Google Scholar]
- 61.Etayo J, Sancho LG. 2008. Hongos Liquenicolas del Sur de Sudamerica, Especialmente de Isla Navarino (Chile). Bibliotheca Lichenologica series vol. 98. J. Cramer, Berlin, Germany. [Google Scholar]
- 62.Fleischhacker A, Grube M, Kopun T, Hafellner J, Muggia L. 2015. Community analyses uncover high diversity of lichenicolous fungi in alpine habitats. Microb Ecol 70:348–360 10.1007/s00248-015-0579-6. [DOI] [PubMed] [Google Scholar]
- 63.Zhang T, Wei XL, Wei YZ, Liu HY, Yu LY. 2016. Diversity and distribution of cultured endolichenic fungi in the Ny-Ålesund region, Svalbard (high Arctic). Extremophiles 20:461–470 10.1007/s00792-016-0836-8. [DOI] [PubMed] [Google Scholar]
- 64.Döbbeler P, Hertel H. 2013. Bryophilous ascomycetes everywhere: distribution maps of selected species on liverworts, mosses and Polytrichaceae. Herzogia 26:361–404 10.13158/heia.26.2.2013.361. [DOI] [Google Scholar]
- 65.Davey ML, Kauserud H, Ohlson M. 2014. Forestry impacts on the hidden fungal biodiversity associated with bryophytes. FEMS Microbiol Ecol 90:313–325 10.1111/1574-6941.12386. [DOI] [PubMed] [Google Scholar]
- 66.Hirose D, Hobara S, Matsuoka S, Kato K, Tanabe Y, Uchida M, Kudoh S, Osono T. 2016. Diversity and community assembly of moss-associated fungi in ice-free coastal outcrops of continental Antarctica. Fungal Ecol 24:94–101 10.1016/j.funeco.2016.09.005. [DOI] [Google Scholar]
- 67.Kohlmeyer J, Kohlmeyer E. 2013. Marine Mycology: The Higher Fungi. Academic Press, San Diego, CA. [Google Scholar]
- 68.Furbino LE, Godinho VM, Santiago IF, Pellizari FM, Alves TM, Zani CL, Junior PA, Romanha AJ, Carvalho AG, Gil LH, Rosa CA, Minnis AM, Rosa LH. 2014. Diversity patterns, ecology and biological activities of fungal communities associated with the endemic macroalgae across the Antarctic peninsula. Microb Ecol 67:775–787 10.1007/s00248-014-0374-9. [DOI] [PubMed] [Google Scholar]
- 69.Arnold AE. 2007. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev 21:51–66 10.1016/j.fbr.2007.05.003. [DOI] [Google Scholar]
- 70.Rodriguez RJ, White JF Jr, Arnold AE, Redman RS. 2009. Fungal endophytes: diversity and functional roles. New Phytol 182:314–330 10.1111/j.1469-8137.2009.02773.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Higgins KL, Arnold AE, Coley PD, Kursar TA. 2014. Communities of fungal endophytes in tropical forest grasses: highly diverse host-and habitat generalists characterized by strong spatial structure. Fungal Ecol 8:1–11 10.1016/j.funeco.2013.12.005. [DOI] [Google Scholar]
- 72.Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, Hyde KD. 2014. The sooty moulds. Fung Div 66:1–36 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- 73.Griffith GW, Baker S, Fliegerova K, Liggenstoffer A, van der Giezen M, Voigt K, Beakes G. 2010. Anaerobic fungi: Neocallimastigomycota. IMA Fungus 1:181–185 10.5598/imafungus.2010.01.02.11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suh SO, McHugh JV, Pollock DD, Blackwell M. 2005. The beetle gut: a hyperdiverse source of novel yeasts. Mycol Res 109:261–265 10.1017/S0953756205002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lichtwardt RW. 2012. Trichomycete gut fungi from tropical regions of the world. Biodiv Conserv 21:2397–2402 10.1007/s10531-011-0146-5. [DOI] [Google Scholar]
- 76.Gouba N, Raoult D, Drancourt M. 2013. Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PLoS One 8:e59474 10.1371/journal.pone.0059474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Tretter ED, Johnson EM, Kandel P, Lichtwardt RW, Novak SJ, Smith JF, White MM. 2014. Using a five-gene phylogeny to test morphology-based hypotheses of Smittium and allies, endosymbiotic gut fungi (Harpellales) associated with arthropods. Mol Phylogenet Evol 79:23–41 10.1016/j.ympev.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 78.Weir A, Hammond PM. 1997. Laboulbeniales on beetles: host utilization patterns and species richness of the parasites. Biodiv Conserv 6:701–719 10.1023/A:1018318320019. [DOI] [Google Scholar]
- 79.Weir A. 2004. The Laboulbeniales: an enigmatic group of arthropod-associated fungi. Symbiosis 4:611–620. [Google Scholar]
- 80.Ruibal C, Gueidan C, Selbmann L, Gorbushina AA, Crous PW, Groenewald JZ, Muggia L, Grube M, Isola D, Schoch CL, Staley JT, Lutzoni F, de Hoog GS. 2009. Phylogeny of rock-inhabiting fungi related to Dothideomycetes. Stud Mycol 64:123–133, S7 10.3114/sim.2009.64.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Egidi E, de Hoog GS, Isola D, Onofri S, Quaedvlieg W, de Vries M, Verkeley GJM, Stielow JB, Zucconi L, Selbmann L. 2014. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothideomycetes based on multi-locus phylogenies. Fung Div 65:127–165 10.1007/s13225-013-0277-y. [DOI] [Google Scholar]
- 82.Le Calvez T, Burgaud G, Mahé S, Barbier G, Vandenkoornhuyse P. 2009. Fungal diversity in deep-sea hydrothermal ecosystems. Appl Environ Microbiol 75:6415–6421 10.1128/AEM.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hawksworth DL, Rossman AY. 1997. Where are all the undescribed fungi? Phytopathology 87:888–891 10.1094/PHYTO.1997.87.9.888. [DOI] [PubMed] [Google Scholar]
- 84.Bebber DP, Carine MA, Wood JR, Wortley AH, Harris DJ, Prance GT, Davidse G, Paige J, Pennington TD, Robson NKB, Scotland RW. 2010. Herbaria are a major frontier for species discovery. Proc Natl Acad Sci USA 107:22169–22171 10.1073/pnas.1011841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brock PM, Döring H, Bidartondo MI. 2009. How to know unknown fungi: the role of a herbarium. New Phytol 181:719–724 10.1111/j.1469-8137.2008.02703.x. [DOI] [PubMed] [Google Scholar]
- 86.Larsson E, Jacobsson S. 2004. Controversy over Hygrophorus cossus settled using ITS sequence data from 200 year-old type material. Mycol Res 108:781–786 10.1017/S0953756204000310. [DOI] [PubMed] [Google Scholar]
- 87.Begerow D, Nilsson H, Unterseher M, Maier W. 2010. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl Microbiol Biotechnol 87:99–108 10.1007/s00253-010-2585-4. [DOI] [PubMed] [Google Scholar]
- 88.Staats M, Cuenca A, Richardson JE, Vrielink-van Ginkel R, Petersen G, Seberg O, Bakker FT. 2011. DNA damage in plant herbarium tissue. PLoS One 6:e28448 10.1371/journal.pone.0028448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Särkinen T, Staats M, Richardson JE, Cowan RS, Bakker FT. 2012. How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS One 7:e43808 10.1371/journal.pone.0043808. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blaalid R, Kumar S, Nilsson RH, Abarenkov K, Kirk PM, Kauserud H. 2013. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol Ecol Resour 13:218–224 10.1111/1755-0998.12065. [DOI] [PubMed] [Google Scholar]
- 91.Crous PW, Giraldo A, Hawksworth DL, Robert V, Kirk PM, Guarro J, Robbertse B, Schoch CL, Damm U, Trakunyingcharoen T, Groenewald JZ. 2014. The genera of Fungi: fixing the application of type species of generic names. IMA Fungus 5:141–160 10.5598/imafungus.2014.05.01.14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ariyawansa HA, Hawksworth DL, Hyde KD, Jones EBG, Maharachchikumbura SSN, Manamgoda DS, Thambugala KM, Udayanga D, Camporesi E, Daranagama A, Jayawardena R, Liu J-K, McKenzie EHC, Phookamsak R, Senanayake IC, Shivas RG, Tian Q, Xu J-C. 2014. Epitypification and neotypification: guidelines with appropriate and inappropriate examples. Fung Div 69:57–91 10.1007/s13225-014-0315-4. [DOI] [Google Scholar]
- 93.Bass D, Richards TA. 2011. Three reasons to re-evaluate fungal diversity ‘on Earth and in the ocean’. Fungal Biol Rev 25:159–164 10.1016/j.fbr.2011.10.003. [DOI] [Google Scholar]
- 94.Schmit JP, Mueller GM. 2007. An estimate of the lower limit of global fungal diversity. Biodivers Conserv 16:99–111 10.1007/s10531-006-9129-3. [DOI] [Google Scholar]
- 95.Hammond PM. 1995. Described and estimated species numbers: an objective assessment of current knowledge, p 29–71. In Allsopp D, Colwell RR, Hawksworth DL (ed), Microbial Diversity and Ecosystem Function. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 96.Chapman AD. 2009. Numbers of Living Species in Australia and the World, 2nd ed. Australian Biological Resources Survey, Canberra, ACT, Australia. [Google Scholar]
- 97.Joppa LN, Roberts DL, Pimm SL. 2011. How many species of flowering plants are there? Proc Biol Sci 278:554–559 10.1098/rspb.2010.1004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.May RM. 1994. Conceptual aspects of the quantification of the extent of biological diversity. Philos Trans R Soc Lond B Biol Sci 345:13–20 10.1098/rstb.1994.0082. [DOI] [PubMed] [Google Scholar]
- 99.Hawksworth DL. 2012. Global species numbers of fungi: are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers Conserv 21:2425–2433 10.1007/s10531-012-0335-x. [DOI] [Google Scholar]
- 100.Dal-Forno M, Lücking R, Bungartz F, Yánez-Ayabaca A, Marcelli MP, Spielmann AA, Coca LF, Chaves JL, Aptroot A, Sipman HJM, Sikaroodi M, Gillevet P, Lawrey JD. 2016. From one to six: unrecognized species diversity in the genus Acantholichen (lichenized Basidiomycota: Hygrophoraceae). Mycologia 108:38–55 10.3852/15-060. [DOI] [PubMed] [Google Scholar]
- 101.Geiser DM, Pitt JI, Taylor JW. 1998. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci USA 95:388–393 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pringle A, Baker DM, Platt JL, Wares JP, Latgé JP, Taylor JW. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886–1899 10.1111/j.0014-3820.2005.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 103.Koufopanou V, Burt A, Szaro T, Taylor JW. 2001. Gene genealogies, cryptic species, and molecular evolution in the human pathogen Coccidioides immitis and relatives (Ascomycota, Onygenales). Mol Biol Evol 18:1246–1258 10.1093/oxfordjournals.molbev.a003910. [DOI] [PubMed] [Google Scholar]
- 104.Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Brown EM, McTaggart LR, Zhang SX, Low DE, Stevens DA, Richardson SE. 2013. Phylogenetic analysis reveals a cryptic species Blastomyces gilchristii, sp. nov. within the human pathogenic fungus Blastomyces dermatitidis. PLoS One 8:e59237 10.1371/journal.pone.0059237. (Erratum, 11:e0168018. doi:10.1371/journal.pone.0168018.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Walker AS, Gautier AL, Confais J, Martinho D, Viaud M, Le Pêcheur P, Dupont J, Fournier E. 2011. Botrytis pseudocinerea, a new cryptic species causing gray mold in French vineyards in sympatry with Botrytis cinerea. Phytopathology 101:1433–1445 10.1094/PHYTO-04-11-0104. [DOI] [PubMed] [Google Scholar]
- 107.Lombard L, Crous PW, Wingfield BD, Wingfield MJ. 2010. Multigene phylogeny and mating tests reveal three cryptic species related to Calonectria pauciramosa. Stud Mycol 66:15–30 10.3114/sim.2010.66.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Douhan GW, Rizzo DM. 2005. Phylogenetic divergence in a local population of the ectomycorrhizal fungus Cenococcum geophilum. New Phytol 166:263–271 10.1111/j.1469-8137.2004.01305.x. [DOI] [PubMed] [Google Scholar]
- 109.Kauserud H, Svegården IB, Decock C, Hallenberg N. 2007. Hybridization among cryptic species of the cellar fungus Coniophora puteana (Basidiomycota). Mol Ecol 16:389–399 10.1111/j.1365-294X.2006.03129.x. [DOI] [PubMed] [Google Scholar]
- 110.Alamouti SM, Wang V, Diguistini S, Six DL, Bohlmann J, Hamelin RC, Feau N, Breuil C. 2011. Gene genealogies reveal cryptic species and host preferences for the pine fungal pathogen Grosmannia clavigera. Mol Ecol 20:2581–2602 10.1111/j.1365-294X.2011.05109.x. [DOI] [PubMed] [Google Scholar]
- 111.Queloz V, Grünig CR, Berndt R, Kowalski T, Sieber TN, Holdenrieder O. 2011. Cryptic speciation in Hymenoscyphus albidus. For Pathol 41:133–142 10.1111/j.1439-0329.2010.00645.x. [DOI] [Google Scholar]
- 112.Alves A, Crous PW, Correia A, Phillips AJL. 2008. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fung Div 28:1–13. [Google Scholar]
- 113.Kroken S, Taylor JW. 2001. A gene genealogical approach to recognize phylogenetic species boundaries in the lichenized fungus Letharia. Mycologia 93:38–53 10.2307/3761604. [DOI] [Google Scholar]
- 114.Moncada B, Lücking R, Betancourt-Macuase L. 2013. Phylogeny of the Lobariaceae (lichenized Ascomycota: Peltigerales), with a reappraisal of the genus Lobariella. Lichenologist 45:203–263 10.1017/S0024282912000825. [DOI] [Google Scholar]
- 115.Bidochka MJ, Small CLN, Spironello M. 2005. Recombination within sympatric cryptic species of the insect pathogenic fungus Metarhizium anisopliae. Environ Microbiol 7:1361–1368 10.1111/j.1462-5822.2005.00823.x. [DOI] [PubMed] [Google Scholar]
- 116.Pavlic D, Slippers B, Coutinho TA, Wingfield MJ. 2009. Multiple gene genealogies and phenotypic data reveal cryptic species of the Botryosphaeriaceae: a case study on the Neofusicoccum parvum/N. ribis complex. Mol Phylogenet Evol 51:259–268 10.1016/j.ympev.2008.12.017. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Roberto TN, Rodrigues AM, Hahn RC, de Camargo ZP. 2016. Identifying Paracoccidioides phylogenetic species by PCR-RFLP of the alpha-tubulin gene. Med Mycol 54:240–247 10.1093/mmy/myv083. [DOI] [PubMed] [Google Scholar]
- 118.Divakar PK, Leavitt SD, Molina MC, Del-Prado R, Lumbsch HT, Crespo A. 2016. A DNA barcoding approach for identification of hidden diversity in Parmeliaceae (Ascomycota): Parmelia sensu stricto as a case study. Bot J Linn Soc 180:21–29 10.1111/boj.12358. [DOI] [Google Scholar]
- 119.Molina MC, Del-Prado R, Divakar PK, Sánchez-Mata D, Crespo A. 2011. Another example of cryptic diversity in lichen-forming fungi: the new species Parmelia mayi (Ascomycota: Parmeliaceae). Organ Div Evol 11:331–342 10.1007/s13127-011-0060-4. [DOI] [Google Scholar]
- 120.Del-Prado R, Divakar PK, Lumbsch HT, Crespo AM. 2016. Hidden genetic diversity in an asexually reproducing lichen forming fungal group. PLoS One 11:e0161031 10.1371/journal.pone.0161031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grünig CR, Duò A, Sieber TN, Holdenrieder O. 2008. Assignment of species rank to six reproductively isolated cryptic species of the Phialocephala fortinii s.1.-Acephala applanata species complex. Mycologia 100:47–67 10.1080/15572536.2008.11832498. [DOI] [PubMed] [Google Scholar]
- 122.Leavitt SD, Fankhauser JD, Leavitt DH, Porter LD, Johnson LA, St Clair LL. 2011. Complex patterns of speciation in cosmopolitan “rock posy” lichens: discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Mol Phylogenet Evol 59:587–602 10.1016/j.ympev.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 123.Cruse M, Telerant R, Gallagher T, Lee T, Taylor JW. 2002. Cryptic species in Stachybotrys chartarum. Mycologia 94:814–822 10.1080/15572536.2003.11833175. [DOI] [PubMed] [Google Scholar]
- 124.Sato H, Yumoto T, Murakami N. 2007. Cryptic species and host specificity in the ectomycorrhizal genus Strobilomyces (Strobilomycetaceae). Am J Bot 94:1630–1641 10.3732/ajb.94.10.1630. [DOI] [PubMed] [Google Scholar]
- 125.Sato H, Murakami N. 2008. Reproductive isolation among cryptic species in the ectomycorrhizal genus Strobilomyces: population-level CAPS marker-based genetic analysis. Mol Phylogenet Evol 48:326–334 10.1016/j.ympev.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 126.Carriconde F, Gardes M, Jargeat P, Heilmann-Clausen J, Mouhamadou B, Gryta H. 2008. Population evidence of cryptic species and geographical structure in the cosmopolitan ectomycorrhizal fungus, Tricholoma scalpturatum. Microb Ecol 56:513–524 10.1007/s00248-008-9370-2. [DOI] [PubMed] [Google Scholar]


