Abstract
The tomato leaf miner (TLM), Phthorimaea absoluta Meyrick, 1917 (Lepidoptera: Gelechiidae) is a destructive invasive insect that has expanded its global distribution. Rapid and accurate identification of invasive pests is essential to support subsequent management and devise control measures. To accurately diagnose P. absoluta, a Loop Mediated Isothermal Amplification (LAMP) assay (TLM-LAMP) was developed to amplify the target region of mitochondrial cytochrome oxidase subunit I (COI) gene. The TLM-LAMP assay can identify the P. absoluta within 60 min at 65 °C after sample extraction. Cross-reactivity analysis against three closely related non-target species, Phthorimaea operculella (Zeller, 1873), Pectinophora gossypiella (Saunders, 1844), and Aproaerema modicella (Deventer, 1904) confirmed species specificity. The TLM-LAMP assay showed high sensitivity to P. absoluta DNA up to 1 × 10− 8 ng/µL and in plasmid DNA template up to 1 × 10–14 ng/µL. In addition, the TLM-LAMP assay was successful in laboratory detection of larvae, pupa, and adult stages of P. absoluta. We have tested the TLM-LAMP assay for field application with quick and simple crude insect extraction procedures and found double distilled water (ddH2O) as an effective extraction solution. The new TLM-LAMP assay was validated in the field and polyhouse using moths collected from pheromone traps followed by ddH2O crude insect extract preparation and incubation. The assay could successfully detect the P. absoluta within 45 min at 65 °C. Sensitivity, specificity, repeatability, and field compatibility of the TLM-LAMP highlights the novelty of the developed method. TLM-LAMP assay is a novel molecular tool for detection of P. absoluta in the laboratory and field which will help in monitoring and aiding biosecurity responses.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84288-1.
Keywords: Phthorimaea absoluta, Cytochrome oxidase subunit 1, Detection, LAMP, Crude insect-extract
Subject terms: Entomology, Invasive species
Introduction
Accurate organism identification is imperative in the arena of biology for various reasons1–3. Consequently, the practice of morphology-centric identification techniques is constrained by its time-consuming nature and significant reliance on the knowledge of experienced taxonomists, thereby leading to a higher probability of errors4,5. Molecular identification methods provide more accurate and efficient inference. Polymerase chain reaction (PCR) coupled with Sanger sequencing is most common technique used for insect detection6–8. Also, advanced molecular detection methods have emerged, such as Multi-Locus Sequence Typing (MLST)9, real-time and digital droplet PCR10–12, high-throughput sequencing (HTS)13–16, single nucleotide polymorphism (SNP)17, Increased Plexing Efficiency and flexibility for the Mass ARRAY assay (iPLEXTM)17 and CRISPR assay18. However, these methods are time-consuming and require complex laboratory setups. Alternatively, Loop-mediated isothermal amplification (LAMP) diagnosis19 and Recombinase polymerase amplification diagnosis (RPA)20 are isothermal amplification methods becoming popular for quick detection of insect pests without the need for complex laboratory setup.
In contrast to other amplification methods, LAMP works in a temperature-stable setting, removing the necessity for a costly thermal cycler21. Moreover, this approach provides the benefit of visual detection and eliminates the necessity of gel electrophoresis22. LAMP assay can be applied for field-based diagnostic testing and is amenable to different rapid DNA extraction methods compared to conventional PCR23. LAMP is frequently employed for the detection of insects such as Mythimna loreyi24, Helicoverpa armigera19, Spodoptera frugiperda25,26, Spodoptera exigua26, Zeugodacus scutellatus27, Bactrocera tryoni28, Bactrocera trivialis29, Thecodiplosis japonensis30, Bactrocera tsuneonis31, Aethina tumida32, Agrilus planipennis33, Nilaparvata lugens34, Laodelphax striatellus34, Sogatella furcifera34 and Diaphorina citri35.
The invasive pest causes significant economic damage in both natural and man-made environments36,37. According to the International Union for Conservation of Nature and Natural Resources (IUCN), invasive insects are arthopods, that invade natural or semi-natural ecosystems or habitats and threaten native biodiversity38. The South American tomato leaf miner, Phthorimaea absoluta Meyrick, 1917 (TLM), which belongs to the lepidopteran family Gelechiidae, is a major invasive pest of tomato39. Since the 1950s, P. absoluta has become a major problem for tomato growers in South America, causing significant declines in productivity due to damage to fruits and leaves40. The $87.9 billion global tomato industry faces a threat from this insect, which directly consumes leaves, buds, calyxes, and fruits. The impact can range from 50 to 100 per cent destruction41. P. absoluta shows an average annual range expansion of 600 km42, that enhances its ability to invade new areas. In addition to the main host plant, tomato, P. absoluta also attacks several secondary hosts40,43,44. P. absoluta was accidentally introduced into Spain in 2006 and subsequently migrated to the Netherlands and east-west Iran40,45. In 2009, it invaded Turkey in Asia and started its invasion in other Asian countries like India46, Nepal47, China48, and other countries causing significant damage to tomato crops49.
To reduce the effect of invasive species, proper identification, subsequent implementation of monitoring, and integrated pest management programs is crucial. Detection of invasive insect pests is a challenge that must be done rapidly to avoid delays in management response. In ports of entry and surveys, morphological identification is a commonly used method. Depending on the stages of the pest, the morphological identification becomes complicated, time-consuming, and increases the chances of misidentifications, which can result in unnecessary control measures and even in the release of invasive species4.
In this study, we developed a LAMP diagnostic assay for the identification of P. absoluta (TLM-LAMP) and tested its species-specificity and limit of detection using DNA, plasmid, and crude extract. The assay was validated in the laboratory for detection of larvae, pupa, and adult stages of P. absoluta. The field deplorability of DNA extraction method and validated field applicability of the TLM-LAMP assay.
Results
Identification of specimens and primer optimization in standard PCR
All the specimens used in this study were documented using morphological characters46,50,51 (Supplementary Fig. 1) and DNA Brcoding (Accession Numbers: PQ451969, PP816326, PQ452113, PQ455498). Conserved sequence of the COI gene served as the basis for designing the primers proposed in this work. A high level of conservation was observed between the sequences from different parts of the world in the multiple alignments that include sequences (Supplementary Table 1 for NCBI accession number) (Supplementary Fig. 2) of the Phthorimaea absoluta, Phthorimaea operculella, Pectinophora gossypiella, and Aproaerema modicella COI gene. The relationship between the target and non-target species shown in Neighbour-Joining tree (Supplementary Fig. 3).
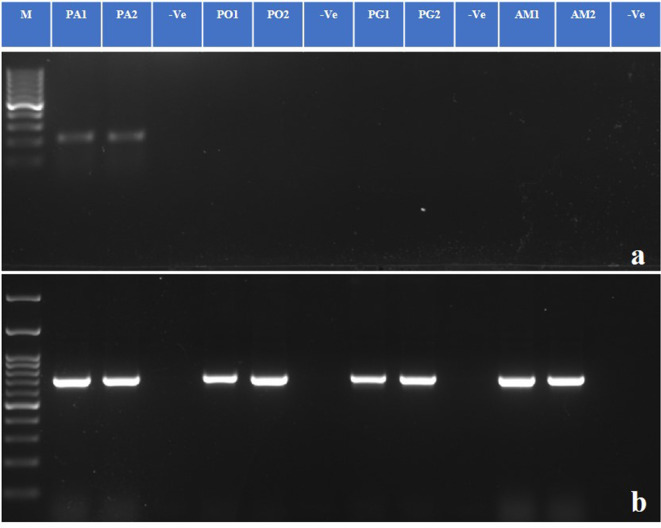
Two sets (P1L1 and P3) of LAMP primers (Supplementary Table 3) targeting 210 bp of the COI gene of P. absoluta were designed. These primer sets consist of two outer primers (F3 and B3) and two inner primers (FIP and BIP). Using outer primers, genomic DNA from 15 samples of P. absoluta was amplified in PCR. Out of these 15 samples, 14 samples were amplified by the primer set (P3), and all 15 samples were amplified by the primer set (P1L1) (Supplementary Fig. 4a, b). Further, the PCR with F3 and B3 of the primer pair (P1L1) yields a 210 bp amplicon for P. absoluta only (Fig. 1a) showing species specificity. These amplicons were sequenced to confirm the specificity. In contrast, universal COI PCR generates 700 bp amplicon from all positive samples (Fig. 1b). However, when we used the primer set (P3), we observed cross-amplification for P. operculella. Therefore, we proceeded further with the primer set (P1L1) in LAMP.
Fig. 1.
Cross reactivity test in PCR (a) primer set (P1L1) specific to Phthorimaea absoluta. (b) LCO1490 and HCO2198 primers as loading controls. M is 100 bp DNA ladder (GeneDireX). Moth species of this study were- PA- P. absoluta, PO- Phthorimaea operculella, PG- Pectinophora gossypiella, AM- Aproaerema modicella. (-Ve)- non-template control. Note: It is a clubbed figure and un-cropped figure of the same has been uploaded as Supplementary file.
Standardized TLM-LAMP
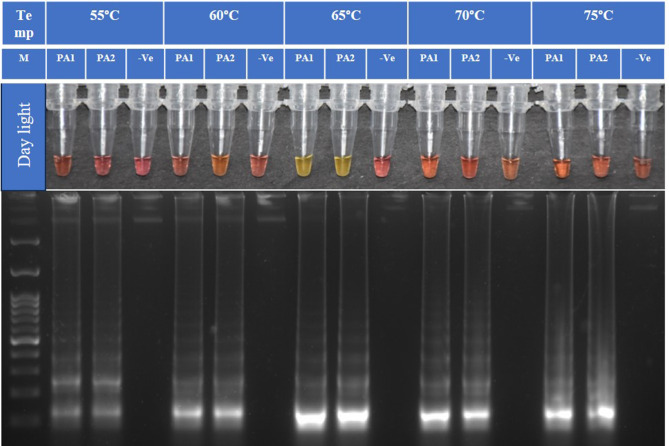
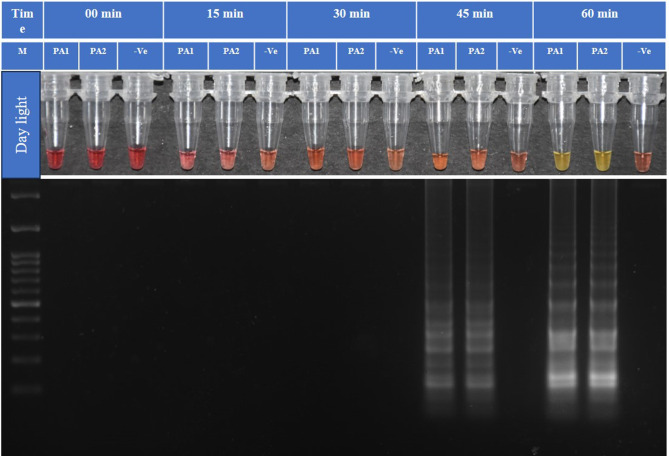
The primer ratio 1:12:6 (F3/B3: FIP: BIP) was found optimum with primer concentrations of 1 µM for F3 and B3, 12 µM for FIP, and 6 µM for BIP. The LAMP assay was standardized using two P. absoluta DNA samples and a non-template control. The LAMP assay was performed in a heating block (Biochem-Life Sciences) at 55 °C, 60 °C, 65 °C, 70 °C, 75 °C for an incubation time of 60 min. A visual change of the color at 65 °C for 60 min showed a transition from a pink negative response to a yellow positive response, as per the manufacturer (WarmStart). The final result was consistent with isothermal amplification analysis by electrophoresis in 2% agarose gel (Fig. 2) showing positive color change and a ladder-like DNA band, which is evidence of LAMP amplification52. When LAMP is executed with four different time intervals (15, 30, 45, 60 min), we found significant amplification exhibited by the color change and ladder-like band after 60-minute interval (Fig. 3).
Fig. 2.
Evaluation of amplification temperature of COI gene of P. absoluta (PA) with two DNA samples and one non-template control (-Ve) using loop-mediated isothermal amplification (LAMP) technique. Isothermal amplification analysis of the COI by Colorimetry and 2% gel electrophoresis. M is 100 bp ladder (GeneDireX). Incubation temperatures 55 °C, 60 °C,65 °C, 70 °C, and 75 °C. Optimum amplification temperature for TLM-LAMP is 65 °C. Note: It is a clubbed figure and un-cropped figure of the same has been uploaded as Supplementary file.
Fig. 3.
Colorimetric test analysis of the isothermal amplification of the COI gene of P. absoluta. LAMP reaction incubated for different times with two DNA samples of P. absoluta (PA) and one Non-template control (-Ve) at 65 °C. M is 100 bp ladder (GeneDireX). Optimum amplification time for TLM-LAMP is 60 min. Note: It is a clubbed figure and un-cropped figure of the same has been uploaded as Supplementary file.
Evaluation of TLM-LAMP with P. absoluta DNA
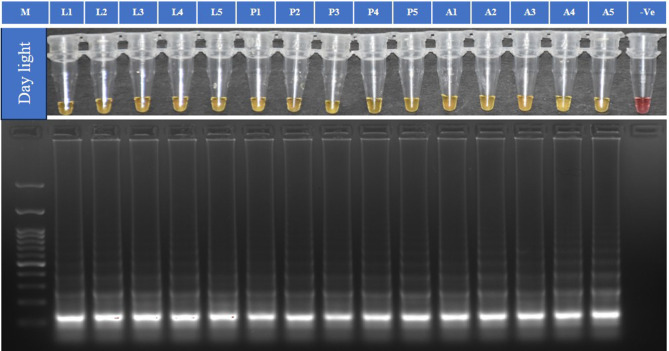
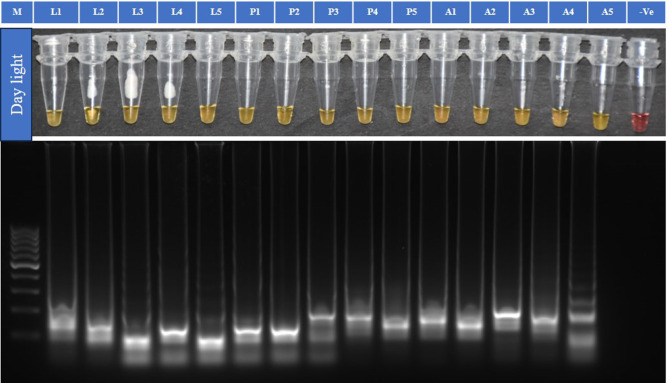
The TLM-LAMP assay with primer ratio 1:12:6 (F3/B3: FIP: BIP) was performed using 15 P. absoluta samples with genomic DNA from three different stages (five from each larval, pupal, and adult stage) at 65 °C for 60 min. The TLM-LAMP assay could identify P. absoluta accurately with results showing the characteristic amplification of the target in all positive samples analysed (Fig. 4).
Fig. 4.
LAMP assay validation with DNA template from different stages of P. absoluta. Isothermal amplification analysis, by Colorimetric test (WarmStart) and 2% gel electrophoresis. M is 100 bp ladder (GeneDireX). L 1–5 DNA from larval stage, P 1–5 DNA from pupal stage, A 1–5 DNA from adult stage of P. absoluta. (-Ve)- non-template control. Note: It is a clubbed figure and un-cropped figure of the same has been uploaded as Supplementary file.
Specificity and sensitivity of TLM-LAMP assay
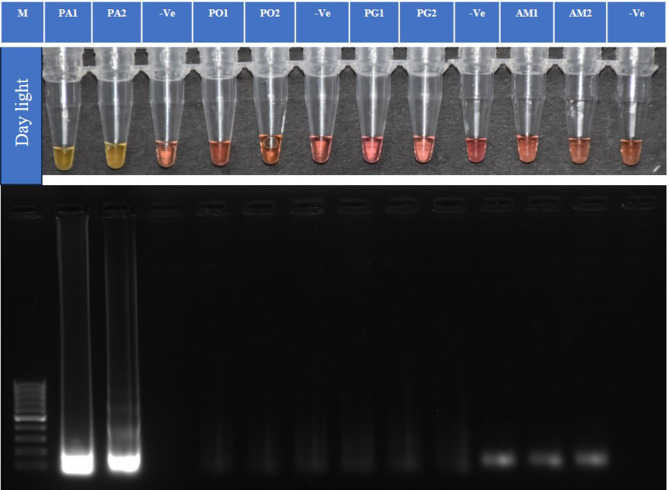
The specificity of the test showed that the primer pair (P1L1) was specific for P. absoluta, since no amplification or color changes were observed in the reactions with the DNA of the non-target insects P. operculella, P. gossypiella, and A. modicella (Fig. 5).
Fig. 5.
Specificity analysis of TLM-LAMP. Amplification for P. absoluta without any cross-amplification. M is 100 bp ladder (GeneDireX). (PA1, PA2)- P. absoluta, (PO1, PO2)- P. operculella, (PG1, PG2)- P. gossypiella, (AM1, AM2)- A. modicella. (-Ve)- Non-template control. Note: It is a clubbed figure and un-cropped figure of the same has been uploaded as Supplementary file.
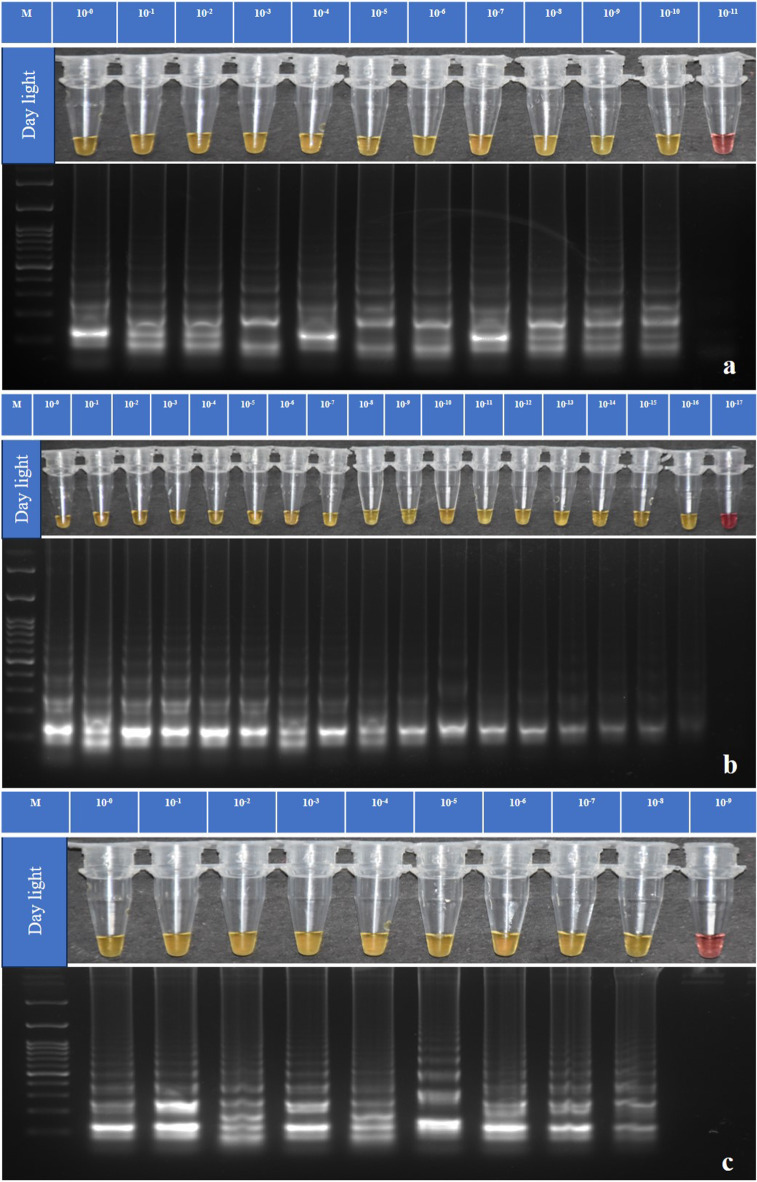
The sensitivity of TLM-LAMP assay was determined for target genomic DNA and plasmid DNA containing respective gene insert in twenty times serial dilution. We detected positive results visually by a gradual color change from pink to light yellow up to a detection limit of 10–10, i.e., 1 × 10− 8 ng/µL (Fig. 6a) and as low as 10–16, i.e., 1 × 10–14 ng/µL (Fig. 6b) with genomic DNA and plasmid DNA, respectively, indicating the high sensitivity of the test. The assay can identify the amplified product at similar dilutions as supported by the similar results obtained with electrophoresis and colorimetric data.
Fig. 6.
Sensitivity analysis of TLM-LAMP for detection of P. absoluta in Colorimetric test (WarmStart) and 2% agarose gel electrophoresis using serial dilution of templates. - (a) for genomic DNA sensitivity up to 10− 10, (b) for Plasmid DNA sensitivity up to 10− 16, and (c) for Crude extract sensitivity up to 10− 8. M is 100 bp (GeneDireX). Note: It is a clubbed figure and un-cropped figure of the same has been uploaded as Supplementary file.
TLM-LAMP assay for field application
LAMP assay with simplified template preparation (crude insect extract)
In the TLM-LAMP assay, we evaluated P. absoluta larvae for crude DNA extraction with four different lysis solutions i.e., 0.02 M EDTA20, double distilled water (ddH2O)20, NAOH: EDTA (1:1)53, and NAOH: EDTA (1:2)53 and the lysis solutions were used directly as template. LAMP assay with the primer set (P1L1) showed significant positive amplification with crude insect extract preapred in two lysis solutions, i.e. 0.02 M EDTA and ddH2O by incubation at 100 °C for 10 min followed by cooling down to room temperature. Based on the simplicity of template preparation using lysis solution, the distinct and specific color change, and the observation of a ladder-like DNA band in the gel (Supplementary Fig. 5), the ddH2O extraction was selected for field application of the test. To validate the ddH2O crude extraction (incubation at 100 °C for 10 min) with TLM-LAMP assay, five specimens from three different life stages (larva, pupa, and adult) i.e. 15 samples of P. absoluta were tested along with a non-template control and the target insect can be accurately identified in all three stages of the P. absoluta just in 45 min of incubation at 65 °C (Fig. 7).
Fig. 7.
LAMP assay for the detection of P. absoluta using crude extract in double distilled water as template. M is 100 bp ladder (GeneDireX). L 1–5 DNA from larval stage, P 1–5 DNA from pupal stage, A 1–5 DNA from adult stage of P. absoluta. (-Ve)- non-template control. Note: It is a clubbed figure and un-cropped figure of the same has been uploaded as Supplementary file.
Sensitivity of crude insect extract-based TLM-LAMP assay
To evaluate the analytical sensitivity of TLM-LAMP assay with crude insect extract of P. absoluta larvae, crude extract template was serially diluted in ddH2O up to 20 times. A LAMP assay was then performed using 1 µL of serial dilution product as a template at 65 °C for 45 min with visualization of results by color change and gel electrophoresis. Results showed positive amplification by color change and ladder-like band up to 10− 8 serial dilution for P. absoluta samples (Fig. 6c).
Geographical validation of TLM-LAMP
The developed TLM-LAMP assay was rigorously validated against the target insect population collected from Bengaluru, Tamil Nadu, Uttarakhand, and Delhi. This validation aimed to assess the assay’s robustness across the genetic variations among populations. The desired result of positive color change was observed after incubation at 65 °C for 45 min and no change of color in non-template control (Supplementary Fig. 6). The results demonstrated consistent sensitivity and specificity, confirming its applicability for reliable detection across diverse geographic regions.
Field and greenhouse validation
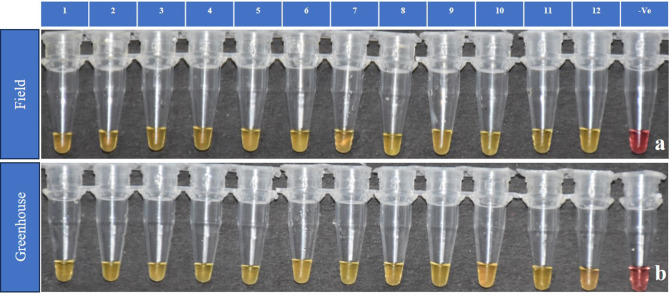
TLM-LAMP assay was validated both in field and greenhouse conditions with 24 adult moths captured in pheromone traps. We confirmed the identity of the P. absoluta samples attracted to the pheromone trap using morphological (genitalia) characters (Supplementary Fig. 8). The P. absoluta adult moth DNA was extracted using the ddH2O extraction method, followed by LAMP assay as mentioned above. Both in field (Fig. 8a) and greenhouse conditions (Fig. 8b) 100% positive amplification with a color change to yellow was evident in the P. absoluta samples, whereas no corresponding change was observed in negative samples (Fig. 8). In addition to this, We completed the identity of the P. absoluta samples attracted to the pheromone trap by universal COI PCR using the primer pair LCO-1490 and HCO-2198 followed by sequencing and BLAST analysis (Accession Number: PQ451970).
Fig. 8.
TLM-LAMP assay validation (a) in Field conditions, (b) in Greenhouse conditions. 1–12 Crude extraction of P. absoluta. (-Ve)- non-template control.
Discussion
The advantageous LAMP test led to its acceptance as an innovative and field-applicable molecular detection tool. The specificity of LAMP is considerably greater than that of conventional detection techniques, for the substantial number of primers required, four or six primers were designed specially to identify six or eight target DNA regions21. LAMP amplification requires at least four primers to hybridize which prevents non-specific amplification22. As majority of the DNA amplification techniques require a sophisticated thermocycler, none of them are appropriate for field conditions23,54. CRISPR-based pest identification of insects is a promising approach under laboratory conditions and till now not been applied in the field with crude extractions; because an intricate extraction of DNA is required to successfully complete the assay18. As crude extract can be used as a template and LAMP reaction can be carried out on a dry bath, it is suitable for field and semi-field conditions54–56.
A specific and sensitive molecular detection assay is necessary for the rapid identification of invasive species12. P. absoluta, is one of the most invasive insect pests and has a rapid invasion history throughout the world46. To curtail the spread and management of this invasive pest there is a need of rapid molecular diagnostic tools for P. absoluta detection. In this study, we developed a diagnostic assay for P. absoluta detection based on LAMP technology. A conserved portion of the COI gene was identified and amplified in order to determine the molecular detection of P. absoluta using the LAMP assay. A significant degree of similarity was observed between the nucleotide sequences accessible in the Genbank, indicating a high degree of conservation among isolates and variation with non-target insects proving specificity in molecular identification. COI gene sequences are an effective alternative to species-specific detection technologies since they are extremely pertinent, especially for closely related species present7. LAMP reaction mix made by adding a set of four primers [1:12:6 (F3/B3: FIP: BIP)], LAMP Colorimetric master mix (WarmStart), and DNA template was incubated for 60 min at 65 °C was found effective in detection of P. absoluta and we named this test as TLM-LAMP. When this assay was evaluated against closely related non-target insects, it didn’t generate any cross-amplification proving our assay is accurate. In order to perform the LAMP test in the field, an on-site crude insect extract-based template preparation procedure from a single insect was standardized. This overcomes the requirement for a prolonged DNA extraction process and enhances its field applicability. Our assay could provide visual detection for crude extracts in 45 min for incubation at 65 °C. Also, it is capable of accurately identifying P. absoluta at larval, pupal, and adult stages. Our study used genomic DNA, plasmid DNA as templates, with sensitivity values of 1 × 10− 8 ng/µL and 1 × 10–14 ng/µL respectively. Correspondingly, the real-time PCR and droplet-digital PCR could distinguish P. absoluta with a sensitivity of DNA concentration of more than 1 × 10− 2 ng/µL and 1 × 10− 3 ng/µL, respectively11,12, showing our assay is more sensitive. As this LAMP assay possesses high sensitivity for the target organism with genomic DNA, Plasmid DNA, and crude extract, this assay will be able to detect the fauna under extreme conditions which will be very beneficial for detecting the mentioned organism. The cost-benefit analysis was done for this TLM-LAMP assay. Cost of TLM-LAMP was compared with standard PCR (Detailed breakup in Supplementary Table 6). TLM-LAMP assay has a slightly higher per-reaction reagent cost compared to PCR, but it compensates with lower equipment costs and quicker turnaround. TLM-LAMP assay requires a simple equipment cost of ($150–200). In PCR the thermal cycler costs around $4,000–6,000. It is difficult to install a thermocycler in the field. But a dry-bath is easy to install in the field. Therefore, TLM-LAMP can be used to detect the target organism under field conditions. Furthermore, we validated the assay in both field and greenhouse setup, where we obtained specific positive results without any false positive and false negative results. Therefore, we are able to refer to our TLM-LAMP assay as a unique molecular method for P. absoluta detection. Also, several published LAMP assays are available for the detection of invasive insects like Spodoptera fruigiperda52, Mythimna loreyi24, Bactrocera tryoni28, and Bactrocera trivialis29 within 30 to 60 min.
In conclusion, we have developed a simplified TLM-LAMP assay which is an accurate, sensitive, and portable diagnostic method that combines crude DNA extraction with the LAMP reaction to identify P. absoluta. The whole procedure, from DNA extraction to detection, can be completed within 1 h. Therefore, this quick, sensitive, specific, and on-site method for field-based detection of P. absoluta can be used without costly laboratory equipment, and it will be helpful in quarantine stations and for the adoption of appropriate pest control strategies.
Materials and methods
Insect collection and rearing
Four moth species from the family Gelechiidae, order Lepidoptera, were used in this study: Phthorimaea absoluta (tomato leaf miner), Phthorimaea operculella (potato tuber moth), Pectinophora gossypiella (pink bollworm), and Aproaerema modicella (groundnut leaf miner) (Supplementary Table 2). Morphological identification was conducted at National Pusa Collection, Division of Entomology, IARI, New Delhi for each species. In addition to this, DNA barcoding was done for each species identification. The target insect, P. absoluta, was obtained from National Bureau of Agricultural Insect Resources, Bengaluru, and reared on tomato plants and 10% honey solution under temperature 27 ± 1 °C and 65 ± 5% relative humidity57 in acrylic jars maintained in Insect Proof Climate Control Chamber at Division of Entomology, ICAR-Indian Agricultural Research Institute, New Delhi. Three other insect pests viz. the groundnut leaf miner, the pink bollworm, and the potato tuber moth were used for specificity validation of the assay.
DNA extraction
DNA isolated from larval, pupal, and adult stages of P. absoluta, and larval stages of P. operculella, P. gossypiella, and A. modicella. DNA isolation was performed using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, USA), following the manufacturer’s instructions with slight modifications to improve DNA yield. Genomic DNA was extracted from an entire specimen for P. absoluta, P. operculella, and A. modicella, as well as from partial larval tissue of P. gossypiella. Specimens were homogenized twice: first using 60 µL of ATL buffer and once more using 120 µL of ATL buffer. The samples were incubated for three hours at 56 °C, to lyse the cells 20 µL proteinase K was added. Later, the spin column filter was filled with 50 µL of AE Buffer to elute the DNA, and it was incubated for 15 min at room temperature. After repeating this process, 100 µL of eluent was produced18. NanoDrop1000 Spectrophotometer (Genova Nano, Jenway Company) was used to quantify the isolated DNA. Isolated DNA was stored at 4 °C for short-term storage and at − 20 °C for long-term preservation.
Designing of primers for LAMP assay
A vital step while developing a LAMP assay is primer designing. Previously, mitochondrial cytochrome oxidase I (COI) has been the target gene for LAMP assays in several insects, including Bactrocera trivialis29, Bactrocera tryoni28, Daktulosphaira vitifoliae23, and Spodoptera frugiperda25,58,59. For its wide utility as a DNA identification ‘barcode’ for insect detection, we selected COI as our target gene. The conserved sequence of the COI gene served as the basis to design the primers proposed in this work. COI region of the target insect P. absoluta was amplified by PCR with primer pair LCO-1490, and HCO-219860, and the amplified DNA product was sequenced by sanger-sequencing by Barcode Biosciences (Karnataka, India). Obtained sequence was submitted to NCBI with accession number: PP506477.
We checked the sequence in NCBI and selected sequences from different parts of the world to observe variation among the sequences using multiple alignments, which include sequences of the COI gene of P. absoluta, P. operculella, P. gossypiella, and A. modicella (Supplementary Table 2). Using the NEB LAMP Primer Design Tool (https://lamp.neb.com/) default settings, two primer sets were generated, and Supplementary Table 3 provides a detailed description of primers selected for this study. The primers were checked using NCBI-BLAST. The BLAST-based amplification of the target COI gene showed the primers were species-specific. The primers were synthesized by Barcode Biosciences (Karnataka, India).
Validation of LAMP primer using conventional PCR protocol
To verify the functionality of the primers, first, we checked the two pairs (P1L1 and P3) of outer primers using PCR. The PCR was done in a 25 µL volume, comprising 12.5 µL of PCR master mix (Dream-taq master mix, Thermo-scientific), 0.5 µL of each primer, 9.5 µL of nuclease-free water, and 2 µL of DNA template. Each reaction included a positive and a negative control. The PCR protocol utilized was as follows: initial denaturation at 94 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 49 °C for 30 s, extension at 65 °C for 1 min, and final extension at 72 °C for 5 min. The reaction was stopped by an indefinite hold at 4 °C. After that, DNA fragments were separated in 2% agarose gel mixed with ethidium-bromide and visualized in Gel Doc XR+ (Bio-rad Laboratories Inc. USA).
Also, we used outer primers to evaluate primer specificity using PCR in order to decide which primer set to use further for the LAMP assay by following the PCR conditions as mentioned in the previous paragraph. The universal COI primers LCO1490 and HCO21982460 were used as positive PCR controls to verify the quality of the extracted genomic DNA (Supplementary Table 4). The products were separated with 2% gel electrophoresis stained with ethidium bromide and viewed in Gel Doc XR+ (Bio-rad Laboratories Inc., USA) using the 100 bp Plus DNA ladder (GeneDireX).
TLM-LAMP assay optimization
An important step in developing a LAMP assay is to standardize the reaction mixture and its parameters, such as temperature and time. The LAMP assay is carried out in a reaction volume of 25 µL with the following components: 10 µL of primer master mix, 1 µL of DNA template, and 14 µL of isothermal master mix (WarmStart Colorimetric LAMP 2x Master Mix - New England BioLabs)58. The primer ratio (F3/B3: FIP: BIP) of this assay was tested and optimized as published28. The primers were tested in the following ratios: 1:6:3, 1:8:4, 1:10:5, and 1:12:6 for outer primers: inner primers: loop primers. However, these ratios didn’t show any positive results. Later, we used outer primers: forward inner primer: backward inner primer (F3/B3: FIP: BIP) 1:6:6, 1:9:6, and 1:12:6.
We found standardized primer ratio for this study was 1:12:6 (F3/B3: FIP: BIP). With a total volume of 100 µL, the primer master mix is prepared using 10 µL of 10 pmol/µL F3 and B3, 12 µL of 100 pmol/µL FIP, 6 µL of 100 pmol/µL BIP, and 62 µL of nuclease-free water.
Samples were incubated at 55 °C, 60 °C, 65 °C, 70 °C, and 75 °C for 60 min which is constant time for most of the LAMP assays. For different time durations, reaction tubes were incubated at 65 °C and 15, 30, 45, and 60 min respectively. Results were observed by visual color change and 2% gel electrophoresis.
Evaluation of the TLM-LAMP assay for P. absoluta with DNA under laboratory
The optimized TLM-LAMP reaction was completed using 15 DNA samples of P. absoluta from three different stages (five from each larva, pupa, and adult stage) were evaluated at 65 °C for 60 min in triplicate. The results were observed by colorimetric change and also in 2% gel electrophoresis.
Specificity testing of TLM-LAMP assay
Using the insect group indicated earlier, the specificity of the LAMP reaction was evaluated. Two DNA samples per insect species (P. absoluta, P. operculella, P. gossypiella, and A. modicella) were used for LAMP reactions, which were carried out under standardized reaction conditions with one non-template reaction in triplicate. DNA samples having a concentration of 100 ng/µL and a 260/280 value of 1.8 were used for specificity testing. The results were visualized by calorimetry and also by using a 2% gel electrophoresis stained with ethidium bromide and viewed in Gel Doc XR+ (Bio-rad Laboratories Inc., USA) using the 100 bp Plus DNA ladder (GeneDireX).
Sensitivity testing of LAMP assay
Insect genomic DNA and plasmid DNA were serially diluted twenty times to assess the detection limit of the TLM-LAMP assay employing a primer set (P1L1). The isolated DNA from the larval insect was adjusted to 100 ng/µl and subsequently serially diluted using DNA up to 10–20. Similarly, plasmid DNA (Concentration- 100 ng/µL) was serially diluted up to twenty times (10–20). LAMP assay was conducted for each concentration in triplicate. The results were observed by color change and in 2% gel electrophoresis stained with ethidium bromide and viewed in Gel Doc XR+ (Bio-rad Laboratories Inc., USA) using the 100 bp Plus DNA ladder (GeneDireX).
Optimization and validation of TLM-LAMP assay for field application
On-site crude insect extract preparation
To develop a quick method for on-site crude insect extraction for its direct use as template, we evaluated different extraction methods used previously to make our test field-friendly. For testing, crude insect extract was prepared by crushing single larval samples in (a) 0.02 M EDTA20, (b) double distilled water20, (c) NaOH: EDTA, (1:1)53, and (d) NAOH: EDTA (1:2)53. In, 0.02 M EDTA and ddH2O, the larval specimen was carefully transferred using a fine camel hair brush into a 1.5 ml microcentrifuge tube, after adding 50 µl of distilled water, the sample was crushed with a sterile micro-pestle in the microcentrifuge tube, followed by incubation at 100 °C for 10 min followed by cooling down for 2 min. Then 1 µl of this crude extract was added to the LAMP reaction. In the same way, insect specimens were crushed in the appropriate NAOH: EDTA buffer (1:1 and 1:2) using a micro-pestle. The extract was then immediately taken for the LAMP reaction, incubated at 65 °C, and monitored in every 15 min. We used 1 µl of crude extraction in the LAMP assay instead of a DNA template and incubated it at 65 °C while observing it every 15 min intervals for color change.
Sensitivity of TLM-LAMP assay
The sensitivity of the TLM-LAMP assay was determined for crude DNA extract of P. absoluta by serially diluting twenty times (10–20) employing the primer set (P1L1). LAMP assay was used to test each dilution in triplicate. The results were observed by color change under daylight and in 2% gel electrophoresis stained with ethidium bromide and viewed in Gel Doc XR+ (Bio-rad Laboratories Inc., USA) with 100 bp Plus DNA ladder (GeneDireX).
Validation under laboratory setup
Phthorimaea absoluta different stages, larva, pupa, and adults were taken from the insect culture and used in LAMP assay validation in laboratory using crude DNA extraction. Five samples of the larval, pupal, and adult stages were collected and 50 µL of ddH2O was used to extract the crude DNA. Next, the LAMP reaction was done using the 1 µl crude extract. Results were observed through a color change and verified using 2% Gel electrophoresis stained with ethidium bromide and viewed in Gel Doc XR+ (Bio-rad Laboratories Inc., USA) using the 100 bp Plus DNA ladder (GeneDireX).
Geographical validation of TLM-LAMP with target insect populations
The TLM-LAMP assay was validated using the insect samples which were collected from Bengaluru, Tamil Nadu, Uttarakhand, Delhi. We have taken twenty larval samples in each batch. Insects were taken for the LAMP reaction after simplified crude insect extraction in ddH2O, as per the mentioned protocol. Each sample batch was then incubated at 65 °C with one non-template control. Then the results were visualized by colorimetry.
Validation under field and greenhouse setup
To confirm the accuracy of TLM-LAMP assay, we set up a DELTA trap (Del-Ta) in the Division of Vegetable Sciences, ICAR-IARI greenhouse (Supplementary Fig. 7b) and Tomato fields (Supplementary Fig. 7a) at ICAR-IARI, New Delhi using a pheromone lure of P. absoluta (TLM Lure, Pest Control Pvt. Ltd.). After three days of trap setting, we took some essential equipment into the field (Supplementary Fig. 4 and Supplementary Table 5), and carried out the crude DNA extraction followed by LAMP assay in both outdoor and greenhouse conditions. The results were observed by colorimetric change. Further, to confirm the samples, mtCOI of the representative samples were amplified using the universal COI primers60, followed by sequencing through sanger sequencing as commercial facility (Barcode Biosciences, Bengaluru, Karnataka India).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Dr. M. Mohan, Principal Scientist, ICAR-NBAIR, Bengaluru and Mr. Kishor Chandra Sahoo, ICAR-IARI, Assam for helping with the target insect culture. The support received from ICAR-IARI, New Delhi is thankfully acknowledged. We thank Dr. Priyank Hanuman Mhatre, Scientist, ICAR-Central Potato Research Station, Muthorai, The Nilgiris, Tamil Nadu for providing Potato tuber moth samples. We thank Dr. G. T. Behere, Head, Division of Crop Protection, ICAR-Central Institute for Cotton Research for providing pink boll worm samples. We thank Dr. Lakshmi Kant, Director, ICAR- Vivekananda Parvatiya Krishi Anusandhan Sansthan, and Dr. K.K. Mishra, Head (Crop Protection Division), ICAR- Vivekananda Parvatiya Krishi Anusandhan Sansthan for providing Tomato leaf miner samples from Uttarakhand that helped in the geographical validation. We acknowledge Ms. Jessa Joseph, PhD Research Scholar, ICAR-Indian Institute of Horticultural Research, Bengaluru for providing Tomato leaf miner samples from Bengaluru that helped in the geographical validation. We acknowledge The Graduate School, ICAR-IARI, New Delhi for providing ICAR-PG Fellowship to first author. P.R.S acknowledges ICAR-IARI funded project titled “Genome editing for improving resource use efficiency, quality, stress tolerance and yield of crops (Flagship Project)” for supporting the work.
Author contributions
A.K., P.R.S. and S.K.S designed the research. Laboratory experiments done by A.K. The LAMP primers were designed by D.D. Non-target insects were sourced and identified by N.D., who also processed the evaluation of male-genitalia of the target insect. P.R.S. and S.K.S planned and assisted with the implementation of this study. A.K. drafted the initial manuscript which was revised by all the authors. P.R.S., S.K.S., S.R. and N.G. critically reviewed the manuscript. P.R.S., conceptualized and supervised the research. P.R.S. and M.K.D. involved in funding the work. All authors read and approved the manuscript.
Data availability
NCBI GenBank, Accession number: PP506477, PQ451969, PP816326, PQ452113, PQ455498, PQ451970.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Balajee, S. A., Nickle, D., Varga, J. & Marr, K. A. Molecular studies reveal frequent misidentification of aspergillus fumigatus by morphotyping. Eukaryot. Cell.5, 1705–1712 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison, T. A., Yoshizaki, J., Nichols, J. D. & Bolger, D. T. Estimating survival in photographic capture–recapture studies: Overcoming misidentification error. Methods Ecol. EvoL.2, 454–463 (2011). [Google Scholar]
- 3.Paredes-Esquivel, C., Donnelly, M. J., Harbach, R. E. & Townson, H. A molecular phylogeny of mosquitoes in the Anopheles Barbirostris Subgroup reveals cryptic species: Implications for identification of disease vectors. Mol. Phylogenet Evol.50, 141–151 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Lyal, C. H. & Miller, S. E. Capacity of United States federal government and its partners to rapidly and accurately report the identity (taxonomy) of non-native organisms intercepted in early detection programs. Biol. Invasions. 22, 101–127 (2020). [Google Scholar]
- 5.McCullough, D. G., Work, T. T., Cavey, J. F., Liebhold, A. M. & Marshall, D. Interceptions of nonindigenous plant pests at US ports of entry and border crossings over a 17-year period. Biol. Invasions. 8, 611–630 (2006). [Google Scholar]
- 6.Hebert, P. D., Cywinska, A., Ball, S. L. & DeWaard, J. R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci.270, 313–321 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebert, P. D., Ratnasingham, S. & De Waard, J. R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. Ser. B Biol. Sci.. 270, S96-S99 (2003). [DOI] [PMC free article] [PubMed]
- 8.Sint, D. et al. A two-dimensional pooling approach towards efficient detection of parasitoid and pathogen DNA at low infestation rates. Methods Ecol. Evol.7, 1548–1557 (2016). [Google Scholar]
- 9.Enright, M. C. & Spratt, B. G. Multilocus sequence typing. Trends Microbiol.7, 482–487 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Doi, H. et al. Droplet digital polymerase chain reaction (PCR) outperforms real-time PCR in the detection of environmental DNA from an invasive fish species. Environ. Sci. Technol.49, 5601–5608 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Zink, F. A., Tembrock, L. R., Timm, A. E. & Gilligan, T. M. A droplet digital PCR (ddPCR) assay to detect Phthorimaea absoluta (Lepidoptera: Gelechiidae) in bulk trap samples. J. Econ. Entomol.115, 2125–2129 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Zink, F. A., Tembrock, L. R., Timm, A. E. & Gilligan, T. M. A real-time PCR assay for rapid identification of Tuta absoluta (Lepidoptera: Gelechiidae). J. Econ. Entomol.113, 1479–1485 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Garzón-Orduña, I. J., Geib, S. M., Ledezma, L., Bremer, F. T. & Barr, N. B. Implementing low-cost, high accuracy DNA barcoding from single molecule sequencing to screen larval tephritid fruit flies intercepted at ports of entry. Ann. Entomol. Soc. Am.113, 288–297 (2020). [Google Scholar]
- 14.Hatzenbuhler, C., Kelly, J. R., Martinson, J., Okum, S. & Pilgrim, E. Sensitivity and accuracy of high-throughput metabarcoding methods for early detection of invasive fish species. Sci. Rep.7, 46393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malacrinò, A. et al. Fungal communities associated with bark and ambrosia beetles trapped at international harbours. Fungal Ecol.28, 44–52 (2017). [Google Scholar]
- 16.Westfall, K. M., Therriault, T. W. & Abbott, C. L. A new approach to molecular biosurveillance of invasive species using DNA metabarcoding. Glob. Change Biol.26, 1012–1022 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Tabuloc, C. A. et al. Sequencing of Tuta absoluta genome to develop SNP genotyping assays for species identification. J. Pest Sci.92, 1397–1407 (2019). [Google Scholar]
- 18.Shashank, P. R. et al. CRISPR-based diagnostics detects invasive insect pests. Mol. Ecol. Resour.24, e13881 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amano, T. & Nomura, M. A diagnostic loop-mediated isothermal amplification method to distinguish Helicoverpa armigera (Lepidoptera: Noctuidae) from other related species in the New World. J. Insect Sci.20, 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priti, J., Baranwal, S., Dietzgen, V. K., Ghosh, A. & R. G., & A rapid field-based assay using recombinase polymerase amplification for identification of Thrips palmi, a vector of tospoviruses. J. Pest Sci.94, 219–229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notomi, T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res.28, e63–e63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notomi, T., Mori, Y., Tomita, N. & Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol.53, 1–5 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Agarwal, A., Cunningham, J. P., Valenzuela, I. & Blacket, M. J. A diagnostic LAMP assay for the destructive grapevine insect pest, phylloxera, Daktulosphaira vitifoliae. Sci. Rep.10, 21229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam, H. Y., Kwon, M., Kim, H. J. & Kim, J. Development of a species diagnostic molecular tool for an invasive pest, Mythimna Loreyi, using LAMP. Insects11, 817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Congdon, B. S., Webster, C. G., Severtson, D. & Spafford, H. In-field capable loop-mediated isothermal amplification detection of Spodoptera frugiperda (Lepidoptera: Noctuidae) larvae using a rapid and simple crude extraction technique. J. Econ. Entomol.114, 2610–2614 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Nam, H. Y., Kim, J. H., Lee, S. H., Heckel, D. G. & Kim, J. Development of a LAMP-Based molecular species diagnosis method for four major agricultural pests in the genus Spodoptera (Lepidoptera: Noctuidae). Insects. 12, 883 (2021). [DOI] [PMC free article] [PubMed]
- 27.Kitano, D. & Takakura, K. I. Simple and on-site DNA purification for LAMP reaction applicable to non‐adult tephritid fruit fly (Diptera: Tephiritidae). J. Appl. Entomol.144, 824–829 (2020). [Google Scholar]
- 28.Blacket, M. J. et al. A LAMP assay for the detection of Bactrocera tryoni Queensland fruit fly (Diptera: Tephritidae). Sci. Rep.10, 9554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starkie, M. L. et al. Loop-mediated isothermal amplification (LAMP) assays for detection of the New Guinea fruit fly Bactrocera trivialis (Drew) (Diptera: Tephritidae). Sci. Rep.12, 12602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao, J., Ren, L., Chen, R., Tao, J. & Luo, Y. A LAMP assay for the detection of Thecodiplosis japonensis, an alien Gall Midge Species Pest of Pine Trees. Insects13, 540 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, W. et al. LAMP assay as a Rapid Identification technique of Chinese Citrus fly and Japanese Orange fly (Diptera: Tephritidae). J. Econ. Entomol.116, 956–962 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Ponting, S., Tomkies, V. & Stainton, K. Rapid identification of the invasive small hive beetle (Aethina tumida) using LAMP. Pest Manag. Sci.77, 1476–1481 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Peterson, D. L. et al. Specificity and sensitivity of a rapid LAMP assay for early detection of emerald ash borer (Agrilus planipennis) in Europe. Forests. 14, 436 (2023).
- 34.Rahman, M. M., Nam, H., Choi, N. & Kim, J. Development of molecular-based species identification and optimization of reaction conditions for molecular diagnosis of three major Asian planthoppers (Hemiptera: Delphacidae). Insects. 14, 124 (2023). [DOI] [PMC free article] [PubMed]
- 35.Agarwal, A., Martoni, F., Eow, L., Rodoni, B. C. & Blacket, M. J. LAMP assay for the detection of the Asian citrus psyllid, Diaphorina citri Kuwayama (Hemiptera: Psylloidea: Psyllidae). Sci. Rep.13, 10895 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis, M. A. Invasion Biology 288 (Oxford University Press, 2009).
- 37.Pimentel, D. Biological Invasions: Economic and Environmental Costs of Alien Plant, Animal, and Microbe Species (No. 10131) (CRC, 2003).
- 38.The International Union for Conservation of Nature. (2024). https://iucn.org/our-work/topic/invasive-alien-species [DOI] [PubMed]
- 39.Han, P. et al. Tuta absoluta continues to disperse in Asia: Damage, ongoing management and future challenges. J. Pest Sci.92, 1317–1327 (2019). [Google Scholar]
- 40.Desneux, N. et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci.83, 197–215 (2010). [Google Scholar]
- 41.Shashank, P. R. et al. Genetic homogeneity in south American tomato pinworm, Tuta absoluta: A new invasive pest to oriental region. 3 Biotech.8, 1–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campos, M. R., Biondi, A., Adiga, A., Guedes, R. N. & Desneux, N. From the Western Palaearctic region to beyond: Tuta absoluta 10 years after invading Europe. J. Pest Sci.90, 787–796 (2017). [Google Scholar]
- 43.Vargas, H. C. Observaciones sobre la biologı´a y enemigos naturales de la polilla del tomate, gnorimoschema absoluta (Meyrick) (Lepidoptera: Gelechiidae). Idesia1, 75–110 (1970). [Google Scholar]
- 44.Larrain, P. Plagas Del tomate. I parte: Descripción, fluctuación poblacional, daño, plantas hospederas, enemigos naturales de las plagas principales. IPA La Platina. 39, 30–35 (1987). [Google Scholar]
- 45.Desneux, N., Luna, M. G., Guillemaud, T. & Urbaneja, A. The invasive south American tomato pinworm, Tuta absoluta, continues to spread in Afro-Eurasia and beyond: The new threat to tomato world production. J. Pest Sci.84, 403–408 (2011). [Google Scholar]
- 46.Shashank, P. R., Chandrashekar, K., Meshram, N. M. & Sreedevi, K. Occurrence of Tuta absoluta (Lepidoptera: Gelechiidae) an invasive pest from India. Indian J. Entomol.77, 323–329 (2015). [Google Scholar]
- 47.Bajracharya, A. S. R. et al. The first record of south American tomato leaf miner, Tuta absoluta (Meyrick 1917) (Lepidoptera: Gelechiidae) in Nepal. J. Entomol. Zool. Stud.4, 1359–1363 (2016). [Google Scholar]
- 48.Xian, X. et al. The potential invasion risk and preventive measures against the tomato leafminer, Tuta absoluta in China. Entomol. Gen.36, (2017).
- 49.Choi, B. H., Hur, J. H., Heckel, D. G., Kim, J. & Koh, Y. H. Development of a highly accurate and sensitive diagnostic tool for pyrethroid-resistant chimeric P450 CYP337B3 of Helicoverpa armigera using loop‐mediated isothermal amplification. Arch. Insect Biochem. Physiol.99, e21504 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Horváth, D., Fazekas, I. & Keszthelyi, S. Phthorimaea operculella (Ze1873, 1873), first record of an invasive pest in Hungary (Lepidoptera, Gelechiidae). Acta Phytopathol. Entomol. Hung.52, 117–122 (2017). [Google Scholar]
- 51.Ramya, R. S., Mohan, M. & Joshi, S. A simple method for sexing live larvae of pink bollworm, Pectinophora gossypiella (Lepidoptera: Gelechiidae). Anim. Biol.. 70, 97–100 (2020).
- 52.Osabutey, A. F. et al. Identification of a fall armyworm, Spodoptera frugiperda-specific gene and development of a rapid and sensitive loop-mediated isothermal amplification assay. Sci. Rep.12, 874 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diksha, D. et al. Simplified extraction protocol of citrus tissues for rapid detection of ‘Candidatus Liberibacter asiaticus’ using isothermal recombinase polymerase amplification assay and its application for prevalence studies. S. Afr. J. Bot.165, 517–525 (2024). [Google Scholar]
- 54.Kim, Y. H., Hur, J. H., Lee, G. S., Choi, M. Y. & Koh, Y. H. Rapid and highly accurate detection of Drosophila suzukii, spotted wing Drosophila (Diptera: Drosophilidae) by loop-mediated isothermal amplification assays. J. Asia Pac. Entomol.19, 1211–1216 (2016). [Google Scholar]
- 55.Polley, S. D. et al. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J. Clin. Microbiol.48, 2866–2871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reta, D. H. et al. Molecular and immunological diagnostic techniques of medical viruses. Int. J. Microbiol. (2020). [DOI] [PMC free article] [PubMed]
- 57.Joshi, R., Gaur, N. & Mathpal, S. Biochemical mechanism of Insecticide Resistance in Spodoptera litura (F) populations from Uttarakhand. Indian J. Entomol.84, 892–897 (2023). [Google Scholar]
- 58.Agarwal, A. et al. A diagnostic LAMP assay for rapid identification of an invasive plant pest, fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). Sci. Rep.12, 1116 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinha, T. et al. Development of a loop-mediated isothermal amplification assay for accurate and rapid identification of Spodoptera frugiperda in maize from India. Cereal Res. Commun.51, 1–11 (2023). [Google Scholar]
- 60.Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplifcation of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol.3, 294–299 (1994). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NCBI GenBank, Accession number: PP506477, PQ451969, PP816326, PQ452113, PQ455498, PQ451970.