Abstract
Chronic kidney disease affects ~10% of people worldwide and there are no disease modifying therapeutics that address the underlying cause of any form of kidney disease. Genome wide association studies have identified the G1 and G2 variants in the apolipoprotein L1 (APOL1) gene as major contributors to a subtype of proteinuric kidney disease now referred to as APOL1-mediated kidney disease (AMKD). We hypothesized that inhibition of APOL1 could have therapeutic potential for this genetically-defined form of kidney disease. Here we describe the development of preclinical assays and the discovery of potent and specific APOL1 inhibitors with drug-like properties. We provide evidence that APOL1 channel activity drives podocyte injury and that inhibition of this activity stops APOL1-mediated cell death and kidney damage in a transgenic mouse model. These preclinical data, combined with clinical data from our previously published phase 2 proof-of-concept study, support the potential of APOL1 channel inhibition for the treatment of AMKD.
Subject terms: Focal segmental glomerulosclerosis, End-stage renal disease, Target validation
Apolipoprotein L1 genetic variants contribute to a subtype of proteinuric kidney disease referred to as APOL1-mediated kidney disease (AMKD). Here the authors report the discovery and characterization of potent and selective APOL1 ion channel inhibitors for the potential treatment of AMKD.
Introduction
Chronic kidney disease (CKD) represents a significant global health burden, and the discovery of disease modifying treatments that address the underlying cause of kidney disease has proven difficult. Genome-wide association studies identified that individuals with recent ancestry from sub-Saharan Africa and chronic kidney disease are more likely to be homozygous or compound heterozygous for the G1 (S342G and I384M) or G2 (N388del:Y389del) variants in the apolipoprotein L1 (APOL1) gene. Proteinuric kidney disease in the setting of two APOL1 alleles has been termed APOL1-mediated kidney disease (AMKD)1,2. Patients with AMKD have accelerated disease progression relative to patients with similar clinical features who lack APOL1 risk variants3–5, and have higher rates of end stage renal disease, transplant, dialysis and death1,6. Moreover, inheriting two APOL1 risk variants carries increased risk of proteinuric kidney disease both in the absence of any other condition, as well as in the setting of inflammatory conditions such as interferon treatment, systemic lupus erythematosus, sickle cell disease, HIV, and COVID-197,8.
APOL1 is expressed in multiple tissues, most predominantly the liver9, where APOL1 is secreted into the bloodstream as part of high-density lipoprotein. Circulating APOL1 protects against Human African Trypanosomiasis, caused by Trypanosoma brucei brucei (T.b.b.). The mechanism whereby APOL1 confers resistance is through the formation of ion channels in the parasite membrane resulting in swelling and lysis of T.b.b.1,10. The APOL1 G1 and APOL1 G2 variants evolved from the ancestral allele, APOL1 G0, and rose to high frequency in sub-Saharan Africa due to positive selection: they protect against evolved strains of Trypanosoma brucei that evade APOL1 G011.
APOL1 is also expressed in the podocyte12, a specialized kidney cell involved in glomerular function13. Cell-specific podocyte APOL1 variant expression in mice leads to proteinuric nephropathy, providing evidence that renal APOL1 expression is intrinsic to the causal biology of disease progression14. In response to inflammation, expression levels of APOL1 in the podocyte increase15. The relationship between inflammation and APOL1 expression in the kidney offers a potential explanation both as to why not all people carrying two APOL1 G1/G2 variants develop kidney disease, and why numerous inflammatory conditions are associated with AMKD.
The cellular and molecular mechanisms whereby APOL1 G1/G2 expression leads to podocyte cytotoxicity remains unclear. Proposed mechanisms include effects on autophagy, lysosomal permeability, mitochondrial dysfunction, endoplasmic reticulum stress, cation channel formation10,16–18, pyroptosis and activation of stress kinases15,19–21. While these various cellular processes may be consequential for downstream pathophysiology, the key to addressing the underlying cause of AMKD is to define the root cause whereby the G1 and G2 variants trigger podocyte cell damage.
We hypothesized that APOL1 ion channel activity in the podocyte membrane was the source of APOL1-mediated podocyte toxicity and thus AMKD. We based this hypothesis on the high frequency of G1 and G2 variants due to protection from Trypanosoma infection, and that the biological mechanism of APOL1 protection is known to involve formation of ion channels in the parasite membrane. We further hypothesized that a highly specific APOL1 ion channel inhibitor would be effective in AMKD. We reasoned an APOL1 ion channel inhibitor would be likely to have limited on-target safety risk, because there is a published report of an individual with no detectable APOL1 protein expression due to homozygous truncation mutations. This individual was healthy except for a sensitivity to trypanosome infection22. Further support for this hypothesis comes from the observation that the APOL1 gene is found only in certain higher primates and is absent in other species, suggesting that APOL1 is not an essential gene in mammals23,24.
To test the hypothesis that APOL1 ion channel activity drives kidney disease in AMKD, we set out to discover and develop a specific inhibitor of channel function, and then to evaluate the impact of APOL1 inhibition in a clinical trial of patients with AMKD. Here we present the development of assays and discovery of a novel series of oral, small molecule APOL1 inhibitors, exemplified by the clinical candidate inaxaplin (VX-147)25. These molecules are potent and specific inhibitors of APOL1 channel function in multiple cellular models. They both prevent proteinuria in an acute mouse model of AMKD and resolve established proteinuria and glomerular histopathology in a chronic mouse model of AMKD. As recently published, a 13-week clinical trial of VX-147 in patients with two APOL1 risk alleles and proteinuric non-diabetic kidney disease demonstrated a 47.6% reduction in proteinuria25.
Results
Development of assays to identify small molecule inhibitors of APOL1 channel activity
To identify small molecule APOL1 channel inhibitors and characterize their properties and biological consequence, we generated a suite of in vitro assays and then characterized compounds in in vivo transgenic mouse models26. First, using tetracycline (tet)-inducible HEK293 cell lines expressing APOL1 G0, G1 or G2, we assessed APOL1 channel function by monitoring APOL1-mediated ion flux in a fluorescence-based thallium flux assay. Second, to confirm that inhibition of ion flux seen in the thallium flux assay was directly related to APOL1 ion channel inhibition, we also established a patch clamp electrophysiology assay to directly monitor APOL1-mediated ionic current. Third, to assess the impact of APOL1 ion channel function on cell viability, we generated an APOL1-mediated cell death assay in the same tet-inducible cell lines. Fourth, we rationalized that an APOL1 inhibitor should be able to prevent the only known biological function for APOL1, namely trypanolysis, so we established a trypanosome cell death assay. Finally, to investigate the effect of APOL1 channel inhibition in vivo, we utilized transgenic mice homozygous for the APOL1 G2 variant26 to assess the impact of APOL1 inhibition on APOL1-dependent proteinuria and renal pathology. Our expectation was that an APOL1 ion channel inhibitor would show efficacy against all these preclinical endpoints.
Discovery of small molecule inhibitors of APOL1 channel activity
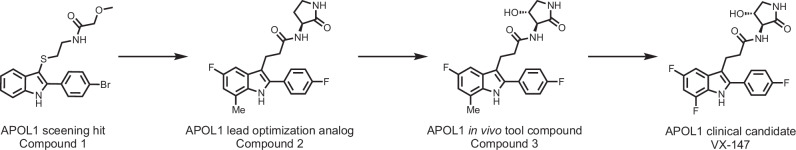
Using the tet-inducible APOL1 G1 cell line, we initiated a high throughput screening campaign using a fluorescence-based thallium flux assay, which monitored APOL1-mediated passage of thallium ions (a surrogate for potassium ions [K+]) across the cell membrane. Compound 1 was our initial hit from the screen, with modest potency but full efficacy at inhibiting APOL1-mediated flux. We embarked on a medicinal chemistry campaign, which brought us to the lead optimization candidate, Compound 2. Further optimization of this candidate gave rise to a series of compounds that included Compound 3 and the clinical candidate, VX-147 (Fig. 1). Characterization of Compounds 1-3 and VX-147 are described in the methods, in Table 1, Supplementary Tables 1 and 2 and Supplementary Figs. 1–8. Importantly, in vitro characterizations of Compound 3 and VX-147 in both the thallium flux and HEK cell rescue assay indicate this series of APOL1 inhibitors display comparable inhibitory potential across the APOL1 variants.
Fig. 1.
Medicinal chemistry progression of the screening hit to VX-147.
Table 1.
Medicinal chemistry progression of the screening hit 1 to VX-147
| Compound 1 | Compound 2 | Compound 3 | Compound 4 | |
|---|---|---|---|---|
| LogD7.4 | 5.0 | 2.8 | 2.7 | 2.7 |
| APOL1 G1/G2 Thallium flux IC50 (nM) | 580/ND (4) | 40/ND (5) | 4.2/5.4 (13/7) | 2.0/1.2 (32 /4) |
| APOL1 G1/G2 HEK Cell Rescue EC50 (nM) | 1100/ND (5) | 37/ND (8) | 3.6/10 (21/9) | 2.0/2.2 (4/1) |
| Rat/human Clint (mL/min/106 cells) | > 400/259 | 8.2/15.4 | 5.1/6.3 | 4.7 / < 2.5 |
| Rat Cl (mL/min/kg) | 61 | 15 | 6.0 | 4.8 |
| Rat t1/2 | 0.81 | 4.1 | 4.2 | 2.5 |
| Rat Vd (L/kg) | 1.6 | 2.1 | 1.4 | 0.9 |
| Rat oral bioavailability (%) | 11 | 34 | 28 | 57 |
Reversal of the amide and installation of the aminolactam afforded compound 2 with increased potency and metabolic stability. Oxidation of the lactam to compound 3 further increased potency and metabolic stability. Further Structure-Activity-Relationship (SAR) investigation of the indole core gave VX-147 with optimized properties. In vitro efficacy replicates are shown in brackets. ND not determined, Vd volume of distribution, T1/2 half-life, Cl clearance.
APOL1 risk variants lead to increased channel activity
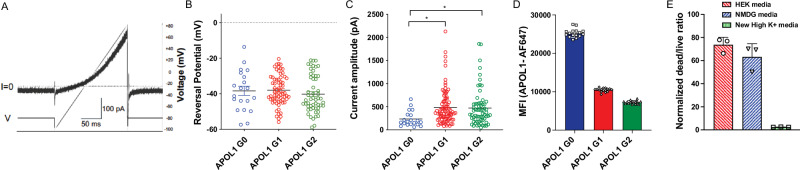
We hypothesized that the enhanced toxicity related to APOL1 G1 and G2 was the result of increased ion flux. Indeed, in the thallium flux assay, we saw increased flux activity in cells expressing risk variants relative to APOL1 G025. Using these APOL1-inducible cell lines, we assessed the ability of APOL1 to function as an ion channel by performing automated whole-cell patch clamp recordings. We found that all forms of APOL1 are permeable to monovalent cations (Na+, K+, Cs+) and had a negative reversal potential when isolating APOL1-mediated outward currents by Na+ replacement extracellularly with NMDG and by Cs+ addition intracellularly to block voltage-gated potassium channels (Fig. 2A, B). In addition, the reversal potential is close to 0 mV when recording with extracellular Na+ and intracellular Cs+ (Supplementary Fig. 9) consistent with previous reports supporting non-selective permeability of APOL1 channels27. Together these results demonstrate that the APOL1 ion channel is permeable to Cs+ ions and that Cs+ can act as surrogate for K+, which is traditionally applied intracellularly to inactivate voltage-gated potassium channels. In addition, we discovered that the outward current amplitude by APOL1 G1 and APOL1 G2 was at least two-fold greater, compared to APOL1 G0-mediated currents (Fig. 2C) similar to previous reports27,28. To ensure the increased current was not due to differential cell surface APOL1 expression, we monitored expression by flow cytometry. Cells expressing APOL1 G0 showed the highest cell surface expression, while cells expressing APOL1 G1 and G2 had significantly lower levels (~50% or less) on the surface (Fig. 2D) demonstrating that this increase outward current amplitude is not the result of elevated expression levels.
Fig. 2. APOL1 G1 and G2 toxicity is associated with enhanced outward currents on the cell surface.
A Representative voltage ramp (V)-current (I) response after 16 h tetracycline induction in APOL1 G1 expressing HEK293 cells. ms, millisecond; pA, picoampere. B Average reversal potential of APOL1-G0, -G1, -G2 -mediated outward currents. The compositions of the intracellular and extracellular solutions are described in the methods section. Results are presented as arithmetic mean ± SEM. mV, millivolt. C Average peak current amplitude of APOL1 G0-, G1- and G2- mediated outward currents at a holding potential of -80mV with the outward currents presented measured at +80 mV at the end of the voltage ramp. Results are presented as arithmetic mean ± SEM. One-way ANOVA with a post-hoc Dunnet’s multiple comparison test with APOL1 G0 as the control. Significance was set at p ≤ 0.05. *p ≤ 0.05. p = 0.011 for APOL1 G0 vs. APOL1 G1, and p = 0.02 for APOL1 G0 vs. APOL1 G2. The extracellular solution contains 137 mM NMDG, 5.4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Glucose, 10 mM Sucrose, 10 mM HEPES, 100 μM Ouabain (pH 7.2; mOsm 300-305); and the intracellular solution contains 124 mM CsF, 2 mM MgCl2, 1 mM CaCl2, 11 mM EGTA, 10 mM HEPES, 1 mM ATP (pH 7.2; mOsm 285-290). D HEK293 cells stably expressing tetracycline-inducible APOL1 G0, G1 or G2 variants. APOL1 protein surface levels were measured using an anti-APOL1 antibody (Abcam, ab252218) by fluorescence activated cell sorting (FACS) analysis and the Median Fluorescence Intensity (MFI) of the AF647 channel was reported. Three independent experiments were performed with 16 replicates per condition. Representative experiment is shown. Results are presented as arithmetic mean ± SEM. E Normalized cell death of HEK293 cells expressing APOL1 G1 in different media compositions 24 h after tet induction. Three independent experiments were performed with 32 replicates per condition. Aggregate mean and SEM is shown. HEK media is DMEM (Catalog # 11960077) with 2% FBS; NMDG media contains 109.50 mM N-Methyl-D-Glucamine, 5.4 mM NaCl, 1.8 mM CaCl2, 0.81 mM MgSO4, 25 mM glucose, 4.5 nM Ferric Nitrate, 10 mM HEPES; and High K+ media contains 115 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 25 mM glucose, 4.5 nM Ferric Nitrate, 10 mM HEPES. Source data are provided as a Source Data file.
Next, we were interested to understand whether K+ efflux or Na+ influx is the initiating event of APOL1-mediated toxicity. Therefore, we performed a cell death rescue assay in three different media conditions. We saw robust cell death in standard assay media allowing for Na+ and K+ flux as well as media only allowing K+ efflux, however, when in media where K+ efflux was inhibited, cell death was almost completely abrogated (Fig. 2E). The near total prevention of cell death was greater than reported previously but is consistent with the necessity for cell K+ efflux to trigger cell death21. These differences may be attributed to the different media compositions used between the studies. Taken together, these experiments support the hypothesis that the cellular toxicity of APOL1 risk variants is due to increased channel activity of the APOL1 protein and associated increases in K+ efflux.
In vitro characterization of VX-147
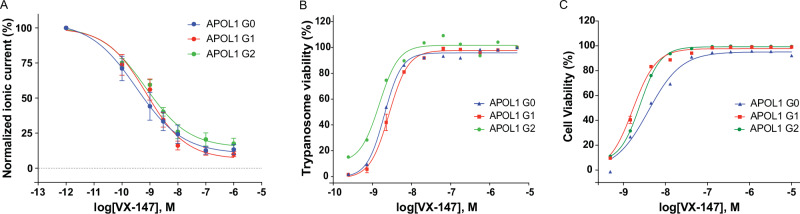
To confirm direct inhibition of APOL1 ion channel activity by APOL1 inhibitors, we performed automated whole-cell patch recordings as above with increasing concentrations of VX-147. Using the average peak outward current before and after application, we determined VX-147 potently inhibited the APOL1 ion channel with IC50 values of 2.3 nM for G0, 1.3 nM for APOL1 G1, and 1.1 nM for APOL1 G2 (Fig. 3A). Next, we evaluated the functional consequence of APOL1 inhibition by the ability of APOL1 to lyse T.b.b. We found that VX-147 prevented APOL1-mediated T.b.b. death with essentially equivalent potencies to ion channel inhibition, with EC50 values of 2.0 nM for APOL1 G0, 2.1 nM for APOL1 G1, and 1.2 nM for APOL1 G2 (Fig. 3B). We further explored whether APOL1 inhibition could rescue cell death in the same HEK293 cells. We saw complete inhibition of APOL1-dependent cell death using VX-147 with EC50 values of 4.3 nM for APOL1 G0, 2.0 nM for APOL1 G1, and 2.2 nM for APOL1 G2 (Fig. 3C). In conclusion, the fact that VX-147 is equipotent across all of these in vitro functional assays underscores the importance of channel activity as the mechanism of APOL1-mediated cell death.
Fig. 3. VX-147 inhibits APOL1-mediated flux and cell death.
A The graph shows baseline normalized APOL1-mediated ionic current measured for HEK293 cell lines expressing APOL1 G0, APOL1 G1, and APOL1 G2 variants, as a function of compound concentration. Whole cell currents were measured for these experiments. The first concentration point is no VX-147 and was artificially assigned as 2 orders of magnitude smaller than the lowest concentration tested for graphical representation. Multiple cells were recorded for each APOL1 variant, n = 8 for APOL1 G0, n = 11 for APOL1 G1 and n = 14 for APOL1 G2. B Assessment of Trypanosoma brucei brucei cell viability in the presence of recombinant APOL1 of the indicated variant using an alamarBlue™ assay. Experiments were performed in duplicate. Experimental replicates performed were n = 1 for APOL1 G0, n = 4 for APOL1 G1 and n = 1 for APOL1 G2. C Assessment of cell viability for HEK293 cell lines expressing APOL1 G0, APOL1 G1, and APOL1 G2 variants using the Promega MultiTox fluorescence assay. Experiments were performed in duplicate. Experimental replicates performed were n = 1 for APOL1 G0, n = 4 for APOL1 G1 and n = 1 for APOL1 G2. Representative traces are shown for all experiments. Results are presented as mean ± SEM when more than 3 replicates or as mean only for replicates less than 3. Source data are provided as a Source Data file.
In vivo characterization of APOL1 channel inhibitors
Given the robust inhibition of APOL1 in vitro, we sought to explore the functional effect of APOL1 inhibition in vivo. Since model organisms lack endogenous APOL1 expression, we licensed and maintained colonies of transgenic mice expressing human APOL1 G2. The APOL1 G2 transgene was inserted multiple times in tandem, and therefore mice were classified as APOL1 G2 multicopy mice (APOL1 G2mc)26. Homozygous APOL1 G2mc were viable, healthy, lived a normal lifespan, and had no baseline phenotype. However, a single injection of interferon-γ (IFNγ), which induces APOL1 kidney expression (Supplementary Fig. 10A), led to significant APOL1-dependent proteinuria in APOL1 G2mc compared to control Friend leukemia B virus (FVB) mice (Supplementary Fig. 10B, C and 11). We recently reported prophylactic treatment with VX-147 reduced APOL1-dependent proteinuria in APOL1 G2mc25. Here we present a close structural analog of VX-147, Compound 3, with comparable potency (Table 1), was able to prevent APOL1-mediated proteinuria in this model at 30 mg/kg bid (Supplementary Fig. 10D, E, F and 12). The efficacious dose used for Compound 3 is elevated relative to the dose we previously reported for VX-147 (3 mg/kg tid) due to the pharmacokinetic properties of compound 3 in mice and the less frequent dosing interval.
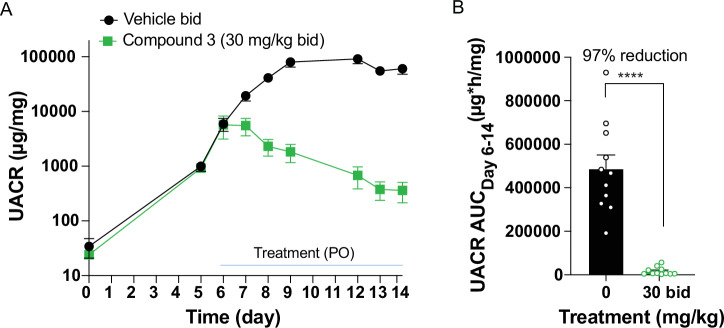
Our prophylactic model demonstrated that APOL1 inhibitors could prevent proteinuria, but their impact on pre-established proteinuria and kidney histology was unknown. Therefore, we utilized a more aggressive published proteinuria model to assess the effect of APOL1 inhibition on established proteinuria and kidney pathology. This model entails the hydrodynamic injection of an IFNγ-expressing plasmid into single copy, homozygous APOL1 G2 mice (APOL1 G2sc)26. These mice develop proteinuria approximately 5 days after plasmid injection, peaking around day 8 and remaining elevated through the study time course (Fig. 4A). Mice were assigned into two groups based on day 5 urinary albumin-to-creatinine ratio (UACR), to establish groups with similar starting mean UACR. All mice developed proteinuria. IFNγ levels were monitored across timepoints of the experiment and are reported in Supplementary Fig. 14. Treatment with vehicle or 30 mg/kg bid Compound 3 began on day 6. Compound 3 had a rapid effect at lowering the UACR (Fig. 4A and Supplementary Fig. 13). Analysis of the UACR AUC following compound intervention revealed a 97% reduction over the 8 days of compound administration (Fig. 4B). Exposure levels of Compound 3 are found in Supplementary Table 3.
Fig. 4. Assessment of Compound 3 efficacy in therapeutic APOL1 G2sc mouse model of APOL1-mediated kidney disease.
A Assessment of urinary albumin-to-creatinine ratio (UACR) in response to hydrodynamic injection of IFNγ plasmid in APOL1 G2sc transgenic mice over 14 days. n = 12 (Vehicle) and n = 11 (Compound 3). Vehicle group had 5 F and 7 M; Compound 3 group had 6 F and 5 M. B UACR area under the curve (AUC) showed reduced proteinuria in Compound 3-treated mice. One male Vehicle mouse was excluded from UACR AUC analysis due to premature removal from study because of disease progression. n = 11 mice in both groups. Percent reduction in UACR AUC with compound 3 treatment was calculated. UACR AUC data were log-transformed and analyzed by unpaired t-test. Significance was set at p ≤ 0.05. ****p < 0.0001. p < 0.0001 for Vehicle vs. treatment group. Results are presented as arithmetic mean ± SEM with each symbol representing an individual mouse. Representative data of n = 2 experiments. Source data are provided as a Source Data file.
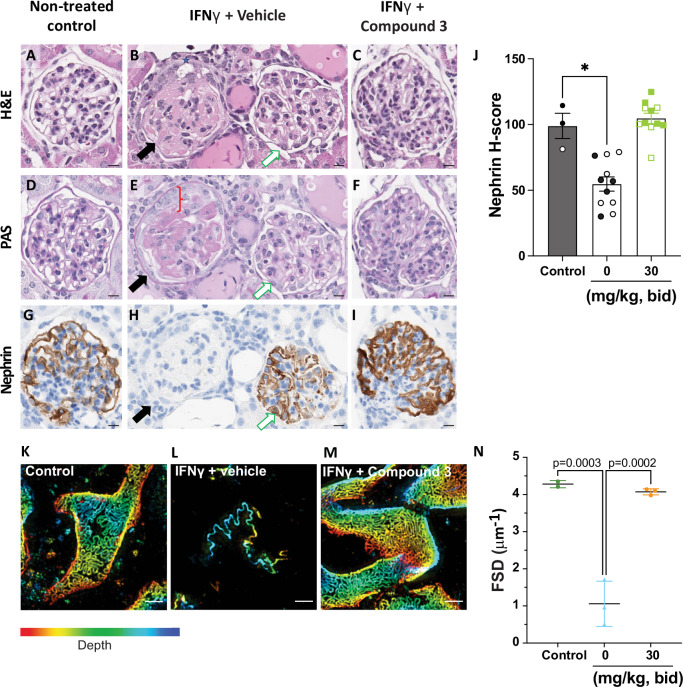
Microscopic evaluations of kidney tissues showed that mice not injected with the IFNγ plasmid (Fig. 5A, D, G) had no evidence of glomerular or tubular damage. In vehicle-treated APOL1 G2sc, approximately 60% of glomeruli exhibited histomorphologic changes and nephrin loss (Figs. 3, 5B, H, black arrow) while remaining glomeruli were morphologically normal (Fig. 5B, H, white arrow). Glomerulopathy was characterized by segmental to global collapse of capillary lumina (Fig. 5B), mesangial hyalinization (Fig. 5E), synechiae between capillary basement membranes and parietal epithelium, and vacuolation and reduplication of parietal epithelial cells (PEC). Tubules were often dilated, lined by attenuated epithelium, and contained eosinophilic proteinaceous fluid (Fig. 5B, E). In contrast, no microscopic abnormalities or nephrin loss were noted in the animals treated with Compound 3 (Fig. 5C, F, I). A larger field of view with multiple glomeruli can be found in Supplementary Fig. 15.
Fig. 5. Compound 3 prevents APOL1-mediated morphological and histopathological damage of kidney glomeruli.
A, D, G No microscopic findings were present in control-group. B, E, H Glomerulopathy was frequent and severe in vehicle-treated mice, however all glomeruli were not affected. The affected glomerulus on left (B, E, H, black-arrow) was characterized by synechia (B, grey-star) and duplication of parietal epithelium (E, red-bracket), collapsed capillary lumina, mesangial hyalinization) and accumulation of PAS+ve (E) material in the mesangium. The adjacent glomerulus on right (B, E, H, white-arrow) was morphologically normal. Near total loss of nephrin in the affected glomerulus (H, black-arrow), while the adjacent glomerulus (H, white-arrow) has preserved nephrin staining. C, F, I No findings were noted in Compound 3-treated mice. Scale bar is 10 μm for all images. J Quantitative image analysis of nephrin immunolabeling using H-score. H-score (label intensity * % area); weak (+1), moderate (+2) strong (+3). Nephrin H-score data was analyzed by a Kruskal-Wallis test with post-hoc Dunn’s multiple comparison test. Significance was set at p ≤ 0.05. *p ≤ 0.05. p = 0.0231 for Control vs. Vehicle mice. Results presented as arithmetic mean ± SEM with each symbol representing an individual mouse (closed are females, open are males) with n = 11 mice per group. K–M Maximum intensity projection of kidney filtration slit diaphragm in control mice (K), in mice treated with IFNγ and vehicle (L), and in mice treated with IFNγ and Compound 3 (M). Kidneys were immunolabeled for podocin and integrin α3. Images were acquired with 3D-Structured Illumination Microscopy. Scale bar is 1 μm. The depth color scale goes from −2 μm (red) to 2 μm (blue). N The filtration slit density (FSD) of n = 2−3 mice/group. 20 glomeruli per mouse were randomly selected and analyzed. FSD was calculated as the ratio between filtration slit length and surface area. Data were analyzed with One-way ANOVA. The Benjamini-Hochberg procedure was applied post-hoc to control for false discovery rate. Significance was set at p ≤ 0.05. p = 0.0003 for control vs. Vehicle and p = 0.0002 Vehicle vs. Compound 3. Results presented as arithmetic mean ± SEM with each symbol representing an individual mouse. Representative data of n = 2 experiments shown for all panels. Source data are provided as a Source Data file.
Serial sections of kidneys were immunolabeled for nephrin29,30 to investigate the effect of systemic inflammation (IFNγ) on glomerular filtration barrier function. Whole slide imaging and an artificial intelligence (AI) based classifier31,32 were used to quantify glomerular expression of nephrin. The quantitative scoring was expressed as an H-score, a cumulative assessment of area and intensity. The nephrin H-scores were comparable between the no IFNγ control and Compound 3-treated groups, while the vehicle-treated group had significantly lower nephrin H-scores (Fig. 5G).
Based on the microscopic data, we employed a recently described quantitative analysis of kidney health using the podocyte exact morphology measurement procedure (PEMP)33. This technique uses fluorescence immunolabeling for podocin and integrin α3 and super-resolution microscopy to assess podocyte health based on the filtration slit density (FSD). The FSD represents the ratio of length of the filtration slit for a given glomeruli (podocin staining) relative to its surface area (integrin staining) with high FSD values indicating healthy, intact glomeruli. Representative maximum intensity projections of the kidney filtration slit are shown for naïve, vehicle-treated and Compound 3-treated mice (Fig. 5H, I, J and Supplementary Fig. 16). Vehicle-treated mice showed significantly reduced FSD when compared to Compound 3-treated mice or naïve mice as summarized in Fig. 5K.
Discussion
Individuals with proteinuric CKD and two APOL1 risk variants (G1 or G2) have a genetically defined form of kidney disease referred to as AMKD. AMKD occurs in the presence of inflammatory conditions such as interferon treatment, lupus, SCD, HIV and COVID, but may also be observed in the absence of an overt inflammatory comorbidity. People with CKD and two APOL1 risk variants progress more rapidly to end-stage kidney disease relative to those with one or no APOL1 risk variants. While the genetic evidence demonstrates a key role of APOL1 G1 and G2 genetic variants in the human causal biology of proteinuric non-diabetic kidney disease, the molecular and cellular mechanism whereby APOL1 variants lead to kidney damage has been less clear. APOL1 is expressed in multiple tissues, and various cellular and molecular mechanisms have been proposed as relevant to AMKD.
Based on the known mechanism of human biology whereby APOL1 variants protect from Trypanosoma Brucei infection, we hypothesized that APOL1 ion channel function leads to kidney damage. To test this hypothesis we developed multiple assays, conducted a high throughput screen to identify small molecule inhibitors of APOL1-mediated ion flux, and optimized those molecules to identify compounds with the potency, specificity, and drug-like properties suitable to evaluate the impact of APOL1 inhibition in cells, animal models, and, ultimately, AMKD patients.
Here we present data that show the highly specific channel blocker, VX-147, directly inhibits APOL1-mediated ion flux and prevents both APOL1-dependent trypanosome and mammalian cell death. VX-147 was previously shown to bind directly to purified, recombinant APOL1 and broad profiling across a large panel of potential off-targets did not identify any significant activity, indicating that the mechanism of action is due to direct inhibition of the APOL1 ion channel25. We report that the mechanism by which APOL1 G1 and G2 variants cause cell death is likely a result of their enhanced channel activity in human cells. We demonstrate that efflux of K+, and not influx of Na+, is responsible for APOL1-mediated cell death in our recombinant cell system. A report previously indicated that reducing the extracellular sodium concentration in HEK293 cells using choline+ or K+ ions reduced APOL1-mediated cell death significantly34. However, based on their published methodology, it is not possible to differentiate between a prevention of sodium influx or potassium efflux as the extracellular media composition may have been altering the ability for potassium efflux as well. There seem to be many downstream mechanisms by which APOL1-mediated ion flux leads to cell death10,15–21, but the root cause, and thus ideal therapeutic target, appears to be the ion channel function of APOL1. In fact, a recent publication highlighted the many proximal and distal events caused by APOL1-mediated ion flux and demonstrated that VX-147 treatment prevented or reversed these downstream events35.
In transgenic APOL1 mouse models, we used Compound 3, a close analog of VX-147 with comparable potency and selectivity, to show that blockage of the APOL1 channel can both prevent and reverse APOL1-dependent proteinuria as well as preserve glomerular integrity. This is the first reported evidence demonstrating that functional inhibition of APOL1 can significantly reduce established proteinuria in an in vivo preclinical model.
Further evidence in support of the ion channel hypothesis comes from a recent human genetics study demonstrating that the N264K substitution in the protein sequence of APOL1 both decreased APOL1 ion flux and reduced kidney disease incidence in APOL1 G2 carriers36.
The most important evaluation of the ion channel hypothesis is to perform clinical trials of potent and specific APOL1 inhibitors in AMKD patients. We recently reported Phase 2a proof-of-concept data with VX-147 in patients with two APOL1 risk alleles and proteinuric non-diabetic kidney disease25. This 13-week study in 16 individuals demonstrated a 47.6% reduction in proteinuria with stable glomerular filtration rate. An ongoing Phase 2b/3 clinical trial of inaxaplin in patients with two APOL1 risk variants and proteinuric non-diabetic kidney disease will further define the clinical potential of APOL1 inhibition for patients with AMKD.
Methods
Cell line generation and growth
Human embryonic kidney (HEK) cell lines containing a tetracycline (tet)-inducible expression system (T-REx™; Invitrogen, Carlsbad, CA) and pAAVS1-Puro-APOL1 G0, pAAVS1-Puro-APOL1 G1 or pAAVS1-Puro-APOL1 G2 were generated. The physiological haplotype sequence of APOL1 was used for all 3 APOL1 sequences with E150/I228/K255. Individual clones of each cell line were generated and clones G0 DC2.13, G1 DC3.25, and G2 DC4.44 were used in this study. Cell growth medium was Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Gaithersburg, MD; Catalog # 11960-077) supplemented with 10% (v/v) tet-free fetal bovine serum (FBS; Takara Bio USA, Mountain View, CA; Catalog # 631368), 2 mM L-glutamine (Gibco; Catalog # 25030-081), 5 µg/mL Blasticidin S HCl (Gibco; Catalog # A11139-03), 1 µg/mL puromycin dihydrochloride (Gibco; Catalog # A11138-03), and 100 U/mL penicillin-streptomycin (Gibco; Catalog # 15140-122).
Electrophysiology
HEK293 APOL1-inducible cells were induced with 15 ng/mL of tet for 16–20 h. Automated electrophysiology was performed by Qpatch48 (Sophion Biosciences). Cells were perfused with extracellular solution containing: 137 mM NMDG (Sigma Aldrich; Catalog # M2004), 5.4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Glucose, 10 mM HEPES. The pH was adjusted to 7.2 with NaOH and adjusted to 300–305 mOsm with sucrose. The intracellular solution contained: 124 mM CsF, 2 mM MgCl2, 1 mM CaCl2, 11 mM EGTA, 10 mM HEPES, 1 mM ATP pH 7.2, 285–290 mOsm. Ouabain (100 μM) was added in the extracellular solution for blockade of Na+/K+ ATP-ase. NMDG replaced Na+ in the extracellular solution to isolate Cs+ efflux. We used Cs+ in the internal solution because we identified APOL1 channels to be permeable to Cs+. Cs+ was selected also to block endogenous voltage-gated K+ channels. Specifically, the reversal potential of the APOL1 ionic current in the presence of NaCl extracellularly and CsF intracellularly was near to 0 mV, supporting nonspecific monovalent cationic flow (Supplementary Fig. 2). Resistance of the QPatch plate chambers (holes) was 1.5–2.5 MΩ. Steady state inhibition of APOL1 current was measured using a ramp protocol in response to six consecutive compound concentrations (100 pM, 1 nM, 3 nM, 10 nM, 100 nM, 1 μM) with an exposure duration of 5 min to each concentration, 5 min after initial current recording (control). Each solution exchange of 5 mL volume was performed twice. The voltage protocol applied to the cells was as follows: Resting step: −80 mV for 100 ms; test pulse: −100 mV to 80 mV for 200 ms; resting step: −80 mV for 100 ms repeated at 10 s intervals, from a holding potential of −90 mV. Peak outward current was measured at the end of the 200 ms test pulse at +80 mV. Data acquisition and analysis was performed using the Sophion QPatch Assay Software (Sophion Biosciences). We continuously monitored the series resistance (MΩ) and the capacitance (pF). Cells were removed from compound analysis if the series resistance exceeded 15 MΩ, if the peak current amplitude was smaller than 100 pA, and if the reversal potential was smaller than −20 mV. The average peak outward currents before and after test compound application were used to calculate the percentage of current inhibited at each concentration. Outward currents in response to each concentration of VX-147 were normalized to its initial control current without VX-147. Concentration-response data were fit by GraphPad Prism using non-linear regression analysis with variable slope (four parameters).
Flow cytometry
HEK293 APOL1-inducible cells were grown in DMEM with 2% (v/v) doxycycline (dox)-free FBS in 96 well plates (Corning, 3904) at a seeding density of 50,000 cells/well in 100 μL of media and treated +/− 15 ng/mL doxycycline to induce APOL1 expression for 16–24 h. Cells were washed with 100 μL DPBS and detached with 100 μl of 1x Versene (Gibco, 15040066) at 37 °C for 15 min. Cells were then resuspended in complete media and transferred to a 96-well plate round bottom plate (Corning, 3799) and pelleted in a tabletop centrifuge at 300 × g for 5 mins. All staining reactions were performed in FACS buffer (DPBS supplemented with 2% FBS and 2 mM EDTA). Cells were stained for surface APOL1 expression in 50 μL of a 1:100 dilution of anti-APOL1 rabbit monoclonal antibody (Abcam, ab252218) in FACS buffer on ice for 30 min. Cells were washed in 200 μL of FACS buffer twice. Secondary staining was performed in 50 μL of a 1:2000 dilution of secondary goat anti-rabbit Alexa Fluor 647 antibody (ThermoFisher, A32733) on ice for 20 min. Cells were washed in 200 μL of FACS buffer twice. Cells were fixed in 50 μl of 4% paraformaldehyde for 15 min at room temperature. 4% PFA was washed off and replaced with 200 μL of FACS buffer for FACS analysis. Cells were immediately analyzed on a Beckman Coulter CytoFLEX instrument. The Alexa Fluor 647 signal was excited using the 638 nm laser and 660/10 nm BP filter. The Beckman coulter CytExpert software was used for data collection and FlowJo software was used for analysis. Live cells gated using FSC-A and SSC-A, then Alexa Fluor 647 intensity was analyzed.
Thallium assay methods
HEK293 APOL1-inducible cells were plated in DMEM with 2% (v/v) tet-free FBS with 15 ng/mL tet to induce APOL1 expression for 17–19 h. Cells were seeded into 384 well BioCoat™ Poly-D-Lysine coated, black/clear, flat-bottom, tissue culture-treated microplates at 2.6 × 104 cell/well. Loading buffer (HBSS, 20 mM HEPES, 5 mM Probenecid, 0.2 mM Ouabain, and components A and C from the FLIPR potassium assay bulk kit (Molecular Devices; Catalog # R8223) was added to the wells of the cell-containing assay plate. The cells were incubated for 30 min at room temperature (23 °C) and then compounds were added to the assay plate wells. Compounds were tested with a top concentration of 10 μM followed by a quarter-log dilution series with 20 total points down to 0.312 nM. An active control was used in each assay and the DMSO concentration was kept fixed at 0.2%. The cells were incubated with compound at room temperature (23 °C) for 30 min then the assay plate was placed on a FLIPR Tetra or Penta high-throughput cellular screening system to measure sample fluorescence intensity. Fluorescence readings were recorded every second for 10 s before the addition of thallium sulfate solution and an additional 60 seconds after the addition of thallium sulfate solution. Data were imported into Genedata for analysis and IC50 generation.
Trypanosome assay methods
Trypanosoma brucei brucei were obtained from ATCC and grown in ATCC medium 2834 modified HMI-9 medium. Trypanosomes in phenol red/FBS free media containing Serum Plus medium supplement (Sigma, 14008 C) were plated at a concentration of 120,000 parasites/well in 384-well plates containing compound and recombinant APOL1. The final concentration of APOL1 used in this assay was sufficient to kill approximately 90% of the parasites. AlamarBlue™ was added to the plates following 16 h incubation at 37 °C with APOL1 and compound, and then returned to the incubator for an additional 2.5 h37. The plate was then read on a SpectraMax plate reader with excitation/emission wavelengths of 555/585 nm. Data was imported into Genedata for analysis and dose response curves generated using the Smart Fit algorithm.
Multitox assay methods
HEK293 APOL1-inducible cells were plated in DMEM with 2% (v/v) tet-free FBS with 50 ng/mL tet to induce APOL1 expression for 24 h. Cells were plated into a 384-well Poly-D-Lysine coated plate at a density of 23,400 cells/well. Compounds were added to the plates for the full 24 h with a 10 point 3-fold dilution curve (top concentration, 10 μM) run in duplicate. The next day, assay buffer containing 2x MultiTox reagent from the MultiTox Multiplex cytotoxicity assay was added to each well and incubated an additional 30 min at 37 °C. The plates were read on an EnVision plate reader for viability (excitation: 400 nm, emission: 486 nm) and cytotoxicity (excitation: 485 nm, emission: 535 nm). A ratio of dead (cytotoxicity) to live (viability) cells was used to normalize data. The data was exported and analyzed in Genedata. Data was normalized using two controls, no tet treatment (100% viability) and 50 ng/mL tet treatment (0% viability) and dose response curves generated using the Smart Fit algorithm. NMDG media (109.50 mM N-Methyl-D-Glucamine, 5.4 mM NaCl, 1.8 mM CaCl2, 0.81 mM MgSO4, 25 mM glucose, 4.5 nM Ferric Nitrate, 10 mM HEPES) or High K+ media (115 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 25 mM glucose, 4.5 nM Ferric Nitrate, 10 mM HEPES) were used to isolate the ionic flux to assess the critical ion for cell death. The MultiTox Multiplex cytotoxicity assay was performed as described above with standard media.
Mouse model methods
To evaluate the pharmacodynamic effects of APOL1 inhibitors in vivo, we licensed transgenic APOL1 G2 mice from Beth Israel Deaconess Medical Center26. The mice were re-derived to allow for housing of the lines in pathogen-free facilities. Strains were propagated and maintained at Charles River Laboratories. The transgenic APOL1 G2 mice have a lysine at amino acid 150 in the APOL1 sequence. The mice were bred to homozygosity as hemizygous mice do not have a phenotype. Experiments were conducted in transgenic male and female FVB APOL1 multi-copy G2-expressing homozygous mice (APOL1 G2mc) (8–15 weeks old, Charles River, Wilmington, MA, USA). Mice were group-housed in a temperature-controlled environment (22 ± 1.5 °C, 30–70% relative humidity, 12 h light/dark) and were acclimatized in the animal facility for at least 3 days prior to use. FVB APOL1 G2 single copy (APOL1 G2sc) homozygous transgenic mice (with glutamate at amino acid 150) were generated at the Beth Israel Deaconess Medical Center (BIDMC) transgenic core facility26. The mice were re-derived to allow for housing of the lines in pathogen-free facilities. APOL1 G2sc mice were group-housed in a temperature-controlled environment (22 ± 1.5 °C, 30 to 70% relative humidity, 12 h light/dark) and were acclimatized in the animal facility for at least 3 days prior to use. Studies were conducted under a protocol that has been approved by Vertex Pharmaceuticals, Inc. Animal Care and Use Committee. Experiments were performed at the Vertex Boston site, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Prophylactic assessment of Compound 3
In APOL1 G2hom copy mice, increased proteinuria (mean urinary albumin-to-creatinine ratio, UACR) was induced by a single intraperitoneal (IP) injection of 150 µg/kg of murine IFNγ (R&D Systems, Catalog# 485-MI/CF) in a volume of 10 mL/kg. Blood (20 µL) was collected 1 h post IFNγ administration to evaluate serum IFNγ levels. Blood was allowed to clot for 30 min at room temperature and then centrifuged at 1500 relative centrifugal force (RCF) for 15 min. Serum IFNγ levels were quantified using the ProQuantum Mouse IFNγ Immunoassay (Invitrogen, Catalog# A41150) to confirm successful administration of IFNγ. Urine was collected at various time points throughout the experiment, which lasted no more than 72 h. For proteinuria analysis, urine was diluted appropriately, and urine albumin and creatinine were measured using a mouse albumin ELISA and creatinine companion kit (Bethyl Laboratories, Catalog# E99-134 and Ethos Biosciences, Catalog# 1012). Compound 3 (30 mg/kg, dosed as a crystalline suspension) or vehicle, which is composed of 1% Hydroxypropyl methylcellulose acetate succinate, type H, 0.25% of Polyvinylpyrrolidone K 30 and 2% of d-α-Tocopheryl polyethylene glycol 1000 succinate (1%HPMCAS-H/0.25%PVP-K30/2%TPGS, Shin-Etsu, MP Biomedicals and Antares manufacturers, respectively) was administered orally twice daily (bid) at 12 h interval in a volume of 10 mL/kg. Compound 3 was administered 1.5 h before IFNγ injection. UACR was calculated by dividing the average of albumin replicate values (expressed in ng/mL) to the average of creatinine replicate values (expressed to mg/dL) and then converted to the unit µg/mg. Area under the curve (AUC) was calculated per animal for UACR values from 0 to 48 h or 72 h, using GraphPad Prism, with baseline set to the mean of all animals of the same strain at baseline (day 0). Percent reduction of UACR AUC was calculated using the following equation:
((Meanvehicle - Meancompound 3)/(MeanVehicle)) * 100. Statistical analysis was performed using GraphPad Prism. UACR AUC data was log-transformed and analyzed with two tailed t-test comparing Vehicle vs IFNγ within a mouse strain or to compound 3. Statistical significance was set at p ≤ 0.05.
Therapeutic assessment of Compound 3 in plasmid chronic G2 single copy mice
Murine IFNγ cDNA was cloned in a CpG-free plasmid (Catalog #pcpgf-mcs) at Vertex laboratories26. The day before the hydro dynamic injection (HDI) individual body weight was collected. The day of the injection, CpG-free IFNγ plasmid was thawed on wet ice and diluted in sterile saline to administer a total of 0.3 μg in a total volume corresponding to 10% body weight in grams. Mice were left in their cage under a heat lamp for 2 min, transferred to a restrainer where the area of injection was cleaned with ethanol wipes. Syringes (3 mL) with a 25-gauge needle were used for the tail vein injection. The full volume was injected in a continuous motion over approximately 5–10 s. Mice were placed in a cage on a heating pad and monitored for 30 min immediately following injection.
Serum IFNγ levels were quantified using the ProQuantum Mouse IFNγ Immunoassay (Invitrogen, Catalog# A41150), to confirm successful administration of IFNγ plasmid in the chronic therapeutic mouse model. Blood (20 µL) was collected on days 1, 5, 9, 14 to evaluate serum IFNγ levels. Blood was allowed to clot for 30 min at room temperature and then centrifuged at 1500 relative centrifugal force (RCF) for 15 min. The serum fraction was collected and used to monitor IFNγ levels. For the ProQuantum Mouse IFNγ Immunoassay, the manufacturer’s protocol to quantify IFNγ was followed except for the following modifications to sample dilution: for day 1 dilution was 1:1000, for day 5 dilution was 1:50, for day 9 and 14 dilution was at 1:20. The standard curve was fit using a 4-parameter fit and sample values were interpolated using GraphPad Prism.
UACR and UACR AUCDay6-14 data were graphed as mean ± standard error of the mean (SEM). UACR AUCDay6-14 data was log-transformed and analyzed with One-way ANOVA followed by Tukey’s comparison test. Statistical significance was set at p ≤ 0.05.
Histopathology and immunohistochemistry
At the end of the study, animals were euthanized as per the IACUC-approved protocol. Euthanized animals were necropsied immediately, the left kidney was resected and fixed in 10% neutral buffered formalin (48 h), followed by transfer to 70% histology grade ethanol (Cat # R3154-1GA, Millipore Sigma) and submitted to the pathology core laboratory. Formalin-fixed kidneys were longitudinally bisected through the mid-sagittal plane to include cortex, medulla, and pelvis. Bisected kidney samples were routinely processed (Tissue-Tek tissue processing system, Sakura-Fintek), and paraffin-embedded. Serial sections from paraffin-embedded blocks were stained for a) hematoxylin and eosin (H&E), and b) periodic acid Schiff’s (PAS) using standard staining protocols as per Ventana Discovery Ultra autostainer protocols (Ventana Medical Systems, Roche). Specific protocols for APOL1 and nephrin immunohistochemistry are described below and were performed using Ventana Discovery Ultra autostainer (Ventana Medical Systems, Roche).
APOL1 and nephrin immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were cut at 3 µm thickness, dried overnight at 37 °C, and baked at 60 °C for 1 h before use. All subsequent steps were performed on the Ventana Discovery Ultra platform (Ventana Medical Systems, Roche). The protocol for nephrin and APOL1 immunohistochemistry were similar with minor differences. FFPE sections were deparaffinized at 69 °C, followed by epitope retrieval (APOL1 – 91 °C, CC2 buffer for 32 min; nephrin – 95 °C, CC1 buffer for 64 min), peroxide block for 8 min (inhibitor conditioning medium), and primary antibody incubation for 1 h (APOL1 – rabbit anti APOL1 monoclonal antibody, Cat # ab252218, 1:250, Abcam; nephrin – rabbit anti-nephrin monoclonal antibody, Cat# ab216341, 1:1000, Abcam). Signals were amplified using OmniMap anti-Rabbit HRP kit (Cat # 760-4311) for 16 min and detected using DAB chromogen (Chromomap DAB Kit; Cat #760-159) for 8 min. Slides were counterstained for 8 min (Hematoxylin II, Cat # 970-2208), followed by bluing for 4 min (Bluing reagent, Cat # 760-2037) and coverslipped. Rabbit monoclonal IgG (clone DA1E, Cat # 3900, Cell Signaling Technologies) was used as isotype control for both APOL1 and nephrin primary antibodies. All reagents unless specified otherwise were provided by the manufacturer (Ventana Medical Systems). Coverslipped and immunolabeled glass slides were scanned using Aperio AT2 whole slide scanner (Leica Biosystems) at 40x objective magnification and were visualized using image scope/web scope software (Leica Biosystems).
Immunolabeled glass slides were scanned using Aperio AT2 whole slide scanner (Leica Biosystems) at 40x objective magnification and were visualized using image scope/web scope software (Leica Biosystems). All antibodies were diluted using the antibody diluent (Cat# ADB250, Ventana Medical Systems). All reagents unless specified otherwise were provided by the manufacturer (Ventana Medical Systems).
Morphometric analysis nephrin expression in glomeruli
Immunolabelled Stained glass slides were scanned at 40x magnification (Aperio AT2, Leica Biosystems) and quality checked for any artifacts (i.e., damaged tissue, wrinkles, particles, scanning lines). Scanned whole slide images (WSI) were imported into HALO v3.4 (Indica Labs), where images were manually annotated to include the renal cortex and exclude artifacts. An Artificial Intelligence- Convolution Neural Network I - CNN (DenseNet from HALO, Indica Labs V3.2) based classifier (“Kidney_Glomerulus_APOL1 DenseNet”), was trained to automatically identify glomeruli within kidney sections. The images were manually curated to remove any abnormalities or non-glomerular structures from further analysis. Morphometric image analysis was performed using an Area Quantification (v1.0) module in HALO (“APOL1- Kidney_Glomerulus_APOL1 DenseNet”). Intensity thresholds were set to detect weak, moderate, and strong intensity of DAB chromogen (brown) staining for APOL1 and nephrin with pathologist’s guidance. All the available glomeruli detected by the algorithm in a one longitudinal section of the kidney per animal were analyzed from each sample and a cumulative output (H-score) was obtained. H-score was calculated as the following:
Statistical analysis was performed in GraphPad Prism 9 using the Kruskall-Wallis test with multiple comparisons. Graphs were generated in Microsoft Excel and Prism.
Podocyte exact morphology measurement procedure
A subset of mice was selected for a morphological assessment of the glomerular slit diaphragm. Mice were injected with a cocktail of 40 mg/mL ketamine and 2 mg/mL Xylazine. Level of anesthesia was then assessed with firm toe pinch. Once the subject was fully anesthetized, it was transferred to the working station for the perfusion. Area of incision was cleaned with an alcohol pad and tweezers were used to cut through the skin and expose the chest cavity. The diaphragm and the ribcage were cut to expose the heart. The ribcage was lifted, and a 25-gauge needle connected was inserted in the apex of the left ventricle. The right atrium was then cut, and the subject was perfused with 1X PBS/15,000-unit heparin at the rate of 20 mL/min for 3 min to washout the blood from the organs using a standard programmable infusion pump (Harvard Apparatus, Cat # 70-4500). To restrict fixation to the right kidney, the left kidney renal artery was clamped. Right kidney was perfused with 4% PFA at the rate of 20 mL/min for 5 min using an infusion pump (New Era Pump Systems, Inc. Cat # NE-9000). Right kidneys were then cut in half and left rotating at room temperature for 1 h inside a 15 mL conical tube. Sample were transferred to 4 °C before shipment to NIPOKA GmbH.
After deparaffinization and rehydration, formalin-fixed and paraffin-embedded kidney sections (3 µm) were boiled in Tris-EDTA buffer (10 mmol/l Tris, 1 mmol/l EDTA, pH 9) in a pressure cooker for 5 min, followed by the blocking step (1% FBS, 1% BSA, 0.1% fish gelatin, 1% normal goat serum) for 1 h. The following primary antibodies were incubated overnight at 4 °C: rabbit anti‐podocin 1:150 (IBL International, Hamburg, Germany, JP-29040) and mouse anti‐integrin alpha3 1:500 (Santacruz, Dallas, Texas, USA, sc-374242). After three washing steps in PBS, secondary antibodies were incubated for 1 h at room temperature (anti-rabbit Alexa Fluor 488‐conjugated IgG 1:600 (ChromoTek, Planegg, Germany, srbAF488-1-100) and anti-mouse Cy3‐conjugated IgG antibody 1:600 (Jackson ImmunoResearch, West Grove, PA, 115-166-006) for 1 h at room temperature. DAPI (1:100) was added to the slides for 5 min, followed by a washing step in PBS. Finally, the slides were incubated in H2O and mounted in Mowiol (Carl Roth, Karlsruhe, Germany) using high precision coverslips (Paul Marienfeld, Lauda-Königshofen, Germany). The evaluation of the filtration slit density (FSD) was performed using a recently established super-resolution microscopy-based methodology termed podocyte exact morphology measurement procedure (PEMP)33. For three-dimensional structured illumination microscopy (3D-SIM), z-stacks of 19 planes of both channels (488 and 561 nm) were acquired from the stained kidney section using an N-SIM super-resolution microscope (Nikon, Tokyo, Japan) equipped with a 100x silicone objective. The images were reconstructed into 3D-SIM images using NIS-Elements AR 5.30 (Nikon, Tokyo, Japan). The z-stacks were converted into a maximum intensity projection followed by the automatized identification of podocyte foot process areas that were selected based on the foot process positive antibody. Inside these areas, the filtration slit diaphragm length was automatically determined. The FSD was expressed as the ratio of the total filtration slit diaphragm length per podocyte foot process area. The FSD of 20 randomly selected glomeruli was determined for every animal.
Synthesis and characterization of chemical compounds
The chemical compounds (2 – 3, VX-147) in this manuscript were prepared according to the synthetic procedures detailed below. Purity assessment for final compounds was based on analytical UPLC: 2.1 mm × 50 mm Waters CSH C18 column, 1.7 μM, 130 Å. Mobile phases were as follows: A, H2O with 0.1% trifluoroacetic acid; B, acetonitrile with 0.1% trifluoroacetic acid; gradient, 10–60% B in 0.6 min with a 1.4 min run time. The flow rate was 0.6 mL/min. All compounds were ≥ 95% pure. Mass samples were analyzed on a Waters 3100 single quad mass spectrometer operated in positive MS mode with electrospray ionization. The mobile phase for all mass analysis consisted of acetonitrile–water mixtures with either 0.1% trifluoroacetic acid or ammonium formate. 1H NMR, 13C NMR, and 19F NMR spectra were recorded using either a Bruker Avance 400 (400 MHz) or a Bruker Avance II-300 (300 MHz) instrument. The screening hit, 1, is commercially available and was purchased from an external vendor. Compound 1 has a Chemical Abstracts Service Registry Number of 687574-42-1.
Synthetic route to (3S,4 R)-3-amino-4-hydroxypyrrolidin-2-one (S5)
Synthesis of Methyl (2S,3 R)-2,4-dibromo-3-hydroxy-butanoate (S1)
Potassium (2 R,3 R)-2,3,4-trihydroxybutanoate (280 g) was stirred with a 33% solution of HBr in acetic acid (1 L) at ambient temperature for 24 h. The reaction mixture was then poured into MeOH (5 L). The mixture was stirred at room temperature for 8 h, then at 65 °C for 4 h. The mixture was concentrated, and the crude residue was dissolved in MeOH (1.2 L) and then concentrated sulfuric acid (30 mL) was slowly added. The mixture was heated under reflux for 6 h, then concentrated. The crude residue was taken up with EtOAc (400 mL). The resulting solution was washed with water (250 mL), dried over Na2SO4, filtered, and concentrated to provide the product methyl (2S,3 R)-2,4-dibromo-3-hydroxy-butanoate as an oil which solidified upon storage at 4 °C. (375 g, 74%). 1H NMR (400 MHz, CDCl3) δ 4.71 (d, J = 3.4 Hz, 1H), 4.17-4.14 (m, 1H), 3.82 (s, 3H), 3.53-3.44 (m, 2H).
Synthesis of Methyl (2 R,3S)-3-(bromomethyl)oxirane-2-carboxylate (S2)
To a solution of methyl (2S,3 R)-2,4-dibromo-3-hydroxy-butanoate (200 g, 0.73 mol) in acetone (2.0 L) was added anhydrous K2CO3 (151.1 g, 1.1 mol) while the reaction temperature was maintained at 0 °C. The reaction was stirred at 0 °C for 2 h, then gradually warmed to ambient temperature over 4 h. The reaction mixture was filtered, and the filtrate was concentrated. The residue was distilled under vacuum 75–80 °C/200-300 Pa to provide the product as a colorless liquid (105 g, 74%). 1H NMR (300 MHz, CDCl3) δ 3.83 (s, 3H), 3.71–3.61 (m, 2H), 3.61–3.53 (m, 1H), 3.46 (dd, J = 9.9, 6.6 Hz, 1H) ppm. 13C NMR (75 MHz, CDCl3) δ 167.58, 55.89, 53.52, 52.77, 26.83 ppm.
Synthesis of Methyl (2 R,3 R)-3-(azidomethyl)oxirane-2-carboxylate (S3)
Methyl (2 R,3S)-3-(bromomethyl)oxirane-2-carboxylate (52.6 g, 269.7 mmol) was dissolved in DMF (500 mL) in a 3 L round-bottomed flask equipped with a magnetic stir bar. NaN3 (25.3 g, 388.4 mmol) was added, and the mixture was stirred at room temperature for 1 h. The reaction mixture was poured into water and extracted with EtOAc. The extract was washed with water, dried over MgSO4, and concentrated to afford a dark red oil. The oil residue was dissolved in CH2Cl2 and filtered over a plug of silica gel, eluting with CH2Cl2. The filtrate was concentrated to afford the product as a clear, light red oil (40.8 g, 96%). 1H NMR (300 MHz, CDCl3) δ 3.87-3.74 (m, 3H), 3.67-3.55 (m, 2H), 3.47 (dd, J = 13.3, 5.1 Hz, 1H), 3.38 (ddd, J = 6.3, 5.0, 4.4 Hz, 1H). 13C NMR (75 MHz, CDCl3) δ 167.76, 54.81, 52.67, 51.32, 48.74.
Synthesis of (1 R,5 R)-6-oxa-3-azabicyclo[3.1.0]hexan-2-one (S4)
A 2 L 3-neck flask with overhead stirrer was charged with methyl (2 R,3 R)-3-(azidomethyl)oxirane-2-carboxylate (67 g, 402.5 mmol) in toluene (500 mL), stirred for 10 min, and then warmed to 80 °C. Bu3SnH (220 mL, 817.8 mmol) and AIBN (2 g, 12.2 mmol) were dissolved in toluene (500 mL) and then added to the reaction over 3 h using an addition funnel. The resulting reaction mixture was stirred at 80–87 °C for 1 h, then cooled to ambient temperature, and concentrated under reduced pressure. The residue was partitioned between acetonitrile (2 L) and pentane (1 L), stirred for 10 min, and then the acetonitrile phase (bottom) was separated. The acetonitrile phase was washed with pentane (2 x 500 mL) and concentrated in vacuo to afford a light-yellow solid. The solid residue was triturated with pentane (200 mL) to afford the product as a yellow solid which was used without further purification (52 g, 98%). 1H NMR (300 MHz, CDCl3) δ 5.89 (s, 1H), 4.00 (q, J = 2.5 Hz, 1H), 3.74–3.50 (m, 2H), 3.44 (dd, J = 12.4, 2.4 Hz, 1H). 13C NMR (75 MHz, CDCl3) δ 173.24, 53.28, 52.18, 44.00.
Synthesis of (3S,4 R)-3-amino-4-hydroxypyrrolidin-2-one (S5)
A Parr vessel containing (1 R,5 R)-6-oxa-3-azabicyclo[3.1.0]hexan-2-one (60 g, 605.5 mmol) and NH3 (1.5 L, 58.6 mol) was pressurized to 200 psi and allowed to stir at 18 °C for 2 days. NH3 was released from the vessel to provide a grey solid. Heptanes was added and the mixture stirred for 30 min. The solid was filtered, and then the filter cake was isolated, and EtOAc and heptanes were used to dissolve the solid. The mixture was concentrated in vacuo to afford the product (55 g, 78%). 1H NMR (300 MHz, D2O) δ 4.13 (q, J = 7.2 Hz, 1H), 3.53 (dd, J = 10.4, 7.4 Hz, 1H), 3.36 (d, J = 7.5 Hz, 1H), 3.05 (dd, J = 10.4, 6.8 Hz, 1H).
Synthetic route to Compound 2
Synthesis of 4-fluoro-2-[2-(4-fluorophenyl)ethynyl]-6-methyl-aniline (S6)
To a stirred solution of 2-bromo-4-fluoro-6-methyl-aniline hydrogen bromide salt (25 g, 0.09 mol) and 1-ethynyl-4-fluoro-benzene (26.1 g, 0.22 mol) in toluene (500 mL) was added triethylamine (500 mL) at room temperature. The reaction was degassed for 20 min with nitrogen. To the reaction mixture was added copper iodide (1.67 g, 0.008 mol) and tetrakis(triphenylphosphine)palladium(0) (4.97 g, 0.004 mol), and the reaction was degassed for another 5 min. The reaction mixture was then stirred at 100 °C for 16 h. The reaction mixture was filtered through a pad of celite and washed with EtOAc (50 mL), then the filtrate was concentrated. The crude product was purified by flash column chromatography column using silica gel eluted with 3% EtOAc in heptanes, and concentrated to afford 4-fluoro-2-[2-(4-fluorophenyl)ethynyl]-6-methyl-aniline (12 g, 46%) as dark brown semi-solid. 1H NMR (400 MHz, CDCl3) δ: 7.48-7.52 (m, 2H), 7.02-7.07 (m, 2H), 6.80-6.94 (m, 2H), 4.11 (s, 2H), 2.18 (s, 3H). ESI-MS m/z calc. 243.09, found 244.06 (M + 1)+.
Synthesis of 5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indole (S7)
A solution of 4-fluoro-2-[2-(4-fluorophenyl)ethynyl]-6-methyl-aniline (6.09 g, 24.3 mmol) and palladium(II) chloride (434.9 mg, 2.43 mmol) was prepared in acetonitrile (243 mL) under argon, and stirred at 80 °C for 3 h. The reaction mixture was concentrated, and the crude product was purified by flash column chromatography column using silica gel eluted with 5% ethyl acetate in heptanes, then concentrated to afford 5-fluoro-2-(4- fluorophenyl)-7-methyl-1H-indole (4 g, 66%) as an off-white solid. 1H NMR (300 MHz, DMSO-d6) δ 11.16 (s, 1H), 8.01 – 7.91 (m, 2H), 7.37 – 7.27 (m, 2H), 7.09 (dd, J = 9.7, 2.5 Hz, 1H), 6.84 (d, J = 2.1 Hz, 1H), 6.76 (dd, J = 10.4, 2.4 Hz, 1H), 2.53 (s, 3H). ESI-MS m/z calc. 243.1, found 244.2 (M + 1)+.
Synthesis of Methyl 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]propanoate (S8)
To 5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indole (502 mg, 2.06 mmol) in 10 mL toluene was added methanesulfonic acid (208 μL, 3.21 mmol) and triethylsilane (1.1 mL, 6.89 mmol), followed by methyl 3,3-dimethoxypropanoate (370 μL, 2.61 mmol). The reaction mixture was heated to 70 °C for 2 h. The reaction was quenched with water, extracted with ethyl acetate, and concentrated. The crude residue was dissolved in methanol and isopropanol (1:1, 30 mL), and 50 mg of 5% Pd/C added. The reaction mixture was hydrogenated at 50 psi for 4 h. The reaction mixture was filtered through celite, concentrated, and the crude residue purified by flash column chromatography column using silica gel eluted with 0-25% EtOAc/heptanes gradient to afford methyl 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H- indol-3-yl]propanoate (478 mg, 70%) as a white solid. 1H NMR (300 MHz, CDCl3) δ 7.86 (s, 1H), 7.63 - 7.45 (m, 2H), 7.25 - 7.15 (m, 2H), 7.12 (dd, J = 9.5, 2.4 Hz, 1H), 6.81 (ddd, J = 9.9, 2.4, 0.9 Hz, 1H), 3.65 (s, 3H), 3.22 - 3.07 (m, 2H), 2.74 - 2.57 (m, 2H), 2.51 (d, J = 0.8 Hz, 3H). ESI-MS m/z calc. 329.12, found 330.27 (M + 1)+.
Synthesis of 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]propanoic acid (S9)
To a solution of methyl 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]propanoate (475 mg, 1.43 mmol) and LiOH (344 mg, 14.36 mmol) in 4 mL MeOH, was added 4 mL THF and 2 mL water. The reaction was stirred at 60 °C for 16 h. The reaction mixture was concentrated, then CH2Cl2 (60 mL) and water (40 mL) added, followed by acidification with aqueous HCl to pH = 1. The organic layer was washed with saturated aqueous NaCl and concentrated to provide 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]propanoic acid (451 mg, 90%) 1H NMR (300 MHz, CDCl3) δ 7.88 (s, 1H), 7.63 - 7.44 (m, 2H), 7.26 - 7.02 (m, 4H), 6.82 (ddd, J = 9.8, 2.4, 0.9 Hz, 1H), 3.22 - 3.10 (m, 2H), 2.75 - 2.63 (m, 2H), 2.51 (d, J = 0.7 Hz, 3H). ESI-MS m/z calc. 315.11, found 316.23 (M + 1)+.
Synthesis of 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]-N-[(3S)-2-oxopyrrolidin-3-yl]propanamide (Compound 2)
To a solution of 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]propanoic acid, (19 mg, 0.05 mmol), (3S)-3-aminopyrrolidin-2-one (8 mg, 0.08 mmol) and HATU (28 mg, 0.07 mmol) in 1 mL DMSO was added triethylamine (30 μL, 0.22 mmol). The reaction mixture was stirred for 12 h at ambient temperature. The reaction mixture was filtered, then directly purified by reverse phase chromatography (C18 ACN/Water with TFA modifier) to afford the gave product 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]-N-[(3S)-2-oxopyrrolidin-3-yl]propanamide (16 mg, 74%) as a white solid. 1H NMR (300 MHz, MeOD) δ 7.71 – 7.58 (m, 2H), 7.21 (t, J = 8.8 Hz, 2H), 7.13 (dd, J = 9.8, 2.4 Hz, 1H), 6.70 (dd, J = 10.2, 2.4 Hz, 1H), 4.45 (dd, J = 10.3, 8.7 Hz, 1H), 3.31 (m, 2H *obscured by MeOD solvent peak), 3.11 (t, J = 8.0 Hz, 2H), 2.64 – 2.52 (m, 2H), 2.49 (s, 3H), 2.44 – 2.31 (m, 1H), 1.84 (dq, J = 12.4, 9.4 Hz, 1H). 19F NMR (282 MHz, MeOD) δ -116.62, -127.29. 13C NMR (75 MHz, MeOD) δ 177.68, 175.81, 165.40, 162.14, 160.63, 157.55, 137.01, 133.65, 131.63, 131.53, 130.87, 130.82, 129.77, 129.64, 123.42, 123.30, 116.59, 116.30, 112.72, 112.65, 111.57, 111.23, 102.00, 101.69, 51.63, 40.02, 38.26, 29.34, 22.06, 17.00, 16.98. ESI-MS m/z calc. 397.16, found 398.18 (M + 1)+. Optical rotation: [α] D 19.6 = −21.05 (c = 1.0, 15 mg in 1 mL of MeOH).
Synthetic route to Compound 3
Synthesis of 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]-N-[(3S,4 R)-4-hydroxy-2-oxo-pyrrolidin-3-yl]propanamide (Compound 3)
To a solution of 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]propanoic acid (6.06 g, 16.34 mmol) and (3S,4 R)-3-amino-4-hydroxy-pyrrolidin-2-one (3.25 g, 21.30 mmol) in DMF (41 mL) was added 2-Chloro-4,6-dimethoxy-1,3,5-triazine (3.7 g, 21.07 mmol) and N-methylmorpholine (7.2 mL, 65.49 mmol). The reaction mixture was stirred for 16 h at ambient temperature. The reaction was quenched with water, extracted with EtOAc, washed with brine, dried over anhydrous MgSO4, and concentrated in vacuo. The crude product was recrystallized from 10% MeOH in water to provide 3-[5-fluoro-2-(4-fluorophenyl)-7-methyl-1H-indol-3-yl]-N-[(3S,4 R)-4-hydroxy-2-oxo-pyrrolidin-3-yl]propanamide (4.32 g, 62%) as a white solid. 1H NMR (300 MHz, MeOD) δ 7.69 – 7.59 (m, 2H), 7.27 – 7.17 (m, 2H), 7.18 – 7.11 (m, 1H), 6.71 (ddd, J = 10.1, 2.5, 1.0 Hz, 1H), 4.35 (td, J = 7.6, 6.8 Hz, 1H), 4.21 (d, J = 7.7 Hz, 1H), 3.56 (dd, J = 9.9, 7.6 Hz, 1H), 3.18 – 3.05 (m, 3H), 2.65 – 2.54 (m, 2H), 2.50 (s, 3H). 19F NMR (282 MHz, MeOD) δ -116.51, -127.10. 13C NMR (75 MHz, MeOD) δ 176.17, 174.97, 165.35, 162.09, 160.62, 157.54, 136.94, 133.65, 131.59, 131.48, 130.81, 130.76, 129.69, 129.55, 123.46, 123.34, 116.58, 116.29, 112.63, 112.57, 111.60, 111.25, 101.85, 101.54, 73.47, 60.00, 47.71, 38.02, 21.93, 17.01, 16.99. ESI-MS m/z calc. 413.16, found 414.01 (M + 1)+. Optical rotation: [α] D 19.6 = −40.80 (c = 1.0, 100 mg in 10 mL of MeOH).
Synthetic route to VX-147
Synthesis of 2,4-difluoro-6-[2-(4-fluorophenyl)ethynyl]aniline (S10)
To a flask containing 2,4-difluoro-6-iodo-aniline (134 g, 525.5 mmol) was added Et3N (1.3 L), followed by DMF (250 mL), 1-ethynyl-4-fluoro-benzene (83.5 g, 695.1 mmol), CuI (20.5 g, 107.6 mmol), and PdCl2(PPh3)2 (25 g, 35.6 mmol). The mixture was allowed to stir at room temperature for 2 h. Solvent was removed under reduced pressure and water (500 mL) was added. The mixture was extracted with ethyl acetate, filtered and concentrated. The product mixture was filtered through a silica gel plug (Eluent: CH2Cl2), followed by a second silica plug filtration (Eluent: 30-40% EtOAc in Heptane). The crude product was purified by flash column chromatography column using silica gel eluted with a gradient of 0-20% EtOAc in heptane to afford the product 2,4-difluoro-6-[2-(4-fluorophenyl)ethynyl]aniline as a pale-yellow solid. (87 g, 60%). 1H NMR (300 MHz, CDCl3) δ 7.58 - 7.45 (m, 2H), 7.14 - 7.02 (m, 2H), 6.92 (ddd, J = 8.8, 2.8, 1.7 Hz, 1H), 6.87 - 6.71 (m, 1H), 4.15 (s, 2H). ESI-MS m/z calc. 247.1, found 248.0 (M + 1)+.
Synthesis of 5,7-difluoro-2-(4-fluorophenyl)-1H-indole (S11)
To a solution of 2,4-difluoro-6-[2-(4-fluorophenyl)ethynyl]aniline (46 g, 167.5 mmol) in DMF (600 mL) was added CuI (1.9 g, 10.0 mmol) and the reaction was heated at reflux. Water (800 mL) was added and the mixture extracted with MTBE. The mixture was then washed with sat. NaCl solution, dried over Na2SO4 and then concentrated in vacuo to afford the product, which was used in subsequent steps without further purification (41 g, 87%). 1H NMR (300 MHz, CDCl3) δ 8.43 (s, 1H), 7.72 - 7.58 (m, 2H), 7.27 - 7.15 (m, 2H), 7.09 (dd, J = 9.0, 2.1 Hz, 1H), 6.85 - 6.63 (m, 2H). ESI-MS m/z calc. 247.1, found 248.0 (M + 1)+.
Synthesis of methyl (E)-3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]prop-2-enoate (S12)
A 12 L flask with overhead stirrer was charged with 5,7-difluoro-2-(4-fluorophenyl)-1H-indole (300 g, 1.2 mol), CH2Cl2 (3 L), methyl 3,3-dimethoxypropanoate (195 mL, 1.4 mol) and TFA (300 mL, 3.9 mol). The reaction was heated to reflux for 4 h. Additional CH2Cl2 was added to facilitate stirring. Upon cooling to room temperature, the solid product was filtered, washed with minimal CH2CI2 and dried to afford the product methyl (E)-3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]prop-2-enoate (388 g, 96%). 1H NMR (400 MHz, DMSO-d6) δ 12.66 (s, 1H), 7.77 - 7.57 (m, 4H), 7.56 - 7.37 (m, 2H), 7.19 (ddd, J = 11.0, 9.7, 2.1 Hz, 1H), 6.47 (d, J = 16.1 Hz, 1H), 3.69 (s, 3H). ESI-MS m/z calc. 331.1, found 332.4 (M + 1)+.
Synthesis of methyl 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]propanoate (S13)
To a suspension of methyl (E)-3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]prop-2-enoate (80 g, 236.5 mmol) in EtOH (1.5 L) under a nitrogen atmosphere was added Pd(OH)2 (6 g of 20 % w/w, 8.5 mmol) and ammonium formate (160 g, 2.5 mol). The mixture was heated at reflux for 3 h, then filtered to remove catalyst. The filtrate was concentrated in vacuo to afford the product methyl 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3- yl]propanoate as an off-white solid, which was used without further purification (82 g, 100%). 1H NMR (300 MHz, CDCl3) δ 8.18 (s, 1H), 7.65 - 7.47 (m, 2H), 7.27 - 7.14 (m, 2H), 7.14 - 7.00 (m, 1H), 6.76 (ddd, J = 10.8, 9.4, 2.2 Hz, 1H), 3.65 (s, 3H), 3.27 - 3.04 (m, 2H), 2.75 - 2.49 (m, 2H). ESI-MS m/z calc. 333.1, found 334.3 (M + 1)+.
Synthesis of 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]propanoic acid (S14)
LiOH (67 g, 2.8 mol) was added to a solution of methyl 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]propanoate (217 g, 651.1 mmol) in THF (1 L) and water (100 mL). The mixture was heated at reflux for 2 h, and then allowed to cool overnight. THF was removed by concentration under reduced pressure, and water was added (approx. 1 L). The mixture was cooled on an ice bath and HCl (250 mL of 11.7 M, 2.9 mol) was added to adjust to pH = 4. EtOAc (300 mL) was added, and the aqueous layer extracted with further EtOAc (100 mL). The combined organic extracts were dried over Na2SO4, and filtered through a plug of silica gel, rinsing with EtOAc. The filtrate was concentrated in vacuo to afford an orange oil (50-75 mL). Heptanes (50 mL) were added and the mixture chilled on dry ice. Upon agitation, a crystalline solid formed. The mixture was allowed to stir on an ice-bath until to allow completion of the crystallization process. The solid was filtered, washed with heptane and air dried to afford the product 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]propanoic acid (208 g, 96%). 1H NMR (300 MHz, CDCl3) δ 8.15 (s, 1H), 7.60 - 7.46 (m, 2H), 7.27 - 7.15 (m, 2H), 7.09 (dd, J = 9.1, 2.2 Hz, 1H), 6.77 (ddd, J = 10.8, 9.4, 2.2 Hz, 1H), 3.26 - 3.05 (m, 2H), 2.78 - 2.57 (m, 2H). ESI-MS m/z calc. 319.1, found 320.0 (M + 1)+.
Synthesis of 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]-N-[(3S,4 R)-4-hydroxy-2-oxo-pyrrolidin-3-yl] propanamide (VX-147)
A 2 L 3-neck RB flask with magnetic stirrer, temperature probe and nitrogen inlet was charged with 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]propanoic acid (90.5 g, 283.5 mmol) and (3S,4 R)-3-amino-4-hydroxy-pyrrolidin-2-one (39.9 g, 343.6 mmol) in DMF (1.65 L), and stirred for 15 min. CDMT (61.1 g, 348 mmol) was added. The mixture was then cooled to 2 °C in an ice bath. N-methylmorpholine was added (131 mL, 1.2 mol) dropwise over 20 min and the mixture was heated at 30 °C for 16 h. The reaction mixture was added into 4.5 L of ice water, and extracted with EtOAc (1.2 L x 4). The combined organic layers were washed with 1.2 L of 1 M HCl (x 3) and then water (1.2 L) and brine (1.2 L). The combined organic layers were dried over Na2SO4, filtered and concentrated. The mixture was washed through a silica gel plug (1.8 L of silica gel), first eluting with 25% EtOAc in dichloromethane (8 L) to remove impurities, followed by hot EtOAc (8 L), to elute the product. The EtOAc filtrate was concentrated in vacuo. TBME was then added (400 mL), and the mixture allowed to stir for overnight. Filtration of the resulting solid afforded the product 3-[5,7-difluoro-2-(4-fluorophenyl)-1H-indol-3-yl]-N-[(3S,4 R)-4-hydroxy-2-oxo-pyrrolidin-3-yl] propenamide as a white solid. 62 g, 52%). 1H NMR (300 MHz, MeOD) δ 7.70 – 7.58 (m, 2H), 7.29 – 7.13 (m, 3H), 6.73 (ddd, J = 11.1, 9.6, 2.2 Hz, 1H), 4.34 (q, J = 7.5 Hz, 1H), 4.22 (d, J = 7.8 Hz, 1H), 3.57 (dd, J = 9.9, 7.5 Hz, 1H), 3.20 – 3.04 (m, 3H), 2.65 – 2.53 (m, 2H). 19F NMR (282 MHz, MeOD) δ -115.93, -124.40, -133.12. 13C NMR (75 MHz, MeOD) δ 174.55, 173.56, 164.21, 160.94, 158.22, 158.09, 155.11, 154.98, 150.15, 147.08, 146.89, 136.72, 131.34, 131.25, 131.20, 131.10, 130.21, 130.10, 128.74, 128.69, 120.93, 120.76, 115.40, 115.11, 111.75, 99.18, 99.13, 98.87, 98.82, 96.37, 96.09, 95.97, 95.69, 72.07, 58.58, 46.32, 36.40, 20.41. ESI-MS m/z calc. 417.13, found 418.20 (M + 1)+. Optical rotation: [α] D 19.6 = −13.46 (c = 1.0, 100 mg in 10 mL of MeOH).
Small Molecule X-ray Structure of VX-147
The single crystal X-ray diffraction studies were carried out on a Bruker Kappa diffractometer with a CPAD PHOTON II detector, utilizing Cu Kα radiation (λ = 1.54178(1)). A 0.380 × 0.128 × 0.050 mm colorless blade was mounted on a MiTeGen kapton Micromount with Paratone oil. Crystals were grown via evaporation of VX-147 from 1:1 dixoane:water at 40 °C. Data were collected at low temperature (100 K), under a nitrogen cold stream generated by an Oxford Cryosystems Cryostream 800 Plus, using ϖ and φ scans. Crystal-to-detector distance was 50 mm using variable exposure times depending on θ with a scan width of 1.0°. Data collection was 100% complete to 67.679° in θ. A total of 60490 reflections were collected covering the indices, −5 < =h < =5, −22 < =k < =22, −16 < =l < =16. 4443 reflections were found to be symmetry independent, with a Rint of 0.0402. Indexing and unit cell refinement indicated a primitive, monoclinic lattice, containing one molecule of API and one molecule of dioxane in the asymmetric unit. The space group was found to be P21. The absolute structure was determined to be correctly assigned by anomalous dispersion with a refined flack parameter of -0.06(4). The Bijvoet analysis conducted using PLATON produced Bayesian Statistics consistent with the correct assignment of absolute structure, P2(true) = 1.000 P3(true) = 1.000 P3(rac-twin) = 0.1 × 10−43 P3(false) = 0.3 × 10−161 with a final Hooft parameter of −0.04(4). The data were integrated using the Bruker SAINT software program and scaled using the SADABS software program. Solution by intrinsic phasing (SHELXT) produced a complete phasing model consistent with the proposed structure.
All nonhydrogen atoms were refined anisotropically by full-matrix least-squares (SHELXL-2014). All carbon bonded hydrogen atoms were placed using a riding model. Their positions were constrained relative to their parent atom using the appropriate HFIX command in SHELXL-2014. Crystallographic data are summarized in Supplementary Table 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
Thank you to all the APOL1 project team members, past and present, who helped further our understanding around APOL1 biology and contributed to the Vertex APOL1 program. We would also like to thank Dr. David Altshuler for advice on the project and comments on the manuscript.
Author contributions
Conceptualization: B.Z., L.A.D., A.F., T.S., B.F., M.E.B.; Methodology: B.Z., L.A.D., A.F., E.N., A.B., N.H., S.K., J.P., T.S., B.F.; Investigation: B.Z., L.A.D., E.N., A.B., J.M., H.M., M.F., T.W., S.N., G.M., C.M., P.M., G.W., P.S., K.D., H.S., K.S., F.B., A.P., K.J.G., F.L., S.P., S.K.P., L.H., M.H., F.D., O.N., N.H., S.K., J.P., T.S.; Writing – original draft: B.Z., E.N., B.F., M.E.B.; Writing – review & editing: B.Z., L.A.D., E.N., J.P., B.F., M.E.B.
Peer review
Peer review information
Nature Communications thanks Alessia Fornoni, who co-reviewed with Matthew Tolerico; Katalin Susztak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. CIF file and crystallographic details are available in the supplementary information and have been deposited at the CCDC; deposition number: CCDC-2367433. Source data are provided with this paper.
Competing interests
The authors declare the following competing financial interest(s): All authors listed are current or past employees of Vertex Pharmaceuticals, Incorporated and may hold stock in the company. Compound 2, Compound 3 and VX-147 are disclosed and claimed in granted U.S. Patent No. 11,618,746 B2, among other places, where Vertex Pharmaceuticals Incorporated is the applicant. The inventors on the patent are as follows: Jingrong Cao, Jon H. Come, Leslie A. Dakin, Francois Denis, Warren A. Dorsch, Anne Fortier, Martine Hamel, Elaine B. Krueger, Brian Ledford, Suganthini S. Nanthakumar, Olivier Nicolas, Camil Sayegh, Timothy J. Senter, Tiansheng Wang, Michael Brodney, Kan-Nian Hu, Peter Rose, Kevin Gagnon, Yi Shi, Muna Shrestha, Ales Medek, Faith Witkos. The aspects covered in the manuscript that are found in the patent application are Compound 2, Compound 3 and VX-147, in addition to the thallium flux and trypanosome rescue assay and their use in discovering these inhibitors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Brandon Zimmerman, Leslie A. Dakin.
Contributor Information
Brandon Zimmerman, Email: brandon_zimmerman@vrtx.com.
Mark E. Bunnage, Email: mark_bunnage@vrtx.com
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55408-2.
References
- 1.Friedman, D. J. & Pollak, M. R. APOL1 Nephropathy: From Genetics to Clinical Applications. Clin. J. Am. Soc. Nephrol.16, 294–303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipkowitz, M. S. et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int83, 114–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp, J. B. et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J. Am. Soc. Nephrol.22, 2129–2137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsa, A. et al. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med.369, 2183–2196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D. P. et al. Kidney Disease Progression in Membranous Nephropathy among Black Participants with High-Risk APOL1 Genotype. Clin. J. Am. Soc. Nephrol.18, 337–343 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genovese, G. et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science329, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langefeld, C. D. et al. Genome-wide association studies suggest that APOL1-environment interactions more likely trigger kidney disease in African Americans with nondiabetic nephropathy than strong APOL1-second gene interactions. Kidney Int.94, 599–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husain, S. A. & Chang, J. H. Searching for Second Hits for the Development of APOL1-Associated Kidney Disease. Kidney Int. Rep.4, 911–913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanhamme, L. et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature422, 83–87 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Perez-Morga, D. et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science309, 469–472 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Thomson, R. et al. Evolution of the primate trypanolytic factor APOL1. Proc. Natl Acad. Sci. USA111, E2130–E2139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duchateau, P. N., Pullinger, C. R., Cho, M. H., Eng, C. & Kane, J. P. Apolipoprotein L gene family: tissue-specific expression, splicing, promoter regions; discovery of a new gene. J. Lipid Res.42, 620–630 (2001). [PubMed] [Google Scholar]
- 13.Bazzi, C., Bakoush, O. & Gesualdo, L. Proteinuria: from molecular to clinical applications in glomerulonephritis. Int J. Nephrol.2012, 424968 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckerman, P. et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat. Med.23, 429–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichols, B. et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int.87, 332–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson, R. & Finkelstein, A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: relevance to trypanosome lysis. Proc. Natl Acad. Sci. USA112, 2894–2899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaub, C. et al. Cation channel conductance and pH gating of the innate immunity factor APOL1 are governed by pore-lining residues within the C-terminal domain. J. Biol. Chem.295, 13138–13149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta, N. et al. Apolipoprotein L1 (APOL1) renal risk variant-mediated podocyte cytotoxicity depends on African haplotype and surface expression. Sci Rep.14, 3765 (2024). [DOI] [PMC free article] [PubMed]
- 19.Ma, L. et al. APOL1 Renal-Risk Variants Induce Mitochondrial Dysfunction. J. Am. Soc. Nephrol.28, 1093–1105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granado, D. et al. Intracellular APOL1 Risk Variants Cause Cytotoxicity Accompanied by Energy Depletion. J. Am. Soc. Nephrol.28, 3227–3238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olabisi, O. A. et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc. Natl Acad. Sci. USA113, 830–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanhollebeke, B. et al. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-I. N. Engl. J. Med.355, 2752–2756 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Lugli, E. B., Pouliot, M., Portela Mdel, P., Loomis, M. R. & Raper, J. Characterization of primate trypanosome lytic factors. Mol. Biochem. Parasitol.138, 9–20 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Seed, J. R., Sechelski, J. B. & Loomis, M. R. A survey for a trypanocidal factor in primate sera. J. Protozool.37, 393–400 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Egbuna, O. et al. Inaxaplin for Proteinuric Kidney Disease in Persons with Two APOL1 Variants. N. Engl. J. Med388, 969–979 (2023). [DOI] [PubMed] [Google Scholar]
- 26.McCarthy, G. M. et al. Recessive, gain-of-function toxicity in an APOL1 BAC transgenic mouse model mirrors human APOL1 kidney disease. Dis. Model Mech.14, 10.1242/dmm.048952 (2021). [DOI] [PMC free article] [PubMed]
- 27.Vandorpe, D. H. et al. Apolipoprotein L1 (APOL1) cation current in HEK-293 cells and in human podocytes. Pflug. Arch.475, 323–341 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Toole, J. F. et al. ApoL1 Overexpression Drives Variant-Independent Cytotoxicity. J. Am. Soc. Nephrol.29, 869–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, V. & Singhal, P. C. APOL1 and kidney cell function. Am. J. Physiol. Ren. Physiol.317, F463–F477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruotsalainen, V. et al. Nephrin is specifically located at the slit diaphragm of glomerular podocytes. Proc. Natl Acad. Sci. USA96, 7962–7967 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akatsuka, A., Horai, Y. & Akatsuka, A. Automated recognition of glomerular lesions in the kidneys of mice by using deep learning. J. Pathol. Inf.13, 100129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouteldja, N. et al. Deep Learning-Based Segmentation and Quantification in Experimental Kidney Histopathology. J. Am. Soc. Nephrol.32, 52–68 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegerist, F. et al. Structured illumination microscopy and automatized image processing as a rapid diagnostic tool for podocyte effacement. Sci. Rep.7, 11473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giovinazzo, J. A. et al. Apolipoprotein L-1 renal risk variants form active channels at the plasma membrane driving cytotoxicity. Elife9, 10.7554/eLife.51185 (2020). [DOI] [PMC free article] [PubMed]
- 35.Datta, S. et al. APOL1-mediated monovalent cation transport contributes to APOL1-mediated podocytopathy in kidney disease. J. Clin. Invest.10.1172/jci172262 (2024). [DOI] [PMC free article] [PubMed]
- 36.Hung, A. M. et al. Genetic Inhibition of APOL1 Pore-Forming Function Prevents APOL1-Mediated Kidney Disease. J. Am. Soc. Nephrol.10.1681/ASN.0000000000000219 (2023). [DOI] [PMC free article] [PubMed]
- 37.Rampersad, S. N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sens. (Basel)12, 12347–12360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information. CIF file and crystallographic details are available in the supplementary information and have been deposited at the CCDC; deposition number: CCDC-2367433. Source data are provided with this paper.