Abstract
In gram-negative organisms, enzymes belonging to the low-molecular-weight protein tyrosine phosphatase (LMPTP) family are involved in the regulation of important physiological functions, including stress resistance and synthesis of the polysaccharide capsule. LMPTPs have been identified also in gram-positive bacteria, but their functions in these organisms are presently unknown. We cloned two putative LMPTPs from Bacillus subtilis, YfkJ and YwlE, which are highly similar to each other in primary structure as well as to LMPTPs from gram-negative bacteria. When purified from overexpressing Escherichia coli strains, both enzymes were able to dephosphorylate p-nitrophenyl-phosphate and phosphotyrosine-containing substrates in vitro but showed significant differences in kinetic parameters and sensitivity to inhibitors. Transcriptional analyses showed that yfkJ was transcribed at a low level throughout the growth cycle and underwent a σB-dependent transcriptional upregulation in response to ethanol stress. The transcription of ywlE was growth dependent but stress insensitive. Genomic deletion of each phosphatase-encoding gene led to a phenotype of reduced bacterial resistance to ethanol stress, which was more marked in the ywlE deletion strain. Our study suggests that YfkJ and YwlE play roles in B. subtilis stress resistance.
In higher eukaryotes, protein phosphorylation on tyrosine residues is a key mechanism for cell-cell communication, signal transduction, vesicle traffic, and regulation of cell proliferation and differentiation (21, 36). Tyrosine phosphorylation also plays an important role in many other aspects of cell physiology and embryonic development (14, 21). The human genome contains 90 genes for protein tyrosine kinases (PTKs) and 107 genes for protein tyrosine phosphatases (PTPs) (1).
The study of protein tyrosine phosphorylation in bacteria presents some technical challenges (13). Nevertheless, a number of reports have demonstrated the presence of tyrosine-phosphorylated intracellular proteins in archaea (47), mycobacteria (7), and gram-negative organisms (23, 27, 39, 51). PTKs and PTPs have also been isolated from bacteria (9, 41), suggesting that this posttranslational modification may play an important role in bacterial physiology.
Genome sequencing projects have shown that enzymes similar to eukaryotic PTPs are encoded by many bacteria and archaea (24, 25). Most of the chromosome-encoded PTPs are class I or class II enzymes (1), also known as dual (serine/threonine and tyrosine)-specificity PTPs and low-Mr PTPs (LMPTPs), respectively. Classical class I PTPs, like the plasmid-encoded YopH, which is a virulence factor of Yersinia pestis and Yersinia pseudotuberculosis, have also been identified (1, 10). LMPTPs are among the most ancient and highly conserved subfamilies of PTPs. The first bacterial LMPTPs were isolated from Erwinia amylovora (5), Acinetobacter johnsonii (15), and Escherichia coli (54). These enzymes show a remarkably high degree of identity (∼30%) with their human homologs.
The physiological functions of bacterial LMPTPs remain largely unknown, although some hints are given by the organization of genes surrounding the PTPs on the chromosome. For example, the gene encoding the AmsI LMPTP of E. amylovora is located within the ams operon, which controls capsular exopolysaccharide synthesis (5). Similarly, the Ptp LMPTP of A. johnsonii can dephosphorylate the transmembrane autokinase Ptk, which regulates the synthesis of colanic acid, a fundamental component of the Acinetobacter capsule (15). In fact, control of polysaccharide capsule composition by an autokinase-LMPTP pair encoded in the cps or cps-like operon has been found to be a conserved feature among gram-negative organisms (55). For example, Wzb, one of the pair of LMPTPs (Wzb and Etp) in E. coli (54), as well as one LMPTP from Klebsiella pneumoniae (42), dephosphorylate a tyrosine autokinase that regulates capsule composition. However, LMPTP functions in gram-negatives are apparently not limited to the regulation of polysaccharide capsule composition: a recent report by Klein et al. (27) showed that the Etp LMPTP of E. coli regulates heat shock resistance by dephosphorylating the sigma factor RpoH and the anti-sigma factor RseA. Interestingly, this function was not shared by the second LMPTP in E. coli, Wzb, providing a first example of nonoverlapping functions of LMPTP pairs in bacteria.
LMPTPs have recently also been found in gram-positive organisms (48), but it is not yet known if these enzymes have functions similar to those of their homologs in gram-negative bacteria. Although the regulation of capsular composition by an autokinase/PTP pair seems to occur in Streptococcus pneumoniae and Staphylococcus aureus (3, 34, 46), it was found that the PTP encoded by the cps operon of S. pneumoniae belongs to an unrelated family of Mn2+-dependent PTPs (35) rather than being an LMPTP. The first PTP isolated from B. subtilis, YwqE, is also a Mn2+-dependent enzyme and is located in a cps-like operon. Within this operon, YwqE is able to dephosphorylate the tyrosine autokinase YwqD and the kinase substrate YwqF, which is a UDP-glucose dehydrogenase (30, 32).
In the present study we report the genetic and biochemical characterizations of two B. subtilis LMPTPs, YfkJ and YwlE. The two enzymes are active as PTPs in vitro but show significant differences in their biochemical properties. Deletion of the corresponding coding genes affected B. subtilis resistance to ethanol stress.
MATERIALS AND METHODS
Bioinformatic and statistical analyses.
BLAST searches and amino acid sequence comparisons were performed using the NCBI site (http://www.ncbi.nlm.nih.gov/BLAST/). Genomic context analysis was performed using SEED (http://theseed.uchicago.edu/FIG/index.cgi), a genomic database and annotation platform provided by the Fellowship for Interpretation of Genomes. This software supports a comparative genome context analysis across a collection of ∼300 complete (or almost complete) genomes (38). Experimental errors were evaluated according to the method of J. R. Taylor (50) using the Excel X for Mac software (Microsoft Corporation).
Reagents.
Pfu Turbo was from Stratagene (La Jolla, CA). BIOMOL GREEN reagent was purchased from Biomol Research Labs (Plymouth Meeting, PA). Tyrosine phosphopeptides were provided by E. Ruoslahti (The Burnham Institute), and the serine and threonine phosphopeptides were purchased from Upstate (Charlottesville, VA). All other reagents were from ICN Biomedicals (Irvine, CA) or Sigma (St. Louis, MO) unless otherwise stated.
B. subtilis culture and physiological assays.
Transformation of B. subtilis was performed as described by Anagnostopoulos and Spizizen (2). Liquid cultures of B. subtilis were performed in Luria-Bertani (LB) medium or in Schaeffer's sporulation medium (SM) (17). Induction of transcription by ethanol (EtOH) stress was analyzed by following the procedure described by Price et al., using 5% EtOH (43). EtOH survival assays were carried out as described by Völker et al. (56) with slight modifications. Cells were grown overnight at 37°C in LB agar and then inoculated in LB medium and grown at 37°C to an optical density at 540 nm of 0.3 to 0.4, at which time EtOH was added to a final concentration of 9% (vol/vol). The specific growth rate of the culture was monitored before and after the stress. Aliquots of the culture were sampled at 30, 60, 90, 180, and 260 min, and appropriate dilutions were plated in duplicate on LB agar to determine cell viability. Survival was calculated as a ratio between the average number of colonies at the indicated times and the number of colonies at time zero (11, 56). For analysis of motility, B. subtilis strains were spotted onto a semisolid plates of tryptone (1.0 g tryptone, 0.5 g NaCl, 0.27 g agar in 100 ml water; after sterilization, 1 ml of 10× Spizizen salts, 5 μg/ml tryptophan, and 5 μg/ml phenylalanine were added to the media) and CK's proline (0.25 g agar was solubilized in 100 ml water and sterilized before the addition of 1 ml of 10× Spizizen salts, 5 μg/ml tryptophan, 5 μg/ml phenylalanine, 0.7 mM sorbitol, and 0.2 mM proline) (28, 59). As the bacteria metabolize nutrients, a chemical gradient is established, resulting in the formation of a ring as the bacteria tax outwards. Differences in the swarm diameters show diversity in strain motility. B. subtilis strains were tested for α-amylase and protease activity by growing them on TBAB (tryptose blood agar base; Difco) starch and TBAB milk plates, respectively, prepared as described previously (17). Antibiotic selection in B. subtilis strains was carried out at the following concentrations: for chloramphenicol, 5 μg/ml; for kanamycin, 2 μg/ml; for spectinomycin, 50 μg/ml; and for erythromycin, 1 μg/ml plus lyncomycin at 25 μg/ml.
Construction of B. subtilis strains and isolation of recombinant proteins.
B. subtilis strains and plasmids used are listed in Table 1. Primers used in the study are listed in Table 2. Site-directed mutagenesis was performed using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's recommendations. All mutations were verified by nucleotide sequencing. Genomic DNA from B. subtilis strain JH642 was used as the template for PCR amplifications. The yfkJ coding sequence was cloned between the BamHI and HindIII sites of the pEGST vector (primers FKJBAM-F and FKJHIN-R), which allows the expression of proteins with an N-terminal glutathione S-transferase (GST) tag (26). The same yfkJ fragment and the ywlE coding sequence were also cloned between the NdeI and XhoI sites of the pET15b vector (Novagen, Madison, WI) (primers KJNDE-F and KJXHO-R for yfkJ and WLENDE-F and WLEXHO-R for ywlE) in order to express the proteins with an N-terminal His6 tag.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL10-Gold | Stratagene | |
| BL21(DE3) codonPlus-RIL | Stratagene | |
| B. subtilis | ||
| JH642 | trpC2 pheA1 | Lab stock |
| JH19000 | JH642asigB::cat | 11 |
| LM58 (ΔyfkJ) | JH642::pLM58 yfkJ::[erm PspaclacZ] | This study |
| LM24 (ΔywlE) | JH642::pLM24aywlE::spc | This study |
| LM02 | JH642::pLM02ayfkJ::cat | This study |
| LM50 | LM02::pLM24ayfkJ::cat ywlE::spc | This study |
| LM08 | JH642::pLM06B amyE::[yfkJp-lacZ::aph] | This study |
| LM14 | JH19000::pLM06B sigB amyE::[yfkJp-lacZ::aph] | This study |
| LM35 | JH642::pLM35 amyE::[ywlEp-lacZ::aph] | This study |
| LM60 | JH19000::pLM35 sigB amyE::[ywlEp-lacZ::aph] | This study |
| Plasmids | ||
| pEGST | GST gene in pET23b (Novagen) | 26 |
| pET15b | Cleavable N terminus His6 tag | Novagen |
| pJM134A | Spectinomycin cassette in pBluescriptIIKS (+) | Unpublished |
| pMUTIN2 | PspaclacZ + erythromycin cassette | 32 |
| pJM115 | lacZ transcriptional fusion that integrates in amyE | 40 |
| pLM58 | Construct for nonpolar deletion of yfkJ in pMUTIN2 | This study |
| pLM24 | Left and right arms of ywlE in pJM134 | This study |
| pLM02 | Left and right arms of yfkJ in pJM105A | This study |
| pLM06B | yfkJ promoter in pJM115 | This study |
| pLM35 | ywlE promoter in pJM115 | This study |
This strain was transformed with linearized plasmids.
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| FKJBAM-F | CGGGATCCATGATAAGCGTGTTATTTGTTTG |
| FKJHIN-R | CCCAAGCTTCACAATTGTTTTTCTTTTTG |
| KJNDE-F | GGAATTCCATATGATAAGCGTGTTATTTGTTTG |
| KJXHO-R | CCGCTCGAGTCACAATTGTTTTTCTTTTTGAATG |
| WLENDE-F | GGAATTCCATATGGATATTATTTTTGTCTGTACTG |
| WLEXHO-R | CCGCTCGAGTTATCTACGGTCTTTTTTCAGC |
| MUTKJHIND-FW | CCCAAGCTTTCTCCGATGGCGGAAGC |
| MUTKJBAM-RV | CGGGATCCCGGGTACGTCAGCCAG |
| JRIGHTHIND-F | CCCAAGCTTGTGAAAGGAGAATTTGCATG |
| JRIGHTKPN-R | CGGGGTACCCCTTCATGAAGGGTGTATC |
| JLEFTSAC-F | GCATGAGCTCGGGAAATGATAAAAAAGACTCG |
| JLEFTPST-R | GCATCTGCAGGATGTGCAACCTCCCTATTC |
| WLEHIND-5′ | CCCAAGCTTGACCGTAGATAAGTTGTCAG |
| WLEKPN-3′ | ACCCGGGTACCCATCGCAAGG |
| WLEBAM-5′ | GGATGGGATCCGGTAACATGCTGTC |
| WLEECO-3′ | AAAAAGAATTCCCATGTCAGTCAC |
| YFKJ5′SAC2 | TGGGAGAGCTCTTCAATCATTTGAGTTAATG |
| JLEFTPST-R | GCATCTGCAGGATGTGCAACCTCCCTATTC |
| WLEPROMECO-FW | CGGAATTCGTTCTTGGGGTTCAAATG |
| WLEPROMBAM-RV | CGGGATCCCGTATTTCCAGTACAGAC |
| H3 | CCGGACGATCGCATTC |
| LFRV | CTACAGGTTTTTCTCTTCAT |
| 64F | AAACAACGCCGCAAGTGATG |
| 63R | GGCGATAATCCCGAATGACA |
| YWLF-FW | GTAGCCATTGCATCGGATC |
| YWLF-RV | CGCGTTTGGTGTCTTCCC |
| KJC8SAVR-F | GATAAGCGTGTTATTTGTTTCCCTAGGTAACATTTGCCG |
| KJC8SAVR-R | CGGCAAATGTTACCTAGGGAAACAAATAACACGCTTATC |
| KJD125ABSP-F | GATCTGGCTGACGTACCTGCTCCTTACTACACAGGGAAC |
| KJD125ABSP-R | GTTCCCTGTGTAGTAAGGAGCAGGTACGTCAGCCAGATC |
| YFKJR14KNDE-F | GTTTGTTTAGGTAACATATGCAAGTCTCCGATGGCG |
| YFKJR14KNDE-R | CGCCATCGGAGACTTGCATATGTTACCTAAACAAAC |
| WLEC8SAGEI-F | ATGGATATTATTTTTGTCTCTACCGGTAATACGTGCCGC |
| WLEC8SAGEI-R | GCGGCACGTATTACCGGTAGAGACAAAAATAATATCCAT |
| WLED118ANHEI-F | CATGGTGATGTGCTAGCTCCGTTCGGCGGCTCAATTGAC |
| WLED118ANHEI-R | GTCAATTGAGCCGCCGAACGGAGCTAGCACATCACCATG |
| WLER13KBSPMI-F | ACTGGAAATACCTGCAAGAGCCCAATGGCTGAGGCGC |
| WLER13KBSPMI-R | GCGCCTCAGCCATTGGGCTCTTGCAGGTATTTCCAGT |
Restriction sites used for cloning purposes are in bold.
A nonpolar deletion of yfkJ (strain LM58 [ΔyfkJ]) was achieved by single-crossover integration of a plasmid constructed using the vector pMUTIN2, carrying a 330-bp yfkJ internal fragment cloned between HindIII and BamHI (primers MUTKJHIND-FW and MUTKJBAM-RV) and an erythromycin resistance cassette (32). Integration of the plasmid caused disruption of yfkJ expression, while expression of the genes 3′ to yfkJ in the same operon (yfkI and yfkH) was placed under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Pspac promoter. A polar deletion of yfkJ (strain LM02) was achieved by double-crossover integration of a plasmid constructed using the vector pJM105A (40), which replaced 446 bp within the yfkJ gene with a chloramphenicol cassette. The right arm was cloned between the HindIII and KpnI sites (primers JRIGHTHIND-F and JRIGHTKPN-R), while the left arm was cloned between the SacI and PstI sites (primers JLEFTSAC-F and JLEFTPST-R). The genomic deletion of ywlE (strain LM24 [ΔywlE]) was achieved by double-crossover integration of a plasmid constructed using the vector pJM134A (M. Perego, unpublished data), which replaced 426 bp within the ywlE gene with a spectinomycin cassette carrying a terminator. The right arm was cloned between the HindIII and KpnI sites (primers WLEHIND5′ and WLEKPN3′), while the left arm was cloned between the PstI and EcoRI sites (primers WLEBAM5′ and WLEECO3′). A B. subtilis strain carrying a double deletion of yfkJ and ywlE was also generated by transformation of the LM02 strain with the DNA construct used for generating the ΔywlE deletion strain. Diagnostic PCRs were carried out to ensure plasmid integration within the yfkJ operon or deletion of yfkJ or ywlE.
The promoter regions of yfkJ (from nucleotide [nt] −255 to nt +12 from the ATG codon) and ywlE (from nt −387 to nt +33 from the ATG codon) were cloned, respectively, in the SmaI site and between the EcoRI and BamHI sites of the vector pJM115 (40) (primers YFKJ5′SAC2 and JLEFTPST-R for yfkJp and primers WLEPROMECO-FW and WLEPROMBAM-RV for ywlEp), which allows the expression of a transcriptional fusion with E. coli β-galactosidase upon integration via double crossover at the amyE gene of the B. subtilis chromosome.
Isolation of GST- or His6-tagged recombinant proteins was performed by single-step affinity chromatography using glutathione Sepharose 4B (Amersham Biosciences, Piscataway, NJ) or Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN Inc., Valencia, CA), respectively. The purity of recombinant proteins was consistently over 95%, as determined by Coomassie blue staining of gels.
Phosphopeptides and phosphosubstrate dephosphorylation assays.
Dephosphorylation of phosphopeptides and other phosphorylated compounds was carried out at 37°C in a 50- or 100-μl total volume. For YfkJ, the reaction buffer was 50 mM sodium citrate, pH 6.0, or 50 mM Bis-Tris, pH 6.0. The substrate hydrolysis rate was measured either by reading the p-nitrophenol absorbance at 405 nm after the addition of 200 μl of 1 M NaOH to the reaction mixture and using the extinction coefficient of 18.000 M−1 cm−1 for p-nitrophenol or by reading the absorbance at 620 nm after the addition of 100 to 200 μl of BIOMOL GREEN reagent to the reaction mixture and calculating the release of inorganic phosphate by comparison with a standard curve of inorganic phosphate. The nonenzymatic hydrolysis of the substrate was corrected by measuring the control without addition of enzyme. For YwlE, the reaction buffer was 50 mM MES (pH 5.5), 1 mM dithiothreitol (DTT) and the hydrolysis rate was always measured by the addition of BIOMOL GREEN reagent. Absorbance readings were carried out on a PowerWaveX340 microplate spectrophotometer (Bio-Tek Instruments, Inc., Winooski, VT). The time of the reaction, amount of enzyme, and concentration of the substrate were optimized to have a linear kinetics. The initial hydrolysis rate (v0) was measured in triplicate for p-nitrophenyl-phosphate (pNPP) and in duplicate for phosphopeptides.
Kinetic parameters were determined by fitting the data to the Michaelis-Menten equation or to the Michaelis-Menten with substrate (S) inhibition equation: v0 = Vmax[S]/{Km + [S](1 + [S]/Ki)} (see http://www.lsbu.ac.uk/biology/enztech/inhibition.html), using nonlinear regression and the Prism software (GraphPad Prism version 4.00 for Mac OS X; GraphPad Software, San Diego, CA). The Ki for vanadate was evaluated by fitting the data to the Michaelis-Menten equation for competitive inhibition: v0 = Vmax[S]/(KMapp + [S]), where KMapp is Km(1 + [I]/Ki) (where [I] is the concentration of the inhibitor, in molar units) using the same software.
Purification and dephosphorylation of Spo0F∼P and Spo0B∼P.
His6-Spo0F (50 μM) was phosphorylated with KinA (0.5 μM) and [γ-32P]ATP (0.3 μM at a specific activity of 6,000 Ci/mmol) in the phosphorelay buffer (50 mM EPPS [4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid], pH 8.5, 20 mM MgCl2, 0.1 mM EDTA, 5% glycerol) for 1 h at room temperature. ATP was added to a final concentration of 1 mM, and the phosphotransfer was continued for another hour (52). Spo0F∼P was purified by using Ni-NTA agarose equilibrated with binding buffer (50 mM Tris-HCl, pH 8, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol). The column was washed with 10 ml of binding buffer, and protein elution was obtained with binding buffer containing 150 mM EDTA. Fractions containing radioactivity were dialyzed in 20 mM HEPES-NaOH, pH 7.0, 0.1 mM EDTA, 10% glycerol. Aliquots were stored at −80°C. The ratio of Spo0F to Spo0F∼P in the purified sample was estimated by analysis on a 10% Tris-glycine native polyacrylamide gel electrophoresis gel stained with Coomassie blue. The concentration of Spo0F/Spo0F∼P used throughout this study refers to the fraction of Spo0F∼P only.
His6-Spo0B (100 μM) was labeled in a 250-μl reaction mixture containing KinA (10 μM), Spo0F (100 μM), and [γ-32P]ATP (0.3 μM at a specific activity of 6,000 Ci/mmol) in the phosphorelay buffer (50 mM EPPS, pH 8.5, 20 mM MgCl2, 0.1 mM EDTA, 5% glycerol) for 1 h at room temperature. ATP was added to a final concentration of 1 mM, and the phosphotransfer was continued for another hour (53). The reaction was mixed with Ni-NTA agarose equilibrated with binding buffer containing 20 mM Tris-HCl, pH 7.4, 50 mM KCl, 5 mM imidazole, and 5 mM β-mercaptoethanol. Two additional washes with binding buffer containing 10 mM imidazole were carried out before the protein was eluted in buffer containing 20 mM Tris-HCl, pH 7.4, 50 mM KCl, 250 mM imidazole, and 5 mM β-mercaptoethanol. Fractions (500 μl) were collected, and 4 μl of each was run on a 15% sodium dodecyl sulfate (SDS)-acrylamide gel. The gel was directly exposed to a PhosphorImager screen (Molecular Dynamics/Amersham Biosciences, Piscataway, NJ) and analyzed after 1 h of exposure. Fractions containing radioactivity were dialyzed in 20 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, 1 mM DTT and concentrated using a Centricon-10 concentrator (Amicon/Millipore, Bedford, MA). Protein concentration was determined with a Bradford-based Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA).
The dephosphorylation assay used to test YwlE and YfkJ activity on Spo0F∼P and Spo0B∼P was carried out at 37°C in a reaction buffer containing 50 mM HEPES-NaOH, pH 7.0, 2 mM EDTA, 1 mM DTT. A time course assay was done in which His-YwlE (1.5 μM) or GST-YfkJ (1.5 μM) was incubated with 1.5 μM Spo0F∼P or Spo0B∼P. Control reactions were performed using Spo0F∼P or Spo0B∼P in the reaction buffer in the absence of the phosphatase. At the time points indicated in the figure, 15 μl of samples was withdrawn and the reactions were stopped by the addition of 3 μl 5× SDS loading buffer (25 mM Tris-HCl, pH 6.8, 1.5% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.02% bromophenol blue). Samples were loaded onto 15% SDS-PAGE. Electrophoresis was carried out at constant voltage (100 V) for 2 h. The gels were dried and exposed overnight to a PhosphorImager screen and then analyzed with Image Quant software (Molecular Dynamics/Amersham Biosciences, Piscataway, NJ).
β-Galactosidase assay.
β-Galactosidase assays were performed as described previously (12). Cultures were grown in LB medium or SM. Samples were taken at the stated intervals, and the activities are expressed in Miller units (33).
RT-PCR.
RNA was extracted from strains JH642, LM58 (ΔyfkJ), and LM24 (ΔywlE) grown in 3 ml LB medium overnight at 37°C using the RNAqueous kit from Ambion (Austin, TX). Turbo-DNase (Ambion, Austin, TX) was added to each RNA preparation at a final concentration of 0.05 U/μg RNA/μl for 1 h at 37°C and then inactivated by addition of EDTA to a final concentration of 5 mM and heating the mixture at 75°C for 10 min. The amount of extracted RNA was determined by measuring its absorbance at 260 nm. Reverse transcription (RT) was performed on 400 ng RNA, starting from nt +337 from the ATG of yfkH or from nt +1060 from the ATG of ywlE, using the primer H3′ or LFRV, respectively, and ImPromII reverse transcriptase (Promega, Madison, WI). PCRs on the RT products were performed using primers KJNDE-FW and KJXHO-RV for yfkJ, 64F and 63R for yfkIH, and YWLF-FW and YWLF-RV for ywlF, with AmpliTaq polymerase (Roche, Basel, Switzerland). PCR performed using equal amounts of nonretrotranscribed RNA was used as a control for genomic DNA contamination of the RNA samples.
RESULTS
Bioinformatic analysis of B. subtilis LMPTPs.
A BLAST analysis of the completed and annotated B. subtilis genome (29) was carried out using the human LMPTP-A tyrosine phosphatase as the query sequence. Two genes were identified, yfkJ and ywlE, predicted to encode proteins with high degrees of amino acid identity to LMPTP-A (39% [E = 10−20] and 26% [E = 2 × 10−6], respectively). Additionally, YfkJ and YwlE showed a 29% amino acid identity and 53% similarity (E = 9 × 10−8) to each other (the matrix used was BLOSUM62). An amino acid sequence alignment of YfkJ and YwlE with other prokaryotic and eukaryotic LMPTPs is shown in Fig. 1. Two amino acid sequence motifs are conserved in all LMPTPs: the CXGNXCR stretch at the N terminus defines the P-loop of LMPTPs and corresponds to the CX5R motif in all PTPs (44), while the DP motif is part of the phosphatase D-loop and is followed by two tyrosines in nearly all LMPTPs (44, 58).
FIG. 1.
Amino acid sequence alignment of B. subtilis YfkJ and YwlE with LMPTPs from prokaryotic and eukaryotic organisms. Amino acid residues that are conserved in all the considered species are in bold. The boxes indicate the catalytic motifs referred to in the text.
Our BLAST search revealed the presence of an additional gene with a lower degree of similarity to the human LMPTPs. This protein is a low-molecular-weight arsenate reductase (arsC/yqcM) and is known to have a similar fold but a quite different catalytic mechanism and extremely weak phosphatase activity in vitro (4). We also found that YfkJ and YwlE are included in a single cluster of orthologs (COG) (49) available at the website http://www.ncbi.nlm.nih.gov/cgi-bin/COG/palox?COG0394. On a similarity tree (also available at the same website), based on multiple sequence alignments of all proteins belonging to the COG, YwlE appears to be distant from YfkJ, and both B. subtilis enzymes are distant from the E. coli LMPTPs.
On the B. subtilis annotated chromosome, yfkJ seems to be the first of a putative three-gene operon (yfkJ, yfkI, and yfkH), while ywlE seems to form an operon with the genes ywlC, ywlD, ywlF, and ywlG (29). We performed for the two genes a genome context analysis, including clustering of genes on the chromosome, domain fusions, and cooccurrence profiles. Such analyses often provide important clues to cellular functions of genes (38). A genome context analysis of yfkJ (illustrated in Fig. S1 of the supplemental material) revealed that homologs of genes encoding the inducible RNase BN (yfkH), described for E. coli (6), and a small conserved protein of unknown function (yfkK) have a strong tendency to cooccur in the genomic neighborhood of yfkJ. This tendency is observed over a broad range of gram-positive bacteria, including most of the sequenced species of Bacillus and Staphylococcus aureus. yfkH and yfkK genes are clustered on the chromosome in all sequenced strains of Listeria spp., where yfkJ orthologs are present in a remote locus. All three genes, yfkJ, yfkH, and yfkK, are conserved but dispersed over the chromosome of Bacillus halodurans. On the other hand, homologs of yfkI (for which there is no known function) are present (and coclustered) only in a compact group, including in Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis.
A genome context analysis performed for ywlE revealed an extensive pattern of conservation and possible functional coupling (illustrated in Fig. S2 of the supplemental material). A conserved chromosomal cluster containing ywlE extends well beyond the boundaries of a possible ywl operon. Distal genes prfA and hemK, as well as ywlC, are the most conserved components of this cluster, as detected in a total of 28 sequenced genomes of rather divergent gram-positive bacteria, including all sequenced species of Bacillus, Staphylococcus, Listeria, and Clostridium and of Moorella thermoacetica, Exiguobacterium sp., and Thermoanaerobacter tengcongensis. Interestingly, all three genes are united by a common “theme”—ribosome and translation. The gene hemK encodes an essential and widely conserved N5-glutamine methyltransferase, which modifies peptide release factors, including the one encoded by its conserved chromosomal neighbor, the essential gene prfA (18). Although a precise function of ywlC (widely conserved in many bacteria) is not known, its homolog SUA5 is an essential gene in Saccharomyces cerevisiae implicated in the regulation of translation (37). Among other less conserved components of the cluster are three metabolic enzymes (YwlF [ribose 5-phosphate isomerase], GlyA [serine hydroxymethyltransferase], and Upp [uracil phosphoribosyltransferase]) and two proteins of unknown function (encoded by ywlD and ywlG). Notably, upp, often located immediately downstream of ywlE, forms a bifunctional fusion, YwlE-Upp, in T. tengcongensis. Finally, the spoIIR gene, involved in the regulation of stage II sporulation, is coclustered with ywlE only in a subset of Bacillus subspp.
Biochemical characterization of recombinant B. subtilis LMPTPs.
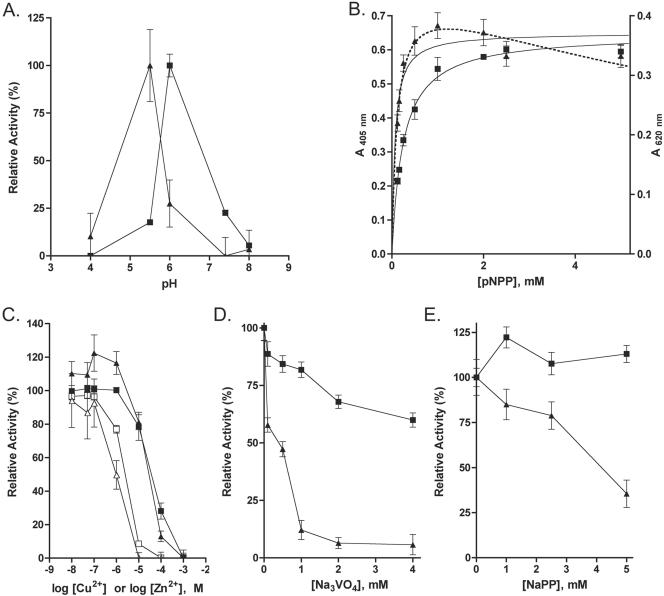
To first characterize the two B. subtilis LMPTPs in vitro, we cloned their genes into E. coli expression vectors. YwlE was purified as a His6-tagged protein, while YfkJ was purified both as a His6 and as an N-terminal GST fusion protein from E. coli lysates. The phosphatase activity of B. subtilis LMPTPs was then measured using pNPP as the substrate. The pH optima for His-YfkJ and His-YwlE were 6.0 and 5.5, respectively (Fig. 2A). Inclusion of 1 mM DTT significantly improved the activity of YwlE (data not shown). Both YfkJ and YwlE dephosphorylated pNPP according to Michaelis-Menten kinetics (Fig. 2B), but YwlE was inhibited by a high concentration of the substrate. The Km values were found to be 0.244 ± 0.009 mM for His-YfkJ and 0.157 ± 0.024 mM for His-YwlE, respectively. These values are comparable to those reported for S. aureus PtpA and PtpB (48) and consistent with those reported for several eukaryotic and prokaryotic Cys-based PTPs and for eukaryotic class IV PTPs (a newly isolated class of metal-dependent and non-Cys-based eukaryotic PTPs) (8, 15, 20, 45). The rate constants (kcat) were calculated to be 0.640 ± 0.006 s−1 for His-YfkJ and 0.010 ± 0.001 s−1 for His-YwlE, which are similar, respectively, to the reported kcat of S. aureus PtpA and PtpB for pNPP (48). We noticed that the activities of S. aureus and B. subtilis LMPTPs on pNPP appear to be several orders of magnitude lower than those of several eukaryotic PTPs or some gram-negative PTPs (8, 15, 20, 45).
FIG. 2.
Biochemical characterization of B. subtilis YfkJ and YwlE enzymatic activity using pNPP as the substrate. (A) Effect of pH on YfkJ (squares) and YwlE (triangles) enzymatic activities. The assays contained 3 mM pNPP as the substrate. Buffers used were sodium citrate (pHs 4.0 and 6.0), MES-NaOH (pH 5.5), and Tris-HCl (pHs 7.4 and 8.0). Data represent relative activities. (B) Analysis of YfkJ (squares) and YwlE (triangles) enzymatic activities using pNPP as the substrate. The graphs represent mean activities and nonlinear fits of the experimental data to the Michaelis-Menten equation (continuous lines). The dotted line is a nonlinear fit of YwlE data to the Michaelis-Menten equation for substrate inhibition (see http://www.lsbu.ac.uk/biology/enztech/inhibition.html). (C, D, E) Effect of CuCl2 (C, open symbols), ZnCl2 (C, filled symbols), Na3VO4 (D), and sodium pyrophosphate (E) on YfkJ (squares) and YwlE (triangles) enzymatic activities. The buffer used for YfkJ was 50 mM sodium citrate or 50 mM Bis-Tris (pH 6.0), and the buffer used for YwlE was 50 mM MES (pH 5.5), 1 mM DTT. The assays contained 3 mM pNPP as the substrate, and data represent relative activities. Error bars in all graphs represent standard errors; if not visible, they are within the resolution of the point.
Both Cu2+ and Zn2+ efficiently inhibited the activity of YfkJ and YwlE (Fig. 2C), while the addition of Mg2+, Ca2+, or Fe3+ to the reaction buffer had minimal effects on both phosphatases even at 5 mM (data not shown). YfkJ and YwlE were differently sensitive to Na3VO4, a classic PTP competitive inhibitor: while it inhibited YwlE in a competitive manner, with a Ki of 224 nM, YfkJ was insensitive to this inhibitor (Fig. 2D and data not shown). The serine phosphatase inhibitors NaF and NaPP did not inhibit YfkJ, while YwlE was insensitive to NaF but was significantly inhibited by 5 mM NaPP (data not shown and Fig. 2E). In contrast, 5 mM NEM completely inhibited both enzymes, while 5 mM iodoacetamide reduced the activity of YfkJ by 65% and of YwlE by 79% (data not shown).
The enzymes were then assayed using phosphoamino acids, phosphopeptides, and other phosphorylated compounds as substrates. For comparison, we included the human LMPTP-A and -B enzymes, which are considered to be tyrosine-specific enzymes in vitro and in vivo (44). YfkJ showed modest activity on phosphotyrosine but was unable to dephosphorylate a phosphotyrosine-containing peptide modeled after the C terminus of the mammalian tyrosine kinase Lck (Table 2). YwlE, on the other hand, was as active against phosphotyrosine as human LMPTP-B and was even more active against several tyrosine phosphopeptides (Table 2). Neither of the two enzymes were active on phosphoserine- or phosphothreonine-containing substrates. The two bacterial LMPTPs and the two human enzymes dephosphorylated other phospho-compounds very similarly (Table 2), with the exception of a moderate activity of YwlE against glucose-1-phosphate. Both enzymes were able to dephosphorylate β-naphthyl phosphate but were inactive against α-naphthyl phosphate, which is consistent with our and other reported data on human LMPTPs (57). Neither YfkJ nor YwlE were able to dephosphorylate phospho-Spo0F (previously phosphorylated on Asp-54 by KinA) or phospho-Spo0B (previously phosphorylated on His-30 by phosphate transfer from Spo0F) over a prolonged period of time, suggesting that N-phosphoamino acids are not substrates for the two enzymes (data not shown). Overall, our data are compatible with YwlE and YfkJ being tyrosine-specific phosphatases. The low activity of YfkJ against the phosphotyrosine-containing peptide included in the screen is likely due to a stricter requirement for certain amino acid residues and/or structural determinants surrounding the phosphorylated tyrosine. Alternatively, YfkJ might be specific for a different phosphosubstrate(s) not included in our screen.
Next, we evaluated the role of amino acid residues predicted to be critical for catalysis. Since the GST-YfkJ protein was kinetically identical to His6-YfkJ against pNPP (data not shown), site-directed mutagenesis was performed on the GST-YfkJ construct. Mutation of Cys-8 to Ser, Arg-14 to Lys, or Asp-125 to Ala completely abolished the enzymatic activity of YfkJ. Corresponding mutations of Cys-7, Arg-13, and Asp-118 in YwlE also completely abolished the enzymatic activity (data not shown). Cys-8/-7 and Arg-14/-13 are critical elements of the P-loop (44) and phosphocysteinyl-substrate intermediate formation, while Asp-125/-118 in the D-loop functions as general base in the catalytic mechanism of LMPTPs (44).
Transcriptional analysis of B. subtilis LMPTPs.
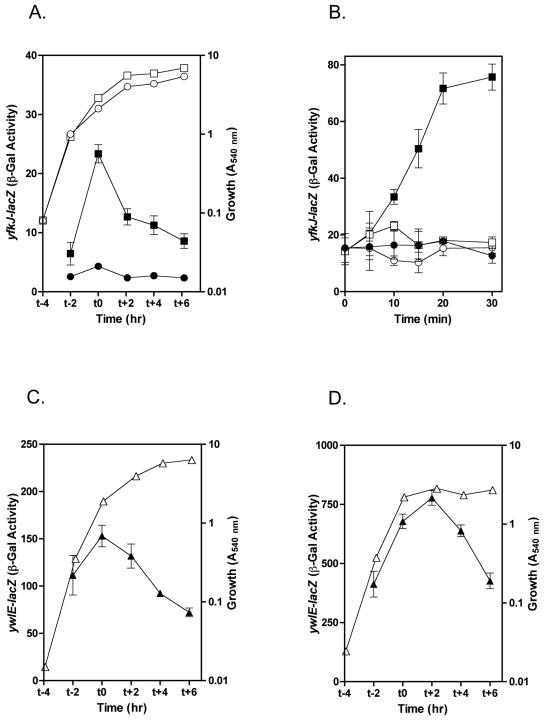
The promoter region of yfkJ includes recognition sequences for both σA- and σB-dependent transcription (43). By direct DNA array analysis, Price et al. (43) detected an upregulation of yfkJ transcription after 5 min of ethanol stress in B. subtilis. By rapid amplification of cDNA ends-PCR they also mapped a σB-dependent 5′ end of the yfkJ mRNA immediately downstream from the putative yfkJ σB promoter (43). In order to assess whether the 5′-end-flanking regions of yfkJ and ywlE contain promoter activity and regulate such activity in various growth phases and conditions, we cloned a 255-bp fragment (from nt −255 from yfkJ ATG) containing the whole yfkK-yfkJ intergenic region and a 420-bp fragment 5′ to ywlE ATG (from nt −387 from ywlE ATG to nt +33 from ywlE ATG, a fragment long enough to include putative ywlE promoter sequences) into the transcriptional fusion vector pJM115 (40), thus creating, respectively, the yfkJ promoter-lacZ and ywlE promoter-lacZ transcriptional fusion plasmids. The B. subtilis strain carrying the yfkJ transcriptional fusion showed an overall low β-galactosidase activity in the absence of stress, and for this reason it was analyzed in comparison to a control B. subtilis strain transformed with the empty vector alone. Transcription from the yfkJ promoter showed a peak of activity in the late exponential phase of growth when bacteria were grown in liquid LB medium (Fig. 3A). When bacteria were grown in sporulation medium, yfkJ activity was almost identical to the control throughout the growth cycle (data not shown). Figure 3B shows that ethanol stress, carried out as described by Price et al. using 5% ethanol (43), caused a rapid induction of yfkJ transcription. When the experiments were performed in a B. subtilis strain that lacks σB, the induction of yfkJ transcription by ethanol stress was absent, confirming that yfkJ transcription is stress inducible and σB regulated. The pattern and rates of induction of β-galactosidase activity from the yfkJ transcriptional fusion during ethanol stress were compatible with the ones obtained by Price et al. for the yfkJ transcript (43).
FIG. 3.
Transcriptional analysis of B. subtilis yfkJ and ywlE. (A, C, D) β-Galactosidase activity from yfkJ (A) and ywlE (C, D) transcriptional fusions during growth in LB medium (A, C) or SM (D). Filled symbols represent β-galactosidase activity expressed in Miller units (33). Open symbols represent average optical densities (540 nm) of bacterial culture at the indicated times. (B) Induction of yfkJ transcription by ethanol stress. Ethanol-induced β-galactosidase activity from the yfkJ transcriptional fusion in wild-type bacteria (strain LM08 [squares]) or bacteria lacking σB (strain LM14 [circles]). Graphs represent β-galactosidase activities expressed in Miller units (33) from bacteria subjected (filled symbols) or not subjected (open symbols) to ethanol treatment. Standard errors have been calculated for all β-galactosidase activity data and reported as error bars. When error bars are not visible, they are within the resolution of the points.
The β-galactosidase activity of the ywlE transcriptional fusion showed a peak in the exponential phase of growth, followed by a significant and gradual reduction in the stationary/sporulation phase (Fig. 3C and D). This pattern was independent from the growth medium, although the activity was much higher when bacteria were grown in SM (Fig. 3D). We did not observe any increase in β-galactosidase activity during ethanol stress (data not shown), suggesting that ywlE and yfkJ transcriptional regulation might differ in their stress sensitivities. We concluded from these experiments that the 5′-end-flanking regions of both LMPTPs contain sequences necessary for promoter activity. Regulation of these two promoter regions shows significant differences between the two enzymes.
Role of B. subtilis LMPTPs in stress resistance.
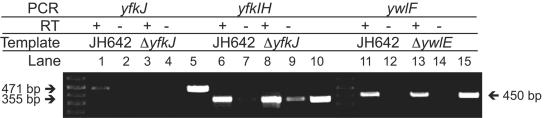
In order to study the physiological role of the two B. subtilis LMPTPs, we generated two B. subtilis strains carrying a deletion of yfkJ or of ywlE. To avoid polar effects on yfkI and yfkH, yfkJ was disrupted by integration of a pMUTIN2-based plasmid, which places the transcription of genes downstream from yfkJ under the control of an IPTG-inducible promoter (32). The absence of polar effects in our LMPTP deletion strain was confirmed by RT-PCR amplification of genes downstream of yfkJ and ywlE. Figure 4 shows that the transcription of yfkI and of ywlF was maintained in the ΔyfkJ and ΔywlE strains, respectively. Considering the strategies used to generate the deletion strains, these results suggest that the yfkJ deletion was nonpolar and that ywlF does not form an operon with ywlE.
FIG. 4.
RT-PCR analysis of JH642, LM58 (ΔyfkJ), and LM24 (ΔywlE) B. subtilis strains. cDNA was synthesized from total RNA using a reverse primer within yfkH (lanes 1 to 9) or within ywlF (lanes 11 to 14) and subsequently amplified by PCR using primers within yfkJ (lanes 1 to 4), yfkIH (lanes 6 to 9), or ywlF (lanes 11 to 14). RNA was extracted from lysates of JH642 (lanes 1, 2, 6, 7, 11, and 12) or a ΔyfkJ (lanes 3, 4, 8, and 9) or ΔywlE (lanes 13 and 14) strain. Each RT-PCR lane is followed by a control lane for PCR performed using equal amounts of nonretrotranscribed RNA (lanes 2, 4, 7, 9, 12, and 14). Control PCRs are also shown, using genomic DNA of the wild-type JH642 strain as the template and primers within yfkJ (lane 5), yfkIH (lane 10), or ywlF (lane 15). Data shown in lanes 1 to 10 are from bacteria grown in the presence of 1 mM IPTG.
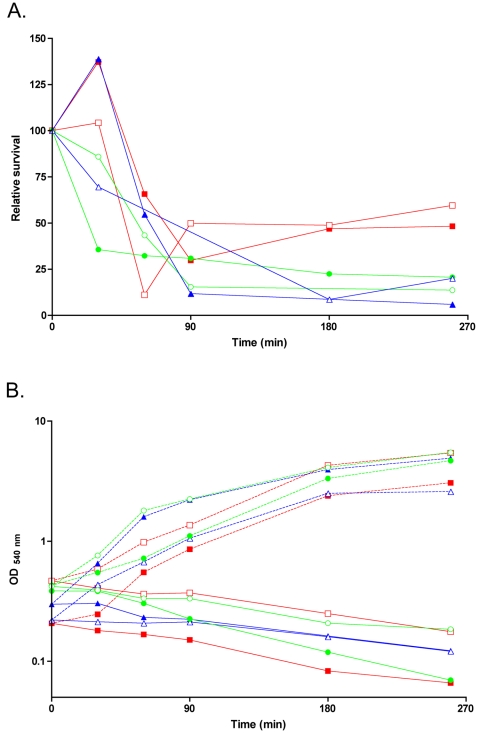
Deletion of one or the other phosphatase did not affect bacterial growth in liquid LB medium or SM at 37°C and in solid SM at 28°C, 37°C, or 50°C (data not shown). The deletion of one or the other phosphatase did not affect protease or amylase production or bacterial motility (data not shown). The σB-dependent upregulation of yfkJ transcription during ethanol stress prompted us to analyze the role of LMPTPs in the general stress response. Thus, the B. subtilis strains carrying deletions of one or the other LMPTPs were analyzed for their ability to respond to ethanol stress, which is known to be an efficient activator of the σB regulon. Figure 5 shows that both deletions were able to affect bacterial resistance to EtOH stress for exposure times of 90 min or longer. The nonpolar deletion of yfkJ led to a moderate reduction (around 50%) of EtOH stress resistance (Fig. 5). An identical phenotype was shown by the B. subtilis strain LM02, carrying a polar deletion of yfkJ, in two independent experiments (data not shown). The deletion of ywlE led to a more marked (more than 75%) decrease of B. subtilis survival (Fig. 5) after EtOH stress. The strain LM50 carrying a deletion of both phosphatases showed a phenotype similar to that of the ΔywlE strain (Fig. 5).
FIG. 5.
Resistance of LM58 (ΔyfkJ), LM24 (ΔywlE), and LM50 (double deletion) B. subtilis strains to ethanol stress. Bacteria were grown in LB medium, and lethal stress was administered by the addition of 9% ethanol to early-log-phase cultures as described by Völker et al. (56). Appropriate dilutions of samples taken at the indicated time points were plated in duplicate on LB solid medium. Survival was calculated as the ratio of the average number of colonies at the indicated times after application of 9% ethanol stress to the number of colonies at time zero. Relative survival was calculated as a ratio between the survival of each strain and the survival of the wild-type JH642 strain under the same experimental conditions. (A) Relative survival after subjection to 9% ethanol stress of the ΔyfkJ (red lines), ΔywlE (blue lines), or double-deletion (green lines) strain. Results for the ΔyfkJ strain have been obtained by growing both the wild type and deletion strains in the presence of 1 mM IPTG. (B) Growth curves of bacteria used in the same experiments and of control bacteria not subjected to EtOH stress. In panels A and B, two independent experiments for each deletion strain (indicated by filled and empty symbols) are shown.
DISCUSSION
In this study we cloned and biochemically characterized two LMPTPs (YfkJ and YwlE) from B. subtilis. The presence of two enzymatically active LMPTPs has been reported for several other bacteria, including E. coli (54) and S. aureus (48). The two enzymes were active on pNPP and tyrosine phosphosubstrates in vitro but showed significant differences in their biochemical properties. The optimal pH in the acidic range and the pattern of enzyme sensitivity to mutations of critical residues suggested that both enzymes dephosphorylate their substrate with a mechanism similar to that of known Cys-based PTPs. Overall the pattern of sensitivity of YfkJ and YwlE to inhibitors is very similar to that reported for eukaryotic and prokaryotic LMPTPs and in general for Cys-based PTPs. The insensitivity of YfkJ to vanadate represents an important exception and, to the best of our knowledge, has not been reported for any other LMPTP. Also the inhibition of YwlE by millimolar concentrations of NaPP that we observe has not been reported for other LMPTPs and might be of physiological relevance, as B. subtilis is known to contain between 1.2 and 6 mM pyrophosphate, depending on the growth conditions (31).
The kinetic parameters of YfkJ and YwlE against pNPP were similar to those of their orthologs in S. aureus, PtpA and PtpB, which are the only other LMPTPs characterized so far from gram-positive bacteria (48). YwlE showed good specific activity against tyrosine-phosphorylated substrates. In contrast, YfkJ was able to dephosphorylate only free O-phosphotyrosine. This suggests that YfkJ may have a stricter requirement for specific sequence determinants surrounding the phosphorylated residue. None of the two enzymes showed significant activity against other O- or N-phosphorylated amino acids.
The present study also showed that there are significant differences in chromosomal and operon structures between LMPTPs in gram-negative organisms and in B. subtilis, and between the two LMPTPs in B. subtilis. Gram-negative LMPTPs are usually located near a gene encoding a tyrosine kinase within cps or cps-like operons (55). No such gene could be identified in the operons containing YfkJ or YwlE, or even in their vicinity, in B. subtilis. Although no exhaustive biochemical comparison between LMPTP pairs isolated from gram-negative organisms has been reported to date, differences in biochemical behavior and chromosomal organization seem to be more common among gram-positive than gram-negative LMPTP pairs. Indeed, YfkJ and YwlE share much less similarity in primary structure to each other than LMPTP pairs encoded by gram-negative genomes, like the E. coli YccY (also called Etp) and Wzb (27, 55). Our phylogenetic analysis showed that LMPTP pairs in gram-positive organisms are evolutionarily distant from each other compared to their gram-negative counterparts. The genome context analysis for the two B. subtilis enzymes did not deliver specific predictions but provided useful guidelines for further experimental studies of functional coupling of each phosphatase gene with coclustered neighbors. It is tempting to speculate that each LMPTP might be involved in the control of the phosphorylation state of one or more of these proteins or their interaction partners.
Transcriptional analyses showed interesting differences between the two B. subtilis LMPTPs. The yfkJ promoter region contains a σB recognition sequence, and yfkJ transcription has been shown by Price et al. (43) and by our data (Fig. 4) to be upregulated in a σB-dependent manner during ethanol stress. The 5′ region of the ywlE gene also showed promoter activity, which drove transcription of a reporter gene in a growth-dependent, but ethanol insensitive, pattern.
The stress-dependent upregulation of yfkJ transcription, together with the recent finding that E. coli Etp plays a critical role in bacterial resistance to heat stress (27), prompted us to analyze the possible role of YfkJ and YwlE in the ethanol stress resistance of B. subtilis. Genomic deletion of each phosphatase led to a phenotype of reduced bacterial resistance to ethanol stress, which was more pronounced in the ywlE deletion strain. Deletion of both phosphatases did not lead to an additive phenotype, suggesting that YwlE is at least partially able to compensate for the loss of YfkJ. The role of each phosphatase and their partial overlap in B. subtilis resistance to ethanol and possibly other stresses needs to be further investigated by complementation and cross-complementation experiments.
In gram-negative organisms tyrosine phosphorylation phenomena are known to be involved in the virulence of Aeromonas spp. (51), and interestingly, the expression of the tyrosine autokinase Etk was found to be restricted to some pathogenic strains of E. coli (22). A possible involvement of LMPTP genes in virulence would not be surprising, considering that stress resistance plays a role in the virulence of several pathogenic bacilli. For example B. anthracis needs to overcome the extreme oxidative stress and very low pH in the macrophage phagosome during outgrowth (16). The role of tyrosine phosphorylation pathways in the σB-dependent and -independent stress resistance of virulent bacilli also warrants further investigation.
In conclusion, we isolated two B. subtilis LMPTPs and found that they behave as bona fide tyrosine phosphatases. We also found that they play a role in ethanol stress resistance. Protein tyrosine phosphorylation has recently been shown to be present in B. subtilis. The first PTK and PTP pair with their substrate have recently been isolated from this organism and characterized (32). The final elucidation of the mechanism of action of YfkJ and YwlE in B. subtilis stress resistance will require the isolation of their physiological substrate(s) and/or interactor(s) during bacterial stress.
Supplementary Material
TABLE 3.
Substrate analysis of YfkJ and YwlE
| Substrateb | Activity of enzyme (Miller units)a
|
|||
|---|---|---|---|---|
| YfkJ | YwlE | LMPTP-A | LMPTP-B | |
| pNPP | 100 | 100 | 100 | 100 |
| FTATEGQpYQPQP | 0 | 18.1 | 61.7 | 17.1 |
| FTATEGQpYQPIP | 0 | 7.6 | 41.3 | 12.6 |
| FTATEGQpYQEIP | 0.4 | 31.8 | 65.3 | 18.8 |
| FTATEGQpYEEIP | 0.1 | 36.9 | 43.2 | 12.1 |
| RRApSVA | 0 | 0 | 2.9 | 4.0 |
| KRpTIRR | 0 | 0 | 3.8 | 1.6 |
| p-Tyr | 2.3 | 13.2 | 82.7 | 26.6 |
| p-Ser | 0 | 3.6 | 6.2 | 3.8 |
| p-Thr | 0 | 0 | 0 | 0 |
| α-Naphthyl phosphate | 0.6 | 3.5 | 4.5 | 1.2 |
| β-Naphthyl phosphate | 21.7 | 69.9 | 102.4 | 98.1 |
| Glucose-1-phosphate | 0 | 20.8 | 7.3 | 5.4 |
| Glucose-6-phosphate | 0 | 0 | 1.4 | 0 |
| Ribose-5-phosphate | 0 | 0 | 8.8 | 0 |
| β-Glycerol phosphate | 0 | 0 | 0.4 | 0 |
| Pyridoxal 5′ Phosphate | 0 | 0 | 0 | 0 |
| IMP | 0.9 | 4.7 | 2.1 | 1.2 |
| ATP | 0 | 0 | 0.5 | 0 |
| ADP | 0 | 0 | 0 | 0 |
| AMP | 0.7 | 2.1 | 2.2 | 0.1 |
| GMP | 0.6 | 2.5 | 4.2 | 0 |
| CMP | 0.7 | 2.3 | 2.8 | 0.5 |
| UMP | 0 | 0 | 1.5 | 1.7 |
Values are relative activities of B. subtilis His-YfkJ and His-YwlE and of human GST-LMPTP-A and GST-LMPTP-B.
The reaction buffer was 50 mM MES (pH 5.5) with 1 mM DTT for His-YwlE or 50 mM Na citrate (pH. 6.0) for the other enzymes. Substrates were present at a 1 mM concentration. pY, phosphotyrosine; pS, phosphoserine; pT, phosphothreonine.
Acknowledgments
This work was partially supported by grants from the National Institutes of Health (to T.M. and NIGMS-GM55594 to M.P.) and the Danish National Research Council (SNF). The Stein Beneficial Trust supported in part oligonucleotide synthesis and DNA sequencing.
We thank James Hoch (The Scripps Research Institute) for helpful discussion.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alonso, A., J. Sasin, N. Bottini, I. Friedberg, I. Friedberg, A. Osterman, A. Godzik, T. Hunter, J. Dixon, and T. Mustelin. 2004. The set of genes encoding protein tyrosine phosphatase family members in the human genome. Cell 117:699-711. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, M. H., and J. Yother. 2001. CpsB is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J. Biol. Chem. 276:47966-47974. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, M. S., Z. Guan, M. Laurberg, and X. D. Su. 2001. Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 98:13577-13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bugert, P., and K. Geider. 1997. Characterization of the amsI gene product as a low molecular weight acid phosphatase controlling exopolysaccharide synthesis of Erwinia amylovora. FEBS Lett. 400:252-256. [DOI] [PubMed] [Google Scholar]
- 6.Callahan, C., D. Neri-Cortes, and M. P. Deutscher. 2000. Purification and characterization of the tRNA-processing enzyme RNase BN. J. Biol. Chem. 275:1030-1034. [DOI] [PubMed] [Google Scholar]
- 7.Chow, K., D. Ng, R. Stokes, and P. Johnson. 1994. Protein tyrosine phosphorylation in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 124:203-207. [DOI] [PubMed] [Google Scholar]
- 8.Cirri, P., T. Fiaschi, P. Chiarugi, G. Camici, G. Manao, G. Raugei, and G. Ramponi. 1996. The molecular basis of the differing kinetic behavior of the two low molecular mass phosphotyrosine protein phosphatase isoforms. J. Biol. Chem. 271:2604-2607. [DOI] [PubMed] [Google Scholar]
- 9.Dadssi, M., and A. J. Cozzone. 1990. Evidence of protein-tyrosine kinase activity in the bacterium Acinetobacter calcoaceticus. J. Biol. Chem. 265:20996-20999. [PubMed] [Google Scholar]
- 10.DeVinney, I., I. Steele-Mortimer, and B. B. Finlay. 2000. Phosphatases and kinases delivered to the host cell by bacterial pathogens. Trends Microbiol. 8:29-33. [DOI] [PubMed] [Google Scholar]
- 11.Dowds, B. C., and J. A. Hoch. 1991. Regulation of the oxidative stress response by the hpr gene in Bacillus subtilis. J. Gen. Microbiol. 137:1121-1125. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, E., D. J. Henner, M. Perego, and J. A. Hoch. 1988. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J. Bacteriol. 170:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, R., J. Thorner, and G. S. Martin. 1989. Nucleotidylation, not phosphorylation, is the major source of the phosphotyrosine detected in enteric bacteria. J. Bacteriol. 171:272-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grangeasse, C., P. Doublet, C. Vincent, E. Vaganay, M. Riberty, B. Duclos, and A. J. Cozzone. 1998. Functional characterization of the low-molecular-mass phosphotyrosine-protein phosphatase of Acinetobacter johnsonii. J. Mol. Biol. 278:339-347. [DOI] [PubMed] [Google Scholar]
- 16.Guidi-Rontani, C. 2002. The alveolar macrophage: the Trojan horse of Bacillus anthracis. Trends Microbiol. 10:405-409. [DOI] [PubMed] [Google Scholar]
- 17.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. John Wiley and Sons, New York, N.Y.
- 18.Heurgue-Hamard, V., S. Champ, A. Engstrom, M. Ehrenberg, and R. H. Buckingham. 2002. The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himanen, J. P., and D. B. Nikolov. 2003. Eph receptors and ephrins. Int. J. Biochem. Cell. Biol. 35:130-134. [DOI] [PubMed] [Google Scholar]
- 20.Howell, L. D., C. Griffiths, L. W. Slade, M. Potts, and P. J. Kennelly. 1996. Substrate specificity of IphP, a cyanobacterial dual-specificity protein phosphatase with MAP kinase phosphatase activity. Biochemistry 35:7566-7572. [DOI] [PubMed] [Google Scholar]
- 21.Hunter, T. 1998. The role of tyrosine phosphorylation in cell growth and disease. Harvey Lect. 94:81-119. [PubMed] [Google Scholar]
- 22.Ilan, O., Y. Bloch, G. Frankel, H. Ullrich, K. Geider, and I. Rosenshine. 1999. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 18:3241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly-Wintenberg, K., T. Anderson, and T. C. Montie. 1990. Phosphorylated tyrosine in the flagellum filament protein of Pseudomonas aeruginosa. J. Bacteriol. 172:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennelly, P. J. 2002. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 206:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Kennelly, P. J. 2003. Archaeal protein kinases and protein phosphatases—insights from genomics and biochemistry. Biochem. J. 370:373-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kholod, N., and T. Mustelin. 2001. Novel tools for studying protein complexes: vectors for co-expression of two proteins in Escherichia coli. BioTechniques 31:322-328. [DOI] [PubMed] [Google Scholar]
- 27.Klein, G., C. Dartigalongue, and S. Raina. 2003. Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol. Microbiol. 48:269-285. [DOI] [PubMed] [Google Scholar]
- 28.Kristich, C. J., G. D. Glekas, and G. W. Ordal. 2003. The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol. Microbiol. 47:1353-1366. [DOI] [PubMed] [Google Scholar]
- 29.Kunst, F., et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 30.Mijakovic, I., L. Musumeci, L. Tautz, D. Petranovic, R. Edwards, P. R. Jensen, T. Mustelin, J. Deutscher, and N. Bottini. 2005. In vitro characterization of B. subtilis protein tyrosine phosphatase YwqE. J. Bacteriol. 187:3384-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mijakovic, I., S. Poncet, A. Galinier, V. Monedero, S. Fieulaine, J. Janin, S. Nessler, J. A. Marquez, K. Scheffzek, S. Hasenbein, W. Hengstenberg, and J. Deutscher. 2002. Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc. Natl. Acad. Sci. USA 99:13442-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mijakovic, I., S. Poncet, G. Boel, A. Maze, S. Gillet, E. Jamet, P. Decottignies, C. Grangeasse, P. Doublet, P. Le Marechal, and J. Deutscher. 2003. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 22:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431-1442. [DOI] [PubMed] [Google Scholar]
- 35.Morona, J. K., R. Morona, D. C. Miller, and J. C. Paton. 2002. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase J. Bacteriol. 184:577-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mustelin, T., G. S. Feng, N. Bottini, A. Alonso, N. Kholod, D. Birle, J. Merlo, and H. Huynh. 2002. Protein tyrosine phosphatases. Front. Biosci. 7:85-142. [DOI] [PubMed] [Google Scholar]
- 37.Na, J. G., I. Pinto, and M. Hampsey. 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osterman, A., and R. Overbeek. 2003. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 7:1-14. [DOI] [PubMed] [Google Scholar]
- 39.Ostrovsky, P. C., and S. Maloy. 1995. Protein phosphorylation on serine, threonine, and tyrosine residues modulates membrane-protein interactions and transcriptional regulation in Salmonella typhimurium. Genes Dev. 9:2034-2041. [DOI] [PubMed] [Google Scholar]
- 40.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 41.Potts, M., H. Sun, K. Mockaitis, P. J. Kennelly, D. Reed, and N. K. Tonks. 1993. A protein-tyrosine/serine phosphatase encoded by the genome of the cyanobacterium Nostoc commune UTEX 584. J. Biol. Chem. 268:7632-7635. [PubMed] [Google Scholar]
- 42.Preneta, R., S. Jarraud, C. Vincent, P. Doublet, B. Duclos, J. Etienne, and A. J. Cozzone. 2002. Isolation and characterization of a protein-tyrosine kinase and a phosphotyrosine-protein phosphatase from Klebsiella pneumoniae. Comp. Biochem. Physiol. B 131:103-112. [DOI] [PubMed] [Google Scholar]
- 43.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 44.Ramponi, G., and M. Stefani. 1997. Structural, catalytic, and functional properties of low Mr phosphotyrosine protein phosphatases. Evidence of a long evolutionary history. Int. J. Biochem. Cell. Biol. 29:279-292. [DOI] [PubMed] [Google Scholar]
- 45.Rayapureddi, J. P., C. Kattamuri, B. D. Steinmetz, B. J. Frankfort, E. J. Ostrin, G. Mardon, and R. S. Hegde. 2003. Eyes absent represents a class of protein tyrosine phosphatases. Nature 426:295-298. [DOI] [PubMed] [Google Scholar]
- 46.Sau, S., N. Bhasin, E. R. Wann, J. C. Lee, T. J. Foster, and C. Y. Lee. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143:2395-2405. [DOI] [PubMed] [Google Scholar]
- 47.Smith, S. C., P. J. Kennelly, and M. Potts. 1997. Protein-tyrosine phosphorylation in the archaea. J. Bacteriol. 179:2418-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soulat, D., E. Vaganay, B. Duclos, A. L. Genestier, J. Etienne, and A. J. Cozzone. 2002. Staphylococcus aureus contains two low-molecular-mass phosphotyrosine protein phosphatases. J. Bacteriol. 184:5194-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, J. R. 1997. An introduction to error analysis. University Science Books, Sausalito, Calif.
- 51.Thomas, S. R., and T. J. Trust. 1995. Tyrosine phosphorylation of the tetragonal paracrystalline array of Aeromonas hydrophila: molecular cloning and high-level expression of the S-layer protein gene. J. Mol. Biol. 245:568-581. [DOI] [PubMed] [Google Scholar]
- 52.Tzeng, Y. L., and J. A. Hoch. 1997. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis J. Mol. Biol. 272:200-212. [DOI] [PubMed] [Google Scholar]
- 53.Tzeng, Y. L., X. Z. Zhou, and J. A. Hoch. 1998. Phosphorylation of the Spo0B response regulator phosphotransferase initiating development in Bacillus subtilis. J. Biol. Chem. 273:23849-23855. [DOI] [PubMed] [Google Scholar]
- 54.Vincent, C., P. Doublet, C. Grangeasse, E. Vaganay, A. J. Cozzone, and B. Duclos. 1999. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J. Bacteriol. 181:3472-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vincent, C., B. Duclos, C. Grangeasse, E. Vaganay, M. Riberty, A. J. Cozzone, and P. Doublet. 2000. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in Gram-negative bacteria. J. Mol. Biol. 304:311-321. [DOI] [PubMed] [Google Scholar]
- 56.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the sigmaB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wo, Y. Y., A. L. McCormack, J. Shabanowitz, D. F. Hunt, J. P. Davis, G. L. Mitchell, and R. L. Van Etten. 1992. Sequencing, cloning, and expression of human red cell-type acid phosphatase, a cytoplasmic phosphotyrosyl protein phosphatase. J. Biol. Chem. 267:10856-10865. [PubMed] [Google Scholar]
- 58.Zhang, M., C. V. Stauffacher, D. Lin, and R. L. Van Etten. 1998. Crystal structure of a human low molecular weight phosphotyrosyl phosphatase. Implications for substrate specificity. J. Biol. Chem. 273:21714-21720. [DOI] [PubMed] [Google Scholar]
- 59.Zimmer, M. A., H. Szurmant, M. M. Saulmon, M. A. Collins, J. S. Bent, and G. W. Ordal. 2002. The role of heterologous receptor in McpB-mediated signaling in Bacillus subtilis chemotaxis. Mol. Microbiol. 45:555-568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.