Abstract
Fluid shear stress (FSS) from blood flow sensed by vascular endothelial cells (ECs) determines vessel behavior, but regulatory mechanisms are only partially understood. We used cell state transition assessment and regulation (cSTAR), a powerful computational method, to elucidate EC transcriptomic states under low shear stress (LSS), physiological shear stress (PSS), high shear stress (HSS), and oscillatory shear stress (OSS) that induce vessel inward remodeling, stabilization, outward remodeling, or disease susceptibility, respectively. Combined with a publicly available database on EC transcriptomic responses to drug treatments, this approach inferred a regulatory network controlling EC states and made several notable predictions. Particularly, inhibiting cell cycle–dependent kinase (CDK) 2 was predicted to initiate inward remodeling and promote atherogenesis. In vitro, PSS activated CDK2 and induced late G1 cell cycle arrest. In mice, EC deletion of CDK2 triggered inward artery remodeling, pulmonary and systemic hypertension, and accelerated atherosclerosis. These results validate use of cSTAR and identify key determinants of normal and pathological artery remodeling.
Computational analysis of EC transcriptomic data revealed cell cycle as a key regulator of flow-dependent artery remodeling.
INTRODUCTION
The vascular system has evolved to provide adequate circulation to the tissues in the face of changing demand due to tissue growth, regression, and changes in metabolic activity. An important aspect of this homeostatic regulation involves sensing of fluid shear stress (FSS) from blood flow by vascular endothelial cells (ECs) (1–3). ECs encode a FSS set point such that sustained FSS above or below this level triggers vessel outward or inward remodeling to increase or decrease lumen diameter and restore FSS to the initial level (4, 5). Available evidence suggests that FSS near the set point activates signaling and gene expression programs that stabilize the vessel, whereas FSS above or below this range destabilizes the vessel to permit remodeling (4). Failure of these homeostatic mechanisms results in tissue ischemia associated with excessive inward remodeling in coronary and peripheral artery disease. Vascular malformations associated with excessive outward remodeling and vessel fragility are much less common but must also be considered diseases of compromised vascular homeostasis.
Regions of arteries that bend sharply, branch, or bifurcate develop FSS with lower magnitude and complex fluctuating changes in direction during the cardiac cycle, termed disturbed shear stress (DSS), generally modeled in vitro by oscillatory shear stress (OSS) (6). Artery segments under DSS are susceptible to metabolic and inflammatory stimuli that induce the formation of atherosclerotic plaques, a form of pathological vessel remodeling. Most plaques are clinically silent due to compensatory outward remodeling by the unaffected parts of the vessel that preserves lumen diameter, often termed Glagov remodeling (7). Only in late stages with more severely inflamed vessels does this mechanism fail, at which point the encroaching plaque decreases lumen diameter to cause tissue ischemia.
These ideas can be conceptualized in terms of a set of interlinked FSS-dependent EC states and transitions between them (4, 8, 9). Physiological FSS [physiological shear stress (PSS)] at or near the set point confers vessel stability with low permeability, low cell proliferation and turnover, and low inflammation (10). Low FSS [low shear stress (LSS)] that triggers inward remodeling is associated with higher EC turnover, higher EC inflammatory activation with recruitment of leukocytes that assist remodeling, and smooth muscle contraction (11). High FSS [high shear stress (HSS)] triggers outward remodeling associated with cell proliferation, inflammatory activation and leukocyte recruitment, and smooth muscle cell relaxation and proliferation (12). DSS alone is insufficient to induce remodeling but renders vessels susceptible to pathological stresses (13). FSS magnitude and patterns thus govern transitions between distinct EC states that drive vessel remodeling, stability, or susceptibility to additional stresses that lead to disease (14).
We have identified multiple pathways that show sharp FSS-dependent regulation and that participate in these processes. For example, Smad2/3 are specifically activated at low FSS (LSS) and required for vessel inward remodeling (15). By contrast, Smad1/5 are specifically activated by physiological FSS (PSS) to signal vessel stability (4, 16) and suppressed by paraphysiological HSS through a Krüppel-like factor 2 bone morphogenetic protein endothelial regulator (KLF2-BMPER) pathway (17). However, our understanding of these crucially important pathways remains highly incomplete. This limited understanding is a major obstacle to developing safe and effective treatments for coronary, peripheral, and cerebral artery disease and vascular malformations, which are characterized by aberrant vessel remodeling that leads to vascular insufficiency and tissue ischemia or to vessel instability, rupture, and bleeding.
Developing improved therapies requires a deep understanding of the regulatory networks that define EC states and transitions between states. Recent efforts applied computational approaches to identify different cell states and predict molecular pathways that govern cell trajectories and cell state transitions (18–20). A systems biology approach, cSTAR, can use and integrate a variety of omics data to identify key regulators of cell state transitions and predict experimental outcomes (21).
FSS potently determines EC phenotype, controlling expression of several thousand genes (22, 23). However, a systematic analysis of the distinct transcriptomic responses to each of these flow patterns has not been reported. Thus, to gain insight into these processes, we assessed responses of cultured ECs to LSS, PSS, HSS, OSS, and static (STAT; no flow) conditions. The use of cSTAR accurately separated the transcriptomic patterns of EC states under these conditions and demonstrated the existence of three orthogonal vector axes that precisely characterize EC state transitions. Combining this information with published EC drug perturbation data [LINCS L1000 dataset (24)] enabled reconstruction of a signaling network that controls EC state transitions and suggested that cell cycle plays a central role in these processes. Experimental studies confirmed these predictions, demonstrating distinct inflammatory mediators under LSS versus OSS and showing that cell cycle position determines pro- and anti-inflammatory susceptibility, with CDK2 as a major determinant of the stable, anti-inflammatory state in quiescent ECs under physiological shear stress.
RESULTS
cSTAR analysis of EC transcriptomic data under flow
Human umbilical vein ECs (HUVECs) were subjected to OSS (0.5 ± 4 dynes/cm2), LSS (3 dynes/cm2), PSS (16 dynes/cm2), and HSS (40 dynes/cm2) for 24 hours, in each case in comparison to STAT as an internal control. The total RNA was extracted and prepared for RNA sequencing (RNA-seq). The fold changes in expression compared to the control static condition were considered as points in the transcriptomic dataspace (see Materials and Methods).
In the initial cSTAR step (21), we used machine learning techniques, such as support vector machines (SVMs), to exploit the high-dimensional omics space and construct separating surfaces between distinct EC states (technically, the SVM algorithm with a linear kernel from the scikit-learn python library was applied to build a maximum margin hyperplane that distinguishes different cell states). SVM is recognized as one of the top-performing algorithms for classification tasks (25). It is particularly effective when the number of features greatly exceeds the number of samples, which is precisely the scenario addressed in this study (26).
Directions in the transcriptomic space are specified by the state transition vectors (STVs), which are unit vectors normal to the hyperplanes that separate cellular phenotypic states. After building the STVs, cSTAR computes quantitative scores of cell phenotypic states termed dynamic phenotype descriptors (DPDs) that describe the phenotypic cell features associated with each STV. These metrics are determined by the Euclidean distances between the separating surface and the cell state, with plus or minus sign depending on the STV directions. If all STVs are orthogonal, then transcriptomics changes along either STV affect distinct phenotypic features that are associated with different DPDs and different molecular processes (27).
We aimed to find the STV directions that describe and determine three different EC functions: (i) FSS sensing (STVFSS) that distinguishes FSS magnitude, (ii) stability (under PSS) versus tendency for inward (LSS) or outward (HSS) vessel remodeling (STVremod), and (iii) abnormal remodeling under OSS (STVOSS). Therefore, we used the SVM to separate different transcriptomic responses that correspond to distinct flow conditions and cell phenotypic responses. Accordingly, the calculated STVs and DPD scores are denoted by different subscripts. The subscript “FSS” corresponds to the separation of LSS and HSS transcriptomic responses, the “remod” subscript corresponds to the separation of PSS versus LSS and HSS taken together, and the “OSS” subscript corresponds to the separation of OSS versus PSS, LSS, and HSS taken together. Grouping of LSS and HSS transcriptomic states together versus PSS state is explained by the fact that remodeling and inflammation pathways are activated in both LSS and HSS states, whereas there is no remodeling/inflammation in PSS state, which is stable. Likewise, OSS state is analyzed versus combined other FSS states because OSS is used to model EC pathological conditions (6). The resulting STVFSS, STVremod, and STVOSS (tables S1 and S2) are mutually orthogonal, suggesting distinct EC transcriptomic and phenotypic responses to perturbations along these axes. Robustness of the EC state classification and the DPD score variability was rigorously tested using cross validation, as we did previously (21).
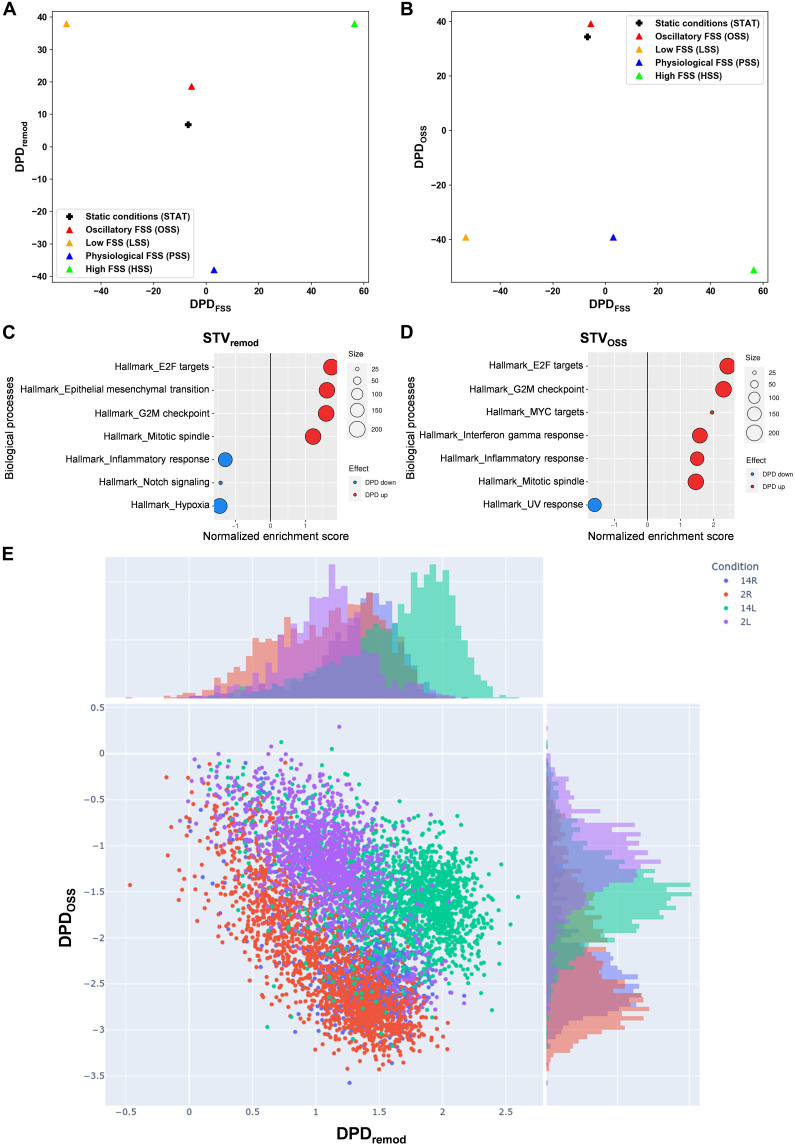
DPD scores calculated along each STV (Fig. 1, A and B) reveal that the remodeling scores, as quantified by DPDremod, for EC transcriptomes under both OSS and STAT conditions substantially differ from other EC states (Fig. 1A). These remodeling scores are higher than the scores under PSS, where vessels are stable, but lower than under LSS and HSS that induce strong vessel remodeling. We conclude that OSS and STAT induce a state susceptible to remodeling, although to a lesser extent than LSS and HSS. The findings for OSS correspond well to behaviors in vivo, where regions of arteries under OSS are stable if unperturbed but show high susceptibility to inflammatory and metabolic stresses, which result in preferential inflammatory gene expression and ultimately formation of atherosclerotic plaques (28). ECs under STAT do not exist in vivo, but our result aligns with general observations that PSS reduces inflammatory gene expression and cell proliferation to levels below those observed under static conditions (29).
Fig. 1. cSTAR analysis of EC states under different flow conditions.
(A to D) RNA-seq data from ECs under OSS, LSS, PSS, HSS, or STAT conditions for 24 hours were analyzed, and DPD scores were calculated. (A) Two-dimensional (2D) plot of phenotypic scores in the DPDFSS and DPDremod plane. (B) 2D plot of phenotypic scores in the DPDFSS and DPDOSS plane. (C and D) Top GSEA Hallmark (HM) gene sets that contribute to the STVremod [(C)] and STVOSS [(D)]. The components of STVs were used as the GSEA input for the GSEA hallmark gene set. Positive-normalized enrichment score (red) reflects an increase in a molecular process while moving along the corresponding STV, and negative-normalized enrichment score (blue) reflects a decrease. Size of circles corresponds to the number of genes in STV corresponding to the specific GSEA term. (E) Scatter plot of the DPDremod and DPDOSS scores calculated for in vivo single-cell RNA-seq data (33) from the left carotid artery partial ligation mice model: 2L, 2 days left carotid artery; 2R, 2 days right carotid; 14L,14 days left carotid; 14R, 14 days right carotid. Histograms on the top and right present distributions of DPD values across one axis.
Molecular features described by the DPDFSS scores that distinguish LSS from HSS (Fig. 1, A and B) are involved in inward versus outward remodeling. The DPDOSS scores of OSS and STAT states are also notably close to each other and significantly higher than the scores under all other FSS conditions (Fig. 1B). Figure 1 (A and B) demonstrates that the DPDFSS scores of EC responses under STAT and OSS conditions closely resemble the DPDFSS scores under PSS condition. This suggests that static and OSS conditions predispose vessels to remodeling (i.e., reduce stability) but without a preference for inward versus outward directions. Again, this result is consistent with the observation that artery regions under disturbed flow are stable but susceptible to inflammatory and metabolic stimuli (28). The similarity in the DPDFSS and DPDOSS scores does not indicate that OSS and STAT states are identical but rather that they have some patterns of gene expression in common. For DPDFSS, it implies that neither OSS nor STAT condition induces patterns of gene expression associated with responses to the FSS magnitude. Likewise, the similar DPDOSS scores between OSS and STAT imply similar responses of gene expression associated with abnormal remodeling. Although the OSS state is closer to the STAT state than to the PSS, HSS, and LSS conditions, these two states are by no means identical, as can be seen for the difference in their DPDremod scores.
While both LSS and OSS are often considered atherogenic (28), our analysis indicates that these two states are distinct, as evident from their distinct positions within the planes representing the phenotypic features of EC states (Fig. 1). To gain initial insight into the differences between physiological and pathological vessel remodeling, we applied the gene set enrichment analysis (GSEA) (30) algorithm to analyze the components of the STVremod and the STVOSS (Fig. 1, C and D). As described (31), this approach identifies the transcriptomic features that characterize these two remodeling programs, irrespective of whether the remodeling is directed inward or outward. We observed that both physiological (characterized by the STVremod components) and pathological (depicted by the STVOSS) remodeling are strongly associated with changes in cell cycle genes (Fig. 1, C and D). The STVremod and STVOSS were distinguished by (i) the presence of epithelial to mesenchymal transition genes in the STVremod but not in the STVOSS, (ii) suppression of inflammation response genes in STVremod and their activation in STVOSS, (iii) suppression of NOTCH signaling in the STVremod but not in the STVOSS, and (iv) distinct patterns of chemokine expression. Specifically, chemokines induced by OSS promote macrophage recruitment, whereas LSS-induced chemokines favor neutrophil recruitment (fig. S1A) (32). These results thus predict differential immune cell recruitment under LSS versus OSS. To test this notion in vivo, we created LSS and OSS in the mouse carotid artery using different ligation protocols (fig. S1, B and C). Both protocols increased infiltration of both neutrophils and macrophages, consistent with their inflammatory nature, but neutrophil infiltration was more pronounced under LSS (fig. S1, D and E), whereas macrophage infiltration was greater under OSS (fig. S1, F and G). The increase in immune cell infiltration indicates that inflammatory processes are indeed activated during both physiological vessel remodeling (guided by STVremod) and pathological (guided by STVOSS) remodeling. This suggests that the GSEA-based conclusion of suppressed inflammatory processes during physiological remodeling (Fig. 1C) is overly simplistic, as certain inflammatory processes are still activated in this context.
To further test the in vivo relevance of the DPD scores derived from in vitro data, we computed these scores for single-cell RNA-seq data from a published carotid ligation protocol that induces OSS (left carotid ligated, right control) (33). Figure 1E displays the computed DPDremod and DPDOSS scores for individual ECs on a scatter plot. Notably, ECs from the operated left carotid artery at 2 and 14 days after surgery (2L and 14L) exhibit the significantly higher DPDOSS scores compared to control right carotids (2R and 14R). DPDOSS scores were slightly higher for ECs from the left carotid at 2 days than at 14 days, whereas the DPDremod scores showed the opposite pattern. This suggests that exposure to OSS initiates negative feedback that partially dampens inflammation, consistent with in vivo observations (34, 35). Our in vitro–derived STVs and DPDs from different flow conditions are thus supported by in vivo results.
Integrating high-throughput LINCS perturbation data
The derived STVs and DPDs aid in comprehending the differences between EC states but do not reveal the causal connections that regulate these states. To infer these connections, cSTAR relies on perturbation data, which are responses measured in the same omics data space as the STVs and DPDs (21). Perturbations can be genetic, such as small interfering RNA (siRNA), or with small-molecule inhibitors or activators. The LINCS datasets report responses to drug perturbations but only under STAT condition and thus are insufficient for constructing the STVs/DPDs and gaining insights into the core network that determines EC phenotypic transitions. However, merging our data with the LINCS L1000 transcriptomics perturbation data for HUVECs (24) elucidates how drug perturbations affect cell states and facilitates the inference of causal connections within the core network governing EC state transitions (Fig. 2A). This data integration will enable us to infer how core pathways control EC phenotypic features that will be included as modules in the core network and expressed by different DPD metrics.
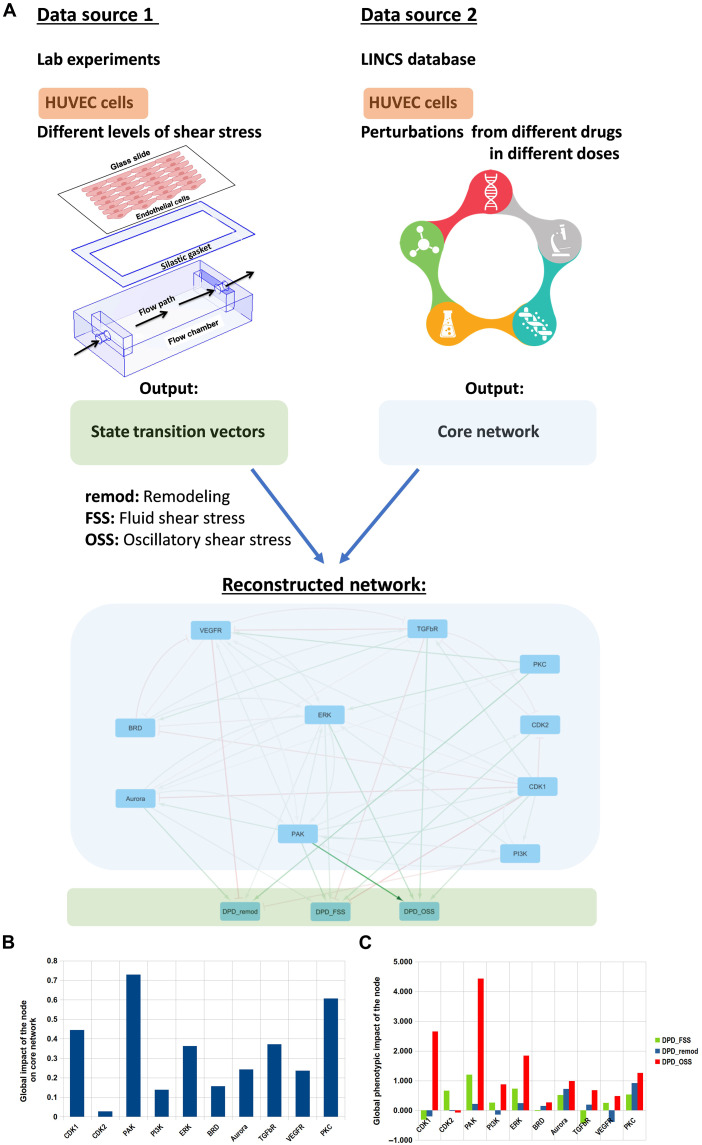
Fig. 2. cSTAR reconstruction of the core network driving EC cell fate decisions.
(A) Schematic illustrating the data analysis pipeline and the reconstructed core network. RNA-seq data from different flow conditions were used to obtain the STVs, and perturbation data from LINCS database (https://clue.io/data/CMap2020#LINCS2020) were used to infer the wiring of the core network and its connection to the phenotypic modules (DPDremod and DPDOSS). The resulting inferred network is presented at the bottom. Arrowheads indicate activation, and blunt ends indicate inhibition. Line widths indicate the absolute values of interaction strengths. Flow chamber was adapted from (81). (B) Global impact scores of the effect of perturbation of each core network module on the rest of core network components (blue) and on the cells’ phenotype (red) calculated as L2 norm of the relevant column in the global response matrix (Materials and Methods) (C) Global impact of the core network modules on the direction of vessel diameter change (calculated using the DPDFSS) and vessel remodeling program regardless of the direction (calculated using the DPDremod).
The core network components are chosen on the basis of two key criteria. First, these components must significantly influence the STVs. Second, their perturbations should substantially affect DPDs scores, as outlined in Materials and Methods. As transcriptomics patterns for the OSS state are close to those for the STAT state (Fig. 1), we calculated log fold changes (LFCs) of transcriptomic responses to drug perturbations with respect to the dimethyl sulfoxide controls using LINCS data. Combining these transcriptomic responses with the STV based on our data enabled us to compute the changes in the DPD scores following drug perturbations (Fig. 2A). This protocol identified components of the core regulatory network controlling both OSS and STAT states, as well as EC transitions to other states. These components include vascular endothelial growth factor receptor (VEGFR), transforming growth factor–β receptor (TGFβR), bromodomain (BRD) family, MAPK kinase (MEK)/extracellular signal–regulated kinase (ERK) pathway, phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR pathway, protein kinase C (PKC), p21-activated kinase (PAK) kinase family, cell cycle-dependent kinase (CDK1/2), and Aurora kinase family.
Inferring causal connections in the core network that controls EC state transitions and phenotypes
The LINCS L1000 datasets contain only transcriptomic responses to drug perturbations. However, to infer causal connections within the core network pathways regulating both OSS and STAT states and EC transitions, it is imperative to ascertain the responses in pathway activities, including the kinases and BRD proteins that were affected by drug perturbations. The existing methods for identifying enzyme transcriptional signatures and estimating enzyme activities are founded on database knowledge and cover only a few well-studied pathways (21, 36). Therefore, we developed a data-driven identification of enzyme activity signatures. It enabled us to deduce not only the drug-induced alterations in the activities of well-known core network modules, such as the MEK/ERK or PI3K/AKT/mTOR pathways, but also for enzymes whose transcriptional signatures are not well-established, such as Aurora kinases and PAK (Materials and Methods).
We previously developed modular response analysis (MRA) (37, 38) and its Bayesian reformulation (BMRA) (39) to precisely reconstruct causal connections between network nodes, including feedback loops, from perturbation data. Each node in the network represents a reaction module, which could be a single protein, gene, or pathway or any entity defined by its input-output relationships. This enables us to incorporate EC phenotypes, quantified through the DPDs, as network modules. The DPD modules contain information about the performance of the cell-wide network, which is controlled by the core regulatory network that plays a key role in determining EC phenotypic features.
MRA quantifies the network topology in terms of connection coefficients, i.e., connection strengths, that quantify the immediate impact of a change in a node (A) on another node (B), assuming that the activities of all other nodes are held constant to prevent indirect influence. The connection coefficients cannot be directly measured because perturbations to any node rapidly propagate through the network, masking direct, causal connections (40). We use BMRA because of its enhanced resilience to noise and its reduced demand for perturbation data, as compared to MRA. Moreover, BMRA offers the flexibility to incorporate pre-existing knowledge about the network in the form of a prior distribution. In BMRA, the inputs consist of the changes in the activities of core network components induced by drug perturbations, along with a prior distribution, which may contain all zeros if no prior knowledge is available. The outputs of BMRA encompass the inferred connections, the associated connection strengths, and their respective confidence intervals (21, 39). Critically, BMRA calculates the confidence intervals for each inferred connection, which estimates robustness of the inferred causal connections, including connections to the phenotypic DPD modules (21).
The inferred network is detailed in table S3 and Fig. 2A. While its causal connections and strengths quantify the local impact of each node on others, they do not offer insights into the overall or global changes (37). The forward MRA approach allows us to calculate how perturbation of each module propagates through the network and enables prediction of the global impact of each node (see Materials and Methods), highlighting the significance of each direct causal connection for the global changes in cell state (41, 42).
To gain insight into the control of EC remodeling programs, we analyzed both the inferred connection coefficients characterizing direct effects and the global system impacts of each node on the regulatory network and EC phenotype. Quantified by DPDremod, Aurora kinase and ERK emerge as the sole immediate drivers, while the VEGFR and PI3K pathways are the sole immediate suppressors of physiological vessel remodeling. Other nodes affect the DPDremod scores via activation or inhibition of these nodes. Top three nodes exerting strongest global impacts on the regulatory network are PAK, PKC, and CDKs (Fig. 2B). The causal, local connections reveal multiple mutual positive and negative feedback loops between the CDKs and other network nodes, and same for PAK (Fig. 2A). Summarizing, we found that cell cycle–related kinases (CDKs and Aurora kinase) and PAK play key role in computing cell fate decisions. PAK, CDKs, and ERK have the strongest impact on different EC phenotypic features (Fig. 2C). The positive impact of the CDK2 module on the DPDFSS score and its negative impact on DPDremod score suggest that CDK2 inhibition will induce inward vessel remodeling (where the DPDFSS decreases and DPDremod increases). Figure 2C predicts that inhibition of CDKs, PI3K, and VEGFR will stimulate vessel remodeling by decreasing DPDremod, whereas inhibition of all other core network modules will suppress vessel remodeling.
The impact of signaling network modules on the DPDremod underscores the importance of these nodes in normal vessel stability versus remodeling, while their impact on the DPDOSS suggests anti-atherosclerotic effects (Fig. 2C). cSTAR calculations suggest that core network modules except CDK2 could facilitate atherosclerosis, with PAK being the most potent. Critically, cSTAR predicts that inhibiting CDK2 promotes atherosclerosis by activating the TGFβ pathway, an essential driver of plaque formation (Fig. 2A) (43).
Summarizing, the cSTAR analysis predicts that cell cycle is pivotal in regulating various shear stress–induced EC states (Fig. 2). Particularly, our analysis predicts that CDK2 inhibition will decrease the DPDFSS score and increase the DPDremod and DPDOSS scores, thus promoting both inward vessel remodeling and atherogenesis. While the roles that core network modules play in shear stress signaling must be inferred with caution, they are consistent with numerous studies linking shear stress to cell cycle regulation such that physiological shear stress inhibits while OSS/DSS promotes proliferation (44–50). We therefore experimentally assessed the role of CDK2 in shear stress-dependent artery remodeling.
Effects of FSS on cell cycle
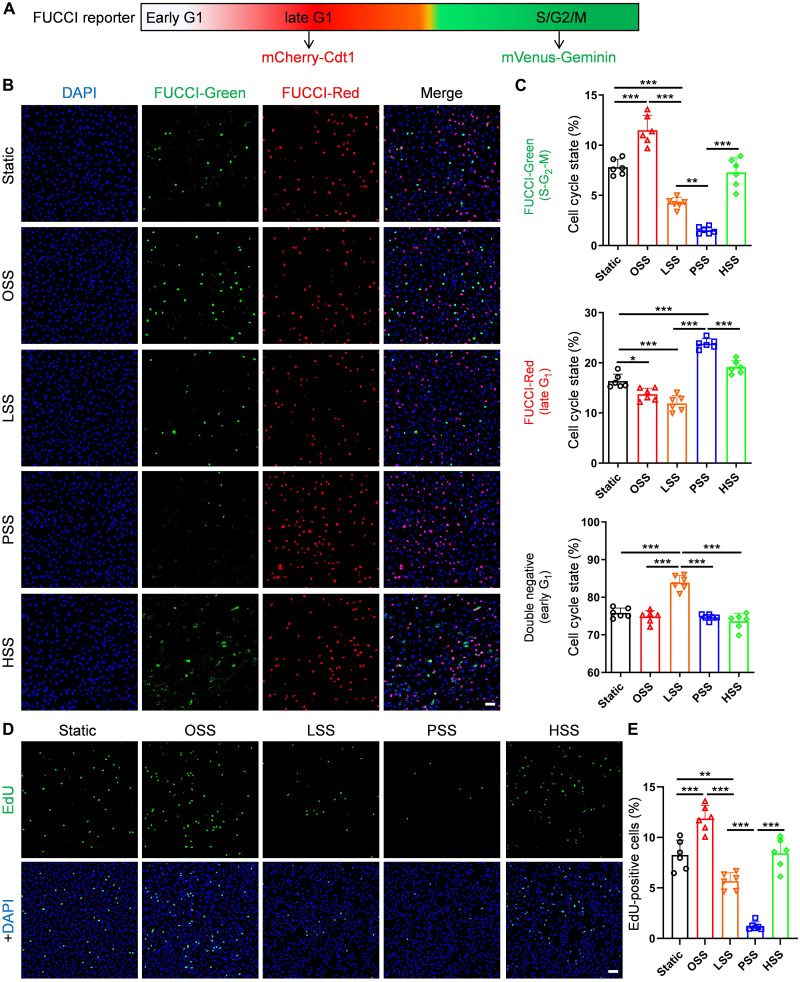
We first assessed cell cycle state under the different FSS conditions using the FUCCI reporter system that distinguishes cells in early G1, late G1 and S-G2-M. FUCCI is based on expression of the fusion proteins mCherry-hCdt1 and mVenus-hGem that are differentially ubiquitinated and degraded at different points in the cell cycle (51). During S, G2, and M phases (S-G2-M), only mVenus-Geminin is expressed, resulting in cytoplasmic green fluorescence (52, 53). Following mitosis, geminin is degraded, and cells are nonfluorescent (G0/early G1). As cells progress to late G1, mCherry-Cdt1 is expressed, and red fluorescence accumulates. We generated stable FUCCI-expressing HUVECs, subjected these cells to different FSS conditions for 24 hours, and scored cells as unlabeled, red, or green. We found that OSS increases cell cycle progression above the level seen under STAT as indicated by an increase in green cells (S-G2-M phases). LSS induces early G1 arrest (increased FUCCI negative, decreased S-G2-M) to a moderate extent. PSS induces much stronger late G1 arrest (FUCCI red with very few in S-G2-M). HSS induces a higher fraction of cells in S-G2-M compared to PSS, about the same as STAT (Fig. 3, A and B). Cell cycle progression is thus ordered OSS > STAT = HSS > LSS > PSS. To confirm these findings, cells were labeled with the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) during the last 2 hours of the 24-hour flow regimen and stained for this marker to identify cells that synthesized DNA in this interval. EdU labeling demonstrated that OSS induced the highest DNA synthesis, and LSS suppressed, PSS strongly suppressed, and HSS had little effect on DNA synthesis compared to static (Fig. 3, C and D). These results confirm the findings from the FUCCI reporter and identify an unexpected difference in the nature of cell cycle arrest that depends on flow magnitude. They are also consistent with published findings that cell proliferation is increased under OSS and reduced by PSS (44, 47–50).
Fig. 3. Characterization of cell cycle state under different FSS.
(A) Diagram of the FUCCI reporter. (B and C) FUCCI HUVECs were subjected to OSS, LSS, PSS, HSS, or static for 24 hours. Cells were fixed, stained with 4′,6-diamidino-2-phenylindole (DAPI), and imaged as described in Materials and Methods. Representative images for these conditions are shown in (B). Scale bar, 100 μm. (C) Quantification of FUCCI-red, FUCCI-green–positive cells, and double-negative cells. n = 6 experiments. (D and E) HUVECs were subjected to flow patterns as in (A), labeled, and stained for EdU as described in Materials and Methods. Percent of EdU-positive cells was quantified. n = 6 experiments. Scale bar, 100 μm. Data are presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001. ns, not significant, calculated by one-way analysis of variance (ANOVA) with Tukey’s multiple comparison tests.
CDK2 activity and function in ECs
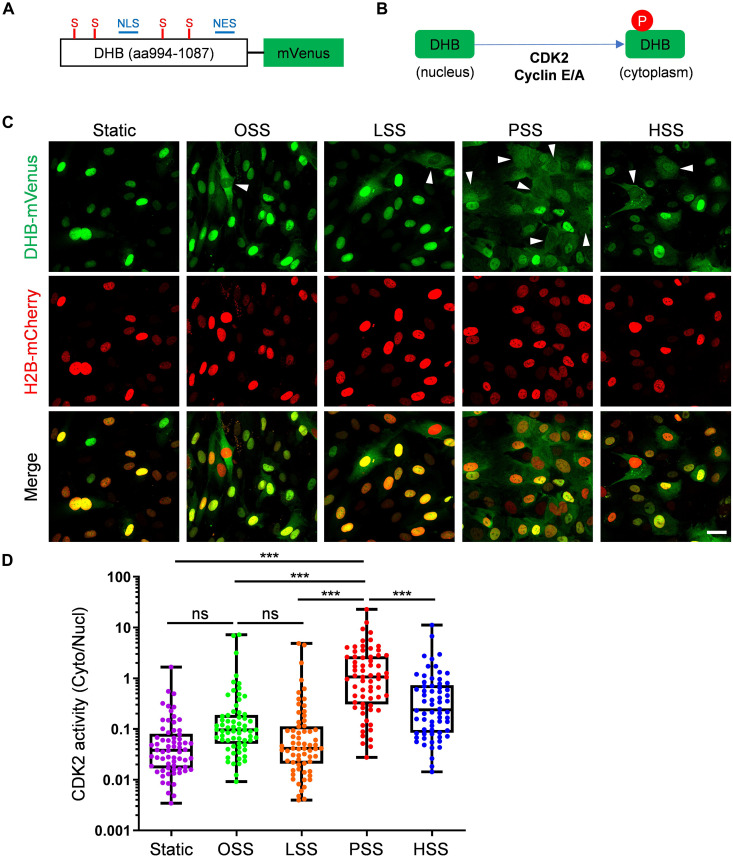
Active CDK2 complexed with cyclin E is thought to be the major determinant of the late G1-to-S transition (54). Our previous work showed that the Smad2/3 pathway, an important mediator of endothelial inflammatory gene expression during atherosclerosis and low flow inward remodeling in vascular ECs, is directly inhibited by CDK2 (15). We therefore assayed CDK2 activity under these FSS patterns, mainly to determine whether the late G1 arrested state under PSS is associated with active CDK2. For this purpose, we used a reporter construct, DHB-mVenus (55), whose phosphorylation by CDK2 triggers its translocation out of the nucleus (Fig. 4, A and B). HUVECs were cotransfected with DHB-mVenus plus histone 2B (H2B)–mCherry as a nuclear marker; cells were then subjected to OSS, LSS, PSS, HSS, and STAT conditions. We found that PSS is the most effective among all conditions to increase the cytoplasm/nucleus ratio, indicating CDK2 activity (Fig. 4, C and D). Therefore, CDK2 is most active in the PSS and late G1-arrested state. The fact that CDK2 activity is higher under PSS than in cells under OSS that are cycling more rapidly is because in cycling cells, CDK2 is active in a short window before entering S phase, whereas under PSS, CDK2 activity is sustained as long as cells remain in late G1 arrest.
Fig. 4. CDK2 activity under FSS.
(A) Schematic of CDK2 sensor. NLS, nuclear localization signal; NES, nuclear export signal; S, serines that are CDK consensus phosphorylation sites. (B) Schematic of CDK2 phosphorylation-mediated translocation of DHB-mVenus. (C and D) HUVECs expressing the DHB sensor were subjected to OSS, LSS, PSS, HSS, or STAT conditions for 6 hours. Cells were fixed and imaged as described in Methods, representative images shown in (C). Scale bar, 25 μm. Arrowheads are CDK2 high activity cells. Cell nuclei were identified using fluorescent H2B images to obtain a mask. Ratio of cytoplasmic/nuclear signal was quantified (D). n = 66 cells for each group from three independent experiments. Data showing all points from minimum to maximum. **P < 0.01 and ***P < 0.001, calculated by one-way ANOVA with Tukey’s multiple comparison.
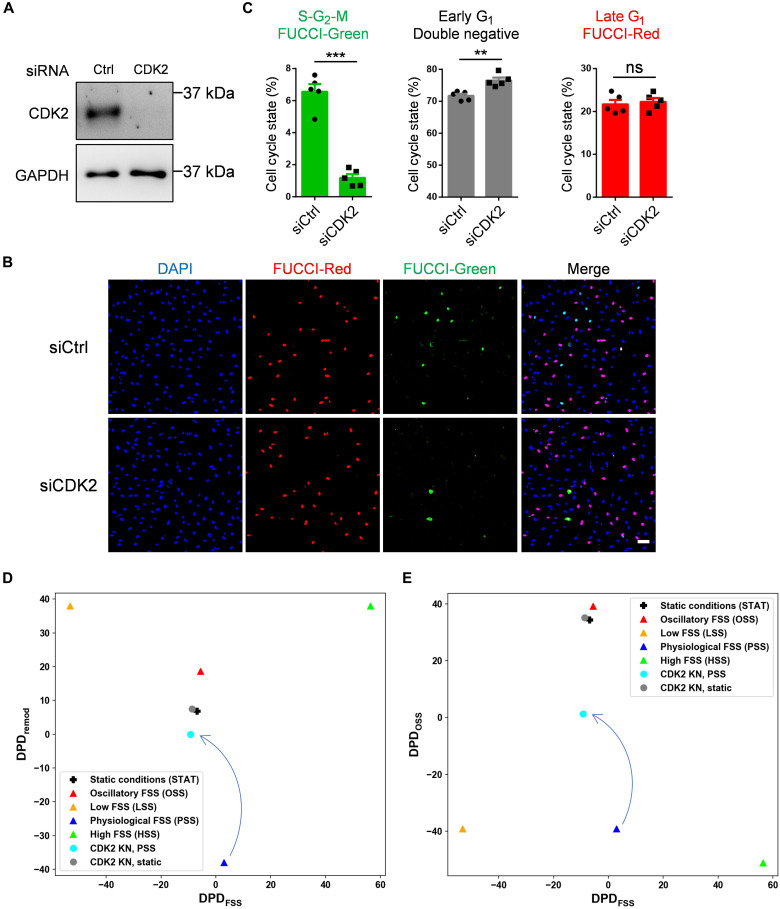
To further evaluate how CDK2 affects cell cycle state, we performed CDK2 knockdown in FUCCI HUVECs (Fig. 5A). CDK2 depletion strongly reduced the fraction of cells in S-G2-M and increased early G1 arrest (Fig. 5, B and C), a hallmark feature of the LSS state (Fig. 3C). This result was consistent under OSS (fig. S2). CDK2 is thus required not only for cell cycle progression but also for the specific late G1 arrest. We next carried out RNA-seq on CDK2 knockdown (KN) cells under STAT and PSS conditions for 24 hours. CDK2 knockdown decreased the DPDFSS and increased the DPDremod and DPDOSS scores (Fig. 5, D and E) with greater changes under PSS than STAT conditions. These results support computational cSTAR predictions (Fig. 2C), indicating that CDK2 knockdown can initiate inward vessel remodeling and atherogenesis in vivo. In addition, different extents of cell state changes after CDK2 knockdown under PSS and STAT conditions suggest that CDK2 activity is essential for the PSS state, and the disruption of CDK2 activity abolishes PSS state, but not STAT state.
Fig. 5. CDK2 depletion induces early G1 arrest.
FUCCI HUVECs were transfected with control (siCtrl) or CDK2 (siCDK2) siRNA and analyzed on day 4. (A) CDK2 knockdown confirmed by Western blotting. (B) Cells were fixed and mounted with DAPI. Representative images are showed. Scale bar, 100 μm. (C) Quantification of cell cycle state. n = 5 experiments. Data are presented as means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, calculated by two-tailed unpaired t tests. (D and E) 2D plots of DPDFSS and DPDremod [(D)] and DPDFSS and DPDOSS [(E)] for CDK2 KN in STAT and PSS conditions. DAPI, 4′,6-diamidino-2-phenylindole.
Furthermore, cSTAR predictions are corroborated by the FUCCI data (Fig. 5, B and C), where CDK2 knockdown induced early G1 arrest, resembling the LSS state. However, the CDK2 knockdown state differs from the bona fide LSS state, which has a low DPDOSS score. The CDK2 knockdown thus induces characteristics of both the LSS and OSS states, activating both normal and pathological vessel remodeling processes.
Deletion of CDK2 in ECs in vivo
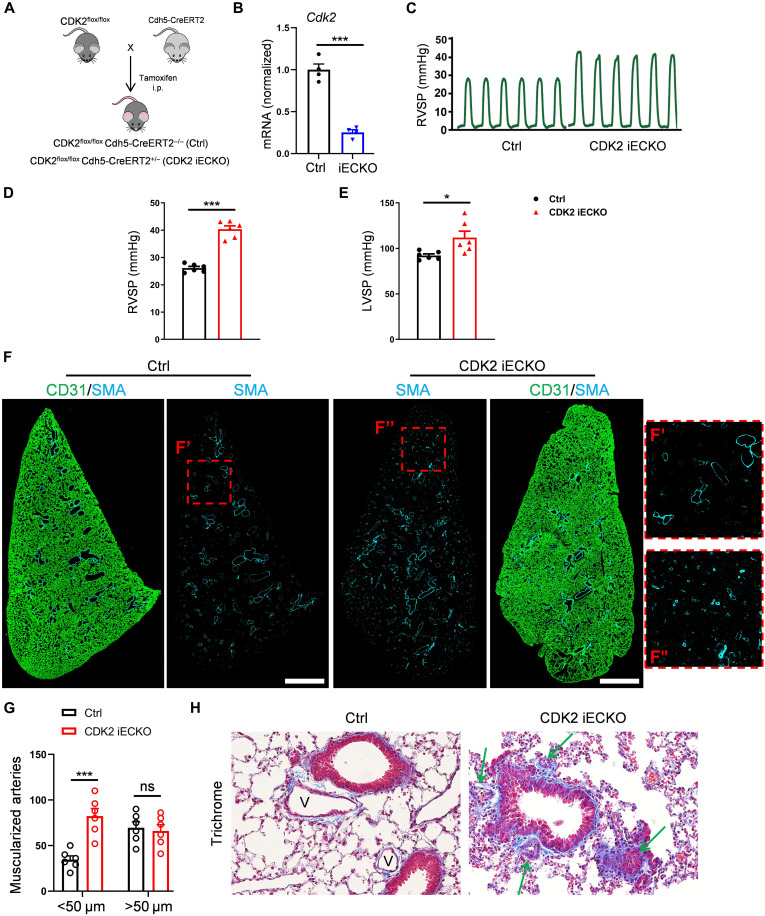
We previously found that a pharmacological inhibitor of CDK2 and related CDKs induced artery inward remodeling and pulmonary arterial hypertension (15), although this protocol was limited by general toxicity. To investigate the role of CDK2 in vivo using a more specific approach, we crossed CDK2 floxed mice with Cdh5-CreERT2 mice to induce endothelial-specific CDK2 knockout (CDK2 iECKO; Fig. 6A). Mice at 8 weeks of age were injected with tamoxifen and Cdk2 deletion confirmed (Fig. 6B). Mice at 2 months after CDK2 iECKO showed markedly higher right ventricular systolic pressure (RVSP) and less dramatic but significant left ventricular systolic pressure (LVSP) (Fig. 6, C to E). CDK2 iECKO thus induces pulmonary arterial hypertension and to a lesser extent systemic hypertension. To investigate mechanisms, we focused on the pulmonary vasculature where effects were stronger. Immunostaining for smooth muscle actin (SMA) in whole lung sections showed an increase in muscularized small vessels, typical of pulmonary hypertension and indicating artery inward remodeling (Fig. 6, F and G). Masson’s trichrome and hematoxylin and eosin (H&E) staining of lung sections confirmed that arteries in CDK2 iECKO lung became narrowed and even occlusive (Fig. 6H and fig. S3). Immunostaining of phospho-Smad2 and 3 in lung sections corroborated the cSTAR prediction that TGFβR signaling is hyperactivated after CDK2 iECKO (fig. S4). CDK2 iECKO thus leads to artery inward remodeling and hypertension.
Fig. 6. Deletion of endothelial CDK2.
(A) Schematic for production of CDK2 iECKO mice. Cdh5-CreERT2; CDK2flox/flox (CDK2 iECKO) and CDK2flox/flox (ctrl) mice at 6 weeks of age were injected with tamoxifen. (B) Cdk2 deletion efficiency in isolated lung ECs confirmed by quantitative PCR. n = 4 mice per group. (C and D) Representative trace and quantification of right ventricle systolic pressure (RVSP) from control and CDK2 iECKO mice. (E) Quantification of left ventricle systolic pressure (LVSP) from control and CDK2 iECKO mice. n = 6 mice per group. (F) Representative SMA immunostaining in Ctrl and CDK2 iECKO entire lung sections. Scale bars, 1 mm. (G) Quantification of SMA+ muscularized arteries (diameter < 50 μm and > 50 μm) in Ctrl and CDK2 iECKO lungs. n = 6 mice per group. (H) Representative Masson’s trichrome staining of Ctrl and CDK2 iECKO lung sections. V, vessels. Arrows indicate narrowed and occlusive vessels. Data are presented as means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, calculated by two-tailed unpaired t tests [(B), (D), and (E)] and two-way ANOVA with Tukey’s multiple comparison [(G)]. i.p., intraperitoneal.
Deletion of CDK2 in ECs accelerates atherosclerosis plaque formation
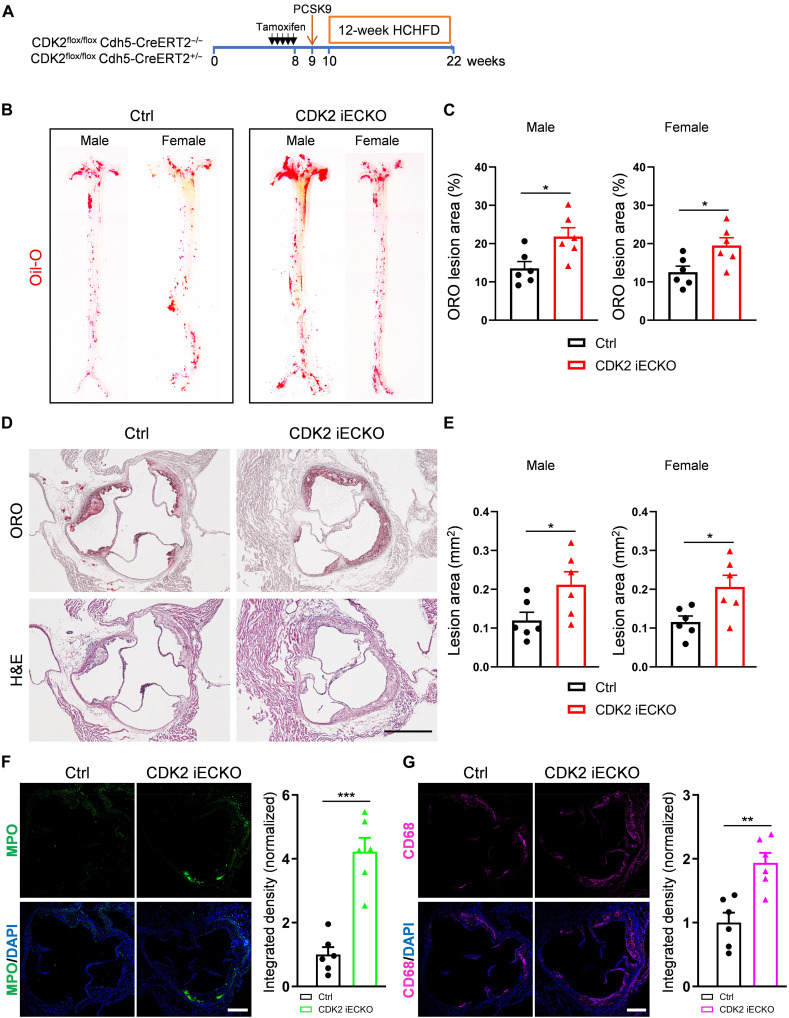
The cSTAR model predicted that activation of the TGFβR pathway due to CDK2 suppression will promote atherogenesis. Previous studies also linked both Smad2/3 hyperactivation and inward remodeling to atherosclerosis (56). We therefore hypothesized that the loss of endothelial CDK2 might enhance atherosclerotic plaque formation. To test this hypothesis, CDK2 iECKO and Ctrl mice were injected intraperitoneally with PCSK9 adeno-associated virus and fed a high-cholesterol/high-fat diet (HCHFD) for 12 weeks (Fig. 7A). Body weight, plasma total cholesterol, and triglyceride levels were unchanged between CDK2 iECKO and Ctrl mice (fig. S5). En face staining of the whole aorta with Oil-Red O (ORO) was used to assess total atherosclerotic burden. This analysis showed that CDK2 iECKO mice developed larger atherosclerosis plaques compared to control mice (Fig. 7, B and C). Histological analysis of aortic root sections also showed larger plaques (Fig. 7, D and E). Immunostaining showed that loss of endothelial CDK2 increased infiltration of both neutrophils and macrophages (Fig. 7, F and G). Together, the deletion of endothelial CDK2 accelerates atherosclerosis plaque formation.
Fig. 7. Atherosclerosis in CDK2 iECKO mice.
(A) Experiment timeline: CDK2 iECKO and Ctrl mice at ~6 weeks were treated with tamoxifen for five consecutive days. At 9 weeks, mice were injected with PCSK9 virus and 1 week later fed with HCHFD for an additional 12 weeks. (B and C) Representative images and quantification of ORO staining for male and female Ctrl and CDK2 iECKO whole aorta. (D) ORO and H&E staining of aortic root sections from Ctrl and CDK2 iECKO mice. Scale bar, 500 μm. (E) Quantification of ORO lesion area for both male and female Ctrl and CDK2 iECKO mice. n = 6 mice per group per gender. (F) Representative images and quantification of anti-myeloperoxidase (MPO; neutrophil marker) on aortic root sections from Ctrl and CDK2 iECKO mice. Scale bar, 250 μm. (G) Representative images and quantification of CD68 (macrophage marker) on aortic root sections from Ctrl and CDK2 iECKO mice. Scale bar, 250 μm. n = 6 mice per group. Data are presented as means ± SEM. *P < 0.05 and **P < 0.01, calculated by two-tailed unpaired t tests.
DISCUSSION
These results lead to three main conclusions. First, cSTAR analysis of transcriptomic data for ECs under low, physiological, high, and oscillatory FSS identifies three distinct, orthogonal axes that describe endothelial states (Fig. 1). In keeping with published studies that show that PSS confers vessel stability (2, 28), ECs under PSS have a low value on the remodeling axis (DPDremod) and an intermediate value on the FSS axis (DPDFSS) indicating no propensity to remodel in either direction. ECs under both STAT and OSS show intermediate values on the remodeling axis, indicating a less stable state than PSS but well below the actively remodeling LSS and HSS conditions. OSS and STAT conditions have low values on the DPDFSS axis, similar to PSS, again indicating no propensity for inward or outward vessel remodeling. OSS and STAT are also in close proximity on the OSS axis (DPDOSS).
These data should be interpreted in light of the concept that in vivo remodeling processes are assisted by immune cells (8, 57). The EC expression of cytokines and leukocyte recruitment receptors bring in monocyte/macrophages that secrete growth factors, degrade, and synthesize extracellular matrix. These processes are essential for high FSS-induced collateral remodeling in the heart for example (58). Pathological remodeling clearly differs in many respects that are only partially understood but both physiological and pathological remodeling associate with inflammation (59).
These findings correspond well to behaviors in vivo, where artery walls under disturbed, atherogenic flow are stable over decades in the absence of additional perturbations but are specifically susceptible to inflammatory and metabolic stresses that lead to atherosclerosis. Consistent with reduced stability, these regions also show amplified rapid responses to inflammatory stimuli, so-called inflammatory priming, quantified by DPDOSS, whereas regions of arteries under PSS are resistant (60). However, these similarities in gene expression patterns and remodeling behaviors for OSS and STAT do not in any way indicate identity. Numerous studies including this one identifies extensive differences in EC gene expression (22, 44, 61). In vivo, only angiogenic ECs exist without shear stress, but this state shares elevated inflammatory status and endothelial-mesenchymal transition with OSS, underscoring both their similarities and differences (62). The cSTAR analysis of transcriptional signatures thus offers a notable perspective on FSS-driven EC states.
Second, using cSTAR to integrate our transcriptomic data with the LINCS L1000 datasets, which describe responses of ECs without flow to a library of compounds, identified causal connections between multiple pathways that determine vessel stability, remodeling and disease susceptibility. This analysis identified a causal network linking VEGFR, TGFβR, CDK1/2, ERK MAP kinases, PI3K/AKT, PAK, BRD, PKC, and Aurora kinases that is likely to play a major role in determining EC state. In this model, CDKs suppress physiological remodeling, i.e., confer stability, whereas PAK, TGFβR, and Aurora kinase promote remodeling. All core network modules except CDK2 promoted the OSS state, associated with pathological remodeling, with PAK being top contributor. These outcomes fit well with the known pro-inflammatory role for PAK (63–67).
Third, based on the prediction that CDKs suppress DPDremod, the FUCCI reporter revealed that LSS suppressed cell proliferation via early G1 arrest, while PSS suppressed more strongly but via late G1 arrest. In this state, CDK2 is active, but cells do not progress into S phase. The nature of the block into S phase is not fully understood, but p53 and the CDK inhibitor p21/WAF1 have been implicated (45, 48). It may be related to the so-called cell growth checkpoint that is later in G1 and distinct from the restriction checkpoint (68). Knockdown of CDK2 in cells or under OSS decreased cell cycle progression but shifted cells into early G1. DPD analysis of transcriptomic data after CDK2 knockdown confirmed similarities to the LSS state, although with features of the atherogenic state. In vivo EC deletion of CDK2 induced artery inward remodeling and pulmonary and systemic hypertension, providing strong evidence that CDK2 is essential for stabilizing arteries under PSS. Together, these data argue that early versus late G1 arrest associate with highly distinct states. CDKs appear to do so not simply by controlling cell proliferation but rather by controlling activities of other signaling modules (Fig. 2), for example, by inactivating Smad2/3. Both in vitro transcriptomic analysis and in vivo results identify CDK2 as a major determinant of the stabilized PSS state.
The emerging concept that CDKs control EC phenotype apart from rates of proliferation or cell cycle per se fits with a recent study examining cell cycle regulation in post-natal retinal angiogenesis (53). They reported that late G1 arrest associates with arterial differentiation, while early G1 arrest associates with venous differentiation in vivo. Their results, however, differed from ours in that the arterial, late G1 state had higher TGFβ signaling. This discrepancy suggests that regulatory networks likely vary depending on developmental stage (adult versus developing vasculature) or location (systemic versus central nervous system vasculature), where different CDKs may be involved.
A second surprising finding is the distinction between LSS and OSS. Numerous publications treat these as comparable features of atherogenic flow patterns that predict lesion formation and vulnerability (69, 70). cSTAR analysis predicted that the overall states are highly distinct, and while both promote expression of inflammatory mediators, even these are different, correlating with differential recruitment of leukocyte subsets. Further evaluation of these differences and their roles in vessel inward remodeling under LSS versus atherogenesis under OSS will be an important direction for future work.
Together, these results strongly validate the cSTAR approach for elucidating the effect of FSS on EC state as a driver of vessel remodeling. They also lead to three important conclusions concerning the nature of FSS-regulated EC states, elucidation of a potential regulatory network (Fig. 2), and the role of CDKs in determining vessel remodeling. Future work to fully elucidate the detailed FSS-dependent regulatory network will require a perturbation analysis under the different FSS conditions and experimental validation of predictions. Doing so offers the opportunity to both advance our basic understanding of vascular regulation and to identify therapeutic targets for treatment of vascular disease.
Limitations of this study
This study examined effects of shear stress on HUVECs cultured on tissue culture plastic with conclusions validated in arteries in mice; effects on different substrates or in microvascular and lymphatic ECs are likely to differ. EC responses to pulsatile versus steady flow show statistically significant but modest differences (71–73); thus, steady flow was used for LSS, PSS, and HSS conditions. The simplified shear stress profiles used in this study may not fully reflect the responses to complex pulsatile FSS in vivo. Because cSTAR is a data-driven approach, its results are tailored to the specific cell types from which the data have been originated. In addition, since cSTAR is a data-driven approach, and most of perturbation data were obtained under static conditions, the cSTAR model predicts cell state responses under nonstatic conditions qualitatively rather than quantitatively. To achieve quantitatively accurate predictions of vessel remodeling processes, a set of perturbation omics experiments under FSS conditions would be needed.
MATERIALS AND METHODS
Animals
CDK2fl/fl mice and Cdh5-CreERT2 mice, all on the C57BL/6 background, were previously described (74, 75). Gene deletion was initiated by intraperitoneal injection with 1.5 mg of tamoxifen (Sigma-Aldrich, T5648) for five consecutive days. Sex-matched 6- to 10-week-old mice were used to induce gene deletion for experiments. All mice were housed in a pathogen–free (SPF) facility with a light (12-hour light cycle), 50% humidity, and ambient temperature (69°F) controlled environment. They were fed a pellet rodent diet and had free access to water. All mouse protocols and experimental procedures were approved (approval number 2023-20148) by Yale University Institutional Animal Care and Use Committee.
RV and LV hemodynamic measurements
RVSP was measured with a 1.4F pressure transducer catheter (Millar Instruments) and LabChart software (ADInstruments) as described (56). Briefly, mice were anesthetized with 2% isoflurane, and the catheter was inserted through the right jugular vein into the right ventricle. For LVSP, the catheter was inserted through the carotid artery into the left ventricle. RVSP and LVSP were all recorded and analyzed with LabChart software (ADInstruments).
Mouse atherosclerosis model
Control and CDK2 iECKO mice at 9 weeks old were intraperitoneally injected with murine PCSK9 adeno-associated virus (pAAV-mPCSK9, produced by the Gene Therapy Program Vector Core at the University of Pennsylvania School of Medicine, Philadelphia, PA). Beginning at 10 weeks, mice were fed a HCHFD (Clinton/Cybulsky high-fat rodent diet with regular casein and 1.25% added cholesterol; Research Diet, D12108) for the indicated times.
Analysis of atherosclerotic lesions
Mice were euthanized and immediately perfused with phosphate-buffered saline (PBS) for 5 min. Whole aortas—including the aortic arch, the thoracic, and abdominal segments—were dissected, gently cleaned of adventitial tissue, and fixed with 4% paraformaldehyde (Electron Microscopy Sciences) overnight at 4°C. The whole aorta was longitudinally opened and stained with 0.6% ORO (Sigma-Aldrich, O0625) for 60 min at room temperature. The aorta then was washed with 60% isopropanol for 20 min and rinsed three times with dH2O for 5 min. Last, the whole aorta was flat-mounted and imaged with Nikon Digital Slight DS-Fi1c camera. For analysis of lesions in the aortic root, the heart and the proximal aorta were excised, and the bottom of the heart was removed. The heart was then fixed with 4% paraformaldehyde overnight at 4°C, dehydrated with 30% sucrose overnight, embedded in optimal cutting temperature compound (OCT), and frozen at −80°C. OCT blocks were sectioned at 6 μm in thickness using a microm cryostat. Sections were further stained with ORO and hematoxylin. Lesion areas were quantified with ImageJ (National Institutes of Health) software.
Plasma lipid analysis
Blood samples were collected into tubes with lithium heparin and centrifuged at 3000g at 4°C for 10 min, and plasma was carefully transferred to a sterile tube for lipid measurement or stored at −80°C. Total cholesterol was measured using the Total Cholesterol Assay Kit (MyBioSource, MBS168179) according to the manufacturer’s instructions. Triglyceride levels were measured using the Triglyceride Colorimetric Assay Kit (Cayman, 10010303) according to the manufacturer’s instructions. Optical density was determined using a microplate reader (BioTek).
Cell culture and siRNA transfection
HUVECs pooled from multiple deidentified donors were obtained from Yale Vascular Biology and Therapeutics tissue-culture core laboratory at passage 1 (P1). Cells were maintained in endothelial growth medium 2 (EGM2) Endothelial Cells Growth Media (Lonza, CC-3162) and used for experiments between P2 and P5. The siRNA transfection was performed with Opti-MEM medium (Thermo Fisher Scientific) and Lipofectamine RNAiMAX (Invitrogen). ON-TARGET plus Smartpool siRNAs from Dharmacon were used against human CDK2 (L-003236-00-0005).
Generation of FUCCI HUVECs
HEK293T cells were transfected with pBOB-EF1-FastFUCCI-Puro (Addgene plasmid #86849) and packaging plasmids using Lipofectamine 2000 (Thermo Fisher Scientific, 11668019) according to the manufacturer’s instructions. Supernatants containing lentivirus were collected 48 hours after transfection and passed through a 0.22-μm filter. Primary HUVECs were infected with lentivirus for 24 hours and then replaced with EMG2 Endothelial Cells Growth Media. Cells were selected and passaged in puromycin (1 μg/ml; Sigma-Aldrich P9620).
Shear stress
HUVECs were seeded on tissue culture plastic slides coated with fibronectin (20 μg/ml) for 2 hours at 37°C and grown to confluence. Shear stress with the calculated intensities indicated in manuscript and each figure was applied in parallel flow chambers as described (15, 76). Shear stress values were chosen with reference to HUVECs based on both in vitro data demonstrating that physiological responses (alignment and suppression of inflammatory pathways) are maximal between 10 and 20 dynes/cm2, which corresponds well to in vivo FSS in the umbilical vein at ~15 dynes/cm2 [(4) and references therein]. Shear stress at 1 to 6 dynes/cm2 activates inflammatory pathways; thus, 3 dynes/cm2 was used to model low FSS. Levels above 30 dynes/cm2 disrupted alignment and activated inflammatory pathways (4); thus, 40 dynes/cm2 was used to model high FSS. For OSS, flow reversal rather than magnitude (or time averaged magnitude) has been identified as the critical variable that influences EC phenotype (59, 77–79); we chose 0.5 ± 4 dynes/cm2, a standard condition that elicits responses similar to those seen in regions of disturbed FSS in vivo.
EdU cell proliferation assay
HUVECs were seeded on tissue culture slides, grown to confluence in EGM2 medium, and subjected to shear stress as indicated. After 22 hours, a 2X working solution of EdU in complete EGM2 medium was added to cells, a final concentration of 10 μM. Two hours later, cells were collected and fixed. EdU DNA synthesis was detected following the manufacturer’s Click-iT EdU imaging kits (Invitrogen, C10337).
Immunofluorescence
For cells, samples were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 10 min at room temperature. For tissues, samples were fixed in 4% paraformaldehyde overnight at 4°C and incubated with 30% sucrose (Sigma-Aldrich) solution in PBS overnight at 4°C. Specimens were embedded in OCT medium (SAKURA), and 8- to 10-μm sections were cut in a cryostat (Leica). For immunofluorescence (IF), samples were incubated in blocking buffer [5% donkey serum, 0.2% bovine serum albumin (BSA), and 0.3% Triton X-100 in PBS], followed by incubation with primary and secondary antibodies diluted in a blocking buffer. Images were taken using the SP8 confocal microscope (Leica).
RNA isolation and quantitative real-time PCR
RNA was extracted from cells with the RNeasy Plus Mini Kit (QIAGEN) according to the manufacturer’s instructions, and reverse transcription was performed with the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad). Then, cDNA was amplified by real-time polymerase chain reaction (PCR) with iQ SYBR Green Supermix (Bio-Rad). The expression of target genes was normalized to expression of housekeeping gene GAPDH. Primer sequences were listed as below: mGapdh (5′-AGGTCGGTGTGAACGGATTTG-3′, 5′-TGTAGACCATGTAGTTGAGGTCA-3′) and mCdk2 (5′-CCTG-CTTATCAATGCAGAGGG-3′, 5′-GTG CTGGGTACACACTAGG-TG-3′).
Computational methods and analysis
Total RNA was extracted from HUVECs subjected to OSS, LSS, PSS, HSS, and static for 24 hours. Total RNA was quantitated by NanoDrop, and RNA integrity number value was measured with an Agilent Bioanalyzer. Samples were subjected to RNA-seq using Illumina NextSeq 500 sequencer (75–base pair paired end reads). The base calling data from sequencer were transferred into FASTQ files using bcl2fastq2 conversion software (version 2.20, Illumina). To analyze EC responses to FSS, the control condition is selected as no treatment, static condition (STAT). Using DeSeq2 (R package for the analysis of RNA-seq data), we determined the differentially expressed genes comparing the conditions LSS, PSS, OSS, HSS, and STAT.
STVs and cell states
Using cSTAR, we determined STVs that govern FSS sensing of ECs using the data we generated. In the cSTAR approach, the STV determines the direction in the data space that corresponds to a cell state transition. To generate the STVFSS, which describes the direction of vessel remodeling, we separated LSS and HSS states. To obtain the STVremod, which describes the initiation of vessel remodeling program regardless of the direction, we separated LSS and HSS states from PSS state. To generate the STVOSS, which describes pathological vessel remodeling, we separated PSS, LSS, and HSS states from OSS state. Mutual orthogonality of these STVs suggests that these three processes activate different groups of genes. The components of STVs are presented in tables S1 and S2.
We used LINCS L1000 perturbation data that contains HUVECs transcriptomic responses to a large number of drug perturbations, enabling us to perform inference of causal connections between the drug targets. To make the STV transferrable between different datasets, these STVs must be built in the space of LFCs. The reference state, with respect to which LFCs are calculated, is STAT for HUVECs in the LINCS database. To estimate phenotypic changes brought about by drug perturbations, for the j-th drug perturbation, we calculated the dot products of LFCs from the LINCS data () and the STV () from our generated data to find the element of the global response matrix () for the corresponding DPD module, as follows
| (1) |
Global responses of core network modules
If a direct measurement of the core network module activity can be done, then the LFC in the expression of the gene i in response to the j-th perturbation are used for value. When direct measurements of the activity of the i-th node are unavailable, such as in transcriptomic assessments of responses to enzyme inhibitor treatments, we apply the following procedure. A relative activity of the of the i-th module after j-th perturbation () is calculated as a linear function of the transcriptomic LFCs () as follows
| (2) |
Coefficients , which are used to infer activity of i-th module, can be found using linear regression over function , which describes response of enzyme activity to an inhibitor. In the simplest case of monomeric target, this function is expressed as follows
| (3) |
Here, is the inhibitor concentration and is the median inhibitory concentration (IC50) dose for this inhibitor, taken from the literature data. If multiple inhibitors of the same target are applied, then the coefficients are found by regressing the function over all doses of all inhibitors . The coefficient is set to 1 for all modules because for the control (no inhibitors) data points, all LFC, , are close to zero, while the value of the function is equal to 1.
To satisfy the MRA modular insulation condition (80), the vectors of coefficients (excluding the intercept coefficient ) for different modules must be mutually orthogonal. In addition, we have penalized the negative pathway activities and added Lasso regularization to minimize the number of potential solutions and ideally find the unique solution. In brief, we infer the coefficients by finding the global minimum of the following function
| (4) |
Here, the first term describes the linear regression over enzyme activity for each module, the second term introduces the orthogonality condition, the third term penalizes the negative pathway activities, and the forth term introduces Lasso regularization. The hyperparameters, , , , and , provide a means to balance the priority among different terms when it is not possible to satisfy all conditions simultaneously.
Taking into account that , expression 4 reads
| (5) |
When coefficients are determined, the elements of the global response matrix for core network modules can be calculated from Eq. 2 as follows
| (6) |
Here, we imply that for control data point .
It is important to highlight that while the STV () is normalized to a unit length, the vectors do not have unit lengths and must not be normalized, as doing so would violate the regression conditions. Completing the entries, , for both the core network modules and the DPD module(s) marks the completion of the transcriptomic data preparation for the BMRA inference in cSTAR. Note that for the CDK2 module, we integrated our transcriptomics siRNA perturbation data together with LINCS drug perturbation data. All codes for computational analysis are available on Zenodo (https://zenodo.org/records/14218021).
MRA-based predictions of the global responses based on the inferred network
The local response matrix, , which is inferred during the BMRA step of cSTAR, enables us to make predictions of system responses to different perturbations. The local response matrix is intimately related to the ODE system that describes the network dynamics. Mathematically, the matrix is the Jacobian matrix normalized by its diagonal elements (41, 42).
The predictive models can vary in complexity. Using both the inferred local response matrix and phosphoproteomics time-course data, we have built nonlinear ODE models of kinase networks using cSTAR (21). However, because the transcriptomics LINCS data only permit indirect measures of the activities of signaling network modules and lack time-course data, attempting to develop a dynamic nonlinear ODE model would be inappropriate. Such a model would be challenging to identify, suggesting that a simpler level of description is needed. Therefore, we employ MRA framework, which facilitates predicting drug responses using a linear function of applied perturbations, as follows (41, 42)
| (7) |
Here is a vector of systems-level responses to a drug perturbation (), is a BMRA-inferred local response matrix, and vector quantifies the local responses of primary targets to a drug . Equation 7 demonstrates that the -th column of matrix (table S4) encapsulates the system-level responses of all modules within the network to a 1% activation of module . Thus, to quantify the phenotypic impact of each signaling core network module on the phenotypic DPD modules in Fig. 2C and to quantify impact of all core network modules on the activation of the TGFβR module in fig. S3C, we used elements of as described above. To quantify the overall effect of the -th module on all other core network modules at Fig. 2B, we used the L2-norm of the vector of core network responses to the activation of module .
Western blotting
Cells were harvested and lysed for 30 min on ice in radioimmunoprecipitation assay buffer (Roche) containing complete mini protease inhibitors (Roche) and phosphatase inhibitors (Roche). Cells were centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was transferred to new 1.5-ml EP tubes and boiled with 4X loading buffer [250 mM tris-HCl (pH 6.8), 8% SDS, 40% glycerol, 20% β-mercaptoethanol, and 0.008% bromophenol blue] at 99°C for 5 min. Cell lysates were resolved by SDS-polyacrylamide electrophoresis on 4 to 15% CriterionTM TGXTM Precast Gels (Bio-Rad) and blotted onto a polyvinylidene difluoride membrane (Millipore). The blotted membranes were blocked with 5% non-fat milk and then incubated with specific antibodies diluted in 5% BSA, followed by horseradish peroxidase (HRP)–conjugated secondary antibodies. Protein bands were imaged using Immobilon Western Chemiluminescent HRP substrate (Millipore). ImageJ was used for densitometry quantification of western blot bands.
Mouse lung ECs isolation
Mouse lungs were collected and digested in a solution of collagenase (2 mg/ml; Sigma-Aldrich). Cell suspension was filtered through a 70-μm sterile cell strainer (Falcon). ECs were isolated using magnetic beads anti-rat immunoglobulin G (Invitrogen) coated with rat antimouse CD31 antibody (BD Biosciences). Cells were lysed for RNA extraction with PicoPure RNA isolation kit (Applied Biosystems) according to the manufacturer’s instructions.
Mouse carotid artery ligation model
C57BL/6J mice (12 weeks) were purchased from the Jackson Laboratory (stock no. 000664). Mice were anesthetized with ketamine/xylazine (100 and 10 mg/kg), and surgical procedures were performed under 0.5 to 1% isoflurane anesthesia condition delivered via a precision vaporizer. Mice received pre-emptive analgesia of buprenorphine (Ethiqa XR) and local anesthetic bupivacaine, and surgical procedures were performed using aseptic and microsurgery techniques. The cervical skin was cut in the midline. Then, the left common carotid artery (LCA) and its bifurcation were bluntly dissected to expose all four distal branches: external carotid artery (ECA), internal carotid artery (ICA), occipital artery (OA), and superior thyroid artery (STA). To generate low shear stress (LSS) in the region of LCA, ECA and STA were ligated using 6-0 silk suture, leaving ICA and OA open. To generate OSS in the region of LCA, four distal branches were ligated using 6-0 silk suture. Last, the skin was closed with a 6-0 Prolene suture. Shear stress profiles after surgery were characterized in previous studies (15, 61).
Antibodies
We used the following antibodies for immunofluorescence (IF) and immunoblotting (IB): Rat antimouse CD31 (BD 550274, clone MEC13.3; IF, 1:200), glyceraldehyde-3-phosphate dehydrogenase (Cell Signaling Technology, 5174S; IB, 1:2000), p-Smad2 Ser465/467 (Millipore, AB3849-I; IF, 1:200), p-Smad3 Ser423/425 (Abcam, ab52903; IF, 1:200), α–smooth muscle actin (Sigma-Aldrich, A2547; IF, 1:400), CDK2 (Cell Signaling Technology, 2546S; IB, 1:1000), myeloperoxidase (R&D Systems, AF3667; IF, 1:400), F4/80 (Invitrogen, 14-4801-82; IF 1:400), and CD68 (Thermo Fisher Scientific, 14-0681-82; IF, 1:400). Secondary antibodies are listed in table S5.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (GraphPad software Inc.). Data were analyzed for normality and equal variance using the Shapiro-Wilk test and Brown-Forsythe test, respectively. If both tests were passed, then statistical significance was further analyzed by two-tailed unpaired t test for two groups comparison or one-way analysis of variance (ANOVA) with Tukey’s post hoc test for multiple groups comparison. Statistical significance between two groups plus treatment was calculated by two-way ANOVA with Tukey’s multiple comparison tests. A P value less than 0.05 was considered significant (*P < 0.05, **P < 0.01, and ***P < 0.001). Statistical details and experimental n are described in the legend of each figure.
Acknowledgments
We thank P. Kaldis (Lund University, Sweden) for CDK2-floxed mice and J. Zhang (Yale University Animal Core, USA) for carotid artery ligation surgery.
Funding: This work was supported by National Institutes of Health (NIH) Grants R01 HL135582 (to M.A.S.) and R01HL171773 (to M.A.S. and B.N.K.), American Heart Association (AHA) career development award 24CDA1268658 (to H.D.), NIH/NCI grant R01CA244660 and EU grant 101136926 MULTIR (to B.N.K.), and SFI grant 22/PATH-S/10875 (to O.S.R.).
Author contributions: H.D.: Conceptualization, investigation, methodology, resources, funding acquisition, data curation, validation, supervision, formal analysis, visualization, and writing (original draft, review, and editing). O.S.R.: Conceptualization, methodology, formal analysis, software, and writing (original draft, review, and editing). D.J.: Investigation, resources, data curation, and validation. X.H.: Investigation and validation. P.J.: Investigation, methodology, data curation, formal analysis, and software. A.T.: Formal analysis and software. B.N.K.: Conceptualization, methodology, funding acquisition, validation, supervision, project administration, visualization, and writing (original draft, review, and editing). M.A.S.: Conceptualization, funding acquisition, supervision, project administration, and writing (original draft, review, and editing).
Competing interests: Patent application (no. UK2107576.7) related to this work was filed (to O.S.R. and B.N.K.). All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data are available from the NCBI Gene Expression Omnibus (GEO) under accession number GSE276195. LINCS L1000 dataset for EC transcriptomic responses to drug treatments are available from CLUE platform (https://clue.io/data/CMap2020#LINCS2020). The codes used in this paper for computational analysis are available at Zenodo (https://zenodo.org/records/14218021). The experimental materials can be provided by reasonable request and a completed material transfer agreement.
Supplementary Materials
The PDF file includes:
Figs. S1 to S5
Legends for tables S1 to S5
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.Hahn C., Schwartz M. A., Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 10, 53–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeyens N., Bandyopadhyay C., Coon B. G., Yun S., Schwartz M. A., Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Invest. 126, 821–828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitken C., Mehta V., Schwartz M. A., Tzima E., Mechanisms of endothelial flow sensing. Nat. Cardiovasc. Res. 2, 517–529 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Baeyens N., Nicoli S., Coon B. G., Ross T. D., Van den Dries K., Han J., Lauridsen H. M., Mejean C. O., Eichmann A., Thomas J. L., Humphrey J. D., Schwartz M. A., Vascular remodeling is governed by a VEGFR3-dependent fluid shear stress set point. eLife 4, e04645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiezzi M., Deng H., Baeyens N., Endothelial mechanosensing: A forgotten target to treat vascular remodeling in hypertension? Biochem. Pharmacol. 206, 115290 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Chien S., Effects of disturbed flow on endothelial cells. Ann. Biomed. Eng. 36, 554–562 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glagov S., Weisenberg E., Zarins C. K., Stankunavicius R., Kolettis G. J., Compensatory enlargement of human atherosclerotic coronary arteries. N. Engl. J. Med. 316, 1371–1375 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Humphrey J. D., Schwartz M. A., Vascular mechanobiology: Homeostasis, adaptation, and disease. Annu. Rev. Biomed. Eng. 23, 1–27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baeyens N., Schwartz M. A., Biomechanics of vascular mechanosensation and remodeling. Mol. Biol. Cell 27, 7–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien S., Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. Am. J. Physiol. Heart Circ. Physiol. 292, H1209–H1224 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Ward M. R., Tsao P. S., Agrotis A., Dilley R. J., Jennings G. L., Bobik A., Low blood flow after angioplasty augments mechanisms of restenosis: Inward vessel remodeling, cell migration, and activity of genes regulating migration. Arterioscler. Thromb. Vasc. Biol. 21, 208–213 (2001). [DOI] [PubMed] [Google Scholar]
- 12.De Mey J. G. R., Schiffers P. M., Hilgers R. H. P., Sanders M. M. W., Toward functional genomics of flow-induced outward remodeling of resistance arteries. Am. J. Physiol. Heart Circ. Physiol. 288, H1022–H1027 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Chiu J. J., Chien S., Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev. 91, 327–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamargo I. A., Baek K. I., Kim Y., Park C., Jo H., Flow-induced reprogramming of endothelial cells in atherosclerosis. Nat. Rev. Cardiol. 20, 738–753 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng H., Min E., Baeyens N., Coon B. G., Hu R., Zhuang Z. W., Chen M., Huang B., Afolabi T., Zarkada G., Acheampong A., McEntee K., Eichmann A., Liu F., Su B., Simons M., Schwartz M. A., Activation of Smad2/3 signaling by low fluid shear stress mediates artery inward remodeling. Proc. Natl. Acad. Sci. U.S.A. 118, e2105339118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baeyens N., Larrivee B., Ola R., Hayward-Piatkowskyi B., Dubrac A., Huang B., Ross T. D., Coon B. G., Min E., Tsarfati M., Tong H., Eichmann A., Schwartz M. A., Defective fluid shear stress mechanotransduction mediates hereditary hemorrhagic telangiectasia. J. Cell Biol. 214, 807–816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng H., Zhang J., Wang Y., Joshi D., Pi X., De Val S., Schwartz M. A., A KLF2-BMPER-Smad1/5 checkpoint regulates high fluid shear stress-mediated artery remodeling. Nat. Cardiovasc. Res. 3, 785–798 (2024). [DOI] [PubMed] [Google Scholar]
- 18.Schiebinger G., Shu J., Tabaka M., Cleary B., Subramanian V., Solomon A., Gould J., Liu S., Lin S., Berube P., Lee L., Chen J., Brumbaugh J., Rigollet P., Hochedlinger K., Jaenisch R., Regev A., Lander E. S., Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 928–943.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu X., Zhang Y., Martin-Rufino J. D., Weng C., Hosseinzadeh S., Yang D., Pogson A. N., Hein M. Y., Min K. H., Wang L., Grody E. I., Shurtleff M. J., Yuan R., Xu S., Ma Y., Replogle J. M., Lander E. S., Darmanis S., Bahar I., Sankaran V. G., Xing J., Weissman J. S., Mapping transcriptomic vector fields of single cells. Cell 185, 690–711.e45 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeo G. H. T., Saksena S. D., Gifford D. K., Generative modeling of single-cell time series with PRESCIENT enables prediction of cell trajectories with interventions. Nat. Commun. 12, 3222 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rukhlenko O. S., Halasz M., Rauch N., Zhernovkov V., Prince T., Wynne K., Maher S., Kashdan E., MacLeod K., Carragher N. O., Kolch W., Kholodenko B. N., Control of cell state transitions. Nature 609, 975–985 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks A. R., Lelkes P. I., Rubanyi G. M., Gene expression profiling of vascular endothelial cells exposed to fluid mechanical forces: Relevance for focal susceptibility to atherosclerosis. Endothelium 11, 45–57 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Nakajima H., Mochizuki N., Flow pattern-dependent endothelial cell responses through transcriptional regulation. Cell Cycle 16, 1893–1901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A., Narayan R., Corsello S. M., Peck D. D., Natoli T. E., Lu X., Gould J., Davis J. F., Tubelli A. A., Asiedu J. K., Lahr D. L., Hirschman J. E., Liu Z., Donahue M., Julian B., Khan M., Wadden D., Smith I. C., Lam D., Liberzon A., Toder C., Bagul M., Orzechowski M., Enache O. M., Piccioni F., Johnson S. A., Lyons N. J., Berger A. H., Shamji A. F., Brooks A. N., Vrcic A., Flynn C., Rosains J., Takeda D. Y., Hu R., Davison D., Lamb J., Ardlie K., Hogstrom L., Greenside P., Gray N. S., Clemons P. A., Silver S., Wu X., Zhao W. N., Read-Button W., Wu X., Haggarty S. J., Ronco L. V., Boehm J. S., Schreiber S. L., Doench J. G., Bittker J. A., Root D. E., Wong B., Golub T. R., A Next Generation Connectivity Map: L1000 Platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernández-Delgado M., Cernadas E., Barro S., Amorim D., Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 15, 3133–3181 (2014). [Google Scholar]
- 26.Lu Z. Q. J., The elements of statistical learning: Data mining, inference, and prediction. J. R. Stat. Soc. Ser. A Stat. Soc. 173, 693–694 (2010). [Google Scholar]
- 27.Kholodenko B. N., Kolch W., Rukhlenko O. S., Reversing pathological cell states: The road less travelled can extend the therapeutic horizon. Trends Cell Biol. 33, 913–923 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou M., Yu Y., Chen R., Liu X., Hu Y., Ma Z., Gao L., Jian W., Wang L., Wall shear stress and its role in atherosclerosis. Front. Cardiovasc. Med. 10, 1083547 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boon R. A., Horrevoets A. J. G., Key transcriptional regulators of the vasoprotective effects of shear stress. Hamostaseologie 29, 39–43 (2009). [PubMed] [Google Scholar]
- 30.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yabaji S. M., Rukhlenko O. S., Chatterjee S., Bhattacharya B., Wood E., Kasaikina M., Kholodenko B. N., Gimelbrant A. A., Kramnik I., Cell state transition analysis identifies interventions that improve control of Mycobacterium tuberculosis infection by susceptible macrophages. Sci. Adv. 9, eadh4119 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohli K., Pillarisetty V. G., Kim T. S., Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 29, 10–21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andueza A., Kumar S., Kim J., Kang D. W., Mumme H. L., Perez J. I., Villa-Roel N., Jo H., Endothelial reprogramming by disturbed flow revealed by single-cell RNA and chromatin accessibility study. Cell Rep. 33, 108491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies P. F., Civelek M., Fang Y., Guerraty M. A., Passerini A. G., Endothelial heterogeneity associated with regional athero-susceptibility and adaptation to disturbed blood flow in vivo. Semin. Thromb. Hemost. 36, 265–275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies P. F., Civelek M., Fang Y., Fleming I., The atherosusceptible endothelium: endothelial phenotypes in complex haemodynamic shear stress regions in vivo. Cardiovascular Res. 99, 315–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert M., Klinger B., Klunemann M., Sieber A., Uhlitz F., Sauer S., Garnett M. J., Bluthgen N., Saez-Rodriguez J., Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kholodenko B. N., Kiyatkin A., Bruggeman F. J., Sontag E., Westerhoff H. V., Hoek J. B., Untangling the wires: A strategy to trace functional interactions in signaling and gene networks. Proc. Natl. Acad. Sci. U.S.A. 99, 12841–12846 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kholodenko B. N., Untangling the signalling wires. Nat. Cell Biol. 9, 247–249 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halasz M., Kholodenko B. N., Kolch W., Santra T., Integrating network reconstruction with mechanistic modeling to predict cancer therapies. Sci. Signal 9, ra114 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Kholodenko B. N., Hoek J. B., Westerhoff H. V., Brown G. C., Quantification of information transfer via cellular signal transduction pathways. FEBS Lett. 414, 430–434 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Kholodenko B. N., Rauch N., Kolch W., Rukhlenko O. S., A systematic analysis of signaling reactivation and drug resistance. Cell Rep. 35, 109157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruggeman F. J., Westerhoff H. V., Hoek J. B., Kholodenko B. N., Modular response analysis of cellular regulatory networks. J. Theor. Biol. 218, 507–520 (2002). [PubMed] [Google Scholar]
- 43.Chen P. Y., Qin L., Baeyens N., Li G., Afolabi T., Budatha M., Tellides G., Schwartz M. A., Simons M., Endothelial-to-mesenchymal transition drives atherosclerosis progression. J. Clin. Invest. 125, 4514–4528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurya M. R., Gupta S., Li J. Y., Ajami N. E., Chen Z. B., Shyy J. Y., Chien S., Subramaniam S., Longitudinal shear stress response in human endothelial cells to atheroprone and atheroprotective conditions. Proc. Natl. Acad. Sci. U.S.A. 118, e2023236118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akimoto S., Mitsumata M., Sasaguri T., Yoshida Y., Laminar shear stress inhibits vascular endothelial cell proliferation by inducing cyclin-dependent kinase inhibitor p21(Sdi1/Cip1/Waf1). Circ. Res. 86, 185–190 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Sun H. Y., Kumar S., Puerta M. D. M., Jo H., Rezvan A., ZBTB46 is a shear-sensitive transcription factor inhibiting endothelial cell proliferation via gene expression regulation of cell cycle proteins. Lab. Invest 99, 305–318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee D. Y., Lee C. I., Lin T. E., Lim S. H., Zhou J., Tseng Y. C., Chien S., Chiu J. J., Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proc. Natl. Acad. Sci. U.S.A. 109, 1967–1972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin K., Hsu P. P., Chen B. P., Yuan S., Usami S., Shyy J. Y., Li Y. S., Chien S., Molecular mechanism of endothelial growth arrest by laminar shear stress. Proc. Natl. Acad. Sci. U.S.A. 97, 9385–9389 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denis J. F., Diagbouga M. R., Molica F., Hautefort A., Linnerz T., Watanabe M., Lemeille S., Bertrand J. Y., Kwak B. R., KLF4-Induced Connexin40 Expression Contributes to Arterial Endothelial Quiescence. Front. Phys. 10, 80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maleszewska M., Vanchin B., Harmsen M. C., Krenning G., The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis 19, 9–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mort R. L., Ford M. J., Sakaue-Sawano A., Lindstrom N. O., Casadio A., Douglas A. T., Keighren M. A., Hohenstein P., Miyawaki A., Jackson I. J., Fucci2a: A bicistronic cell cycle reporter that allows Cre mediated tissue specific expression in mice. Cell Cycle 13, 2681–2696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oki T., Nishimura K., Kitaura J., Togami K., Maehara A., Izawa K., Sakaue-Sawano A., Niida A., Miyano S., Aburatani H., Kiyonari H., Miyawaki A., Kitamura T., A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci. Rep. 4, 4012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chavkin N. W., Genet G., Poulet M., Jeffery E. D., Marziano C., Genet N., Vasavada H., Nelson E. A., Acharya B. R., Kour A., Aragon J., McDonnell S. P., Huba M., Sheynkman G. M., Walsh K., Hirschi K. K., Endothelial cell cycle state determines propensity for arterial-venous fate. Nat. Commun. 13, 5891 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai L. H., Lees E., Faha B., Harlow E., Riabowol K., The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene 8, 1593–1602 (1993). [PubMed] [Google Scholar]
- 55.Spencer S. L., Cappell S. D., Tsai F. C., Overton K. W., Wang C. L., Meyer T., The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng H., Xu Y., Hu X., Zhuang Z. W., Chang Y., Wang Y., Ntokou A., Schwartz M. A., Su B., Simons M., MEKK3-TGFbeta crosstalk regulates inward arterial remodeling. Proc. Natl. Acad. Sci. U.S.A. 118, e2112625118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Medrano-Bosch M., Simon-Codina B., Jimenez W., Edelman E. R., Melgar-Lesmes P., Monocyte-endothelial cell interactions in vascular and tissue remodeling. Front. Immunol. 14, 1196033 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allahwala U. K., Khachigian L. M., Nour D., Ridiandres A., Billah M., Ward M., Weaver J., Bhindi R., Recruitment and maturation of the coronary collateral circulation: Current understanding and perspectives in arteriogenesis. Microvasc. Res. 132, 104058 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Feaver R. E., Gelfand B. D., Blackman B. R., Human haemodynamic frequency harmonics regulate the inflammatory phenotype of vascular endothelial cells. Nat. Commun. 4, 1525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajra L., Evans A. I., Chen M., Hyduk S. J., Collins T., Cybulsky M. I., The NF-kappa B signal transduction pathway in aortic endothelial cells is primed for activation in regions predisposed to atherosclerotic lesion formation. Proc. Natl. Acad. Sci. U.S.A. 97, 9052–9057 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S., Sur S., Perez J., Demos C., Kang D. W., Kim C. W., Hu S., Xu K., Yang J., Jo H., Atorvastatin and blood flow regulate expression of distinctive sets of genes in mouse carotid artery endothelium. Curr. Top. Membr. 87, 97–130 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Welch-Reardon K. M., Wu N., Hughes C. C., A role for partial endothelial-mesenchymal transitions in angiogenesis? Arterioscler. Thromb. Vasc. Biol. 35, 303–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng S., Bowden N., Fragiadaki M., Souilhol C., Hsiao S., Mahmoud M., Allen S., Pirri D., Ayllon B. T., Akhtar S., Thompson A. A. R., Jo H., Weber C., Ridger V., Schober A., Evans P. C., Mechanical activation of hypoxia-inducible factor 1alpha drives endothelial dysfunction at atheroprone sites. Arterioscler. Thromb. Vasc. Biol. 37, 2087–2101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jhaveri K. A., Debnath P., Chernoff J., Sanders J., Schwartz M. A., The role of p21-activated kinase in the initiation of atherosclerosis. BMC Cardiovasc. Disord. 12, 55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orr A. W., Stockton R., Simmers M. B., Sanders J. M., Sarembock I. J., Blackman B. R., Schwartz M. A., Matrix-specific p21-activated kinase activation regulates vascular permeability in atherogenesis. J. Cell Biol. 176, 719–727 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh N. K., Kotla S., Dyukova E., Traylor J. G. Jr., Orr A. W., Chernoff J., Marion T. N., Rao G. N., Disruption of p21-activated kinase 1 gene diminishes atherosclerosis in apolipoprotein E-deficient mice. Nat. Commun. 6, 7450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yurdagul A. Jr., Chen J., Funk S. D., Albert P., Kevil C. G., Orr A. W., Altered nitric oxide production mediates matrix-specific PAK2 and NF-kappaB activation by flow. Mol. Biol. Cell 24, 398–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foster D. A., Yellen P., Xu L., Saqcena M., Regulation of G1 cell cycle progression: Distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s). Genes Cancer 1, 1124–1131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barakat A., Lieu D., Differential responsiveness of vascular endothelial cells to different types of fluid mechanical shear stress. Cell Biochem. Biophys. 38, 323–343 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Souilhol C., Serbanovic-Canic J., Fragiadaki M., Chico T. J., Ridger V., Roddie H., Evans P. C., Endothelial responses to shear stress in atherosclerosis: A novel role for developmental genes. Nat. Rev. Cardiol. 17, 52–63 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Yee A., Bosworth K. A., Conway D. E., Eskin S. G., McIntire L. V., Gene expression of endothelial cells under pulsatile non-reversing vs. steady shear stress; comparison of nitric oxide production. Ann. Biomed. Eng. 36, 571–579 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Lum R. M., Wiley L. M., Barakat A. I., Influence of different forms of fluid shear stress on vascular endothelial TGF-beta1 mRNA expression. Int. J. Mol. Med. 5, 635–641 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Conway D. E., Sakurai Y., Weiss D., Vega J. D., Taylor W. R., Jo H., Eskin S. G., Marcus C. B., McIntire L. V., Expression of CYP1A1 and CYP1B1 in human endothelial cells: Regulation by fluid shear stress. Cardiovasc. Res. 81, 669–677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jayapal S. R., Wang C. Q., Bisteau X., Caldez M. J., Lim S., Tergaonkar V., Osato M., Kaldis P., Hematopoiesis specific loss of Cdk2 and Cdk4 results in increased erythrocyte size and delayed platelet recovery following stress. Haematologica 100, 431–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fisher O. S., Deng H., Liu D., Zhang Y., Wei R., Deng Y., Zhang F., Louvi A., Turk B. E., Boggon T. J., Su B., Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex. Nat. Commun. 6, 7937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng H., Schwartz M. A., High fluid shear stress inhibits cytokine-driven Smad2/3 activation in vascular endothelial cells. J. Am. Heart Assoc. 11, e025337 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X., Shen Y., Shang M., Liu X., Munn L. L., Endothelial mechanobiology in atherosclerosis. Cardiovasc. Res. 119, 1656–1675 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conway D. E., Williams M. R., Eskin S. G., McIntire L. V., Endothelial cell responses to atheroprone flow are driven by two separate flow components: low time-average shear stress and fluid flow reversal. Am. J. Physiol. Heart Circ. Physiol. 298, H367–H374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Godbole A. S., Lu X., Guo X., Kassab G. S., NADPH oxidase has a directional response to shear stress. Am. J. Physiol. Heart Circ. Physiol. 296, H152–H158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lill D., Rukhlenko O. S., Mc Elwee A. J., Kashdan E., Timmer J., Kholodenko B. N., Mapping connections in signaling networks with ambiguous modularity. NPJ Syst. Biol. Appl. 5, 19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu Q., Smith D. O., Hoo K. A., Shear stress effect on endothelial nitric oxide synthase in cultured human umbilical vein endothelial cells. J. Biomed. Sci. Eng. 06, 982–986 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Legends for tables S1 to S5
Tables S1 to S5