Abstract
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), decreases quality of life and causes disability. The underlying processes are not fully understood. This study uses Mendelian randomization (MR) analysis to identify cytokines that may be associated with UC and CD, aiding in early diagnosis and treatment decisions. Methods Genome-wide association study (GWAS) data for inflammatory cytokine levels were obtained from a cohort of 14,824 individuals of European descent. The outcome data were then analyzed using summary-level GWAS data for UC and CD from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). The analysis was primarily conducted using inverse-variance weighted (IVW) methods, with MR-Egger and weighted median serving as supplementary analyses. Sensitivity analyses included Cochran’s Q test, MR-Egger intercept test, MR-PRESSO, and leave-one-out analysis.The inflammatory cytokines were subjected to additional scrutiny through the application of the Steiger test and reverse Mendelian randomization analysis. Subsequently, multivariable Mendelian randomization (MVMR) was employed to examine the associations of metabolites on UC and CD, in conjunction with linkage disequilibrium score regression (LDSC) and colocalization analysis. After FDR correction, we identified significant genetic associations of two inflammatory proteins (CXCL5 and CXCL9) with UC, and CXCL5 and IL-18R1 with CD. These findings were further validated by MVMR. Colocalization analyses demonstrated substantial genetic overlap between inflammatory proteins and IBD, with CXCL5 showing strong evidence of shared genetic variants with UC, and CXCL9 exhibiting genetic colocalization with CD, suggesting common genetic determinants underlying these inflammatory protein-IBD relationships. The current work presents evidence that presents evidence of significant associations between seven inflammatory protein factors and UC, as well as three inflammatory protein factors and CD. These findings provide novel insights into the biological mechanisms of IBD, and have implications for the screening, prevention, and treatment of IBD.
Keywords: Inflammatory protein factors, Colocalization analysis, Ulcerative colitis, Crohn’s disease, Mendelian randomization
Subject terms: Gastrointestinal diseases, Predictive markers
Introduction
Inflammatory bowel disease (IBD)1, encompassing ulcerative colitis (UC)2 and Crohn’s disease (CD)3, represents a significant contributor to diminished quality of life and disability4,5, imposing a substantial burden on public health6,7. The prevalence of these conditions is on the rise8, and the precise pathogenesis of IBD remains elusive9,10. Therefore, enhancing efforts in the prevention and early detection of IBD is imperative from a strategic standpoint.
Cytokines, small protein molecules secreted and synthesized by various immune and non-immune cells, are essential for regulating immunity, stimulating cell activation and proliferation, and maintaining human cell function11. Additionally, cytokines are implicated in inflammation12, autoimmune diseases13, and tumorigenesis14. Emerging research indicates that inflammatory cytokines contribute to the pathogenesis and advancement of IBD15–17.
A diverse array of inflammatory cytokines, including interleukin-1 receptor antagonist (IL1R), IL12, IL23, IL17, tumor necrosis factor-alpha (TNF-α), interleukin-18 (IL-18), C-C motif chemokine ligand (CCL) 2, and CCL3, have been identified in observational studies as being associated with the risk of UC and CD18,19. These findings underscore the complex nature of intestinal inflammation in UC and CD and suggest that targeting cytokines may hold promise for primary prevention and treatment strategies.The existing evidence robustly indicates the participation of TNF-mediated pathways, as well as non-TNF pathways including IL-1, IL-12/23, and IL-18, in the pathogenesis of UC and CD20–24. This highlights the promise of these cytokines as targets for therapy in current and forthcoming treatment regimens for UC and CD.The precise role of particular inflammatory pathways in the pathogenesis of UC and CD remains uncertain, largely as a result of the constraints inherent in observational studies, such as residual confounding and reverse causation, as well as the absence of robust data from randomized trials. These gaps in research and limitations impede the differentiation between the roles of inflammatory cytokines in the development of UC and CD and their influence on the advancement of systemic inflammation.
In contemporary research, Mendelian randomization (MR) studies have become prevalent in investigations of disease etiology25. In the absence of randomized controlled trials (RCTs), MR stands out as a particularly persuasive approach for investigating correlational associations between exposures and outcomes26. Through the utilization of genetic variants as instrumental variables (IVs) for exposures, such as the levels of a specific inflammatory cytokine, MR analysis can enhance the robustness of associative inference by mitigating unobserved confounding and attenuating reverse causation27,28.The IVs approach employed in this study closely resembles RCTs by randomly assigning genetic variations at conception. Under specific assumptions, the MR framework also emulates a randomized controlled trial29. In contrast to traditional epidemiological methods, this approach minimizes the impact of confounding factors such as gender and age, thereby facilitating more reliable associative inference30. Additionally, the risk of reverse causation is reduced in MR studies due to the formation of genotypes before the onset of disease.
Further research is necessary to elucidate the association between inflammatory protein factors and IBD. In this study, we conducted MR analysis utilizing genome-wide association study (GWAS) summary data to comprehensively investigate the associations of 91 inflammatory protein factors on UC and CD.Furthermore, we utilized linkage disequilibrium score regression(LDSC) and colocalization analyses to investigate the genetic and protein-level mechanisms driving the pathogenesis of UC and CD.
Methods and materials
Study design
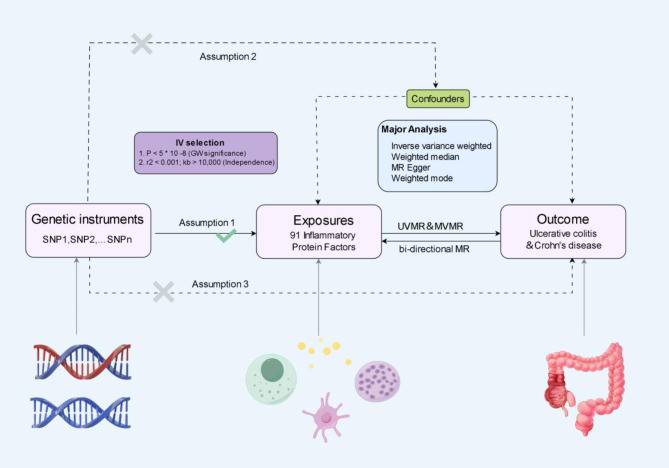
This study utilized comprehensive two-sample bidirectional Mendelian randomization analyses and multivariable Mendelian randomization(MVMR) analyses to investigate potential associations between inflammatory protein factors and UC and CD. The MR analysis and design schematic are depicted in Fig. 1.In order to ensure the validity of MR studies, adherence to three key criteria is essential. Firstly, the single nucleotide polymorphisms (SNPs) of the selected IVs must exhibit a significant association with the inflammatory protein factors under investigation. Secondly, the IVs should demonstrate independence from other potential confounding factors. Lastly, the IVs must exert their influence on the outcome through the exposure factor rather than through their own effects31. Given that this study made use of publicly available GWAS data, no additional ethical approval was necessary. The MR analyses conducted in this study were carried out using the Two Sample MR, MRPRESSO, and coloc MR packages within the R software platform (version 4.2.1).
Fig. 1.
The flowchart presented in this study delineates the fundamental assumptions underlying MR analysis. The principal objective of conducting two-sample bidirectional MR and MVMR analyses is to investigate the potential genetic associations between 91 inflammatory protein factors and the predisposition to UC and CD.
GWAS data for 91 inflammatory protein factors
The most recent GWAS findings have identified a total of 91 cytokines related to inflammation32. This extensive dataset offers increased possibilities for examining the connections between inflammatory factors and both UC and CD. To compile this comprehensive collection of inflammatory protein factors, data from GWAS on 91 inflammation-related cytokines were gathered from 11 cohorts, involving a combined sample of 14,824 individuals of European ancestry.Comprehensive information regarding the aggregated GWAS data can be accessed in the primary publication and the GWAS Catalog. Table S1 contains a complete list of the 91 inflammatory protein factors, along with their respective names, the GWAS Catalog IDs, and abbreviations.
GWAS data for UC and CD
Summary-level GWAS data from the IBDGC was utilized for the UC and CD datasets. The IIBDGC dataset includes genotyping and whole-genome sequencing data for more than 75,000 individuals diagnosed with IBD, based on radiological, endoscopic, and histopathological evaluations. Genetic associations were assessed through logistic regression models, controlling for age, sex, and principal components of genetic ancestry33. Our analysis specifically focused on summary statistics for individuals of European descent, encompassing UC (6,968 cases and 20,464 controls) and CD (5,956 cases and 14,927 controls) data.In order to prevent duplication of study cohorts, we exclusively utilized the GWAS data pertaining to UC and CD from the IIBDGC33. Both the cohorts for exposure and outcome were restricted to individuals of European ancestry to minimize the impact of potential bias resulting from population stratification.
IVs selection
In order to test hypothesis (1), we employed a rigorous selection process to identify instrumental variables (IVs) associated with inflammatory protein factors from various perspectives. To ensure robust and reliable results, we applied the genome-wide significance threshold of p < 5 × 10 − 8 for the selection of relevant SNPs. Subsequently, we pruned the SNPs by excluding those in linkage disequilibrium (LD), defined as R2 > 0.001 within a 10,000 kb range, a criterion commonly utilized in prior research34,35. To address potential bias stemming from weak IVs, we computed the R2 and F-statistics for each SNP36.SNPs with F < 10 were defined as weak IVs and were subsequently excluded from the analysis30.Subsequently, SNPs linked to metabolites were isolated from the findings while SNPs correlated with the outcomes (p < 1 × 10− 5) were excluded. The SNPs were then standardized for both exposures and outcomes by eliminating palindromic SNPs and those with allele discrepancies. In accordance with hypothesis (3), SNPs associated with outcomes (p < 1 × 10− 5) were omitted from the IVs. Ultimately, MR analysis was performed on cytokines with a minimum of two SNPs37.
Statistical analysis and secondary analysis
The associations of 91 inflammatory protein factors on UC and CD were initially assessed using the IVW method. In cases where IVW results indicated no significant heterogeneity, a fixed-effects model was applied, while a random-effects model was used for cases with heterogeneity38. The IVW estimate was derived from a meta-analysis of Wald ratios for all genetic variants. IVW relies on the assumption of no horizontal pleiotropy across all SNPs to yield the most precise estimates of relationships39,40.Therefore, we utilized IVW-based estimates to conduct an initial screening of inflammatory protein factors for their potential statistical relationships on UC and CD.In order to enhance the robustness of our findings, we employed two supplementary methods, the MR-Egger method and the weighted median (WM) method, to assess metabolites exhibiting significant IVW estimates (p < 0.05). These additional analyses were conducted to improve the reliability of our estimates in less stringent conditions.The WM method permits the inclusion of a maximum of 50% of SNPs that may be invalid, whereas MR-Egger provides capabilities for testing horizontal pleiotropy and detecting heterogeneity in the presence of pleiotropy41,42. MR-Egger regression is able to yield unbiased estimates under the InSIDE assumption (Instrument Strength Independent of Direct Effect)43,44.To account for multiple comparisons and maintain statistical rigor, we implemented false discovery rate (FDR) correction to control the type I error rate and ensure the reliability of our findings.
The secondary analyses in MR involve assessing heterogeneity, pleiotropy, and sensitivity. Heterogeneity among SNPs associated with exposures was examined through the application of Cochran’s Q test30. The Q statistic and I² (%) value were utilized to quantify heterogeneity, with I² defined as I² = [Q - (K − 1)] / Q, where K represents the number of SNPs, and Q is the Q statistic. Horizontal pleiotropy was assessed using the MR-Egger intercept method45 and the global test in MR-PRESSO46. An intercept close to zero suggests a reduced likelihood of horizontal pleiotropy.Following the identification of outliers through MR-PRESSO, a repeat MR analysis was conducted after excluding heterogeneous SNPs. Subsequently, MR-PRESSO was utilized once more to ascertain the presence of heterogeneous SNPs. Sensitivity assessment was carried out through leave-one-out(LOO) analysis47, systematically removing each SNP to assess its impact on the overall correlational estimate. Furthermore, sensitivity analyses involved comparing results from different MR methods to ensure the stability and reliability of the conclusions48.
To strengthen the exclusion restriction assumption in MR analyses, the Steiger test was employed. This test serves to confirm that the genetic variants utilized as IVs exhibit a stronger association with the exposure variable compared to the outcome variable, thereby upholding the integrity of the instrumental variable assumptions. This supplementary validation procedure enhances the reliability of our MR investigations by affirming the associative inference’s directionality49.
In conclusion, we conducted a thorough examination of inflammatory protein factors that may have associative effects on UC and CD by applying several criteria: (1) Significance at a p-value of less than 0.05 derived from the IVW method for the primary analysis. (2) Consistency in both direction and magnitude across several MR methods. (3) Absence of heterogeneity or horizontal pleiotropy in the MR results. (4) Minimal influence from any individual SNP on the MR estimates.
Subsequently, utilizing the aforementioned methodologies and protocols, reverse MR analyses were performed on the identified positive findings. The exposures under investigation were UC and CD, while the outcomes focused on inflammatory protein factors.
Confounding analysis and multivariable MR analysis
We conducted sensitivity analyses to evaluate the horizontal pleiotropy of MR results, identifying potential SNPs that may violate the assumptions of MR. Despite these efforts, it is possible that a limited number of residual confounding SNPs remain. We examined instrumental variables linked to inflammatory protein factors in the GWAS Catalog website( https://www.ebi.ac.uk/gwas/) to determine their associations with established risk factors for UC and CD, including smoking, alcohol consumption, education level, postappendectomy status2, and the diseases themselves.If any SNP were found to be linked with the aforementioned confounding variables and the outcome itself (p < 1 × 10− 5), MR analyses would be repeated following the exclusion of these SNPs to confirm the validity of the findings.
In order to adhere to the assumptions 2 and 3 of MR, it is imperative to confirm the association of genetic variants with a singular risk factor. Nonetheless, certain genetic variants exhibit associations with multiple risk factors, a concept referred to as pleiotropy. In instances of pleiotropy, multivariable MR (MVMR) can effectively account for the interactions among genetic variants linked to various exposures that may impact one another50. MVMR offers a solution to the challenges of independence, dominance, and comparability that single-variable MR is unable to address.Essentially, single-variable MR analyzes the overall impact of exposure on the outcome, whereas MVMR examines the specific impact of each exposure on the outcome, without considering other exposures.In this research, we utilized MVMR to account for the interactions of the inflammatory protein factors identified. MVMR was implemented through the IVW51, MR-PRESSO46, and LASSO regression52,53 techniques. The IVW method in MVMR entails regressing SNPs for all exposures against the outcome, with weights determined by the inverse variance of the outcome. MR-PRESSO was employed to identify and remove outliers in order to address pleiotropy among IVs. LASSO regression was utilized to eliminate exposures exhibiting collinearity.
Evaluation of genetic correlation
Based on the genetic correlation between exposure and outcome, estimates from MR studies may potentially compromise associative inference54. Despite the exclusion of SNPs associated with UC and CD during IVs selection, it is possible that SNPs lacking an obvious association could still play a role in mediating the genetic predisposition to UC and CD.Linkage Disequilibrium Score (LDSC) regression is utilized to estimate shared heritability by conducting chi-square statistics on SNP-based traits55. In order to mitigate potential confounding effects of shared heritability on exposures and outcomes, LDSC was employed to investigate the genetic correlation between the selected inflammatory protein factors and UC and CD.
Colocalization analysis
In order to explore the potential relationship between the inflammatory protein factors identified in UC and CD, we conducted a colocalization analysis utilizing the coloc R package56. This analysis enabled us to identify a shared associative variant locus within a specific genomic region that may drive the association between these factors and the two diseases.The Coloc method assessed the posterior probabilities (H0, H1, H2, H3, H4) of five hypotheses within a Bayesian framework for each variant locus: (1) no association with either trait; (2) association with trait 1 only; (3) association with trait 2 only; (4) both traits are associated with different associative variants specific to each trait; (5) both traits are correlated and share the same associative variant57. The colocalization analyses utilized default priors (p1 = 1 × 10− 4, p2 = 1 × 10− 4, p12 = 1 × 10− 5).pp.H4 > 80% of the results of the colocalization analyses ( H4 ) provide strong evidence in support of the existence of shared associative variants affecting gene expression and the risk of UC versus CD in specific genomic regions58.
Results
Preliminary analysis
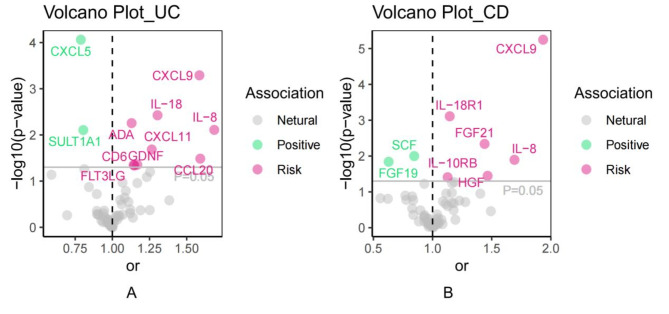
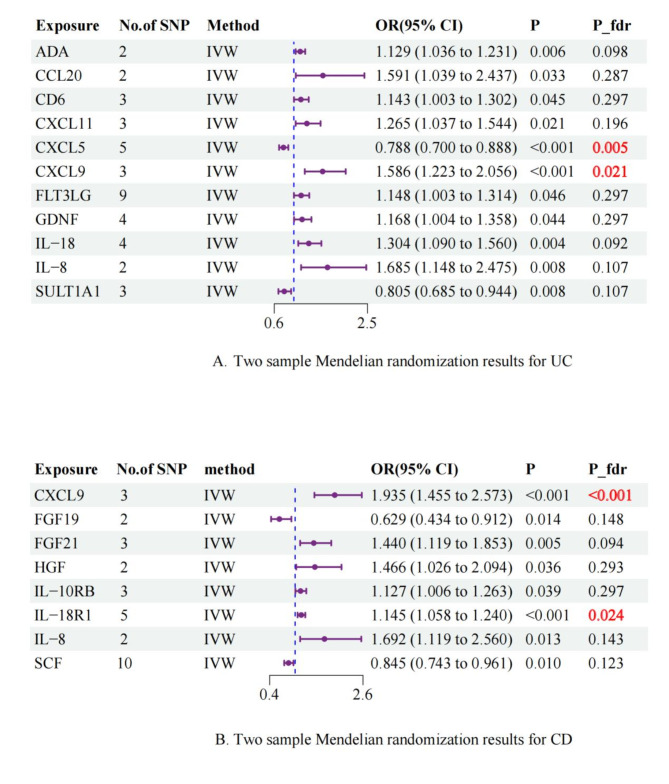
Utilizing a significance criterion of P < 5 × 10− 8, this study discovered 256 SNPs linked to 91 inflammatory protein factors. The F-statistics of these SNPs varied from 32.17 to 1,064.62, affirming the statistical soundness and dependability of the chosen cytokines as IVs in the MR analysis. Detailed information regarding the IVs can be found in Table S2. Prior to conducting the formal MR analysis, all outliers were identified and eliminated through the screening of confounders and MR-PRESSO (Table S3).In this comprehensive MR analysis of 91 inflammatory proteins, the IVW method identified significant associations for 11 and 9 inflammatory proteins with UC and CD, respectively. After FDR correction, two inflammatory proteins remained significantly associated with each of UC and CD (Supplementary Table 4; Figs. 2 and 3). The IVW analysis identified 11 inflammatory proteins significantly associated with UC (p < 0.05), including CXCL5 (odds ratio [OR]: 0.788, 95% confidence interval [CI]: 0.700-0.888, p < 0.001), CXCL9 (OR: 1.586, 95% CI: 1.223–2.056, p < 0.001), IL-18 (OR: 1.304, 95% CI: 1.090–1.560, p = 0.004), and others. After FDR correction, only CXCL5 (p_fdr = 0.005) and CXCL9 (p_fdr = 0.021) remained significantly associated with UC.For CD, 8 inflammatory proteins showed initial associations (p < 0.05), with CXCL9 (OR: 1.935, 95% CI: 1.455–2.573, p < 0.001) and IL-18R1 (OR: 1.145, 95% CI: 1.058–1.240, p < 0.001) demonstrating the strongest associations. Following FDR correction, CXCL9 (p_fdr < 0.001) and IL-18R1 (p_fdr = 0.024) maintained statistical significance(Figs. 2 and 3).The alignment of the MR Egger, Weighted Median, and Weighted Mode methodologies with the IVW approach in the MR analysis underscores the robustness of the findings.
Fig. 2.
Volcano plot of 91 inflammatory protein factors associated with the risk of developing UC and CD.
Fig. 3.
Forest plot of inflammatory protein factors versus UC and CD risk.
In the repeated sensitivity analyses, no SNP exhibiting pleiotropy was identified in either the level pleiotropy or MR-PRESSO analyses (Supplementary Table 5), suggesting the reliability of our instrumental variables. Moreover, the heterogeneity test revealed no heterogeneity in the MR results (Supplementary Table 6), thus bolstering the validity of our findings. Additionally, the LOO method did not detect any notable sources of bias (Supplementary Fig. 1).All checks successfully passed the Steiger test, suggesting the absence of reverse association in the instrumental variables (Supplementary Table 7).The remaining visualizations are shown in Supplementary Figs. 2–4.
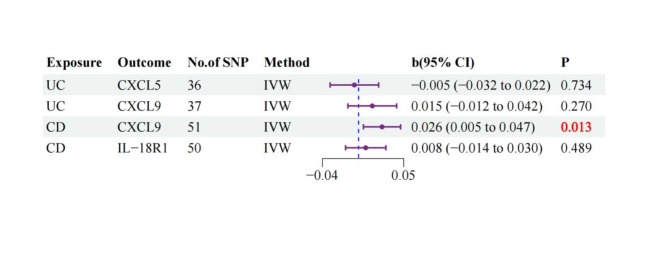
Reverse MR analysis
To assess potential reverse causation, we performed reverse MR analyses examining the associative effects of UC and CD on the identified inflammatory proteins. For UC, no significant reverse effects were observed on either CXCL5 (β = -0.005, 95% CI: -0.032 to 0.022, p = 0.734) or CXCL9 (β = 0.015, 95% CI: -0.012 to 0.042, p = 0.270). For CD, while no significant effect was found on IL-18R1 (β = 0.008, 95% CI: -0.014 to 0.030, p = 0.489), there was evidence of reverse causation for CXCL9 (β = 0.026, 95% CI: 0.005 to 0.047, p = 0.013), suggesting a potential bidirectional relationship between CD and CXCL9 levels (Supplementary Table 8, Fig. 4).
Fig. 4.
Inverse Mendelian randomization of forest maps.
MVMR analysis
After adjusting for inflammatory protein factor interactions, MVMR analyses using IVW, MR-PRESSO, and LASSO regression methods demonstrated consistent associations. For UC, CXCL5 showed an inverse association (OR: 0.808, 95% CI: 0.717–0.910, p < 0.001), while CXCL9 exhibited a positive association (OR: 1.745, 95% CI: 1.312–2.320, p < 0.001). For CD, both CXCL9 (OR: 1.933, 95% CI: 1.531–2.442, p < 0.001) and IL-18R1 (OR: 1.148, 95% CI: 1.061–1.241, p < 0.001) showed significant positive associations. These associations remained consistent across multiple sensitivity analyses.(Supplementary Table 9, Fig. 5).
Fig. 5.
Multivariate Mendelian randomization of forest maps.
Evaluation of genetic correlation
LDSC analysis revealed no significant genetic correlations between inflammatory proteins and IBD. For UC, genetic correlations were not significant with either CXCL5 (Rg = -0.229, SE = 0.163, p = 0.161) or CXCL9 (Rg = -0.006, SE = 0.128, p = 0.962). Similarly for CD, no significant genetic correlations were observed with CXCL9 (Rg = 0.167, SE = 0.119, p = 0.159) or IL-18R1 (Rg = -0.111, SE = 0.148, p = 0.942). These findings suggest minimal genetic overlap between these inflammatory proteins and IBD phenotypes(Supplementary Table 10).
Colocalization analysis
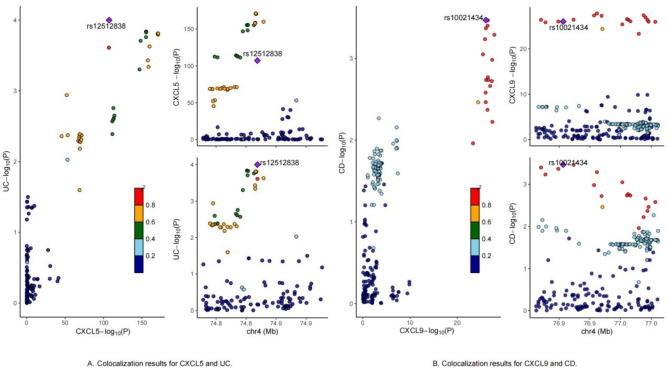
The colocalization analysis for CXCL5 (PP.H4 = 93.07%, lead SNP = rs1251252838) provided strong evidence for a shared genetic basis between CXCL5 expression and UC risk. These findings are consistent with previous studies32. Specifically, key loci identified in genome-wide association studies may contribute to UC risk through biological regulation of CXCL5 expression. Similarly, the colocalization analysis for CXCL9 (PP.H4 = 73.57%, lead SNP = 10021434) provided moderate evidence for a shared genetic basis between CXCL9 expression and CD risk. This suggests that the identified key loci may influence CD risk through mechanisms involving the regulation of CXCL9 expression. In contrast, other inflammatory proteins did not show supporting evidence for colocalization hypotheses related to UC and CD. The complete colocalization results are presented in Supplementary Tables 11 and Fig. 6.
Fig. 6.
Colocalization results.
Discussion
IBD is a complex autoimmune disorder characterized by a multifaceted and intricate pathogenesis. An increasing body of research underscores the significance of inflammatory protein factors in the development and advancement of UC and CD. Nevertheless, the precise associations and underlying mechanisms connecting inflammatory protein factors to UC and CD remain largely ambiguous.In this research, we employed a comprehensive collection of SNPs related to inflammatory protein factors, along with a thorough examination of loci and the use of the IIBDGC database, as outcome for conducting MR to evaluate the potential associative influence of inflammatory protein factors on UC and CD.
This study demonstrates that individuals with genetically predicted elevated levels of ADA, CCL20, CD6, CXCL11, CXCL9, FLT3LG, GDNF, IL-18, and IL-8 showed higher associations with UC risk, while those with elevated CXCL5 and SULT1A1 levels showed lower associations. After FDR correction and MVMR analysis, CXCL5 showed a protective association with UC, while CXCL9 was associated with increased UC risk. Specifically, one standard deviation (SD) increase in CXCL5 was associated with a 19.2% decrease in UC risk, while a similar increase in CXCL9 was associated with a 74.5% increase in UC risk. In the context of CD, CXCL9, FGF21, HGF, IL-10RB, IL-18R1, and IL-8 were found to be associated with increased risk, while FGF19 and SCF were associated with decreased risk. After FDR correction and MVMR analysis, CXCL9 and IL-18R1 showed associations with increased CD risk. Specifically, each SD increase in CXCL9 and IL-18R1 was associated with 93.3% and 14.8% increased risk of CD, respectively, with evidence of bidirectional association between CD and CXCL9. Investigation of the potential mechanisms of these factors may provide new perspectives and potential therapeutic targets for IBD management. Colocalization analysis suggests that certain genetic loci identified in GWAS studies may contribute to UC and CD susceptibility through regulation of CXCL5 and CXCL9 expression.
C-X-C motif chemokine 5(CXCL5), also known as epithelial-derived neutrophil-activating peptide 78 (ENA-78), is a member of the CXC chemokine family and plays a crucial role in neutrophil chemotaxis, activation, and inflammation promotion59. It is secreted by a variety of cell types, including epithelial cells, fibroblasts, and macrophages, especially in response to inflammatory triggers such as IL-1 and TNF-α. Upon binding to the CXCR2 receptor on neutrophils, CXCL5 facilitates their migration to inflammatory sites.The chemotactic activity plays a critical role in the immune response by facilitating the migration and accumulation of neutrophils at sites of tissue damage or infection, allowing them to carry out essential functions such as pathogen clearance and tissue repair. CXCL5 has been associated with a range of inflammatory and autoimmune conditions, including UC.A study revealed that CXCL5 is significantly upregulated in the inflamed colon60, which contrasts with our genetic findings, indicating a potentially intricate and context-specific involvement in disease pathogenesis. Our genetic findings suggest a protective association with UC. This apparent paradox might be explained by the complex and context-dependent functions of CXCL5 in intestinal homeostasis.During inflammatory responses, elevated CXCL5 levels may represent more than just a pathological factor, but rather a compensatory protective response. This response likely functions through enhanced neutrophil recruitment, promoting pathogen clearance, tissue repair, and mucosal healing61. As the first line of defense in the innate immune system, neutrophils can exert multiple beneficial effector functions in acute inflammation, helping maintain tissue homeostasis. However, while neutrophils play a crucial role in tissue repair, their activation may also lead to tissue damage, particularly in cases of chronic inflammation or non-infectious injury62. CXCL5 might also play a crucial role in maintaining intestinal barrier integrity and mucosal homeostasis through regulation of epithelial cell renewal and local immune responses63.Our colocalization analysis provides strong genetic evidence supporting the relationship between CXCL5 and UC, though reverse MR analysis did not establish a direct association. These findings highlight the complex nature of inflammatory mediators in UC pathogenesis, where their effects may vary depending on the disease stage, local environment, and genetic context.
Studies suggest that CXCL9 may play a crucial role in the pathogenesis of IBD. While UC and CD have distinct pathological features, they both involve abnormal immune responses and dysregulation of the gut microbiota64. In CD, aberrant interactions between microbes and the host immune system during early life may be a key factor in its development, while UC can be affected by intestinal dysregulation at any life stage65. Furthermore, elevated CXCL9 expression has been associated with extraintestinal manifestations and long-term disease in ulcerative colitis patients66. This aligns with our MR findings, and the bidirectional association between CD and CXCL9 observed in our reverse MR analysis. This suggests that CXCL9 not only functions within the intestine but may also influence systemic inflammatory responses, potentially exacerbating disease progression. Our colocalization analysis provided moderate evidence for a shared genetic basis between CXCL9 expression and CD risk, with the identified lead SNP (rs10021434) potentially representing a functional genetic variant that influences both CXCL9 expression and CD susceptibility. This variant might function through multiple mechanisms, including regulation of transcription factor binding sites, modulation of enhancer or promoter activity, alteration of chromatin accessibility, and influence on CXCL9 expression patterns in relevant tissues. The SNP may contribute to CD pathogenesis by affecting immune cell recruitment and activation, modifying inflammatory responses, and potentially impacting intestinal barrier function. Therefore, CXCL9 expression levels could serve as an important biomarker for monitoring and evaluating both diseases, while understanding the functional role of this lead SNP could provide valuable insights into the molecular mechanisms underlying the relationship between CXCL9 and CD, potentially leading to more targeted therapeutic approaches.
The IL-18 receptor 1 (IL-18R1) serves as the principal signaling receptor for IL-18, facilitating IL-18-induced pro-inflammatory reactions67. Studies have shown that IL-18 plays a crucial role in various chronic inflammatory diseases, including CD and UC68. IL-18R1 polymorphisms have been associated with CD susceptibility, particularly in specific populations69. Furthermore, IL-18 and its receptor play a critical role in regulating intestinal immune responses by influencing T cell differentiation and function70. In the intestine, IL-18R1 expression is enhanced in both effector and regulatory CD4(+) T cells, particularly within the intestinal lamina propria[1]. Our MR study findings demonstrate consistency with these studies and clearly indicate IL-18R1 as a risk factor for CD, while IL-18 is identified as a risk factor for UC. This suggests that IL-18 and its receptor may play significant roles in the pathogenesis of CD, and further research could contribute to the development of therapeutic strategies targeting IL-18R1 and IL-18 to improve outcomes for CD and UC patients.
Although some inflammatory factors did not pass strict FDR correction, several results still warrant our attention. IL-8 plays a crucial role in IBD, with its elevated levels closely correlating with disease activity. In one study, researchers found significantly increased IL-8 mRNA expression in the colonic mucosa of active UC and CD patients, suggesting IL-8’s potential importance in IBD pathogenesis71. IL-8’s relative stability and longer half-life give it a central function in the sustained attraction and activation of granulocytes, which may contribute to the persistent inflammation in IBD72. In UC patients, The enzyme adenosine deaminase (ADA) expression levels correlate with disease activity and may influence intestinal immune responses73. Additionally, CXCL11, a chemokine, has been found to be associated with the pathological process of ulcerative colitis, with its high expression potentially related to intestinal inflammatory responses and extraintestinal manifestations74.These findings are all consistent with our MR study results, indicating the significant roles of these inflammatory factors in IBD research. Our analyses support that these inflammatory factors are not merely markers of disease activity but potentially contributing factors in disease development, where factors like IL-8, ADA, and CXCL11 may interact synergistically to promote disease progression, suggesting their potential value as therapeutic targets and biomarkers for IBD.
Nonetheless, further investigation is required to thoroughly examine the precise impact of the aforementioned inflammatory protein factors on UC and CD within controlled experimental settings.
This MR demonstrates multiple strengths. Firstly, it represents the most comprehensive and methodical investigation to date into the genetic association association between inflammatory protein factors and UC and CD, encompassing an analysis of 91 inflammatory protein factors. Secondly, a rigorous MR analysis was employed to mitigate inherent limitations such as reverse association and confounding variables. There are several limitations to the current study. One limitation is the restricted number of SNPs available for identifying SNPs of interest at the genome-wide level. To mitigate this limitation, a more lenient threshold for MR analysis was employed, a practice commonly observed in other studies. Nonetheless, all selected SNPs exhibited F-statistic values exceeding 10, suggesting the robustness of our IVs. Additionally, to minimize the impact of ethnic diversity, only individuals of European descent from the IIBDGC were included in the MR analysis.Hence, additional research is warranted to investigate the applicability of our findings to diverse databases and populations. Another constraint of this study is the reliance on sample size for the accuracy of MR estimation. Therefore, it is imperative to increase the sample size to ensure the credibility of our outcomes. Furthermore, while MR analysis offers valuable insights into association, it is essential to underscore the importance of validating our results through rigorous RCTs and fundamental research prior to clinical application.
Conclusion
In conclusion, this current study has identified genetic susceptibility inflammatory protein factors associated with UC and CD. CXCL5, CXCL9, and IL-18R1 warrant further exploration as promising therapeutic targets for UC and CD. The identification of these inflammatory protein factors has significant implications for the early detection, prevention, and management of UC and CD, while also providing important insights for the design of upcoming clinical trials. Furthermore, this integrated analysis utilizing genomics and proteomics provides a guiding framework for investigating the potential causes and mechanisms of IBD.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our gratitude to the GWAS Catalog and IIBDGC for generously providing GWAS data. Additionally, We extend our thanks to the drawing services offered by www.figdraw.com and www.chiplot.online.
Author contributions
Qiang Su: Formal analysis (lead); methodology (lead); software (lead); validation (lead); writing – original draft (lead). Yun Lu: Resources (lead); software (lead); validation (lead); writing – original draft (lead). Song He: Formal analysis (equal); validation (equal); visualization (equal). Song Huang: Data curation (equal); investigation (equal). Jiang Liang: Conceptualization (equal); project administration(equal); supervision (equal); writing – review and editing (equal). Zhenxiang An and Yuanli He: Conceptualization (lead); funding acquisition (lead); methodology (lead); project administration (lead); supervision (lead); writing – review and editing (lead).
Funding
This study was funded by the National Administration of Traditional Chinese Medicine of China, Fifth Batch of the National Clinical Excellence Training Program for Traditional Chinese Medicine Practitioners (Program Number: State TCM Education Letter [2022] No. 1); the Traditional Chinese Medicine Spleen and Stomach Disease Scientific and Technological Innovation Talent Team Construction at Guizhou University of Traditional Chinese Medicine (Program Number: Gui TCM TD Hezi [2022] No. 005);the National Administration of Traditional Chinese Medicine’s 2022 Project for the Construction of National Veteran TCM Experts Inheritance Studios: Wu Zhengshi National Veteran TCM expert inheritance studio(Program Number: National Medical Education Letter [2022] No. 75).
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qiang Su and Yun Lu these two authors contributed equally to this work.
Contributor Information
Jiang Liang, Email: truekingboy-999@163.com.
Yuanli He, Email: 407206115@qq.com.
Zhenxiang An, Email: anzhenxiang057@gzy.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84447-4.
References
- 1.Bruner, L. P., White, A. M. & Proksell, S. Inflamm. Bowel Disease Prim. Care50, 411–427. 10.1016/j.pop.2023.03.009. (2023). [DOI] [PubMed] [Google Scholar]
- 2.Le Berre, C., Honap, S. & Peyrin-Biroulet, L. Ulcerative colitis. Lancet402, 571–584. 10.1016/s0140-6736(23)00966-2 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Dolinger, M., Torres, J. & Vermeire, S. Crohn’s disease. Lancet403, 1177–1191. 10.1016/s0140-6736(23)02586-2 (2024). [DOI] [PubMed] [Google Scholar]
- 4.van Erp, L. W. et al. Improvement of fatigue and quality of life in patients with quiescent inflammatory bowel Disease following a personalized Exercise Program. Dig. Dis. Sci.66, 597–604. 10.1007/s10620-020-06222-5 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Knowles, S. R. et al. Quality of life in inflammatory bowel disease: a systematic review and Meta-analyses-part I. Inflamm. Bowel Dis.24, 742–751. 10.1093/ibd/izx100 (2018). [DOI] [PubMed] [Google Scholar]
- 6.The global. regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a. Lancet Gastroenterol. Hepatol.5, 17–30. 10.1016/s2468-1253(19)30333-4 (2020). systematic analysis for the Global Burden of Disease Study, (2017). [DOI] [PMC free article] [PubMed]
- 7.Ng, S. C. et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet390, 2769–2778. 10.1016/s0140-6736(17)32448-0 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Kaplan, G. G. & Windsor, J. W. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol.18, 56–66. 10.1038/s41575-020-00360-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Mahony, C., Amamou, A. & Ghosh, S. Diet-Microbiota Interplay: An Emerging Player in Macrophage Plasticity and Intestinal Health. Int J Mol Sci 23. (2022). 10.3390/ijms23073901 [DOI] [PMC free article] [PubMed]
- 10.Tran, A. et al. Estrogen-related receptor alpha (ERRα) is a key regulator of intestinal homeostasis and protects against colitis. Sci. Rep.1110.1038/s41598-021-94499-5 (2021). [DOI] [PMC free article] [PubMed]
- 11.Liu, C. et al. Cytokines: from clinical significance to quantification. Adv. Sci. (Weinh). 8, e2004433. 10.1002/advs.202004433 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins, J. M. & Pillai, V. TAFRO syndrome or Castleman-Kojima syndrome: a variant of multicentric Castleman disease. Blood126, 2163. 10.1182/blood-2015-07-662122 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Mosser, D. M. & Zhang, X. Interleukin-10: new perspectives on an old cytokine. Immunol. Rev.226, 205–218. 10.1111/j.1600-065X.2008.00706.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Propper, D. J. & Balkwill, F. R. Harnessing cytokines and chemokines for cancer therapy. Nat. Rev. Clin. Oncol.19, 237–253. 10.1038/s41571-021-00588-9 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Wang, L. et al. Targeting JAK/STAT signaling pathways in treatment of inflammatory bowel disease. Inflamm. Res.70, 753–764. 10.1007/s00011-021-01482-x (2021). [DOI] [PubMed] [Google Scholar]
- 16.Bourgonje, A. R., Ungaro, R. C., Mehandru, S. & Colombel, J. F. Targeting the IL-23 pathway in inflammatory bowel disease. Gastroenterology10.1053/j.gastro.2024.05.036 (2024). [DOI] [PubMed] [Google Scholar]
- 17.Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol.14, 329–342. 10.1038/nri3661 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Strober, W. & Fuss, I. J. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology140, 1756–1767. 10.1053/j.gastro.2011.02.016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honap, S. et al. JAK inhibitors for inflammatory bowel disease: recent advances. Frontline Gastroenterol.15, 59–69. 10.1136/flgastro-2023-102400 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neurath, M. F., Fuss, I., Kelsall, B. L., Stüber, E. & Strober, W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J. Exp. Med.182, 1281–1290. 10.1084/jem.182.5.1281 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silvagni, E. et al. From Bed to Bench and back: TNF-α, IL-23/IL-17A, and JAK-Dependent inflammation in the Pathogenesis of Psoriatic Synovitis. Front. Pharmacol.12, 672515. 10.3389/fphar.2021.672515 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atreya, R. & Neurath, M. F. IL-23 blockade in Anti-TNF refractory IBD: from mechanisms to clinical reality. J. Crohns Colitis. 16, ii54–ii63. 10.1093/ecco-jcc/jjac007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, Y., Chen, J. & Zhou, H. Antibody-based Biotherapeutics in Inflammatory diseases. In Pharmaceutical Biotechnology: Fundamentals and Applications, (eds Crommelin, D. J. A., Sindelar, R. D. & Meibohm, B.) (Springer International Publishing), 591–644. 10.1007/978-3-031-30023-3_24. (2024).
- 24.Eltantawy, N. et al. A review article of inflammatory bowel disease treatment and pharmacogenomics. Beni-Suef Univ. J. Basic. Appl. Sci.1210.1186/s43088-023-00361-0 (2023).
- 25.Smith, G. D. & Ebrahim, S. Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol.32, 1–22. 10.1093/ije/dyg070 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Zuccolo, L. & Holmes, M. V. Commentary: mendelian randomization-inspired causal inference in the absence of genetic data. Int. J. Epidemiol.46, 962–965. 10.1093/ije/dyw327 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading mendelian randomisation studies: a guide, glossary, and checklist for clinicians. Bmj362, k601. 10.1136/bmj.k601 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skrivankova, V. W. et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. Bmj375 (n2233). 10.1136/bmj.n2233 (2021). [DOI] [PMC free article] [PubMed]
- 29.Richmond, R. C. & Davey Smith, G. Mendelian Randomization: Concepts and Scope. Cold Spring Harb Perspect Med 12. (2022). 10.1101/cshperspect.a040501 [DOI] [PMC free article] [PubMed]
- 30.Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol.37, 658–665. 10.1002/gepi.21758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boef, A. G., Dekkers, O. M. & le Cessie, S. Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int. J. Epidemiol.44, 496–511. 10.1093/ije/dyv071 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Zhao, J. H. et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat. Immunol.24, 1540–1551. 10.1038/s41590-023-01588-w (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet.47, 979–986. 10.1038/ng.3359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, J. et al. Assessing the Causal effects of human serum metabolites on 5 Major Psychiatric disorders. Schizophr Bull.46, 804–813. 10.1093/schbul/sbz138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi, K. W. et al. Assessment of Bidirectional relationships between physical activity and depression among adults: a 2-Sample mendelian randomization study. JAMA Psychiatry. 76, 399–408. 10.1001/jamapsychiatry.2018.4175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lv, X. et al. Causal relationship between ischemic stroke and its subtypes and frozen shoulder: a two-sample mendelian randomization analysis. Front. Neurol.14, 1178051. 10.3389/fneur.2023.1178051 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gill, D. et al. Effects of genetically determined Iron status on risk of venous thromboembolism and carotid atherosclerotic disease: a mendelian randomization study. J. Am. Heart Assoc.8, e012994. 10.1161/jaha.119.012994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, F. et al. Causality between heart failure and epigenetic age: a bidirectional mendelian randomization study. ESC Heart Fail.10, 2903–2913. 10.1002/ehf2.14446 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgess, S., Dudbridge, F. & Thompson, S. G. Combining information on multiple instrumental variables in mendelian randomization: comparison of allele score and summarized data methods. Stat. Med.35, 1880–1906. 10.1002/sim.6835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yavorska, O. O. & Burgess, S. MendelianRandomization: an R package for performing mendelian randomization analyses using summarized data. Int. J. Epidemiol.46, 1734–1739. 10.1093/ije/dyx034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce, B. L. & Burgess, S. Efficient design for mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol.178, 1177–1184. 10.1093/aje/kwt084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren, Z., Simons, P., Wesselius, A., Stehouwer, C. D. A. & Brouwers, M. Relationship between NAFLD and coronary artery disease: a mendelian randomization study. Hepatology77, 230–238. 10.1002/hep.32534 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, M., Xie, C. R., Shi, Y. Z., Tang, T. C. & Zheng, H. Gut microbiota and major depressive disorder: a bidirectional mendelian randomization. J. Affect. Disord. 316, 187–193. 10.1016/j.jad.2022.08.012 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Burgess, S. & Thompson, S. G. Interpreting findings from mendelian randomization using the MR-Egger method. Eur. J. Epidemiol.32, 377–389. 10.1007/s10654-017-0255-x (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol.44, 512–525. 10.1093/ije/dyv080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat. Genet.50, 693–698. 10.1038/s41588-018-0099-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flatby, H. M., Ravi, A., Damås, J. K., Solligård, E. & Rogne, T. Circulating levels of micronutrients and risk of infections: a mendelian randomization study. BMC Med.21, 84. 10.1186/s12916-023-02780-3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mbutiwi, F. I. N., Dessy, T. & Sylvestre, M. P. Mendelian randomization: a review of methods for the Prevention, Assessment, and discussion of Pleiotropy in studies using the Fat Mass and obesity-Associated Gene as an instrument for Adiposity. Front. Genet.13, 803238. 10.3389/fgene.2022.803238 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemani, G., Tilling, K. & Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet.13, e1007081. 10.1371/journal.pgen.1007081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanderson, E. Multivariable mendelian randomization and mediation. Cold Spring Harb Perspect. Med.1110.1101/cshperspect.a038984 (2021). [DOI] [PMC free article] [PubMed]
- 51.Burgess, S. & Thompson, S. G. Multivariable mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol.181, 251–260. 10.1093/aje/kwu283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao, S. et al. A Six-microRNA signature Nomogram for Preoperative Prediction of Tumor deposits in Colorectal Cancer. Int. J. Gen. Med.15, 675–687. 10.2147/ijgm.S346790 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grant, A. J. & Burgess, S. Pleiotropy robust methods for multivariable mendelian randomization. Stat. Med.40, 5813–5830. 10.1002/sim.9156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reay, W. R. et al. Genetic estimates of correlation and causality between blood-based biomarkers and psychiatric disorders. Sci. Adv.8, eabj8969. 10.1126/sciadv.abj8969 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet.47, 1236–1241. 10.1038/ng.3406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu, B., Gloudemans, M. J., Rao, A. S., Ingelsson, E. & Montgomery, S. B. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet.51, 768–769. 10.1038/s41588-019-0404-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foley, C. N. et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat. Commun.12, 764. 10.1038/s41467-020-20885-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yun, Z. et al. Genetically predicted 486 blood metabolites in relation to risk of colorectal cancer: a mendelian randomization study. Cancer Med.12, 13784–13799. 10.1002/cam4.6022 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, K. J. et al. (eds) Editor’s Highlight: Ah Receptor Activation Potentiates Neutrophil Chemoattractant (C-X-C Motif) Ligand 5 Expression in Keratinocytes and Skin. Toxicol Sci 160, 83–94. (2017). 10.1093/toxsci/kfx160 [DOI] [PMC free article] [PubMed]
- 60.Li, Q. et al. mRNA-engineered mesenchymal stromal cells expressing CXCR2 enhances cell migration and improves recovery in IBD. Mol. Ther. Nucleic Acids. 26, 222–236. 10.1016/j.omtn.2021.07.009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papayannopoulos, V. Neutrophils stepping through (to the other side). Immunity49, 992–994. 10.1016/j.immuni.2018.12.006 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Phillipson, M. & Kubes, P. The Healing Power of neutrophils. Trends Immunol.40, 635–647. 10.1016/j.it.2019.05.001 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Matthews, J. D., Weight, C. M. & Parkos, C. A. Leukocyte-epithelial interactions and mucosal homeostasis. Toxicol. Pathol.42, 91–98. 10.1177/0192623313511336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tontini, G. E. et al. Confocal laser endomicroscopy for the differential diagnosis of ulcerative colitis and Crohn’s disease: a pilot study. Endoscopy47, 437–443. 10.1055/s-0034-1391226 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Beaugerie, L., Langholz, E., Nyboe-Andersen, N., Pigneur, B. & Sokol, H. Differences in epidemiological features between ulcerative colitis and Crohn’s disease: the early life-programmed versus late dysbiosis hypothesis. Med. Hypotheses. 115, 19–21. 10.1016/j.mehy.2018.03.009 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Pierce, E. S. Ulcerative colitis and Crohn’s disease: is Mycobacterium avium subspecies paratuberculosis the common villain? Gut Pathog 2, 21. (2010). 10.1186/1757-4749-2-21 [DOI] [PMC free article] [PubMed]
- 67.Thomas, J. M., Huuskes, B. M., Sobey, C. G., Drummond, G. R. & Vinh, A. The IL-18/IL-18R1 signalling axis: diagnostic and therapeutic potential in hypertension and chronic kidney disease. Pharmacol. Ther.239, 108191. 10.1016/j.pharmthera.2022.108191 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Harrison, O. J. et al. Epithelial-derived IL-18 regulates Th17 cell differentiation and Foxp3⁺ Treg cell function in the intestine. Mucosal Immunol.8, 1226–1236. 10.1038/mi.2015.13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim, S. W. et al. Genetic polymorphisms of IL-23R and IL-17A and novel insights into their associations with inflammatory bowel disease. Gut60, 1527–1536. 10.1136/gut.2011.238477 (2011). [DOI] [PubMed] [Google Scholar]
- 70.Kaplanski, G. Interleukin-18: Biological properties and role in disease pathogenesis. Immunol. Rev.281, 138–153. 10.1111/imr.12616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Izutani, R. et al. Increased expression of interleukin-8 mRNA in ulcerative colitis and Crohn’s disease mucosa and epithelial cells. Inflamm. Bowel Dis.1, 37–47 (1995). [PubMed] [Google Scholar]
- 72.Ha, H., Debnath, B. & Neamati, N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory diseases. Theranostics7, 1543–1588. 10.7150/thno.15625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li, M. et al. IL-6 downregulates hepatic carboxylesterases via NF-κB activation in dextran sulfate sodium-induced colitis. Int. Immunopharmacol.99, 107920. 10.1016/j.intimp.2021.107920 (2021). [DOI] [PubMed] [Google Scholar]
- 74.Bradford, K. L., Moretti, F. A., Carbonaro-Sarracino, D. A., Gaspar, H. B. & Kohn, D. B. Adenosine Deaminase (ADA)-Deficient severe combined Immune Deficiency (SCID): Molecular Pathogenesis and Clinical manifestations. J. Clin. Immunol.37, 626–637. 10.1007/s10875-017-0433-3 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- The global. regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a. Lancet Gastroenterol. Hepatol.5, 17–30. 10.1016/s2468-1253(19)30333-4 (2020). systematic analysis for the Global Burden of Disease Study, (2017). [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].