Abstract
Mitogen-activated protein kinase (MAPK) cascades are central components of the intracellular signaling networks used by eukaryotic cells to respond to a wide spectrum of extracellular stimuli. An MAPK is activated by an MAPK kinase, which in turn is activated by an MAPK kinase kinase (MAP3K). However, little is known about the molecular aspects of the regulation and activation of large numbers of MAP3Ks that are crucial in relaying upstream receptor-mediated signals through the MAPK cascades to induce various physiological responses. In this study, we identified a novel MEKK2-interacting protein, Mip1, that regulates MEKK2 dimerization and activation by forming a complex with inactive and nonphosphorylated MEKK2. In particular, Mip1 prevented MEKK2 activation by blocking MEKK2 dimer formation, which in turn blocked JNKK2, c-Jun N-terminal kinase 1 (JNK1), extracellular signal-regulated kinase 5, and AP-1 reporter gene activation by MEKK2. Furthermore, we found that the endogenous Mip1-MEKK2 complex was dissociated transiently following epidermal growth factor stimulation. In contrast, the knockdown of Mip1 expression by siRNA augmented the MEKK2-mediated JNK and AP-1 reporter activation. Together, our data suggest a novel model for MEKK2 regulation and activation.
Mitogen-activated protein kinase (MAPK) cascades are central components of the intracellular signaling networks that eukaryotic cells use to respond to a wide spectrum of extracellular stimuli. An MAPK is activated by an MAPK kinase (MAPKK), which in turn is activated by an MAPKK kinase (MAP3K). A growing number of MAP3Ks have been identified that are capable of activation of the extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38 MAPKs, including MEKK1, MEKK2, MEKK3, MEKK4, ASK1, TAK1, MLKs, DLK, and Tpl2 (1, 15, 18, 22, 23, 35, 36, 40). These kinases have in common a conserved kinase domain with substantial homology to that of the Saccharomyces cerevisiae MAP3K STE11 (28). There is increasing evidence from biochemical and genetic studies showing that the MAP3Ks are crucial in relaying distinct cell surface signals through various downstream MAPK pathways to their cytoplasmic and nuclear effectors.
Despite extensive efforts in the past, it is still largely unknown how each MAP3K is activated and which upstream molecules activate each MAP3K at the molecular level. One major hurdle preventing us from understanding these processes is that when expressed in transient transfection, most of the MAP3Ks become constitutively activated in the absence of any agonist stimuli toward their downstream MAPKs, such as the JNKs, ERKs, and p38. To complicate the matter further, overexpression of many MAP3Ks could also activate the IκB kinase-NF-κB pathway in addition to the MAPK cascades (20, 42). These transfection studies suggest a likely possibility that these protein kinases are negatively regulated by inhibitors or through intracellular compartmentalization under normal conditions to maintain their inactive status. Yet, direct evidence supporting this hypothesis remains to be shown. In addition, it is still unclear whether these kinases require an activating kinase (an MAP3K kinase) for their activation in a mammalian system. If not, how are these MAP3Ks activated once the cells are stimulated?
MEKK2 is a Ser/Thr protein kinase belonging to the MEKK/STE11 subgroup of the MAP3K family (1, 30). MEKK2 is expressed in multiple tissues (1, 30), but its physiological functions in different tissues are largely unstudied. Ample in vitro studies show that MEKK2 activates the JNK1/2, ERK1/2, p38, and ERK5 MAPKs (1, 5, 9, 14, 26). The strong and specific JNK activation by MEKK2 is mediated by JNK-activating kinase 2 (JNKK2) through the formation of a tripartite molecular complex consisting of MEKK2, JNKK2, and JNK1 (5). Activation of the ERK1/2, p38, and ERK5 MAPKs by MEKK2 is mediated by MEK1/2, MKK3/6, and MEK5, respectively (1, 4, 9). MEKK2 has been suggested to play a role in T-cell receptor signaling and may participate in immune synapse formation during antigenic stimulation (26, 30). MEKK2-deficient T cells were shown to hyperproliferate suggesting a negative role for MEKK2 in T-cell receptor signaling (16). Consistent with this finding, T cells deficient in JNKK2/MKK7, a major target MAPKK for MEKK2, also displayed hyperproliferative phenotypes (5, 25). MEKK2 has also been shown to regulate cytokine gene expression in mast cells (4, 14), mediate epidermal growth factor receptor (EGFR) and fibroblast growth factor 2 receptor signals (21, 34), and play a role in rheumatoid arthritis (17).
In the study reported here, we investigated the molecular mechanisms of MEKK2 activation and regulation. We identified and cloned an MEKK2 interacting protein (Mip), Mip1, that interacts preferentially with the nonphosphorylated and inactive MEKK2. Mip1 expression led to decreased MEKK2 activity, which inhibited JNKK2, JNK1, ERK5, and AP-1 activation by MEKK2. Stimulation of cells with epidermal growth factor, a known MEKK2 agonist that activates the MEKK2-dependent JNK-AP-1 pathway (34), led to a transient dissociation of MEKK2 from Mip1. In contrast, the knockdown of Mip1 expression by small interfering RNA (siRNA) resulted in the augmented activation of the MEKK2-regulated JNK/AP-1 pathway.
MATERIALS AND METHODS
Cell culture and transient transfection.
COS-1, 293T, and mouse embryonic fibroblast cells were cultured in Dulbecco's modified Eagle's medium, supplemented with 5% fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. Plasmid DNA was transfected with Lipofectamine (Invitrogen, La Jolla, CA).
Plasmids, proteins, and antibodies.
Flag-tagged JNK1; HA-tagged JNK1, JNKK2(DD), MEKK2 (1-342), MEKK2 (342-619), MEKK2 (342-424), MEKK2 (1-619), MEKK2(342-619)KM, MEKK2(1-619)KM, and MEKK1; and glutathione transferase (GST)-tagged MEKK2 and MEKK2 (342-619) mammalian expression plasmids, Gal4-luc, Gal4-c-Jun, and GST-c-Jun have been previously described (5, 30, 31, 41, 43). Standard cloning procedures were used to construct hemagglutinin (HA)-tagged Mip1 and Mip1-GFP expression vectors; GST-tagged Mip1 mammalian expression plasmids; and GST-fused Mip1 (1-184), Mip1 (1-457), Mip1 (1-486), Mip1 (152-486), and Mip1 (313-486) bacterial expression plasmids. Anti-HA antibody 12CA5 was prepared from a 12CA5 hybridoma and further purified by using a protein A-Sepharose column. Anti-Flag antibody M2 was purchased from IBI-Kodak (New Haven, CT). MEKK2-specific antibody 1128 was described before (5), and 8384 was prepared by immunizing rabbits with peptide RPALSLQETRKAKSSSPKKQN, and further-affinity-purified. Mip1-specific antibody K87 was prepared by immunizing rabbits with peptide CKNIQWKERSKQSA and further affinity purified.
Protein purification.
The whole-cell lysates of 293T cells transfected with either a GST empty vector or GST-MEKK2 (342-619) were prepared by using lysis buffer (50 mM HEPES, pH 7.6, 300 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, and 10% glycerol). After being precleared with protein A-Sepharose beads, the lysates were incubated with reduced glutathione (GSH)-Sepharose beads at 4°C for 4 h on a rotator. The beads were washed six times with lysis buffer, and the precipitates were eluted with sample buffer, resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by silver staining. The putative MEKK2 interacting protein bands were excised from the gel and analyzed by mass spectrometry.
Immunoprecipitation, in vitro kinase assay, and GST pull-down assay.
Cell lysate preparation, immunoprecipitation, and in vitro kinase assays were performed as described in our previous reports (5, 31). For the GST pull-down assay, COS-1 cells were transfected with HA-, Flag-, or GST-tagged expression vectors and they were lysed 40 h later using low-salt lysis buffer (50 mM HEPES, pH 7.6, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, and 10% glycerol). Nuclear and cellular debris was removed from the lysates by centrifugation for 20 min at 4°C. GST fusion proteins were precipitated from the clarified lysates with GSH-Sepharose at 4°C during a 4-h incubation in a rotator. The beads were washed four times with low-salt lysis buffer, the precipitates were eluted with a sample buffer and resolved by SDS-PAGE, and the interacting proteins were analyzed by immunoblotting with appropriate antibodies.
RNA interference studies.
Oligonucleotides 5′-ACCGATTCATCCTCCTTCAATGTTCAAGAGACATTGAAGGAGGATGAATCTTTTTC-3′ and 3′-TAAGTAGGAGGAAGTTACAAGTTCTCTGTAACTTCCTCCTACTTAGAAAAAGAGCT-5′ (mip1-siRNA1) and 5′-ACCGATTAGAACGACTCCGAAATTCAAGAGATTTCGGAGTCGTTCTAATCTTTTTC-3′ and 3′-TAATCTTGCTGAGGCTTTAAGTTCTCTAAAGCCTCAGCAAGATTAGAAAAAGAGCT-5′ (mip1- siRNA2) containing 19-nucleotide sequences matching two independent mip1 cDNA sequences (underlined, corresponding to amino acids 40 to 46 and 89 to 95, respectively) in reverse orientation were synthesized (Sigma Chemical Company, St. Louis, MO). The mip1 and nonspecific siRNA oligonucleotides (matching the lacZ sequence) (a gift from X. Qin) were inserted into a pBS-U6 vector (24). 293T cells grown in a six-well plate were transfected with siRNA expression plasmids using Lipofectamine (Invitrogen, La Jolla, CA). Knockdown of the endogenous Mip1 expression was analyzed by immunoblotting with K87 antibody.
Nucleotide sequence accession numbers.
The sequences of the genes encoding Mip1α, Mip1β, and Mip1γ have been assigned GenBank accession numbers AY633624, AY633625, and AY633626, respectively.
RESULTS
Cloning of Mip1.
We and other groups have shown previously that MEKK2, like many other members in the MAP3K family, when transiently expressed, is capable of self-activation without the need for any agonist stimulation (1, 5, 6, 12, 43). Thus, an intriguing question is how MEKK2 activation is regulated under normal conditions to prevent unwanted self-activation. One possibility is that MEKK2 may be negatively regulated by inhibitors under normal conditions. To investigate this possibility, we expressed GST-MEKK2 (342-619) as a bait to isolate the Mips using the same GST pull-down assay condition as described before (5).
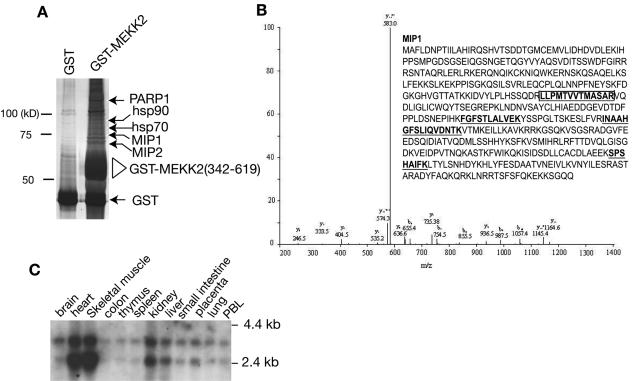
As shown in Fig. 1A, many unique protein bands (putative Mips) were precipitated specifically by GST-MEKK2 (342-619) but not by GST alone. Because of potential problems with the degradation products, we isolated only those Mips larger than GST-MEKK2 (342-619) for peptide sequencing by mass spectrometry. In comparing the sequence data with the gene bank database, we found that the Mips included hsp70, hsp90, kinesin-like protein-1, PARP1, and several novel proteins (Fig. 1A and data not shown). One of the Mips, which we identified by mass spectrometry and named Mip1 (Fig. 1B), with an apparent molecular mass of 65 kDa, was particularly interesting to us because it shared sequence homology with a conserved gene called JC310, which encodes a truncated protein that was characterized as a Ras-inhibitory factor (8) (a full-length cDNA for JC310 was recently deposited in the National Center for Biotechnology Information gene bank during the course of this study). Although the function of the JC310 clone has not been studied in mammalian cells, it was shown that a yeast gene encoding a protein called Sty-1-interacting protein 1 (Sin1) that is involved in the yeast MAPK pathway shared considerable homology with the JC310 clone (38). In addition, the loss of function of the yeast Sin1 (ySin1) could be compensated for by a chicken homolog gene isolated from chicken hindbrain with unknown function (38). These results strongly suggest that Mip1 is a critical regulator of the MAPK cascades in mammalian cells.
FIG. 1.
Identification and cloning of human MEKK2 interacting protein Mip1. (A) Purification and mass-spectrometric analysis of Mips. Cells (293T) were transfected with an empty vector or with GST-MEKK2 (342-619). The total cell lysates were prepared for precipitation with glutathione beads, and the complex was thoroughly washed, resolved by a SDS-polyacrylamide gel, and visualized by silver staining. The MEKK2-interacting protein bands (indicated by arrows) that were specifically precipitated by GST-MEKK2 (342-619) but were not in the control gel were excised from the gel and analyzed by mass spectrometry. Endogenous GST and GST-MEKK2 (342-619) were indicated. (B) The high-performance liquid chromatography/tandem mass spectrometry (HPLC/MS/MS) spectra of one representative Mip1 tryptic peptide. The predicted amino acid sequence of Mip1 is shown in the insert. The peptide sequence corresponding to the HPLC/MS/MS data of Mip1 is boxed and highlighted. Other peptides identified by MS/MS are also shown (underlined in boldface). (C) Northern blot analysis of Mip1 expression in human tissues. A PstI (0.6-kb) fragment of mip1 cDNA was labeled with 32P and used to probe an mRNA blot prepared from human tissues as indicated.
We searched the human expressed sequence tag (EST) cDNA database to obtain the full-length Mip1 cDNA and acquired several EST clones. One clone (IMAGE clone ID 303135) containing a 1.9-kb insert was completely sequenced, which revealed an open reading frame of 1,458 base pairs encoding a polypeptide of 486 amino acids. The predicted polypeptide sequence is shown in Fig. 1B. There is a stop codon upstream of the first methionine codon in the same coding frame, indicating that this EST clone is the full-length cDNA that encodes human Mip1.
Comparing the sequence of human Mip1 with the sequences of chicken Sin1 and JC310 revealed a stretch of 36 amino acids that was missing from the human Mip1, indicating that additional forms of human Mip1 may exist. To obtain these different forms, we designed primers to flank this region and carried out a reverse transcriptase PCR in which human Jurkat cDNA was used as a template. We identified three different isoforms of human Mip1, which we named α, β, and γ; the corresponding genes encode 486, 522, and 323 amino acids, respectively (accession numbers AY633624, AY633625, and AY633626, respectively) (data not shown). A searching of the human genomic DNA database revealed that these isoforms are derived from the alternative splicing of different exons on chromosome 9q34.12, locus ID79109. Although we still do not know the functional differences between these isoforms, the present study was performed with Mip1α. Northern blot analysis showed that Mip1 is ubiquitously expressed; however, it is expressed at the highest levels in the heart and skeletal muscle (Fig. 1C). During the course of this study, Schroder and colleagues also reported the cloning and identification of these differential spliced forms of human Mip1, which they named human Sin1β, Sin1, and Sin1α (29). They also found that Mip1/hSin is ubiquitously expressed with highest expression in heart and skeletal muscle. Interestingly, high MEKK2 expression in heart and skeletal muscle compared to other tissues was also observed (30).
Transiently expressed and endogenous Mip1 interacts with MEKK2.
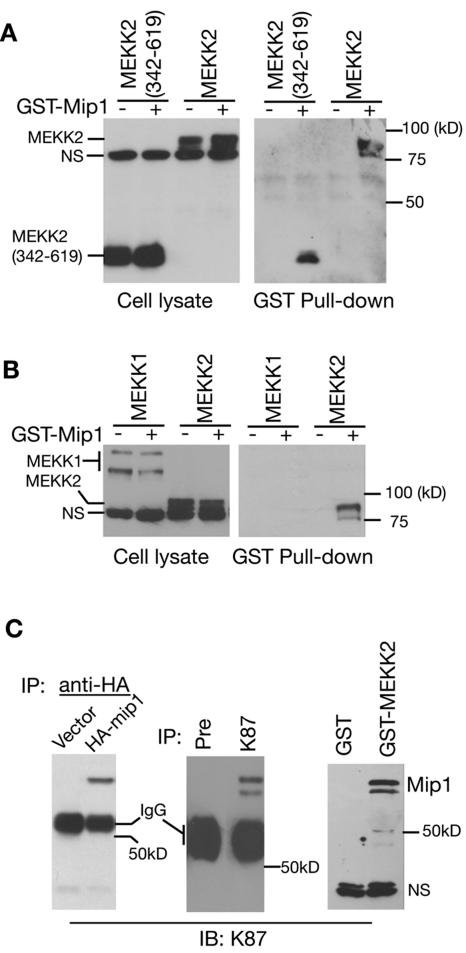
To confirm that the human cDNA indeed encodes the Mip1 that interacts with MEKK2, we constructed expression vectors with HA and GST tags with this cDNA and carried out GST pull-down and coimmunoprecipitation assays. As shown in Fig. 2A, the transiently expressed Mip1 interacted with both the catalytic domain (the bait) and the full-length MEKK2. This interaction appeared to be specific, as Mip1 did not interact with MEKK1, another member of the MEKK/Ste11 family, in the same assay (Fig. 2B). Furthermore, we prepared a Mip1-specific peptide antibody (K87), and using this antibody, we detected the transiently transfected Mip1 (Fig. 2C, left panel) and the endogenous Mip1 protein with an apparent molecular mass of 65 kDa (Fig. 2C, middle panel). The interaction of MEKK2 and Mip1 was further confirmed by using the K87 antibody to detect the endogenous Mip1 protein that was pulled down by GST-MEKK2 (342-619) (Fig. 2C, right panel), thereby confirming that the cDNA is indeed the Mip1 gene.
FIG. 2.
mip1 gene product specifically interacts with MEKK2. (A) Mip1 binds to transfected MEKK2. HA-tagged MEKK2 and MEKK2 (342-619) were expressed alone or with GST-Mip1 in COS-1 cells for a GST pull-down assay (right panel). The total lysates were analyzed by immunoblotting (IB) with an anti-HA antibody (left panel). NS, nonspecific band. (B) Mip1 binds to MEKK2 but not MEKK1. HA-tagged MEKK1 or MEKK2 were transfected alone or with GST-Mip1 into COS-1 cells for a GST pull-down assay. (C) MEKK2 interacts with endogenous Mip1 protein detected by a Mip1-specific antibody. An empty vector or HA-Mip1 expression vector was transfected into COS-1 cells. The cell lysates were subjected to immunoprecipitation (IP) with an anti-HA antibody followed by IB analysis with an anti-Mip1 peptide antibody K87 (left panel). The lysates prepared from 293T cells were subjected to immunoprecipitation with preimmune serum (Pre) or K87 followed by IB analysis with K87 (middle panel). 293T cells were transfected with an empty vector or a GST-MEKK2 (342-619) expression vector (right panel). The cell lysates were precipitated with glutathione beads followed by IB analysis with K87 antibody. IgG, immunoglobulin G.
Mip1 is a negative regulator of MEKK2 signaling.
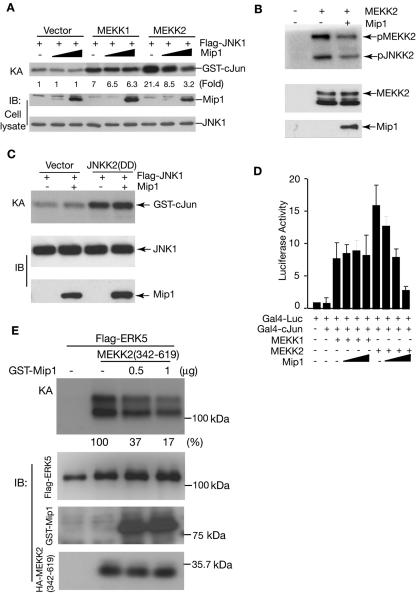
To determine the role of Mip1 in MEKK2 signaling, we next examined its effect on MEKK2-induced JNK activation. As shown in Fig. 3A, the expression of Mip1 by itself had no effect on the basal activity of JNK. However, its expression did significantly inhibit MEKK2-induced JNK1 activation in a dose-dependent manner. Consistent with the observation that Mip1 does not interact with MEKK1, Mip1 did not affect MEKK1-induced JNK1 activation, although both MEKK2 and MEKK1 are potent JNK activators. Since a study of Schizosaccharomyces pombe showed that the Mip1 ortholog ySin1 regulates the yeast Sty-1 MAPK function (38), it is possible that the inhibition we observed is due to a direct blockade of JNK activity rather than to the inhibition of MEKK2 activity. To assess this possibility, we examined the MEKK2 activity in the presence and absence of Mip1. As shown in Fig. 3B, Mip1 inhibited MEKK2 activity directly (as shown by its autophosphorylation) and hence its activation of its substrate JNKK2. We further found that Mip1 did not inhibit the JNKK2-induced JNK1 activation, thereby confirming that Mip1 is a direct regulator of MEKK2 in the control of the MEKK2 signaling pathway (Fig. 3C). Consistent with the inhibition of MEKK2-induced JNK activation, we found that Mip1 blocked the MEKK2-induced, but not the MEKK1-induced, AP-1 reporter gene expression (Fig. 3D). Thus, these results establish that Mip1 is a negative regulator of MEKK2 signaling in the JNK-AP-1 pathway. Since MEKK2 has also been shown to regulate the ERK5 pathway (14, 33), we further determined if Mip1 inhibits the MEKK2-mediated ERK5 activation and found that Mip1 was able to block ERK5 activation by MEKK2 (Fig. 3E).
FIG. 3.
Mip1 inhibits MEKK2-mediated JNK1 activation, ERK5 activation, and AP-1 reporter gene expression. (A) Flag-tagged JNK1 was cotransfected into COS-1 cells with an empty vector, GST-MEKK1, or GST-MEKK2 in the absence or presence of HA-tagged Mip1, as indicated. JNK1 was immunoprecipitated for an in vitro kinase assay (KA). The relative induction of JNK activity was determined by a PhosphorImager (Bio-Rad FX) and normalized to the JNK1 expression level. IB, immunoblotting. (B) Mip1 blocks MEKK2 phosphorylation and kinase activity toward JNKK2. HA-tagged MEKK2 expressed with an empty vector or Mip1 in COS-1 cells was immunoprecipitated with anti-HA antibody for an in vitro kinase assay using JNKK2(KR) as a substrate. The expression levels of MEKK2 and Mip1 were determined by IB with an anti-HA antibody (middle and bottom panels). (C) Mip1 does not block JNKK2-mediated JNK activation. Flag-tagged JNK1 was cotransfected with an empty vector or HA-tagged JNKK2(DD) (an active form of JNKK2) in the absence or presence of GST-Mip1, as indicated. JNK1 activity was determined by an in vitro kinase assay as described for panel A. The expression levels of Flag-JNK1 and GST-Mip1 were determined by IB with an anti-Flag antibody and K87, respectively. (D) Mip1 inhibits the AP-1 reporter gene expression induced by MEKK2 but not MEKK1. Gal4-Luc reporter plasmid and Gal4-cJun plasmid were cotransfected with an empty vector or with MEKK1 or MEKK2 expression vectors in the absence or presence of Mip1 plasmid as indicated. An actin-β-galactosidase (β-gal) plasmid was cotransfected in every transfection as a control for transfection efficiency. The reporter gene expression was determined 36 h later and normalized to the β-gal activity. The results shown are the averages of the results of three independent experiments. (E) Flag-tagged ERK5 was cotransfected into COS-1 cells with an empty vector or MEKK2 (342-619) in the absence or presence of the Mip1 expression vector as indicated. ERK5 was immunoprecipitated for an in vitro kinase assay. Expression levels of Mip1, ERK5, and MEKK2 (342-619) were determined by IB as indicated (middle and bottom panels). The relative ERK5 activity was determined by a PhosphorImager (Bio-Rad FX) and normalized to the ERK5 expression level.
Mip1 binds to the same region required for MEKK2 dimerization.
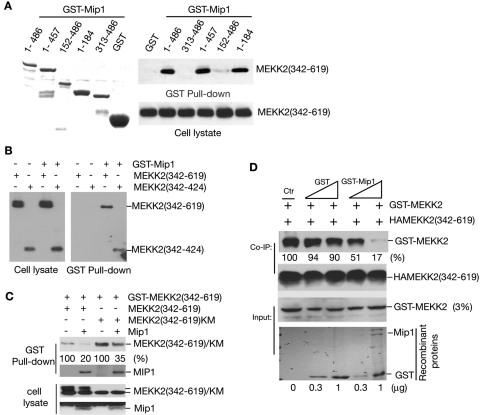
To understand how the interaction of Mip1 and MEKK2 negatively regulates MEKK2 activation, we performed a GST pull-down assay to map the domains in both Mip1 and MEKK2 that were required for their interaction. As shown in Fig. 4A, we found that the 184 amino acids at the N terminal of Mip1 were sufficient to confer MEKK2 binding. The carboxyl terminal was not required for this binding and did not interact with MEKK2.
FIG. 4.
Mapping the Mip1-MEKK2 interaction motif. (A) Recombinant GST and GST fused with various regions of Mip1 were purified from bacteria (left panel) for a GST pull-down assay by using COS-1-expressed HA-tagged MEKK2 (342-619) (right panel). (B) Mip1 binds to the MEKK2 dimerization motif. HA-tagged MEKK2 (342-619) or MEKK2 (342-424) was cotransfected into COS-1 cells with an empty vector or GST-Mip1. The cell lysates were prepared 36 h later for a GST pull-down assay, as described for Fig. 2. The expression levels of MEKK2 (342-619) and MEKK2 (342-424) in cell lysates were determined by immunoblotting (IB) with an anti-HA antibody (left panel). (C) Mip1 disrupts MEKK2 dimers. GST-MEKK2 was cotransfected with HA-tagged MEKK2 (342-619) or MEKK2(342-619)KM into COS-1 cells with or without Mip1. The cell lysates were prepared 36 h later for a GST pull-down assay as described in Fig. 2. The expression levels of MEKK2 (342-619), MEKK2(342-619)KM, and Mip1 were determined by IB with an anti-HA antibody (bottom two panels). (D) Recombinant Mip1 disrupts MEKK2 dimer formation. HA-MEKK2 (342-619) expressed in COS-1 cells was immunoprecipitated with an anti-HA antibody and incubated with equal amount of GST-MEKK2 in the presence of GST or GST-Mip1 recombinant proteins for a coprecipitation assay as indicated. MEKK2 dimers were determined by IB with anti-MEKK2 antibody 8483. Co-IP, coimmunoprecipitation.
We recently mapped a small region (dimerization motif; amino acids 342 to 424) in the catalytic domain of MEKK2 that is required for MEKK2 dimer formation and demonstrated that MEKK2 dimer formation is an essential step for MEKK2 activation (6). As shown in Fig. 2A, Mip1 interacted with MEKK2 through the catalytic domain of MEKK2. Since Mip1 appears to act as an inhibitor for MEKK2 activation, one possibility is that Mip1 may interact with the MEKK2 dimerization motif thereby preventing MEKK2 from forming dimers, hence activation. Indeed, as shown in Fig. 4B, the MEKK2 dimerization motif MEKK2 (342-424) was sufficient to interact with Mip1. This interaction appeared to be as strong as that with the entire catalytic domain, suggesting that this is the binding motif in MEKK2 to interact with Mip1.
Thus the above results suggested that Mip1 negatively regulated MEKK2 activation by preventing MEKK2 from forming dimers. If this were the case, we would expect that Mip1 expression should disrupt MEKK2 dimer formation. To further confirm that Mip1 interferes with MEKK2 dimer formation, we performed a GST pull-down assay to examine the dimer formation between GST-MEKK2 (342-619) and either MEKK2 (342-619) or MEKK2 (342-619)(KM) in the presence or absence of Mip1. As shown in Fig. 4C, Mip1 expression significantly inhibited MEKK2 dimer formation with either MEKK2 (342-619) or MEKK2 (342-619)(KM). Concomitantly, GST-MEKK2 (342-619) formed a complex with Mip1 (Fig. 4C). We also observed similar results when we used GST-MEKK2 (342-619) to pull down the full-length MEKK2 protein (data not shown). To further confirm that Mip1 is able to directly disrupt MEKK2 dimer formation, we purified GST-MEKK2, HA-MEKK2 (342-619), and GST-Mip1 and then examined if Mip1 could disrupt MEKK2 dimer formation. As shown in Fig. 4D, addition of GST-Mip1 but not control GST recombinant proteins significantly blocked MEKK2 dimer formation.
Mip1 interacts with nonphosphorylated and inactive MEKK2.
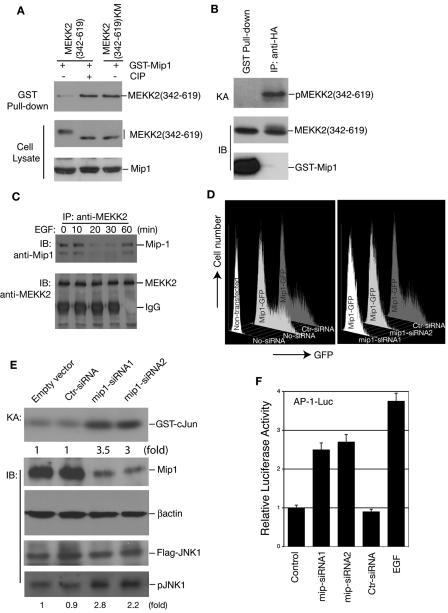
Transiently expressed MEKK2 (342-619) was a phosphor protein with a retarded mobility on an SDS-PAGE gel, whereas the kinase-inactive MEKK2 (342-619)(KM) mutant was nonphosphorylated, with only the band of fastest mobility (Fig. 4C). Protein phosphatase treatment of MEKK2 (342-619) led to its dephosphorylation so that it regained the fast mobility on an SDS-PAGE gel like the mutant MEKK2 (342-619)(KM) (Fig. 5A). Thus we were able to distinguish if the transiently expressed MEKK2 (342-619) protein was phosphorylated or not phosphorylated according to its mobility. Interestingly, we found that the interaction between Mip1 and MEKK2 (342-619) was significantly increased when MEKK2 (342-619) had been pretreated with a protein phosphatase compared to the untreated sample (Fig. 5A). In addition, more MEKK2 (342-619)(KM) than MEKK2 (342-619) was coprecipitated by Mip1 (Fig. 5A). Most importantly, we found that almost all the MEKK2 protein pulled down by Mip1 corresponded to the nonphosphorylated MEKK2 as shown by their fastest mobility on the SDS-PAGE gel, suggesting that Mip1 preferentially interacted with the nonphosphorylated, and likely inactive, MEKK2 (Fig. 5A).
FIG. 5.
Mip1 is a negative regulator of the MEKK2 signaling pathway. (A) Mip1 preferentially interacts with nonphosphorylated MEKK2. GST-Mip1 was cotransfected into COS-1 cells with expression vectors for HA-tagged MEKK2 (342-619) and MEKK2(342-619)KM for a GST pull-down assay, as indicated (top panel). For calf intestine phosphatase (CIP) treatment, cell lysates were prepared in the absence of phosphatase inhibitors and incubated with 1 unit/ml CIP at 37°C for 30 min before immunoblotting (IB) analysis. The levels of the MEKK2 and Mip1 proteins in the lysates were determined by IB (middle and bottom panels). (B) Mip1-associated MEKK2 is inactive. HA-tagged MEKK2 (342-619) was cotransfected with an empty vector or GST-Mip1 into COS-1 cells. The cells were lysed 36 h later and subjected to precipitation with glutathione beads or with an anti-HA antibody. The amounts of HA-tagged MEKK2 (342-619) precipitated by immunoprecipitation and by glutathione beads were determined by IB (middle panel). Equal amounts of HA-tagged MEKK2 (342-619) from each precipitation were used for an in vitro kinase assay (KA) (top panel). (C) EGF stimulation dissociates the endogenous Mip1-MEKK2 complex. Ten million 293T cells were either untreated or stimulated with EGF (25 ng/ml) for the times indicated before being lysed for immunoprecipitation (IP) with anti-MEKK2 antibody 8384. The immunocomplexes were thoroughly washed and separated on an SDS-PAGE gel for IB with anti-Mip1 antibody K87 (top panel) or anti-MEKK2 antibody 8384 (bottom panel). (D) mip1 siRNA specifically knocks down Mip1 gene expression. A Mip1-GFP expression vector was cotransfected with control siRNA (Ctr-siRNA) or with mip1-siRNA1 or mip1-siRNA2 expression vectors into 293T cells as indicated. Cells were harvested 48 h later and analyzed by flow cytometry for GFP-positive cells. Nontransfected cells with no siRNA (No-siRNA) were used as a baseline control whereas the Mip1-GFP transfections with No-siRNA or Ctr-siRNA were used as a positive control for GFP expression for the flow cytometry analysis (left graph). (E) The knockdown of Mip1 expression by siRNA induces JNK activity. Flag-JNK1 and HA-MEKK2 were cotransfected into 293T cells with Ctr-siRNA or mip1-siRNA1 or mip1-siRNA2 as indicated. The endogenous Mip1 protein levels were determined by IB with K87, and anti-β-actin IB was used as a loading control as indicated. The JNK1 activity was determined by an in vitro KA and by anti-phospho-JNK IB (pJNK1). The JNK1 level was determined by IB with an anti-Flag antibody (Flag-JNK1). (F) The knockdown of Mip1 expression activates AP-1 reporter gene expression. AP-1-Luc reporter plasmid and HA-MEKK2 were cotransfected into 293T cells with an empty vector (control) or with mip1-siRNA1 or mip1-siRNA2 or Ctr-siRNA as indicated. AP-1-Luc transfected alone stimulated with EGF was used as positive control. Relative luciferase activity was determined 48 h later and normalized to Renilla luciferase activity by using Promega's dual-luciferase system. The results shown are the averages of the results of three independent experiments.
If this were true, the MEKK2 that was associated with Mip1 should be inactive, thus unable to undergo autophosphorylation. To test this possibility, MEKK2 (342-619) was either coprecipitated by GST-Mip1 or immunoprecipitated with an anti-HA antibody, before being used for an in vitro kinase assay to determine its activity. As shown in Fig. 5B, although a significant amount of MEKK2 was pulled down by Mip1, the ability of this MEKK2 to autophosphorylate itself was significantly decreased, suggesting that the MEKK2 associated with Mip1 was inactive. In contrast, the MEKK2 precipitated by an anti-HA antibody was active and phosphorylated. These results thus further indicate that Mip1 inhibited MEKK2 signaling by binding to the catalytic domain of MEKK2, which prevented its dimerization and activation.
Epidermal growth factor stimulation dissociates the MEKK2-Mip1 complex.
If MEKK2 is negatively regulated through its binding to Mip1, Mip1 may need to dissociate from MEKK2 in order for MEKK2 to be activated during cell stimulation. To test this possibility, we examined the association of MEKK2 and Mip1 in nonstimulated cells and in cells treated with EGF for different times since EGF is a potent MEKK2 activator (13). Using an MEKK2-specific antibody, we then immunoprecipitated the endogenous MEKK2 from the unstimulated and stimulated cells and examined the Mip1 that was coprecipitated with MEKK2 by immunoblotting. In the absence of EGF treatment, we were able to detect the endogenous MEKK2 and Mip1 complex (Fig. 5C). However, after 10 min of EGF treatment, there was a significant decrease of the MEKK2-Mip1 complex. Interestingly, the MEKK2-Mip1 complex appeared to reform after 60 min of EGF stimulation, indicating that Mip1 may also be involved in the negative feedback regulation of MEKK2 signaling following cell stimulation (Fig. 5C).
Knockdown Mip1 expression in vivo activates the JNK-AP-1 pathway.
Although the above studies strongly suggest that Mip1 is a negative regulator of the MEKK2 signaling pathway, it is still possible that Mip1 might act as an adaptor molecule to positively control the MEKK2 signal transduction. However, if Mip1 could acts as an endogenous inhibitor, it is conceivable that, under normal conditions, most endogenous MEKK2 is either sequestered in specific intracellular compartments or associated with Mip1, thus preventing MEKK2 from forming dimers and activating itself. When MEKK2 is ectopically expressed, there may not be enough Mip1 in the cells to inhibit MEKK2 dimer formation and activation, thus allowing the nonspecific activation of the downstream JNK-AP-1 pathway. If this were the case, we would expect that the knockdown of Mip1 might allow MEKK2 to activate the JNK-AP-1 pathway. To test this possibility, we constructed mip1 siRNA expression vectors that target two independent mip1 coding sequences. As shown in Fig. 5D, the mip1 siRNAs but not the control siRNA specifically knocked down the expression of a cotransfected Mip1-GFP fusion protein. Using these two mip1 siRNAs, we knocked down the endogenous Mip1 protein expression (Fig. 5E) and examined how it would affect the JNK and AP-1 activation by an in vitro kinase assay and an AP-1 reporter assay. As shown in Fig. 5E and 5F, transfection of the mip1-specific siRNA but not control siRNA led to the augmented activation of both JNK and the AP-1 reporter gene. Together, these results confirm that Mip1 is a negative regulator of MEKK2 in vivo.
DISCUSSION
The existence of multiple MAP3Ks that are capable of activating multiple MAPKs within a single cell raises the question of how a specific extracellular cue can activate distinct repertoires of MAPK effectors (19, 28, 32). Different models based on large numbers of biochemical studies have been proposed to explain the specificity of the MAPK modules. The common feature of these models is that each MAPK module is able to form a unique multimolecular signaling complex through protein-protein interaction (7, 37). Such signaling complexes have been demonstrated in both yeast and mammalian cells (5, 11, 27, 39). However, one of the key questions regarding the specific activation of each MAPK module is how a MAP3K within a given MAPK module is first activated and regulated. Although ample evidence has suggested that MAP3Ks respond to numerous upstream stimuli, precisely how they are activated at the molecular level remains largely speculative. In yeast, there is genetic evidence of MAP3K kinase (such as Ste20) that appears to act upstream of MAP3Ks (10). However, such evidence remains elusive in the mammalian system. One of the major hurdles in studying the MAP3Ks in the mammalian system is that, unlike MAP2Ks or MAPKs, once over expressed, most MAP3Ks become active, thus preventing further analysis of how they are activated.
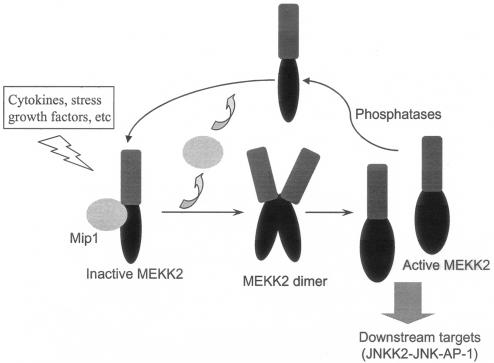
In this study, in which we attempted to elucidate the underlying molecular mechanisms governing the activation of the MAP3K MEKK2, we identified a MEKK2-interacting protein, Mip1, that interacts with the inactive MEKK2 and negatively regulates MEKK2 activation by preventing MEKK2 dimer formation. We recently found that MEKK2 dimer formation is a critical step for MEKK2 activation (6). The MEKK2 and Mip1 interaction essentially blocks the MEKK2 dimerization motif that is required for MEKK2 to form a dimer thus keeping it from being activated. Mip1 expression decreased MEKK2 activity and inhibited JNKK2, JNK1, ERK5, and AP-1 activation by MEKK2. In contrast, EGF stimulation led to a transient dissociation of MEKK2 from Mip1 suggesting that Mip-1 may control the basal activity of MEKK2. Furthermore, we found that the knockdown of Mip1 expression by siRNA resulted in the augmented activation of the MEKK2 downstream target JNK/AP-1 pathway. We have thus identified a novel mechanism for the negative regulation of MEKK2 involving an intracellular inhibitors as depicted in Fig. 6.
FIG. 6.
Model of MEKK2 regulation and activation.
Mip1 has not been characterized in mammalian cells before. A partial human sequence clone called JC310 was reported to suppress the defect resulting from a RAS2Val19 mutation in Saccharomyces cerevisiae (8). In addition, a chicken full-length cDNA sharing about 98% homology with JC310 with an unknown function in mammalian cells was isolated from the chicken hindbrain (38). The chicken cDNA was able to partially compensate for the defects caused by deletion of the yeast Mip1 ortholog Sin1 in S. pombe suggesting that Mip1 has a conserved function (38). The function of ySin1 was studied in more detail in yeast (38). ySin1 was found to be required for the function of yeast MAPK Sty-1 possibly through physical interaction although it is not a substrate of Sty-1. How ySin1 affects the Sty-1-mediated phosphorylation of Atf1 and the activation of yeast AP-1-responsive gene expression remains unclear. ySin-1 appeared to work at the MAPK Sty-1 level and did not interact with yeast MAP3K Wak1 (38). Yet, whether Sin1 interacts with and regulates other MAP3Ks such as Ste11, Ssk2p, and Ssk22p remains to be shown. In our study, we found that Mip1 plays a major role in MEKK2 signaling suggesting that it is capable of functioning at the MAP3K level in mammals. In S. cerevisiae, JC310 acted as a ras inhibitor, suggesting that Mip1 is capable of regulating the MAPK module at distinct levels.
Interestingly, the C terminal of chicken Sin1 was required to complement the loss of function of ySin-1 in yeast MAPK Sty-1 signaling, whereas, in our study, the N-terminal region was involved in regulating MEKK2 activation. Sequence analysis of Mip1 homologues from different species revealed several conserved regions in both the N and C termini of Mip1 suggesting that different regions of Mip1 have distinct functions in regulating the MAPK module (29). It is possible that in addition to its function as a MEKK2 regulator, Mip1 may also act as a scaffold protein to modulate the MAPK cascade similar to that of JIP1 in the MLK-MKK7-JNK module (44). In this regard, we found that Mip1 was also capable of interacting with JNK1/2 although this interaction seems not to facilitate JNK activation by MEKK2 nor does it affect the MEKK2-Mip1 interaction (data not shown). In addition, Mip1 appeared not to affect JNKK2-induced JNK activation. Future investigation on this issue may reveal a distinct function of Mip1 in the MAPK cascade regulation.
We mapped the motifs in MEKK2 that are required for MEKK2 dimerization and for Mip1 binding and discovered that the same motif in MEKK2 confers the ability both for MEKK2 to form dimers and for MEKK2 to interact with Mip1. These data may partially explain how Mip1 regulates MEKK2 activation; that is, an MEKK2-Mip1 complex prevents MEKK2 from being activated by occupying the motif required for MEKK2 dimer formation, an essential step for MEKK2 activation. At the molecular level, the interaction motif in Mip1 contains a unique coiled-coil structure, as predicted by computer modeling. Such a coiled-coil structure has been found in large numbers of proteins and is suggested to form a specific protein-protein interaction surface (2, 3). Thus, Mip1 may use this putative coiled coil structure to interact with MEKK2.
Although our present study established Mip1 as an important regulator of MEKK2 signaling, it is likely that Mip1 is also a critical regulator of MEKK3 since the Mip1 interaction motif in MEKK2 is conserved between MEKK2 and MEKK3. Indeed, a preliminary study confirmed that Mip1 and MEKK3 interact (data not shown). The use of mip1-specific siRNAs to knock down Mip1 in nonstimulated cells led to augmented JNK and AP-1 reporter gene expression, further suggesting that Mip1 is a naturally existing inhibitor of the MEKK2 and perhaps also the MEKK3 signaling pathways. Future studies on how Mip1 regulates the MEKK2 and MEKK3 pathways under normal physiological conditions should prove fruitful.
Acknowledgments
We thank Buna Wang and Guizi Sun for technical assistance, Tony Hunter for critical reading of the manuscript, X. Qin for providing pBS-U6 plasmid, and Beth Notzon for editing the manuscript.
This study was supported partially by grants AI44016 (NIH), HL070225 (NIH), and ARP (Texas Higher Education) (to B.S.) and Cancer Center Core grant CA16672 (M. D. Anderson Cancer Center).
REFERENCES
- 1.Blank, J. L., P. Gerwins, E. M. Elliott, S. Sather, and G. L. Johnson. 1996. Molecular cloning of mitogen-activated protein/ERK kinase kinases (MEKK) 2 and 3. Regulation of sequential phosphorylation pathways involving mitogen-activated protein kinase and c-Jun kinase. J. Biol. Chem. 271:5361-5368. [DOI] [PubMed] [Google Scholar]
- 2.Burkhard, P., J. Stetefeld, and S. V. Strelkov. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82-88. [DOI] [PubMed] [Google Scholar]
- 3.Chang, J. F., B. E. Hall, J. C. Tanny, D. Moazed, D. Filman, and T. Ellenberger. 2003. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure (Cambridge) 11:637-649. [DOI] [PubMed] [Google Scholar]
- 4.Chayama, K., P. J. Papst, T. P. Garrington, J. C. Pratt, T. Ishizuka, S. Webb, S. Ganiatsas, L. I. Zon, W. Sun, G. L. Johnson, and E. W. Gelfand. 2001. Role of MEKK2-MEK5 in the regulation of TNF-alpha gene expression and MEKK2-MKK7 in the activation of c-Jun N-terminal kinase in mast cells. Proc. Natl. Acad. Sci. USA 98:4599-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, J., J. Yang, Y. Xia, M. Karin, and B. Su. 2000. Synergistic interaction of MEK kinase 2, c-Jun N-terminal kinase (JNK) kinase 2, and JNK1 results in efficient and specific JNK1 activation. Mol. Cell. Biol. 20:2334-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J., L. Yu, D. Zhang, Q. Huang, D. M. Spencer, and B. Su. 2005. Dimerization through the catalytic domain is essential for MEKK2 activation. J. Biol. Chem. 280:13477-13482. [DOI] [PubMed] [Google Scholar]
- 7.Cobb, M. H. 1999. MAP kinase pathways. Prog. Biophys. Mol. Biol. 71:479-500. [DOI] [PubMed] [Google Scholar]
- 8.Colicelli, J., C. Nicolette, C. Birchmeier, L. Rodgers, M. Riggs, and M. Wigler. 1991. Expression of three mammalian cDNAs that interfere with RAS function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:2913-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deacon, K., and J. L. Blank. 1997. Characterization of the mitogen-activated protein kinase kinase 4 (MKK4)/c-Jun NH2-terminal kinase 1 and MKK3/p38 pathways regulated by MEK kinases 2 and 3. MEK kinase 3 activates MKK3 but does not cause activation of p38 kinase in vivo. J. Biol. Chem. 272:14489-14496. [DOI] [PubMed] [Google Scholar]
- 10.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3:573-581. [DOI] [PubMed] [Google Scholar]
- 11.Elion, E. A. 2001. The Ste5p scaffold. J. Cell Sci. 114:3967-3978. [DOI] [PubMed] [Google Scholar]
- 12.Ellinger-Ziegelbauer, H., K. Brown, K. Kelly, and U. Siebenlist. 1997. Direct activation of the stress-activated protein kinase (SAPK) and extracellular signal-regulated protein kinase (ERK) pathways by an inducible mitogen-activated protein kinase/ERK kinase kinase 3 (MEKK) derivative. J. Biol. Chem. 272:2668-2674. [DOI] [PubMed] [Google Scholar]
- 13.Fanger, G. R., N. L. Johnson, and G. L. Johnson. 1997. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 16:4961-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrington, T. P., T. Ishizuka, P. J. Papst, K. Chayama, S. Webb, T. Yujiri, W. Sun, S. Sather, D. M. Russell, S. B. Gibson, G. Keller, E. W. Gelfand, and G. L. Johnson. 2000. MEKK2 gene disruption causes loss of cytokine production in response to IgE and c-Kit ligand stimulation of ES cell-derived mast cells. EMBO J. 19:5387-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerwins, P., J. L. Blank, and G. L. Johnson. 1997. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J. Biol. Chem. 272:8288-8295. [DOI] [PubMed] [Google Scholar]
- 16.Guo, Z., G. Clydesdale, J. Cheng, K. Kim, L. Gan, D. J. McConkey, S. E. Ullrich, Y. Zhuang, and B. Su. 2002. Disruption of Mekk2 in mice reveals an unexpected role for MEKK2 in modulating T-cell receptor signal transduction. Mol. Cell. Biol. 22:5761-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammaker, D. R., D. L. Boyle, M. Chabaud-Riou, and G. S. Firestein. 2004. Regulation of c-Jun N-terminal kinase by MEKK-2 and mitogen-activated protein kinase kinase kinases in rheumatoid arthritis. J. Immunol. 172:1612-1618. [DOI] [PubMed] [Google Scholar]
- 18.Ichijo, H., E. Nishida, K. Irie, P. ten Dijke, M. Saitoh, T. Moriguchi, M. Takagi, K. Matsumoto, K. Miyazono, and Y. Gotoh. 1997. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science 275:90-94. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911-1912. [DOI] [PubMed] [Google Scholar]
- 20.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 21.Kesavan, K., K. Lobel-Rice, W. Sun, R. Lapadat, S. Webb, G. L. Johnson, and T. P. Garrington. 2004. MEKK2 regulates the coordinate activation of ERK5 and JNK in response to FGF-2 in fibroblasts. J. Cell. Physiol. 199:140-148. [DOI] [PubMed] [Google Scholar]
- 22.Lange-Carter, C. A., C. M. Pleiman, A. M. Gardner, K. J. Blumer, and G. L. Johnson. 1993. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science 260:315-319. [DOI] [PubMed] [Google Scholar]
- 23.Patriotis, C., A. Makris, S. E. Bear, and P. N. Tsichlis. 1993. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc. Natl. Acad. Sci. USA 90:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki, T., T. Wada, H. Kishimoto, J. Irie-Sasaki, G. Matsumoto, T. Goto, Z. Yao, A. Wakeham, T. W. Mak, A. Suzuki, S. K. Cho, J. C. Zuniga-Pflucker, A. J. Oliveira-dos-Santos, T. Katada, H. Nishina, and J. M. Penninger. 2001. The stress kinase mitogen-activated protein kinase kinase (MKK)7 is a negative regulator of antigen receptor and growth factor receptor-induced proliferation in hematopoietic cells. J. Exp. Med. 194:757-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer, B. C., M. F. Ware, P. Marrack, G. R. Fanger, J. W. Kappler, G. L. Johnson, and C. R. Monks. 1999. Live cell fluorescence imaging of T cell MEKK2: redistribution and activation in response to antigen stimulation of the T cell receptor. Immunity 11:411-421. [DOI] [PubMed] [Google Scholar]
- 27.Schaeffer, H. J., A. D. Catling, S. T. Eblen, L. S. Collier, A. Krauss, and M. J. Weber. 1998. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281:1668-1671. [DOI] [PubMed] [Google Scholar]
- 28.Schlesinger, T. K., G. R. Fanger, T. Yujiri, and G. L. Johnson. 1998. The TAO of MEKK. Front. Biosci. 3:D1181-D1186. [DOI] [PubMed] [Google Scholar]
- 29.Schroder, W., N. Cloonan, G. Bushell, and T. Sculley. 2004. Alternative polyadenylation and splicing of mRNAs transcribed from the human Sin1 gene. Gene 339:17-23. [DOI] [PubMed] [Google Scholar]
- 30.Su, B., J. Cheng, J. Yang, and Z. Guo. 2001. MEKK2 is required for T-cell receptor signals in JNK activation and interleukin-2 gene expression. J. Biol. Chem. 276:14784-14790. [DOI] [PubMed] [Google Scholar]
- 31.Su, B., E. Jacinto, M. Hibi, T. Kallunki, M. Karin, and Y. Ben-Neriah. 1994. JNK is involved in signal integration during costimulation of T lymphocytes. Cell 77:727-736. [DOI] [PubMed] [Google Scholar]
- 32.Su, B., and M. Karin. 1996. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr. Opin. Immunol. 8:402-411. [DOI] [PubMed] [Google Scholar]
- 33.Sun, W., K. Kesavan, B. C. Schaefer, T. P. Garrington, M. Ware, N. L. Johnson, E. W. Gelfand, and G. L. Johnson. 2001. MEKK2 associates with the adapter protein Lad/RIBP, and regulates the MEK5-BMK1/ERK5 pathway. J. Biol. Chem. 276:5093-5100. [DOI] [PubMed] [Google Scholar]
- 34.Sun, W., X. Wei, K. Kesavan, T. P. Garrington, R. Fan, J. Mei, S. M. Anderson, E. W. Gelfand, and G. L. Johnson. 2003. MEK kinase 2 and the adaptor protein Lad regulate extracellular signal-regulated kinase 5 activation by epidermal growth factor via Src. Mol. Cell. Biol. 23:2298-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teramoto, H., O. A. Coso, H. Miyata, T. Igishi, T. Miki, and J. S. Gutkind. 1996. Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase/stress-activated protein kinase pathway. A role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J. Biol. Chem. 271:27225-27228. [DOI] [PubMed] [Google Scholar]
- 36.Tibbles, L. A., Y. L. Ing, F. Kiefer, J. Chan, N. Iscove, J. R. Woodgett, and N. J. Lassam. 1996. MLK-3 activates the SAPK/JNK and p38/RK pathways via SEK1 and MKK3/6. EMBO J. 15:7026-7035. [PMC free article] [PubMed] [Google Scholar]
- 37.Weston, C. R., D. G. Lambright, and R. J. Davis. 2002. Signal transduction. MAP kinase signaling specificity. Science 296:2345-2347. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson, M. G., T. S. Pino, S. Tournier, V. Buck, H. Martin, J. Christiansen, D. G. Wilkinson, and J. B. Millar. 1999. Sin1: an evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 18:4210-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia, Y., Z. Wu, B. Su, B. Murray, and M. Karin. 1998. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 12:3369-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi, K., K. Shirakabe, H. Shibuya, K. Irie, I. Oishi, N. Ueno, T. Taniguchi, E. Nishida, and K. Matsumoto. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science 270:2008-2011. [DOI] [PubMed] [Google Scholar]
- 41.Yang, J., M. Boerm, M. McCarty, C. Bucana, I. J. Fidler, Y. Zhuang, and B. Su. 2000. Mekk3 is essential for early embryonic cardiovascular development. Nat. Genet. 24:309-313. [DOI] [PubMed] [Google Scholar]
- 42.Yang, J., Y. Lin, Z. Guo, J. Cheng, J. Huang, L. Deng, W. Liao, Z. Chen, Z. Liu, and B. Su. 2001. The essential role of MEKK3 in TNF-induced NF-κB activation. Nat. Immunol. 2:620-624. [DOI] [PubMed] [Google Scholar]
- 43.Yang, J., L. New, Y. Jiang, J. Han, and B. Su. 1998. Molecular cloning and characterization of a human protein kinase that specifically activates c-Jun N-terminal kinase. Gene 212:95-102. [DOI] [PubMed] [Google Scholar]
- 44.Yasuda, J., A. J. Whitmarsh, J. Cavanagh, M. Sharma, and R. J. Davis. 1999. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 19:7245-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]