Abstract
The genetic investigation of Campylobacter jejuni, an important gastrointestinal pathogen, has been hampered by the lack of an efficient system for introduction of exogenous genetic information, as commonly used vectors designed for Escherichia coli and other bacteria cannot be maintained in Campylobacter cells. Additionally, gene expression in Campylobacter requires the presence of species-specific promoters. In this study we exploited the availability of several conserved copies of rRNA gene clusters for insertion of various genes into the chromosome by homologous recombination. The high conservation of the rRNA sequences means that the procedure can be applied to other Campylobacter strains. The presence of a Campylobacter-derived promoter in this vector ensures expression of exogenous genes in target cells. The efficiency of the procedure was demonstrated by complementation of mutations in two strains of Campylobacter. In addition, we applied the system for introduction and expression of a green fluorescent protein (GFP). GFP-expressing Campylobacter allowed visualization of sessile bacteria attached to a glass surface in stationary liquid culture. The study demonstrated that the attached bacteria contained an assemblage of coccoid and spiral forms with liquid channels preserving viable highly motile cells. We demonstrate a novel universal procedure for gene delivery and expression that can be used as an efficient tool to study this poorly understood pathogen. The principles developed in this study could be more widely applied for the manipulation of other bacteria that are refractory to genetic analysis.
Despite some reduction in the number of cases linked to infection with Campylobacter, it remains one of the major causes of gastrointestinal disease worldwide (3). Genetic studies on the physiology and virulence of Campylobacter have become more efficient since the publication of the complete genomic sequence of Campylobacter jejuni strain NCTC 11168 (25). Subsequent studies resulted in the discovery of heterogeneity in gene content and genetic variations among various strains of C. jejuni (10). One of the key mechanisms of such diversity is a remarkable propensity of the bacterium to undergo phase variation and recombination (13, 15), combined with many strains being naturally transformable with exogenous DNA (31). The marker rescue technique developed for Campylobacter cells (20) has now become the most common approach used in mutagenesis of these bacteria.
Although insertional mutagenesis is now routine, the introduction and expression of exogenous genes into Campylobacter cells remain a major problem, as many strains, including the sequenced strain NCTC 11168, are refractive to complementation and gene expression. Although some reports described shuttle vectors for the complementation of mutations in trans and for introduction and expression of a gfp gene, their use is often inefficient and is limited to certain strains (23, 24). For example we have been unable to introduce shuttle vectors pRY112 (34), pMW10 (32), pGUO0202 (1), and pMEK91 (24) into various C. jejuni strains, including the sequenced strain NCTC 11168 and its derivative 11168H.
In this study we describe an efficient procedure for gene delivery and expression, which overcomes shuttle vector-related limitations. The method has been successfully tested in a number of applications that have facilitated our understanding of this important pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The following C. jejuni strains were used in this study: 11168H (HS:2), which is a hypermotile derivative of strain NCTC 11168 (15), and 81-176 (HS:23/36, enteritis isolate used in human challenge studies) (4). Escherichia coli strain XL2-MRF′ (Stratagene, La Jolla, CA) was used in cloning experiments. C. jejuni was grown under microaerobic conditions in an incubator (85% N2, 10% CO2, 5% O2) for 2 days at 37°C on Columbia base agar (Oxoid, Basingstoke, United Kingdom) supplemented with 6% horse blood. E. coli was grown on Luria-Bertani agar (Oxoid, Basingstoke, United Kingdom). Where necessary, kanamycin or ampicillin was added to a concentration of 50 μg ml−1 or 100 μg ml−1, respectively. Motility was tested on 0.4% Mueller-Hinton agar (Oxoid, Basingstoke, United Kingdom). For investigation of bacteria attached to glass cover slides in stationary cultures, a 11168H strain expressing a green fluorescent protein (GFP) was first grown in brucella broth medium (Oxoid, Basingstoke, United Kingdom) supplemented with chloramphenicol at 15 μg ml−1 on a rotatory platform at 90 rpm for 2 days and then diluted 1:20 with the same medium and incubated for 4 days without shaking in flasks containing glass coverslips. The coverslips were removed and mounted on a glass slide for visualization under a confocal microscope.
Electroporation.
The electroporation procedure was carried out as follows. A 2-day bacterial plate culture was resuspended in buffer containing 272 mM sucrose and 15% glycerol at 0oC and washed three times with the same buffer, and 50-μl aliquots were used for each transformation. After addition of DNA (0.5 μg in 5 μl), the mixture was transferred to an ice-cold electroporation cuvette. Electroporation was performed at 2.5 kV, 200 Ω, and 25 μF, after which 100 μl of SOC buffer were added to the cuvette, and the bacterial suspension was transferred onto a nonselective blood agar plate. After overnight incubation at 37°C under microaerobic conditions, bacteria were spread onto a selective plate and incubated for a further 3 to 4 days.
Growth competition test.
The growth competition index (CI) was determined as described previously (8). Two-day agar cultures of the Campylobacter Camr derivative 11168H/pRRC4 and the wild-type recipient 11168H, grown with or without chloramphenicol (10 μg/ml), respectively, were inoculated into separate tissue culture flasks containing 10 ml of brucella broth without chloramphenicol. After incubation for a further 2 days on a rotary platform at 90 rpm under microaerobic conditions, the optical density at 600 nm was measured and serial dilutions were plated onto agar plates with (11168H/pRRC4) or without (11168H) chloramphenicol. A 100-μl aliquot of liquid culture of the Camr derivative (2.8 × 104 CFU) was mixed with an equal volume of wild-type strain 11168H (3.5 × 104 CFU) in a flask with fresh brucella broth (10 ml), which corresponded to an input Camr/Cams ratio of 1.25. After incubation for two days, serial dilutions were plated onto blood agar plates with and without chloramphenicol in triplicates. The colonies were counted, and the output Camr/Cams ratio was calculated. The CI was determined as output Camr/Cams ratio divided by input Camr/Cams ratio.
General cloning techniques.
Plasmids used in this study are listed in Table 1. Restriction enzymes were purchased from either Promega (Southampton, United Kingdom) or New England Biolabs (Hitchin, United Kingdom). T4 DNA ligase, T4 DNA polymerase, and calf thymus alkaline phosphatase were purchased from Promega (Southampton, United Kingdom). Oligonucleotides (Table 2) were from Sigma-Genosys (Pampisford, United Kingdom). Standard restriction digestion, DNA ligation, and cloning procedures were essentially as described elsewhere (26). T4 DNA polymerase treatment for generation of blunt ends was performed according to the manufacturer's protocol (Promega, Southampton, United Kingdom).
TABLE 1.
Plasmids
| Plasmid | Description | Source (reference) |
|---|---|---|
| pRY112 | Shuttle vector | P. Guerry (34) |
| pMW10-13 | Shuttle vector containing wild-type gfp gene | E. Gaynor (unpublished) |
| pMEK91 | Shuttle vector containing egfp-pMEK under control of ompE promoter | M. E. Konkel (24) |
| pAV35 | Source of Camr gene cassette | A. van Vliet (29) |
| pJMK30 | Source of Kanr gene cassette | A. van Vliet (29) |
| pGUO0202 | Shuttle vector | V. Korolik (1) |
| pGEM-T Easy | Cloning vector | Promega, Southampton, United Kingdom |
| pEGFP | Contains egfp gene | BD Biosciences Clontech, Palo Alto, CA |
| pRR | Fragment of rRNA gene cluster cloned into pGEM-T Easy | This study |
| pRRC | Camr cassette cloned into pRR | This study |
| pRPGL | pglH gene in pRRC | This study |
| pRMAF | maf5 gene in pRRC | This study |
| pRGW | Wild-type gfp gene in pRRC | This study |
| pRED | egfp gene from pEGFP with added optimal SD sequence cloned into pRRC | This study |
| pREM | PompE-egfp fusion from pMEK91 in pRRC | This study |
TABLE 2.
Primers useda
| Primer | Sequencea | Descriptionb |
|---|---|---|
| ak231 | CTGGAACTCAACTGACGCTAAG | rrs (16S rRNA) (dir) |
| ak232 | CTCTTGCACATTGCAGTCCTAC | rrl (23S rRNA) (rev) |
| ak233 | GCAAGAGTTTTGCTTATGTTAGCAC | Cj0029 (dir) |
| ak234 | GAAATGGGCAGAGTGTATTCTCCG | Cj0431 (dir) |
| ak235 | GTGCGGATAATGTTGTTTCTG | Cj0742 (dir) |
| ak237 | TCCTGAACTCTTCATGTCGATTG | Camr gene cassette (up) |
| ak248 | GCTCTAGACTTAAAGAGGAGAAATGATGAAAATAAGC | pglH (dir) |
| ak249 | GCTCTAGATCATTAGGCATTTTTAACCTCGGCTATAAGC | pglH (rev) |
| ak261 | GGACTAGTAGGAGATTTAAATGGTGAGCAAGGGCGAGGAGCTGTTCAC | egfp from pEGFP (dir) |
| ak262 | GCTCTAGAAGGCCTTTACTTGTACAGCTCGTCCATGCCGAGAGTG | egfp from pEGFP (rev) |
| ak244M | GCTCTAGAAAGGAAGATAAATGGATGGAAAGGGTGAGAAGGTG | maf5 (dir) |
| ak245M | GCTCTAGATTAAAGTGCTTTTTTCTTTTCTAAGAAG | maf5 (rev) |
| ak238 | GCTTGCATCTGATAAAGCACCTG | Cj0863 (xerD) primer, reverse |
| DL3 | ACCCAGCGAACCATTTGAGG | Kanr gene cassette-specific primer |
| ak290 | GCTTTTAGTTTATAAGACAAAAACCAAAAGAG | rrs (16S rRNA) gene-derived primer for sequencing of insertion sites |
The start codons are shown in boldface; the regions complementary to Campylobacter 16S rRNA (SD sequences) are underlined.
dir, direct primer; rev, reverse primer; up, upward primer.
Construction of the delivery vector pRRC.
The rRNA region containing fragments of genes encoding 16S and 28S rRNAs (rrs and rrl genes, respectively) was PCR amplified using primers ak231 and ak232 (Fig. 1) and cloned into pGEM-T Easy vector (Promega, Southampton, United Kingdom) to produce pRR. The pRR plasmid was digested with XbaI, made blunt ended using T4 DNA polymerase, and ligated with a blunt-ended BamHI fragment of plasmid pAV35 (29) containing a Camr gene cassette. After transformation into E. coli, a recombinant plasmid containing a chloramphenicol resistance gene inserted in the same orientation as the rRNA genes was selected and designated pRRC.
FIG. 1.
A, Organization of the three rRNA gene clusters in strain NCTC 11168; the location of the intergenic XbaI site and positions of primers ak231 and ak232 used for PCR amplification are shown. B, A fragment of plasmid pRR containing the ak231/ak232 PCR product cloned into pGEM-T Easy vector. C, A fragment of plasmid pRRC derived from pRR via insertion of the Camr gene cassette. D, Three possible products of allelic replacement resulting from recombination of pRRC with the genome; the primers used for localization of insertion sites (ak233, ak234, ak235, and ak237) are shown. E, Genes inserted into the chromosome in this study. Blunt-ended DNA fragments are indicated by a “b” in the name of a restriction site; the arrowhead in the egfp-pMEK construct represents and additional promoter (PompE). The genes are shown as thick black arrows (not to scale). Open arrows represent the Camr gene. Open boxes indicate vector regions. Locations and directions of PCR primers and promoters are indicated by solid and open arrowheads, respectively.
Construction of delivery vectors for gene expression in C. jejuni. (i) pglH gene.
The pglH gene was PCR amplified using primers ak248 and ak249, and the product was digested with XbaI enzyme and cloned into XbaI-digested pRRC plasmid to produce pRPGL in such a way that pglH gene was transcribed in the same orientation as the Camr gene (Table 1; Fig. 1), which was verified via restriction analysis.
(ii) maf5 gene.
The maf5 gene was PCR amplified using primers ak244 M and ak245 M, and the product was digested with XbaI enzyme and cloned into XbaI-digested pRRC plasmid to produce pRMAF in such a way that the maf5 gene was transcribed in the same orientation as the Camr gene, which was verified via restriction analysis. The forward primer (ak244 M) contained a Shine-Dalgarno (SD) region optimized for Campylobacter (Fig. 1; Table 1).
(iii) Wild-type gfp gene.
Plasmid pMW10-13 (Table 1) containing the wild-type gfp gene was digested with XbaI and ClaI. The fragments were made blunt ended using T4 DNA polymerase and ligated to blunt-ended vector pRRC after digestion with XbaI (Fig. 1; Table 1). A recombinant clone containing pRRC with the wild-type gfp gene transcribed in the same direction as the Camr gene cassette was selected. A plasmid with the correct orientation of the wild-type gfp gene was selected via restriction analysis. Expression of GFP was confirmed via visualization of colonies with a fluorescence microscope.
(iv) egfp-SD gene.
Primers ak261 and ak262 were used to PCR amplify the enhanced GFP gene (egfp) from the pEGFP plasmid (Fig. 1; Table 1). The primer ak261 contained a Campylobacter SD sequence. The PCR product was digested with XbaI enzyme and cloned into vector pRRC so that the gene encoding the enhanced green fluorescent protein with the Campylobacter-specific SD sequence (egfp-SD) was under the control of Camr gene promoter as verified by restriction analysis.
(v) egfp-pMEK gene.
The EcoRI fragment of plasmid pMEK91 (24) containing the efgp gene (23) under the control of the PompE promoter of C. jejuni was blunt ended and inserted into XbaI-digested and blunt-ended vector pRRC (Fig. 1; Table 1). By using restriction analysis, a recombinant plasmid containing the gene encoding enhanced green fluorescent protein from plasmid pMEK91 (egfp-pMEK) under the control of two promoters, Pcam and PompE, was selected. E. coli colonies harboring this plasmid appeared yellow when visualized by eye under daylight and appeared bright green under the fluorescence microscope (excitation at 488 nm).
Construction of xerD mutant.
Plasmid cam71a9, constructed during the course of the C. jejuni genome sequencing project (25), contains a 1.5-kb DNA insert with a fragment of gene xerD (Cj0863). After insertion of a blunt-ended BamHI fragment of pJMK30 (29) containing a Kanr gene cassette into the unique SwaI site within the xerD gene on plasmid cam71a9 and transformation into 11168H, Kanr clones were selected. Insertion of the Kanr gene cassette in a nonpolar orientation was confirmed by PCR with primers ak238 and DL3 (Table 2).
Gel electrophoresis and Western blotting.
Bacteria were resuspended in sample buffer and incubated at 100°C for 10 min, and the lysate was analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 12.5% precast polyacrylamide gels (Invitrogen, Paisley, United Kingdom). Gels were blotted onto a polyvinylidene difluoride membrane (Millipore, Watford, United Kingdom), blocked in phosphate-buffered saline containing 0.5% Tween 20 (PBST) for at least 30 min, and incubated with biotinylated soybean agglutinin (Vector Laboratories, Burlingame, CA) at a concentration of 10 to 20 μg ml−1 in PBST for 1 hour. Following three brief washes in PBST, blots were incubated with extravidin peroxidase (Sigma-Aldrich, Poole, United Kingdom) diluted 1 in 1,000 in PBST for 30 min. Following a further three brief washes in PBST, blots were developed using a diaminobenzidine staining kit with nickel enhancement according to the manufacturer's instructions (Vector Laboratories, Burlingame, CA). Alternatively, the blots were probed with Penner 6 typing antiserum (1:100 dilution), followed by treatment with anti-rabbit immunoglobulin G peroxidase conjugate (Sigma-Aldrich, Poole, United Kingdom) at a 1:1000 dilution. All antibody dilutions were made using Tris-buffered saline containing 0.01% Tween 20 and supplemented with 1% bovine serum albumin (Sigma). The blots were developed using the diaminobenzidine staining kit with nickel enhancement according to the manufacturer's instructions (Vector Laboratories, Burlingame, CA). Broad-range molecular weight standards were supplied by New England Biolabs (Hitchin, United Kingdom).
Confocal microscopy.
Bacterial cells were visualized using a Carl Zeiss LSM 510 confocal microscope according to the manufacturer's manual. The laser excitation wavelength was 488 nm, and the detection wavelength was 505 nm.
RESULTS
Transformation of C. jejuni by using shuttle vectors.
The shuttle vectors pMW10-13, pRY112, pMEK91, and pGUO0202 (Table 1) were used for transformation into C. jejuni NCTC 11168 via natural transformation and electroporation, using standard protocols (31). Both electroporation and natural transformation protocols are very efficient tools for making insertional mutants via allelic replacement in many strains of C. jejuni, including NCTC 11168. However, repeated attempts with any of the available shuttle vectors and strain NCTC 11168 (or its derivative 1118H) resulted in no colonies on selective media.
Construction of the delivery vector.
In order to minimize the disruption of rRNA genes, a spacer region between the 16S and 28S rRNA genes was selected. The genome of C. jejuni strain NCTC 11168 carries three identical clusters of rRNA genes. An XbaI site located immediately downstream from 16S rRNA genes was selected as an insertion site (Fig. 1A). A region containing this site with long flanking regions was PCR amplified and cloned into pGEM-T Easy vector to produce plasmid pRR (Fig. 1B). Insertion of the blunt-ended BamHI fragment containing the Camr gene into the blunt-ended XbaI site of pRR resulted in plasmid pRRC (Fig. 1C). A unique XbaI site of pRRC downstream from the Camr gene was used for cloning other genes, so that, if inserted in the correct orientation, these genes would be under the control of the constitutively expressed Camr gene promoter.
The Camr gene can be inserted into any of the three rRNA clusters.
The pRRC vector was transformed into C. jejuni strain 11168H via electroporation, and Camr colonies were selected. The transformants were analyzed using the Camr gene-specific PCR primer ak237 and three other primers (ak233, ak234, and ak235) corresponding to the regions adjacent to the three potential insertion sites on the chromosome (Fig. 1D). The results of the analysis demonstrated random recombination with any of the three rRNA clusters (Fig. 2).
FIG. 2.
Insertion of the Camr cassette into various rRNA clusters. PCR analysis with primers ak233/ak237, ak234/ak237, and ak235/ak237 of 10 insertion derivatives (lanes 1 to 10) resulting from transformation of strain 11168H with pRRC is shown. Lane 11, negative control (no DNA); lane 12, molecular size standards.
The minor bands present in some insertion derivatives possibly result from the migration of the cassette between the rRNA clusters.
As can be seen in Fig. 2, in some cases PCR products with more than one primer pair could be detected. In order to find out whether the additional bands resulted from the simultaneous presence of the Camr gene cassette in two or more clusters in the genome or from the heterogenous population of the cell culture, we selected three typical derivatives and performed subcloning. Three subclones of these derivatives were analyzed by PCR with three primer pairs (Fig. 1), each specific to one of the possible integration sites.
The subclones of isolate A (Fig. 3) produced the same band patterns as the original strains, with one major band with the primer pair ak234/ak237 and minor bands of variable yields with the two other primer pairs. The presence of the minor bands suggested occasional migration of the Camr gene cassette from the primary insertion site into one of the two other rRNA clusters after subcloning.
FIG. 3.
PCR analysis of three insertion derivatives A, B, and C (lanes O) and respective subclones (lanes 1 to 3) with primers ak233/ak237, ak234/ak237, and ak235/ak237. Lane 4, molecular size standards.
In the case of isolate B, it was possible to select subclones producing a PCR product with just one primer pair (ak235/ak237) (Fig. 3), indicating that the minor bands were not due to insertion of the Camr gene cassette into multiple sites of the same cell but were due to heterogeneity of bacterial population.
In contrast, all subclones of isolate C produced strong bands with two primer pairs, ak234/ak235 and ak235/ak237, indicating that both the original isolate and its subclones contained the cassette integrated into two rRNA clusters on the same chromosome. The presence of minor bands with the ak233/ak237 primer pair also suggested migration of the cassette into another rRNA cluster (Fig. 1) in a subpopulation of bacterial cells, similarly to what was observed with derivatives A and B (see above). The authenticity of these minor bands to the respective rRNA cluster was confirmed by gel extraction followed by sequencing with Camr primer ak290 (Table 2) (data not shown).
Allelic replacement in C. jejuni is not affected by the product of the xerD gene.
Many strains of C. jejuni are capable of highly efficient double recombination events; the mechanism involved is unknown. One of the candidate genes that might be involved in such recombination was the xerD (Cj0863) gene. In E. coli XerD acts in concert with XerC and is involved in segregation of chromosome strands after replication (12). It was also shown that the XerC and XerD proteins of E. coli are required for filamentous phage integration (14).
The xerD mutant of 11168H was transformed via electroporation by plasmid pRRC, and Kanr clones were selected and allelic replacement confirmed by PCR. The mutant was then transformed with pRRC and, following selection of Camr Kanr clones, the chromosomal DNA was analyzed for the presence of Camr gene- and vector-specific sequences using PCR. The Camr gene was found to be inserted into an rRNA gene cluster near gene Cj0029 (Fig. 1D). However, no vector-derived sequences could be detected (data not shown), indicating that the allelic replacement does not require expression of the XerD protein.
Integration of the Camr gene cassette into the spacer region of the rRNA gene cluster does not affect growth in liquid culture.
Preliminary experiments demonstrated no difference in growth rates in liquid cultures (brucella broth) or in colony sizes on solid medium (blood agar plates) between the recipient strain 11168H and the Camr derivatives. For a more detailed comparative analysis of growth rates, in vitro competition studies were carried out as described elsewhere (8). One randomly selected Camr derivative, 11168H/pRRC4, with the Camr gene inserted into cluster III (Fig. 1) was grown in a mixture with the wild-type strain 11168H. The CI determined after 2 days of incubation (CI = 0.89) indicated that the insertion of the Camr gene cassette into the XbaI site of the rRNA cluster does not have a dramatic effect on cell viability under the conditions used. Similar results were obtained with the derivatives containing the Camr gene cassette inserted into clusters I and III (Fig. 1) and even with the derivative containing a Camr-gfp (wild-type gfp) fusion inserted into two clusters (II and III) simultaneously (data not shown).
Complementation of the pglH mutation in strain 11168H.
It was shown previously that insertional inactivation of the pglH gene affects protein glycosylation (21). In order to check whether the integration system described in this study can restore protein glycosylation in this mutant, the pglH gene was PCR amplified and inserted into the pRRC vector to produce pRPGL (Table 1). After transformation of the 11168H/pglH::Kanr mutant with plasmid pRPGL, Kanr Camr clones were selected. PCR analysis confirmed integration of the pglH gene into one of the rRNA gene clusters. Protein glycosylation in the derivatives was found to be completely restored (Fig. 4), indicating expression of a functionally active PglH protein.
FIG. 4.
Restoration of protein glycosylation after complementation of the pglH mutation. Lanes 1, wild-type strain 11168H; lanes 2, 11168H/pglH::Kanr mutant; lanes 3, complementation derivative 11168H/pglH::Kanr/pRPGLH1. After electrophoresis of whole-cell lysates, the gels were stained with Coomassie blue (A) or probed with either Pen2 antiserum (B) or biotinylated soybean agglutinin (C). D, molecular size standards.
Complementation of the maf5 (Cj1337) mutation in strain 11168H.
It was shown previously that mutation of the maf5 gene results in the loss of motility (15). However, the putative Maf5 protein does not show significant sequence similarity to any other protein with a known function, and the role of this protein in motility remains uncertain. Remarkably, in strain NCTC 11168 there are six other genes with close similarity of the respected putative products to Maf5. Some of these genes contain variable poly(G) tracts and are involved in phase variation of motility. Expression of a functionally active Maf5 protein, which could be simply monitored via the restoration of motility in the maf5 mutant, both in the intact form and as His-tagged derivatives, would assist in understanding the function of this new family of motility-related proteins.
Complementation of mutants in motility studies is especially important because of a large number of genes involved in flagellar formation and motility (33). Spontaneous mutations in these genes may also result in reduced motility in a specific mutant, thus skewing the effect of a primary mutation. In our previous study we overcame this problem by analyzing several independent clones of the maf5 mutation in various strains (15). However, a possibility of direct complementation of the mutation would be advantageous.
The maf5 gene was cloned into the pRRC vector, and the resulting plasmid pRMAF (Table 1) was transformed into the maf5 mutant described previously (15). The integration of the maf5 gene into an rRNA cluster was confirmed by PCR. The motility of the Kanr Camr derivatives was restored (although partially [see Discussion]) compared with that of the mutant (Fig. 5), indicating that the cells produced a functionally active Maf5 protein.
FIG. 5.
Restoration of motility after complementation of the maf5 mutation after growth for 3 days on 0.4% Mueller-Hinton agar. 1, 11168H/maf5::Kanr; 2,11168H/maf5::Kanr/pRMAF; 3, 11168H. A similar picture of restoration of motility was observed after complementation of such mutations in strain 81-176.
Complementation of a maf5-like mutation in strain 81-176.
In order to demonstrate the applicability of the integration system to other strains of C. jejuni, we have used complementation of the maf5-like mutation in strain 81-176. The amino acid sequences of the respective gene products from strains NCTC 11168 and 81-176 reveal 55% identity (GenBank accession number AY102622). The maf5 mutant of 81-176 was created using a protocol similar to that for constructing the maf5 mutant of strain 11168H (15) (A. V. Karlyshev et al., unpublished). As expected, the 81-176/maf5::Kanr mutant was nonmotile. Complementation of this mutation using the pRMAF5 delivery vector resulted in partial restoration of the motility, as in the case of strain 11168H (Fig. 5). The results indicate that the integration method for introduction and expression of exogenous genes is not limited to strain 11168H. The finding also demonstrates that despite the sequence difference, the product of the maf5 gene of 11168H strain performs a function similar to the product of the maf5-like gene of strain 81-176.
Expression of wild-type GFP.
The gene encoding wild-type GFP was PCR amplified and cloned into pRRC to produce pRGW (Table 1). Integration of the Camr-wild-type gfp cassette into various rRNA gene clusters was confirmed by PCR. No fluorescence of the Camr colonies could be detected with the fluorescence microscope (excitation at 488 nm). Similarly, only very weakly fluorescent cells could be seen using a confocal microscope. Importantly, this fluorescence was the same as that in the recipient strain 11168H, indicating some background fluorescence. Since both the clones and cells of the control strain E. coli/pRGW were strongly fluorescent, the lack of fluorescence in C. jejuni could be explained by a low level of expression of the wild-type GFP. This could be because in E. coli the wild-type gfp gene is present on a multicopy vector, whereas in C. jejuni it is present at only one copy per cell due to integration into the chromosome. In order to circumvent the problem, we tried expressing an enhanced protein (EGFP) in C. jejuni cells.
Expression of the egfp-SD gene.
The commercially available plasmid pEGFP was used as a source of the egfp gene (Table 1). Although both E. coli/pEGFP clones and cells were highly fluorescent, the expression vector had to be modified for expression in Campylobacter. The egfp gene was PCR amplified using a primer containing an SD sequence optimal for expression in Campylobacter, which resulted in generation of the vector pRED. The E. coli cells and clones containing pRED (Table 1) revealed bright fluorescence. After transformation into C. jejuni, the integration of the Camr-egfp-SD cluster into an rRNA cluster was confirmed by PCR as described above. Compared with wild-type gfp derivatives, the egfp-SD derivatives of C. jejuni revealed enhanced fluorescence under the confocal microscope. However, no fluorescence could be detected using a conventional fluorescence microscope. A relatively low fluorescence level in this case could be explained by a suboptimal codon usage in the egfp gene. Indeed, pEGFP was designed for expression in eukaryotic cells, particularly in human cell lines. Due to a low AT content in the Campylobacter genome, its codon preference is dramatically different from that in E. coli and human cells. Therefore, we decided to use the same technique for expression of the egfp gene from plasmid pMEK91, which as a codon content more optimal for C. jejuni.
Expression of EGFP from pMEK91.
The source of this type of egfp gene was plasmid pMEK91 (24). The plasmid contains a version of the egfp gene (egfp-pMEK) under the control of the Campylobacter ompE promoter (PompE) (Fig. 1). The EcoRI fragment containing the egfp-pMEK gene was cloned into pRRC, and fluorescence of E. coli transformants was tested. Two types of colonies with different levels of fluorescence were detected. The highest level of fluorescence corresponded to both the Camr and egfp-pMEK genes transcribed in the same direction. A plasmid from one such clone was used in transformation into C. jejuni, and integration of the Camr-egfp-pMEK cassette into an rRNA cluster was confirmed as described above. The colonies of the Campylobacter derivatives were fluorescent but were not as bright as E. coli colonies containing the delivery plasmid. However, very strong fluorescence of the C. jejuni cells could be detected using a confocal microscope. Therefore, a better codon composition and the use of an additional promoter allowed efficient expression of GFP in C. jejuni even when the gene was present as just a single copy per cell.
Use of the GFP derivative of C. jejuni for visualization of bacterial forms in situ.
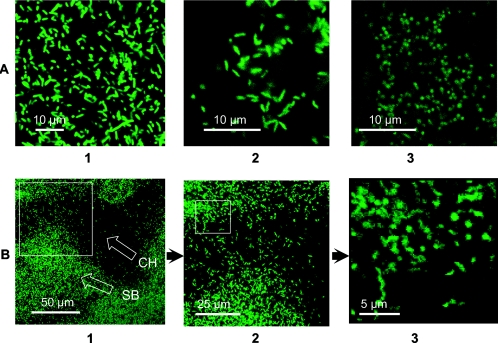
The GFP derivative of C. jejuni strain 11168H (11168H/egfp-pMEK) was used for investigation of sessile bacteria attached to a coverslip in a stationary liquid culture. The sessile bacteria grew on glass coverslips immersed in flasks with stationary liquid cultures as indicated in Materials and Methods. Figure 6 demonstrates intense fluorescence of E. coli/pREM donor cells (Fig. 6A, panel 1) and 11168H/egfp-pMEK derivatives (Fig. 6A, panel 2). Similarly to the recipient 11168H cells (16), prolonged incubation (e.g., 4 days) of 11168H/egfp-pMEK on the solid medium resulted in conversion of spiral cells to coccoid forms (Fig. 6A, panel 3). Such a conversion was significantly reduced in bacteria attached to a coverslip in the stationary liquid culture (Fig. 6B, panels 1 to 3). The aggregates of sessile bacteria were found to contain extensive channels with highly motile darting rod- and spiral-shaped cells (Fig. 6B, panel 3).
FIG. 6.
Confocal microscope images of bacteria expressing EGFP. A, agar cultures. 1, E. coli/pREM5; 2, 11168H/egfp-pMEK 2-day culture; 3, 11168H/egfp-pMEK 4-day culture. B, C. jejuni biofilm formed on a glass coverslip placed at the bottom of a flask with stationary liquid culture (four day incubation, brucella broth). 1 to 3, images of the same area of the coverslip with increased magnification. Magnified areas are shown as squares. The open arrows point to the areas of sessile bacteria (SB) and channels (CH).
DISCUSSION
In this study we describe the design and application of an efficient procedure for the introduction and expression of selected genes in Campylobacter cells. The procedure is based on insertion of a gene expression cassette into a noncoding conserved spacer region of one of several rRNA gene clusters present in the bacterial genome. The insertion occurs via a double recombination event that is common for C. jejuni (31). This is in contrast to the case for many other bacteria, including E. coli and Yersinia, in which insertions via single recombination events are more common and allelic replacement is achieved by either using an additional selection step, e.g., with the help of vectors containing sucrose sensitivity genes (5), or via introduction of exogenous genes required for allelic replacement (7).
Despite routine use of allelic replacement in Campylobacter, at present little is known about its mechanism and the genes involved. One of the candidate genes that could be responsible for allelic replacement was xerC (Cj0863). Indeed, the gene shows extensive similarity with other genes involved in resolution of cointegrants (data not shown). In order to check the possibility that this gene is also involved in allelic replacement in C. jejuni, we used our integration system as a model in the investigation of recombination in the 11168H/xerC::Kanr mutant. Transformation of the pRRC delivery plasmid into this mutant resulted in insertion of the Camr gene cassette into different rRNA clusters. However, in all cases no vector-derived sequences could be detected, indicating double recombination and elimination of the vector sequences even in the xerC-negative strain. Therefore, genes other than xerC are likely to be responsible for allelic replacement.
The number of rRNA clusters in bacterial genomes varies significantly (18). The reason for such variation is not completely clear. There is no strong correlation between the rRNA gene copy number per genome and the bacterial growth rate. The reduction of the number of rRNA genes in E. coli, which is normally seven, has a moderate effect on the growth rate (2, 9). Overexpression of the rRNA gene may actually decrease the bacterial growth rate (28). A link between the rRNA gene copy number and ecological strategies of bacteria has been reported (17). Those authors showed that depending on environmental conditions, either a higher or a lower copy number of rRNA gene clusters may be preferable. It was suggested that the multiple copies of rRNA clusters may in some circumstances provide an advantage when a quick response to varying environmental conditions is required (9). However, for slowly growing bacteria, usually isolated from a low-nutrient environment, a low copy number of rRNA clusters is adequate (11). Our results indicated little or no effect of insertion of the Camr gene cassette on growth of C. jejuni. One should bear in mind, however, that our integration system was designed in such a way that only a minimal disruption (if any) of the rRNA cluster affected would occur.
Analysis of the insertion products revealed that the Camr gene cassette could be inserted randomly into any of the rRNA gene clusters. Interestingly, in some cases the PCR analysis indicated integration into two rRNA clusters simultaneously. This was not due to the presence of two different kinds of cells in the same sample, since subcloning resulted in colonies still producing PCR products with different primers. Simultaneous integration of the Camr gene into two rRNA clusters did not have a significant effect on the colony sizes or on the bacterial growth in a liquid culture. The additional minor PCR products produced by some derivatives suggest a possibility of migration of the Camr gene cassette between the rRNA clusters.
Various Campylobacter shuttle vectors have been described previously. However, these vectors have not been proven versatile. In our hands, none of the available shuttle vectors could be introduced into the sequenced strain C. jejuni strain NCTC 11168 or its derivative 11168H. In addition, even in a few cases of successful introduction of shuttle vectors into a Campylobacter strain as described by other authors, additional steps were required to overcome host-specific restriction. For example, in order to introduce pMW10-based vectors into C. jejuni RM1221 via electroporation, Miller and colleagues had to first transform these vectors into an Strr derivative of this strain via conjugation (23). Only plasmids extracted from C. jejuni could then transform the same strain of C. jejuni. However, strains carrying other restriction-modification systems would be refractory to acquiring this DNA. Another limitation of the shuttle vectors may be a requirement for the presence of residential plasmids in the recipient strains, which may be required for plasmid rescue (30). An additional disadvantage of the shuttle vectors used in other studies is the requirement for using a complementing gene with its own promoter (19, 22, 27). However, this is not always feasible, since a promoter is often located at quite a significant distance from a gene. In our study we overcame the shuttle vector-related limitations by putting an exogenous gene under the control of a constitutive Camr promoter, followed by integration of this cassette into a spacer region within an rRNA gene cluster.
The use of the rRNA gene cluster as a gene insertion target has a number of other advantages. First, due to redundancy of the rRNA genes in Campylobacter, even if a certain rRNA gene cluster is affected by insertion, it would result in only a minor effect on cell functioning. Indeed, we could detect no significant difference in growth rates even after simultaneous insertion of the Camr cassette into two different rRNA clusters. Second, due to the very high conservation of rRNA genes between Campylobacter spp., a recombination vector designed for one strain can be effectively used for most other Campylobacter strains. The vector contains long conserved flanking regions of DNA, ensuring high efficiency of recombination. Using a total DNA preparation extracted from an already-constructed derivative, an inserted gene can be transferred to other Campylobacter strains with even higher transformation efficiency.
In this study we demonstrated the complementation of two knockout mutants. However, while the complementation completely restored the glycosylation phenotype in the pglH mutant, the complementation of the maf5 mutation only partially restored motility. This could be due do tight requirements for the regulation, timing, and level of expression of the genes required for motility. However, even partial phenotype restoration confirms that the phenotype changes are really associated with a particular gene. Moreover, partial phenotype restoration after expressing a His tag fusion protein would provide a tool for isolating functional protein complexes, thus assisting functional analysis. We have demonstrated the same restoration of motility in a 11168H/maf5::Kanr mutant complemented with a His-tagged Maf5 protein (data not shown).
We also investigated expression of three genes of non-Campylobacter origin (gfp) in C. jejuni. Despite extensive differences among the three gfp genes, the derived amino acid sequences are almost identical. Fluorescence properties of the EGFP-SD and EGFP-pMEK proteins are enhanced compared with those of wild-type GFP due to the presence of two mutations, F64L and S65T. The EGFP-pMEK protein also contains a Q80R replacement compared with EGFP. E. coli cells containing either pRED (EGFP-SD) or pREM (EGFP-pMEK) were highly fluorescent. However, the fluorescence level of C. jejuni cells expressing EGFP-pMEK was much higher than that of cells expressing EGFP-SD, which is due to the presence of two tandem promoters and more optimal codon usage (Table 3) in the latter construct.
TABLE 3.
Examples of the difference in codon usage frequencies between two versions of egfp genes, with the highly expressed C. jejuni gene ompE shown for comparison
| Codon (amino acid) | Frequency in:
|
||
|---|---|---|---|
| egfp | egfp-pMEK | ompE | |
| TTT (Phe) | 0 | 8 | 9 |
| TTC (Phe) | 12 | 4 | 14 |
| CTT (Leu) | 0 | 11 | 17 |
| CTC (Leu) | 3 | 1 | 0 |
| CTC (Leu) | 0 | 3 | 4 |
| CTG (Leu) | 18 | 1 | 0 |
| AAA (Lys) | 1 | 15 | 22 |
| AAG (Lys) | 19 | 5 | 0 |
The construction of the fluorescent derivative of the sequenced strain of C. jejuni allows a wide variety of studies involving situ detection of bacterial cells in various forms both in vivo and in vitro. One advantage is the real-time observation of bacterial cells, which does not require any subsequent fixation or staining steps. This is particularly important when visualizing fragile communities of C. jejuni sessile cells attached to solid surfaces. Similarly to the recipient strain 11168H (16), the cells of its GFP derivative were mostly spiral or rod-like after 2 days incubation on blood agar plates (Fig. 6A, panel 2) but were mostly coccoid after 4 days of incubation (Fig. 6A, panel 3). The coccoid cells are considered to be a degenerate bacterial form accumulated in cultures during prolonged incubation or are induced under unfavorable environmental conditions (6). We were interested to investigate if attached aggregates (biofilms) of bacteria provide a mechanism of protection from such conversion. The results demonstrated much higher preservation of live rod-like or spiral forms, indicating that aggregation/immobilization is indeed advantageous for bacterial survival. Detailed analysis of the conglomerates of sessile bacteria under the confocal microscope revealed the presence of extensive internal liquid channels containing a large number of highly motile cells (Fig. 6B). It appears that the formation of the sessile form serves as a protective mechanism increasing bacterial survival under unfavorable conditions.
In summary, we demonstrated an efficient procedure of introduction and expression of various genes in Campylobacter cells. It is noteworthy that the method is applicable to the sequenced strain NCTC 111168, which is used by the majority of researchers. The fact that this strain was particularly difficult to complement was hindering functional genomics studies on this pathogen. This study therefore is a breakthrough for the research community. The method allows complementation of mutations and expression of other genes of non-Campylobacter origin, assisting better understanding of the physiology of this microorganism, including the mechanisms of survival of bacteria in the environment, which may be important for the design of intervention strategies to reduce the presence of Campylobacter in the food chain.
Acknowledgments
We thank M. E. Konkel for the kind gift of plasmid pMEK91, P. Guerry for plasmid pRY112, E. Gaynor for plasmid pMW10-13, and V. Korolik for plasmid pGUO0202.
Financial support for this research was from the BBSRC and the Leverhulme Trust.
REFERENCES
- 1.Alfredson, D. A., and V. Korolik. 2003. Sequence analysis of a cryptic plasmid pCJ419 from Campylobacter jejuni and construction of an Escherichia coli-Campylobacter shuttle vector. Plasmid 50:152-160. [DOI] [PubMed] [Google Scholar]
- 2.Asai, T., C. Condon, J. Voulgaris, D. Zaporojets, B. Shen, M. Al-Omar, C. Squires, and C. L. Squires. 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bereswill, S., and M. Kist. 2003. Recent developments in Campylobacter pathogenesis. Curr. Opin. Infect. Dis. 16:487-491. [DOI] [PubMed] [Google Scholar]
- 4.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, S. N., E. R. Slater, A. H. Chamberlain, and M. R. Adams. 1994. Production and viability of coccoid forms of Campylobacter jejuni. J. Appl. Bacteriol. 77:303-307. [DOI] [PubMed] [Google Scholar]
- 7.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 9.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grainge, I., and D. J. Sherratt. 1999. Xer site-specific recombination. DNA strand rejoining by recombinase XerC. J. Biol. Chem. 274:6763-6769. [DOI] [PubMed] [Google Scholar]
- 13.Guerry, P., C. M. Szymanski, M. M. Prendergast, T. E. Hickey, C. P. Ewing, D. L. Pattarini, and A. P. Moran. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber, K. E., and M. K. Waldor. 2002. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417:656-659. [DOI] [PubMed] [Google Scholar]
- 15.Karlyshev, A. V., D. Linton, N. A. Gregson, and B. W. Wren. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473-480. [DOI] [PubMed] [Google Scholar]
- 16.Karlyshev, A. V., and B. W. Wren. 2001. Detection and initial characterization of novel capsular polysaccharide among diverse Campylobacter jejuni strains using alcian blue dye. J. Clin. Microbiol. 39:279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labigne-Roussel, A., P. Courcoux, and L. Tompkins. 1988. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J. Bacteriol. 170:1704-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linton, D., E. Allan, A. V. Karlyshev, A. D. Cronshaw, and B. W. Wren. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 43:497-508. [DOI] [PubMed] [Google Scholar]
- 22.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 23.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mixter, P. F., J. D. Klena, G. A. Flom, A. M. Siegesmund, and M. E. Konkel. 2003. In vivo tracking of Campylobacter jejuni by using a novel recombinant expressing green fluorescent protein. Appl. Environ. Microbiol. 69:2864-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Siegesmund, A. M., M. E. Konkel, J. D. Klena, and P. F. Mixter. 2004. Campylobacter jejuni infection of differentiated THP-1 macrophages results in interleukin 1 beta release and caspase-1-independent apoptosis. Microbiology 150:561-569. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson, B. S., and T. M. Schmidt. 1998. Growth rate-dependent accumulation of RNA from plasmid-borne rRNA operons in Escherichia coli. J. Bacteriol. 180:1970-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Vliet, A. H. M., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131-135. [DOI] [PubMed] [Google Scholar]
- 32.Wösten, M. M., M. Boeve, M. G. Koot, A. C. van Nuene, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wösten, M. M., J. A. Wagenaar, and J. P. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 34.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]