Abstract
Peptide sequencing via tandem mass spectrometry (MS/MS) is essential in proteomics. Unlike traditional database searches, deep learning excels at de novo peptide sequencing, even for peptides missing from existing databases. Current deep learning models often rely on autoregressive generation, which suffers from error accumulation and slow inference speeds. In this work, we introduce π-PrimeNovo, a non-autoregressive Transformer-based model for peptide sequencing. With our architecture design and a CUDA-enhanced decoding module for precise mass control, π-PrimeNovo achieves significantly higher accuracy and up to 89x faster inference than state-of-the-art methods, making it ideal for large-scale applications like metaproteomics. Additionally, it excels in phosphopeptide mining and detecting low-abundance post-translational modifications (PTMs), marking a substantial advance in peptide sequencing with broad potential in biological research.
Subject terms: Proteome informatics, Machine learning, Proteomics
Peptide sequencing is critical to the advancement of proteomics research. Here, the authors present π-PrimeNovo, a non-autoregressive deep learning model that achieves high accuracy and up to 89x faster sequencing. This enables large-scale sequencing and multiple downstream applications.
Introduction
Protein identification is essential in proteomics, with shotgun proteomics via mass spectrometry recognized as the primary method1. This approach involves enzymatically digesting proteins into peptides for tandem mass spectrometry analysis, providing spectra that reveal peptide sequences and structures. Decoding amino acid sequences from these spectra is key to protein identification2. Currently, database searching is the main method, with tools like SEQUEST3, Mascot4, MaxQuant/Andromeda2, PEAKS DB5, and pFind6. However, these methods depend on comprehensive sequence databases, limiting their applicability in areas like monoclonal antibody sequencing7, novel antigen identification8, and metaproteome analysis without established databases9.
Over the past two decades, various de novo peptide sequencing tools have advanced the field8,10–21. These algorithms infer amino acid compositions and modifications by analyzing mass differences between fragment ions in spectra. Early methods like PepNovo11 and PEAKS10 used the graph theory and dynamic programming approach. DeepNovo12 introduced a deep learning-based model, integrating CNNs for spectral peak analysis with LSTMs for sequence processing. PointNovo13 enhanced prediction precision with an order-invariant network, while Casanovo15 applied Transformer architecture, treating sequencing as a translation task. Casanovo V216 was later trained on a 30 million spectra dataset to further scale up the model performance. Recent innovations like PepNet19 use fully convolutional networks for speed, and GraphNovo22 uses graph neural networks to address missing-fragmentation issues. Despite these advances15–22, deep learning-based de novo sequencing in shotgun proteomics still achieves low peptide recall rates of 30–50% on standard benchmark.
Currently, all deep learning models for de novo peptide sequencing are based on the autoregressive framework23, meaning the generation of each amino acid is heavily reliant on its predicted predecessors, resulting in a unidirectional generation process. However, the significance of bidirectional information is paramount in peptide sequencing, as the presence of an amino acid is intrinsically linked to its neighbors in both directions21. In autoregressive models, any errors in early amino acid predictions can cascade, affecting subsequent generations. Autoregressive decoding algorithms such as beam search lack the capability to retrospectively modify previously generated content, making it challenging to control the total mass of the generated sequence. This limitation arises because each token is produced based on its predecessor, meaning that altering any previously generated token would consequently shift the distribution of subsequent tokens and, therefore, require a re-generation of the whole sequence24.
In this research, we introduce π-PrimeNovo (shortened as PrimeNovo) (Fig. 1), representing a significant departure from conventional autoregressive approaches by adopting a non-autoregressive approach to effectively address the unidirectional problems of autoregressive methods. This innovation stands as the pioneer non-autoregressive transformer-based model in this field. Such design enables a simultaneous sequence prediction, granting each amino acid a comprehensive bidirectional context. Another key advancement in PrimeNovo is the integration of a precise mass control (PMC) unit, uniquely compatible with the non-autoregressive framework, which utilizes precursor mass information to generate controlled and precise peptide sequences. This precise mass control, coupled with bidirectional generation, significantly enhances peptide-level performance.
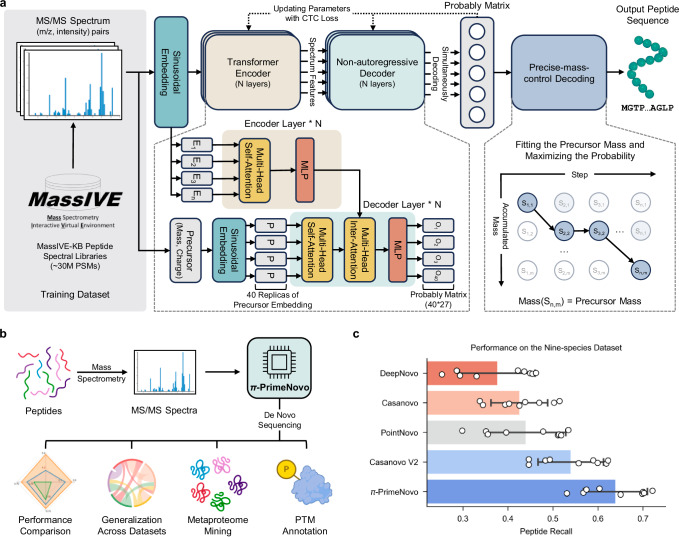
Fig. 1. PrimeNovo stands as the pioneering biological non-autoregressive Transformer model, delivering precise peptide sequencing.
a Model architecture overview: Our model takes MS/MS spectra as input and generates the predicted peptide sequence. It comprises two key components: (1) a non-autoregressive Transformer model backbone optimized with connectionist temporal classification (CTC) loss, enabling simultaneous amino acid prediction at all positions. (2) The precise mass control (PMC) decoding unit, which utilizes predicted probabilities to precisely optimize peptide generation to meet mass requirements. b Applications and biological insights: PrimeNovo’s capabilities extend to downstream tasks and offer valuable insights for various biological investigations. c Average performance comparison: This chart illustrates the average performance of PrimeNovo alongside four other top-performing models on the widely utilized nine-species benchmark dataset (93,750 tested spectrum samples across all 9 species). Each bar represents the mean peptide recall for the respective approach. The black line indicates the 95% confidence interval (n = 9). Notably, results for DeepNovo, Casanovo, and Casanovo V2 are based on model weights released by the original authors, while PointNovo’s results are cited from the published work, as the original model weights were not shared by PointNovo’s authors. Source data are provided as a Source Data file. Some figures were created in BioRender56.
PrimeNovo consistently demonstrates impressive peptide-level accuracy, achieving an average peptide recall of 64% on the widely used nine-species benchmark dataset. This performance significantly surpasses the existing best model, which achieves a peptide recall of 54%16. Across a diverse range of other MS/MS datasets, PrimeNovo consistently maintains a notable advantage in peptide recall over the state-of-the-art model, achieving relative improvements from 16% to even doubling the accuracy, which highlights its exceptional performance and reliability. Moreover, by avoiding the sequential, one-by-one generation process inherent in autoregressive models, PrimeNovo also substantially increases its inference speed. This acceleration is further enhanced through the use of dynamic programming and CUDA-accelerated computation, allowing PrimeNovo to surpass the existing autoregressive models by up to 89 times. This speedup advantage enables PrimeNovo to make accurate predictions on large-scale spectrum data. We have demonstrated that PrimeNovo excels in large-scale metaproteomic research by accurately identifying a significantly greater number of species-specific peptides compared to previous methods, reducing the processing time from months, as required by Casanovo V2 with beam search, to just days. Furthermore, PrimeNovo’s versatility extends to the identification of PTMs, showcasing its potential as a transformative tool in proteomics research.
Results
PrimeNovo sets a benchmark with 64% peptide recall, achieving over 10% improvement in widely used nine-species dataset
Echoing the approach of Casanovo V2, we utilized the large-scale MassIVE-KB dataset25, featuring around 30 million peptide-to-spectrum matches (PSMs), as our training data. PrimeNovo was then evaluated on the nine-species testing benchmark directly. It is crucial to note, however, that baseline models like PointNovo, DeepNovo, and Casanovo were originally trained using the leave-one-species-out cross-validation (CV) strategy26 on the nine-species dataset. This strategy involves training on eight species and evaluating on the ninth each time. To facilitate a fair comparison, we also trained PrimeNovo on the nine-species dataset using the same CV strategy, following the data split used by all other baseline models. As shown in Fig. 2a, PrimeNovo CV outperformed other baseline models trained with this strategy by a large margin. Notably, even when trained solely on the nine-species benchmark dataset, PrimeNovo CV already matched the performance of Casanovo V2, which is the model trained on the large-scale MassIVE-KB dataset. When trained on the MassIVE-KB dataset, PrimeNovo set state-of-the-art results across all species in the nine-species benchmark (Fig. 2b and Supplementary Fig. 6). The average peptide recall improved significantly, increasing from 45% with Casanovo to 54% with Casanovo V2, and further to 64% with PrimeNovo. This marks a 10% improvement over Casanovo V2 and a 19% increase over Casanovo. In the recall-coverage curve (Fig. 2a), PrimeNovo consistently held the top position across all coverage levels and species, reaffirming its status as a leading model in de novo peptide sequencing. At the amino acid (AA) level, PrimeNovo demonstrates significantly higher accuracy, as measured by both AA recall and AA precision, compared to Casanovo V2. As shown in Fig. 2c, PrimeNovo outperforms Casanovo V2 in AA recall across all nine species, with an improvement ranging from 3% to 6%. This performance advantage is consistent in AA precision, with a detailed comparison provided in the Supplementary Information. Additionally, we tested PrimeNovo on a revised nine-species test set introduced by Casanovo V216, which featured higher data quality and a larger quantity of spectra, covering a wider range of data distributions for each species. In this updated test, PrimeNovo’s average peptide recall soared to 75% across all species, from the previous 65% by Casanovo V2. A detailed comparison of these results is available in Supplementary Fig. 4. The outcomes from both the original and revised nine-species benchmark datasets highlight PrimeNovo’s capability to accurately predict peptides across various species, demonstrating its effectiveness and versatility.
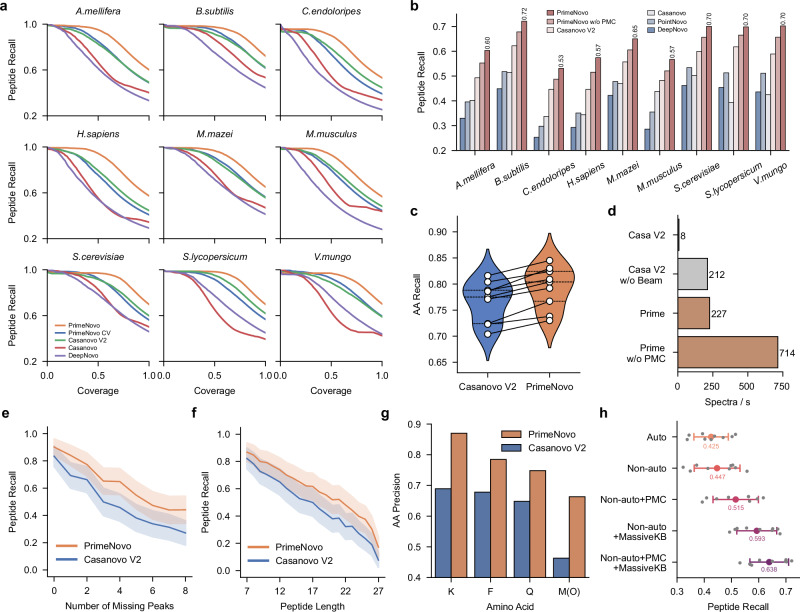
Fig. 2. A detailed comparison between PrimeNovo and previous deep learning-based approaches on the nine-species benchmark dataset.
a The performance comparison between PrimeNovo and other de novo algorithms for recall-coverage curves on the nine-species benchmark dataset. These curves illustrate recall (the averaged peptide recall)—coverage (the proportion of the predicted spectra to all annotated spectra ranked by the model’s confidence) relationships across all confidence levels for each test species. PrimeNovo CV represents our model trained on the nine-species benchmark dataset using a cross-validation strategy. PrimeNovo represents our model trained on the MassIVE-KB dataset. b The average prediction performance on each individual species for PrimeNovo and comparison models. PrimeNovo w/o PMC presents results obtained using CTC beam search decoding without PMC. c Comparison of Amino Acid level prediction recall across nine different species between Casanovo V2 and PrimeNovo. d Inference Speed Comparison: A comparison of inference speeds, measured in the number of spectra decoded per second, between PrimeNovo and Casanovo V2. The speed tests were conducted on the same computational hardware (single A100 NVIDIA GPU) and averaged over data from all test species. e and (f) Influence of Missing Peaks and Peptide Length: These plots reveal how the degree of missing peaks (less or equal to 8 for missing peaks and length ranging from 7 to 27) and the length of true labels affect the predictions of PrimeNovo and Casanovo V2. We plot a central curve that connects the mean values of the data points (n = 9714), with a light background representing the s.d. (scale factor=0.2) g. Performance on Amino Acids with Similar Masses: A comparison of Casanovo V2 and PrimeNovo in predicting amino acids with very similar molecular masses, such as K (128.094963) with Q (128.058578) and F (147.068414) with Oxidized M (147.035400). h Ablation study: An analysis of the impact of adding each module of our approach on the overall performance (of the nine-species benchmark dataset. (n = 9, data are presented as mean values ± sd). Source data are provided as a Source Data file.
PrimeNovo, leveraging its bi-directional information integration and parallel generation process as a non-autoregressive model, convincingly establishes its superiority across various facets of sequencing tasks, transcending mere high prediction accuracy. Firstly, our non-autoregressive model offers a substantial improvement in the inference speed compared to the autoregressive models of similar sizes, thanks to its concurrent generation process. As depicted in Fig. 2d, PrimeNovo, even without the Precise Mass Control (PMC) unit, achieves a staggering speed advantage of 3.4 times faster over Casanovo V2 without beam search decoding under identical testing conditions (i.e., using the same machine with identical CPU and GPU specifications). Upon incorporating post-prediction decoding strategies (PMC for PrimeNovo and beam search for Casanovo V2), PrimeNovo’s advantage in inference speed becomes even more pronounced, making it over 28 times faster than Casanovo V2. Notably, considering that PrimeNovo without PMC can already outperform Casanovo V2 with beam search by an average of 6% on the nine-species benchmark dataset (as demonstrated in Fig. 2b), users can experience a maximum speedup of 89 times while making only minimal sacrifices in prediction accuracy when PMC is not deployed. We further investigated other factors, such as batch size on the speed and the results are included in Supplementary Information.
Furthermore, PrimeNovo exhibits exceptional prediction robustness across various challenges, including different levels of missing peaks in the spectrum, varying peptide lengths, and amino acid combinations that are prone to confusion. To illustrate this robustness, we categorized predictions on the nine-species benchmark dataset based on the degree of missing peaks in the input spectrum and the number of amino acids in the target peptide. The calculation of missing peaks in each spectrum follows the methodology outlined in a previous study by Beslic et al.7, where we compute all the theoretical m/z values for potential y ions and b ions based on the true label and determine how many of these theoretical peaks are absent in the actual spectrum. As presented in Fig. 2e, it is not surprising to observe a decline in prediction accuracy as the number of missing peaks in the spectra increases. However, PrimeNovo consistently indicates superior performance across all levels of missing peaks and consistently outperforms Casanovo V2. Similarly, Fig. 2f illustrates that PrimeNovo maintains its higher accuracy compared to Casanovo V2, irrespective of the length of the peptide being predicted. In Fig. 2g, we further observe that PrimeNovo excels in accurately predicting amino acids that are challenging to identify due to their closely similar mass (<0.05 Da) to other amino acids. Specifically, the aa precision of all four similar amino acids is more than 10% more accurate on average compared to that of Casanovo V2. Specifically, the precision advantage is more than 18% on both K and Oxidized M amino acids.
We then conducted an ablation study to investigate the performance gains achieved by each component of our model on the nine-species benchmark dataset. From Fig. 2h, we observe a 2% improvement in peptide recall when transitioning from an autoregressive model to a non-autoregressive model. The gain in performance is magnified by a large amount (7%) when PMC is introduced, as controllable generation is important in such tasks and improves the accuracy of our generated sequence. Remarkably, the performance boost from the non-autoregressive model is most pronounced when transitioning from the CV training data to the MassIVE-KB dataset, as the substantial increase in training data proves invaluable for learning the underlying bi-directional patterns in the sequencing task. Lastly, we see that utilizing PMC with augmented training data achieves the highest prediction accuracy, which further demonstrates PMC’s importance under different data availability situations.
PrimeNovo exhibits strong generalization and adaptability capability across a wide array of MS/MS data sources
As MS/MS data can vary significantly due to differences in biological samples, mass spectrometer parameters, and post-processing procedures, there is often a substantial degree of distributional shift across various MS/MS datasets. To demonstrate PrimeNovo’s ability to generalize effectively across a wide spectrum of distinct MS/MS data for diverse downstream tasks, we conducted an evaluation of PrimeNovo’s performance on some of the most widely used publicly available MS/MS datasets. We then compared the results with those of the current state-of-the-art models, Casanovo and Casanovo V2. In addition to the nine-species benchmark dataset discussed earlier, we selected three prominent MS/MS datasets that represent varying data sources and application settings: the PT27, IgG1-Human-HC26, and HCC28 datasets, and the details of these datasets are included in the Supplementary Information.
We start by evaluating PrimeNovo’s ability to perform well in a zero-shot scenario, which means the model is tested without any specific adjustments to match the characteristics and distribution of the target dataset. As depicted in Fig. 3a and Supplementary Fig. 8, PrimeNovo exhibits significant performance superiority over both Casanovo V2 and Casanovo in terms of peptide recall when directly tested on three distinct datasets. Specifically, PrimeNovo outperforms Casanovo V2 by 13%, 14%, and 22% on PT, IgG1-Human-HC, and HCC datasets, respectively. This performance gap widens to 30%, 43%, and 38% when compared to Casanovo. For the IgG1-Human-HC dataset, following7, we present the evaluation results for each human antigen type, as illustrated in Fig. 3b. PrimeNovo consistently outperforms Casanovo V2 across all six antigen types, achieving increased peptide recall ranging from 9% to 20%. We further examine the amino acid level accuracy on the unseen dataset. From Fig. 3c, it’s notable that PrimeNovo has a dominant AA level precision advantage over Casanovo V2 across all confidence levels of the model output. This indicates PrimeNovo’s better prediction of amino acids’ presence and locations.
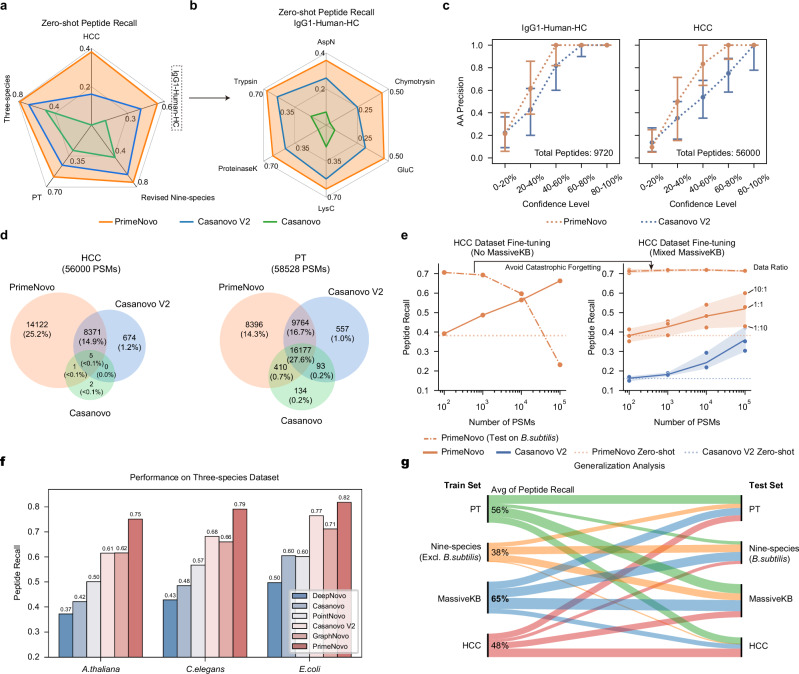
Fig. 3. PrimeNovo’s exceptional performance extends to unseen spectra from various biological sample sources.
a Average peptide recall: This section details the average peptide recall of PrimeNovo compared to baseline models across four distinct large-scale MS/MS datasets. b Enzyme-specific performance: Performance breakdown among six different proteolytic enzymes in the IgG1-Human-HC dataset. c Amino acid-level precision: The chart depicts the amino acid-level precision for PrimeNovo and Casanovo V2 on the IgG1-Human-HC (9719 tested spectrum samples) and HCC datasets (56,000 tested spectrum samples). The x-axis shows the coverage rate of predicted peptides based on each model’s confidence score. For instance, 20%-40% indicate the 20%-40% least confident predictions based on confidence scores. AA precision is then calculated within each coverage range. Note that data are presented as median values of each confidence level with interquartile range (50% percentile interval). d A Venn diagram illustrates the number of overlapping peptides among three de novo sequencing models and a traditional database searching algorithm. Each count represents identical peptides identified by both MaxQuant and the respective model for the same spectrum. e Model fine-tuning results: This chart demonstrates how performance on the HCC test dataset changes with the addition of more HCC training data during fine-tuning. The left side shows fine-tuning with only the HCC dataset, leading to catastrophic forgetting of the original data distribution (nine-species benchmark dataset). The right side shows fine-tuning with a mix of HCC and MassIVE-KB training data. The data points in the right figure show the performance of three different data ratios during the fine-tuning stage. We plot a central curve that connects the mean values of the data points, with a light background representing the s.d. f A comparison of performance between PrimeNovo and five other de novo models on a 3-species test dataset. g This diagram demonstrates the model’s generalization capability when trained exclusively with each training dataset. The left-hand side indicates each one of the four training data PrimeNovo is trained on. The thickness of each line indicates the performance on each of the four testing sets on the right-hand side, with a thicker line being better performance. The numbers on the stem indicate the averaged peptide recall over all four testing sets, highlighting the distributional transferability of each training data. The model trained on MassIVE-KB exhibited the highest average peptide recall, 65% (bolded). Source data are provided as a Source Data file.
To further assess the performance disparities under the zero-shot setting, we leveraged identified PSMs from MaxQuant in each dataset as the benchmark. Then we compared the number of overlapping PSMs between the predicted PSMs generated by each de novo algorithm and the PSMs identified by MaxQuant. As displayed in Fig. 3d, Casanovo performed poorly on the HCC dataset, with only 8 PSMs overlapping with MaxQuant. In contrast, Casanovo V2 identified 9050 overlapping PSMs, while PrimeNovo predicted up to 22499 PSMs that perfectly matched those identified by MaxQuant. On the PT dataset, PrimeNovo, Casanovo V2, and Casanovo had 34747, 26591, and 16814 overlapping PSMs with MaxQuant search results, respectively. PrimeNovo demonstrates a much more consistent prediction behavior, aligning closely with high-quality traditional database-searching peptide identification software.
Next, we examine how well PrimeNovo generalizes under the fine-tuning setting, which involves quickly adapting the model to new training data from the target distribution without starting the training process from scratch. This approach allows the model to leverage its previously acquired knowledge from the large dataset it was originally trained on and apply it to a more specific task or domain with only a minimal amount of additional training. We fine-tuned PrimeNovo on both the PT and HCC training datasets to assess the model’s adaptability. In order to gauge the impact of the quantity of additional data on fine-tuning performance, we conducted the fine-tuning with 100, 1000, 10,000, and 100,000 additional data points, respectively. We also fine-tuned Casanovo V2 under identical settings to compare the adaptability of the two models fairly. As depicted on the right side of Fig. 3e, augmenting the amount of additional data for fine-tuning does indeed enhance the model’s prediction accuracy on the corresponding test set, as the model gains a better understanding of the distributional nuances within the data. In comparison, PrimeNovo demonstrates a more robust ability to adapt to new data distributions and achieves higher accuracy after fine-tuning compared to the zero-shot scenario. It consistently outperforms Casanovo V2 when subjected to the same fine-tuning conditions, with 18% and 12% higher peptide level recall on HCC and PT test sets respectively when the fine-tuning reaches the best performance (Fig. 3e). It is noteworthy that a noticeable improvement in prediction accuracy is only observed after incorporating 10,000 additional MS data points during the fine-tuning process, indicating a recommended data size for future fine-tuning endeavors involving other data distributions.
It’s important to note that the fine-tuning process can lead the model to forget the original data distribution from the training set, which is referred to as catastrophic forgetting. As illustrated in the left part of Fig. 3e, when fine-tuning is conducted exclusively with the target data, the performance in the nine-species benchmark dataset experiences a significant and gradual decline as more data samples are included (indicated by the dashed line). However, when the target data is mixed with the original training data, catastrophic forgetting is mitigated, as evident from the dashed line in the right part of Fig. 3e. Indeed, fine-tuning exclusively with the target data does introduce a relatively higher performance gain in the target test set compared to fine-tuning with mixed data (solid line in Fig. 3e), where the difference can be as much as 15% when the amount of the new data used for fine-tuning is large.
By fine-tuning the model using a single dataset and then testing it on others, we can explore the similarities and disparities in data distributions among different pairs of datasets. This approach provides valuable insights into how closely related each MS/MS dataset is to the others and the extent to which a model’s knowledge can be transferred when trained on one dataset. In Fig. 3g, it’s not surprising to observe that the model exhibits the strongest transferability when the training and testing data share the same data source. Notably, MassIVE-KB, the training set for both our model and Casanovo V2, demonstrates the highest average peptide recall of 65% across all other test sets. This can be attributed to the diverse range of MS/MS data sources encompassed within the MassIVE-KB dataset, covering a wide spectrum of distinct MS/MS data. The PT dataset, with an average peptide recall of 56%, is also considered a high-quality dataset with robust transferability. It has been employed in the training of numerous other de novo models21. However, the models trained on the HCC and nine-species benchmark datasets do not generalize well to other testing datasets. The nine-species benchmark exclusively covers MS/MS data for the included nine species and has a relatively small data size, while the HCC dataset is specific to human hepatocellular carcinoma. Additionally, we observe that models trained with the nine-species benchmark dataset and MassIVE-KB datasets exhibit relatively poor performance when applied to the HCC dataset, suggesting a notable disparity in their data distribution.
Finally, we conduct a comparative analysis between PrimeNovo and concurrent approaches in de novo sequencing to illustrate the advancements and effectiveness of our method. Our comparative models, namely GraphNovo, a graph-based neural network, and PepNet, a CNN-based neural network, approach the problem from distinct angles, utilizing the latest deep learning techniques. It’s worth noting that both GraphNovo22 and PepNet19 are trained on their own designated training and testing datasets for their respective model versions. Consequently, we adopt a zero-shot evaluation approach, testing PrimeNovo on each of their test sets and comparing the results with their reported performances. We carefully examined the used data and ensured that there was no overlap between our training dataset and the test sets used by GraphNovo and PepNet. For the 3-species test set employed by GraphNovo, PrimeNovo demonstrates remarkable improvements in peptide recall, surpassing GraphNovo by 13%, 13%, and 11% in the A. thaliana, C. elegans, and E. coli species, respectively (see Fig. 3f). Furthermore, when tested on the PepNet test set, PrimeNovo exhibits a notable advantage of 14% and 24% in peptide recall over PepNet when predicting the peptide with charges of 2 and 3 respectively, detailed results of which are in Supplementary Fig. 13.
PrimeNovo’s behavior analysis reveals an effective error correction mechanism behind non-autoregressive modeling and PMC unit
To gain a comprehensive understanding of the model’s behavior and to analyze how PrimeNovo utilizes the spectrum data to arrive at its final results, we employ some of the most recent model interpretability techniques, examining each component of our model in detail. We commence by visualizing the attention behavior of the encoder network in PrimeNovo and comparing it to that of Casanovo V2. The encoder’s role is critical, as it is responsible for feature extraction from the spectrum, significantly influencing how well the model utilizes input spectrum data. As depicted in the attention map in Fig. 4a, it is evident that Casanovo V2 predominantly assigns most of its attention weights to the first input position (the special token added at the beginning of the peak tokens). Attention weights for the remaining tokens are sparse, insignificant, and primarily concentrated along the diagonal direction. This behavior suggests that Casanovo V2 encodes information primarily within its special token, with limited utilization of other peak positions. In contrast, PrimeNovo exhibits a well-distributed attention pattern across different input peaks, each with varying levels of information density. Furthermore, we observe that the attention of PrimeNovo is more heavily allocated to peaks corresponding to the b-y ions of the true label, which are among the most crucial pieces of information for decoding the spectrum (as detailed in Supplementary Fig. 19). This highlights PrimeNovo’s capacity to extract information more effectively from tokens it deems essential, and this behavior remains consistently active across all nine layers.
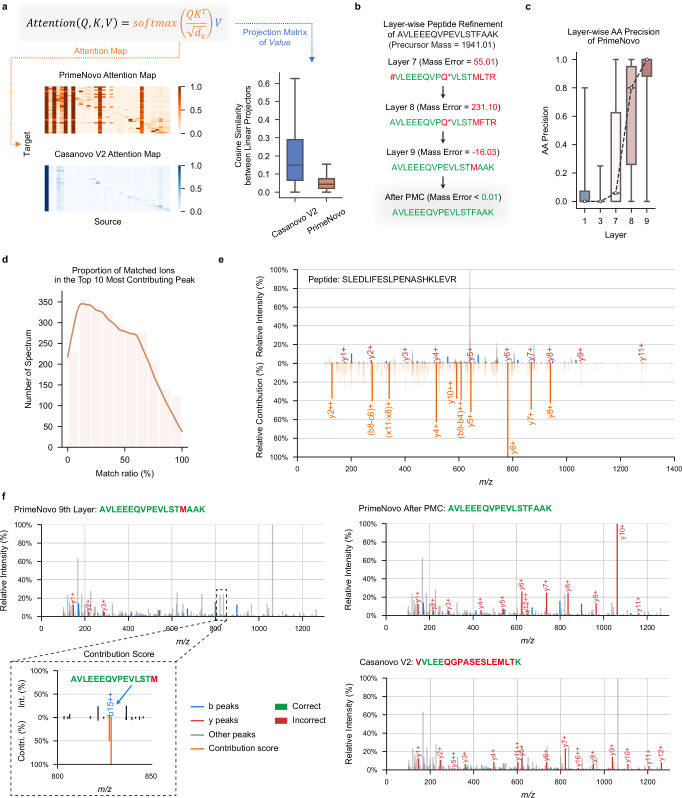
Fig. 4. Error analysis and model explainability offer valuable insights into the performance of PrimeNovo.
a Attention map and feature vector similarity: This section showcases the visualization of attention maps between the Transformer encoders of Casanovo V2 and PrimeNovo. It also includes a detailed similarity analysis of each column in the feature vector from the value matrix projection. The boxplot displays the minimum, maximum, median, and quartiles of the similarities scores (n = 421,232, outliers omitted). b Layerwise prediction refinement: A case study demonstrates how PrimeNovo’s non-autoregressive model progressively refines predictions layer by layer, highlighting the model’s capacity for self-correcting its predictions as a whole. Note that * represents the Glutamine deamidation modification on amino acid Q. c The points display the average prediction accuracy at the amino acid level across each layer in PrimeNovo, with the boxplot showing the minimum, maximum, median, and quartiles of the prediction accuracy (n = 88,236). d This diagram illustrates the proportion of peaks corresponding to b-y ions, as determined from predictions, based on all peaks within the PT test set ranked within the top 10 by their contribution scores. e Alignment between the model’s contribution scores and the theoretical b-y ion peaks derived from predictions is presented. The diagram’s lower half shows the magnitude of all contribution scores, emphasizing those matching the b-y ions. The upper half provides a comparison with the original spectrum. f A case study on how the theoretical ions, calculated from the predicted peptide, align with the input spectrum. The matched theoretical b-y ions are distinctly marked in red and blue for predictions made by PrimeNovo and Casanovo, respectively. This comparison seeks to identify potential sources of error in incorrect predictions. The diagram’s bottom left section highlights a high contribution score assigned to an incorrect peak, corresponding to a b-ion peak linked to an erroneous amino acid prediction in PrimeNovo’s final layer. Source data are provided as a Source Data file.
Furthermore, we conducted a numerical comparison of the Value matrices learned by the encoder networks of both models29. Each column in the Value matrix projection represents a hidden feature. To assess the diversity of features present in the Value matrix, we calculated the average cosine similarity between every pair of columns. As illustrated in the bar plot in Fig. 4a, it is evident that PrimeNovo’s feature vectors exhibit lower similarity to each other, as indicated by the lower average cosine similarity values in the plot. This suggests that our model’s Value matrix encompasses a broader spectrum of information and a more diverse set of features30,31. This finding could provide an additional explanation for our model’s superior performance. For a more comprehensive assessment of the orthogonality of the Value matrix projection, which is evaluated by measuring the norm of the Gram matrix29–31 (see Supplementary Fig. 18).
Since our non-autoregressive model predicts the entire sequence at once, we can examine how each of the nine model layers progressively improves the overall sequence prediction. We decode the whole sequence from each layer of our model and observe how the amino acids evolve over time. As illustrated in Fig. 4c, amino acid-level accuracy experiences a significant surge from layer seven to nine, with a consistent increasing trend across each layer. This signifies a continual improvement in prediction accuracy at each layer. By examining the case study presented in Fig. 4b, we discern that this increase in accuracy is achieved through a layer-wise self-correction mechanism. In this process, each layer gradually adjusts the erroneously predicted amino acids throughout the entire sequence, making them more reasonable and closer to the true answer. The non-autoregressive model’s capability of enabling each amino acid to reference the surrounding amino acids for information facilitates accurate and effective correction across its layers. PMC, acting as the final safeguard against errors, rectifies model prediction errors by selecting the most probable sequence that adheres to the mass constraint. This process yields a slightly modified sequence compared to the output from the last layer, ultimately leading to the correct answer.
We also employed the feature contribution technique saliency maps32 to analyze the impact of each peak on the prediction results. This technique generates contribution scores that provide a quick view of the impact of each peak on the prediction. A higher contribution score for a peak indicates a larger impact on the results. On the test set of PT, the contribution scores for all peaks in each spectrum. Subsequently are calculated, we sorted all peaks in descending order based on their contribution scores and selected the top 10 peaks. Using the known peptide sequences associated with these spectra, all possible fragment ions considering only 1+ and 2+ ions, are generated using the in-house script (see Supplementary Note 7 for more details). We then compared the m/z values of the top 10 peaks with the m/z values of all possible fragment ions, considering a match if the difference was within 0.05 Da. Finally, the percentage of the top 10 peaks that could be matched is calculated. As shown in Fig. 4d, ~40% of the spectra had a matched percentage of above 50%. Importantly, our model not only focused on the major peaks but also considered internal fragment ions. For example (Fig. 4e), in the spectrum corresponding to the peptide sequence SLEDLIFESLPENASHKLEVR, among the top 10 peaks with the highest contribution scores, seven were b ions, while the remaining three corresponded to intermediate fragment ions FE ((b8−c6)+), LIFES ((b9−b4)+), and PEN ((x11−x8)+), respectively. These results demonstrate that our model learned a few informative peaks from the spectra, which are useful for peptide inference.
To analyze which peak in the spectrum led to the erroneous generation of the model, we visualized the spectrum by highlighting b-y ion peaks corresponding to the model’s predictions. As shown in Fig. 4f, Casanovo V2’s predicted sequence predominantly aligns its y-ions with input spectrum peaks, with very few calculated b-ions aligning with input peaks. This behavior is a consequence of the autoregressive model’s prediction direction from right to left, making it more natural to choose y-ion peaks for forming predictions. However, given the presence of noise in the spectrum, this prediction approach can lead to errors when y-ions are inaccurately selected, as demonstrated in Fig. 4f. In contrast, PrimeNovo’s predictions exhibit an alignment with both b-ions and y-ions in the input spectrum. This is due to our model’s prediction process, which leverages information from both directions, allowing it to effectively utilize the peak information from both ends of the sequence. Furthermore, we conducted a detailed analysis to identify the specific peak responsible for prediction errors in the last layer. This is achieved by calculating a gradient-based contribution score for each input peak, serving as a robust indicator of which input has a greater impact on the output, determined by the magnitude of the gradient. As observed in the left corner of Fig. 4f, the highest contribution scores across the entire spectrum coincide precisely with the peak corresponding to PrimeNovo’s incorrectly predicted b-ion, and this critical information is captured and corrected by our PMC unit.
PrimeNovo demonstrates exceptional performance in taxon-resolved peptide annotation, enhancing metaproteomic research
We conducted an evaluation to gauge PrimeNovo’s proficiency in enhancing the identification of taxon-unique peptides, particularly in the context of metaproteomic research. The field of metaproteomics poses significant challenges when it comes to taxonomic annotation, primarily due to the vast diversity within microbiomes and the presence of closely related species that share high protein sequence similarity. Consequently, increasing the number of unique peptides represents a crucial approach for achieving precision in taxonomic annotations. In our assessment, we turned to a metaproteomic dataset33 obtained from gnotobiotic mice, hosting a consortium of 17 pre-defined bacterial strains (as summarized in Supplementary Table 2). Within this dataset, we applied PrimeNovo and Casanovo V2 to sequence unidentified MS/MS spectra through database search, all without the need for fine-tuning33. It’s worth noting that we are using Casanovo V2 without Beam Search (BS) due to the estimated inference time with BS exceeding 4000 A100 GPU hours on this large-scale dataset, which amounts to more than 21 days of inference with 8 A100 GPUs.
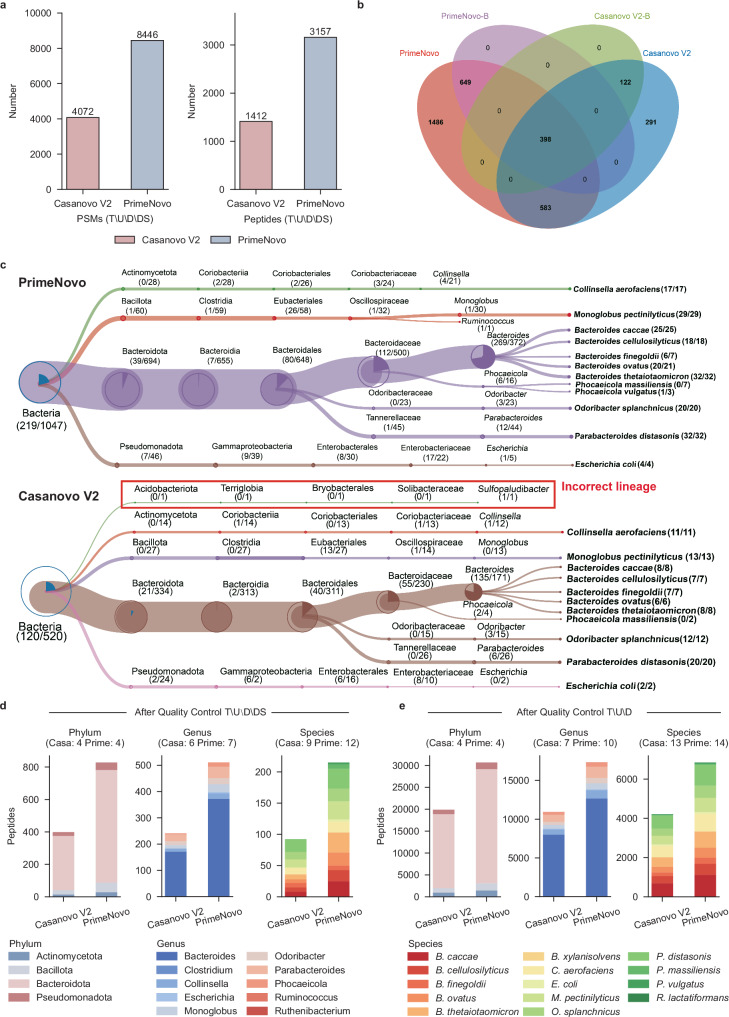
As illustrated in Fig. 5a, PrimeNovo exhibits superior performance compared to Casanovo V2, identifying a significantly higher number of PSMs (8446 vs. 4072) and peptides (3157 vs. 1412) following the rigorous quality control process T\U\D\DS, resulting in a relative increase of 107% and 124%, respectively. Furthermore, PrimeNovo excels in enhancing taxonomic resolution, outperforming Casanovo V2 in the detection of taxon-specific peptides. Notable increases are observed in bacterial-specific (1047 vs. 520), phylum-specific (828 vs. 399), genus-specific (511 vs. 241), and species-specific (215 vs. 92) peptides (Fig. 5b–d). Particularly noteworthy is the high identification accuracy achieved by PrimeNovo, where all identified peptides are correctly matched to known species, while Casanovo V2 exhibits one incorrect matching at the genus level (Fig. 5c).
Fig. 5. The advantages of PrimeNovo in metaproteomic analysis.
a Identification of PSMs and peptides through the quality control process T\U\D\DS, which involves the following steps: first, we identify sequences present in the target database. Then, we filter out results that are (1) unmatched with the precursor mass (mass error >0.1 Da); (2) found within the decoy database; (3) identified in database search results. Both the target and decoy databases were provided in the original study33. Additionally, the T\U\D approach is similar but does not entail a comparison with the database search results. b The Venn diagram illustrates the overlap between peptides identified by PrimeNovo and Casanovo V2, as well as the bacterial-specific peptides (PrimeNovo-B and Casanovo V2-B). c The treeview representation of species-level identification. d The number of peptides identified at the phylum, genus, and species levels, with the note that taxa identified by fewer than three unique peptides are excluded. e The number of peptides at the phylum, genus, and species levels after the quality control process T\U\D.
We further conducted an analysis of high-confidence identification results under the quality control process T\U\D. PrimeNovo demonstrated a significant increase in both PSM and peptide identifications, with a 66% increase (513,590 vs. 308,499) in PSMs and a 46% increase (58,392 vs. 39,866) in peptides. This result is further emphasized by the higher identifications of taxon-unique peptides achieved by PrimeNovo, surpassing Casanovo V2 in several categories, including bacterial-specific (36,704 vs. 24,349), phylum-specific (30,652 vs. 19,866), genus-specific (17,332 vs. 10,906), and species-specific (6848 vs. 4209) peptides (Fig. 5e). Subsequently, we assessed the models’ performance in taxonomic annotation at the protein level, which is crucial for enhancing the taxonomic resolution and contributing to subsequent research in the taxon-function network. As depicted in Supplementary Fig. 23, proteins identified by PrimeNovo and Casanovo were correctly assigned to 10 genera, 14 species, and 20 COG (Clusters of Orthologous Groups of proteins) categories. On the genus level, PrimeNovo identified a total of 6,883 proteins assigned to the 10 genera, with 6709 of them annotated to specific COG functions. In contrast, Casanovo V2 identified only 5028 proteins, with 4896 of them annotated. Thus, PrimeNovo achieved a 36.89% and 37.03% increase over Casanovo V2 in taxon and functional annotations.
Furthermore, a detailed examination at the genus level revealed that PrimeNovo increased the number of proteins assigned to each genus compared to Casanovo V2: Bacteroides (4926 vs. 3623), Clostridium (3 vs. 2), Collinsella (486 vs. 383), Escherichia (91 vs. 62), Monoglobus (294 vs. 197), Odoribacter (297 vs. 204), Parabacteroides (576 vs. 425), Phocaeicola (204 vs 130), Ruminococcus (3 vs. 1), Ruthenibacterium (3 vs. 1). Similarly, PrimeNovo exhibited significant potential for taxonomic annotation at the species level. Compared to Casanovo V2, PrimeNovo identified an additional 45.32% (3136 vs. 2158) of proteins assigned to the 14 species, with 45.03% (3034 vs. 2092) of these proteins annotated to specific COG functions. These results demonstrate that PrimeNovo significantly enhances taxonomic resolution at both the peptide and protein levels, highlighting its substantial potential in metaproteomic research.
PrimeNovo enables accurate prediction of a wide range of different post-translation modifications
PTMs play a crucial role in expanding the functional diversity of the proteome34, going well beyond the inherent capabilities of the genetic code. The primary challenge lies in the underrepresentation of modified peptides within the dataset, especially those that have not been enriched for certain modifications. The detection of such peptides is often overshadowed by the more prevalent unmodified peptides. Moreover, the distinct physical properties of modified residues—namely their mass and ionization efficiency—further complicate the detection35–39. The capabilities of current database search engines are limited, permitting the consideration of only a select few modifications. This scarcity leads to a low presence of modified peptides in the training data, thereby making it difficult for models to accurately identify diverse PTMs from spectral data.
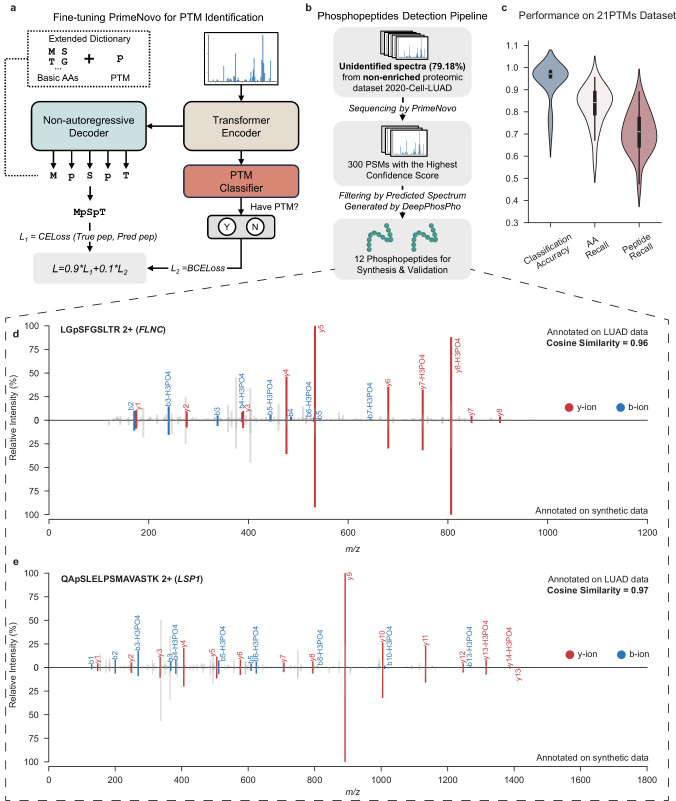
To address these challenges, PrimeNovo has been advanced in predicting peptide sequences with multiple PTMs, establishing itself as a foundational model divergent from conventional methods that start anew for each PTM type. By fine-tuning enriched PTM data, PrimeNovo gains extensive exposure to multiple PTM types while retaining its ability to recognize standard peptides. Architectural adjustments, as illustrated in Fig. 6a, including the addition of a classification head above the encoder to identify specific PTMs and a newly initialized linear layer above the decoder, enhance PrimeNovo’s ability to decode peptides with PTMs, broadening the model’s token repertoire. The final loss is formulated in a multi-task setting, combining the peptide decoding loss with a binary classification task for PTM identification loss.
Fig. 6. De novo sequencing of peptides with PTMs.
a A fine-tuning pipeline for PrimeNovo’s PTM prediction. b The methodology for selecting high-quality phosphopeptides predicted by PrimeNovo. c Performance metrics on the 21PTMs dataset (n = 21), including classification accuracy, amino acid-level recall, and peptide-level recall. d and (e) A comparative analysis of the actual input spectrum and the spectrum of the synthesized peptide predicted by PrimeNovo. The diagrams' upper sections display the original input spectrum, whereas the lower sections illustrate the spectrum generated from the predicted peptide sequence. Overlapping peaks are highlighted in red and blue for b-y ions. The cosine similarity is calculated based on spectrum encoding using the GLEAMS package. Source data are provided as a Source Data file. Some figures were created in BioRender56.
Our training methodology employed a dataset encompassing 21 distinct PTMs, referred to as the 21PTMs dataset, as detailed in ref. 40. We fine-tuned PrimeNovo for each PTM to ascertain its proficiency in peptide generation and PTM classification, in accordance with previously described methods. To ensure dataset balance, we included an approximately equal number of peptides with and without PTMs, culminating in a total of 703,606 PSMs for the dataset. The comprehensive fine-tuning endeavor across the 21 PTMs allows PrimeNovo to discern a broad spectrum of PTMs, a capability evidenced by the exemplary performance metrics for each PTM category depicted in Fig. 6c. Specifically, the classification accuracies for all PTMs exceeded 95%, except asymmetric and symmetric Dimethylation at Arginine (R), and Monomethylation at Arginine (R), which have classification accuracies of 77%, 77%, and 69%, respectively. Excluding Monomethylation at Arginine (R), which recorded a peptide recall rate of 48%, the de novo sequencing recall for peptides with the other 20 PTMs exceeded 61%. Such peptide recall levels are on par with performance in other datasets without special PTMs, such as an average peptide recall of 64% across nine-species datasets. Detailed insights into the classification accuracy and peptide recall for each PTM are provided in the supplementary Fig. 20.
To assess PrimeNovo’s inference performance on PTMs within a more applied context, we selected a phosphorylation dataset from Xu et al.41 (denote as the 2020-Cell-LUAD dataset), which concentrates on Human Lung Adenocarcinoma with 103 LUAD tumors and their corresponding non-cancerous adjacent tissues. It offers both phosphorylation-enriched and non-enriched data. We randomly selected a portion (3389 PSMs) of the enriched data for testing and the rest for training, checking of no overlapping peptide sequence between the training and testing sets. We fine-tuned PrimeNovo on such training data and the test results demonstrate that PrimeNovo distinguishes between phosphorylated and non-phosphorylated spectra with a classification accuracy of 98% and achieves a peptide recall rate of 66% on both cancer tissue data and non-cancerous adjacent tissues test data, as detailed in Supplementary Table 9.
To assess PrimeNovo’s capability to identify modified peptides within non-enriched proteomic datasets, we deployed it for the analysis of unidentified MS/MS spectra from the non-enriched 2020-Cell-LUAD dataset, notably without conducting dataset-specific fine-tuning. Given the absence of peptide identifications from existing databases in this dataset, we relied on the model’s confidence scores to select 300 high-quality predicted peptides. We then undertook a comparative analysis between the theoretical spectrum, as generated by DeepPhosPho42, and the original input spectrum corresponding to these peptides, as illustrated in Fig.6b. Through this process, we pinpointed 12 peptides as candidates for synthesis validation and further functional investigation. The details of the selection methodology are elaborated upon in Supplementary Note 8.
All 12 phosphopeptides predicted by PrimeNovo from non-enriched data were validated using their synthetic counterparts, as depicted in Fig. 6 and Supplementary Figs. 21 and 22. In Fig. 6d, e, they showcase the alignment between theoretical and experimental spectra for two representatives of 12 synthesized phosphorylated peptides. The comparison reveals a strong correspondence between the predicted b-ions and y-ions peaks and the experimental spectrum’s signal peaks, evidenced by a Pearson correlation exceeding 0.90 for nine paired spectra, and 0.70, 0.72, and 0.86 for the remaining three pairs. This correlation underscores the model’s high predictive precision. Further investigation into the proteins associated with these phosphopeptides highlighted their relevance to lung adenocarcinoma (LUAD). For example, the peptide LGpSGFSLTR (2+) (Fig. 6d) from Filamin-C (FLNC) aligns with findings that the ITPKA and Filamin C interaction fosters a dense F-actin network, enhancing LUAD cell migration43. Another identified peptide, HGpSDPAFAPGPR (2+) from FAM83H (Fig. 6e), is noted for being upregulated in LUAD, indicating a potential prognostic marker of LUAD44,45. Additionally, peptides WLDEpSDAEMELR, GPAGEAGApSPPVR, and AQpTPPGPSLSGSK reveal proteins (HACD3, SNTB2, and SRRM2) not previously associated with LUAD, but there are studies suggesting potential relevance between these three proteins and other cancer types. This offers directions for potential biological research on the disease by examining the above-relevant proteins. For detailed results concerning the remaining peptides and the comprehensive experimental methodologies used for their synthesis and analysis, please see Supplementary Note 8.
These results demonstrate that PrimeNovo has a high sensitivity in detecting PTMs from proteomic datasets, especially those non-enriched ones, which provides a solution for low-abundance PTM discovery.
Peptide sequencing is vital for understanding protein structures and functions. This work introduces PrimeNovo, a Transformer-based model for fast, accurate de novo peptide sequencing. Using a non-autoregressive architecture46 and a precise PMC decoding unit, PrimeNovo achieves state-of-the-art performance across spectrum datasets. Its speed and adaptability make it ideal for large-scale sequencing, with robust performance in zero-shot and fine-tuning scenarios. PrimeNovo excels in metaproteomic peptide annotation, aiding microorganism identification and functional analysis, while its PTM detection capability after finetuning enables the discovery of peptides beyond traditional methods.
Methods
Training datasets
The dataset used for training our model is the MassIVE Knowledge Base spectral library version 1 (MassIVE-KB)25, which we obtained from the MassIVE repository. This extensive dataset comprises over 2.1 million precursors originating from 19,610 proteins. These precursors were distilled from a vast pool of human data, amounting to more than 31 terabytes, gathered from 227 public proteomics datasets within the MassIVE repository.
Overview and notation
In the de novo sequencing task, we are provided with a spectrum instance denoted as S = {I, c, m}, which is generated by a mass spectrometer when analyzing biological samples. Here, I = {(m/z1, i1), (m/z2, i2), ⋯ , (m/zk, ik)} represents a set of mass-to-charge ratio and corresponding intensity pairs. These pairs are retained after being filtered by the mass spectrometer threshold. Additionally, c denotes the measured charge of the peptide (precursor), and m represents the measured total mass of this peptide. Our primary objective in this context is to derive the correct amino acid sequence denoted as A = {a1, a2, ⋯ , an} from the information contained within S.
Non-autoregressive transformer backbone
We adopt the transformer encoder–decoder network as our foundational model, following the work of Casanovo16. In the encoder network, we handle the mass-to-charge ratio m/z and the intensity information i from set I separately before merging them. To represent each m/z value, we employ a sinusoidal embedding function, which effectively captures the relative magnitude—an essential factor in determining the peptide fragments:
Here, j signifies the position in the d-dimensional hidden embedding. The parameters and define the wavelength range for this embedding. In contrast, we handle intensity values through a linear projection layer.
In the non-autoregressive model, the only architectural distinction between the encoder and decoder lies in the cross-attention mechanism. Therefore, we employ identical notations for both components. In a formal sense, each layer computes a representation R, based on the preceding feature embeddings. For the kth layer, the representation is
| 1 |
Here, R(0) signifies the spectrum embedding for the encoder, while for the decoder, it represents the summation of positional and precursor embeddings. To maintain consistency, we keep the generation length fixed as t for the decoder. Consequently, the output of the final decoder layer undergoes a softmax operation, which calculates the probability distribution over tokens for each position.
Peptide reduction strategy for our non-autoregressive modeling
Our strategy for non-autoregressive modeling deviates from conventional autoregressive generation, which predicts each token’s probability as P(a(i+1)∣a1). This approach, however, restricts bidirectional information, contrasting with protein structures where each amino acid is informed by both neighbors. To address this, we propose a non-autoregressive model where all amino acids are generated simultaneously, allowing each position to access bidirectional context. In this framework, each amino acid probability, P(a), is independently modeled, but this independence can lead to weak global coherence, resulting in nonsensical sequences despite locally accurate regions. For instance, a phrase like “au revoir" might ambiguously split into “see bye" in non-autoregressive translation with cross-entropy loss due to a lack of sequence-level cohesion. To mitigate this, we employ CTC loss47, which improves global consistency by enhancing sequence-level coherence, leading to more accurate and cohesive peptide generation.
To address cases where the generated token sequence, with a maximum length t, exceeds the target length, we introduce a reduction function, Γ( ⋅ ), in non-autoregressive generation. This function merges consecutive identical amino acids, for example:
| 2 |
However, simple reduction is unsuitable for sequences with consecutive identical amino acids. Inspired by Graves et al.47, we use a blank token ϵ during generation. Identical amino acids separated by ϵ are not merged, and ϵ is later removed, resulting in
| 3 |
For a visual representation of this process, please refer to the Supplementary Fig. 1.
Definition of CTC loss
Following the CTC reduction rule described above, it’s possible to obtain multiple decoding paths denoted as y, which can all be reduced to the target sequence A. For instance, both CCGT and CGϵT, among many others, can be transformed into the target sequence CGT. Consequently, the probability of generating the target sequence A is the sum of the probabilities associated with all paths y that can be reduced to A:
| 4 |
Here, y = (y1, y2, ⋯ , yt) represents a single decoding path in the non-autoregressive model output, satisfying the condition Γ(y) = A. The overall probability of generating the target sequence A, denoted as P(A∣S), is then computed as the sum of the probabilities of generating each y, with yi at each position. Since the probability is modeled independently, the probability of each y can be calculated as the multiplication of the probabilities of generating all yi ∈ y. This multiplication can be expressed as the sum of the logarithm of the probabilities of each yi.
During the training process, our objective is to maximize the total probability of generating the target sequence A for each input spectrum S. Since we are utilizing gradient descent to optimize our model, this goal is equivalent to minimizing the negative total probability. Therefore, our loss function is simply defined as:
| 5 |
One could theoretically enumerate all possible paths y for each target sequence A in order to calculate the total probability (loss) for training our network. However, this approach becomes impractical as the number of paths grows exponentially with respect to the maximum generation length. This would result in an unmanageable amount of computation time. Instead, we adopt a dynamic programming method, as detailed in the Supplementary Information, to optimize the calculation of this loss efficiently. This approach allows us to train our model effectively without the computational burden of exhaustively enumerating all possible paths.
Knapsack-like dynamic programming decoding algorithm for precise mass control
The generated de novo peptide sequence should be strictly grounded by molecular mass measured by the mass spectrometer. Specifically, the molecular mass of the ground truth peptide, mtr falls in the range of [m−σ, m + σ], where m is precursor mass given by mass spectrometer, and σ is measurement error, usually at 10−3 level, of used mass spectrometer. However, neural network models are of low explainability and controllability, making it difficult to control the generated results to cater to certain desires. To allow accurate generation, we reformulate the non-auto regressive generation as a knapsack-like optimization problem48, where we are picking items (amino acids) to fill the bag with a certain weight constraint, while the value (predicted log probability) is maximized. Such optimization problem can be formulated as:
| 6 |
where and are the desired lower bound and upper bound for decoded peptide mass. We denote and where tol is decoding tolerance within which we think the true mass mtr falls in, after taking into measurement error.
Inspired by a similar idea by Liu et al.48, we propose a dynamic programming method to solve such an optimization task. We denote e as the decoding precision to construct a two-dimensional DP table. For each time step, we would have cells with being the ceiling function. The lth cell can only store the peptide with mass precisely within [e*(l−1), e*l]. Specifically, the lth cell at τth time step dτ,l stores the most probable, calculated by the sum of log probability by non-autoregressive model, τ tokens sequence y1:τ satisfying the mass constraint of .
We first initialize our DP table by filling the first time step, τ = 1, as follows:
| 7 |
In the first case, d1,1 stores the one-token sequence with the total mass in the range of [0, e], where e is usually a very small number (e < 1) for higher decoding accuracy, therefore no amino acid other than ϵ can fall under this mass limit. On the other hand, when l ≠ 1, there might be multiple amino acids whose mass falls within . We store all of them in lth cell to avoid overlooking of any possible starting amino acid.
We then divide the recursion steps into three cases, , each storing its corresponding set of sequences following the rules below:
- When yτ = ϵ, we know Γ(y1:τ−1) = Γ(y1:τ) due to CTC reduction, therefore the mass stays the same. This gives the set of candidate sequences :
where ⊕ is the concatenation.8 - When the newly decoded non-ϵ token is the repetition of the last token, the reduced sequence still remains the same with the mass unchanged, due to the CTC rule. We get the second set of potential sequences:
9 - When the newly decoded non-ϵ token is different from the last token in the already generated sequence, the mass will be increased. We select the potential sequence by examining the total mass that falls in the mass constraint:
The we update the cell dτ,l using all candidates from the above three sets:10
where topB is taking the top B most probable sequences according to generated probability. We then select the most probable sequence at dt,∣A∣ cell as our final result.11
CUDA acceleration for proposed mass control decoding algorithm
The time complexity of our proposed mass control dynamic programming algorithm, when executed sequentially, is O(Nat(U/e)2), where Na represents the total number of tokens (which, in our case, corresponds to the number of amino acids plus one).
To implement the parallel algorithm for the PMC unit, we employ the compute unified device architecture (CUDA). CUDA is a parallel computing programming framework developed by NVIDIA, which allows programs to leverage the computational power of NVIDIA graphics processing units (GPUs) for a wide range of general-purpose computing tasks. Detailed information regarding our CUDA algorithm is provided in the Supplementary Information.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This research was supported by grant from National Key Research and Development Program of China (2021YFA1301603) to C.C. and F.H., Shanghai Artificial Intelligence Laboratory and the Focus Project of AI for Science of Comprehensive Prosperity Plan for Disciplines of Fudan University to S.S., the National Natural Science Foundation of China (32088101) to C.C. and F.H., the Natural Sciences and Engineering Research Council of Canada (RGPIN-2020-05408) to L.V.S.L., and the Canada Research Chairs Program and the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2018-04267) to M.A.M. We also thank the authors of PepNet19 in providing their processed training and testing data to help complete our additional experiments during reviewing stage.
Author contributions
S.S. and C.C. co-supervised and conceived the study. T.L., Z.G., and N.D. collected and preprocessed all the datasets for training and testing. S.X., Z.J., and X.Z. replicated the baseline models. X.Z. proposed and implemented the non-autoregressive model pipeline and the precise mass control unit. S.S., L.V.S.L., and M.A.-M. provide guidance on algorithm designs. Z.J. implemented the CUDA code for the precise mass control unit. X.Z. and Z.J. ran the training and inference for de novo sequencing on all datasets. Z.Q., Z.G., Z.X., and Z.J. conducted the interpretation analysis. T.L. performed the phosphopeptides identification analysis and drafted this section. G.W. performed the LC–MS/MS experiment analysis of the synthetic phosphopeptides. T.L., B.S., and L.L. performed the metaproteomic data analysis and drafted this section. S.X. and T.L. conducted data analysis and visualization. X.Z. drafted the manuscript, and S.S., C.C., W.O., T.L., S.X., Z.G., N.D., L.V.S.L., and M.A.-M. edited it. J.W. ran and analyzed the speed comparison of all approaches. W.O., L.V.S.L., M.A.-M., and F.H. coordinated the study and provided suggestions for the study. All authors read and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks Lei Xin, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The nine-species benchmark dataset12 was directly downloaded as Mascot Generic Format (MGF) files from the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository (identifier: MSV000081382), shared by the authors of the DeepNovo paper. The dataset was searched using PEAKS DB5 software [version 8.0] with a false discovery rate (FDR) of 1%. The MassIVE-KB dataset25 was obtained by downloading the raw files and the filtered identification results from the All Candidate Library Spectra section of the MassIVE Knowledge Base spectral library v1 (https://massive.ucsd.edu/ProteoSAFe/static/massive-kb-libraries.jsp). The PT27, 21PTMs40, and PXD01948349 datasets were obtained by downloading the raw files and MaxQuant2 identification results from the PRIDE50 repository by PXD00473227, PXD00944940, and PXD01948349, respectively. The HCC28 and 2020-Cell-LUAD41 datasets were obtained by downloading the raw files and MaxQuant identification results from the iProX51 repository (identifier: IPX0000937000 and IPX0001804000, respectively). The IgG1-Human-HC26 dataset was obtained by downloading the combined identification results of the database algorithms MS-GF+52 and X!Tandem53 with an FDR rate of 1% from the MassIVE repository (identifier: MSV000079801). The three-species dataset22 was obtained by downloading the SEQUEST3 search results with a 1% false positive rate for these three species datasets from the data shared by the GraphNovo authors on Zenodo (identifier: zenodo.8000316). The revised nine-species benchmark dataset16 was obtained by downloading the raw files and Crux54 identification results from the MassIVE repository (identifier: MSV000090982). The cell-metaproteome dataset33 was obtained by downloading the raw files and MyriMatch55 identification results from the MassIVE repository (identifier: MSV000082287). Source data are provided with this paper.
Code availability
We have open-sourced the codebase and trained model weights for π-PrimeNovo on GitHub: https://github.com/PHOENIXcenter/pi-PrimeNovo and https://github.com/BEAM-Labs/pi-PrimeNovo. Future updates and new releases will also be available at this link.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiang Zhang, Tianze Ling, Zhi Jin, Sheng Xu.
Contributor Information
Wanli Ouyang, Email: ouyangwanli@pjlab.org.cn.
Cheng Chang, Email: changcheng@ncpsb.org.cn.
Siqi Sun, Email: siqisun@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55021-3.
References
- 1.Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature422, 198–207 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p. Nat. Biotechnol.26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom.5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis20, 3551–3567 (1999). [DOI] [PubMed]
- 5.Zhang, J. et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteom.11, M111.010587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi, H. et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol.36, 1059–1061 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Beslic, D., Tscheuschner, G., Renard, B. Y., Weller, M. G. & Muth, T. Comprehensive evaluation of peptide de novo sequencing tools for monoclonal antibody assembly. Brief. Bioinform.5, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karunratanakul, K., Tang, H.-Y., Speicher, D. W., Chuangsuwanich, E. & Sriswasdi, S. Uncovering thousands of new peptides with sequence-mask-search hybrid de novo peptide sequencing framework. Mol. Cell. Proteom.18, 2478–2491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hettich, R. L., Pan, C., Chourey, K. & Giannone, R. J. Metaproteomics: Harnessing the power of high-performance mass spectrometry to identify the suite of proteins that control metabolic activities in microbial communities. Anal. Chem.85, 4203–4214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma, B. et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom.17, 2337–2342 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Frank, A. & Pevzner, P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem.77, 964–973 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Tran, N.H., Zhang, X., Xin, L., Shan, B. & Li, M. De novo peptide sequencing by deep learning. Proc. Natl Acad. Sci. USA114, 8247–8252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao, R. et al. Computationally instrument-resolution-independent de novo peptide sequencing for high-resolution devices. Nat. Mach. Intell.3, 420–425 (2021). [Google Scholar]
- 14.Yang, H., Chi, H., Zeng, W.-F., Zhou, W.-J. & He, Si-Min pNovo 3: Precise de novo peptide sequencing using a learning-to-rank framework. Bioinformatics35, i183–i190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz, M., Fondrie, W., Bittremieux, W., Oh, S. & Noble, W. S. De novo mass spectrometry peptide sequencing with a transformer model. In Proc. 39th International Conference on Machine Learning 25514–25522 (ICML, 2022).
- 16.Yilmaz, M. et al. Sequence-to-sequence translation from mass spectra to peptides with a transformer model. Nat. Commun.15, 6427 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, T. et al. Introducing π-helixnovo for practical large-scale de novo peptide sequencing. Brief. Bioinform.25, bbae021 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, Z. et al. ContraNovo: a contrastive learning approach to enhance de novo peptide sequencing. In AAAI Conference on Artificial Intelligence 144–152 (AAAI, 2024).
- 19.Liu, K., Ye, Y., Li, S. & Tang, H. Accurate de novo peptide sequencing using fully convolutional neural networks. Nat. Commun.14, 7974 (2023). [DOI] [PMC free article] [PubMed]
- 20.Klaproth-Andrade, D. et al. Deep learning-driven fragment ion series classification enables highly precise and sensitive de novo peptide sequencing. Nat. Commun.15, 151 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eloff, K. et al. De novo peptide sequencing with instanovo: accurate, database-free peptide identification for large scale proteomics experiments. bioRxiv, 10.1101/2023.08.30.555055 (2023).
- 22.Mao, Z., Zhang, R., Xin, L. & Li, M. Mitigating the missing-fragmentation problem in de novo peptide sequencing with a two-stage graph-based deep learning model. Nat. Mach. Intell.5, 1250–1260 (2023). [Google Scholar]
- 23.Brown, T. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst.33, 1877–1901 (2020). [Google Scholar]
- 24.Arora, K., Asri, Layla El. Bahuleyan, H. & Cheung, J. Why exposure bias matters: an imitation learning perspective of error accumulation in language generation. In Findings of the Association for Computational Linguistics: ACL 2022, Dublin, Ireland, 2022, (eds Muresan, S. Nakov, P. & Villavicencio, A.) 700–710 (Association for Computational Linguistics, 2022).
- 25.Wang, M. et al. Assembling the community-scale discoverable human proteome. Cell Syst.7, 412-421.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran, N.H. et al. Complete de novo assembly of monoclonal antibody sequences. Sci. Rep.6, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zolg, D. P. et al. Building ProteomeTools based on a complete synthetic human proteome. Nat. Methods14, 259–262 (2017). [DOI] [PMC free article] [PubMed]
- 28.Jiang, Y. et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature567, 257–261 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Zhang, A. et al. On orthogonality constraints for transformers. In Proc. 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing 375–382 (Association for Computational Linguistics, 2021).
- 30.Xie, D., Xiong, J. & Pu, S. All you need is beyond a good init: exploring better solution for training extremely deep convolutional neural networks with orthonormality and modulation. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 6176–6185 (IEEE, 2017).
- 31.Wang, J., Chen, Y., Chakraborty, R. & Yu, S. X. Orthogonal convolutional neural networks. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 11505–11515 (IEEE, 2020).
- 32.Simonyan, K., Vedaldi, A. & Zisserman, A. Deep inside convolutional networks: visualising image classification models and saliency maps. In Workshop at International Conference on Learning Representations (ICLR, 2014).
- 33.Patnode, M. L. et al. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell179, 59–73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barber, K. W. & Rinehart, J. The abcs of PTMS. Nat. Chem. Biol.14, 188–192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carr, S. et al. The need for guidelines in publication of peptide and protein identification data: working group on publication guidelines for peptide and protein identification data. Mol. Cell. Proteom.3, 531–533 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Andersen, J. S. & Mann, M. Organellar proteomics: turning inventories into insights. EMBO Rep.7, 874–879 (2006). [DOI] [PMC free article] [PubMed]
- 37.Wilkins, M. R. et al. Guidelines for the next 10 years of proteomics. Proteomics6, 4–8 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Shen, Y. et al. Proteome-wide identification of proteins and their modifications with decreased ambiguities and improved false discovery rates using unique sequence tags. Anal. Chem.80, 1871–1882 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duncan, M. W., Aebersold, R. & Caprioli, R. M. The pros and cons of peptide-centric proteomics. Nat. Biotechnol.28, 659–664 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Paul, D. et al. ProteomeTools: systematic characterization of 21 post-translational protein modifications by liquid chromatography tandem mass spectrometry (lc-ms/ms) using synthetic peptides. Mol. Cell. Proteom.17, 1850–1863 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, J. Y. et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell182, 245–261.e17 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Lou, R. et al. DeepPhospho accelerates DIA phosphoproteome profiling through in silico library generation. Nat. Commun.12, 1–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Windhorst, S. et al. Inositol 1, 4, 5-trisphosphate 3-kinase-a is a new cell motility-promoting protein that increases the metastatic potential of tumor cells by two functional activities. J. Biol. Chem.285, 5541–5554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, J.-Y. et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell182, 245–261 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Uhlén, M. et al. Tissue-based map of the human proteome. Science347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Gu, J., Bradbury, J., Xiong, C., Li, V. O. K. & Socher, R. Non-autoregressive neural machine translation. In International Conference on Learning Representations (ICLR, 2018).
- 47.Graves, A., Fernández, S., Gomez, F. & Schmidhuber, J. Connectionist temporal classification: labelling unsegmented sequence data with recurrent neural networks. In Proc. 23rd International Conference on Machine Learning 369–376 (ICML, 2006).
- 48.Liu, P., Zhang, X. & Mou, L. A character-level length-control algorithm for non-autoregressive sentence summarization. Adv. Neural Inf. Process. Syst.35, 29101–29112 (2022). [Google Scholar]
- 49.Müller, J. B. et al. The proteome landscape of the kingdoms of life. Nature582, 592–596 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Vizcaíno, J. A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res.44, D447–D456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res.47, D1211–D1217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim, S. & Pevzner, P. A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun.5, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langella, O. et al. X!TandemPipeline: a tool to manage sequence redundancy for protein inference and phosphosite identification. J. Proteome Res.16, 494–503 (2017). [DOI] [PubMed] [Google Scholar]
- 54.McIlwain, S. et al. Crux: rapid open source protein tandem mass spectrometry analysis. J. Proteome Res.13, 4488–4491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabb, D. L., Fernando, C. G. & Chambers, M. C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res.6, 654–661 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beam, L. Biorender 2024, https://www.biorender.com (accessed 5 November 2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nine-species benchmark dataset12 was directly downloaded as Mascot Generic Format (MGF) files from the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository (identifier: MSV000081382), shared by the authors of the DeepNovo paper. The dataset was searched using PEAKS DB5 software [version 8.0] with a false discovery rate (FDR) of 1%. The MassIVE-KB dataset25 was obtained by downloading the raw files and the filtered identification results from the All Candidate Library Spectra section of the MassIVE Knowledge Base spectral library v1 (https://massive.ucsd.edu/ProteoSAFe/static/massive-kb-libraries.jsp). The PT27, 21PTMs40, and PXD01948349 datasets were obtained by downloading the raw files and MaxQuant2 identification results from the PRIDE50 repository by PXD00473227, PXD00944940, and PXD01948349, respectively. The HCC28 and 2020-Cell-LUAD41 datasets were obtained by downloading the raw files and MaxQuant identification results from the iProX51 repository (identifier: IPX0000937000 and IPX0001804000, respectively). The IgG1-Human-HC26 dataset was obtained by downloading the combined identification results of the database algorithms MS-GF+52 and X!Tandem53 with an FDR rate of 1% from the MassIVE repository (identifier: MSV000079801). The three-species dataset22 was obtained by downloading the SEQUEST3 search results with a 1% false positive rate for these three species datasets from the data shared by the GraphNovo authors on Zenodo (identifier: zenodo.8000316). The revised nine-species benchmark dataset16 was obtained by downloading the raw files and Crux54 identification results from the MassIVE repository (identifier: MSV000090982). The cell-metaproteome dataset33 was obtained by downloading the raw files and MyriMatch55 identification results from the MassIVE repository (identifier: MSV000082287). Source data are provided with this paper.
We have open-sourced the codebase and trained model weights for π-PrimeNovo on GitHub: https://github.com/PHOENIXcenter/pi-PrimeNovo and https://github.com/BEAM-Labs/pi-PrimeNovo. Future updates and new releases will also be available at this link.