Abstract
The COP9 signalosome (CSN) is a highly conserved protein complex in eukaryotes, with CSN5 serving as its critical catalytic subunit. However, the role of CSN5 in plant immunity is largely unexplored. Here, we found that suppression of OsCSN5 in rice enhances resistance against the fungal pathogen Magnaporthe oryzae and the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo) without affecting growth. OsCSN5 is ubiquitinated and degraded by the E3 ligase OsPUB45. Overexpression of OsPUB45 increased resistance against M. oryzae and Xoo, while dysfunction of OsPUB45 decreased resistance. In addition, OsCSN5 stabilized OsCUL3a to promote the degradation of a positive regulator OsNPR1. Overexpression of OsPUB45 compromised accumulation of OsCUL3a, leading to stabilization of OsNPR1, whereas mutations in OsPUB45 destabilized OsNPR1. These findings suggest that OsCSN5 stabilizes OsCUL3a to facilitate the degradation of OsNPR1, preventing its constitutive activation without infection. Conversely, OsPUB45 promotes the degradation of OsCSN5, contributing to immunity activation upon pathogen infection.
Ubiquitination of COP9 signalosome subunit 5 confers plant broad-spectrum disease resistance without growth penalties.
INTRODUCTION
Pathogenic organisms can cause severe damages to crop plants, posing a threat to global food security. Plants have developed complex immune systems to protect against these pathogens (1, 2). However, activating defense mechanisms in the absence of pathogens can be costly and harmful to plant growth and overall fitness (3). For example, removing the Mildew Resistance Locus O (MLO) gene in barley, Arabidopsis (Arabidopsis thaliana), and wheat (Triticum aestivum) can provide broad-spectrum resistance to powdery mildew, but it can also lead to unintended consequences such as premature aging (4). Similarly, knocking out SPOTTED LEAF 11 (SPL11) and ENHANCED BLIGHT AND BLAST RESISTANCE 1 (EBR1) in rice can enhance resistance to Magnaporthe oryzae, but it can also cause notable cell death (5, 6), making it challenging to use these defense genes in practical agricultural settings. Recent studies have shown that some gene knockouts can provide disease resistance without adversely affecting plant growth (7). For instance, knocking-out PUCCINIA STRIIFORMIS-INDUCED PROTEIN KINASE 1 (TaPsIPK1) in wheat, BROAD-SPECTRUM RESISTANCE DIGU 1 (BSR-D1) and valine-glutamine (VQ) motif-containing protein OsVQ25 in rice, or nicotinate N-methyltransferase (ZmNANMT) in maize can enhance resistance to fungal and/or bacterial pathogens without affecting plant growth (8–11). Therefore, it is essential to identify genes that offer resistance without causing yield penalties.
The COP9 signalosome (CSN) is a highly conserved protein complex found in higher eukaryotes, consisting of eight subunits known as CSN1 to CSN8 (12). Among these subunits, CSN5 plays a crucial role in removing “Related to Ubiquitin” (RUB) modification from the cullin subunit in Cullin (CUL)-RING ubiquitin ligase (CRL) complexes (13). In mammals, CSN5 positively regulates the Cul3/Keap1-mediated degradation of the nuclear factor E2-related factor 2 to control innate immune responses in macrophages (14). In Arabidopsis, mutations in either CSN5A or CSN5B lead to the inactivation of CSN and a loss of deRUBylation by CUL1 and CUL4 (15). Arabidopsis CSN5A interacts with NB-LRR proteins, RLKs, and 29 distinct effectors from Hyaloperonospora arabidopsidis (Hpa) and Pseudomonas syringae (Psy). Dysfunction of CSN5A enhances resistance to Hpa and Psy (16), indicating a critical role of CSN5 proteins in immunity. Furthermore, silencing or mutation of TaCSN5 enhances wheat resistance against Puccinia triticina and multiple Puccinia striiformis f. sp. tritici isolates (17, 18). Transient silencing of VvCSN5 in grapevine (Vitis vinifera) boosts resistance to powdery mildew (19). These studies have shown that plant CSN5 proteins are crucial for immunity. However, the regulation of CSN5 and its related signaling pathway in plant immunity remains not fully understood.
Ubiquitination-mediated protein degradation is a universal mechanism that regulates the amount of key immune regulators to maintain immune system function (20–22). For instance, the U-box type E3 ligases PUB25 and PUB26 target nonactivated BIK1 for polyubiquitination and degradation in Arabidopsis (23). In rice, SDS2 and OsBAG4 positively regulate resistance against M. oryzae and their overexpression, leading to autoimmunity and cell death. However, the E3 ligases SPL11 and EBR1 degrade SDS2 and OsBAG4, respectively, to coordinate the trade-off between defense and growth (5, 6). Rice plants overexpressing OsNPR1, an ortholog of Arabidopsis salicylic acid (SA) and immune signaling master regulator NPR1, have shown enhanced resistance to M. oryzae and Xoo, although this overexpression affects plant growth and development (24–26). While OsCUL3a interacts with OsRBX1a or OsRBX1b to form a CRL complex, which mediates the degradation of OsNPR1, thereby negatively regulating resistance against the aforementioned pathogens (27). In mammals, phosphorylated CSN5 undergoes ubiquitination through the 26S proteasome pathway to activate nuclear factor κB (NF-κB) activity (28). CSN5 plays crucial roles in plant immunity regulation, while whether it can be modified by ubiquitination, and the related E3 ubiquitin ligase remains largely unexplored in plants.
In this study, we found that silencing of OsCSN5 in rice enhanced disease resistance against fungus M. oryzae and the bacterial Xoo without growth penalties. We found that OsCSN5 is ubiquitinated and degraded by the E3 ligase OsPUB45 via the 26S proteasome pathway. Conversely, OsPUB45 positively regulates disease resistance against M. oryzae and Xoo. OsCSN5 interacts with OsCUL3a, which is proven to degrade OsNPR1 to negatively regulate immunity. Overall, our results indicate that OsCSN5 contributes to the stabilization of OsCUL3a, which facilitates the degradation of OsNPR1, while OsPUB45 degrades OsCSN5 to inhibit OsCUL3a and activate OsNPR1-mediated immunity.
RESULTS
Silencing of OsCSN5 in rice leads to broad-spectrum disease resistance without growth penalties
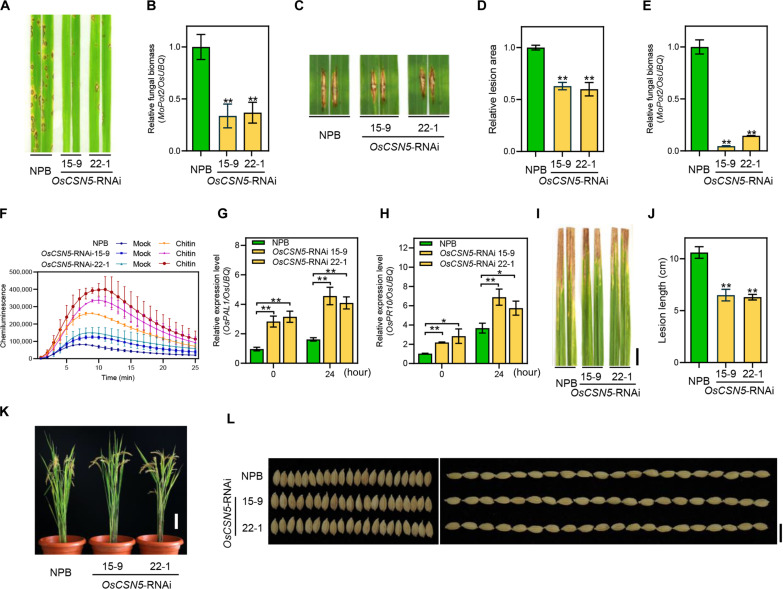
To determine the role of OsCSN5 in blast disease resistance, we examined its expression pattern in Nipponbare (NPB) plants following infection with the compatible M. oryzae isolate RB22. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) analysis showed that OsCSN5 was rapidly induced at 12 and 24 hours after inoculation with M. oryzae (fig. S1A). To further explore the disease resistance phenotype of OsCSN5, OsCSN5 knockout mutants (oscsn5) were generated by CRISPR-Cas9–mediated genome editing approach (fig. S1B). Various editing types in the T0 generation, including 1–, 2–, and 4–base pair (bp) deletions leading to early termination occurred. However, we were unable to obtain loss-of-function oscsn5 homozygous mutants in T1 generation (fig. S1C), suggesting potential embryo mortality. As an alternative, we generated OsCSN5 knockdown lines via RNA interference (RNAi) approach. Two OsCSN5 RNAi constructs, OsCSN5 RNAi (I) and OsCSN5 RNAi (II), targeting the regions of 594 to 891 bp and 822 to 1083 bp of OsCSN5 coding sequence, respectively, were built and introduced into NPB plants (fig. S1D), resulting in two independent OsCSN5-RNAi lines with significantly reduced expression of OsCSN5 as estimated by qRT-PCR (fig. S1E). The spray inoculation assay at the seedling stage demonstrated that the OsCSN5-RNAi plants exhibited enhanced resistance with lower fungal biomass relative to the wild type NPB plants (Fig. 1, A and 1B). Subsequent punch inoculation of OsCSN5-RNAi lines at the tillering stage also developed smaller lesion area and reduced fungal biomass (Fig. 1, C to E), indicating that OsCSN5 negatively regulates M. oryzae resistance at both seedling and tillering stages.
Fig. 1. Silencing of OsCSN5 in rice enhances disease resistance against M. oryzae and Xoo without growth penalties.
(A and B) Disease symptoms (A) and relative fungal biomass (B) of representative leaves from OsCSN5-RNAi and NPB plants after spray inoculation with M. oryzae isolate RB22. (C to E) Disease symptoms (C), the relative leaf area with lesions as measured by ImageJ (D), and relative fungal biomass (E) of representative leaves of OsCSN5-RNAi and NPB plants after punch inoculation with M. oryzae isolate RB22. (F) Chitin-induced ROS accumulation dynamics in OsCSN5-RNAi and NPB plants. Water treatment (Mock) was the negative control. (G and H) Relative transcript levels of the defense-related genes OsPAL1 (G) and OsPR10 (H) in OsCSN5-RNAi and NPB plants at 0 and 24 hours after spray inoculation with M. oryzae isolate RB22 as determined by qRT-PCR. (I and J) Phenotypes of the leaves of OsCSN5-RNAi plants inoculated with the Xoo isolate PXO99A (I) and lesion length (J). Scale bar, 2 cm. (K) Morphological phenotypes of OsCSN5-RNAi plants. Scale bar, 10 cm. (L) Grain width (left) and grain length (right) of OsCSN5-RNAi and NPB plants. Scale bar, 1 cm. All the statistical analysis data are shown as mean ± SE, and significance was determined at *P < 0.05 and **P < 0.01 (n = 3) with a two-tailed Student’s t test.
To investigate the resistance mechanism mediated by OsCSN5, we measured the accumulation of reactive oxygen species (ROS) in response to chitin treatment. The OsCSN5-RNAi lines exhibited a higher ROS burst compared to the NPB control (Fig. 1F). Moreover, the transcript levels of defense marker genes, OsPAL1 and OsPR10, were significantly up-regulated in the OsCSN5-RNAi plants at 0 and 24 hours after inoculation with M. oryzae isolate RB22 (Fig. 1, G and 1H). To further assess the impact of OsCSN5 suppression on resistance against bacterial pathogen, the OsCSN5-RNAi plants were inoculated with a compatible Xoo isolate. Two weeks after inoculation, the OsCSN5-RNAi plants displayed enhanced resistance to Xoo, as evidenced by shorter lesions and lower bacterial growth compared to NPB plants (Fig. 1, I and J, and fig. S1F). These findings indicated that OsCSN5 plays a negative regulatory role in resistance to fungal and bacterial diseases.
The broad-spectrum resistance displayed by OsCSN5-RNAi lines prompted us to investigate their agronomic traits in field conditions. Unexpectedly, the OsCSN5-RNAi plants showed normal growth in the field (Fig. 1K). To further evaluate their performance, various agronomic traits such as plant height, grain length, grain width, effective panicle numbers per plant, grain number per panicle, seed setting rate, and thousand-grain weight were analyzed. These traits were comparable to those of the NPB plants (Fig. 1L and fig. S1, G to K). These findings demonstrated that suppression of OsCSN5 can improve broad-spectrum disease resistance without obvious growth penalties.
OsCSN5 interacts with and is ubiquitinated by OsPUB45
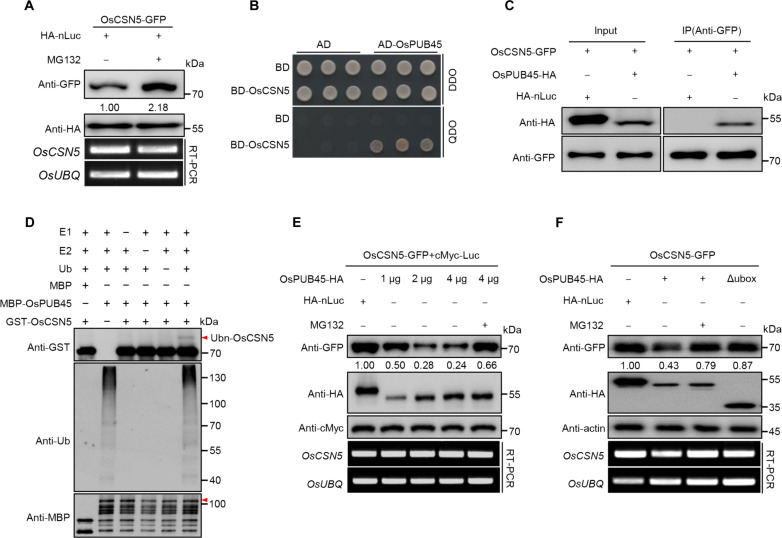
To explore the regulation mechanism of the OsCSN5 protein, transiently experiments were conducted in rice protoplasts through expressing OsCSN5-GFP and treatment with the 26S proteasome inhibitor MG132. Immunoblotting analysis showed a notable increase of the OsCSN5 protein level upon MG132 treatment. In addition, RT-PCR analysis indicated that the transcript levels of OsCSN5 remained consistent (Fig. 2A), suggesting that OsCSN5 protein may be targeted for degradation through the 26S proteasome pathway. To further explore the proteins involved in the degradation of OsCSN5, we screened the ubiquitin E3 ligase (UbE3)–ORFeome library using OsCSN5 as bait (29), leading to the identification of a U-box domain-containing protein known as OsPUB45 (encoded by LOC_Os02g33680). Subsequent yeast two-hybrid assay confirmed the interaction between OsCSN5 and OsPUB45 (Fig. 2B). To examine their interaction in vivo, we conducted a co-immunoprecipitation (co-IP) assay by coexpressing OsCSN5-GFP and OsPUB45-HA or HA-nLuc in rice protoplasts. Immunoblot analysis with an anti-GFP antibody showed that OsCSN5–green fluorescent protein (GFP) coprecipitates with OsPUB45–hemagglutinin (HA), but not with the control HA-nLuc (Fig. 2C). Moreover, a bimolecular fluorescence complementation (BiFC) assay demonstrated the reconstitution of yellow fluorescent protein when OsCSN5-nYFP [OsCSN5 fused to the N-terminal half of yellow fluorescent protein (YFP)] and OsPUB45-cYFP (OsPUB45 fused to the C-terminal half of YFP) were coexpressed, supporting the interaction between these proteins in cytoplasm and nucleus (fig. S2, A and B). The collective evidence solidifies the conclusion that OsCSN5 interacts with OsPUB45 in planta.
Fig. 2. OsPUB45 ubiquitinates and targets OsCSN5 for degradation via the 26S proteasome pathway.
(A) OsCSN5-GFP abundance was detected in transfected protoplasts after treatment with dimethyl sulfoxide or MG132. OsCSN5-GFP was then detected by immunoblotting with an anti-GFP antibody. HA-nLuc serves as a loading control. The relative transcript levels of OsCSN5 and OsUBQ were detected by RT-PCR. (B) OsPUB45 interacts with OsCSN5 in yeast. AD, pGADT7 AD vector; BD, pGBKT7 BD vector. Yeast were grown on DDO (SD-Leu-Trp) or QDO (SD-Leu-Trp-His-Ade) medium. (C) Co-IP assay for the interaction between OsCSN5 and OsPUB45 in rice protoplasts. OsCSN5-GFP was coexpressed with HA-nLuc and OsPUB45-HA in rice protoplasts. (D) Ubiquitination assay of OsCSN5 by OsPUB45 in vitro. GST-OsCSN5 from Escherichia coli was incubated with E1, E2, Ub, and rice total extract preincubated MBP-OsPUB45 in the reactions. Ubiquitination of GST-OsCSN5 was detected by anti-GST antibody. Ubn-OsCSN5 denotes ubiquitinated OsCSN5 band. (E) Degradation of OsCSN5 is OsPUB45 dosage dependent. OsCSN5-GFP was coexpressed with HA-nLuc, OsPUB45-HA in rice protoplasts, respectively. Numbers under the bands indicate relative OsCSN5-GFP abundance, normalized to cMyc-Luc as loading control. The relative transcript levels of OsCSN5 and OsUBQ were detected by RT-PCR. Similar results were obtained from three independent biological experiments. (F) OsPUB45 promotes the degradation of OsCSN5 via 26S proteasome pathway in vivo. OsCSN5-GFP was cotransfected with HA-nLuc, OsPUB45-HA, or OsPUB45-Δubox-HA in rice protoplasts, respectively. Actin serves as an internal control. The relative transcript levels of OsCSN5 and OsUBQ were detected by RT-PCR. Similar results were obtained from three independent biological experiments.
We then investigated whether OsPUB45 ubiquitinates OsCSN5. In the presence of ubiquitin, E1, and E2, the auto-ubiquitination activity of MBP-OsPUB45 was detected using an anti-ubiquitin antibody (Fig. 2D). A high–molecular weight polyubiquitination band of glutathione S-transferase (GST)–OsCSN5 was evident when incubated with MBP-OsPUB45, E1, E2, and ubiquitin (Fig. 2D), indicating that OsCSN5 can be polyubiquitinated by OsPUB45. To determine whether OsPUB45 promotes the degradation of OsCSN5 in vivo, we coexpressed the same amount of OsCSN5-GFP either alone or together with increasing amounts of OsPUB45-HA in rice protoplasts. The OsCSN5-GFP protein decreased substantially with higher levels of OsPUB45 protein (Fig. 2E). Treatment with MG132 markedly inhibited the degradation of OsCSN5 by OsPUB45, and RT-PCR analysis showed consistent transcript levels of OsCSN5 in all conditions (Fig. 2E). As OsPUB45 exhibits E3 ligase activity, we investigated whether OsPUB45-mediated OsCSN5 degradation depended on its U-box domain. We first tested the self-ubiquitination activity with deletion of the U-box domain. The auto-ubiquitination activity lost in MBP-OsPUB45-Δubox compared to MBP-OsPUB45 (fig. S2C), indicating the importance of the U-box domain for self-ubiquitination. In vivo degradation assays revealed that the decreased OsCSN5-GFP levels by OsPUB45-HA was partially blocked when coexpressing with OsPUB45-Δubox-HA, with consistent OsCSN5 transcript levels in all conditions (Fig. 2F). These findings suggested that OsPUB45 ubiquitinates OsCSN5, leading to its proteasomal degradation. To determine whether the ubiquitination and degradation of OsCSN5 by OsPUB45 is influenced by chitin treatment. We transiently coexpressed OsCSN5-GFP and OsPUB45-HA in rice protoplasts with or without 80 nM chitin treatment for 1 hour. Immunoblot analysis revealed that chitin treatment enhanced the ubiquitination and degradation of OsCSN5 by OsPUB45 (fig. S2, D and E).
OsPUB45 positively regulates disease resistance against M. oryzae and Xoo
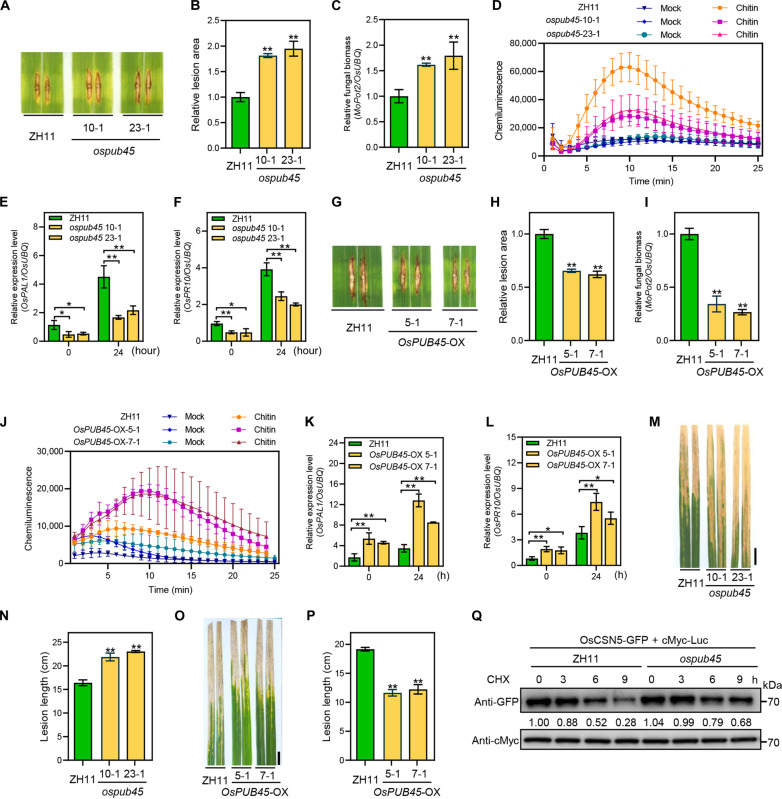
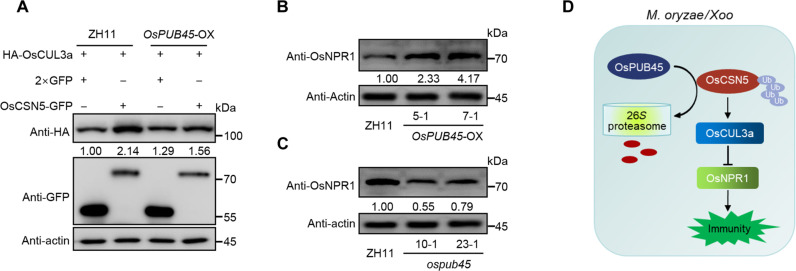
To determine the role of OsPUB45 in rice blast disease resistance, the expression levels of OsPUB45 were examined in NPB plants infected with compatible M. oryzae isolate. OsPUB45 was significantly up-regulated at 48 hours after inoculation compared to mock-treated plants (fig. S3A), suggesting a potential involvement of OsPUB45 in rice blast resistance. Next, ospub45 knockout mutants were generated using the CRISPR-Cas9 approach in the japonica rice variety Zhonghua 11 (ZH11), which is highly conserved with NPB at the genome level (30). Two independent homozygous lines, i.e., 10-1 and 23-1 in which a 1-bp insertion and 2-bp deletion occurred, respectively, leading to early termination (fig. S3B). The spray inoculation assay at the seedling stage showed that the ospub45 mutants displayed increased susceptibility to M. oryzae infection, as evidenced by higher fungal biomass compared to the wild-type ZH11 plants (fig. S3, C and D). We subsequently also performed the punch inoculation with the same isolate at tillering stage. The ospub45 mutants developed much larger disease lesion area and more fungal biomass than that in ZH11 plants (Fig. 3, A to C). Corresponding to reduced resistance, the ospub45 mutants showed reduced chitin-triggered ROS burst and lower expression levels of defense-related genes OsPAL1 and OsPR10 at 0 and 24 hours after inoculation with M. oryzae isolate RB22 (Fig. 3, D to F), these findings demonstrated that dysfunction of OsPUB45 compromised rice blast responses. The OsPUB45 overexpressing transgenic rice plants driven by maize UBIQUITIN promoter (OsPUB45-OX) were then generated and identified by qRT-PCR analysis (fig. S3E), two independent lines were selected for resistance evaluation inoculated with M. oryzae isolate. Contrast with the ospub45 mutants, the OsPUB45-OX plants showed enhanced resistance to M. oryzae infection, with smaller lesion size and reduced fungal biomass compared with the wild-type ZH11 (Fig. 3, G to I). Moreover, the chitin-triggered ROS burst and the expression of defense-associated genes OsPAL1 and OsPR10 were largely increased in OsPUB45-OX plants at 0 and 24 hours after inoculation with M. oryzae isolate RB22 compared to the wild type (Fig. 3, J to L). To explore whether OsPUB45 was also involved in bacterial disease resistance, we evaluated the resistance of the ospub45 mutants and OsPUB45-OX lines to infection with the Xoo isolate PXO99A. Two weeks after inoculation, the Xoo disease lesions developed longer, and bacterial growth was higher on the ospub45 mutants than on ZH11 plants (Fig. 3, M and N, and fig. S3F), whereas the OsPUB45-OX plants showed shorter lesion length and lower bacterial growth (Fig. 3, O and P, and fig. S3G). Together, these data further supported the role of OsPUB45 as a positive regulator of rice immunity. Next, we determined the stability of OsCSN5 in ospub45 mutants through transiently expressing OsCSN5-GFP in rice protoplasts isolated from ospub45 mutant and ZH11 plants. The protoplasts were treated with protein synthesis inhibitor cycloheximide (CHX). The results revealed that the degradation of OsCSN5-GFP protein was slower in ospub45 protoplasts compared to ZH11 protoplasts (Fig. 3Q). Conversely, the OsCSN5-GFP protein exhibited decreased stability when expressed in OsPUB45-OX protoplasts (fig. S3H). These results indicate that OsPUB45 facilitates the degradation of OsCSN5 in rice cells.
Fig. 3. OsPUB45 positively regulates rice disease resistance to M. oryzae and Xoo.
(A to C) Disease symptoms (A), the relative leaf area with lesions as measured by ImageJ (B), and relative fungal biomass (C) of representative leaves of ospub45 mutants and ZH11 plants after punch inoculation with M. oryzae isolate RB22. (D) Chitin-induced ROS accumulation dynamics in ospub45 mutants and ZH11 plants. Water treatment (Mock) was the negative control. (E and F) Relative transcript levels of defense-related genes OsPAL1 (E) and OsPR10 (F) in ospub45 mutants and ZH11 plants at 0 and 24 hours after spray inoculation with M. oryzae isolate RB22 as determined by qRT-PCR. (G to I) Disease symptoms (G), the relative leaf area with lesions as measured by ImageJ (H), and relative fungal biomass (I) of representative leaves of OsPUB45-OX and ZH11 plants after punch inoculation with M. oryzae isolate RB22. (J) Chitin-induced ROS accumulation dynamics in OsPUB45-OX and ZH11 plants. Water treatment (Mock) was the negative control. (K and L) Relative transcript levels of defense-related genes OsPAL1 (K) and OsPR10 (L) in OsPUB45-OX and ZH11 plants at 0 and 24 hours after spray inoculation with M. oryzae isolate RB22 as determined by qRT-PCR. (M and N) Phenotypes of ospub45 mutants inoculated with the Xoo isolate PXO99A (M) and lesion length (N). Scale bar, 2 cm. (O and P) Phenotypes of OsPUB45-OX plants inoculated with the Xoo isolate PXO99A (O), and lesion length (P). Scale bar, 2 cm. (Q) Time-course degradation of OsCSN5-GFP in rice protoplasts from ZH11 or ospub45 mutants. Cotransfected rice protoplasts were treated with 100 μM CHX to block protein synthesis for the indicated duration of time, and OsCSN5-GFP levels were monitored by immunoblotting. All the statistical analysis data are shown as mean ± SE, and significance was determined at *P < 0.05 and **P < 0.01 (n = 3) with a two-tailed Student’s t test.
OsCSN5 interacts with and stabilizes OsCUL3a
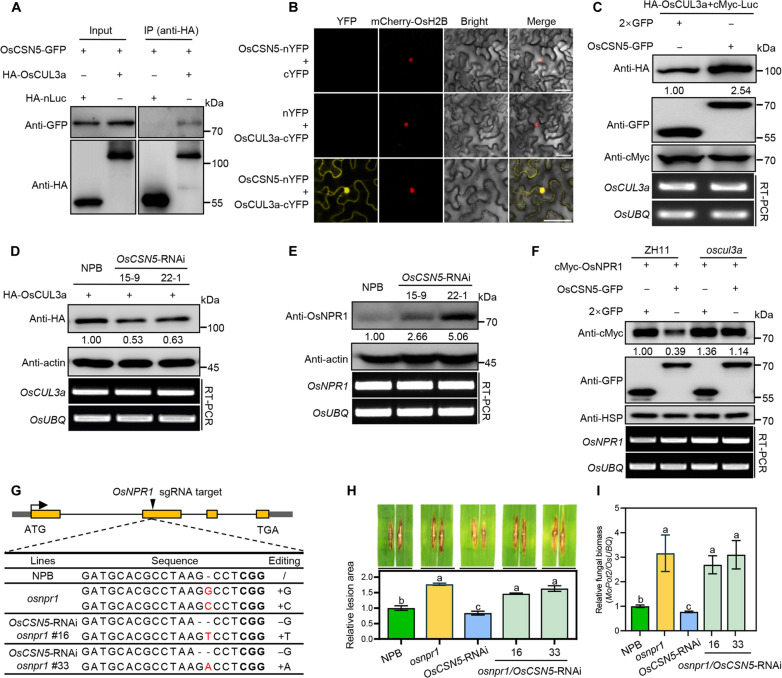
In Arabidopsis, csn5a-2 cul3a double mutants exhibit substantially enhanced disease resistance against the virulent Oomycete Hpa and overaccumulation of PR1 protein compared to cul3a single mutants, suggesting that CUL3a and CSN5 coordinately negatively regulate resistance (16). In rice, OsCUL3a has been found to negatively regulate cell death and resistance against M. oryzae and Xoo (27). To confirm the interaction between OsCSN5 and OsCUL3a in rice cell, we performed a co-IP assay by transfecting rice protoplasts with OsCSN5-GFP and either HA-OsCUL3a or HA-nLuc plasmids. The HA-OsCUL3a fusion protein was immunoprecipitated from the rice extracts using an anti-HA antibody, and OsCSN5-GFP was specifically detected in the immunocomplex (Fig. 4A). To further verify the location of the interaction between OsCSN5 and OsCUL3a in the cell, Agrobacterium strains containing OsCSN5-nYFP and OsCUL3a-cYFP constructs were co-infiltrated into Nicotiana benthamiana leaves. The BiFC assay showed that reconstituted YFP fluorescence occurred in the cytoplasm and nucleus, while no signal was observed in the negative control combinations (Fig. 4B and fig. S4A). These findings collectively demonstrate that OsCSN5 interacts with OsCUL3a in planta.
Fig. 4. OsCSN5-mediated resistance through OsCUL3a-OsNPR1 pathway.
(A) Co-IP assay for the interaction between OsCSN5 and OsCUL3a in rice protoplasts. (B) BiFC assay to confirm the interaction between OsCSN5 and OsCUL3a in N. benthamiana. Scale bars, 50 μm. mCherry-OsH2B was coexpressed to indicate the nucleus. (C) Stability of OsCUL3a promoted by OsCSN5 in rice protoplasts. HA-OsCUL3a was cotransfected with 2×GFP, OsCSN5-GFP in rice protoplasts, followed by immunoblotting. cMyc-Luc protein was coexpressed as a loading control. The relative transcript levels of OsCUL3a and OsUBQ were detected by RT-PCR. There were three biological replicates with similar results. (D) Stability of OsCUL3a in OsCSN5-RNAi rice protoplasts. Plasmid of HA-OsCUL3a was transfected into NPB and OsCSN5-RNAi rice protoplasts. Actin protein was served as the internal control. The relative transcript levels of OsCUL3a and OsUBQ were detected by RT-PCR. (E) Immunoblot analysis of OsNPR1 protein levels in the NPB and OsCSN5-RNAi plants using an anti-OsNPR1 antibody. Three independent experiments were carried out with similar results. (F) Degradation of OsNPR1 by OsCSN5 in ZH11 and oscul3a rice protoplasts. The relative transcript levels of OsNPR1 and OsUBQ were detected by RT-PCR. (G) Diagram of the mutations in osnpr1 mutants generated by the CRISPR-Cas9 technology. The yellow boxes represent exons, and gray boxes represent untranslated regions (UTRs). (H and I) Disease symptoms, the relative leaf area with lesions as measured by ImageJ (H), and relative fungal biomass (I) of representative leaves of osnpr1, OsCSN5-RNAi, osnpr1/OsCSN5-RNAi, and NPB plants after punch inoculation with M. oryzae isolate RB22. Values are means ± SD (n = 3, technical repeats).
To understand how OsCSN5 interacts with OsCUL3a to regulate rice resistance, we conducted experiments by cotransfecting HA-OsCUL3a with either OsCSN5-GFP or 2×GFP in rice protoplasts. Immunoblotting analysis revealed a substantial increase of OsCUL3a protein level when coexpressed with OsCSN5-GFP compared to the 2×GFP control while similar transcript levels of OsCUL3a in both combinations (Fig. 4C), demonstrating that OsCSN5 can promote the accumulation of OsCUL3a protein. Subsequently, we transfected HA-OsCUL3a plasmid into protoplasts isolated from NPB and OsCSN5-RNAi lines. The intensity of the HA-OsCUL3a band was notably weaker in OsCSN5-RNAi protoplasts compared to that in NPB protoplasts (Fig. 4D). These results demonstrated that OsCSN5 plays a role in stabilizing OsCUL3a protein in planta.
OsCSN5-mediated resistance through downstream OsCUL3a-OsNPR1 pathway
OsCUL3a plays a crucial role in regulating cell death and immunity by promoting the degradation of OsNPR1 (27). The stabilization of OsCUL3a by OsCSN5 prompted us to determine the impact of suppressing OsCSN5 on OsCUL3a-mediated OsNPR1 degradation. Total proteins were extracted from OsCSN5-RNAi and NPB plants, and immunoblotting analyses revealed higher levels of OsNPR1 protein in OsCSN5-RNAi lines compared to that in NPB plants despite similar transcript levels of OsNPR1 in both (Fig. 4E). Conversely, when coexpressing with OsCSN5-GFP, a lower level of OsNPR1 protein was detected using anti-cMyc antibody compared to coexpress with 2×GFP in rice protoplasts (fig. S4B). To determine whether OsCSN5-mediated OsNPR1 degradation is dependent on OsCUL3a, we transiently coexpressed cMyc-OsNPR1 with OsCSN5-GFP or the control 2×GFP in rice protoplasts from oscul3a mutant and ZH11. Immunoblotting analysis showed a rapid decrease in OsNPR1 levels when coexpressed with OsCSN5-GFP in ZH11 plants (Fig. 4F, lane 1 and lane 2). In contrast, cMyc-OsNPR1 protein levels remained stable in the oscul3a mutant (Fig. 4F, lane 3 and lane 4), with consistent transcript levels of OsNPR1 in all combinations as confirmed by RT-PCR analysis (Fig. 4F). These findings demonstrate that OsCSN5 stabilizes OsCUL3a to promote OsNPR1 degradation.
To determine whether OsCSN5-mediated resistance through OsNPR1, we utilized CRISPR-Cas9 technology to generate osnpr1 knockout mutants in both NPB and OsCSN5-RNAi backgrounds (Fig. 4G and fig. S4C). Subsequently, we assessed their resistance phenotypes against M. oryzae. The osnpr1 mutants exhibited decreased resistance compared to NPB, as evidenced by larger lesions and higher fungal biomass (fig. S4, D to F). The resistance of OsCSN5-RNAi plants was compromised in plants harboring both osnpr1 mutation and OsCSN5 suppression (Fig. 4, H and I). These results indicate that OsCSN5-mediated resistance likely achieved through the downstream action of OsNPR1.
OsPUB45 interferes with the effects of OsCSN5 on OsCUL3a-mediated OsNPR1 degradation
To further explore the impact of OsPUB45 on the stability of OsCUL3a mediated by OsCSN5, we conducted an experiment by coexpressing HA-OsCUL3a with OsCSN5-GFP or 2×GFP in OsPUB45-OX and ZH11 protoplasts. Immunoblotting analysis revealed that the increased amount of HA-OsCUL3a by OsCSN5-GFP in ZH11 was remarkably reduced in OsPUB45-OX plants (Fig. 5A), indicating that OsPUB45-mediated degradation of OsCSN5 leads to a decrease in OsCUL3a protein abundance. Given that OsCSN5 negatively regulates OsNPR1 accumulation, we also investigated the impact of OsPUB45 on OsNPR1 protein accumulation. Upon isolating total RNA and proteins from OsPUB45-OX lines and ZH11, we observed comparable transcript levels of OsNPR1 (fig. S5A). However, the protein levels of OsNPR1 were increased in two OsPUB45-OX lines when analyzed using an anti-OsNPR1 antibody (Fig. 5B). Conversely, the transcript level of OsNPR1 remained consistent (fig. S5B), but the OsNPR1 protein level was reduced in ospub45 mutants (Fig. 5C). These findings indicate that OsPUB45 degrades OsCSN5 to positively regulate immunity through the OsCUL3a-OsNPR1 pathway. Upon chitin treatment, OsPUB45 was found to promote the ubiquitination and degradation of OsCSN5 (fig. S2E). To further explore the impact of chitin treatment on OsNPR1 degradation by OsCUL3a, we coexpressed cMyc-OsNPR1 and HA-nLuc or HA-OsCUL3a in rice protoplasts with or without 80 nM chitin treatment for 1 hour. Immunoblot analysis revealed that chitin treatment attenuated the ubiquitination and degradation of OsNPR1 by OsCUL3a (fig. S6). These findings suggest that chitin treatment enhances the ubiquitination and degradation of OsCSN5, leading to reduced OsCSN5 levels. This reduction in OsCSN5 subsequently lowers OsCUL3a level, weakening the ubiquitination and degradation of OsNPR1 and ultimately activating immunity.
Fig. 5. OsPUB45 interferes with the effects of OsCSN5 on OsCUL3a and OsNPR1 abundance.
(A) Stability of OsCUL3a by OsCSN5 in ZH11 and OsPUB45-OX rice protoplasts. Plasmid of HA-OsCUL3a was cotransfected with 2×GFP or OsCSN5-GFP into ZH11 and OsPUB45-OX rice protoplasts. Actin protein was served as the internal control. OsCUL3a protein levels were determined by immunoblotting. (B) Immunoblot analysis of OsNPR1 protein levels in ZH11 and OsPUB45-OX plants by an anti-OsNPR1 antibody. Actin was used as the internal control. (C) Immunoblot analysis of OsNPR1 protein levels in ZH11 and ospub45 mutants by an anti-OsNPR1 antibody. Actin was used as the internal control. (D) OsCSN5 interacts with and stabilizes OsCUL3a to promote the degradation of OsNPR1 for rice normal growth in the absent of infection. The expression of OsPUB45 is induced by pathogen infection. OsPUB45 promotes OsCSN5 degradation to inhibit OsCUL3a and activate OsNPR1-mediated immunity.
DISCUSSION
Suppression of OsCSN5 in rice endows enhanced resistance without any yield penalties
Developing resistant materials against multiple pathogens is a desirable goal in crop breeding programs. However, it is often observed that strong immune responses can come at the cost of reduced growth (31, 32). Therefore, identifying resistant materials that confer immunity without impacting growth is of great value in crop breeding (33–35). While completely loss of susceptibility (S) genes or negative regulators can lead to growth defects and reduced yield, recent studies have shown that specific gene mutations can strike a balance between immunity and growth (36). For example, knocking out the RESISTANCE OF RICE TO DISEASES1 (ROD1) gene in rice confers resistance to multiple pathogens but results in reduced yield. A naturally ROD1 allele with a single-base substitution in the coding sequence maintains resistance while alleviating the yield penalty of the rod1 mutant (37). Similarly, knockout of RESISTANCE TO BLAST1 (RBL1) provides broad-spectrum resistance against rice blast but caused a considerable reduction in yield. However, a mutation lacking of 12-bp segment of RBL1 produces an elite allele that confers broad-spectrum resistance without compromising yield (38).
It has been shown that mutation of CSN5a in Arabidopsis and dysfunction or silencing of its orthologs in wheat or grapevine largely enhances resistance to respective fungal pathogens (16–19). We found that knockout of OsCSN5 in rice resulted in embryo mortality, but the suppression of OsCSN5 by RNAi approach substantially boosted disease resistance against M. oryzae and Xoo, chitin-induced ROS production, and the expression of defense-related genes (Fig. 1). Silencing of OsCSN5 did not have a noticeable impact on major agronomic traits (Fig. 1), indicating that reducing the expression of OsCSN5 could be beneficial for rice resistance breeding. Recent studies has indicated that knockdown of OsCSN5 leads to stronger symptoms of rice black-streaked dwarf virus (39); in addition, a positive role of CSN5 in immunity have been reported in which down-regulation of SlCSN5-1 and SlCSN5-2 in tomato results in reduced resistance against the necrotrophic fungal pathogen Botrytis cinerea and stunted growth (40). The suppression of SlCSN5-1 and SlCSN5-2 did not affect resistance against tobacco mosaic virus (40). These results imply that CSN5 may play distinct roles in mediating immunity against different pathogens.
Ubiquitination of OsCSN5 by OsPUB45 modulates plant immunity
In mammals, the CSN complex inhibits constitutive NF-κB activity in nonactivated cells. Knocking down CSN5 enhances basal NF-κB activity and improves cell survival under stress (28). The IκB kinase 2 interacts with and phosphorylates CSN5, leading to its ubiquitination via the 26S proteasome pathway to activate NF-κB activity (28). However, the specific E3 ubiquitin ligase responsible for targeting CSN5 has not been identified. In the current study, we revealed that OsCSN5 protein can be degraded by an E3 ligase, and the U-box–type E3 ligase OsPUB45 was initiatively identified from a rice UbE3 library (29). In vitro ubiquitination assay revealed that OsCSN5 was directly ubiquitinated by OsPUB45 (Fig. 2). Moreover, OsPUB45 positively regulated resistance against M. oryzae and Xoo (Fig. 3), resemble to the phenotype observed in OsCSN5-RNAi plants, indicating that ubiquitination of OsCSN5 triggered immunity. Given the high conservation of CSN5 protein across species and the importance of U-box–type E3 ligase in coordinating plant immunity, it is likely that CSN5 protein is targeted for degradation by OsPUB45 orthologs in other species.
OsCSN5-mediated resistance through OsCUL3a-OsNPR1 module
Cullin-RING ligases (CRLs) are a major class of E3 ligases, comprising approximately 63% of all E3s in rice (41). CRLs can be categorized on the basis of different CULs and the substrate adapters (42). Loss-of-function mutations of CUL1 in Arabidopsis are lethal, while a weak mutation, Atcul1–7, results in a dwarf phenotype and constitutive defense response (43, 44). Emerging studies have highlighted the crucial role of CUL3 E3 ligases in plant immunity (41). For instance, OsCUL3a has been shown to negatively regulate resistance against fungal M. oryzae and bacterial Xoo in rice (27). The canonical function of CSN5 is to catalyze the removal of NEDD8 (de-neddylation) from CUL proteins to regulate E3 ligase activity (45). In Arabidopsis, csn5a csn5b double mutants exhibit a notable increase in neddylation levels of CUL1 and CUL4, but not CUL3s (15). The barley yellow striate mosaic virus (BYSMV) P6 protein interacts with CSN5 of barley plants and negatively affects CSN5-mediated deRUBylation of CUL1 (46). Our research revealed that the expression of OsCSN5 promotes the accumulation of OsCUL3a, while the suppression of OsCSN5 reduces OsCUL3a protein abundance (Fig. 4, C and D), indicating that OsCSN5 plays a role in regulating OsCUL3 protein stability. This finding is similar to previous studies conducted in Drosophila melanogaster, where the CUL3 protein was lower in CSN5 null mutants and exhibited a shorter half-life in CSN5-RNAi cells, indicating that CUL3 protein degradation maybe a prominent mechanism (47). The degradation of CUL3 in CSN5 null mutants was prevented by coexpressing CSN5 protein, suggesting that CSN5 protects CUL3 from degradation (47). It is possible that CSN5 recycles unstable CUL3 into a more stable form and thereby promoting CUL3-organized E3 activity in vivo. We infer that this protective effect of CSN5 is likely also occur with OsCUL3a in rice. csn5a-2 cul3a double mutants in Arabidopsis displayed notably enhanced immunity compared to cul3a mutant (16). However, it is still unknown whether CUL3a and CSN5a in Arabidopsis share similar regulatory mechanisms. Notably, a substantial decrease in OsCUL3a and increase in OsNPR1 protein levels were detected in OsCSN5-RNAi transgenic plants (Fig. 4, D and E), with the decrease in OsNPR1 by OsCSN5 being compromised in oscul3a mutants (Fig. 4F). This suggests that OsCSN5 promotes the accumulation of OsCUL3a, preventing overactivation of OsNPR1 due to reduced OsCUL3a protein level. The relationship between protein regulation can also be confirmed through disease resistance phenotypes. We further found that the enhanced blast resistance in OsCSN5-RNAi plants was lost in osnpr1/OsCSN5-RNAi mutants (Fig. 4, H and I), supporting the role of OsCSN5 in negatively regulating rice disease resistance through OsNPR1. Therefore, OsCSN5 stabilizes OsCUL3a protein levels and maintains OsNPR1 homeostasis to balance defense response and plant growth in rice.
On the basis of our research findings, we have developed a working model that illustrates how OsPUB45, OsCSN5, and OsCUL3a-OsNPR1 interact to regulate immunity in rice (Fig. 5D). Our results show that OsCSN5 plays a key role in stabilizing OsCUL3a, which destabilizes the OsNPR1 protein, preventing its constitutive activation in noninfected plants. In addition, pathogens such as M. oryzae induce the accumulation of OsPUB45, which interacts with and promotes the degradation of OsCSN5. This leads to a reduction in OsCUL3a protein levels and an increase in OsNPR1 accumulation, ultimately activating immunity in rice.
MATERIALS AND METHODS
Plant materials and growth conditions
The rice (Oryza sativa L.) cultivars NPB and ZH11 were used for disease evaluation in this study. The OsCSN5-RNAi lines were generated in NPB background. In the OsCSN5-RNAi constructs, a fragment of 594 to 891 bp and a fragment of 822 to 1083 bp in the OsCSN5 CDS region were targeted to suppress its expression. The oscsn5 and ospub45 mutants were generated in NPB and ZH11 backgrounds by CRISPR-Cas9. The OsPUB45-OX (driven by the maize Ubiquitin promoter) transgenic plants were generated in the ZH11 background. All the resulting constructs were introduced into Agrobacterium (Agrobacterium tumefaciens) strain EHA105 for rice transformation. T0 progeny were screened by PCR, sequencing, or qRT-PCR. Rice seedlings were grown in a growth chamber at 26°C and 70% relative humidity with a 12-hour light/12-hour dark photoperiod.
N. benthamiana plants
N. benthamiana were cultivated in soil under a 12-hour light/12-hour dark photoperiod at 25°C. Four-week-old N. benthamiana leaves were used in bimolecular fluorescence complementation assays.
Blast fungus inoculation and phenotypic analysis
M. oryzae isolate RB22 was cultivated on oat solid medium at 28°C for 14 to 21 days to produce spores. For punch inoculation, a spore suspension of 3.0 × 105 spores/ml was used on the second leaf (from the top) of 6-week-old plants as previously described (48). Disease symptoms on leaves were scored 14 days after inoculation; lesion areas were measured with ImageJ. For spray inoculation, 3-week-old rice seedlings were sprayed with spore suspension (2.0 × 105 spores/ml). At 7 days after inoculation, leaves with representative lesions were photographed. A segment of rice leaf 3 cm in length with a lesion was then cut and subjected to DNA extraction with the cetyltrimethyl ammonium bromide method. Relative fungal biomass was measured as previously described with DNA-based qPCR.
Xanthomonas oryzae pv. oryzae (Xoo) inoculation and disease symptom evaluation
Xoo PXO99A was grown on prostate-specific antigen (PSA) medium plates at 30°C for 2 days before it was suspended in sterilized water. The bacterial suspension was adjusted to optical density at 600 nm (OD600) = 0.5 and then used to inoculate rice leaves. Leaves of 6-week-old rice plants were cut with a scissors that had been dipped into the bacterial suspension (48). The lesion lengths were measured at 14 days after inoculation. For bacterial growth analysis, inoculated leaves (n = 3) were thoroughly ground, and the ground tissue was suspended in 1 ml of sterilized water. The leaf suspensions were then diluted and transferred to PSA medium plates containing cephalexin (15 mg/liter). The plates were incubated at 30°C, and colonies were counted within 2 to 3 days (49).
Expression pattern analysis
Rice leaves were collected at different time points after spray inoculation with M. oryzae isolates RB22. Water with 0.1% (v/v) Tween 20 was used as a mock inoculation control (Mock). Total RNA was extracted from rice seedlings and converted to first-strand cDNA using the HiScript II First Strand cDNA Synthesis Kit (Vazyme, R212-01). qRT-PCR was performed with 2×RealStar Fast SYBR qPCR Mix (GenStar, A301-01) on an ABI QuantStudio 6 instrument (10). The rice Ubiquitin gene (OsUBQ, LOC_Os03g13170) was used as the reference genes for rice. Gene expression levels were calculated using data from three technical repeats, and all experiments were repeated thrice.
Measurement of ROS
Pathogen-associated molecular pattern (PAMP)-triggered oxidative burst in OsCSN5-RNAi, ospub45, and OsPUB45-OX seedlings was detected as described previously. Leaf punches collected from the second leaves of 4-week-old plants were immersed in ddH2O overnight and were then transferred into 100 μl of luminol containing 1.0 μl of horseradish peroxidase (1 μg/μl), and elicitor (8 μM hexaacetylchitohexaose or water as a control). Chemiluminescence was measured using a GloMax 20/20 luminometer (50). Three technical replicates were included for each sample.
Yeast two-hybrid assays
For E3 library screening, the OsCSN5 coding region was cloned in-frame into the pGBKT7 vector as the bait for the screening of interacting proteins from the rice E3 ubiquitin ligase library (29). After screening the yeast transformants on SD-Leu-Trp selected plates, positive clones were transferred to SD-Leu-Trp-His-Ade medium to confirm the interaction. The yeast cells were grown on selective medium for 3 days at 30°C.
Co-IP assays
For co-IP assay, the desired constructs were cotransfected into rice protoplasts. After being kept in an incubator for 20 hours, total protein was extracted in native buffer [50 mM tris-MES, (pH 8.0), 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM dithiothreitol (DTT), and protease inhibitor cocktail]. A total of 300 μl of supernatants of cell lysate was used for immunoprecipitation with 2 μl of GFP or HA antibody at 4°C for 2 hours with gentle shaking before the addition of 35 μl of Protein G agarose beads (Millipore) followed by 2 hours of incubation at 4°C. The beads were washed six times with 1× phosphate-buffered saline (PBS) with 1% Tween 20 (PBST) buffer and subsequently separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) (51).
BiFC assays
For BiFC assay, full-length coding sequences of OsCSN5, OsPUB45, and OsCUL3a were individually cloned into the p2YN (nYFP) or p2YC (cYFP) vectors to produce the fusion to the N- or C-terminal of YFP, including OsCSN5-nYFP, OsPUB45-cYFP, and OsCUL3a-cYFP. The desired plasmids were separately transformed into Agrobacterium strain EHA105 and then transiently infiltrated in N. benthamiana leaves. Fluorescent signals were observed using a laser scanning confocal microscope (Zeiss LSM880) at 48 hours after infiltration (48). Total proteins were extracted from 4-week-old N. benthamiana leaves in ice-cold protein extraction buffer. After centrifuge at 15,000g for 10 min at 4°C, the protein extracts were mixed with 4×SDS loading buffer and separated by SDS-PAGE. Proteins were detected by immunoblotting using anti-HA (ABclonal, AE008) or anti-GFP antibody (Abcam, ab290).
In vivo degradation assay
The in vivo degradation assays were conducted in rice protoplasts. The desired combinations of plasmids were transfected into NPB, ZH11, or the indicated rice protoplasts. The proteins were extracted using protein denaturing buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, 4 M urea, and 1 mM phenylmethylsulfonyl fluoride]. Appropriate amounts of 4×protein loading buffer was added and separated by SDS-PAGE before immunoblotting. Protein abundance was detected by immunoblotting and normalized to the indicated protein in each sample by ImageJ software (51). In addition, the total RNA was isolated and the relative transcript levels of each gene were determined by RT-PCR (10).
Anti-OsNPR1 antibody
Immunoblotting for OsNPR1 protein was performed with anti-OsNPR1 antibody (ABclonal, A20582) according to the manufacturer’s instructions.
In vitro E3 ligase ubiquitination assay
In vitro OsPUB45 E3 ligase activity and in vitro ubiquitination of OsCSN5 by OsPUB45 was done by following the method described previously (29). Briefly, crude extract containing E1 (wheat E1), E2 (AtUBC8), ubiquitin (2 μg/μl), MBP, or MBP-OsPUB45 were mixed in 1× reaction buffer [50 mM tris-HCl (pH 7.4), 10 mM MgCl2, 5 mM adenosine 5′-triphosphate, and 2 mM DTT]. The reactions were incubated at 30°C for 2 hours, and in vitro E3 ligase activity was determined using an anti-Ub antibody and anti-MBP antibody. For the substrate ubiquitination assay, equal amounts of GST-OsCSN5 were added to the reaction mixture, and ubiquitination was measured by immunoblotting.
Accession numbers
The accession numbers of major genes mentioned in this study are as follows: OsCSN5, LOC_Os04g56070; OsPUB45, LOC_Os02g33680; and OsCUL3a, LOC_Os02g51180.
Quantification statistical analysis
Statistical analysis was performed with GraphPad Prism. Data were analyzed by two-tailed Student’s t test or one-way analysis of variance (ANOVA) to test the significance of the changes as indicated in the figure legends. Different lowercase letters or asterisks (*P < 0.05 and **P < 0.01; ns, not significant) representing significance levels are given in the figures.
Acknowledgments
We thank Dr. Lei Wang from Institute of Botany, Chinese Academy of Sciences for BiFC vectors.
Funding: This study was supported by the National Key Research and Development Program of China (2022YFD1401400), National Natural Science Foundation of China (32001858 and U20A2021, and 32161143009), Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-CSCB-202301), Central Public-interest Scientific Institution Basal Research Fund (Y2023PT18), and China Postdoctoral Science Foundation (2021M703556 and 2022T150715).
Author contributions: C.Z., L.F., R.W. and Y.N. designed the experiments and wrote the paper. C.Z., L.F., F.H., X.Y., M.W., T.Z., Y.H. and N.X. performed the experiments. R.W., Y.N., G.-L.W., J.R., F.F., J.Y. and A.L. revised the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Table S1
REFERENCES AND NOTES
- 1.Jones J. D., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Zhou J.-M., Zhang Y., Plant immunity: Danger perception and signaling. Cell 181, 978–989 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Brown J. K. M., Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 5, 339–344 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Li S., Lin D., Zhang Y., Deng M., Chen Y., Lv B., Li B., Lei Y., Wang Y., Zhao L., Liang Y., Liu J., Chen K., Liu Z., Xiao J., Qiu J.-L., Gao C., Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460 (2022). [DOI] [PubMed] [Google Scholar]
- 5.You Q., Zhai K., Yang D., Yang W., Wu J., Liu J., Pan W., Wang J., Zhu X., Jian Y., Liu J., Zhang Y., Deng Y., Li Q., Lou Y., Xie Q., He Z., An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20, 758–769 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Fan J., Bai P., Ning Y., Wang J., Shi X., Xiong Y., Zhang K., He F., Zhang C., Wang R., Meng X., Zhou J., Wang M., Shirsekar G., Park C. H., Bellizzi M., Liu W., Jeon J. S., Xia Y., Shan L., Wang G. L., The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe 23, 498–510.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao M., Hao Z., Ning Y., He Z., Revisiting growth-defence trade-offs and breeding strategies in crops. Plant Biotechnol. J. 22, 1198–1205 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang N., Tang C., Fan X., He M., Gan P., Zhang S., Hu Z., Wang X., Yan T., Shu W., Yu L., Zhao J., He J., Li L., Wang J., Huang X., Huang L., Zhou J. M., Kang Z., Wang X., Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185, 2961–2974.e19 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Li W., Zhu Z., Chern M., Yin J., Yang C., Ran L., Cheng M., He M., Wang K., Wang J., Zhou X., Zhu X., Chen Z., Wang J., Zhao W., Ma B., Qin P., Chen W., Wang Y., Liu J., Wang W., Wu X., Li P., Wang J., Zhu L., Li S., Chen X., A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170, 114–126.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Hao Z., Tian J., Fang H., Fang L., Xu X., He F., Li S., Xie W., Du Q., You X., Wang D., Chen Q., Wang R., Zuo S., Yuan M., Wang G. L., Xia L., Ning Y., A VQ-motif-containing protein fine-tunes rice immunity and growth by a hierarchical regulatory mechanism. Cell Rep. 40, 111235 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Li Y. J., Gu J. M., Ma S., Xu Y., Liu M., Zhang C., Liu X., Wang G. F., Genome editing of the susceptibility gene ZmNANMT confers multiple disease resistance without agronomic penalty in maize. Plant Biotechnol. J. 21, 1525–1527 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng X. W., Dubiel W. G., Wei N., Hofmann K., Mundt K., Colicelli J., Kato J., Naumann M., Segal D., Seeger M., Unified nomenclature for the COP9 signalosome and its subunits: An essential regulator of development. Trends Genet. 16, 202–203 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Hu Q., Chen H., Zhou Z., Li W., Wang Y., Li S., He Q., Role of individual subunits of the neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins. PLOS Genetics 6, e1001232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Z., Pardi R., Cheadle W., X. Xiaoyu, Zhang S., Shah S. V., Grizzle W., Miller D., Mountz J., Zhang H.-G., Plant homologue constitutive photomorphogenesis 9 (COP9) signalosome subunit CSN5 regulates innate immune responses in macrophages. Blood 117, 4796–4804 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Gusmaroli G., Figueroa P., Serino G., Deng X. W., Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19, 564–581 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukhtar M. S., Carvunis A.-R., Dreze M., Epple P., Steinbrenner J., Moore J., Tasan M., Galli M., Hao T., Nishimura M. T., Pevzner S. J., Donovan S. E., Ghamsari L., Santhanam B., Romero V., Poulin M. M., Gebreab F., Gutierrez B. J., Tam S., Monachello D., Boxem M., Harbort C. J., McDonald N., Gai L., Chen H., He Y., European Union Effectoromics Consortium, Vandenhaute J., Roth F. P., Hill D. E., Ecker J. R., Vidal M., Beynon J., Braun P., Dangl J. L., Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333, 596–601 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H., Wang X., Giroux M. J., Huang L., A wheat COP9 subunit 5-like gene is negatively involved in host response to leaf rust. Mol. Plant Pathol. 18, 125–133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai X., Huang X., Tian S., Peng H., Zhan G., Goher F., Guo J., Kang Z., Guo J., RNAi-mediated stable silencing of TaCSN5 confers broad-spectrum resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 22, 410–421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui K.-C., Liu M., Ke G.-H., Zhang X.-Y., Mu B., Zhou M., Hu Y., Wen Y.-Q., Transient silencing of VvCSN5 enhances powdery mildew resistance in grapevine (Vitis vinifera). Plant Cell Tiss. Org. 146, 621–633 (2021). [Google Scholar]
- 20.Langin G., González-Fuente M., Üstün S., The plant ubiquitin–Proteasome system as a target for microbial manipulation. Annu. Rev. Phytopathol. 61, 351–375 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Wang G., Chen X., Yu C., Shi X., Lan W., Gao C., Yang J., Dai H., Zhang X., Zhang H., Zhao B., Xie Q., Yu N., He Z., Zhang Y., Wang E., Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice. Nature 629, 1158–1164 (2024). [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y. T., Li X., Ubiquitination in NB-LRR-mediated immunity. Curr. Opin. Plant Biol. 15, 392–399 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Grubb L. E., Wang J., Liang X., Li L., Gao C., Ma M., Feng F., Li M., Li L., Zhang X., Yu F., Xie Q., Chen S., Zipfel C., Monaghan J., Zhou J. M., A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69, 493–504.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Yuan Y., Zhong S., Li Q., Zhu Z., Lou Y., Wang L., Wang J., Wang M., Li Q., Yang D., He Z., Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Feng J.-X., Cao L., Li J., Duan C.-J., Luo X.-M., Le N., Wei H., Liang S., Chu C., Pan Q., Tang J.-L., Involvement of OsNPR1/NH1 in rice basal resistance to blast fungus Magnaporthe oryzae. Eur. J. Plant Pathol. 131, 221–235 (2011). [Google Scholar]
- 26.Li X., Yang D. L., Sun L., Li Q., Mao B., He Z., The systemic acquired resistance regulator OsNPR1 attenuates growth by repressing auxin signaling through promoting IAA-Amido synthase expression. Plant Physiol. 172, 546–558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q., Ning Y., Zhang Y., Yu N., Zhao C., Zhan X., Wu W., Chen D., Wei X., Wang G. L., Cheng S., Cao L., OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 29, 345–359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orel L., Neumeier H., Hochrainer K., Binder B. R., Schmid J. A., Crosstalk between the NF-kappaB activating IKK-complex and the CSN signalosome. J. Cell Mol. Med. 14, 1555–1568 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R., You X., Zhang C., Fang H., Wang M., Zhang F., Kang H., Xu X., Liu Z., Wang J., Zhao Q., Wang X., Hao Z., He F., Tao H., Wang D., Wang J., Fang L., Qin M., Zhao T., Zhang P., Xing H., Xiao Y., Liu W., Xie Q., Wang G. L., Ning Y., An ORFeome of rice E3 ubiquitin ligases for global analysis of the ubiquitination interactome. Genome Biol. 23, 154 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin H., Xia P., Wing R. A., Zhang Q., Luo M., Dynamic intra-japonica subspecies variation and resource application. Mol. Plant 5, 218–230 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Ning Y., Liu W., Wang G.-L., Balancing immunity and yield in crop plants. Trends Plant Sci. 22, 1069–1079 (2017). [DOI] [PubMed] [Google Scholar]
- 32.He Z., Webster S., He S. Y., Growth-defense trade-offs in plants. Curr. Biol. 32, R634–R639 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Li W., Deng Y., Ning Y., He Z., Wang G. L., Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Nelson R., Wiesner-Hanks T., Wisser R., Balint-Kurti P., Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 19, 21–33 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Boutrot F., Zipfel C., Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Zhou X., Liao H., Cherne M., Yin J., Chen Y., Wang J., Zhu X., Chen Z., Yuan C., Zhao W., Wang J., Li W., He M., Ma B., Wang J., Qin P., Chen W., Wang Y., Liu J., Qiang Y., Wang W., Wu X., Li P., Zhu L., Li S., Ronald P. C., Chen X., Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. U.S.A. 115, 3174–3179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao M., He Y., Yin X., Zhong X., Yan B., Wu Y., Chen J., Li X., Zhai K., Huang Y., Gong X., Chang H., Xie S., Liu J., Yue J., Xu J., Zhang G., Deng Y., Wang E., Tharreau D., Wang G.-L., Yang W., He Z., Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 184, 5391–5404.e17 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Sha G., Sun P., Kong X., Han X., Sun Q., Fouillen L., Zhao J., Li Y., Yang L., Wang Y., Gong Q., Zhou Y., Zhou W., Jain R., Gao J., Huang R., Chen X., Zheng L., Zhang W., Qin Z., Zhou Q., Zeng Q., Xie K., Xu J., Chiu T.-Y., Guo L., Mortimer J. C., Boutté Y., Li Q., Kang Z., Ronald P. C., Li G., Genome editing of a rice CDP-DAG synthase confers multipathogen resistance. Nature 618, 1017–1023 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He L., Chen X., Yang J., Zhang T., Li J., Zhang S., Zhong K., Zhang H., Chen J., Yang J., Rice black-streaked dwarf virus-encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol. 225, 896–912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hind S. R., Pulliam S. E., Veronese P., Shantharaj D., Nazir A., Jacobs N. S., Stratmann J. W., The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. Plant J. 65, 480–491 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Ban Z., Estelle M., CUL3 E3 ligases in plant development and environmental response. Nat. Plants 7, 6–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hua Z., Vierstra R. D., The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Shen W.-H., Parmentier Y., Hellmann H., Lechner E., Dong A., Masson J., Granier F., Lepiniec L., Estelle M., Genschik P., Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13, 1916–1928 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilkerson J., Hu J., Brown J., Jones A., Sun T.-p., Callis J., Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics 181, 945–963 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwechheimer C., Serino G., Callis J., Crosby W. L., Lyapina S., Deshaies R. J., Gray W. M., Estelle M., Deng X.-W., Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Gao D.-M., Zhang Z.-J., Qiao J.-H., Gao Q., Zang Y., Xu W.-Y., Xie L., Fang X.-D., Ding Z.-H., Yang Y.-Z., Wang Y., Wang X.-B., A rhabdovirus accessory protein inhibits jasmonic acid signaling in plants to attract insect vectors. Plant Physiol. 190, 1349–1364 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J.-T., Lin H.-C., Hu Y.-C., Chien C.-T., Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 7, 1014–1020 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Zhang C., Fang H., Wang J., Tao H., Wang D., Qin M., He F., Wang R., Wang G.-L., Ning Y., The rice E3 ubiquitin ligase-transcription factor module targets two trypsin inhibitors to enhance broad-spectrum disease resistance. Dev. Cell 59, 2017–2033.e5 (2024). [DOI] [PubMed] [Google Scholar]
- 49.Liu M., Shi Z., Zhang X., Wang M., Zhang L., Zheng K., Liu J., Hu X., Di C., Qian Q., He Z., Yang D. L., Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat. Plants 5, 389–400 (2019). [DOI] [PubMed] [Google Scholar]
- 50.You X., Zhu S., Sheng H., Liu Z., Wang D., Wang M., Xu X., He F., Fang H., Zhang F., Wang D., Hao Z., Wang R., Xiao Y., Wan J., Wang G.-L., Ning Y., The rice peroxisomal receptor PEX5 negatively regulates resistance to rice blast fungus Magnaporthe oryzae. Cell Rep. 42, 113315 (2023). [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Wang R., Fang H., Zhang C., Zhang F., Hao Z., You X., Shi X., Park C. H., Hua K., He F., Bellizzi M., Xuan Vo K. T., Jeon J. S., Ning Y., Wang G. L., Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice. Mol. Plant 14, 253–266 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1