Abstract
The novel pathogen, Elizabethkingia anophelis, has gained attention due to its high mortality rates and drug resistance facilitated by its inherent metallo-β-lactamases (MBLs) genes. This study successfully identified and outlined the functions of the B3-Q MBLs variant, GOB-38, in a clinical sample of E. anophelis. The T7 expression system was employed to stimulate the expression of recombinant protein in Escherichia coli, followed by an analysis of the biochemical properties of purified GOB-38. Our findings indicate that the enzyme GOB-38 displays a wide range of substrates, including broad-spectrum penicillins, 1–4 generation cephalosporins, and carbapenems, potentially contributing to in vitro drug resistance in E. coli through a cloning mechanism. It is important to highlight that GOB-38 exhibits a distinct active site composition compared to GOB-1/18, featuring hydrophilic amino acids Thr51 and Glu141 at both ends of its active center instead of hydrophobic alanine, potentially indicating a preference for imipenem. Furthermore, the co-isolation of Acinetobacter baumannii and E. anophelis, two opportunistic pathogens, from a single lung infection is noteworthy. Our in vitro co-culture experiments suggest that E. anophelis, carrying two MBL genes, may have the ability to transfer carbapenem resistance to other bacterial species through co-infection.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-82748-2.
Keywords: Metallo-β-lactamases, GOB-38, Antibiotic resistance, Elizabethkingia anophelis, Acinetobacter baumannii
Subject terms: Antimicrobials, Bacteria, Clinical microbiology, Pathogens
Introduction
Currently, there is a growing prevalence of multidrug-resistant (MDR) bacteria, leading to a significant global health concern regarding antibiotic resistance1. In developed nations, the annual mortality rate attributed to MDR bacteria surpasses the combined mortality rates of Parkinson’s disease, emphysema, AIDS, and homicides2. Acinetobacter baumannii, as a highly successful pathogen in hospital settings, has been extensively researched for its resistance mechanisms in various scenarios. A. baumannii has been designated as an ESKAPE pathogen by the World Health Organization3 owing to its capacity to circumvent the effects of antibiotics via enzymatic degradation, target modification, multidrug efflux pump activity, and alterations in permeability4,5. The emergence of metallo-β-lactamases (MBLs) in A. baumannii, notably the IMP6, NDM7, and VIM8 variants, is regarded as a noteworthy issue. MBLs, belonging to the Class B beta-lactamases, utilize Zn2+-activated hydroxides to initiate the inactivation of antibiotics. MBLs possess the capability to hydrolyze a broader spectrum of β-lactam substrates, such as penicillins, cephalosporins, and carbapenems, in comparison to serine-β-lactamases (SBLs) found in classes A, C, and D9,10. Additionally, MBLs demonstrate resistance to a variety of inhibitors commonly employed in clinical settings, including clavulanic acid and avibactam11.
In addition to being clinically relevant, many of these enzymes are produced by a diverse range of environmental bacteria12–17. The genus Elizabethkingia is an emerging species that is increasingly associated with life-threatening infections and outbreaks in humans. Recent studies have identified six species within Elizabethkingia, with E. anophelis emerging as the predominant pathogen, accounting for 59–99% of all isolates, while E. meningoseptica is less commonly implicated18. The escalating prevalence rates of E. anophelis infections, with a range of 0.01 to 0.6 cases per 1000 hospital admissions19, coupled with the high mortality rates of 24–60% 19–21, have garnered increased attention. The genus Elizabethkingia is characterized by its environmental origins and intrinsic multidrug resistance, rendering it impervious to most β-lactams, β-lactam/β-lactam inhibitor combinations, carbapenems, and aminoglycosides20,22,23. Notably, Elizabethkingia is unique in that it is the only microorganism reported to possess two chromosomally encoded MBL genes, namely blaB and blaGOB24.
In the present study, both A. baumannii and E. anophelis were identified in a single case of pulmonary infection. Given the frequent occurrence of A. baumannii co-infections in clinical contexts25,26, this finding is not unexpected. Furthermore, our research delved into the biochemical properties of GOB-38 produced by E. anophelis, as well as explored the potential interactions between E. anophelis and A. baumannii through in vitro co-culture experiments. Additionally, we conducted DNA sequencing and genomic analysis to characterize the evolution, plasmid profiles, and resistance profiles of these bacterial strains.
Results
Genomic and evolutionary analysis of Ab6-2 and EA-1
The A. baumannii Ab6-2 strain was isolated directly from sputum samples of a 38-year-old female patient diagnosed with severe myocarditis. The patient underwent 33 days of intensive medical care in the intensive care unit of the First Affiliated Hospital of Jiangxi Medical College, Nanchang University, and was subsequently discharged following various interventions, including endotracheal intubation and ventilator support. Following the detection of E. anophelis EA-1 trace in the whole genome sequencing (WGS) data of Ab6-2, EA-1 was successfully isolated using a blood agar plate, and its genome was fully sequenced. A comparative analysis of 417 MDR strains of A. baumannii collected between 2022 and 2023 revealed that strain Ab6-2 exhibited a MBL phenotype; however, WGS analysis did not identify a common MBL gene associated with strain Ab6-2. Ab6-2 was discovered to have a 3.9 Mb chromosome and two plasmids of 72.3 kb and 8.7 kb (see Table 1). According to the AB-PBRT scheme, the plasmids of Ab6-2 were classified as incompatible types GR6 (plasmid 1) and GR2 (plasmid 2). Plasmid 1 harbored a Type IV secretion system (T4SS), while neither plasmid carried any resistance genes (refer to Fig. S1). In contrast, EA-1 lacked plasmids and contained only a 4.0 Mb chromosome (Table 1).
Table 1.
Genomic data for E. Anophelis EA-1 and A. Baumannii Ab6-2.
| Strain | Genome size (bp) | GC (%) | *No. plasmids: size (kp) |
No. gene | No. tRNA | No. ARGs |
|---|---|---|---|---|---|---|
| EA-1 | 4,030,207 | 36% | - | 3719 | 44 | 9 |
| Ab6-2 | 3,989,450 | 39% | 2:72.3, 8.7 | 3848 | 73 | 15 |
*Number of plasmids are denoted in bold text.
ARGs, antibiotics resistance genes antibiotics resistance genes.
EA-1 harbored class A β-lactamases blaCME-1, B1 MBL blaB-29, and B3 MBL blaGOB-38. EA-1, along with the closely related E6809 (isolated from a blood culture in the USA) and SEA01 (isolated from a blood culture in India) strains, were grouped together in cluster II (Fig. 1A). These strains exhibited distinct variants of blaCME, blaB, and blaGOB. Ab6-2, a prevalent ST2 (Pastur) strain, was a member of the international clone II (IC2) clonal complex and shared the highest genetic similarity with AB021-A001 (isolated from an oral swab of a French patient) (Fig. 1B). Ab6-2 harbored β-lactamase genes, specifically blaTEM-1, blaOXA-66, and blaOXA-23-like. The insertion sequence (IS)Aba1, a member of the IS4 family translocase, was identified upstream of the blaOXA-23-like gene in Ab6-2. In contrast, AB021-A001 only contained AmpC and blaOXA-64 genes, with no adjacent ISs detected.
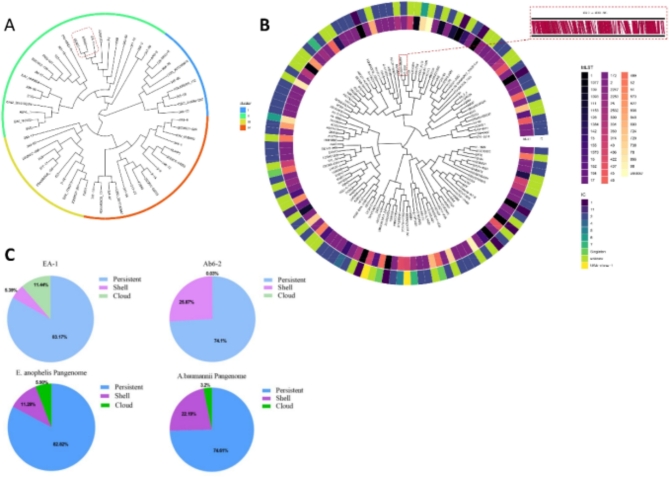
Fig. 1.
Phylogenetic tree and pangenomes of A. baumannii Ab6-2 and E. anopheles EA-1. Based on the average nucleotide identity (ANI), phylogenetic trees of (A), 60 complete genomes of E.anophelis (including EA-1 and 59 reference strains), and (B), 120 complete genomes of A. baumannii (including Ab6-2 and 119 reference strains) were constructed. (C), The distribution of persistent (softcore), shell (core/shell), and cloud (accessory/dispensable) genes, as well as the pangenome composition of EA-1 and 59 E. anophelis strains, and the pangenomes of Ab6-2 and 119 A. baumannii strains. Pangenome was calculated by PPanGGOLiN.
The pangenomes of Ab6-2 and EA-1, along with their corresponding reference strains, were determined using PPanGGOLiN. The proportion of cloud genes in the Ab6-2 genome was found to be a mere 0.03%, significantly lower than the average cloud gene content of 3.2% observed in 119 complete A. baumannii genome sequences obtained from GenBank. In contrast, EA-1 exhibited a notably higher cloud gene content at 11.44%, which was substantially greater than the average cloud gene content of 5.9% observed in 59 complete E. anophelis genome sequences (Fig. 1C).
Detection of drug resistance in coculture ofE. anophelisandA. baumannii
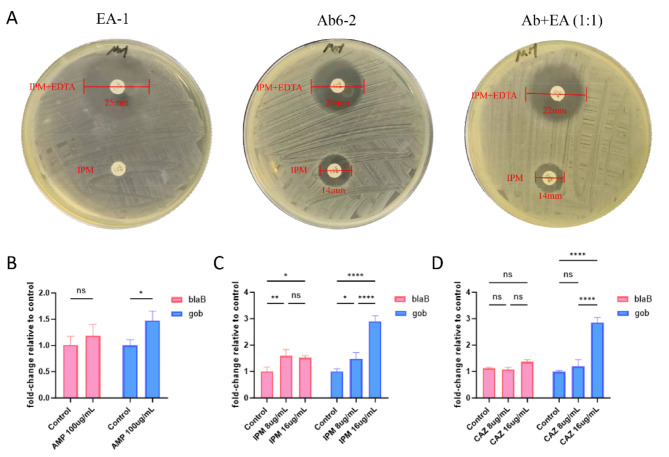
ECT was employed to assess the MBL phenotype. Pure cultures of Ab6-2 and EA-1, along with cocultures at varying ratios (1:9–9:1), were inoculated onto Mueller-Hinton (MH) agar plates. Following incubation at 37 ℃ for 18 h, variations in bacteriostatic circle sizes were noted. Ab6-2 exhibited a negative response (inhibition zone increased by 6 mm), EA-1 showed a positive response (increased by 20 mm), and various proportions of cocultures demonstrated positive responses (inhibition zone increased by 8–11 mm) (Fig. 2A).
Fig. 2.
MBL phenotype examine and RT-qPCR. (A) MBL phenotype of Ab6-2 and EA-1 pure cultures and cocultures (1:1 ratio). (B–D) RT-qPCR analysis of blaB-29 and blaGOB-38 expression in coculture. Data are expressed as the mean and standard error of three independent biological replicates. (*, p < 0.05; **, p < 0.01 and ****, p < 0.0001; ns, not significant).
AMP, Ampicillin; IPM, Imipenem; CAZ, Ceftazidime.
The minimum inhibitory concentrations (MICs) of Ab6-2 and EA-1 monocultures and cocultures (at a 1:1 ratio) were assessed against various antibiotics, including ampicillin, azlocillin, ceftazidime, cefepime, and imipenem, using the broth microdilution method (refer to Table 2). Subsequently, antibiotic-treated cocultures were plated on blood agar to validate that the MIC values were influenced by the presence of cocultures rather than monocultures. The larger size of individual colonies of A. baumannii compared to those of E. anopheles (Fig. S2) allowed for the distinction between Ab6-2 and EA-1 based on morphological characteristics, a distinction later confirmed by 16SrRNA analysis. The presence of viable cells of both A. baumannii and E. anophelis in the coculture was observed. Given the disparity in MIC values between the two strains, ampicillin, ceftazidime, and imipenem emerged as the most probable antibiotics for cross-protection.
Table 2.
Minimum inhibitory concentrations for mono-cultures and co-cultures (1:1) of E. Anophelis EA-1 and A. Baumannii Ab6-2 (µg/mL).
| Culture | Ampicillin(ug/mL) | Azlocillin(ug/mL) | Ceftazidime(ug/mL) | Cefepime(ug/mL) | Imipenem(ug/mL) |
|---|---|---|---|---|---|
| EA-1 | 128 | 512 | 128 | 512 | 512 |
| Ab6-2 | 1024 | 512 | 512 | 512 | 256 |
| *Ab + EA(1:1) | 1024 | 512 | 512 | 512 | 512 |
*Ab + EA = Ab6-2 + EA-1.
Due to CME’s incapacity to hydrolyze carbapenem, we conducted a study to assess the gene copy numbers of two MBLs, specifically blaB-29 and blaGOB-38, in order to identify which MBL gene amplification led to resistance to ampicillin, ceftazidime, and imipenem in previous co-cultures. Quantitative polymerase chain reaction (qPCR) was employed to measure the expression levels of blaB-29 and blaGOB-38 in co-cultures exposed to various antibiotic conditions, with normalization to 16SrRNA levels. The study found that blaGOB transcription significantly increased with 100 µg/ml ampicillin, 8 µg/ml imipenem, and 8 µg/ml cefotaxime, and increased further with 16 µg/ml of imipenem and cefotaxime. In contrast, blaB transcription only significantly increased with imipenem exposure, but to a lesser extent than blaGOB (Fig. 2B–D).
Nucleotide sequence analysis
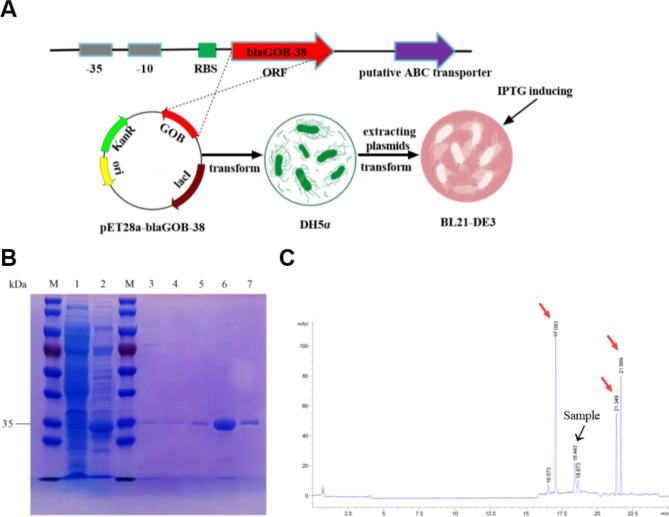
The blaGOB-38 gene was characterized through amplicon sequencing utilizing primers detailed in Table S1. This gene exhibited a total length of 1956 bp and a GC content of 34%. The open reading frame (ORF) of the gene spanned 873 bp with a GC content of 36%, a proportion closely resembling the G + C ratio found in Elizabethkingia genes (ranging from 36.1 to 41.6%)27,28 and comparable to that of A. baumannii (averaging approximately 39.6%)29. Conserved promoter regions − 35 (TTGAAA) and − 10 (TTTATT) were identified upstream, alongside a mutation at the final base (A→G) in the ribosome binding site (RBS) sequence (AAAACG) in comparison to blaGOB-1. Nevertheless, the mutation did not impede protein expression in subsequent cloning experiments conducted in Escherichia. coli (Fig. S3A). Furthermore, a putative ABC transporter gene resembling blaGOB-1 was identified downstream of the ORF (Fig. 3A).
Fig. 3.
DNA cloning, the purification of proteins, and the determination of pI. (A) Description of the upstream and downstream sequences of blaGOB-38 and the experimental steps for overexpression of the GOB-38 protein. (B) SDS-PAGE after the different purification steps. M, molecular mass standards; lane 1, crude extract from BL21-DE3; lane 2, Ni-NTA agarose resin eluent; lane 3, STarm Streptactin beads flow through liquid; lane 4, STarm Streptactin beads detergent; lane 5–7, STarm Streptactin beads eluent. (C) GOB-38 pI determination. Red arrow, standard; black arrow, sample.
DNA cloning and susceptibility profiles ofE. coliand ATCC17978
Utilizing pK-18mob, the ORF of blaGOB-38 and its putative promoter region were cloned to validate its resistance function. The recombinant plasmid was then introduced into E. coli DH5α recipient cells. The transformed cells were cultured on columbia blood agar plates supplemented with kanamycin (50 µg/mL) and amoxicillin (30 µg/mL). Subsequent susceptibility testing, as outlined in Table 3, revealed that the transformant displayed resistance to ticarcillin/clavulanic acid and ceftazidime, with MIC levels increasing by 16-fold and 64-fold, respectively, in comparison to the control strain (pK-18/DH5α). The production of GOB-38 resulted in a twofold increase in the MIC of piperacillin-tazobactam. However, the transformants remained fully susceptible to piperacillin-tazobactam and did not demonstrate resistance to cefepime, consistent with the findings observed in transformants producing SMB-130 and THIN-B31 subclass B3 MBLs. Additionally, the transformants exhibited sensitivity to carbapenems, with the MIC of imipenem (1) slightly higher than that of meropenem (0.5), a pattern akin to that observed in GOB-127.
Table 3.
Result of susceptibility testing.
| Antibiotic | MIC (µg/ml) against strain: | ||||||
|---|---|---|---|---|---|---|---|
| Ab6-2 | EA-1 | pK-blaGOB-38/DH5α | pK-18/DH5α | pK-blaGOB-38(A) /ATCC17978 | pK-blaGOB-38(G) /ATCC17978 | pK-18/ATCC17978 | |
| Ticarcillin/clavulanic acid | ≥ 128 | ≥ 128 | ≥ 128 | ≤ 8 | ≤ 8 | ≤ 8 | ≤ 8 |
| Piperacillin tazobactam | ≥ 128 | ≥ 128 | 8 | ≤ 4 | ≤ 4 | ≤ 4 | ≤ 4 |
| Cefoperazone sulbactam | ≥ 64 | ≥ 64 | ≤ 8 | ≤ 8 | ≤ 8 | ≤ 8 | ≤ 8 |
| Ceftazidime | ≥ 64 | ≥ 64 | 16 | 0.25 | 2 | 2 | 2 |
| Cefepime | ≥ 32 | ≥ 32 | ≤ 0.12 | ≤ 0.12 | 16 | 2 | 4 |
| Ciprofloxacin | ≥ 4 | ≥ 4 | ≤ 0.25 | ≤ 0.25 | ≤ 0.25 | ≤ 0.25 | ≤ 0.25 |
| Levofloxacin | 4.0 | ≥ 8 | ≤ 0.12 | ≤ 0.12 | ≤ 0.12 | ≤ 0.12 | ≤ 0.12 |
| Meropenem | ≥ 16 | ≥ 16 | 0.5 | ≤ 0.25 | 8 | 1 | ≤ 0.25 |
| Imipenem | ≥ 16 | ≥ 16 | 1 | ≤ 0.25 | ≤ 0.25 | ≤ 0.25 | ≤ 0.25 |
| Tigecycline | ≤ 0.5 | 4 | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 |
| Tobramycin | ≥ 16 | ≥ 16 | ≤ 1 | ≤ 1 | ≤ 1 | ≤ 1 | ≤ 1 |
| SMZ-TMP | ≥ 320 | ≤ 20 | ≤ 20 | ≤ 20 | ≤ 20 | ≤ 20 | ≤ 20 |
| Colistin | ≤ 0.5 | ≥ 16 | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 |
Furthermore, we conducted mutations in the RBS region and subsequently introduced the engineered recombinant plasmids pK-blaGOB-38 (A) and pK-blaGOB-38 (G) into A. baumannii ATCC17978. Notably, the transformation of ATCC17978 with pK-blaGOB-38 (G) resulted in minimal alteration in resistance levels; however, upon mutation of the RBS region (AAAACG→AAAACA), the ATCC17978 transformant exhibited a significant increase in resistance to meropenem (32-fold higher MIC).
Purification and physicochemical properties of GOB-38
A yield of approximately 4 mg of purified protein per 1 L of bacterial culture was achieved through a double-tags two-step purification method, resulting in a purity of ≥ 95% as confirmed by SDS-PAGE analysis (Fig. 3B). The molecular weight of the purified enzyme was determined to be approximately 33 kDa, in line with ExPASy analysis32 following the removal of an 18-residue N-terminal signal peptide predicted by the SignalP 4.1 algorithm33. The specific activity of the purified enzyme was found to be 26.7 µM ·min− 1·mg of protein− 1, representing a 146-fold increase compared to the crude enzyme. The results of CIEF analysis indicated that the isoelectric point (pI) of GOB-38 was 7.2, as shown in Fig. 3C. This value differed from that of GOB-1 (pI = 8.7)27 but was more similar to SMB-1 (pI = 7.4)30.
Additionally, ICP-MS analysis demonstrated that recombinant GOB-38 contained a significant amount of zinc, with the total metal content consistently below one metal ion per protein molecule (0.8–0.84 zinc/ GOB-38), consistent with the findings for GOB-1834.
Kinetic parameters of the GOB-38 enzyme
The kinetic analysis of GOB-38 revealed a broad hydrolytic capacity, particularly towards imipenem (Table 4). GOB-38 exhibited the ability to hydrolyze a variety of broad-spectrum penicillins and most cephalosporins, with the exception of five-generation ceftaroline. Notably, GOB-38 displayed the capability to hydrolyze cefepime, a fourth generation cephalosporin previously considered a poor substrate for most B3 MBLs such as CAU-135. Interestingly, the catalytic efficiency of GOB-38 (0.17 µM− 1·s− 1) surpassed that of other B3-Q MBLs such as FEZ-136, L137, BJP-138, THIN-B31, and CPS-139, which exhibit a catalytic efficiency of 0.08 µM− 1·s− 1. Moreover, the efficiency of GOB-38 was found to be similar to that of IMP-140 and VIM-241 in B1 MBLs. In contrast to the notable catalytic efficiency observed with imipenem, the reduced catalytic efficiency of meropenem was attributed to its lower substrate turnover rate (Kcat 140.8 s− 1). This observation diverged from the catalytic behavior of GOB-127 and GOB-1834, as well as the majority of B3 MBLs (including SIE-1, a representative enzyme of B3-E MBL)31,36,37,42, but was similar to that of CPS-139.
Table 4.
Kinetic parameters and inhibition profile of GOB.
| Substrate or chelating agent | Km(µM) | Kcat(s− 1) | Kcat/Km(µM− 1·s− 1) | Relative Kcat/Km | IC50(mM) |
|---|---|---|---|---|---|
| Azlocillin | 437.7 | 25.1 | 0.06 | 15 | |
| Mezlocillin | 195 | 23.7 | 0.12 | 29 | |
| Caphalothin | 797 | 143 | 0.18 | 44 | |
| Cefoxitin | 1719.3 | 208.1 | 0.12 | 29 | |
| Cefotaxime | 814.6 | 136 | 0.17 | 41 | |
| Ceftazidime | 180.8 | 48.4 | 0.27 | 66 | |
| Ceftriaxone | 313.9 | 64 | 0.2 | 49 | |
| Cefoperazone | 821.5 | 109.4 | 0.13 | 32 | |
| Cefepime | 1049 | 125 | 0.12 | 29 | |
| Ceftaroline | NDa | - | - | ||
| Aztreonam | ND | - | - | ||
| Meropenem | 531 | 140.8 | 0.27 | 66 | |
| Imipenem | 626.9 | 256.3 | 0.41 | 100 | |
| EDTA | 21.2 | ||||
| Dipicolinic acid | 0.05 | ||||
| Potassium clavulanate | >200 | ||||
| Avibactam | >100 |
aND, not determined.
The GOB-38 enzyme demonstrated increased resistance to EDTA, as indicated by its notably higher IC50 value (21.2 mM) in comparison to GOB-1 (25 µM)27, GOB-18 (approximately 2 mM)34, and other B3 MBLs30,43,44. Notably, GOB-38 exhibited a comparable level of resistance to its distant relatives BJP-1 (40% inactivation after exposure to 50 mm EDTA for 30 min)38 and IMP-1 (10% inactivation after exposure to 10 mm EDTA for 1 h)40. Dipicolinic acid, a metal chelating agent, demonstrated significant efficacy in deactivating GOB-38, as evidenced in Table 4. In contrast, the SBL inhibitors clavulanic acid and avibactam were found to be ineffective in inhibiting GOB-38, as expected.
Structural features of GOB-38
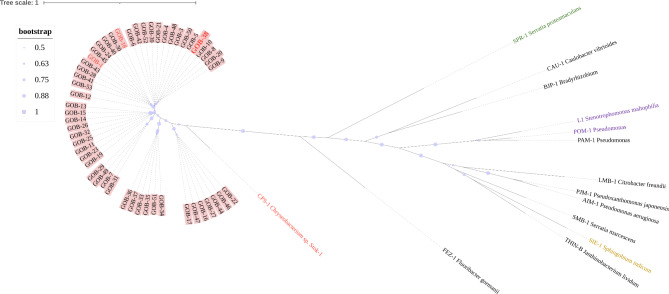
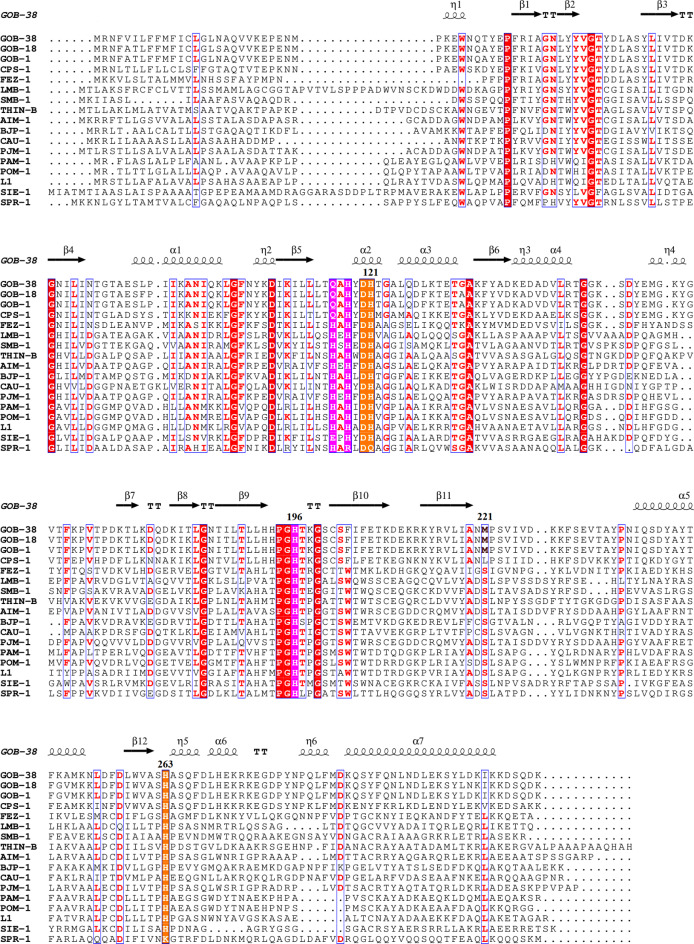
The comparative analysis of evolutionary relationships among functionally characterized B3-β-lactamases revealed that GOB forms a distinct branch in the phylogenetic tree, separate from other B3 MBLs with the exception of CPS-1, which also falls within the B3-Q MBL group (Fig. 4). Within the GOB family, GOB-38 demonstrated the highest amino acid (AA) identity with GOB-1 (93.8%) and GOB-18 (95.2%). Additionally, GOB-38 displayed the highest level of AA sequence identity with CPS-1 (68.3%), followed by FEZ-1 (36.7%), and the lowest similarity with SMB-1 (23.3%) (Table S2). In the AA sequence of GOB-38, the active site residues B3-Q (QHH/DHH) were found to be conserved, while eight AA substitutions (Ala51Thr, Phe128Leu, Ala133Gly, Ala141Glu, Lys182Thr, Thr187Ile, Gly249Lys, Ile258Leu) were detected when compared to other members of the GOB family (GOB-1 and GOB-18), excluding the initial peptide sequence (Figs. 5 and 6A).
Fig. 4.
The maximum likelihood tree illustrated the relationship between GOB and other functionally characterized B3-MBLs. Bootstrap support for internal nodes was indicated by purple dots, with values ranging from 0.5 to 1. The GOB family was depicted with a pink background, while the B3-Q MBLs, including GOB-1/18/38 and CPS-1, were highlighted in red font. Additionally, the tetramer members L1 and POM-1 were emphasized in purple, while the representative enzyme SPR-1 of B3-RQK was highlighted in green, and the representative enzyme SIE-1 of B3-E was highlighted in yellow.
Fig. 5.
The amino acid alignment of GOB-38 (NCBI Reference protein Sequence number: WP_058879139.1), GOB-1 (WP_063860523.1), GOB-18 (WP_063860560.1), FEZ-1 (WP_058468956.1), CPS-1 (WP_063857696.1), LMB-1 (WP_108361548.1), SMB-1 (WP_063864723.1), THIN-B (WP_063842948.1), BJP-1 (WP_011088970.1), AIM-1 (WP_063857820.1), CAU-1 (WP_063859390.1), PJM-1 (WP_213603971.1), PAM-1 (WP_043245728.1), POM-1 (WP_044403015.1), L1 (WP_063842689.1), SIE-1 (WP_007683232.1), SPR-1 (WP_012145975.1) with the secondary structure of GOB-38 was depicted. Conserved residues were indicated by red boxes, while similar residues were indicated by white boxes. The QHH site (Zn1) was highlighted in pink, and the DHH site (Zn2) was highlighted in yellow. The secondary structure elements of GOB-38 were displayed above the alignments. This figure was generated using ESPript.
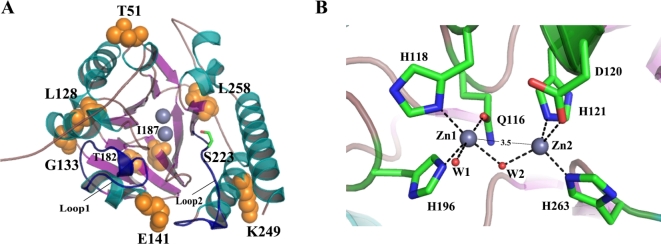
Fig. 6.
Overall and active-site structure of GOB-38. (A) The GOB-38 protein was depicted in a cartoon representation, where the helices were colored in dark green, the strands in pink, and the non-structured loops in dark yellow. Specifically, the two loops, referred to as loop1 and loop2, which covered the active site, were highlighted in dark blue. The presence of zinc atoms was represented by gray spheres. The residues that were the subject of discussion in this paper were visually represented using orange spheres and stick models, with carbon atoms depicted in green, nitrogen atoms in blue, and oxygen atoms in red. (B) The active-site structure of GOB-38 was depicted, with gray representing the zinc atoms, red representing the water molecules (Wat1, Wat2), and dashed lines indicating the coordination bonds. The estimated distance between the two Zn2+ ions was 3.5 Å.
The enzyme GOB-38 underwent molecular modeling based on the established structure of the GOB-18 enzyme, resulting in the discovery of several significant attributes. Notably, GOB-38 displayed the expected αβ/βα sandwich fold, characterized by two central β-sheets composed of strands β1-β7 and β8-β12, respectively. Furthermore, seven α-helices were identified surrounding the two exposed β-sheets, which were in direct contact with the surrounding solvent. The central active site cavity of GOB-38 was shielded by loop1, which connected the α4-helix and β7-strand, as well as loop2, which connected the β11-strand and α5-helix. The conformation of the zinc ligand at the active site of GOB-38 was stabilized by a hydrogen bond network (Figs. 5 and 6A). Additionally, the two Zn2+ ions were connected by a water molecule positioned centrally, creating an angular configuration, with an approximate separation of 3.5 Å between the ions. Zn1 displayed a unique association with an additional water molecule (Fig. 6B), diverging from established findings on B3 MBLs45–49. It was essential to consider that the reliability of this observation was contingent upon the accuracy of our initial hypothesis. Another significant finding was the modification of eight AAs within the mature protein, all located in the outer layer of the active site. Of particular interest are residues T51 and E141, situated at opposite ends of the active center. These residues experienced a change from the hydrophobic alanine in GOB-1/18 to the currently hydrophilic AAs threonine and glutamate, respectively. This change can be compared to the formation of a hydrophilic pathway within the hydrophobic core, potentially leading to differences in substrate specificity.
Discussion
Polymicrobial infections are common in various human diseases50–52, where microbial communities interact through metabolites53, signaling systems54, direct contact55, and regulation of host immune responses56. In response to antibiotics, some community members can release compounds that degrade antibiotics or activate tolerance in others for cross protection57. When carbapenem resistant A. baumannii (CRAB) was co-cultured with Staphylococcus aureus, the effectiveness of meropenem against S. aureus decreased58. CRAB released class D β -lactamases (CHDL) outside the cell to hydrolyze carbapenem and protect against sensitive bacteria57, suggesting A. baumannii may shelter rather than kill certain pathogens59.
Ab6-2 and EA-1 showed cross protection against high concentrations of antibiotics, with the coculture having a greater or equal MIC compared to the monoculture. The antibiotic sensitivity of cocultures may be influenced by the medium60. In ECT, EA-1 with two MBLs masked the Ab6-2 MBL negative phenotype. At higher concentrations of antibiotics, EA-1 may provide resistance to cocultures by expressing more active GOB. Both Ab6-2 and EA-1 carried extended-spectrum β-lactamase (ESBL) genes (blaTEM-1 and blaCME-1), but while TEM-1 conferred resistance to certain antibiotics in A. baumannii, it did not affect sensitivity to carbapenem61,62. CME-1, on the other hand, can provide resistance to cephalosporins but is inhibited by certain antibiotics63. These genes are unlikely to provide imipenem resistance in cocultures. Ab6-2 had two OXA type carbapenemase genes, blaOXA-66 and blaOXA-23-like, with ISAba1 located before blaOXA-23-like. The presence of ESBL and OXA type carbapenemase genes gave Ab6-2 broad-spectrum resistance. Imipenem resistance in cocultures was similar to EA-1, likely due to MBL64,65. Previous reports indicated that BlaB was the sole β-lactamase responsible for imipenem resistance in E. meningoseptica24. However, our study found that the expression of blaGOB-38 was higher than blaB-29 in the coculture, possibly due to differences in nutrient availability in the culture medium. Previous research has shown that bacteria have evolved ways to communicate and coordinate their activities. For example, Streptococci produce a peptide signal to enhance biofilm formation54, Haemophilus influenzae produces autoinducer-2 (AI-2) to increase antibiotic resistance in Moraxella catarrhalis66, and Pseudomonas aeruginosa can sense AI-2 from co-infected flora to boost its virulence67. We hypothesized that blaGOB may be influenced by quorum sensing and regulated independently24. We have not yet established the connection between the two, which may be the focus of our future research.
Based on their active site structure, the optimal number of Zn2+ ions necessary for maximal activity, and their substrate profile, these MBLs can be categorized into three subclasses: B1, B2, and B312. BlaB is classified under the B1 MBLs, while GOB falls within the B3 MBLs. Despite displaying less than 20% sequence similarity to both B1 and B2 MBLs, the B3 subgroup demonstrates a substrate spectrum as broad as that of the B1 subgroup68. Within the catalytic center containing two Zn2+ ions, MBLs display similar yet distinct sequence motifs, including B1 (His116, His118, His196, Asp120, Cys221, His263, denoted as HHH/DCH), B2 (NHH/DCH), and classic B3 (HHH/DHH)68. Among these motifs, B3 MBLs exhibit a higher level of active site diversity compared to B1 and B2 MBLs. Pedroso et al.68 conducted a sequence analysis on 1449 B3 MBL proteins, identifying 1150 classic B3 active site residues HHH/DHH, 162 QHH/DHH, 90 EHH/DHH, and 47 rare variants (HRH/DQK) that show variations at three sites. The GOB enzymes derived from Elizabethkingia spp. represent the most extensive family of B3 MBLs, with only two out of 52 members, namely GOB-169 and GOB-1870, having been characterized to date. These two members exhibit significant diversity in their Zn2+ binding properties.
Of the two enzymes in the GOB family, GOB-18 is versatile and prefers mono-Zn (II) or bi-Zn (II) activity, with optimal activity when metal ions bind to the Zn2 (DHH) site70. On the other hand, GOB-1 enhances its hydrolytic capacity by binding two Zn2+ ions, expanding the range of metal sites in MBLs69. To better understand the mechanism of B3-Q MBLs enzymes, we characterized GOB-38 enzymes due to the extensive hydrolysis ability of B3 MBLs and the diversity of GOB family members. In the protein production experiment, two temperatures, 37 ℃ and 16 ℃, were used. After a 4-hour induction at 37 ℃ with 0.1mM IPTG, the culture showed a red color change in nitrocefin. This did not happen after a 12-hour induction at 16 ℃ with the same IPTG concentration. WB analysis showed that higher IPTG concentrations led to decreased expression of recombinant protein. Longer induction times and higher induction intensities also resulted in lower levels of recombinant enzyme in the cell. The preGOB protein was synthesized in the cytoplasm, where it was shielded from degradation by cytoplasmic proteases through the action of the DnaK chaperone. Subsequently, it was translocated to the bacterial periplasm via the Sec translocation pathway. Within the periplasm, GOB underwent folding and incorporated Zn (II) ions to achieve its native conformation, after which it was ultimately secreted extracellularly71. As temperature increases, the accelerated rate of protein synthesis can result in a relative deficiency of chaperone proteins, thereby impeding the correct folding of precursor proteins and leading to an excessive secretion of proteins into the extracellular environment. The secretion characteristics of GOB72 may explain the increase in external environment. Co-cultured bacteria, especially Ab6-2, may have increased resistance to imipenem due to the secretion of GOB by the neighboring EA-1 strain, which likely inactivated the antibiotics in the surrounding environment.
The actual pI value of 7.2 closely matched the predicted value of 6.7 for the mature recombinant protein. ICP-MS data showed Zn (II) concentration in GOB-38 enzyme was below 1, but it didn’t mean GOB-38 only functions as a mono-Zn (II) enzyme. Zn2+ was acquired by the GOB enzyme once in the periplasm, where MBLs in Gram-negative bacteria were located72–74. The production of periplasmic GOB-38 enzyme was not targeted specifically, possibly due to intermixing of proteins from different cellular compartments. MBLs have similar sequence motifs in their catalytic center, which contains two Zn2+ ions. GOB has a weaker metal binding at the Zn1 site due to Gln116, resulting in only one Zn (II) ion being carried by the enzyme. The absence of metal ions in the crystal structure of CSR-1 suggests that mutations in the B3 active site residues, such as H263K, impair the protein’s ability to bind to metals75.
Previous research on the L1 enzyme indicated that loop2 might act like the “flapping loop” in B1 MBLs76. Differences in loop structure and AAs may explain the substrate preference variations in B3 MBLs48,77. The GOB-38 enzyme, with unchanged AAs on its loops compared to GOB-1/18 and slightly shorter loops than other B3 MBLs, could potentially accommodate diverse substrates. Variations in the N-terminal structure of B3 MBLs affect their hydrolytic activity47. BJP-1’s α-helix restricts substrate binding, whereas SMB-1, AIM-1, and L1’s random coil enhances catalytic efficiency. GOB enzymes, with a small helical N-terminal, have moderate catalytic efficiency. The closer proximity of Zn2 + ions in GOB-38, at approximately 3.5 Å, enhances hydrolysis capacity42, comparable to AIM-1 (3.48 Å)49 and GOB-18 (3.52 Å)73, but superior to SIE-1 (3.75 Å)42. Lisa et al.9 suggested that substrate binding occurs via the expansion of the Zn2 coordination range during mono-Zn (II) catalysis. We propose that the two carboxyl oxygen groups in D120 may facilitate this flexibility.
The Zn1 site in the GOB enzyme has little effect on activity but may be crucial for in vivo function78. The binuclear Zn (II) site enhances B1 MBL resistance79, with Zn2 usually binding a water molecule for nucleophilic attacks9. Contrary to this, we found that Zn1 is associated with an additional water molecule, possibly increasing nucleophilic attack opportunities. The model suggested that hydrophilic AAs at the active center ends may alter substrate specificity, enhancing GOB-38’s interaction with β-lactams with less hydrophobic side chains36. This could explain GOB-38’s preference for hydrolyzing cefepime and imipenem. Notably, S223, near M221, was hydrophilic and oriented in a way that may aid in substrate hydrolysis, similar to L145. In the experiment, EDTA did not inhibit the GOB-38 enzyme, while dipicolinic acid did, likely due to sequence differences near the metal coordination site73.
In vitro experiments revealed that most B3-type MBLs, except SPR-180, exhibited strong β-lactamase activity, making E. coli highly resistant, with resistance levels ranked as B3 > B3-E > B3-Q75. E. coli with GOB-38 showed limited resistance, similar to BlaB27 and GOB-127, and no resistance to cefepime15,81–83. Only SMB-1 slightly increased resistance (MIC of 0.5 µg/mL)30, while GOB-38 modestly enhanced carbapenem resistance, akin to B1-type MBLs84,85. E. coli strains with the blaGOB-19 gene had higher MICs for imipenem and meropenem but were less resistant than the parent strain86. In A. baumannii ATCC17978 with pK-blaGOB-38 (G), resistance levels remained unchanged until an RBS mutation increased meropenem resistance 32-fold, indicating the original blaGOB-38 promoter was suboptimal. The RBS mutation enhanced protein translation but didn’t account for the MIC difference between meropenem and imipenem, likely due to outer membrane permeability variations87 and other resistance mechanisms such as altered penicillin-binding protein affinity, permeability barriers, or efflux systems88.
Ab6-2 is part of the ST2 lineage, with phylogenetic analysis indicating EA-1’s close relation to US (1979) and Indian (2014) strains, underscoring its global spread89. E. anophelis has recently caused numerous outbreaks worldwide19,21–23,90, primarily in hospitals and communities, leading to pneumonia, meningitis, bacteremia, and urinary tract infections20,22. These infections are alarming due to multi-drug resistance and high mortality rates18,91. All isolates contained two MBLs, blaB and blaGOB, which may enhance resistance and facilitate spread through conjugative elements (ICEs)92,93, potentially offering protection to other strains.
Conclusion
This study examined the physicochemical properties of B3-Q MBL GOB-38 to understand genetic and structural changes affecting the enzyme’s function. Initial research investigated the cross-protection between A. baumannii and E. anopheles in co-culture, highlighting the GOB enzyme’s exocrine activity as a key factor. The findings offer new insights into the role of these enzymes in drug resistance.
Materials and methods
Bacteria and plasmids
A. baumannii (Ab6-2) and E. anophelis (EA-1) were sourced from the First Affiliated Hospital of Jiangxi Medical College, Nanchang University, China. The identification of these isolates was accomplished utilizing the Vitek-2 automated system (BioMerieux, France) and 16 S rRNA analysis. Cloning and protein expression procedures were carried out using E. coli DH5α, BL21-DE3, and A. baumannii ATCC17978, respectively. The bacterial strains and plasmids utilized in this research can be referenced in Table 5.
Table 5.
Bacterial strains and plasmids used in this study.
| Strains and plasmids | Description | Source |
|---|---|---|
| EA-1 | The wild-type strain of E. anophelis | Isolated from patient sputum |
| Ab6-2 | The wild-type strain of A.baumannii | Isolated from patient sputum |
| DH5α | E. coli DH5α as a host cell for cloning of the blaGOB-38 gene | Our laboratory collection |
| BL21-DE3 | E. coli BL21-DE3 as a host cell for expression of GOB-38 | Our laboratory collection |
| ATCC 25,922 | E. coli ATCC 25,922 as quality control for antimicrobial susceptibility testing | Our laboratory collection |
| Ab5075 | A.baumannii 5075 as nitrocefin chromogenic test positive control | Our laboratory collection |
| ATCC17978 | A. baumannii as a host cell for cloning of the blaGOB-38 gen | As a gift from Professor Yu Yunsong |
| pK-18mob | Cloning vector for the PCR product of the blaGOB-38 gene with its upstream promoter region, Kanr | Our laboratory collection |
| pET28a | Expression vector for the PCR product of the ORF of the blaGOB-38 gene, Kanr | As a gift from Professor Zheng Fanglin |
| pK-blaGOB-38(G) | The recombinant plasmid harbors the blaGOB ORF and putative promoter region | Constructed in this research |
| pK-blaGOB-38(A) | The recombinant plasmid harbors the blaGOB ORF and putative promoter region, RBS was mutated | Constructed in this research |
| pET28a-blaGOB-38 | The recombinant plasmid harbors the blaGOB ORF | Constructed in this research |
r, resistance; Kan, Kanamycin; ORF, open reading frame; RBS, ribosome binding site.
WGS and genome analysis
The complete DNA of Ab6-2 and EA-1 was extracted utilizing the FastPure Bacteria DNA Isolation Mini Kit from Vazyme Biotech. Subsequently, the genomes of Ab6-2 and EA-1 were sequenced through a hybrid approach involving Pacbio long-read and Illumina short-read sequencing technologies. The resultant assembled genomes were then submitted to the GenBank database under the respective BioProject IDs: PRJNA1129009 for Ab6-2 and PRJNA1130100 for EA-1. Resistance gene analysis was conducted by referencing the Comprehensive Antibiotic Resistance Database (CARD)94. The A. baumannii PCR-based replicon typing (AB-PBRT) method, which utilized a multiplex approach to define 19 groups of replicase-encoding genes, was conducted following established protocols95. Plasmid circles were visualized using BRIG software96.
Due to the potential applicability of the Multilocus Sequence Typing (MLST) analysis based on the Pasteur scheme for the current A. baumannii group as opposed to the Oxford scheme97, we performed MLST using the aforementioned Pasteur schemes98. The identification of allelic numbers and sequence types (STs) was achieved by consulting the Pubmlst database (http://pubmlst.org/abaumannii/). Reads were aligned to the reference genomes of specific international clones utilizing smalt (v0.7.6, www.sanger.ac.uk/science/tools/smalt-0).
The genomes of 119 strains of A. baumannii and 59 strains of E. anophelis were obtained from Genbank (Supplementary Data 1–2) and used to construct the pangenome of these species with PPanGGOLiN v 1.1.10899. The average nucleotide identity (ANI) of the whole genome sequences was calculated, and the hclust R function was utilized to generate a distance matrix based on this ANI data100. The resulting matrix was then converted into a dendrogram using Ggtree101.
Phenotypic MBL detection and susceptibility testing
The imipenem-EDTA combined-disk test (ECT) was performed in accordance with established protocols102–104. Pure cultures of Ab6-2 and EA-1 at a concentration of 0.5 McFarland standard, as well as cocultures at ratios ranging from 1:9 to 9:1, were inoculated onto MH agar plates. Specifically, two 10 µg imipenem disks were positioned on the plate with a distance exceeding 4 cm between them. Furthermore, 750 µg EDTA was applied to one of the disks. The plate was then placed in an incubator at 37 ℃ for a duration of 18 h. A positive outcome was determined if the inhibition zones surrounding the imipenem plus EDTA disks exhibited an increase of 7 mm or more.
The MICs were ascertained utilizing the Vitek-2 system and interpreted in accordance with the guidelines outlined in the CLSI M100 (33rd Edition, 2023). E. coli ATCC 25,922 served as the control organism in this investigation. Subsequent analysis was conducted through the broth microdilution method105,106 using cation-regulated Mueller Hinton (CAMH, Solarbio) broth to assess the MIC values of both cocultures and pure cultures.
RNA isolation and RT-qPCR
Ab6-2 and EA-1 were individually cultured in Luria-Bertani (LB) broth overnight, then combined at a 1:1 ratio and transferred to CAMH broth (1:50 ratio) with varying antibiotic concentrations. Following incubation at 37 ℃ for 1 h, the mixed culture was transferred to CAMH broth with corresponding antibiotic concentrations at a 1:100 ratio and continued to grow until reaching an optical density (OD) of 600 nm of 0.6. RNA extraction was performed using the RNA pure bacteria Kit (Tiangen biotech), with cocultures grown without antibiotics serving as controls.
Total RNA extracted from cocultures was processed using the Hiscript III RT Supermix for qPCR commercial kit (+ gDNA wiper, R323-01, Vazyme) to remove DNA and undergo reverse transcription. Subsequent RT-qPCR analysis was conducted on an ABI RT-qPCR system (ABI QuantStudio DX) utilizing ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme). The primer sequences employed for RT-qPCR can be found in Table S1. Each sample underwent three biological replicates, with 16SrRNA genes serving as endogenous controls for normalizing the relative transcript levels of the genes under investigation.
DNA cloning and sequence analysis
The DNA of EA-1 was extracted using the FastPure Bacteria DNA Isolation Mini Kit from Vazyme Biotech and utilized as the subsequent polymerase chain reaction (PCR) template. Primers GOB-Seq/f and GOB-Seq/r were utilized for the amplification of 2163 bp sequences containing the blaGOB ORF and its adjacent regions. Sanger sequencing from Tsingke Biotech was employed for the determination of the nucleotide sequence. Sequence similarities were evaluated using the BLAST program accessible through the BLAST interface of the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST/). The N-terminal region of ORF and the upstream promoter were examined using the methodologies described in SignalP 4.1-DTU Health Tech - Bioinformatic Services and Softberry – BPROM33.
The blaGOB-38 ORF and its potential promoter region were amplified utilizing primers GOB-Clo/f and GOB-Clo/r, with the blaGOB-38 ORF specifically amplified using primers GOB-Exp/f and GOB-Exp/r (see Table S1 for details). Subsequently, the amplified fragments were inserted into the pK-18mob vector and pET28a vector through the utilization of the ClonExpress Ultra One Step Cloning Kit from Vazyme Biotech. The resultant plasmids, pK-blaGOB-38 and pET28a-blaGOB-38, were then individually introduced into DH5α. Furthermore, a single nucleotide mutation (AAAACG→AAAACA, with corresponding primers listed in Table S1) was introduced at the putative ribosome binding site (RBS).
Overexpression and purification of GOB-38
The recombinant plasmid pET28a-blaGOB-38, harboring both His and StrepII double tags, was isolated utilizing the FastPure Plasmid Mini Kit from Vazyme Biotech and subsequently transfected into BL21-DE3 cells. To induce the expression of the GOB-38 protein, a 100 mL overnight preculture in LB broth was utilized to inoculate 1 L of fresh Terrific Broth (TB) supplemented with kanamycin (50 µg/mL) at a dilution ratio of 1:100. Isopropyl-β-D-thiogalactopyranoside (IPTG, Solarbio Life Sciences) was introduced at a final concentration of 0.1 mM once the culture reached an OD600 = 0.5 (Fig. 3A). The culture underwent orbital shaking at 250 revolutions per minute (r.p.m) for an additional 12 h at 16 °C. Prior to this step, the induction conditions were examined. Recombinant protein expression was evaluated using the nitrocefin (0.5 mg/mL, Apexbio) chromogenic assay and Western blot (WB) with StrepII Tag Mouse Monoclonal Antibody (Beyotime Biotechnology) (Fig. S3B and S3C). Cells were harvested via centrifugation at 7000 r.p.m for 15 min at 4 °C. The harvested cells were stored at -20 °C overnight and then resuspended in a lysis buffer containing 50mM NaH2PO4, 500mM NaCl, 10mM imidazole, and a pH of 7.5. A ratio of 2 g of pellets to 10 milliliters of lysis buffer was utilized prior to disruption using the SCIENTZ-IID equipment from China. Centrifugation at 10,000 r.p.m for 30 min at 4 °C was performed to remove any cell debris from the mixture.
Subsequently, the crude extract was combined with pre-balanced nickel-nitrilotriacetic acid (Ni-NTA) agarose resin obtained from Smart-Lifesciences Biotechnology and gently agitated for a duration of 4 h at a temperature of 4 °C. The resulting recombinant protein, with an estimated purity of 80%, was then purified through elution using elution buffer A (consisting of 50 mM NaH2PO4, 500 mM NaCl, 250 mM imidazole, pH 7.5) and subsequently dialyzed overnight against 1×PBS (pH 7.5). The protein was subsequently immobilized on STarm Streptactin beads (Smart-Lifesciences Biotechnology) and eluted using elution buffer B (1×PBS with 5 mM D-biotin, pH 7.5). Subsequently, the protein underwent four rounds of dialysis against 100 volumes of 15 mM Hepes (pH 7.5) with 50 µM ZnCl2. The fraction displaying β-lactamase activity demonstrated a purity level exceeding 95%, as confirmed by SDS-PAGE analysis. Before being stored at -20 ℃, the pure enzymes utilized the BCA method to determine the protein concentration, resulting in an approximate value of 0.5 mg/mL. Glycerol was then added to achieve a final concentration of 20%.
Peptide mapping, protein analysis and capillary isoelectric focusing (CIEF)
An adequate sample size was obtained through reduction alkylation, enzymatic hydrolysis, and desalination treatment. Separation was achieved using the nanoElute ® Nanoflow UHPLC system (Baizhen Biotechnologies), while detection and analysis were performed with the Bruker timsTOF Pro2 mass spectrometer. Data collection was carried out in DDA mode with positive ion detection, utilizing a 60-minute collection time and a scanning range of mass-to-charge ratio of 350–2000 m/z. The resulting mass spectrometry data were analyzed using PEAKS Online software. Finally, the coverage rate of the test sample sequence is 100%.
Database searches were conducted using the resources available on the NCBI website. The ClustalW program, integrated within MEGA11, was utilized for conducting multiple protein sequence alignments, which were then visualized using ESPrint 3.0107. Furthermore, MEGA11 was used to construct a Maximum Likelihood (M-L) phylogenetic tree, incorporating GOB-38 and other functionally characterized β-lactamases. The bootstrap method with 1000 replications was employed for this analysis, and the resulting image was generated using the online tool iTol4108. A homology model of the putative target protein identified in E. anophelis was generated utilizing SWISS-MODEL, with the crystal structure of GOB-18 (PDB ID: 5k0w) employed as a reference template.
Subsequently, CIEF analysis was conducted on GOB-38, employing established methodologies109.
Metal content determination
Zinc concentration was quantified using inductively coupled plasmaatomic mass spectrometry (ICP-MS)34.
Kinetic parameters
The assessment of β-lactamase activity was carried out by measuring the hydrolysis of nitrocefin110,111. One unit of enzyme activity was defined as the quantity of enzyme that can hydrolyze 1 µmol of nitrocefin per minute at a pH of 7.5 and a temperature of 30 ℃.
The enzyme’s efficacy in degrading different β-lactams was evaluated at a temperature of 30 °C in a 15 mM Hepes buffer solution with 50 µM ZnCl2 (pH 7.5), using a multimode plate reader from Perkin-Elmer. The values of Km and Kcat for 96-well plates were determined by analyzing the initial hydrolysis velocity against substrate concentration. The Michaelis-Menten equation was employed in Prism software (version 8.0.1) by GraphPad Software in California to compute all kinetic parameters. The reported kinetic values are representative of the means derived from a minimum of three independent measurements. The kinetic parameters for each substrate were determined based on a minimum of three distinct concentrations.
The enzyme underwent preincubation with different concentrations of EDTA, dipicolinic acid, potassium clavulanate, or avibactam in a 15 mM Hepes buffer (pH 7.5) at 30 °C for 10 min prior to assessing the rate of nitrocefin (0.1 mM) hydrolysis. The 50% inhibitory concentration (IC50) values of these inhibitors were determined through non-linear regression analysis using GraphPad Prism, version 8.0.1, by fitting logs (inhibitor) against responses (four parameters).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are very grateful for the support of the Jiangxi Clinical Research Center for Respiratory Diseases, Nanchang, China, as well as the expert advice on protein purification from Professors Fanglin Zheng and Ye Huang.
Author contributions
Conceptualization, R.L., Y.L. and N.C.; methodology, Y.L.; validation, J.Q., Q.R. and C.W.; formal analysis, R.L., D.P. and J.S.; resources, D.W. and P.L.; writing—original draft preparation, R.L., Y.L. and J.Q.; writing—review and editing, R.L., Y.L., T.X. and N.C.; supervision, T.X. and N.C.; funding acquisition, N.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the National Natural Science Foundation of China (81960121), Jiangxi Natural Science Foundation (20202BAB206008).
Data availability
Sequences analysed during the current study were deposited into the National Center for Biotechnology Information (NCBI) under BioProject Accession Number PRJNA1129009 and PRJNA1130100, and are available at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1129009 and https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1130100.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All research methods were approved by the Medical Ethics Committee of the First Affiliated Hospital of Nanchang University (CDYFYYLK 02–020) and were conducted in accordance with the Declaration of Helsinki and the legal requirements of the country (China) in which the work was performed. Informed consent was obtained from all participants.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ren Liu and Yang Liu contributed equally to this work.
Contributor Information
Tianxin Xiang, Email: txxiangmed@163.com.
Na Cheng, Email: chengnah@sina.com.
References
- 1.Huemer, M., Mairpady Shambat, S., Brugger, S. D. & Zinkernagel, A. S. Antibiotic resistance and persistence-implications for human health and treatment perspectives. EMBO Rep.21, e51034. 10.15252/embr.202051034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laxminarayan, R. et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis.13, 1057–1098. 10.1016/s1473-3099(13)70318-9 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Suay-García, B. & Pérez-Gracia, M. T. Future prospects for Neisseria gonorrhoeae Treatment. Antibiot. (Basel Switzerland). 710.3390/antibiotics7020049 (2018). [DOI] [PMC free article] [PubMed]
- 4.Gordon, N. C. & Wareham, D. W. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents. 35, 219–226. 10.1016/j.ijantimicag.2009.10.024 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Lee, C. R. et al. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol.7, 55. 10.3389/fcimb.2017.00055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arakawa, Y. et al. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemotherapy 39(7), 1612–1615 (1995). [DOI] [PMC free article] [PubMed]
- 7.Chen, Y. et al. Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66(6), 1255-1259 (2011). [DOI] [PubMed]
- 8.Lauretti, L. et al. Cloning and characterization of bla VIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43(7), 1584-1590 (1999). [DOI] [PMC free article] [PubMed]
- 9.Lisa, M. A. O. et al. A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-β-lactamases. [DOI] [PMC free article] [PubMed]
- 10.Crowder, M. W., Spencer, J., Fau - Vila, A. J. & Vila, A. J. Metallo-beta-lactamases: novel weaponry for antibiotic resistance in bacteria. [DOI] [PubMed]
- 11.Walsh, T. R., Weeks, J., Fau - Livermore, D. M. & Toleman, M. A. Livermore Dm Fau - Toleman, M. A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. [DOI] [PubMed]
- 12.Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol.74, 1686–1701. 10.1016/j.bcp.2007.05.021 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Bellais, S., Naas, T. & Nordmann, P. Genetic and biochemical characterization of CGB-1, an Ambler class B carbapenem-hydrolyzing beta-lactamase from Chryseobacterium gleum. Antimicrob. Agents Chemother.46, 2791–2796. 10.1128/aac.46.9.2791-2796.2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boschi, L. et al. The Legionella (Fluoribacter) gormanii metallo-beta-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 beta-lactamases. Antimicrob. Agents Chemother.44, 1538–1543. 10.1128/aac.44.6.1538-1543.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Docquier, J. D. et al. CAU-1, a subclass B3 metallo-beta-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. Antimicrob. Agents Chemother.46, 1823–1830. 10.1128/aac.46.6.1823-1830.2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, X. H., Xu, Y. H., Cheng, J., Li, T. & Wang, Z. X. Heterogeneity of bla(IND) metallo-beta-lactamase-producing Chryseobacterium indologenes isolates detected in Hefei, China. Int. J. Antimicrob. Agents. 32, 398–400. 10.1016/j.ijantimicag.2008.04.028 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Mammeri, H., Bellais, S. & Nordmann, P. Chromosome-encoded beta-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymes. Antimicrob. Agents Chemother.46, 3561–3567. 10.1128/aac.46.11.3561-3567.2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, J. N., Lai, C. H., Yang, C. H. & Huang, Y. H. Elizabethkingia Infections in humans: from Genomics to Clinics. Microorganisms710.3390/microorganisms7090295 (2019). [DOI] [PMC free article] [PubMed]
- 19.Choi, M. H. et al. Risk factors for Elizabethkingia Acquisition and clinical characteristics of patients, South Korea. Emerg. Infect. Dis.25, 42–51. 10.3201/eid2501.171985 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, J. N., Lai, C. H., Yang, C. H. & Huang, Y. H. Comparison of clinical manifestations, Antimicrobial susceptibility patterns, and mutations of Fluoroquinolone Target genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis isolated in Taiwan. J. Clin. Med.710.3390/jcm7120538 (2018). [DOI] [PMC free article] [PubMed]
- 21.Lin, J. N., Lai, C. H., Yang, C. H., Huang, Y. H. & Lin, H. H. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J. Antimicrob. Chemother.73, 2497–2502. 10.1093/jac/dky197 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Lau, S. K. et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep.6, 26045. 10.1038/srep26045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrin, A. et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun.8, 15483. 10.1038/ncomms15483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González, L. J. & Vila, A. J. Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-β-lactamase BlaB. Antimicrob. Agents Chemother.56, 1686–1692. 10.1128/aac.05835-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchaim, D. et al. Swimming in resistance: co-colonization with carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii or Pseudomonas aeruginosa. Am. J. Infect. Control. 40, 830–835. 10.1016/j.ajic.2011.10.013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mammina, C. et al. Co-colonization with carbapenem-resistant Klebsiella pneumoniae and Acinetobacter baumannii in intensive care unit patients. Scand. J. Infect. Dis.45, 629–634. 10.3109/00365548.2013.782614 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Bellais, S. et al. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum. [DOI] [PMC free article] [PubMed]
- 28.Teo, J. et al. Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis. [DOI] [PMC free article] [PubMed]
- 29.Touchon, M. et al. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. [DOI] [PMC free article] [PubMed]
- 30.Wachino, J. et al. SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. [DOI] [PMC free article] [PubMed]
- 31.Docquier, J. D. et al. Biochemical characterization of the THIN-B metallo-beta-lactamase of Janthinobacterium lividum. [DOI] [PMC free article] [PubMed]
- 32.Wilkins, M. R. et al. Protein identification and analysis tools in the ExPASy server. [DOI] [PubMed]
- 33.Kyte, J. F., - Doolittle, R. F. & Doolittle R. F. A simple method for displaying the hydropathic character of a protein. [DOI] [PubMed]
- 34.Morán-Barrio, J. et al. The metallo-beta-lactamase GOB is a mono-Zn(II) enzyme with a novel active site. [DOI] [PubMed]
- 35.Docquier, J. D. et al. CAU-1, a subclass B3 metallo-beta-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. [DOI] [PMC free article] [PubMed]
- 36.Mercuri, P. S. et al. Biochemical characterization of the FEZ-1 metallo-beta-lactamase of Legionella gormanii ATCC 33297T produced in Escherichia coli. [DOI] [PMC free article] [PubMed]
- 37.Felici, A. & Amicosante, G. Kinetic analysis of extension of substrate specificity with Xanthomonas maltophilia, Aeromonas hydrophila, and Bacillus cereus metallo-beta-lactamases. [DOI] [PMC free article] [PubMed]
- 38.Stoczko, M., Frère, J. F., Rossolini, G. M., Docquier, R. G. F. & Docquier, J. D. -, J.-D. Postgenomic scan of metallo-beta-lactamase homologues in rhizobacteria: identification and characterization of BJP-1, a subclass B3 ortholog from Bradyrhizobium japonicum. [DOI] [PMC free article] [PubMed]
- 39.Gudeta, D. D. et al. Biochemical Characterization of CPS-1, a Subclass B3 Metallo-β-Lactamase from a Chryseobacterium piscium Soil Isolate. [DOI] [PMC free article] [PubMed]
- 40.Laraki, N. et al. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-beta-lactamase IMP-1 produced by Escherichia coli. [DOI] [PMC free article] [PubMed]
- 41.Poirel, L. et al. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. [DOI] [PMC free article] [PubMed]
- 42.Wilson, L. A. O. et al. Kinetic and Structural Characterization of the First B3 Metallo-β-Lactamase with an Active-Site Glutamic Acid. [DOI] [PMC free article] [PubMed]
- 43.Yamada, K. A. O., Ishii, Y. A. O. & Tateda, K. Biochemical characterization of the Subclass B3 Metallo-β-Lactamase PJM-1 from Pseudoxanthomonas japonensis. [DOI] [PMC free article] [PubMed]
- 44.Borgianni, L. et al. Biochemical characterization of the POM-1 metallo-β-lactamase from Pseudomonas otitidis. [DOI] [PMC free article] [PubMed]
- 45.Ullah, J. H. et al. The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 a resolution. [DOI] [PubMed]
- 46.García-Sáez, I. et al. Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril. [DOI] [PubMed]
- 47.Docquier, J. D. et al. High-resolution crystal structure of the subclass B3 metallo-beta-lactamase BJP-1: rational basis for substrate specificity and interaction with sulfonamides. [DOI] [PMC free article] [PubMed]
- 48.Wachino, J. et al. Structural insights into the subclass B3 metallo-β-lactamase SMB-1 and the mode of inhibition by the common metallo-β-lactamase inhibitor mercaptoacetate. [DOI] [PMC free article] [PubMed]
- 49.Leiros, H. K. et al. Crystal structure of the mobile metallo-β-lactamase AIM-1 from Pseudomonas aeruginosa: insights into antibiotic binding and the role of Gln157. [DOI] [PMC free article] [PubMed]
- 50.Short, F. L., Murdoch, S. L. & Ryan, R. P. Polybacterial human disease: the ills of social networking. Trends Microbiol.22, 508–516. 10.1016/j.tim.2014.05.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vos, M. G. J., Zagorski, M., McNally, A. & Bollenbach, T. Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc. Natl. Acad. Sci. U.S.A.114, 10666–10671. 10.1073/pnas.1713372114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bottery, M. J. et al. Inter-species interactions alter antibiotic efficacy in bacterial communities. ISME J.16, 812–821. 10.1038/s41396-021-01130-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramsey, M. M., Rumbaugh, K. P. & Whiteley, M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog.7, e1002012. 10.1371/journal.ppat.1002012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cook, L. C., LaSarre, B. & Federle, M. J. Interspecies communication among commensal and pathogenic streptococci. mBio410.1128/mBio.00382-13 (2013). [DOI] [PMC free article] [PubMed]
- 55.Yamada, M., Ikegami, A. & Kuramitsu, H. K. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol. Lett.250, 271–277. 10.1016/j.femsle.2005.07.019 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Kline, K. A., Schwartz, D. J., Gilbert, N. M., Hultgren, S. J. & Lewis, A. L. Immune modulation by group B Streptococcus influences host susceptibility to urinary tract infection by uropathogenic Escherichia coli. Infect. Immun.80, 4186–4194. 10.1128/iai.00684-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao, Y. T. et al. Sheltering effect and indirect pathogenesis of carbapenem-resistant Acinetobacter baumannii in polymicrobial infection. Antimicrob. Agents Chemother.58, 3983–3990. 10.1128/aac.02636-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith, N. M. et al. Interaction of Staphylococcus aureus and Acinetobacter baumannii during in Vitro β-Lactam exposure. Antimicrob. Agents Chemother.6510.1128/aac.02414-20 (2021). [DOI] [PMC free article] [PubMed]
- 59.Weber, B. S. et al. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PloS One. 8, e55142. 10.1371/journal.pone.0055142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adamowicz, E. M., Flynn, J., Hunter, R. C. & Harcombe, W. R. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J.12, 2723–2735. 10.1038/s41396-018-0212-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livermore, D. M., Moosdeen, F., Lindridge, M. A., Kho, P. & Williams, J. D. Behaviour of TEM-1 beta-lactamase as a resistance mechanism to ampicillin, mezlocillin and azlocillin in Escherichia coli. J. Antimicrob. Chemother.17, 139–146. 10.1093/jac/17.2.139 (1986). [DOI] [PubMed] [Google Scholar]
- 62.Krizova, L., Poirel, L., Nordmann, P. & Nemec, A. TEM-1 β-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J. Antimicrob. Chemother.68, 2786–2791. 10.1093/jac/dkt275 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Rossolini, G. M. et al. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaA(CME)) encoding an extended-spectrum class A beta-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER beta-lactamases. Antimicrob. Agents Chemother.43, 2193–2199. 10.1128/aac.43.9.2193 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El-Badawy, M. F., Abdelwahab, S. F., Alghamdi, S. A. & Shohayeb, M. M. Characterization of phenotypic and genotypic traits of carbapenem-resistant Acinetobacter baumannii clinical isolates recovered from a tertiary care hospital in Taif, Saudi Arabia. Infect. drug Resist.12, 3113–3124. 10.2147/idr.S206691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeon, J. H. et al. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci.16, 9654–9692. 10.3390/ijms16059654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Armbruster, C. E. et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 1, doi: (2010). 10.1128/mBio.00102-10 [DOI] [PMC free article] [PubMed]
- 67.Duan, K., Dammel, C., Stein, J., Rabin, H. & Surette, M. G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol.50, 1477–1491. 10.1046/j.1365-2958.2003.03803.x (2003). [DOI] [PubMed] [Google Scholar]
- 68.Pedroso, M. M. et al. Broad spectrum antibiotic-degrading metallo-β-lactamases are phylogenetically diverse. Protein cell.11, 613–617. 10.1007/s13238-020-00736-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horsfall, L. E. et al. Broad antibiotic resistance profile of the subclass B3 metallo-β-lactamase GOB-1, a di-zinc enzyme. [DOI] [PubMed]
- 70.Lisa, M. N., Hemmingsen, L., Fau - Vila, A. J. & Vila, A. J. Catalytic role of the metal ion in the metallo-beta-lactamase GOB. [DOI] [PMC free article] [PubMed]
- 71.Morán-Barrio, J., Limansky, A. S. & Viale, A. M. Secretion of GOB metallo-beta-lactamase in Escherichia coli depends strictly on the cooperation between the cytoplasmic DnaK chaperone system and the Sec machinery: completion of folding and zn(II) ion acquisition occur in the bacterial periplasm. Antimicrob. Agents Chemother.53, 2908–2917. 10.1128/aac.01637-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morán-Barrio, J., Viale, L. A. F. & Viale, A. M. A. M. Secretion of GOB metallo-beta-lactamase in Escherichia coli depends strictly on the cooperation between the cytoplasmic DnaK chaperone system and the Sec machinery: completion of folding and zn(II) ion acquisition occur in the bacterial periplasm. [DOI] [PMC free article] [PubMed]
- 73.Morán-Barrio, J. et al. Crystal Structure of the Metallo-β-Lactamase GOB in the Periplasmic Dizinc Form Reveals an Unusual Metal Site. [DOI] [PMC free article] [PubMed]
- 74.Tottey, S. et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. [DOI] [PubMed]
- 75.Pedroso, M. A. O. X. et al. Broad spectrum antibiotic-degrading metallo-β-lactamases are phylogenetically diverse. [DOI] [PMC free article] [PubMed]
- 76.Moali, C. et al. Analysis of the importance of the metallo-beta-lactamase active site loop in substrate binding and catalysis. [DOI] [PubMed]
- 77.Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. [DOI] [PubMed]
- 78.González, J. M. et al. Metallo-β-lactamases withstand low zn(II) conditions by tuning metal-ligand interactions. [DOI] [PMC free article] [PubMed]
- 79.González, L. J., Moreno, D. M., Bonomo, R. A. & Vila, A. J. Host-specific enzyme-substrate interactions in SPM-1 metallo-β-lactamase are modulated by second sphere residues. [DOI] [PMC free article] [PubMed]
- 80.Yong, D. et al. Genetic and biochemical characterization of an acquired subgroup B3 metallo-β-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. [DOI] [PMC free article] [PubMed]
- 81.Stoczko, M., Frère, J. M., Rossolini, G. M. & Docquier, J. D. Postgenomic scan of metallo-beta-lactamase homologues in rhizobacteria: identification and characterization of BJP-1, a subclass B3 ortholog from Bradyrhizobium japonicum. Antimicrob. Agents Chemother.50, 1973–1981. 10.1128/aac.01551-05 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yong, D. et al. Genetic and biochemical characterization of an acquired subgroup B3 metallo-β-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob. Agents Chemother.56, 6154–6159. 10.1128/aac.05654-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stoczko, M., Frère, J. M., Rossolini, G. M. & Docquier, J. D. Functional diversity among metallo-beta-lactamases: characterization of the CAR-1 enzyme of Erwinia carotovora. Antimicrob. Agents Chemother.52, 2473–2479. 10.1128/aac.01062-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lauretti, L. et al. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother.43, 1584–1590. 10.1128/aac.43.7.1584 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Osano, E. et al. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother.38, 71–78. 10.1128/aac.38.1.71 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu, R., Zhang, Q. & Gu, Z. Whole-genome analysis of the potentially zoonotic Elizabethkingia miricola FL160902 with two new chromosomal MBL gene variants. J. Antimicrob. Chemother.75, 526–530. 10.1093/jac/dkz480 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Bellais, S., Aubert, D., Naas, T. & Nordmann, P. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother.44, 1878–1886. 10.1128/aac.44.7.1878-1886.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walsh, T. R. et al. Metallo-beta-lactamases: the quiet before the storm?. [DOI] [PMC free article] [PubMed]
- 89.Hu, S. et al. Population genomics of emerging Elizabethkingia anophelis pathogens reveals potential outbreak and rapid global dissemination. Emerg. Microbes Infections. 11, 2590–2599. 10.1080/22221751.2022.2132880 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Navon, L. et al. Notes from the field: investigation of Elizabethkingia anophelis Cluster - Illinois, 2014–2016. MMWR Morb. Mortal. Wkly Rep.65, 1380–1381. 10.15585/mmwr.mm6548a6 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Hu, S. et al. Elizabethkingia anophelis isolated from patients with multiple organ dysfunction syndrome and lower respiratory tract infection: report of two cases and literature review. Front. Microbiol.8, 382. 10.3389/fmicb.2017.00382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson, C. M. & Grossman, A. D. Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet.49, 577–601. 10.1146/annurev-genet-112414-055018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wozniak, R. A. & Waldor, M. K. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol.8, 552–563. 10.1038/nrmicro2382 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Alcock, B. P. et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res.48, D517–d525. 10.1093/nar/gkz935 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bertini, A. et al. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother.54, 4168–4177. 10.1128/aac.00542-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alikhan, N. F., Petty, N. K., Zakour, B., Beatson, S. A. & N. L. & BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genom.12, 402. 10.1186/1471-2164-12-402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wareth, G. A. O. et al. Molecular characterization of German Acinetobacter baumannii isolates and Multilocus sequence typing (MLST) analysis based on WGS reveals novel STs. LID – 10.3390/pathogens10060690 [doi] LID – 690. [DOI] [PMC free article] [PubMed]
- 98.Diancourt, L. et al. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. [DOI] [PMC free article] [PubMed]
- 99.Gautreau, G. et al. PPanGGOLiN: Depicting microbial diversity via a partitioned pangenome graph. PLoS computational biology 16, e1007732, doi: (2020). 10.1371/journal.pcbi.1007732 [DOI] [PMC free article] [PubMed]
- 100.Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T. & Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun.9, 5114. 10.1038/s41467-018-07641-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu, S. et al. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 1, e56, doi: (2022). 10.1002/imt2.56 [DOI] [PMC free article] [PubMed]
- 102.Yong, D. et al. Imipenem-EDTA disk Method for Differentiation of metallo-beta-lactamase-producing Clinical Isolates of Pseudomonas spp. and Acinetobacter spp. [DOI] [PMC free article] [PubMed]
- 103.Franklin, C., Liolios, L., Fau - Peleg, A. Y. & Peleg, A. Y. phenotypic detection of carbapenem-susceptible metallo-beta-lactamase-producing gram-negative bacilli in the clinical laboratory. [DOI] [PMC free article] [PubMed]
- 104.Chang, T. Y., Chen, H. Y., Chou, Y. C., Cheng, Y. H. & Sun, J. A.-O. In vitro activities of imipenem, Vancomycin, and rifampicin against clinical Elizabethkingia species producing BlaB and GOB metallo-beta-lactamases. [DOI] [PubMed]
- 105.Chew, K. L., Cheng, B., Lin, R. T. P. & Teo, J. W. P. Elizabethkingia anophelis is the Dominant Elizabethkingia species found in blood cultures in Singapore. J. Clin. Microbiol.5610.1128/jcm.01445-17 (2018). [DOI] [PMC free article] [PubMed]
- 106.Cheng, Y. H. et al. Multicentre study evaluating matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically isolated Elizabethkingia species and analysis of antimicrobial susceptibility. Clin. Microbiol. Infection: Official Publication Eur. Soc. Clin. Microbiol. Infect. Dis.25, 340–345. 10.1016/j.cmi.2018.04.015 (2019). [DOI] [PubMed] [Google Scholar]
- 107.Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. [DOI] [PMC free article] [PubMed]
- 108.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. [DOI] [PubMed]
- 109.Padhan, N. Highly sensitive and Quantitative Detection of Proteins and their isoforms by Capillary Isoelectric focusing Method. LID – 10.3791/56794 [doi] LID – 56794. [DOI] [PMC free article] [PubMed]
- 110.O’Callaghan Ch Fau -, Morris, A., Morris, A., Fau - Kirby, S. M. & Shingler, A. H. Kirby Sm Fau - Shingler, A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. [DOI] [PMC free article] [PubMed]
- 111.Rashamuse, K., Magomani, V., Fau - Ronneburg, T., Ronneburg, T. & Fau - Brady, D. & Brady, D. A novel family VIII carboxylesterase derived from a leachate metagenome library exhibits promiscuous beta-lactamase activity on nitrocefin. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences analysed during the current study were deposited into the National Center for Biotechnology Information (NCBI) under BioProject Accession Number PRJNA1129009 and PRJNA1130100, and are available at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1129009 and https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1130100.