Abstract
Human neuroendocrine (NE) cancers range from relatively indolent to highly aggressive. In this study, we combine functional genomics with metabolomics to identify features of NE cancers associated with a poor outcome. Analysis of GeneChip datasets of primary prostate tumors, as well as lymph node and liver metastases from transgenic mice with a NE cell cancer, plus derived NE cell lines yielded a signature of 446 genes whose expression is enriched in neoplastic mouse prostatic NE cells. This signature was used for in silico metabolic reconstructions of NE cell metabolism, directed liquid chromatography/tandem MS analysis of metabolites in prostatic NE tumors and cell lines, and analysis of GeneChip datasets of human NE tumors with good or poor prognoses. The results indicate that a distinguishing feature of poor-prognosis NE tumors is a glutamic acid decarboxylase-independent pathway for production of GABA and a pathway for production of imidazole-4-acetate that involves dopa decarboxylase and a membrane-associated amine oxidase, amiloride-binding protein 1. Electrophysiological studies disclosed that imidazole-4-acetate can bind and activate GABAA receptors expressed by transformed NE cells, thus providing a previously uncharacterized paradigm for NE tumor cell signaling. Transcriptional, metabolic, and electrophysiologic features of transformed mouse NE cells are also evident in neural progenitor cells.
Keywords: GABA signaling, MS, metastatic neuroendocrine cancers, polyamine metabolism, transcriptome-directed metabolomics
Human neuroendocrine (NE) cancers (NECs) can arise in neuroectodermal tissues or in endoderm-derived epithelia (e.g., pheochromocytomas and small cell lung cancers, respectively). Many types of NECs are aggressive and diagnosed only after metastatic spread (1, 2). In addition, the appearance of NE features in cancers arising from cells that do not normally express neural or endocrine fates is often associated with more aggressive disease (3–5). A panel of biomarkers and mediators of tumorigenesis is needed for better diagnosis and stratification of NECs and new, more efficacious therapeutic approaches. In this report, we present one approach for addressing this issue that uses a combination of transgenic mice with a metastatic cancer arising from their prostatic NE cell lineage, NE cell lines established from this cancer, human NE tumors with varying degrees of aggressiveness, functional genomics, and MS-based metabolomics.
The prostate contains three epithelial lineages: secretory, basal, and NE (6). The secretory or luminal cell is the most abundant type and requires continuous exposure to androgens for its survival (7–11). Basal cells represent the primary proliferating epithelial population and are not androgen-dependent (12, 13). NE cells are rare (<1% of the total), have distinctive projections that allow them to contact epithelial neighbors located several cell diameters away, and secrete a variety of neuropeptides and neurotransmitters (14–17). Conventional adenocarcinoma of the prostate (CaP) is the most commonly diagnosed cancer and the second leading cause of cancer death in males (18): it displays features common to luminal/secretory cells (19–23). Pure NE cell tumors of the prostate are rare but aggressive (2). In ≥30% of cases, conventional CaP displays features of NE differentiation (NED; ref. 14). NED is focal, correlates with androgen-independent growth, and is associated with a poor prognosis (3, 24–28).
We have produced FVB/N transgenic mice (CR2-TAg) where transcriptional regulatory elements from mouse Defcr2 (cryptdin-2) were used to express simian virus 40 large T antigen (TAg) in a subset of prostate NE cells. Males from multiple pedigrees develop a stereotyped pattern of initiation and progression of prostatic NE cell cancer. Transgene expression commences at 7 weeks of age. Within 1 week, foci of transformed NE cells appear that phenocopy human prostatic intraepithelial neoplasia, the postulated precursor of conventional adenocarcinoma of the prostate (17, 29). By 16 weeks of age, 90% of CR2-TAg prostates demonstrate local invasion, with 25% having tumor nodules >3 mm. These nodules are composed of sheets of malignant cells with no glandular features. The majority of mice die by 24 weeks, with transformed NE cells comprising >90% of all cells in their prostates and metastases in their lymph nodes, liver, lungs, and bone.
CR2-TAg tumors and metastases have been used to generate prostate NEC (PNEC) lines. PNECs produce prominent neurite-like processes when grown as attached monolayers and express a variety of gene products that are also produced by NE cells in CR2-TAg prostates (30). When grown in suspension cultures, they form multicellular aggregates that resemble the neurospheres produced when neural stem cells are cultured on noncoated surfaces. Subcutaneous injection of PNEC cells produces rapidly growing tumor xenografts (average doubling time, 7.5 d), whereas i.v. injection results in liver and bone metastases (30).
Below, we describe how GeneChip analyses of primary tumors and metastases from 24-week-old CR2-TAg mice and PNEC lines, combined with (i) in silico metabolic reconstructions of NE cell metabolism from the GeneChip transcriptional profiles, (ii) mass spectrometric analysis of metabolites present in tumors and PNEC extracts, and (iii) hierarchical clustering of GeneChip expression profiles generated from 182 human tumors (127 lung adenocarcinomas, 20 carcinoids, 6 small cell lung carcinomas, 10 medullary thyroid carcinomas, and 19 adrenal pheochromocytomas) have provided a list of mRNA transcripts and metabolites indicative of a poor prognosis for various human NE cancers (NEC).
Materials and Methods
Transcriptome-Directed Metabolomics. Methods for acquiring transcript signatures from mouse and human GeneChip datasets of NE tumors are outlined in Results and Discussion in Supporting Text and Table 1, which are published as supporting information on the PNAS web site. In silico metabolic reconstructions of cellular metabolism were performed by using metaview software (developed in our laboratory) and transcript lists. These reconstructions in turn were used to direct gas chromatography (GC)/MS and liquid chromatography/MS analysis of metabolites in specific pathways, as described below. Methods for sample preparation, chromatography, and MS are described in detail in Supporting Text.
Electrophysiology. PNEC cells were plated on poly-l-lysine- and laminin-coated 35-mm plastic culture dishes (Falcon) at a density of 10,000 cells per dish and grown for ≥60 h as described in Supporting Text. Methods used to record currents in PNEC cells after application of selected metabolites are presented in Supporting Text.
Results and Discussion
Defining a “Core CR2-TAg NE Cell Transcript Signature.” Significance Analysis of Microarrays (sam; www-stat.stanford.edu/∼tibs/SAM; ref. 31) comparisons of GeneChip-derived datasets of mRNAs present in prostates, lymph node metastases, and liver metastases of 24-week old CR2-TAg mice vs. datasets obtained from the prostates of their normal littermates revealed 669 transcripts (identified by 732 probesets) enriched ≥2-fold in CR2-TAg primary tumors and all metastases (Table 2, which is published as supporting information on the PNAS web site).
To remove transcripts originating from non-NE cell populations in CR2-TAg tumors, GeneChip datasets were acquired from PNEC30 cells (derived from a primary CR2-TAg prostate tumor), PNEC30–3 cells (a subclone of PNEC30), PNEC25 cells (from a liver metastasis), and nonattached PNEC30 cells (neurosphere morphotype), plus PNEC30 and PNEC30–3 xenografts. sam comparisons of (i) all PNEC monolayer datasets vs. normal prostate, (ii) PNEC30 neurosphere datasets vs. normal prostate, and (iii) PNEC30/PNEC30–3 xenografts vs. normal prostate yielded a total of 596 transcripts (recognized by 639 probesets) that were enriched ≥2-fold in all three comparisons (Table 3, which is published as supporting information on the PNAS web site). When this PNEC filter of 639 probe sets was applied to the list of 732 probe sets that recognize transcripts enriched in 24-week-old CR2-TAg prostates and metastases vs. normal prostate, 472 probe sets (446 transcripts) were obtained and defined as a “core CR2-TAg NE cell transcript signature” (Table 4, which is published as supporting information on the PNAS web site).
Transcriptome-Directed Metabolomics. Using metaview, we transferred the resulting core CR2-TAg NE cell transcript signature onto metabolic maps present in the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg) so we could make and test predictions about the metabolic properties of malignant prostate NE cells (see http://gordonlab.wustl.edu/metaview/ippolito for output maps based on the core signature, as well as maps derived from “Present” calls in CR2-TAg primary tumor, PNEC25 and PNEC30 GeneChip datasets).
Glutamic acid decarboxylase (Gad1; E.C. 4.1.1.15) exhibited the greatest enrichment among all transcripts that encoded enzymes (219-fold; see Table 2). metaview reconstructions predicted synthesis of GABA in transformed prostate NE cells via decarboxylation of glutamate by Gad1 (glutamate metabolism pathway). GC/MS assays subsequently confirmed that GABA levels were enriched 104- to 136-fold in PNEC30 monolayers (51.8 ± 7.6 nmol/mg dry weight tissue) and primary tumor (39.6 ± 7.8 nmol/mg) vs. normal prostates (0.38 + 0.14 nmol/mg). GABA was also significantly elevated in venous blood sampled from CR2-TAg transgenic mice (27.4 ± 1.0 μM) vs. normal mice (7.3 ± 2.2 μM) (n = three animals/group; P < 0.001).
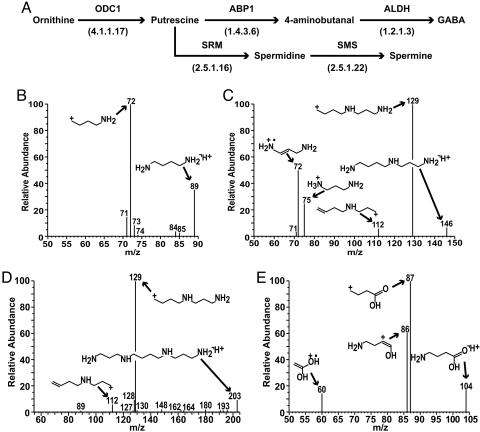
The metaview reconstructions also predicted that primary CR2-TAg tumors and PNEC cells contained transcripts encoding enzymes able to synthesize GABA through an alternative pathway that does not involve Gad1 (Fig. 1A). In this alternate pathway, ornithine decarboxylase (E.C. 4.1.1.17) converts ornithine, a component of the urea cycle, to putrescine, which is then metabolized to 4-aminobutanal by the amine oxidase, amiloride-binding protein 1 (ABP1; E.C. 1.4.3.6). Aminobutanal is then metabolized to GABA by aldehyde dehydrogenases (ALDH; E.C. 1.2.1.3). In addition to GABA, putrescine was predicted to give rise to spermidine and spermine. These latter metabolites have been implicated in many aspects of cancer biology, including tumor progression and metastasis (32, 33). Exact mass identification by Fourier transform mass spectrometry and liquid chromatography/tandem MS analyses confirmed the KEGG reconstructions by detecting putrescine, spermidine, spermine, and GABA in PNEC cells (Fig. 1 B–E).
Fig. 1.
Transcriptome-directed metabolomics of polyamine metabolism in prostate NEC cells. (A) metaview in silico reconstruction of polyamine metabolism in the KEGG arginine/proline metabolic map. Reconstructions are based upon “Present” calls in GeneChip datasets of PNEC30 cell lines and CR2-TAg prostate tumors. ODC1, ornithine decarboxylase; SRM, spermidine synthase; SMS, spermine synthase. E.C. numbers are in parentheses. (B–E) Mass spectral fragmentation of GABA and polyamines in PNEC cell lines: (B) putrescine ([M+H]+ = 89): (C) spermidine ([M+H]+ = 146); (D) spermine ([M+H]+ = 203); (E) GABA ([M+H]+ = 104). [Fragmentation spectra were acquired with an ion trap mass spectrometer and structures assigned to peaks with mass frontier 4.0 (ThermoElectron; see Methods).]
Generation of Metabolic Signatures Indicative of Poor-Prognosis Human NE Tumors. We hypothesized that the presence of these and perhaps other metabolic pathways could be used to distinguish poor-prognosis metastatic NE tumors from benign human NE tumors. To explore this notion, we examined GeneChip datasets representing human NE lung tumors and tumors from patients with multiple endocrine neoplasia (MEN). Our analysis included publicly available GeneChip datasets from three types of lung cancers: adenocarcinomas with NED (n = 12; Fig. 5 and Tables 5 and 6, which are published as supporting information on the PNAS web site, and Results in Supporting Text), carcinoids (n = 20), and small cell carcinomas (n = 6). We also generated GeneChip datasets from metastatic medullary thyroid carcinomas (MEN2B; n = 10), and benign pheochromocytomas (MEN2A, MEN2B, and from sporadic cases; n = 19; available at http://gordonlab.wustl.edu/metaview/ippolito). Lung adenocarcinomas with NED (see Fig. 6, which is published as supporting information on the PNAS web site, for survival analysis), small cell lung carcinomas (5-year survival rate of 1–5%; ref. 34), and medullary thyroid carcinomas (5-year survival rate of 55%; ref. 35) were assigned to a poor-prognosis NE group. Pulmonary carcinoids (5-year survival rate of 92%; ref. 36) and benign pheochromocytomas were defined as the good-prognosis NE group.
A supervised learning approach, Prediction Analysis of Microarrays (pam; www-stat.stanford.edu/∼tibs/PAM; ref. 37), was used to identify transcripts that best distinguished the good and poor-prognosis groups. dchip (38) was then used to visualize their expression patterns among the various datasets (Fig. 7, which is published as supporting information on the PNAS web site). Table 7, which is published as supporting information on the PNAS web site, presents a list of the top 50 genes whose increased expression in poor-prognosis tumors and low expression in good-prognosis tumors best delineate poor-prognosis NECs (see Table 8, which is published as supporting information on the PNAS web site, for an analogous list for good prognosis tumors). As a supplement to the pam analysis, sam was used to generate a dataset of transcripts that were ≥2-fold enriched in poor-prognosis NE tumors (Table 9, which is published as supporting information on the PNAS web site).
Spermidine/spermine N1-acetyltransferase (SAT) and ABP1 (see above) were among the top 50 transcripts whose expression demarcated poor-prognosis NE tumors. SAT, the rate-limiting enzyme in polyamine catabolism, catalyzes N1 acetylation of spermine and spermidine and, together with polyamine oxidase, allows production of putrescine. Based on KEGG pathway maps, we predicted that in poor-prognosis human (and mouse CR2-TAg) NE tumors, ornithine decarboxylase and SAT act in concert to produce putrescine, which is converted by ABP1 to 4-aminobutanal, and finally to GABA via ALDH (ALDH is not up-regulated in poor-prognosis tumors, but ALDH transcripts are “Present” in 100% of human NE tumors; Fig. 2).
Fig. 2.
Metabolic reconstructions of the polyamine pathway based upon pam and sam analyses of poor-prognosis human NE tumors. Transcripts enriched in poor-compared with good-prognosis tumors are indicated in green with the gene symbol. Fold enrichment was obtained from the sam analysis (see Table 9). SAT, spermidine/spermine N1-acetyltransferase; PAO, polyamine oxidase.
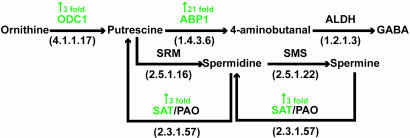
metaview reconstructions of CR2TAg tumors and PNEC cells also revealed that ABP1 is downstream of dopa decarboxylase (DDC; E.C. 4.1.1.28) in the histidine, phenylalanine, tryptophan, and tyrosine metabolic pathways, where it catalyzes the oxidative deamination of histamine, phenylethylamine, tryptamine, and tyramine to the corresponding aldehydes followed by their conversion to acids by ALDH (Fig. 3A). DDC is a component of the core CR2-TAg NE cell transcript signature (35-fold enriched compared with normal prostate; Table 2), and a “top 10” transcript for defining lung adenocarcinomas with NED (see Results in Supporting Text).
Fig. 3.
Biogenic amine metabolism in poor-prognosis mouse and human NE cell cancers. (A) Metabolic reconstructions of aromatic amino acid metabolism in mouse and human tumors. Note that DDC and ALDH transcripts were “Present” in 100% of good- and poor-prognosis human NE tumors examined (n = 67). ABP1 received a “Present” call in 22/28 (78.6%) poor-prognosis NE tumors and only 1/39 (2.6%) good-prognosis NE tumors. (B–E) Mass spectral fragmentation profiles of DDC- and ABP1-generated metabolites in PNEC30 cell monolayers: (B) histamine ([M+H]+ = 112); (C) tryptamine ([M+H]+ = 161); (D) imidazole-4-acetate ([M+H]+ = 127); (E) indole-3-acetate ([M+H]+ = 176). Fragmentation spectra were acquired with an ion trap mass spectrometer and structures assigned to peaks with mass frontier 4.0 (see Methods).
We used Fourier transform MS and liquid chromatography/ tandem MS analyses to profile these pathways. We confirmed tryptophan and histidine metabolism in PNEC cells by identifying the biogenic amines tryptamine and histamine as well as indole-3-acetate, a plant auxin whose role in mammalian cellular physiology remains poorly understood, and imidazole-4-acetate, a GABAA receptor agonist and partial antagonist of GABAC receptors (39) (Fig. 3 B–E).
Inspection of expression values for DDC and ABP1 mRNAs in the various GeneChip datasets revealed that DDC was scored “Present” in all NE tumor samples whether in the poor- or good-prognosis group, indicating this enzyme is a general biomarker for NE tumors. In contrast, a statistically significant demarcation (P = 2.8 × 10–11; Fisher exact probability test) in ABP1 expression was noted between good- and poor-prognosis NECs: ABP1 mRNA was scored Present in 78.6% (22/28) of poor-prognosis tumors, compared to only 2.6% (1/39) of good-prognosis tumors.
GABA derived from putrescine appears to have a role in the normal development and physiology of several tissues, including the small intestine (40), liver (41, 42), and retina (43). Moreover, GABA obtained from this pathway in gastrin-producing enteroendocrine cells is a postulated paracrine modulator within the gastric mucosa (44, 45). These findings, together with the observations that (i) ABP1 is cell membrane-associated (46, 47) and (ii) its mRNA is called “Present” almost exclusively in poor- vs. good-prognosis NE tumors, suggested that metabolites produced from ABP1-generated aldehydes in the polyamine or DDC pathways may not only be useful as prognostic biomarkers but also function as mediators of cellular communications within NE tumor microenvironments. We used PNEC cells to examine this latter hypothesis.
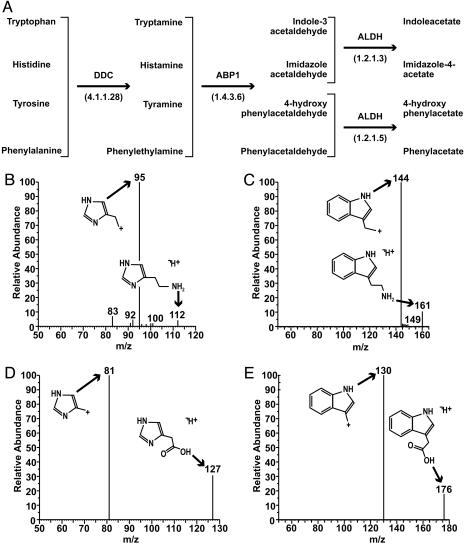
Response of PNEC Cell Lines to GABA. Electrophysiological studies disclosed that PNEC30 cells respond to GABA application in a dose-dependent manner (Fig. 4 A and D). The current induced is consistent with activation of a chloride channel, as demonstrated by comparison of reversal potentials between intracellular solutions containing chloride and methanesulfonate (a membrane-impermeant anion) (Fig. 4B). Picrotoxin, a GABAA antagonist, abolishes the GABA-stimulated current, whereas lorazepam (a benzodiazepine) and pentobarbital (a barbiturate) potentiate the current (Fig. 4 A and C). The findings indicate these metastatic tumor cells express a functional GABAA receptor. Importantly, imidazole-4-acetate acts on PNEC30 cells in a similar manner, producing a dose-dependent response, although it is 30-fold less potent than GABA (Fig. 4 E and F). Indole-3-acetate, the other ABP1 product detected in PNEC30 cells, does not activate a current in PNEC cells at concentrations up to 1 mM.
Fig. 4.
Functional evidence for GABAA receptor expression in PNEC cells. The horizontal bar indicates the duration of drug exposure. (A) Responses to 100 μM GABA ± 100 μM picrotoxin. (B) Reversal potential of GABA currents, obtained from digital subtractions of current responses to voltage ramps in the presence and absence of 30 μM GABA and measured by using the indicated pipette solutions (n = 3 cells for each pipette solution). (C) Responses to 10 μM GABA ± 1 μM lorazepam or 50 μM pentobarbital. (D) Concentration-response curve for GABA. The solid line represents a fit of the plotted data to the Hill equation, which estimated a half-maximum concentration of 17.5 μM for GABA (n = 8 cells per point). (E) Responses to 1 mM imidazole-4-acetate ± 100 μM picrotoxin. (F) Concentration-response curve for imidazole-4-acetate. The solid line represents a fit of the plotted data to the Hill equation, which estimated an EC50 value of 500 μM(n = 5–10 cells per point).
We subsequently surveyed PNEC and human NE tumor GeneChip datasets for transcripts encoding various GABA receptor subunits. GABAA γ2 subunit mRNA was called “Present” in both PNEC30 and PNEC25 cells (derived from a primary CR2-TAg prostate tumor and liver metastasis, respectively). When compared with normal prostate, the γ2 subunit mRNA was ≥10-fold enriched in 24-week-old CR2-TAg prostate tumors and metastases, PNEC30 and PNEC25 monolayers, as well as PNEC30 neurospheres (Tables 2 and 3). GeneChip studies also revealed “Present” calls for GABAA receptor β3 subunit and GABAB receptor 1 mRNAs in PNEC cells and primary CR2-TAg tumors.
Interestingly, the γ2 subunit mRNA was scored “Present” in 17/19 pheochromocytomas but “Absent” in virtually all other human NE tumors. The β3 subunit was “Present” in 18/20 carcinoids, 17/19 pheochromocytomas, and 8/10 metastatic MEN2B medullary thyroid cancers. Another distinctive pattern of subunit expression involved the GABAC ρ1 subunit, which was Present in 100% (10/10) of metastatic MEN2B medullary thyroid cancers.
In the prostates of CR2-TAg mice, neoplastic NE cells synthesizing GABA are juxtaposed next to normal epithelial cells, representing other prostatic lineages that express GABAA and GABAB receptors (17). This observation, together with the findings described above, provide a previously undescribed view of cellular communications in poor-prognosis human NECs. In this view, the presence of a membrane-associated amine oxidase (ABP1) allows for the synthesis of metabolites, such as GABA and imidazole-4-acetate, which act on GABA receptors associated with these and neighboring cells. The repertoire of metabolites produced and the pattern of GABA receptor subunits expressed act in concert to effect the biological behavior of neoplastic NE cells and their neighbors within a given tumor microenvironment through autocrine and paracrine mechanisms. Our findings suggest that noninvasive imaging modalities such as positron emission tomography (48) and magnetic resonance spectroscopy (49) may be useful for phenotyping patient populations with NE tumors based on the activity of key enzymes in these metabolic pathways (e.g., DDC) and their metabolic products (e.g., GABA). Moreover, GABA, imidazole-4-acetate, and indole-3-acetate may only be a subset of products of these pathways; ABP1-derived aldehydes may react with amine-containing compounds in tumor cells to form precursors for alkaloids similar to those found in plants and other organisms (50).
Similarities Between PNEC and Neural Progenitor Cells. PNEC cells, which produce NECs as xenografts, resemble neural progenitors in a number of ways. Both form neurospheres ex vivo. Both produce GABA and GABAA receptors (51, 52). Voltage traces recorded in response to current injections into PNEC cells (Fig. 8, which is published as supporting information on the PNAS web site) are very similar to the “immature action potentials” generated by neural progenitor cells (51). PNEC cells express transcription factors important to neural progenitor cell survival, including SRY-box proteins (Table 4). In fact, when we extracted 170 probe sets representing 158 transcripts encoding products with a “nuclear” Gene Ontology term from the CR2-TAg NE core signature and examined their representation in previously published GeneChip datasets obtained from mouse neural, hematopoietic, and embryonic stem cells (53), 150 of the 158 transcripts (95%) were called “Present” in at least one of the three stem cell populations (Table 10 and Fig. 9, which are published as supporting information on the PNAS web site). Neural stem cells had the greatest overlap with the CR2-TAg core NE signature components having “nuclear” gene ontology terms (146/158 transcripts; 93%), followed by embryonic (140/158 transcripts; 89%) and hematopoietic stem cells (119/158 transcripts; 75%). When metaview was used to determine whether PNEC polyamine and biogenic amine metabolic pathways were present in these stem cell populations, the results indicated that both neural and hematopoietic stem cells are capable of synthesizing GABA: neural stem cells express Gad1 but not ABP1, whereas hematopoietic stem cells express ABP1 but not Gad1. Hematopoietic stem cells are also predicted to synthesize imidazole-4-acetate using histidine decarboxylase and ABP1.
Neural progenitors have been isolated from tissues located outside the central nervous system and have the ability to adopt several different cell fates (54, 55). This raises the intriguing possibility that some NE carcinomas may represent transformed neural stem cells, or that some NE differentiated adenocarcinomas (e.g., prostate, lung) may be more accurately described as adenocarcinomas with a neural progenitor phenotype.
Supplementary Material
Acknowledgments
This work is dedicated to Americo Ippolito. We thank Jeffrey Mil-brandt for generously supplying GeneChip datasets from medullary thyroid carcinomas and pheochromocytomas and Ling Munsell and Julia Gross for advice on liquid chromatography/MS. This work was supported by National Institutes of Health Grants DK59129, DK63483, CA94056, RR00954, AA12952, and DA18109, and funds from the William M. Keck Foundation.
Author contributions: J.E.I., S.M., R.R.T., and J.I.G. designed research; J.E.I., K.M., S.M., and J.R.C. performed research; J.E.I., J.X., and S.J. contributed new reagents/analytic tools; J.E.I., K.M., S.M., and J.R.C. analyzed data; and J.E.I., K.M., S.M., and J.I.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NE, neuroendocrine; NEC, NE cancer; NED, NE differentiation; TAg, T antigen; PNEC, prostate NEC line; KEGG, Kyoto Encyclopedia of Genes and Genomes; DDC, dopa decarboxylase; ABP1, amiloride-binding protein 1; ALDH, aldehyde dehydrogenase; Gad1, glutamic acid decarboxylase; MEN, multiple endocrine neoplasia.
References
- 1.Sorensen, J. B. & Hansen, H. H. (1994) Curr. Opin. Oncol. 6, 162–170. [DOI] [PubMed] [Google Scholar]
- 2.Cohen, R. J., Glezerson, G., Haffejee, Z. & Afrika, D. (1990) Br. J. Urol. 66, 405–410. [DOI] [PubMed] [Google Scholar]
- 3.McWilliam, L. J., Manson, C. & George, N. J. (1997) Br. J. Urol. 80, 287–290. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee, A., Richards, W. G., Staunton, J., Li, C., Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R., Gillette, M., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Syder, A. J., Karam, S. M., Mills, J. C., Ippolito, J. E., Ansari, H. R., Farook, V. & Gordon, J. I. (2004) Proc. Natl. Acad. Sci. USA 101, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonkhoff, H. & Remberger, K. (1996) Prostate 28, 98–106. [DOI] [PubMed] [Google Scholar]
- 7.Aumuller, G. (1983) Prostate 4, 195–214. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs, J. T. & Coffey, D. S. (1989) Prostate Suppl. 2, 33–50. [DOI] [PubMed] [Google Scholar]
- 9.Masai, M., Sumiya, H., Akimoto, S., Yatani, R., Chang, C. S., Liao, S. S. & Shimazaki, J. (1990) Prostate 17, 293–300. [DOI] [PubMed] [Google Scholar]
- 10.de Winter, J. A., Trapman, J., Brinkmann, A. O., Boersma, W. J., Mulder, E., Schroeder, F. H., Claassen, E. & van der Kwast, T. H. (1990) J. Pathol. 160, 329–332. [DOI] [PubMed] [Google Scholar]
- 11.van der Kwast, T. H., Schalken, J., Ruizeveld de Winter, J. A., van Vroonhoven, C. C., Mulder, E., Boersma, W. & Trapman, J. (1991) Int. J. Cancer 48, 189–193. [DOI] [PubMed] [Google Scholar]
- 12.Bonkhoff, H., Stein, U. & Remberger, K. (1994) Prostate 24, 114–118. [DOI] [PubMed] [Google Scholar]
- 13.Bonkhoff, H., Stein, U. & Remberger, K. (1994) Hum. Pathol. 25, 42–46. [DOI] [PubMed] [Google Scholar]
- 14.di Sant'Agnese, P. A. (1992) Hum. Pathol. 23, 287–296. [DOI] [PubMed] [Google Scholar]
- 15.di Sant'Agnese, P. A. & Cockett, A. T. (1996) Cancer 78, 357–361. [DOI] [PubMed] [Google Scholar]
- 16.Abrahamsson, P. A. (1999) Endocr. Relat. Cancer 6, 503–519. [DOI] [PubMed] [Google Scholar]
- 17.Hu, Y., Ippolito, J. E., Garabedian, E. M., Humphrey, P. A. & Gordon, J. I. (2002) J. Biol. Chem. 277, 44462–44474. [DOI] [PubMed] [Google Scholar]
- 18.Jemal, A., Thomas, A., Murray, T. & Thun, M. (2002) CA Cancer J. Clin. 52, 23–47. [DOI] [PubMed] [Google Scholar]
- 19.Wernert, N., Seitz, G. & Achtstatter, T. (1987) Pathol. Res. Pract. 182, 617–626. [PubMed] [Google Scholar]
- 20.Sherwood, E. R., Berg, L. A., Mitchell, N. J., McNeal, J. E., Kozlowski, J. M. & Lee, C. (1990) J. Urol. 143, 167–171. [DOI] [PubMed] [Google Scholar]
- 21.Dhom, G., Seitz, G. & Wernert, N. (1988) Am. J. Clin. Oncol. 11, S37–S42. [DOI] [PubMed] [Google Scholar]
- 22.Nagle, R. B., Brawer, M. K., Kittelson, J. & Clark, V. (1991) Am. J. Pathol. 138, 119–128. [PMC free article] [PubMed] [Google Scholar]
- 23.Okada, H., Tsubura, A., Okamura, A., Senzaki, H., Naka, Y., Komatz, Y. & Morii, S. (1992) Virchows Arch. A. Pathol. Anat. Histopathol. 421, 157–161. [DOI] [PubMed] [Google Scholar]
- 24.Allen, F. J., Van Velden, D. J. & Heyns, C. F. (1995) Br. J. Urol. 75, 751–754. [DOI] [PubMed] [Google Scholar]
- 25.Bohrer, M. H. & Schmoll, J. (1993) Verh. Dtsch. Ges. Pathol. 77, 107–110. [PubMed] [Google Scholar]
- 26.Dema, A., Raica, M. & Tudose, N. (1996) Rom. J. Morphol. Embryol. 42, 83–88. [PubMed] [Google Scholar]
- 27.Speights, V. O., Jr., Cohen, M. K., Riggs, M. W., Coffield, K. S., Keegan, G. & Arber, D. A. (1997) Br. J. Urol. 80, 281–286. [DOI] [PubMed] [Google Scholar]
- 28.Weinstein, M. H., Partin, A. W., Veltri, R. W. & Epstein, J. I. (1996) Hum. Pathol. 27, 683–687. [DOI] [PubMed] [Google Scholar]
- 29.Garabedian, E. M., Humphrey, P. A. & Gordon, J. I. (1998) Proc. Natl. Acad. Sci. USA 95, 15382–15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, Y., Wang, T., Stormo, G. D. & Gordon, J. I. (2004) Proc. Natl. Acad. Sci. USA 101, 5559–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manni, A., Washington, S., Griffith, J. W., Verderame, M. F., Mauger, D., Demers, L. M., Samant, R. S. & Welch, D. R. (2002) Clin. Exp. Metastasis 19, 95–105. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, T. & Thomas, T. J. (2003) J. Cell. Mol. Med. 7, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elias, A. D. (1997) Chest 112, 251S–258S. [DOI] [PubMed] [Google Scholar]
- 35.Cohen, M. S. & Moley, J. F. (2003) J. Intern. Med. 253, 616–626. [DOI] [PubMed] [Google Scholar]
- 36.Mezzetti, M., Raveglia, F., Panigalli, T., Giuliani, L., Lo Giudice, F., Meda, S. & Conforti, S. (2003) Ann. Thorac. Surg. 76, 1838–1842. [DOI] [PubMed] [Google Scholar]
- 37.Tibshirani, R., Hastie, T., Narasimhan, B. & Chu, G. (2002) Proc. Natl. Acad. Sci. USA 99, 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tunnicliff, G. (1998) Gen. Pharmacol. 31, 503–509. [DOI] [PubMed] [Google Scholar]
- 40.Raul, F., Gosse, F., Galluser, M., Hasselmann, M. & Seiler, N. (1995) JPEN J. Parenter. Enteral Nutr. 19, 145–150. [DOI] [PubMed] [Google Scholar]
- 41.Fogel, W. A., Bieganski, T., Bechtold, E. & Maslinski, C. (1982) Agents Actions 12, 49–52. [DOI] [PubMed] [Google Scholar]
- 42.Fogel, W. A., Bieganski, T. & Maslinski, C. (1982) Comp. Biochem. Physiol. C 73, 431–434. [DOI] [PubMed] [Google Scholar]
- 43.Eliasson, M. J., McCaffery, P., Baughman, R. W. & Drager, U. C. (1997) Neuroscience 79, 863–869. [DOI] [PubMed] [Google Scholar]
- 44.Hardt, J., Larsson, L. I. & Hougaard, D. M. (2000) J. Histochem. Cytochem. 48, 839–846. [DOI] [PubMed] [Google Scholar]
- 45.Hougaard, D. M., Houen, G. & Larsson, L. I. (1992) FEBS Lett. 307, 135–138. [DOI] [PubMed] [Google Scholar]
- 46.Barbry, P., Champe, M., Chassande, O., Munemitsu, S., Champigny, G., Lingueglia, E., Maes, P., Frelin, C., Tartar, A., Ullrich, A., et al. (1990) Proc. Natl. Acad. Sci. USA 87, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbry, P., Chassande, O., Marsault, R., Lazdunski, M. & Frelin, C. (1990) Biochemistry 29, 1039–1045. [DOI] [PubMed] [Google Scholar]
- 48.Hoegerle, S., Nitzsche, E., Altehoefer, C., Ghanem, N., Manz, T., Brink, I., Reincke, M., Moser, E. & Neumann, H. P. (2002) Radiology 222, 507–512. [DOI] [PubMed] [Google Scholar]
- 49.Bielicki, G., Chassain, C., Renou, J. P., Farges, M. C., Vasson, M. P., Eschalier, A. & Durif, F. (2004) NMR Biomed. 17, 60–68. [DOI] [PubMed] [Google Scholar]
- 50.Deitrich, R. & Erwin, V. (1980) Annu. Rev. Pharmacol. Toxicol. 20, 55–80. [DOI] [PubMed] [Google Scholar]
- 51.Wang, D. D., Krueger, D. D. & Bordey, A. (2003) J. Physiol. 550, 785–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gago, N., El-Etr, M., Sananes, N., Cadepond, F., Samuel, D., Avellana-Adalid, V., Baron-Van Evercooren, A. & Schumacher, M. (2004) J. Neurosci. Res. 78, 770–783. [DOI] [PubMed] [Google Scholar]
- 53.Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. (2002) Science 298, 597–600. [DOI] [PubMed] [Google Scholar]
- 54.Kabos, P., Ehtesham, M., Kabosova, A., Black, K. L. & Yu, J. S. (2002) Exp. Neurol. 178, 288–293. [DOI] [PubMed] [Google Scholar]
- 55.Fernandes, K. J., McKenzie, I. A., Mill, P., Smith, K. M., Akhavan, M., Barnabe-Heider, F., Biernaskie, J., Junek, A., Kobayashi, N. R., Toma, J. G., et al. (2004) Nat. Cell Biol. 6, 1082–1093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.