Abstract
Detailed evaluation of gene functions in an asexual fungus requires advanced methods of molecular biology. For the generation of targeted gene deletions in the opportunistic pathogen Aspergillus fumigatus we designed a novel blaster module allowing dominant selection of transformants due to resistance to phleomycin as well as dominant (counter)selection of a Cre recombinase-mediated marker excision event. For validation purposes we have deleted the A. fumigatus pabaA gene in a wild-type isolate by making use of this cassette. The resulting pabaA::loxP strain served as the recipient for subsequent targeting of the velvet locus. Homologous reconstitution of the deleted gene was performed by an allele whose expression is driven in a nitrogen source-dependent manner, as validated by Northern analyses. Overexpression of the veA locus in A. fumigatus does not result in any obvious phenotype, whereas the sporulation capacities of the veA null mutant are reduced on nitrate-containing medium, a phenotype that is completely restored in the reconstituted strain.
With the advent of a steadily increasing number of microbial genome sequences, molecular tools to assign gene functions are a necessity in future basic research. Within the plethora of genome sequencing projects, fungal genomes have always been of special relevance: the ascomycete Saccharomyces cerevisiae was the first eukaryote whose genomic content was determined (23), lately complemented by efforts to resolve the complete genetic information from its filamentous ancestor Ashbya gossypii (15). The genome sequence of the model organism Neurospora crassa has been annotated recently (4, 21), and the genomes of three aspergilli, Aspergillus (Emericella) nidulans, A. fumigatus, and A. oryzae, are currently under comparative investigation.
One of the major obstacles in evaluation of any eukaryotic genome sequence lies in the identification of loci encoding potential gene products. Comparative BLAST studies may identify conserved loci that are likely to be expressed. Comparison of stretches of genomic sequences with entries in expressed sequence tag (EST) databases or cDNA sequences serves to identify expressed loci together with their exon/intron architecture (2). Gene functions might be assessed via identification of well-characterized orthologues from comprehensive databases; nevertheless, the function of a large fraction of the annotated genes from any newly determined genome sequence remains putative, as exemplified by the 41% of predicted Neurospora crassa proteins lacking significant similarity to gene products from the public databases (21).
A first glance on gene functions is achieved by the generation of null mutations in the organism under investigation, provided that adequate screening for phenotypes of interest is available. An established molecular biology is a vital prerequisite for controlled manipulation of any organism on the genomic level, and targeted gene manipulation by means of precise gene replacement has become the method of choice in addressing gene functions and characteristics. Genetic manipulation of deuteromycetous fungi is restricted due to the absence of any mode of sexual propagation, which would be accompanied by meiotic recombination. Therefore, genetic markers have to be introduced by transformation or by exploiting any parasexual cycle (20, 59). Furthermore, when addressing factors that might contribute to the virulence of a pathogen, dominance of the genetic markers is desirable to exclude effects that might be solely based on imperfect complementation of the genetic lesion (6).
Saprophytes of the genus Aspergillus have become one of the most relevant opportunistic fungal pathogens in present times (38). Based on advances in immunosuppressive medical treatment, the incidence of aspergilloses has increased steadily over the past decade (34), and the vast majority of them are caused by A. fumigatus spores. The pathogenicity of A. fumigatus is regarded as multifactorial, with a variety of cellular attributes contributing to the virulence potential of this asexual fungus (39). A. fumigatus has advanced to a practical level with standard procedures such as transformation, gene disruption and deletion, or integration of reporter fusions being well established (5). In virulence studies, the replacement of any gene under scrutiny by a dominant marker followed by homologous reconstitution with a silently mutated allele is useful to determine its potential as a virulence factor following the proposed molecular postulates of Koch (17, 18). Although the molecular techniques of genetic marker rescue and precise allelic replacement have been evaluated for Aspergillus (8, 12, 13, 35), a dominant system of selection and counterselection facilitating this task has not been described to date.
The annotation of the A. fumigatus genome sequence has uncovered gene loci formerly characterized in the model representative A. nidulans (46). Of special interest are genes that contribute to the sexual propagation mode or fruit body formation of the latter species (7), as their role in the life cycle of the asexual fungus A. fumigatus remains tentative. One prominent example is represented by the velvet locus (30). The nature of the veA gene determines the developmental capacities of A. nidulans. The Glasgow wild-type strain commonly inherits the wild-type allele of this gene, whereas most laboratory strains express a truncated version encoded by the veA1 allele. In A. nidulans the balance between asexual conidiophore formation and sexual fruit body differentiation is influenced by the nature of the velvet-encoded gene product. A wild-type isolate usually forms conidia on a growth substrate under illumination, and cleistothecium formation is preferred in the dark under conditions of restricted air exchange. For veA1 strains this dependency on distinct environmental conditions is partly diminished, as these strains conidiate profoundly in the absence of light but produce fruiting bodies in a delayed and reduced fashion (11). In its overall appearance, these veA1 mutant strains do not display the velvety appearance of older A. nidulans colonies, and, furthermore, they are blind for red light, which promotes conidiation in a wild-type background (43). Full deletion of the veA locus results in a completely acleistothecial phenotype accompanied by the secretion of a dark pigment, whereas overexpression of the veA gene product induces the formation of sexual structures under inappropriate conditions (32). Based on these and other findings, the VeA protein was designated a positive-acting factor of A. nidulans, promoting the sexual differentiation of this homothallic species. Recent studies have demonstrated a further role of the veA gene product in the regulation of secondary metabolism (10, 31), including the control of genes which directly or indirectly participate in carbohydrate metabolism, such as the mannoprotein-encoding gene mnpA (27) and fructosyl amine induction of the faoA gene, which encodes a fructosyl amino acid oxidase activity (28).
Here we address the role of the A. fumigatus-encoded velvet gene in the lifestyle of this obligate asexual fungus. By designing a suitable fungal marker module for dominant selection as well as counterselection (41) we aimed at the precise exchange of the endogenous A. fumigatus veA locus for a conditional allele by a two-step procedure of gene deletion and marker replacement. For safety reasons these studies were carried out by manipulating a strain of A. fumigatus that carries a complete deletion of the pabaA locus (8, 54) from which the reuseable marker module had been excised by Cre-mediated recombination (55). Inspection of the corresponding A. fumigatus strains revealed that overexpression of the veA gene does not result in any obvious developmental phenotype, whereas VeA is required for full sporulation capacity in the presence of nitrate as the sole nitrogen source.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The fungal strains used throughout this study are listed in Table 1. The clinical isolate D141 (50) served as the wild-type progenitor for all A. fumigatus strains constructed. Escherichia coli strains DH5α (61) and SURE (Stratagene) were employed for preparation of plasmid DNA and were propagated in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) in the presence of ampicillin at 100 μg ml−1. For recombinogenic engineering (recombineering) purposes (12), the bacterial strain KS272 carrying the pKOBEG plasmid was grown in low-salt LB medium with 0.5% NaCl in the presence of 25 μg ml−1 chloramphenicol. Minimal medium (0.52 g liter−1 KCl, 0.52 g liter−1 MgSO4, 1.52 g liter−1 KH2PO4, 0.1% trace element solution [29], pH 6.5) was used for growth of fungal strains, supplemented with appropriate amounts of 4-aminobenzoic acid (PABA, 1 μg ml−1), phleomycin (30 μg ml−1), 5-fluoro-2′-deoxyuridine (FUDR, 100 μM), or pyrithiamine (0.1 μg ml−1); 1% d-glucose was used as the source of carbon with 10 mM nitrogen source such as ammonium, supplemented as tartrate salt, or sodium nitrate.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| D141 | A. fumigatus wild type, clinical isolate | 50 |
| AfS11 | pabaA::loxP-phleo/tk | This study |
| AfS12 | pabaA::loxP | This study |
| AfS13 | veA::loxP-phleo/tk | This study |
| AfS15 | pabaA::loxP veA::loxP-phleo/tk | This study |
| AfS25 | pabaA::loxP pniiA::veA | This study |
Transformation procedures.
Protocols for E. coli were either for calcium- and manganese-treated cells (26) or for electroporation (56) with a Bio-Rad GenePulser at 2.5 kV in 0.2-cm cuvettes. A. fumigatus was transformed by polyethylene glycol-mediated fusion of protoplasts essentially as described (49).
Manipulation of nucleic acids.
Standard protocols of recombinant DNA technology were carried out (53). Recombination of DNA in E. coli employing the Redabγ system was executed following the procedure of Chaveroche et al. (12). Taq and Pfu polymerases were generally used in PCRs (52) and essential cloning steps were confirmed by sequencing on an ABI Prism 310 capillary sequencer. Fungal genomic DNAs were prepared from ground mycelia (33), and Southern analyses were carried out as described (57). Samples of total RNA were isolated using the TRIzol reagent of Invitrogen followed by Northern hybridization according to the protocols cited by Brown and Mackey (9). Random-primed labeling of hybridization probes was carried out with the Stratagene Prime-It II kit in the presence of [α-32P]dATP (19). To generate autoradiographs, the washed membranes were exposed to Kodak X-omat films. Sequence analyses were carried out using the Lasergene Biocomputing software package from DNAStar and alignments were created by the Lipman-Pearson method (40).
Plasmid constructions.
The plasmids used and constructed during the course of this study are listed in Table 2, accompanied by the oligonucleotide sequences recorded in Table 3. Plasmid pME2889 was constructed by inserting an XhoI/NotI fragment from pME2256 in pBluescript II KS (Stratagene). The phleomycin blaster cassette pME2891 was constructed in five steps: an 0.4-kb amplicon generated by PCR on pAN8-1 (49, GenBank accession number Z32751) with primer pair Sv95 (positions 2297 to 2320)/Sv85 (positions 2672 to 2655, plus additional nucleotide to create EcoRV half site) was inserted into the EcoRV site of cloning phagemid pBluescript II KS, followed by insertion of a 1.1-kb fragment amplified from a plasmid containing the coding sequence of herpes simplex virus type 1 (HSV1) thymidine kinase (42) with oligonucleotides Sv86 (positions +4 to +22 of coding sequence) and Sv87 (positions +1131 to +1113). From the resulting plasmid a 1.5-kb NcoI/XbaI fragment was released to replace a 1.1-kb NcoI/XbaI fragment in pAN8-1.
TABLE 2.
Plasmid constructs employed in this study
| Plasmid | Description | Reference |
|---|---|---|
| pBluescript II KS | General cloning plasmid (bla, multiple cloning site) | Stratagene |
| pUG6 | kanMX marker module (bla loxP-TEF2::kanr-loxP) | 25 |
| pSH47 | Yeast Cre expression plasmid (pGAL1::cre URA3 ARS/CEN) | 25 |
| pAN7-1 | Aspergillus marker cassette conferring resistance to hygromycin [pgpdA::hph::trpCt] | 49 |
| pAN8-1 | Aspergillus marker cassette conferring resistance to phleomycin [pgpdA::ble::trpCt] | 49 |
| pPTRII | Autonomously replicating Aspergillus plasmid (ptrA AMA1 bla) | Takara |
| pME2256 | Yeast plasmid pRS316 (URA3, ARS/CEN) with BglII site in polylinker | Collection of AG Braus |
| pME2889 | pBluescript II KS derivative carrying BglII site in polylinker | This study |
| pME2890 | A. nidulans pniaD-niaDt expression module in pBluescript II KS | This study |
| pME2891 | loxP-phleo/tk blaster (loxP-pgpdA::ble/HSV1 tk::trpCt-loxP) | This study |
| pME2892 | Cre expression module in pPTRII (A. nidulans niaD::cre ptrA AMA1) | This study |
| pME2893 | pabaA genomic locus as 9.0-kb BamHI/SacII fragment in pBluescript II KS | This study |
| pME2894 | pabaA deletion cassette (pabaA::loxP-phleo/tk) | This study |
| pME2895 | veA genomic locus as 6.7-kb BglII/NruI fragment in pME2889 (BglII/SmaI) | This study |
| pME2896 | veA deletion cassette (veA::loxP-phleo/tk) | This study |
| pME2897 | veA reconstitution allele for promoter insertion (pniiA::veA) | This study |
TABLE 3.
Oligonucleotides used in this study
| Designation | Sequence |
|---|---|
| Sv85 | 5′-ATC CTG CTC CTC GGC CAC G-3′ |
| Sv86 | 5′-GCT TCG TAC CCC TGC CAT-3′ |
| Sv87 | 5′-TCA GTT AGC CTC CCC CAT C-3′ |
| Sv88 | 5′-TTC ATT ACT TCT AGA CGG GTT CG-3′ |
| Sv89 | 5′-AAG AGG TAT TAC TAG TCT ACA GTG-3′ |
| Sv95 | 5′-TCA CCA TGG CCA AGT TGA CCA GTG-3′ |
| Sv160 | 5′-TCA CTC CCC TTT TGG TGT TCT CTG AGT ACC GTC GGA GCT TGC GGG CCA CAG CAA TTA GGC CAC TAG TGG ATC TG-3′ |
| Sv161 | 5′-TGA AGT ATT CAC CTC GAC GTC CAA AAC ATC ATC CAC AAG AAA GAA GAG ATG CTT GCA GCT GAA GCT TCG TAC GC-3′ |
| Sv233 | 5′-TAC ATT CGA AGA GTA CTA CAG CTG ATA ATG TCG GTA TAG-3′ |
| Sv234 | 5′-ATC GTT CGA AGT CTC AAC GCC AGT ATT GAC C-3′ |
| OLSK58 | 5′-TAT AGA ATT CAT GTC CAA TTT ACT GAC CG-3′ |
| OLSK59 | 5′-ATA TAG ATC TCT AAT CGC CAT CTT CCA GC-3′ |
Next, the trpC terminator sequence was inserted into this plasmid as a 0.7-kb BamHI/XbaI fragment isolated from pAN7-1 (49). From this construct the resulting 4.4-kb BglII/XbaI pgpdA::ble/tk::trpCt cassette was transferred to a derivative of pUG6 (25) from which the kanMX module had been removed after BglII/XhoI digestion, followed by Klenow treatment and religation.
The Cre recombinase expression plasmid pME2892 was assembled as follows. The coding sequence of the cre gene was amplified from pSH47 (25) (accession number AF298782) with primer pair OLSK58 (EcoRI site, positions 3673 to 3655)/OLSK59 (BglII, positions 2642 to 2660) and inserted into the EcoRV site of pME2890, which carries the A. nidulans niiA/niaD intergenic region as a 1.2-kb KpnI fragment (45) and the niaD 3′ region (accession number M58291) as an XbaI/SpeI amplicon from genomic DNA via Sv88 (positions 3303 to 3325) and Sv89 (positions 3588 to 3565). The resulting niaD::cre module was amplified by PCR and inserted into the SmaI site of the autonomously replicating Aspergillus plasmid pPTRII from Takara. Both constructs, pME2891 and pME2892, will be made available at the Fungal Genetics Stock Center (http://www.fgsc.net/).
Plasmids pME2893 and pME2895 were isolated from partial sublibraries created from genomic DNA of strain D141 that had been fragmented by the restriction endonucleases BamHI and SacII or BglII and NruI, respectively. In pME2894 the pabaA coding sequence was exchanged for the loxP-phleo/tk blaster module by recombineering linearized pME2893 with the PCR product of primers Sv160 and Sv161 on template pME2891 (12, 37). For construction of pME2896, the 4.5-kb HpaI/HindIII fragment of pME2891 was ligated to the Csp45I/HindIII backbone of pME2895 to yield the veA::loxP-phleo/tk deletion cassette. The pME2897 reconstitution allele to exchange the endogenous veA promoter sequence for the A. fumigatus niiA 5′ region was generated by insertion of a 1.2-kb PCR product amplified with primers Sv233 (BstBI, positions −1222 to −1194 relative to the niiA coding sequence) and Sv234 (BstBI, positions −10 to −30) from the genomic DNA of strain D141 into the unique BstBI site of plasmid pME2895.
Nucleotide sequence accession numbers.
pME2891 and pME2892 were submitted to GenBank under accession no. DQ023271 and DQ023272.
RESULTS
Novel blaster module facilitates subsequent gene deletions and reconstitution in Aspergillus fumigatus.
The molecular biology of the pathogen Aspergillus fumigatus is hampered by the lack of a sexual cycle. Introduction of precise deletions often calls for the reconstitution of the deleted locus in later steps, which is made difficult by the low frequency of homologous recombination in this filamentous fungus. Furthermore, the limited number of marker genes suitable for gene inactivation by gene replacement restricts the number of genetic lesions that can be introduced in a particular recipient strain. We therefore sought an approach to circumvent these general issues and to develop a generally applicable system suitable for targeted gene inactivation accompanied by the possibility of homologous reconstitution (Fig. 1).
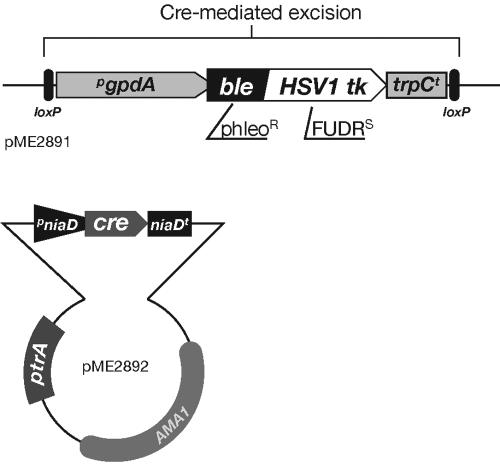
FIG. 1.
Plasmid constructs apt for A. fumigatus gene deletions and homologous reconstitution. Schematically depicted are the elements of plasmids pME2891 (top) and pME2892 (bottom). The marker module is a derivative of pAN8-1 and includes a translational fusion of the ble gene with the thymidine kinase-encoding sequence from herpes simplex virus type 1 (HSV1 tk). This chimeric sequence confers resistance to the antibiotic phleomycin (phleor) as well as sensitivity to nucleoside analogues such as 5-fluoro-2′-deoxyuridine (FUDRs). To accomplish excision by site-specific recombination, loxP acceptor sites were added on either edge of the cassette. Transient expression of the Cre recombinase in a nitrogen source-dependent manner can be achieved from the expression module integrated into the autonomously replicating vector pME2892, which carries the ptrA marker allele for plasmid maintenance in the presence of pyrithiamine.
In general, this task requires a marker suitable for dominant negative selection, and based on earlier studies in filamentous fungi, the suicide gene constituted by the thymidine kinase gene of herpes simplex virus type 1 seemed highly appropriate (24, 48, 51). The enzymatic activities encoded by such genes catalyze the phosphorylation of nucleoside analogues, which in turn interfere with the nucleotide metabolism of the host cell. Conclusively, the coding sequence of the HSV1 thymine kinase gene was combined with the ble gene sequence as a translational fusion to yield a chimeric dominant marker module allowing positive selection against the antibiotic phleomycin as well as negative selection against the antimetabolite 5-fluoro-2′-deoxyuridine (FUDR). To allow constitutive expression in the fungal host, this sequence stretch was positioned between the gpdA promoter sequence and trpC termination region of A. nidulans, as validated in the well-established expression module of pAN8-1 (49).
In order to expand the practicability of the novel marker we made marker rescue possible by the insertion of inverted repeats upstream and downstream of the genetic marker. These loxP sites serve as acceptor sites for the Cre recombinase and mediate excision of sequences sandwiched between them (55). To achieve controlled expression of the loxP-specific recombinase, an expression module placing the encoding cre gene between the A. nidulans niaD 5′ and 3′ regulatory sequences was constructed. This niaD::cre component was cloned into plasmid pPTRII, suitable for autonomous replication in Aspergillus species (36). Both plasmids, pME2891 and pME2892, constitute a novel system for dominant marker selection in a positive as well as negative manner, allowing us to employ the marker in a repeated fashion for subsequent targeting of multiple loci in Aspergillus fumigatus in addition to homologous reconstitution of deletion mutant strains (for a detailed protocol outline, see our website at http://wwwuser.gwdg.de/∼molmibio/organigr.htm).
Generation of an A. fumigatus pabaA::loxP strain.
In order to validate our marker system we targeted the pabaA locus of the wild-type clinical isolate D141. This locus encodes the para-aminobenzoic acid (PABA) synthetase, which converts chorismate, the last common intermediate of aromatic amino acid biosynthesis, into the folate precursor PABA. PABA-requiring Aspergillus mutant strains have been shown in earlier studies to be dramatically impaired in their virulence characteristics, and the pabaA gene product was regarded as an attractive drug target to counteract microbial pathogens (8).
The complete locus as deduced from the genome sequence was cloned in plasmid pME2895 from a genomic D141 sublibrary and served as the template in a recombineering approach, which is genetic engineering based on homologous recombination in an E. coli host strain expressing phage-derived proteins (44). For that purpose, the loxP-phleo/tk module of pME2891 was amplified with oligonucleotides that contained short homology arms of 50 nucleotides complementary to the 5′ and 3′ untranslated regions of the pabaA locus. Cotransformation of linearized plasmid pME2895 with the amplicon into the arabinose-induced E. coli host strain KS272/pKOBEG (12) yielded the recombined construct pME2894, in which the pabaA coding sequence had been replaced completely by the marker component. This replacement cassette with approximately 4.0-kb 5′ and 2.5-kb 3′ homologous flanking regions was introduced into strain D141, and primary transformants were selected on phleomycin-containing medium supplemented with PABA.
Detailed inspection of selected candidates after colony purification revealed two isolates that displayed the predicted phenotype: these strains only grow in the presence of the supplement PABA, exhibit resistance to the antibiotic phleomycin, and, furthermore, are impaired in growth on medium containing the nucleoside analogue FUDR (Fig. 2C). From Southern analysis, homologous replacement of the pabaA locus by the deletion marker cassette could be confirmed in both transformants (not shown), and one of them was chosen as representative AfS11 for further studies. Using AfS11 as the transformation recipient, the autonomously replicating plasmid pME2892 was put into action, and transformants were allowed to grow in the presence of pyrithiamine. To induce pniaD-driven expression of the cre gene, several of them were propagated on medium containing nitrate as the sole source of nitrogen, and clonal isolates from this lineage were screened for acquired FUDR resistance due to pop-out of the marker cassette. Out of 20 segregates, five displayed impaired growth on FUDR-containing medium, and all of these isolates were additionally scored as sensitive to phleomycin.
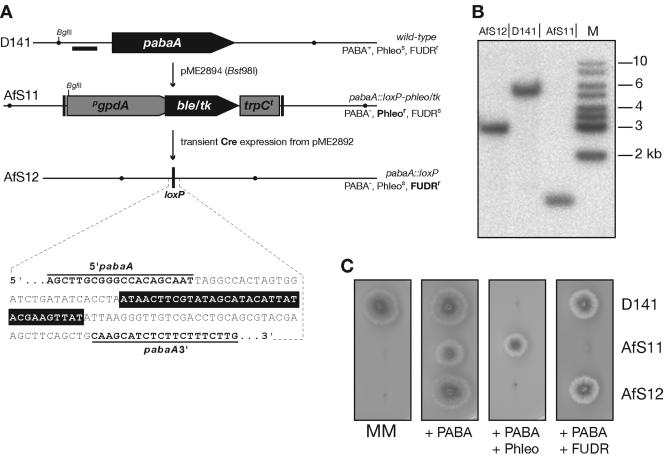
FIG. 2.
Deletion of the A. fumigatus pabaA locus followed by marker rescue. (A) Schematic representation of the pabaA gene as well as the deleted locus prior to and after excision of the loxP-phleo/tk marker module. Complete deletion of the pabaA coding sequence was achieved by transformation of the wild-type isolate D141 with a replacement cassette from pME2894. Recombination between the flanking loxP sites (vertical bars) and rescue of the genetic marker was carried out by transient expression of the Cre recombinase after transformation with pME2892. BglII restriction sites are indicated by dots; the black horizontal bar indicates the position of the probe used in hybridization experiments. The strains' phenotypes corresponding to the resulting genotypes are indicated. The sequence extract from the genomic pabaA::loxP lesion as determined by sequence analyses is given below with pabaA-specific 5′ and 3′ stretches in bold and the loxP inverted repeat boxed in black. (B) Southern analysis of D141, AfS11, and AfS12. Equivalent genomic DNA was subjected to BglII digestion and hybridization signals were detected employing a 5′-specific probe as specified. Fragment sizes of the DNA standard (M) are indicated. (C) Growth phenotypes of the resulting strains D141, AfS11, and AfS12. Conidia were plated on minimal medium (MM) supplemented with 4-aminobenzoic acid (PABA), phleomycin (Phleo), or 5-fluoro-2′-deoxyuridine (FUDR) and grown at 37°C.
When plating conidia of AfS11 at various densities on FUDR-containing medium, no colonies could be grown (not shown), indicating the absence of spontaneous recombination between the flanking loxP sites. Furthermore, the pabaA::loxP lesion from two FUDRs isolates after Cre expression was amplified from the genome and sequenced to validate precise recombination between the loxP sites. Identical sequences from both isolates demonstrated the presence of one loxP site flanked by 5′ and 3′ pabaA sequences, as expected (Fig. 2A). To check for loss of the expression plasmid, conidia from these two isolates were streaked out on supplemented medium without the selective agent, and single colonies from these plates were scored for resistance to pyrithiamine. All of these descendants were sensitive to pyrithiamine, indicating a high rate of plasmid loss when selective pressure is relieved. As a representative, one isolate was chosen, and this strain, AfS12, clearly exhibits the phenotype expected, which is proper growth in the presence of FUDR as well as sensitivity to phleomycin, accompanied by PABA auxotrophy (Fig. 2C). Correspondingly, the correct genotypes of both strains, AfS11 and AfS12, could be confirmed in Southern hybridization experiments (Fig. 2B).
In summary, we were able to replace a target gene of A. fumigatus by the novel dominant marker module and successfully rescued the genetic marker after precise excision mediated by Cre/loxP recombination. The resulting pabaA::loxP strain can serve as the recipient in any subsequent gene targeting procedure.
veA locus is conserved among aspergilli.
To exploit the characteristics of the loxP-phleo/tk marker module further we aimed at the deletion of an additional A. fumigatus gene in the pabaA::loxP background of strain AfS12. As gene of interest the veA locus was chosen, based on its significance for fruiting body formation in the homothallic relative A. nidulans (43). By inspection of the genome sequence of the A. fumigatus isolate Af293, a gene locus could be identified that carries a coding sequence with significant similarity to the veA gene of A. nidulans. As deduced from the BLAST results that were obtained by submission of this sequence stretch, this genetic locus holds the capacity to encode a polypeptide of 570 amino acids, which is encoded by two exons that are separated by one intronic sequence of 79 nucleotides (Fig. 3A).
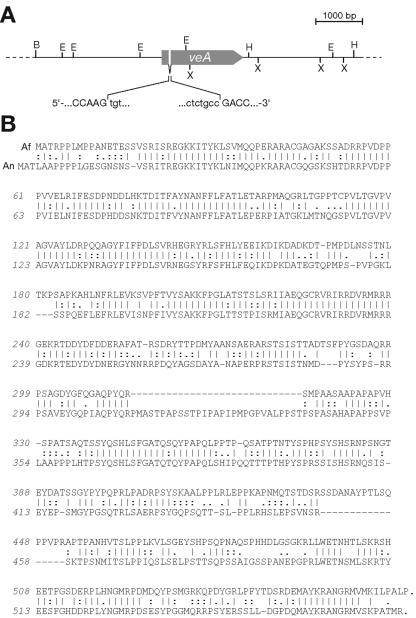
FIG. 3.
A. fumigatus veA gene is conserved. (A) Genomic organization of the A. fumigatus veA open reading frame and its environment. Restriction sites depicted are BglII (B), EcoRV (E), HindIII (H), and XhoI (X); exon/intron boundaries in the veA coding sequence are illustrated. (B) Global alignment of the deduced VeA amino acid sequences from A. fumigatus (Af) and A. nidulans (An). Identical residues are indicated by vertical bars, conservative replacements by colons, and neutral changes by periods.
In accordance with these in silico assumptions, a complete cDNA could be amplified by reverse transcription-PCR that confirmed the absence of this intron in the processed veA transcript of A. fumigatus. The veA gene product shows a high degree of conservation with its A. nidulans counterpart (Fig. 3B). In the alignment based on the Lipman-Pearson algorithm, the similarity index was calculated to be 53.3%. Only two stretches, one in each deduced amino acid sequence, interfere with proper alignment, which may hint at different molecular characteristics of the proteins. Nevertheless, when transformed into an A. nidulans recipient strain deleted of its veA locus, the A. fumigatus gene restored the phenotypes originating from the genetic lesion (not shown): these reconstituted isolates did profoundly form cleistothecia but did not excrete a dark brown pigment, which is highly characteristic of A. nidulans mutants impaired in fruiting body formation. Therefore, the A. fumigatus veA gene represents a true functional orthologue of the A. nidulans velvet locus.
veA gene product is required for proper conidiation of A. fumigatus.
To assess any functional role of the A. fumigatus-encoded VeA in the life style of this asexual fungus, we aimed at inactivation of the encoding gene (Fig. 4A). For this purpose, regions encompassing 2.7 kb of the 5′ and 2.2 kb of the 3′ region of the target locus were cloned to border the loxP-phleo/tk marker. The resulting cassette from pME2896 was then transformed into strain AfS12 with its loxP::pabaA genetic background, and selection was carried out on PABA- as well as phleomycin-containing medium. Inspection of the transformant pool by Southern analysis revealed several isolates in which the endogenous veA gene had been replaced by the dominant marker (Fig. 4B) to yield strain AfS15.
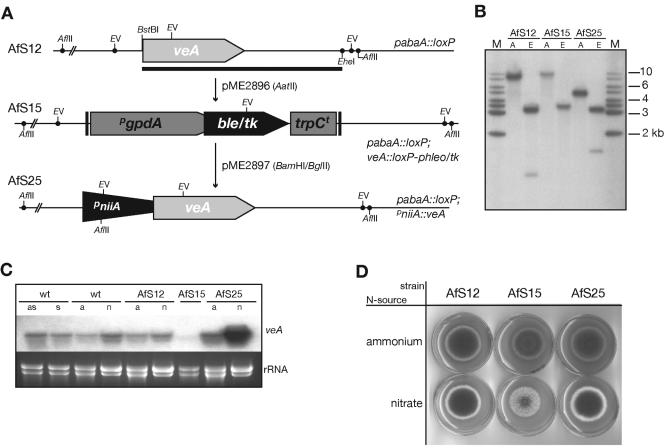
FIG. 4.
Deletion and allelic exchange of the A. fumigatus veA gene. (A) Schematic representation of the veA open reading frame as well as the deleted locus before and after replacement of the loxP-phleo/tk marker module. Complete deletion of the veA coding sequence was achieved by transformation of the pabaAΔ strain AfS12 with a replacement cassette from pME2896. Exchange by the niiA promoter-driven allele from pME2897 could be selected by resistance of strain AfS25 to FUDR. Restriction sites are indicted; the horizontal bar indicates the position of the probe used in hybridization experiments. (B) Southern analysis of strains D141, AfS15, and AfS25. Corresponding genomic DNA was digested with AflII (A) or EcoRV (E) and hybridization signals were detected employing a veA-specific probe as specified. Fragment lengths of the DNA size standard (M) are indicated. (C) Northern analysis of A. fumigatus strains for validation of the allelic exchange. Strains were propagated in the presence of nitrogen sources ammonium (a) or nitrate (n); additionally, induction of asexual (as) or sexual (s) differentiation as established for A. nidulans was applied for wild-type (wt) strain D141 by incubation with illumination or in the absence of light accompanied by restricted air exchange, respectively. Steady-state levels of the veA transcript are shown; ethidium bromide-stained rRNA signal served as a loading control. (D) Developmental characteristics of A. fumigatus strains expressing VeA at various levels. Equal amounts of spores were inoculated on medium supplemented with ammonium or nitrate as the sole nitrogen source and allowed to spread out at 37°C for 3 days.
To demonstrate and exploit the negative selection capacities of the novel marker module, we further intended to reconstitute this strain by integration of an alternative veA construct at the homologous gene locus. Therefore, the niiA promoter sequence of A. fumigatus was inserted upstream of the presumed veA start codon to result in an allele whose expression can be adjusted by the added nitrogen source (1). This reconstitution cassette, covering the same flanking regions as pME2896, was transformed into the pabaA::loxP veAΔ deletion mutant, and transformants were screened for reconstituted strains based on their growth phenotype on FUDR-containing medium. Whereas the recipient strain AfS15 grew poorly on FUDR control plates, several single sporulating colonies were visible among the transformants on this medium. Further inspection of one of these isolates (AfS25) by Southern hybridization confirmed homologous replacement of the veA::loxP-phleo/tk lesion by the pniiA-driven veA allele (Fig. 4B).
The strains generated were scrutinized by Northern analysis to estimate the steady-state levels of the veA transcript (Fig. 4C). For the veAΔ strain AfS15, no signal could be detected, confirming the deletion genotype of this strain. When propagated in liquid minimal medium containing the rich nitrogen source ammonium, the veA transcript is abundant in the wild-type strain D141 as well as in the pabaA::loxP descendant AfS12. When transferred onto solid ammonium-containing medium, a clear veA transcript could be detected for D141, irrespective of whether conditions that support asexual sporulation (aeration and illumination) or fruiting body formation (taping of plates and incubation in the dark) in A. nidulans were applied. Growing the fungus in the presence of the poor nitrogen source nitrate resulted in veA transcript levels similar to those for ammonium-grown strains D141 and AfS12. For the reconstituted strain AfS25, clear regulation of veA expression was detected due to the nitrate-inducible niiA promoter that precedes the veA coding sequence: when propagated in the presence of ammonium; basal expression of the veA transcript could be monitored in Northern experiments, whereas induction of veA transcription was evident when nitrate was the sole source of nitrogen. Conclusively, overexpression of the veA gene under conditions of nitrate feeding had been accomplished by allelic replacement employing the versatile loxP::phleo/tk marker module.
Ultimately, the set of A. fumigatus strains generated were put on solid medium containing either ammonium or nitrate as the nitrogen source to assess any phenotypes linked to altered veA expression levels (Fig. 4D). Whereas no obvious growth phenotype could be observed for strain AfS25, which carries the pniiA::veA allele, in comparison to its progenitor AfS12 on either type of medium, a clear reduction in sporulation capacities was observed for AfS15, which lacks the veA coding sequence. This diminution in the formation of asexual conidia was more pronounced in the presence of nitrate as the sole source of nitrogen. A rough estimation of spore numbers by counting the conidia from equal-sized plugs of mycelia yielded an approximately twofold decrease in spore quantity in the presence of ammonium and a ≈10-fold reduction when nitrate had to be utilized by the fungus, each time compared to the wild-type strain grown on the same nitrogen source (data not shown). Moreover, this cut in sporulation capacity is not based on inappropriate supplementation with PABA or interference of the pabaA::loxP lesion with the veA deletion, as a strain deleted only of its veA locus (AfS13) displayed the same phenotype as strain AfS15 (not shown). This interesting phenotype indicates an influence of the nitrogen source on asexual sporulation of A. fumigatus, which seems to be balanced by the veA gene product. Moreover, our data indicate that sheer overexpression of the gene orthologous to A. nidulans veA is not sufficient to trigger any similar but cryptic developmental pathway in the asexual fungus A. fumigatus.
DISCUSSION
In our aim to enhance the molecular biology of the opportunistic pathogen A. fumigatus we have designed and validated a novel marker system allowing gene replacement accompanied by marker rescue. Alternatively, based on its negative selection capacities, reconstitution or allelic exchange of target genes is facilitated by this marker module. The combination of several genetic functions—positive as well as negative selection together with acceptor sites for recombination—in a single marker module is a novel and applicable concept for the generation of genetic lesions in A. fumigatus. Because genetic markers are limited in this particular host, the concept of multiple marker usage, as proven before in A. nidulans (13, 35), was adapted in our studies.
The basic idea of recombinase-assisted marker rescue has been applied with great success in studying gene families in bakers' yeast (14), e.g., in the construction of Saccharomyces cerevisiae mutants impaired in hexose transport (60). Here, the highly redundant functions of more than 20 loci were eliminated by rounds of gene deletion and marker rescue, resulting in a strain completely devoid of any hexose transporter activity. In putting up the comprehensive Euroscarf (Frankfurt, Germany) yeast mutant collection, a recyclable blaster cassette was employed to facilitate downstream genetic manipulation of any deletion strain. With the genome sequence of A. fumigatus in hand, follow-up functional studies will have to include the generation of comprehensive knockout mutant collections, too.
The marker system developed may serve as a standard tool in these molecular studies for several reasons. (i) Dominance of the marker gene that replaces the gene of interest is important. Selection systems based on complementation of auxotrophies are generally hampered by the risk of incomplete complementation resulting in interfering phenotypes (3, 6). Especially in the in vivo situation of virulence studies, this may lead to biased results. (ii) The option of marker rescue is of great advantage when studying gene families encoding redundant products. Furthermore, the genetic interactions of several gene lesions can be explored, a task that, due to the deficiency of sexual crossing techniques, has been prevented in A. fumigatus until now. (iii) The marker facilitates the homologous reconstitution of any targeted gene irrespective of an expressed strong phenotype that would allow direct selection. Clear reconstitution of a mutant strain is vital for phenotypic characterization and to prove that the nature of any observed phenotype is based solely on the lack of the gene locus and not on additional mutations.
However, we cannot exclude that expression of the viral thymine kinase might result in unwanted side effects that interfere with proper propagation in an infected host animal. On the other hand, the marker enables the direct comparison of a deletion strain carrying the complete marker module with a deletion strain in which the marker has been removed to leave a single loxP site, and future virulence studies will have to address this issue. Also, we seek to downsize the marker module by testing alternative promoter sequences that support expression of the resistance fusion protein at sufficient levels. Additionally, the necessity to aim at high-throughput processing when constructing deletion cassettes for the generation of a comprehensive mutant collection calls for automation of the cloning processes. Recombineering has evolved as the method of choice for purposes such as this, and we will explore the possibility of implementing this option in the novel marker module.
In our aim to provide a proof-of-concept we have deleted the pabaA locus of an A. fumigatus wild-type strain followed by marker excision. As indicated by other gene deletion experiments (our unpublished data), the low frequency of homologous recombination when deleting the pabaA locus is more likely due to the target locus than to the marker module. Precise excision of the marker module by the Cre/loxP system could be confirmed by sequencing of the genomic lesion, and the necessity for expression of the site-specific recombinase was demonstrated. As an alternative to plasmid-delivered Cre expression, electroporation of the recipient strain with purified, recombinant recombinase would facilitate the marker rescue procedure. Nevertheless, due to the likelihood of genetic rearrangements in applying this particular transformation method to A. fumigatus (20), we have not taken this into account. Furthermore, transformation of proteins via electroporation is not validated for aspergilli to our knowledge, so controlled and transient action of Cre appears more feasible by employing the expression plasmid.
When scoring the events of marker rescue, a frequency of 25% (5 out of 20) was observed. As this appears rather low, we have optimized the procedure by selecting primary pME2892 transformants on nitrate-containing medium and plating harvested conidia directly on FUDR plates without focusing on single isolates. Fast-sporulating clones can then clearly be identified on these plates and scrutinized further. Therefore, by screening a large number of descendants in this manner, segregates lacking the marker module can easily be isolated.
The resulting pabaA::loxP strain could facilitate molecular studies in this pathogen, as Aspergillus strains auxotrophic for the vitamin precursor PABA have been shown to be completely avirulent in several virulence testing models. This particular strain might serve as a safety strain when investigating basic features of A. fumigatus besides pathogenicity. One of the topics that have always been of interest is the apparent lack of any sexual cycle in the A. fumigatus lifestyle. Earlier studies stated a close relationship of the anamorph A. fumigatus and the teleomorph Neosartorya fischeri, but these organisms represent distinct species (22). So far, the conditions under which any cryptic sexual mode of propagation is executed by A. fumigatus have not been revealed.
Comparative genome studies have uncovered a variety of genetic features in this fungus that were characterized in other fungal species as required for mating, meiosis, or the formation of fruiting bodies, but their role in the life cycle of A. fumigatus remains enigmatic (16, 47, 58). One of these factors is the veA gene product. In this study we were able to demonstrate that the veA gene of the asexual fungus A. fumigatus is expressed and that it encodes a functional protein. By allelic exchange of the locus in A. fumigatus we were able to induce the expression of the veA gene in its original host under inappropriate conditions. Given the highly conserved genomic structure of the Aspergillus veA loci, the absence of any obvious phenotype when overexpressed in A. fumigatus was unexpected. However, this indicates that the genetic program of A. nidulans cleistothecium formation is not completely mirrored in the asexual relative and that a single factor might not be sufficient to trigger this cryptic developmental process in A. fumigatus.
Detailed examination of the fungal genomes of three aspergilli has uncovered the existence of two mating type idiomorphs, MAT-1 and MAT-2, with either one being present in the A. fumigatus genome, whereas both are encoded by the genome of the homothallic species A. nidulans (58). Accordingly, the presence of both mating type loci in one vegetative thallus might be a strict prerequisite that determines mating ability in aspergilli to ensure proper communication between compatible mating partners. As a logical consequence, the sheer overexpression of positive regulators of sexual development might be insufficient to initiate the cellular program in the absence of activating upstream signals such as pheromone perception.
Furthermore, we could show that VeA is likely to function in the nitrogen metabolism of A. fumigatus, a role that has not been described for its A. nidulans counterpart. The reduced sporulation capacity of an A. fumigatus veAΔ strain in the presence of ammonium was accentuated by feeding nitrate. Chae and coworkers tested the sporulation capacity of their A. nidulans veA deletion strain DVAR1 on different media all containing the rich nitrogen source ammonium (32). However, the A. nidulans veA gene product was shown to influence the ratio of the α and β transcripts synthesized from the brlA locus (31), a function that indicates an influence of the sexual effector on asexual sporulation. A veA deletion strain of the aflatoxin-producing fungus Aspergillus parasiticus was characterized to exhibit reduced conidial production, in particular when grown on its natural substrate peanut seed (10). This could specify a conserved role for the VeA factor in sporulation under adverse nutritional conditions. Comparative studies in the postgenomic era will have to address the nature of the numerous attributes that are directed by the veA gene product in different species of Aspergillus, as this conserved factor constitutes a key regulator of cellular processes in this highly relevant fungal genus.
Acknowledgments
We thank Christian Monnerjahn for the generous gift of an HSV1 thymidine kinase-encoding sequence. The excellent technical assistance of Verena Grosse is highly appreciated, and we value the work of Roland Graf during the initial phase of the project. All members of the department are thanked for assistance, helpful advice, and inspiring discussions.
Funding was received from the Deutsche Forschungsgemeinschaft and its priority program SPP 1160 (KR 2294/1-1) as well as from the Volkswagenstiftung and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Amaar, Y. G., and M. M. Moore. 1998. Mapping of the nitrate-assimilation gene cluster (crnA-niiA-niaD) and characterization of the nitrite reductase gene (niiA) in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Genet. 33:206-215. [DOI] [PubMed] [Google Scholar]
- 2.Ayoubi, P., X. Jin, S. Leite, X. Liu, J. Martajaja, A. Abduraham, Q. Wan, W. Yan, E. Misawa, and R. A. Prade. 2002. PipeOnline 2.0: automated EST processing and functional data sorting. Nucleic Acids Res. 30:4761-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baganz, F., A. Hayes, D. Marren, D. C. Gardner, and S. G. Oliver. 1997. Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast 13:1563-1573. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brakhage, A. A., and K. Langfelder. 2002. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 56:433-455. [DOI] [PubMed] [Google Scholar]
- 6.Brand, A., D. M. MacCallum, A. J. Brown, N. A. Gow, and F. C. Odds. 2004. Ectopic Expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braus, G. H., S. Krappmann, and S. E. Eckert. 2002. Sexual Development in Ascomycetes-Fruit Body Formation of Aspergillus nidulans, p. 215-244. In H. D. Osiewacz (ed.), Molecular Biology of Fungal Development. Marcel Dekker, Inc., New York, NY.
- 8.Brown, J. S., A. Aufauvre-Brown, J. Brown, J. M. Jennings, H. Arst, Jr., and D. W. Holden. 2000. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol. Microbiol. 36:1371-1380. [DOI] [PubMed] [Google Scholar]
- 9.Brown, T., and K. Mackey. 1997. Analysis of RNA by Northern and slot blot hybridization., p. 4.9.1-4.9.16. In Current protocols in molecular biology. John Wiley and Sons Inc., New York, NY. [DOI] [PubMed]
- 10.Calvo, A. M., J. Bok, W. Brooks, and N. P. Keller. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ Microbiol. 70:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champe, S. P., M. B. Kurtz, L. N. Yager, N. J. Butnick, and D. E. Axelrod. 1981. Spore formation in Aspergillus nidulans: competence and other developmental processes, p. 255-276. In H. R. Hohl and G. Turian (ed.), The fungal spores: morphogenic controls. Academic, New York, NY.
- 12.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.d'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 14.Delneri, D., G. C. Tomlin, J. L. Wixon, A. Hutter, M. Sefton, E. J. Louis, and S. G. Oliver. 2000. Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene 252:127-135. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 16.Dyer, P. S., M. Paoletti, and D. B. Archer. 2003. Genomics reveals sexual secrets of Aspergillus. Microbiology 149:2301-2303. [DOI] [PubMed] [Google Scholar]
- 17.Falkow, S. 2004. Molecular Koch's postulates applied to bacterial pathogenicity-a personal recollection 15 years later. Nat. Rev. Microbiol. 2:67-72. [DOI] [PubMed] [Google Scholar]
- 18.Falkow, S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10:S274-276. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 20.Firon, A., A. Beauvais, J. P. Latge, E. Couve, M. C. Grosjean-Cournoyer, and C. d'Enfert. 2002. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics 161:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 22.Girardin, H., M. Monod, and J. P. Latge. 1995. Molecular characterization of the food-borne fungus Neosartorya fischeri (Malloch and Cain). Appl. Environ. Microbiol. 61:1378-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546-567. [DOI] [PubMed] [Google Scholar]
- 24.Grivell, A. R., and J. F. Jackson. 1968. Thymidine kinase: evidence for its absence from Neurospora crassa and some other micro-organisms, and the relevance of this to the specific labelling of deoxyribonucleic acid. J. Gen. Microbiol. 54:307-317. [DOI] [PubMed] [Google Scholar]
- 25.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 27.Jeong, H. Y., H. Kim, D. M. Han, K. Y. Jahng, and K. S. Chae. 2003. Expression of the mnpA gene that encodes the mannoprotein of Aspergillus nidulans is dependent on fadA and flbA as well as veA. Fungal Genet. Biol. 38:228-236. [DOI] [PubMed] [Google Scholar]
- 28.Jeong, H. Y., M. H. Song, J. H. Back, D. M. Han, X. Wu, V. Monnier, K. Y. Jahng, and K. S. Chae. 2002. The veA gene is necessary for the inducible expression by fructosyl amines of the Aspergillus nidulans faoA gene encoding fructosyl amino acid oxidase (amadoriase, EC 1.5.3). Arch. Microbiol. 178:344-350. [DOI] [PubMed] [Google Scholar]
- 29.Käfer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 30.Käfer, E. 1965. Origins of translocations in Aspergillus nidulans. Genetics 52:217-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato, N., W. Brooks, and A. M. Calvo. 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, H., K. Han, K. Kim, D. Han, K. Jahng, and K. Chae. 2002. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37:72-80. [DOI] [PubMed] [Google Scholar]
- 33.Kolar, M., P. J. Punt, C. A. van den Hondel, and H. Schwab. 1988. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62:127-134. [DOI] [PubMed] [Google Scholar]
- 34.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161-172. [DOI] [PubMed] [Google Scholar]
- 35.Krappmann, S., and G. H. Braus. 2003. Deletion of Aspergillus nidulans aroC using a novel blaster module that combines ET cloning and marker rescue. Mol. Genet. Genomics 268:675-683. [DOI] [PubMed] [Google Scholar]
- 36.Kubodera, T., N. Yamashita, and A. Nishimura. 2002. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 66:404-406. [DOI] [PubMed] [Google Scholar]
- 37.Langfelder, K., S. Gattung, and A. A. Brakhage. 2002. A novel method used to delete a new Aspergillus fumigatus ABC transporter-encoding gene. Curr. Genet. 41:268-274. [DOI] [PubMed] [Google Scholar]
- 38.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latge, J. P. 2001. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 9:382-389. [DOI] [PubMed] [Google Scholar]
- 40.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 41.Lupton, S. D., L. L. Brunton, V. A. Kalberg, and R. W. Overell. 1991. Dominant positive and negative selection using a hygromycin phosphotransferase-thymidine kinase fusion gene. Mol. Cell. Biol. 11:3374-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monnerjahn, C., and M. Konrad. 2003. Modulated nucleoside kinases as tools to improve the activation of therapeutic nucleoside analogues. Chem. Biochem. 4:143-146. [DOI] [PubMed] [Google Scholar]
- 43.Mooney, J. L., and L. N. Yager. 1990. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 4:1473-1482. [DOI] [PubMed] [Google Scholar]
- 44.Muyrers, J. P., Y. Zhang, and A. F. Stewart. 2000. ET-cloning: think recombination first. Genet. Eng. (N.Y.) 22:77-98. [DOI] [PubMed] [Google Scholar]
- 45.Osherov, N., J. Mathew, and G. S. May. 2000. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet. Biol. 31:181-188. [DOI] [PubMed] [Google Scholar]
- 46.Pain, A., J. Woodward, M. A. Quail, M. J. Anderson, R. Clark, M. Collins, N. Fosker, A. Fraser, D. Harris, N. Larke, L. Murphy, S. Humphray, S. O'Neil, M. Pertea, C. Price, E. Rabbinowitsch, M. A. Rajandream, S. Salzberg, D. Saunders, K. Seeger, S. Sharp, T. Warren, D. W. Denning, B. Barrell, and N. Hall. 2004. Insight into the genome of Aspergillus fumigatus: analysis of a 922 kb region encompassing the nitrate assimilation gene cluster. Fungal Genet. Biol. 41:443-453. [DOI] [PubMed] [Google Scholar]
- 47.Pöggeler, S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42:153-160. [DOI] [PubMed] [Google Scholar]
- 48.Pratt, R. J., and R. Aramayo. 2002. Improving the efficiency of gene replacements in Neurospora crassa: a first step towards a large-scale functional genomics project. Fungal Genet. Biol. 37:56-71. [DOI] [PubMed] [Google Scholar]
- 49.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447-457. [DOI] [PubMed] [Google Scholar]
- 50.Reichard, U., S. Buttner, H. Eiffert, F. Staib, and R. Rüchel. 1990. Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. J. Med. Microbiol. 33:243-251. [DOI] [PubMed] [Google Scholar]
- 51.Sachs, M. S., E. U. Selker, B. Lin, C. J. Roberts, Z. Luo, D. Vaught-Alexander, and B. S. Margolin. 1997. Expression of herpes virus thymidine kinase in Neurospora crassa. Nucleic Acids Res. 25:2389-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saiki, R. K., T. L. Bugawan, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1986. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature 324:163-166. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Sandhu, D. K., R. S. Sandhu, Z. U. Khan, and V. N. Damodaran. 1976. Conditional virulence of a p-aminobenzoic acid-requiring mutant of Aspergillus fumigatus. Infect. Immun. 13:527-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauer, B. 1987. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song, S., T. Zhang, W. Qi, W. Zhao, B. Xu, and J. Liu. 1993. Transformation of Escherichia coli with foreign DNA by electroporation. Chin. J. Biotechnol. 9:197-201. [PubMed] [Google Scholar]
- 57.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 58.Varga, J. 2003. Mating type gene homologues in Aspergillus fumigatus. Microbiology 149:816-819. [DOI] [PubMed] [Google Scholar]
- 59.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]
- 60.Wieczorke, R., S. Krampe, T. Weierstall, K. Freidel, C. P. Hollenberg, and E. Boles. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123-128. [DOI] [PubMed] [Google Scholar]
- 61.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]