Abstract
To ensure survival, most bacteria must acquire iron, a resource that is sequestered by mammalian hosts. Pathogenic bacteria have therefore evolved intricate systems to sense iron limitation and regulate gene expression appropriately. We used a pan-Neisseria microarray to examine genes regulated in Neisseria gonorrhoeae in response to iron availability in defined medium. Overall, 203 genes varied in expression, 109 up-regulated and 94 down-regulated by iron deprivation. In iron-replete medium, genes essential to rapid bacterial growth were preferentially expressed, while iron transport functions, and predominantly genes of unknown function, were expressed in low-iron medium. Of those TonB-dependent proteins encoded in the FA1090 genome with unknown ligand specificity, expression of three was not controlled by iron availability, suggesting that these receptors may not be high-affinity transporters for iron-containing ligands. Approximately 30% of the operons regulated by iron appeared to be directly under control of Fur. Our data suggest a regulatory cascade where Fur indirectly controls gene expression by affecting the transcription of three secondary regulators. Our data also suggest that a second MerR-like regulator may be directly responding to iron availability and controlling transcription independent of the Fur protein. Comparison of our data with those recently published for Neisseria meningitidis revealed that only a small portion of genes were found to be similarly regulated in these closely related pathogens, while a large number of genes derepressed during iron starvation were unique to each organism.
Iron plays a prominent role in a variety of metabolic pathways, making it essential for life in most organisms. Iron is a cofactor for proteins such as catalase, cytochromes, hemoglobin, metalloflavoproteins, myoglobin, ribonucleotide reductase, and peroxidase (43). Despite its essential importance to metabolism, iron is paradoxically a difficult nutrient to manage. Under biological conditions (oxygenated, aqueous, neutral pH), iron quickly becomes oxidized to the ferric state (Fe3+) (5). Ferric iron reacts rapidly with water to form insoluble oxy-hydroxide complexes that are not metabolizable. Further, the ferric iron is extremely toxic due to the ability to drive free radical production via the Fenton reaction. This can lead to DNA strand breakage, lipid peroxidation, and protein denaturation (73). To avoid these potential problems, mammalian physiology has evolved a variety of mechanisms to sequester iron, suppress its redox reactivity, and maintain solubility. This is achieved by incorporating iron intracellularly in proteins such as ferritin and hemoproteins and binding iron extracellularly by lactoferrin and transferrin (43). This results in free iron concentrations of approximately 10−18 M, a concentration that will not sustain growth for most microorganisms, which typically require an iron concentration of ∼10−6 M to sustain life. Thus, mammalian physiology nonspecifically suppresses the growth of many potential pathogens by withholding iron, commonly referred to as “nutritional immunity” (78).
To be a successful pathogen, bacteria must therefore evolve methods to acquire iron from the host. The sexually transmitted disease pathogen Neisseria gonorrhoeae is typically seen as a pathogen of mucosal surfaces, predominantly associated with symptomatic urethritis in males and an often asymptomatic endocervicitis in women. For women, this can progress to pelvic inflammatory disease and salpingitis, potentially resulting in ectopic pregnancy and sterility (15, 22). Iron is sequestered on the urogenital mucosal surface by lactoferrin and transferrin (1), while during menses hemoglobin is released into the environment (36). The gonococcus, an obligate human pathogen, has evolved several iron transport systems that bind human iron carrier proteins and remove the iron directly from these ligands. Thus, the gonococcus possesses receptors that bind and remove iron from human hemoglobin (HmbR), hemoglobin-haptoglobin complexes (HpuAB), lactoferrin (LbpAB), and transferrin (TbpAB) (58). The gonococcus also can use exogenously produced enterobactin (a siderophore) via the FetA receptor (20); presumably, this may be important for allowing the organism to metabolize catecholate siderophores found in mixed microbial environments, such as the female urogenital tract. The lactoferrin and transferrin receptors are extremely important for gonococcal survival in vivo. In infection of male volunteers, both lbpAB+ ΔtbpAB and ΔlbpAB tbpAB+ strains were capable of initiating disease where a ΔlbpAB ΔtbpAB strain was not (3, 24).
Thus, it is not surprising that iron availability is often found to be a key environmental signal that controls virulence in a variety of pathogens. For instance, iron availability regulates expression of Corynebacterium diphtheriae diphtheria toxin, Shigella dysenteriae Shiga toxin, and Pseudomonas aeruginosa exotoxin A (43). Microarray analysis of Helicobacter pylori revealed iron-regulated expression of the virulence genes cagA, napA, and vacA (47). Early studies demonstrated that iron starvation enhanced capsular polysaccharide biosynthesis in Neisseria meningitidis (45) and was associated with increased virulence in mice (14, 37).
In gram-negative and gram-positive bacteria, transcription of genes involved in iron acquisition and virulence are often under the control of the ferric uptake regulator (Fur) protein. Fur, in the presence of ferrous iron, binds as a dimer to DNA regulatory sequences (Fur boxes), which typically results in the repression of transcription of many iron-repressible genes (25). Several gonococcal iron-repressible promoters in the gonococcus, including fetA, hmbR, fbpABC, hpuAB, lbpAB, and tbpAB, have this consensus Fur box (33, 42). Thomas and Sparling found 32 proteins by two-dimensional gel electrophoresis that were directly controlled by the gonococcal fur gene (72). However, aside from this little is known about the gonococcal iron response regulon. To begin to address this, we have used a pan-Neisseria microarray to analyze the steady-state, mid-log gene expression profiles of gonococci in response to iron availability.
MATERIALS AND METHODS
Bacterial strains and growth.
N. gonorrhoeae strain FA1090 was kept as a freezer stock at −80°C in GC medium (Difco) supplemented with 20% glycerol. Prior to growth experiments, FA1090 was inoculated on GCB agar (Becton Dickinson) supplemented with IsoVitaleX (Becton Dickinson) and grown at 37°C, 5% CO2 for 16 h. Several nonpiliated colonies were transferred to a fresh GCB agar plate and grown for an additional 16 h. Nonpiliated organisms were used in this study, as piliated bacteria clump significantly in broth, making the measurement of growth based on optical density difficult. Piliation was determined based on criteria established by Swanson et al. (71). These bacteria were used to inoculate Chelex-treated (Bio-Rad) chemically defined medium (CDM) broth in acid-washed glassware (27). The organisms were allowed to undergo one doubling in CDM without the addition of 10 μM iron (CDM-0) to deplete the organisms' internal iron pools. The cultures were then diluted threefold (20-ml total volume) in either CDM plus 10 μM FeNO3 (CDM-10) or CDM-0 and grown to stationary phase. Ten-milliliter samples for microarray experiments were taken at mid-log phase (see Fig. 1, below). Each growth experiment was performed four times.
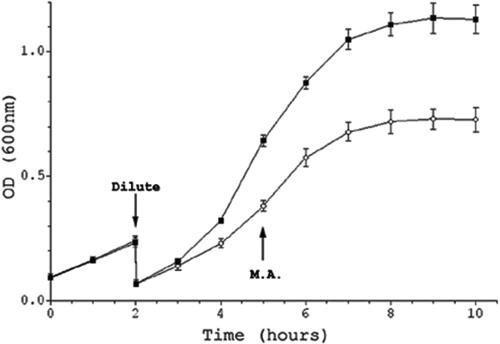
FIG. 1.
Growth of Neisseria gonorrhoeae FA1090 in CDM grown with (CDM-10 [▪]) or without (CDM-0 [○]) 10 μM ferric nitrate. Cultures were diluted after first doubling, and a 10-ml sample for microarray analysis (M.A.) was taken at mid-log as indicated on the graph. This figure is the average of four independent experiments, with standard deviations shown as error bars; at some time points, the error was so small that it cannot be seen on this figure. OD, optical density.
RNA extraction.
An aliquot of bacterial culture was immediately added to an ice-cold 95% phenol-5% ethanol (EtOH) mixture at a 1:10 (vol/vol) ratio and gently inverted to arrest bacterial growth and preserve RNA integrity. Samples were allowed to remain on ice for several minutes and then spun at 5,000 × g for 10 min to pellet the cells. Cells were resuspended in TE (10 mM Tris-HCl, 1 mM EDTA), pH 8.0, lysed by addition of a 1/10 volume of 10% sodium dodecyl sulfate (SDS), and incubated at 64°C for 2 min. A 1/10 volume of 1 M sodium acetate, pH 5.2, was added, and RNA was isolated by extraction, using an equal volume of prewarmed water-saturated phenol followed by an extraction using an equal volume of chloroform. Each extraction mixture was incubated at 64°C for 6 min with inversion every minute, chilled, and centrifuged at 15,000 × g for 10 min, 4°C. After the second extraction, bulk RNA was precipitated from the aqueous layer using a standard EtOH precipitation (80). The RNA was then resuspended in 10 μl distilled water (dH2O) and treated with 20 U RNase inhibitor (Roche) and 5 U RNase-free DNase (Invitrogen) in a 50-μl volume for 15 min, followed by extraction with the RNeasy Mini kit (QIAGEN) according to the manufacturer specifications. The DNase treatment was repeated, and the RNA concentration and integrity were determined by measuring the optical density at 260 nm and examined by 1% agarose gel electrophoresis using Tris-acetate-EDTA buffer (59). PCR against the porI locus of FA1090 was performed to assess DNA contamination. If necessary, DNase treatment was repeated to ensure that no detectable chromosomal DNA was in each RNA preparation.
Construction of pan-Neisseria DNA microarray.
The detailed description of the pan-Neisseria microarray will be published elsewhere (60). Briefly, the published genome sequences of meningococcal strains Z2491 and MC58 and gonococcal strain FA1090 were used to generate a PCR amplicon-based microarray. Open reading frames (ORFs) that shared 90% or greater nucleotide sequence identity between at least two of the three genomes were used to design a set of “core” amplicons using FA1090 DNA. Strain-specific genes were amplified using the respective DNA from each meningococcal or gonococcal strain. Amplified ORF targets ranging from 150 to 450 bp in length were resuspended in 50% (vol/vol) dimethyl sulfoxide and spotted in triplicate onto GAPS slides (Corning). The printed microarray contains 99.6% of all the FA1090 annotated features.
Microarray hybridization and analysis.
Equal concentrations (16 μg) of test RNA extracted from cells grown under high- or low-iron conditions were used to set up standard reverse transcription reactions using random nonamers (Integrated DNA Technologies), SuperScript II (Invitrogen), and aminoallyl-dUTP (Sigma) to generate cDNA. The cDNA was purified using Micron 30 spin columns (Millipore) and indirectly labeled with a monofunctional N-hydroxysuccinimide ester Cy3- or Cy5-containing dye (Amersham). Reaction mixtures were combined, and unincorporated dye was removed using the QIAquick PCR purification kit (QIAGEN) according to the manufacturer specifications. The labeled cDNA was vacuum concentrated without heat prior to suspension in 35 μl of hybridization buffer (25% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS) containing 1 μl of a 10-mg/ml solution of sheared salmon sperm DNA. The probe was heated to 99°C for 2 min, briefly bench top cooled, added to the back of a prewarmed Hybri-slip (Sigma), placed on a prehybridized array (25% formamide, 5× SSC, 0.1% SDS, and 1% bovine serum albumin for 45 min at 42°C), and enclosed in a prewarmed glass array hybridization chamber (Ambion). After an 18-h hybridization, the array was removed from the hybridization chamber and stringently washed in 1-liter volumes with stir bar agitation as follows: 2× SSC and 0.1% SDS for 5 min at 42°C, 0.1× SSC and 0.1% SDS for 10 min at room temperature, and three washes of 0.1× SSC for 1 min at room temperature. The slide was briefly washed with dH2O, dried by spinning in a microcentrifuge with slide adapter (Telechem International), and scanned with a ScanArray ExpressHT microarray scanner (Perkin-Elmer). The brightest spots on each array (rRNA) were used to establish a scan setting (80% of signal maximum) for each individual array. Photomultiplier tube gain and laser settings were varied from array to array to achieve this setting.
After scanning, the data were analyzed using the ArrayVision 7.0 software package (Imaging Research). Actual signal intensity was calculated by subtracting the local mean background intensity of each spot from the mean signal intensity of each spot. Any spot with an adjusted signal intensity lower than its mean background intensity plus two standard deviations in both channels was excluded from further analysis; this removed 445 genes from consideration. Data were formatted for import into GeneSpring version 4.2.1 (Silicon Genetics) and normalized per spot and per chip using Lowess transformation. The data of each spot represent the average of four experiments. The Student t test was then applied to the data to find genes with highly reproducible gene expression. Differentially expressed genes were defined as genes in which the normalized ratios were >1.5-fold (CDM-0/CDM-10 for iron-repressed genes or CDM-10/CDM-0 for iron-responsive genes) (38, 63, 69) with a P value of <0.02. Genes with a P value of >0.02 were excluded from consideration, resulting in the removal of 87 additional genes from further analysis. Raw array data are stored at the National Center for Biotechnology Information's GEO database (7) under the series accession number GSE2147.
Confirmation of pan-Neisseria microarray data.
Verification of gene expression levels was measured by real-time quantitative PCR (qRT-PCR). Five derepressed genes (fetA, hemO, tbpA, tbpB, and cah), four repressed genes (L11, NGO0295, NGO1246, and secY), and two stably expressed genes (NGO1996 and porI), as well as the four expressed, unidentified ligand TonB-dependent genes (NGO0021, NGO0553, NGO0952, and NGO1205), were chosen for this confirmatory study. Oligonucleotide primers (Table 1) were designed with PRIMER EXPRESS software (PE Biosystems) and were tested prior to qRT-PCR analysis to confirm that they resulted in a single amplicon of the proper size when genomic DNA was used as template. After RNA isolation, 2 μg of total RNA was reverse transcribed using SuperScript II with 250 ng of random nonamers according to the manufacturer's specifications (Invitrogen). After reverse transcription, 5 ng of cDNA along with 250 nM of each primer pair was subjected to qRT-PCR using an ABI Prism 7700 sequence detection system (Applied Biosystems) and SYBR Green master mix, according to manufacturer specifications (PE Biosystems). The relative expression level of each gene was normalized to porI, the gene encoding a major outer membrane protein of N. gonorrhoeae whose expression level remained stable irrespective of iron availability (data not shown). Quantitative values were obtained by using the comparative threshold cycle (ΔΔCT) method, as recommended by Applied Biosystems. Each gene was assayed in duplicate from each of the four pools of microarray-isolated RNA, and the mean CT value was used for further comparison. Gene expression was calculated as 2−ΔΔCT, where values for ΔΔCT (ΔCT,Fe− − ΔCT,Fe+) were obtained by subtracting the mean CT value of the specific gene tested from the mean CT value of the porI gene. Controls to ensure that there was no contaminating genomic DNA in the cDNA (cDNA reaction mixtures generated using RNA with no reverse transcriptase) as well as controls to identify any possible primer dimer artifacts (qRT-PCR mixtures containing primers alone) were also run with each set of qRT-PCRs.
TABLE 1.
Oligonucleotide primers for RT-PCR microarray validation
| Gene | Forward primer | Reverse primer |
|---|---|---|
| fetA | ATCCAACACCAATTTGGCGTAC | CATCGGCGGAATAGCGTTT |
| hemO | GAAACTGCAATCCGTGTTCCA | CATCGTATCGCGCCATGTATT |
| tbpA | AGACACTTGGGCGGATTATG | GAACAGTGCGTCTGCTGAAA |
| tbpB | TGGATGCGGTTGAATTGACA | GGACAGGAGCGGAATCATAATG |
| cah | GGCGTATCATGGTTGGTGTTG | CACGTGCATTCAGAGGCTGTAC |
| L11 | TGATGGAACACGGTGCACTCT | TGCTGCCAGATTTGTTCGGT |
| NGO0295 | GCGGTTTATCGGCATTCTGAT | CTGCCTGATTTTCGGTGATGTT |
| NGO1246 | TCTGCCGAAACGCAAAAATC | AAAGCACCGCCGTAATCTCA |
| secY | TGGTGCCCATATACCCGTACCT | TCTAACGACCCACCGGAAAAC |
| NGO1996 | TATCCCGTTGTGCAGATGTCC | ATATGCCTGCCCTGTATCCGT |
| porI | TGTCCGTACGCTACGATTCTCC | AGCCAACGTGGTAAGATTCGC |
| NGO0021 | GGCAGATACAGGCAGGCTAC | GGTAGGCGGTGAAGAGTTTG |
| NGO0553 | AAAGACGACAGGCAGCTGAT | GCCGTCTGTAATGTCGGTTT |
| NGO0952 | GATCAGGACAACGGGCTTTA | GCCCTTTATCGCCCTTAAAC |
| NGO1205 | TCAACGCAAGAACTGTACGC | CCGACTTCAAAGGTGTTGGT |
Bioinformatics.
To examine the potential role of Fur in the transcriptional regulation of each gene, it was first necessary to predict the location of putative operons in the FA1090 genome, so that presumptive promoter regions in the genome could be identified. First, GeSTer was used to predict transcriptional termination sites within the gonococcal genome (75). ORFs were then assigned to a putative operon based on proximity to nearby genes (within 100 bp), the presence of a predicted transcriptional termination site, and the predicted direction of the surrounding ORFs (see Table S3 in the supplemental material). For the first differentially expressed gene of each predicted operon, a Perl script was written to compile the 150 bp immediately upstream of the translational start into a file in FASTA format. These sequences were analyzed by using MEME (6) for both the Escherichia coli (GATAATGATAATCATTATC) (70) and N. gonorrhoeae (GATAAT-ATAATAATTATC-TTT) (33) Fur-binding consensus sequences. Putative Fur-binding sites were then placed in a multiple sequence alignment using ClustalX, and a consensus sequence logo was generated using Seqlogo (26).
The predicted amino acid sequences of the differentially expressed genes were collected in FASTA format in their unprocessed and processed forms. A Perl script was written to generate both SignalP (50) and TMHMM (65) data on each protein sequence. Additionally, each hypothetical and conserved hypothetical protein was subjected to Pfam analysis by comparison of the protein translation to the Pfam database (64). The Institute for Genomic Research Comprehensive Microbial Resource was consulted for functional classifications (53). To compare gene expression between N. gonorrhoeae and N. meningitidis, Perl scripts were used to create protein databases of both the entire genomes and the iron-responsive, differentially regulated genes of N. gonorrhoeae and N. meningitidis. These databases were then compared by BLASTP comparisons (2). Orthologues were identified as having a 70% identity over the entire span of the predicted amino acid sequences of both proteins.
RESULTS AND DISCUSSION
Microarray analysis of N. gonorrhoeae FA1090 steady-state mRNA levels in response to iron availability.
To study the gene expression profile of the gonococcus in response to iron availability, we used Chelex-treated CDM rather than resorting to the use of chelators to withhold iron. We were concerned that chelators might introduce confounding variables that could complicate an attempt to analyze iron-specific responses. Chelators such as 2,2-dipyridyl diffuse into the cell and could affect intracellular events by chelating divalent cations other than iron (62). In addition to having a strong affinity for ferrous iron, 2,2-dipyridyl can also chelate Mg2+ (46), Mn2+ (55), and Zn2+ (66). Similarly, although Desferal probably withholds iron from Neisseria by binding it extracellularly, it also can bind copper, nickel, and zinc (16). Such effects may be enough to trigger noniron, ion-specific regulatory events or otherwise affect gene expression in such a way as to complicate our analysis of iron-specific responses. CDM is a modification of the Morse and Bartenstein medium (49) developed by Dyer et al. to study transferrin-specific iron uptake in the meningococcus (27). This medium has been successfully utilized in the study of iron acquisition of both pathogenic Neisseria species. CDM has been used to study Neisseria iron acquisition from aerobactin (79), heme-bound iron (28), lactoferrin (10, 12), and transferrin (13, 23, 44) and the role of three putative TonB-dependent outer membrane proteins in iron transport (74). CDM-0 supported limited growth (Fig. 1). Bacteria grown in CDM-0 versus CDM-10 had a slower doubling time (approximately 90 min versus 60 min) and lower optical density during mid-log phase (0.38 ± 0.03 versus 0.64 ± 0.03). Cells were taken directly from these broth cultures and mixed with buffer-saturated phenol to freeze the mRNA population. Bulk RNA was then prepared and either labeled for microarray hybridization or used in RT-PCR experiments to validate the microarray data.
The microarray data were processed as described in Materials and Methods. We detected an alteration in steady-state mRNA levels of 203 genes, representing slightly over 10% of the total ORF content of FA1090. Of these 203 genes, 109 genes were repressed by high iron (see Table S1 in the supplemental material) and 94 were induced by growth in high iron (see Table S2 in the supplemental material).
Validation of microarray data by dilution RT-PCR.
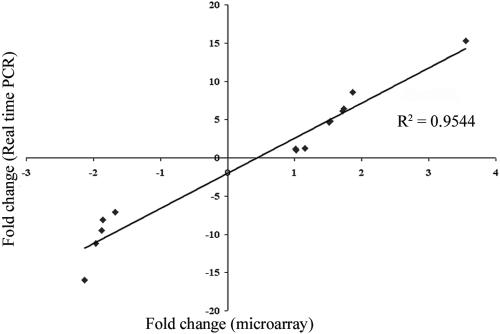
We used qRT-PCR to validate the microarray data (Fig. 2), analyzing transcript levels for 14 genes. These included genes whose transcripts appeared to be derepressed (fetA, hemO, tbpA, tbpB, and cah), repressed (L11, NGO0295, NGO1246, and secY), or unresponsive (NGO1996) in the absence of iron. In each instance, the RT-PCR results correlated well (R2 = 0.95) with those obtained from the microarrays, suggesting that the microarray results were an accurate reflection of the gene expression profile in N. gonorrhoeae FA1090.
FIG. 2.
Validation of microarray results by qRT-PCR. Shown are five genes derepressed, four genes repressed, and one gene unresponsive to iron deprivation, as well as the four expressed, TonB-dependent protein genes of unknown ligand specificity. Fold changes are shown [(iron-depleted expression level)/(iron-replete expression level)].
Response to growth under low-iron conditions.
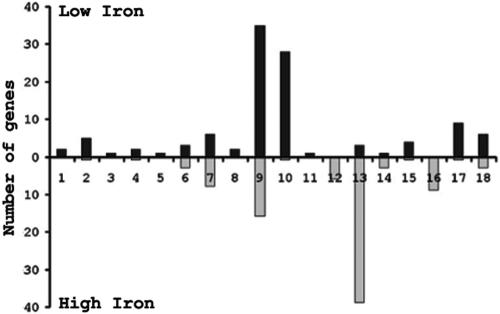
Figure 3 represents the number of differentially expressed genes grouped by functional categories. Interestingly, genes encoding transport functions constituted the largest category of genes with known or predicted function (nine genes total) found to be derepressed under iron-depleted growth conditions (Fig. 3, column 7). Of these nine genes, seven encode the proteins ExbB, essential for TonB-dependent energy transduction (11), FbpA, an iron-binding periplasmic protein (8), FetAB, which is involved in the binding and transport of ferric enterobactin (20), the transferrin receptor TbpAB (4), and TdfG, an iron-repressible TonB-dependent family protein of as-of-yet-undetermined function (74). FbpA, FetAB, TbpAB, and TdfG have been previously reported to be iron-repressible proteins (9, 31, 68, 74). While we observed a derepression of expression of many gonococcal iron-repressible proteins, we did not observe all that may have been anticipated. For instance, we did not observe differences in expression of the hmbR or the hpuAB receptors in response to iron availability. This was expected; the hpuAB operon is phase variable by slipped-strand mispairing in a poly(G) tract in the hpuA gene (41) and is phase varied “off” in FA1090. Similarly, the FA1090 hmbR gene has multiple frameshifts that inactivate protein expression. Since mRNA from untranslated frameshifted genes is less stable than transcripts being translated (51), the apparent lack of hpuAB and hmbR expression was probably masked by mRNA instability.
FIG. 3.
Differentially expressed genes categorized by functional classification according to TIGRFAM designations. Columns: 1, amino acid biosynthesis; 2, biosynthesis of cofactors; 3, cell envelope; 4, cellular processes; 5, central intermediary metabolism; 6, DNA metabolism; 7, energy metabolism; 8, fatty acid metabolism; 9, conserved hypothetical proteins; 10, hypothetical proteins; 11, other; 12, protein fate; 13, protein synthesis; 14, purines, pyrimidines, nucleosides, and nucleotides; 15, regulatory functions; 16, transcription; 17, transport; 18, unknown function. The solid bars and shaded bars represent genes up-regulated and down-regulated in the absence of iron, respectively.
A number of other genes derepressed under low-iron conditions are predicted to encode potential virulence determinants or to be involved in DNA transformation. NGO0788 encodes a putative macrophage infectivity potentiator protein (mip), similar to one found in Legionella pneumophila. Knockouts of mip attenuate Legionella mutants infecting human mononuclear phagocytes and lung epithelial cells (39). NGO1718 encodes a putative MviN-like protein. Deletion of this gene in Salmonella enterica serovar Typhimurium attenuates mouse infectivity (19). The four identical comE genes within the FA1090 genome also were derepressed under low-iron conditions, although we could not discriminate between these in this assay. The comE loci are necessary for DNA-mediated transformation (21). Our observations are consistent with those of Serkin and Seifert, who showed that iron-starved gonococci have an increased transformability and increased, RecA-independent DNA recombination and repair activity (61). Also consistent with this observation is the derepression of NGO0173, which encodes a putative very short patch DNA repair endonuclease.
The largest number of iron-repressed loci encoded hypothetical and conserved hypothetical proteins, accounting for 58% (63 of 109) of the total. This also was observed in H. pylori, N. meningitidis, and Pasteurella multocida (35, 47, 52). SignalP analysis (50) predicted that 13 conserved hypothetical and 2 hypothetical proteins could be exported. Further examination of these 15 genes for the presence of α-helices using TMHMM analysis (65) revealed that NGO0449 and NGO1471, both conserved hypothetical proteins, were predicted to contain three and two transmembrane helices, respectively. This indicates that these genes probably encode proteins that localize to the inner membrane. The remaining 13 genes may possibly encode proteins that are either periplasmic or associated with the outer membrane. In addition to those observations mentioned above, this suggests that iron starvation may significantly affect the antigenic mosaic on the surface of the gonococcus. Pfam analysis of these hypothetical proteins (Table 2) (64) revealed four conserved hypothetical proteins with functional domains. In one particular instance, the conserved hypothetical gene NGO1318 encodes a PigA-like protein which in Pseudomonas aeruginosa functions as a heme oxygenase (56). PigA is responsible for liberating iron from heme, yielding biliverdin and carbon monoxide. NGO1318 shares 84% identity with the experimentally proven heme oxygenase (hemO) of N. meningitidis (81). Cleavage of heme to release iron serves as an iron source for the gonococcus (81).
TABLE 2.
SignalP and Pfam analysis of gonococcal iron-regulated conserved hypothetical proteinsa
| Pfam family | Special characteristic | N. gonorrhoeae CHP ORF(s) |
|---|---|---|
| NA | Contain a signal peptide | NGO0165, NGO0449, NGO0757, NGO0787, NGO0853, NGO0895, NGO1063, NGO1168, NGO1237, NGO1471, NGO1559, NGO1622, NGO1656, NGO1686 |
| PF01551 | Zinc metallopeptidase | NGO1686 |
| PF01430 | Molecular chaperone | NGO1189 |
| PF00582 | Universal stress protein | NGO0959 |
| PF01126 | Heme oxygenase | NGO1318 |
| PF02498 | BRO family protein | NGO1652 |
| PF00691 | OmpA family protein | NGO1559 |
| PF01722 | BolA family protein | NGO1657 |
Abbreviations: NA, not applicable; CHP, conserved hypothetical protein.
Expression profiles for genes encoding TonB-dependent proteins.
In addition to FetA, HmbR, HpuB, and TbpA (see above), strain FA1090 harbors five additional genes encoding TonB-dependent proteins. These ORFs include NGO0021 (tdfF), NGO0553 (tdfG), NGO0560, NGO0952 (tdfH), and NGO1205. Typically, gram-negative TonB-dependent proteins are high-affinity receptors for iron-carrying ligands, and the transcription of these iron transport proteins is generally repressed by iron (58). The lone exception to this is the E. coli BtuB protein, which transports vitamin B12 and whose expression is not iron responsive (18). Recently, Turner et al. examined expression of tdfF, tdfG, and tdfH by Western blotting and found that only TdfG expression was iron repressible. Similarly, we noticed an increase in the transcript levels of the tdfG gene in CDM-0 (see Table S1 in the supplemental material [microarray fold change of 1.58]) (Fig. 2, qRT-PCR fold change of 4.8), and expression of tdfF and tdfH was unaltered in response to iron availability. Assuming that derepression of a TonB-dependent protein in response to iron starvation implies a role in iron transport, then our data conversely suggest that tdfF and tdfH may be high-affinity receptors for important ligands other than iron carriers, since their expression was unresponsive to iron availability. Strikingly, expression of NGO1205 was significantly enhanced by growth under high-iron conditions (see Table S2 in the supplemental material [microarray fold change of −1.67]) (Fig. 2, qRT-PCR fold change of −7.1), suggesting that this protein also is a high-affinity transporter for a noniron ligand. We could not detect expression of NGO0560; NGO0560 contains multiple frameshifts that block translation, most likely rendering the transcript unstable and difficult to detect (51).
Response to growth under high-iron conditions.
In general, genes encoding proteins involved in protein synthesis, energy metabolism, and transcription were preferentially expressed under high-iron conditions, suggesting an overall increase of cellular metabolism (Fig. 3). The expression profile of these genes is consistent with an increased growth rate in iron-containing medium compared to iron-restricted medium (Fig. 1). Members of the translational apparatus accounted for the largest functional category induced in high iron, accounting for almost half of the genes regulated in this manner. This included ribosomal proteins and translation factors, such as initiation factors IF-2 (infB) and IF-3 (infC) and elongation factors G (fusA) and TS (tsf). Consistent with this, secE and secY, encoding components of the protein translocation machinery, and dnaJ, encoding a protein chaperone, were also induced.
This increased level of protein synthesis would dictate a higher energy requirement for the organism. This is reflected in the microarray data. The putative membrane-associated proteins involved in electron transport, nqrC (NGO1415) and nqrF (NGO1418), along with nqrA (NGO1413) were induced by growth in CDM-10. The NQR complex is a sodium pump coupling electron transfer (NADH to ubiquinone) to the transport of Na+ across the periplasmic membrane (54). Complex I, another proton-pumping NADH:ubiquinone oxidoreductase, is a similar system (77) and is likewise regulated in an iron-dependent manner, as seen by the induction of nuoD (NGO1748). NqrF, in addition to containing binding motifs for NADH and FAD, also contains an iron-sulfur center (67). In complex I, NuoB has been shown to contain an iron-sulfur complex (30).
In contrast to the iron-repressible gene set, the number of conserved hypothetical and hypothetical genes induced by high iron was limited. Only 1 hypothetical and 16 conserved hypothetical proteins were induced by high iron. SignalP analysis suggested that only one of these genes, NGO1686, encoded a protein with a putative signal sequence. None of these proteins was predicted to contain transmembrane α-helices by TMHMM analysis. Pfam analysis did reveal that NGO1686, encoding a conserved hypothetical protein, contains a motif similar to other zinc metallopeptidases. Another conserved hypothetical protein, encoded by NGO1189, putatively contains an Hsp33 motif. Hsp33, a redox-regulated chaperone, contains four cysteines (all present in NGO1189) which are oxidized to form disulfide bonds, resulting in its activation. Again, this may be consistent with the increased protein synthetic capacity of the organism under high-iron conditions. Notably, genes involved in amino acid and cofactor biosynthesis were not appreciably induced under high-iron conditions. This is likely due to the fact that CDM is an extremely rich, defined medium that supplies every cofactor and amino acid necessary for growth.
Mechanisms of iron regulation.
We assigned the 203 iron-responsive gonococcal genes to 153 putative operons (mono- or polycistronic) (see Table S3 in the supplemental material). Of these, 64 operons were preferentially expressed under high-iron conditions, while 89 were iron repressed. The predicted promoter regions of these operons were examined for the presence of putative Fur boxes. We used the Fur box consensus derived from analysis of both the E. coli (GATAATGATAATCATTATC) (70) and the Neisseria (GATAAT-ATAATAATTATC-TTT) (33) Fur-responsive genes. This was based on the observation that the DNA-binding regions of the E. coli and N. gonorrhoeae Fur proteins appear to be highly conserved. The DNA-binding motif of the E. coli Fur protein is an atypical helix-turn-helix motif, with amino acid residues 20 to 24 and 55 to 61 responsible for protein-DNA interaction (34). A sequence alignment between the Fur proteins of E. coli and N. gonorrhoeae reveals a high similarity throughout the protein and the DNA-binding motifs (data not shown). Amino acids 20 to 24 (KILEV) of the E. coli Fur protein are 60% identical to N. gonorrhoeae Fur (KILDL) with strong conservation between the last two amino acids. The E. coli Fur protein amino acids 55 to 61 (YRVLNQF) are identical to the gonococcal Fur (YRVLTQF) except for a weakly conserved asparagine-to-threonine difference. The two amino acids absolutely necessary for DNA binding (R56 and F61) are identical in the two Fur proteins (34).
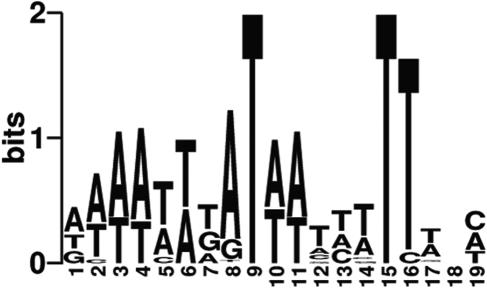
Comparison of putative promoters to these consensus Fur-binding sites demonstrated that 50 operons (33%) were predicted to be under the direct regulatory control of Fur (see Table S3 in the supplemental material). Of these operons, 31 were derepressed in the absence of iron, including tbpAB, fetA, fetB, and fbpABC, as well as tdfG. The remaining 19 were up-regulated in the presence of iron. The predicted Fur boxes contained within the promoters of the iron-regulated operons were used to generate a consensus logo sequence (Fig. 4). This logo reveals an AT-rich sequence common to previously studied Fur boxes (29). Significantly, putative Fur boxes were found in the promoter regions for three of the four regulatory proteins derepressed in low iron, including two AraC-like regulators, NGO0025 and NGO2115, and an unclassified phage-like regulator, NGO1013. This suggests that low-iron conditions may activate a regulatory cascade where some effects of Fur are indirect.
FIG. 4.
Sequence logo of the Fur box of N. gonorrhoeae FA1090. Fur boxes found in the promoters of genes regulated by iron were used to determine nucleotide frequencies. Regulatory sequence determinations were done using the E. coli and N. gonorrhoeae Fur box consensus sequences.
AraC-like transcriptional regulators vary in the types of functions that they control, being involved in controlling carbon metabolism, protein synthesis, regulation of virulence factors, and response to environmental stressors (32). One of the putative AraC-like regulators, NGO0025, shares significant identity (99%) with NMB1879, which is similarly derepressed under low-iron conditions in the meningococcus (35). Unfortunately, the diversity of this regulatory family results in an extremely degenerate DNA-binding motif (57), and so we were unable to reliably predict promoter regions that might contain binding sites for these regulators (data not shown). The phage-like regulator NGO1013 may be responsible for control of some or all of the 19 phage-associated hypothetical proteins derepressed under low-iron conditions (see Table S2 in the supplemental material).
The lone iron-repressible regulator that does not have a putative Fur box in the promoter encodes a MerR-like protein. Some MerR-like proteins respond to heavy metals, such as the product of the E. coli gene zntR, which is responsive to zinc (17). This suggests that two potential regulators, Fur and the MerR-like regulator, may be directly involved in the gonococcal response to iron availability, while three additional regulators indirectly control gene expression in a potential Fur-directed transcriptional cascade.
Comparative genomics of N. gonorrhoeae and N. meningitidis grown in iron-depleted and iron-replete environments.
Grifantini et al. recently published a microarray analysis of the response of meningococcal strain MC48 to iron availability (35). While Grifantini et al. used a chemically defined medium, the formulation of this medium was not explicitly described, and so comparison of the meningococcal data to ours is somewhat problematic. In addition, Grifantini et al. employed 12.5 μM Desferal to iron limit N. meningitidis MC58; as described above, this might complicate the interpretation of these data. Nevertheless, the comparison of our data set with theirs still appears to be instructive. While N. gonorrhoeae and N. meningitidis are closely related at the genetic level, they cause significantly different diseases. Unlike the gonococcus, N. meningitidis typically attaches to the nasopharyngeal mucosa (48) and is transmitted person-to-person via aerosolized respiratory droplets or oral secretions (76). Of the 1,999 gonococcal ORFs, 1,487 (74%) are shared between FA1090 and MC58, 535 of which are conserved hypothetical proteins (40). Approximately 20% of iron-regulated gonococcal genes had iron-regulated orthologues in the meningococcal data set (Fig. 5 and Tables 3 and 4). The majority of genes preferentially expressed in both organisms under high-iron conditions are involved in protein synthesis. Additionally, the three predominant functional categories differentially expressed under high-iron conditions are identical for both organisms, including hypothetical proteins and those involved in protein synthesis and energy metabolism. Of the 26 genes coregulated under high-iron conditions (Table 3), 10 encode ribosomal proteins, with only 2 encoding conserved hypothetical proteins. The genes encoding SecY, NuoD, and Rho also were similarly regulated in both organisms.
FIG. 5.
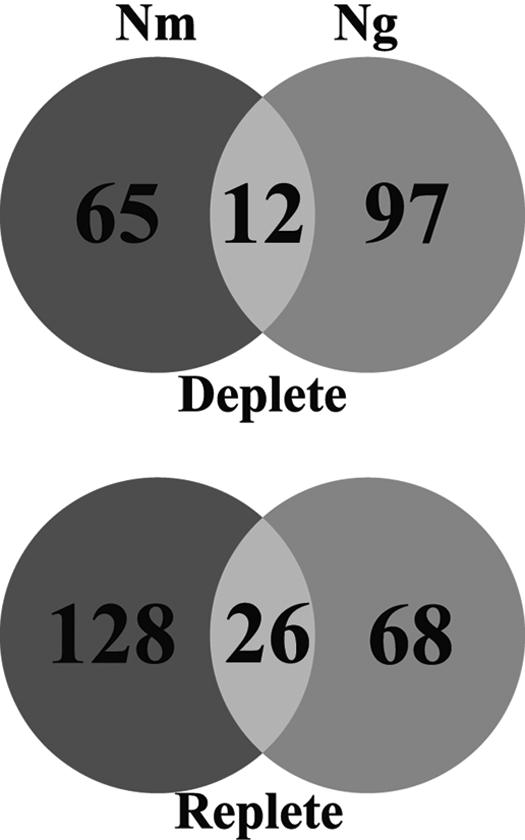
Venn diagram comparing differentially expressed iron-regulated genes of N. gonorrhoeae FA1090 and N. meningitidis MC58.
TABLE 3.
N. gonorrhoeae and N. meningitidis orthologs differentially expressed under high-iron conditions
|
N. gonorrhoeae
|
N. meningitidis
|
% IDa | E valueb | ||
|---|---|---|---|---|---|
| ORF | Gene product | ORF | Gene product | ||
| NGO0037c | Conserved hypothetical protein | NMB1866 | Conserved hypothetical protein | 98.87 | 0 |
| NGO0043c | 50S ribosomal protein L11 | NMB1862 | 50S ribosomal protein L11 | 94.92 | 8.00E-165 |
| NGO0057 | Putative thioredoxin | NMB1845 | Thioredoxin | 93.49 | 3.40E-079 |
| NGO0191 | 30S ribosomal protein S15 | NMB0609 | 30S ribosomal protein S15 | 100 | 4.60E-048 |
| NGO0199c | Transcription termination factor rho | NMB0617 | Transcription termination factor rho | 98.81 | 0 |
| NGO0260 | Putative GTPase | NMB0687 | GTP-binding protein Era | 98.39 | 2.00E-156 |
| NGO0295 | Threonyl-tRNA synthetase | NMB0720 | Threonyl-tRNA synthetase | 98.12 | 0 |
| NGO0296 | Translation initiation factor IF-3 | NMB0721 | Translation initiation factor IF-3 | 99.35 | 9.70E-086 |
| NGO0297 | 50S ribosomal protein L35 | NMB0722 | 50S ribosomal protein L35 | 100 | 2.10E-034 |
| NGO0584 | 50S ribosomal protein L9 | NMB1320 | 50S ribosomal protein L9 | 99.25 | 2.00E-059 |
| NGO0905 | Conserved hypothetical protein | NMB1437 | Conserved hypothetical protein | 97.42 | 2.00E-132 |
| NGO1246c | Serine protease | NMB1587 | Putative protease | 96.86 | 1.00E-171 |
| NGO1413 | Na+-translocating NADH-ubiquinone | NMB0569 | Na+-translocating NADH-ubiquinone | 96.2 | 0 |
| NGO1748c | NADH dehydrogenase I chain D | NMB0244 | NADH dehydrogenase 1 chain D | 98.56 | 0 |
| NGO1776 | Glyceraldehyde 3-phosphate | NMB0207 | Glyceraldehyde 3-phosphate | 99.42 | 0 |
| NGO1818c | DNA-directed RNA polymerase alpha | NMB0168 | DNA-directed RNA polymerase alpha | 98.78 | 0 |
| NGO1822c | Preprotein translocase SecY subunit | NMB0162 | Preprotein translocase SecY subunit | 99.08 | 0 |
| NGO1824.1c | 50S ribosomal protein L18 | NMB0158 | 50S ribosomal protein L18 | 100 | 3.30E-045 |
| NGO1826.1c | 30S ribosomal protein S14 | NMB0155 | 30S ribosomal protein S14 | 100 | 5.40E-043 |
| NGO1828c | 50S ribosomal protein L24 | NMB0153 | 50S ribosomal protein L24 | 98.13 | 6.30E-045 |
| NGO1829c | 50S ribosomal protein L14 | NMB0152 | 50S ribosomal protein L14 | 99.18 | 1.80E-065 |
| NGO1830c | 30S ribosomal protein S17 | NMB0151 | 30S ribosomal protein S17 | 98.85 | 2.00E-045 |
| NGO1836c | 50S ribosomal protein L23 | NMB0144 | 50S ribosomal protein L23 | 91.76 | 1.70E-039 |
| NGO1851c | DNA-directed RNA polymerase, beta subunit | NMB0132 | DNA-directed RNA polymerase, beta subunit | 99.28 | 0 |
| NGO1867c | Two-component signal sensor kinase | NMB0114 | Putative nitrogen regulation protein NtrY | 93.77 | 0 |
| NGO1870c | Methionyl-tRNA formyltransferase | NMB0111 | Methionyl-tRNA formyltransferase | 97.4 | 1.00E-162 |
Abbreviation: % ID, percent identity.
E value as calculated by BLASTP.
TABLE 4.
N. gonorrhoeae and N. meningitidis orthologs differentially expressed under low-iron conditions
|
N. gonorrhoeae
|
N. meningitidis
|
% IDa | E valueb | ||
|---|---|---|---|---|---|
| ORF | Gene product | ORF | Gene product | ||
| NGO1690c | Conserved hypothetical protein | NMB0311 | Hypothetical protein | 96.3 | 1.60E-042 |
| NGO1496c | Transferrin-binding protein B | NMB0460 | Transferrin-binding protein B | 61.59 | 0 |
| NGO1495c | Transferrin-binding protein A | NMB0461 | Transferrin-binding protein A | 93.99 | 0 |
| NGO0217c | ABC transporter, periplasmic-binding protein | NMB0634 | Iron(III) ABC transporter, periplasmic-binding protein | 99.09 | 3.00E-178 |
| NGO0322 | Conserved hypothetical protein | NMB0744 | Hypothetical protein | 96.23 | 7.70E-087 |
| NGO0385 | Delta-aminolevulinic acid dehydratase | NMB0801 | Delta-aminolevulinic acid dehydratase | 98.2 | 1.00E-177 |
| NGO0165c | Conserved hypothetical protein | NMB0861 | Hypothetical protein | 97.79 | 6.20E-091 |
| NGO1442 | Alcohol dehydrogenase | NMB1395 | Alcohol dehydrogenase | 70.5 | 5.50E-023 |
| NGO0108 | Conserved hypothetical protein | NMB1796 | Conserved hypothetical protein | 99.38 | 4.10E-093 |
| NGO1066 | MafI protein | NMB1798 | IS1016 transposase fragment | 96.55 | 4.50E-011 |
| NGO0025 | AraC family transcriptional regulator | NMB1879 | Hypothetical protein | 99.03 | 6.20E-059 |
| NGO2093c | Ferric enterobactin receptor FetA | NMB1988 | Iron-regulated outer membrane protein FrpB | 93.28 | 0 |
Abbreviation: % ID, percent identity.
E value as calculated by BLASTP.
Under low-iron conditions (Table 4), expression of genes encoding four conserved hypothetical proteins was shared by the two organisms, as well as the AraC family transcriptional regulator noted above. Not surprisingly, the genes encoding FbpA, FetA, HemO, TbpA, and TbpB were also regulated similarly in each pathogen. In MC58, the paralogous lactoferrin receptor genes lbpA and lbpB were also derepressed. In FA1090, the lbpAB operon has been inactivated by a large deletion that removes the promoter and a significant portion of the lbpB ORF (40).
While hypothetical proteins and conserved hypothetical proteins were the largest functional category of both organisms derepressed under low-iron conditions, only four of these iron-repressible proteins were shared by N. gonorrhoeae and N. meningitidis. Twenty of the derepressed meningococcal hypothetical and 30 of the gonococcal hypothetical protein-encoding genes are unique to their respective genomes. The gonococcus causes a sexually transmitted disease and primarily inhabits the urogenital tract, whereas the meningococcus colonizes the upper respiratory tract and often invades the bloodstream of susceptible individuals. So, while these two organisms share a high degree of genetic similarity, the diseases they cause are very different; the iron-regulated hypothetical proteins of each organism probably play a significant role in the specific pathogenesis of these diseases.
In summary, use of microarray analysis to examine differences in gene expression of N. gonorrhoeae in response to iron availability resulted in demonstrating that roughly 10% of gonococcal ORFs are involved in this response. Under high-iron conditions, the predominance of up-regulated genes encode proteins involved in protein synthesis, while under low-iron conditions, the predominance of derepressed genes encode proteins of unknown function. Of these ORFs roughly 30% are putatively controlled by the Fur protein, with additional control being provided by a MerR-like regulator. Likewise, a putative Fur-directed transcriptional cascade is suggested. Comparison of the gonococcal data to similar meningococcal data has revealed a surprisingly low degree of overlap between the iron regulons of the two organisms, suggesting that specific responses of each organism to iron availability dictate in part the different diseases that each pathogen causes.
Supplementary Material
Acknowledgments
We thank A. F. Gillaspy for critical review of the manuscript and J. K. Davies for printing the pan-Neisseria arrays.
This work was supported by USPHS grants 5P20-RR-15564 (COBRE) and 5P20-RR-016478 (INBRE).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aisen, P., and A. Leibman. 1972. Lactoferrin and transferrin: a comparative study. Biochim. Biophys. Acta 257:314-323. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. E., M. M. Hobbs, G. D. Biswas, and P. F. Sparling. 2002. Presented at the Thirteenth International Pathogenic Neisseria Conference, Oslo, Norway.
- 4.Anderson, J. E., P. F. Sparling, and C. N. Cornelissen. 1994. Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J. Bacteriol. 176:3162-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagg, A., and J. B. Neilands. 1987. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 51:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey, T. L., and C. Elkan. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:21-29. [PubMed] [Google Scholar]
- 7.Barrett, T., T. O. Suzek, D. B. Troup, S. E. Wilhite, W. C. Ngau, P. Ledoux, D. Rudnev, A. E. Lash, W. Fujibuchi, and R. Edgar. 2005. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 33:D562-D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berish, S. A., T. A. Mietzner, L. W. Mayer, C. A. Genco, B. P. Holloway, and S. A. Morse. 1990. Molecular cloning and characterization of the structural gene for the major iron-regulated protein expressed by Neisseria gonorrhoeae. J. Exp. Med. 171:1535-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beucher, M., and P. F. Sparling. 1995. Cloning, sequencing, and characterization of the gene encoding FrpB, a major iron-regulated, outer membrane protein of Neisseria gonorrhoeae. J. Bacteriol. 177:2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas, G. D., J. E. Anderson, C. J. Chen, C. N. Cornelissen, and P. F. Sparling. 1999. Identification and functional characterization of the Neisseria gonorrhoeae lbpB gene product. Infect. Immun. 67:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas, G. D., J. E. Anderson, and P. F. Sparling. 1997. Cloning and functional characterization of Neisseria gonorrhoeae tonB, exbB and exbD genes. Mol. Microbiol. 24:169-179. [DOI] [PubMed] [Google Scholar]
- 12.Biswas, G. D., and P. F. Sparling. 1995. Characterization of lbpA, the structural gene for a lactoferrin receptor in Neisseria gonorrhoeae. Infect. Immun. 63:2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulton, I. C., M. K. Yost, J. E. Anderson, and C. N. Cornelissen. 2000. Identification of discrete domains within gonococcal transferrin-binding protein A that are necessary for ligand binding and iron uptake functions. Infect. Immun. 68:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brener, D., I. W. DeVoe, and B. E. Holbein. 1981. Increased virulence of Neisseria meningitidis after in vitro iron-limited growth at low pH. Infect. Immun. 33:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Britigan, B. E., M. S. Cohen, and P. F. Sparling. 1985. Gonococcal infection: a model of molecular pathogenesis. N. Engl. J. Med. 312:1683-1694. [DOI] [PubMed] [Google Scholar]
- 16.Brown, D. M., V. Stone, P. Findlay, W. MacNee, and K. Donaldson. 2000. Increased inflammation and intracellular calcium caused by ultrafine carbon black is independent of transition metals or other soluble components. Occup. Environ. Med. 57:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 18.Cadieux, N., P. G. Phan, D. S. Cafiso, and R. J. Kadner. 2003. Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc. Natl. Acad. Sci. USA 100:10688-10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carsiotis, M., B. A. Stocker, D. L. Weinstein, and A. D. O'Brien. 1989. A Salmonella typhimurium virulence gene linked to flg. Infect. Immun. 57:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carson, S. D., P. E. Klebba, S. M. Newton, and P. F. Sparling. 1999. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J. Bacteriol. 181:2895-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, I., and E. C. Gotschlich. 2001. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J. Bacteriol. 183:3160-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen, M. S., and P. F. Sparling. 1992. Mucosal infection with Neisseria gonorrhoeae. Bacterial adaptation and mucosal defenses. J. Clin. Investig. 89:1699-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornelissen, C. N., J. E. Anderson, I. C. Boulton, and P. F. Sparling. 2000. Antigenic and sequence diversity in gonococcal transferrin-binding protein A. Infect. Immun. 68:4725-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornelissen, C. N., M. Kelley, M. M. Hobbs, J. E. Anderson, J. G. Cannon, M. S. Cohen, and P. F. Sparling. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol. Microbiol. 27:611-616. [DOI] [PubMed] [Google Scholar]
- 25.Coy, M., and J. B. Neilands. 1991. Structural dynamics and functional domains of the fur protein. Biochemistry 30:8201-8210. [DOI] [PubMed] [Google Scholar]
- 26.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. Weblogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyer, D. W., W. McKenna, J. P. Woods, and P. F. Sparling. 1987. Isolation by streptonigrin enrichment and characterization of a transferrin-specific iron uptake mutant of Neisseria meningitidis. Microb. Pathog. 3:351-363. [DOI] [PubMed] [Google Scholar]
- 28.Dyer, D. W., E. P. West, and P. F. Sparling. 1987. Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect. Immun. 55:2171-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flemming, D., A. Schlitt, V. Spehr, T. Bischof, and T. Friedrich. 2003. Iron-sulfur cluster N2 of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) is located on subunit NuoB. J. Biol. Chem. 278:47602-47609. [DOI] [PubMed] [Google Scholar]
- 31.Forng, R. Y., C. R. Ekechukwu, S. Subbarao, S. A. Morse, and C. A. Genco. 1997. Promoter mapping and transcriptional regulation of the iron-regulated Neisseria gonorrhoeae fbpA gene. J. Bacteriol. 179:3047-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genco, C. A., and P. J. Desai. 1996. Iron acquisition in the pathogenic Neisseria. Trends Microbiol. 4:179-184. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez de Peredo, A., C. Saint-Pierre, J. M. Latour, I. Michaud-Soret, and E. Forest. 2001. Conformational changes of the ferric uptake regulation protein upon metal activation and DNA binding; first evidence of structural homologies with the diphtheria toxin repressor. J. Mol. Biol. 310:83-91. [DOI] [PubMed] [Google Scholar]
- 35.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallberg, L., and L. Rossander-Hulten. 1991. Iron requirements in menstruating women. Am. J. Clin. Nutr. 54:1047-1058. [DOI] [PubMed] [Google Scholar]
- 37.Holbein, B. E., K. W. Jericho, and G. C. Likes. 1979. Neisseria meningitidis infection in mice: influence of iron, variations in virulence among strains, and pathology. Infect. Immun. 24:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 39.Köhler, R., J. Fanghänel, B. König, E. Lüneberg, M. Frosch, J.-U. Rahfeld, R. Hilgenfeld, G. Fischer, J. Hacker, and M. Steinert. 2003. Biochemical and functional analyses of the Mip protein: influence of the N-terminal half and of peptidylprolyl isomerase activity on the virulence of Legionella pneumophila. Infect. Immun. 71:4389-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis, L. A., A. F. Gillaspy, R. E. McLaughlin, M. Gipson, T. F. Ducey, T. Ownbey, K. Hartman, C. Nydick, M. Carson, J. Vaugh, C. Thomson, L. Song, S. Lin, X. Yuan, F. Najar, M. Zhan, Q. Ren, H. Zhu, S. Qi, S. M. Kenton, H. Lai, J. D. White, S. Clifton, B. A. Roe, and D. W. Dyer. Submitted for publication.
- 41.Lewis, L. A., M. Gipson, K. Hartman, T. Ownbey, J. Vaughn, and D. W. Dyer. 1999. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32:977-989. [DOI] [PubMed] [Google Scholar]
- 42.Lewis, L. A., E. Gray, Y. P. Wang, B. A. Roe, and D. W. Dyer. 1997. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23:737-749. [DOI] [PubMed] [Google Scholar]
- 43.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masri, H. P., and C. N. Cornelissen. 2002. Specific ligand binding attributable to individual epitopes of gonococcal transferrin binding protein A. Infect. Immun. 70:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masson, L., and B. E. Holbein. 1985. Influence of nutrient limitation and low pH on serogroup B Neisseria meningitidis capsular polysaccharide levels: correlation with virulence for mice. Infect. Immun. 47:465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masuda, T., K. Inoue, M. Masuda, M. Nagayama, A. Tamaki, H. Ohta, H. Shimada, and K. Takamiya. 1999. Magnesium insertion by magnesium chelatase in the biosynthesis of zinc bacteriochlorophyll a in an aerobic acidophilic bacterium Acidiphilium rubrum. J. Biol. Chem. 274:33594-33600. [DOI] [PubMed] [Google Scholar]
- 47.Merrell, D. S., L. J. Thompson, C. C. Kim, H. Mitchell, L. S. Tompkins, A. Lee, and S. Falkow. 2003. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect. Immun. 71:6510-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merz, A. J., and M. So. 2000. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16:423-457. [DOI] [PubMed] [Google Scholar]
- 49.Morse, S. A., and L. Bartenstein. 1980. Purine metabolism in Neisseria gonorrhoeae: the requirement for hypoxanthine. Can. J. Microbiol. 26:13-20. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 51.Nilsson, G., J. G. Belasco, S. N. Cohen, and A. von Gabain. 1987. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc. Natl. Acad. Sci. USA 84:4890-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfenninger-Li, X. D., S. P. Albracht, R. van Belzen, and P. Dimroth. 1996. NADH:ubiquinone oxidoreductase of Vibrio alginolyticus: purification, properties, and reconstitution of the Na+ pump. Biochemistry 35:6233-6242. [DOI] [PubMed] [Google Scholar]
- 55.Posey, J. E., J. M. Hardham, S. J. Norris, and F. C. Gherardini. 1999. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. USA 96:10887-10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ratliff, M., W. Zhu, R. Deshmukh, A. Wilks, and I. Stojiljkovic. 2001. Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohde, K. H., and D. W. Dyer. 2003. Mechanisms of iron acquisition by the human pathogens Neisseria meningitidis and Neisseria gonorrhoeae. Front. Biosci. 8:d1186-d1218. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 60.Saunders, N. J., C. M. Kahler, L. A. S. Snyder, E. A. Stohl, I. A. Gunesekere, M. Apicella, J. P. Dillard, T. F. Ducey, D. W. Dyer, D. Entz, J. P. Folster, P. Jordan, D. Powell, H. S. Seifert, W. M. Shafer, D. Williams, and J. K. Davies. Submitted for publication.
- 61.Serkin, C. D., and H. S. Seifert. 2000. Iron availability regulates DNA recombination in Neisseria gonorrhoeae. Mol. Microbiol. 37:1075-1086. [DOI] [PubMed] [Google Scholar]
- 62.Silver, S., and M. Walderhaug. 1992. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol. Rev. 56:195-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sonnhammer, E. L., S. R. Eddy, and R. Durbin. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405-420. [DOI] [PubMed] [Google Scholar]
- 65.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 66.Soong, C. L., J. Ogawa, E. Sakuradani, and S. Shimizu. 2002. Barbiturase, a novel zinc-containing amidohydrolase involved in oxidative pyrimidine metabolism. J. Biol. Chem. 277:7051-7058. [DOI] [PubMed] [Google Scholar]
- 67.Steuber, J., W. Krebs, and P. Dimroth. 1997. The Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio alginolyticus—redox states of the FAD prosthetic group and mechanism of Ag+ inhibition. Eur. J. Biochem. 249:770-776. [DOI] [PubMed] [Google Scholar]
- 68.Stevenson, P., P. Williams, and E. Griffiths. 1992. Common antigenic domains in transferrin-binding protein 2 of Neisseria meningitidis, Neisseria gonorrhoeae, and Haemophilus influenzae type b. Infect. Immun. 60:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 71.Swanson, J., S. Bergstrom, O. Barrera, K. Robbins, and D. Corwin. 1985. Pilus− gonococcal variants. Evidence for multiple forms of piliation control. J. Exp. Med. 162:729-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas, C. E., and P. F. Sparling. 1996. Isolation and analysis of a fur mutant of Neisseria gonorrhoeae. J. Bacteriol. 178:4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 74.Turner, P. C., C. E. Thomas, I. Stojiljkovic, C. Elkins, G. Kizel, D. A. Ala'Aldeen, and P. F. Sparling. 2001. Neisserial TonB-dependent outer-membrane proteins: detection, regulation and distribution of three putative candidates identified from the genome sequences. Microbiology 147:1277-1290. [DOI] [PubMed] [Google Scholar]
- 75.Unniraman, S., R. Prakash, and V. Nagaraja. 2002. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res. 30:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Deuren, M., P. Brandtzaeg, and J. W. van der Meer. 2000. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin. Microbiol. Rev 13:144-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walker, J. E. 1992. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25:253-324. [DOI] [PubMed] [Google Scholar]
- 78.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.West, S. E., and P. F. Sparling. 1987. Aerobactin utilization by Neisseria gonorrhoeae and cloning of a genomic DNA fragment that complements Escherichia coli fhuB mutations. J. Bacteriol. 169:3414-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeugin, J. A., and J. L. Hartley. 1985. Ethanol precipitation of DNA. Focus 7:1-2. [Google Scholar]
- 81.Zhu, W., A. Wilks, and I. Stojiljkovic. 2000. Degradation of heme in gram-negative bacteria: the product of the hemO gene of neisseriae is a heme oxygenase. J. Bacteriol. 182:6783-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.