Abstract
Yeast sex-hormone whole-cell biosensors are analytical tools characterized by long-time storage and low production cost. We engineered compact β-estradiol biosensors in S. cerevisiae cells by leveraging short (20-nt long) operators bound by the fusion protein LexA-ER-VP64—where ER is the human estrogen receptor and VP64 a strong viral activation domain. Our best biosensors showed high accuracy since their recovery concentration ranged between 97.13% and 104.69%. As a novelty, we built on top of them testosterone biosensors that exploit the conversion of testosterone into β-estradiol by the human aromatase enzyme—expressed in S. cerevisiae together with its co-factor CPR. We used our engineered yeast strains to evaluate aromatase activity through fluorescence measurements without the need for protein purification. Besides, we set up an aromatase-inhibitors evaluation assay to measure the IC50 (half-maximal inhibitory concentration) of candidate inhibitory compounds and developed a screening assay for enzymes that metabolize β-estradiol that demands only to measure fluorescence. These two assays allow the screening of a large number of chemicals and proteins in a fast and economic fashion. We think that our work will facilitate considerably high throughput screening for the discovery of new drugs and unknown metabolic processes.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-85022-7.
Keywords: Biosensor, CYP enzyme, β-estradiol, Testosterone, Aromatase
Subject terms: Biotechnology, Breast cancer
Introduction
In recent years, research in synthetic biology has led to the construction of whole-cell biosensors1. However, optimizing the performance of this kind of bio-circuits in terms of low basal output signal (noise), high ON/OFF ratio, and large detection range is still a challenging task2. Several sex-hormone whole-cell biosensors have been developed3–5 in microbes. They can be almost permanently preserved (as cryostocks) and have low production costs6.
The chimeric transcription factor LexA-ER-AD contains—as a DNA-binding domain—the bacterial protein LexA7, the β-estradiol binding domain of the human estrogen receptor (ER)8, and an activation domain (AD) of viral, bacterial, or eukaryotic origin. It was used, initially, as an orthogonal protein to control the expression of target genes in the yeast Saccharomyces cerevisiae9. Later, it was repurposed as a component of a two-gene system that behaves as a highly sensitive biosensor or a steep toggle switch10. Moreover, LexA-AD was reported to effectively increase gene expression by binding a large number (6 to 8) of short operators (16 to 20 nt)3,11.
Cytochrome P450s (CYPs) are a vast family of monooxygenase enzymes whose characteristic absorption peak (in a reduced state and bound to CO) lies at 450 nm12. On one hand, CYP enzymes play an important role in drug metabolism. On the other hand, they are used as targets to discover new pharmaceuticals. To be functional, CYP enzymes require the assistance of redox partner proteins that transfer electrons, mainly, from reduced nicotinamide adenine dinucleotides (such as NADPH or NADH)13. Peroxides14, photosensitizers15, and electrode surfaces16 are other possible (though more expensive) sources of electrons. Studying mammalian CYPs is comparatively difficult since both their purification and storage are complex and costly operations17. The enzyme bags technique18, which demands a strong expression of CYPs and their usage inside permeabilized cells, has become an attractive and economical way to deepen our knowledge of the properties of these enzymes. A luminescence-based assay, which takes about a week, has been developed to evaluate CYP activity via enzyme bags in fission yeast cells19.
The human CYP19A1 (aromatase) enzyme is of particular interest because it is an inhibitor target to treat certain kinds of illnesses such as breast cancer20,21. Human placental microsomes are used for the evaluation of aromatase activity22, even though they present several disadvantages: an overall complicated procedure, high cost for aromatase extraction, and low reproducibility of the experimental results.
The conversion of testosterone into β-estradiol is a reaction carried out by CYP19A123 that permits to evaluate aromatase activity by monitoring the concentration of β-estradiol. Moreover, it allows to screen for aromatase inhibitors24. S. cerevisiaecan be engineered to express aromatase. The long-term cryopreservation of budding yeast maintains CYP19A1 functionality almost unaltered25. Furthermore, the low probability of genetic variation in yeast guarantees that the same strain, used for multiple independent experiments, remains basically unchanged26.
Whole-cell testosterone biosensors have a great potential for practical applications27. They can be an alternative to: (1) immunoassays, which not only require the separation of interfering substances but also have reduced accuracy at low concentrations28; (2) mass spectrometry analysis, which may be difficult and expensive29; (3) optical30, electrochemical31, and microfluidic biosensors32, which are hard to be preserved for a long time and have high production cost. A cell-based testosterone biosensor that relies on tritium3H, a radioactive isotope of hydrogen) has been previously reported33. Moreover, testosterone has been used as an input for genetic circuits34. However, no whole-cell testosterone biosensors that are sufficiently safe and low-cost have been developed so far.
In this work, we constructed, first, a β-estradiol-concentration detection tool in S. cerevisiae. It consists of a library of β-estradiol biosensors characterized by the presence of short (20-nt long) lex operators. On top of it, we built an S. cerevisiae testosterone biosensor that leverages the interaction between the CPR-CYP19A1 system and the male sex hormone to synthesize β-estradiol. Furthermore, we made use of the enzyme bags technique to establish a luminescence-based assay in S. cerevisiae—which has a shorter experimental cycle than in S. pombe—to evaluate CYPs’ activity and utilize yeast strains as ‘off the shelf’ reagents. We also developed an aromatase-activity evaluation assay that does not need enzyme bags but exploits the yeast strains that convert testosterone into β-estradiol and the β-estradiol biosensors. On the same synthetic yeast strains, we set up an aromatase-inhibitors evaluation assay to measure the IC50 (half-maximal inhibitory concentration) of candidate compounds and a screening assay for enzymes that metabolize β-estradiol. These new assays guarantee simple operational procedures, low cost—which allows for large-scale screening—and elevated accuracy.
Results and discussion
Construction of a library of efficient β-estradiol biosensors based on short lex operators
The new β-estradiol biosensors presented in this work were constructed by taking as a reference a previous work from our lab10. Hence, they were organized into two main components: the receptor part that expresses constitutively the chimeric activator LexA-ER-AD9—which is transported into the nucleus only in the presence of the inducer β-estradiol—and the reporter part that contains a synthetic activated promoter, which gathers a variable number of short lex operators in front of a minimal CYC1promoter sequence (pCYC1min) and leads the synthesis of yEGFP35. The whole lex operator (indicated as lex2Op) was divided in the left and right part—termed lexOpL and lexOpR, respectively—to explore whether these short operators could be used to build a more compact reporter transcription unit (TU). Inside the nucleus, LexA-ER-AD binds the short lex operators and recruits RNA polymerase II to the core promoter region to initiate the transcription of yEGFP mRNA.
The various parameters for evaluating the β-estradiol biosensors are listed in Table S8. Among them, we find: the maximal and basal fluorescence, the Hill coefficient (n), the ON/OFF ratio, and the detection range. The Hill coefficient is proportional to the steepness of the fitted dose-response curve. Higher values of n mean that the dose-response curve is closer to a step function, i.e., the biosensor tends to behave like a switch (see Table S9). The ON/OFF ratio is calculated between the maximum and the basal fluorescence value. The limit of detection (LOD) is the lowest β-estradiol concentration whose output is significantly different and at least two-fold higher than the basal fluorescence10. The tolerance is the highest concentration of β-estradiol at which the biosensor works (i.e., returns a fluorescence signal) before the appearance of toxicity effects in the cell culture. The detection range is the interval between LOD and the tolerance.
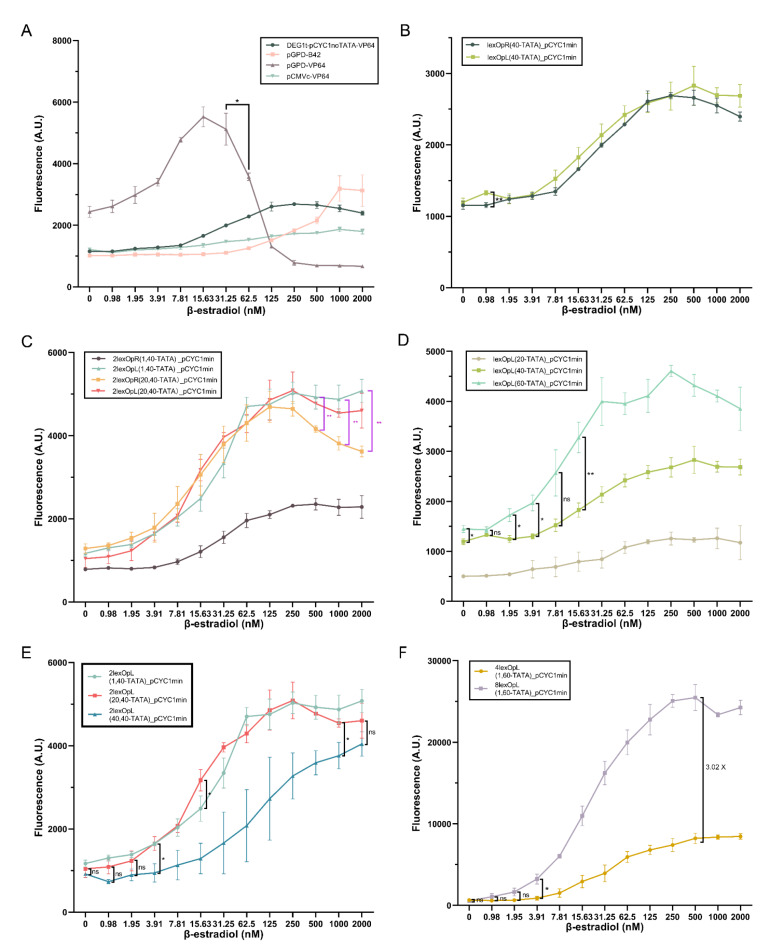
Initially, we tested four receptor parts—made of overall four different promoters and two activation domains—on a single reporter part containing (and referred to as) lexOpR(40-TATA)_pCYC1min (i.e., the short lexOpR was placed 40 nt upstream of the pCYC1min TATA box starting at position − 52—TATA−52—with respect to the transcription start site—TSS). Two receptors shared the strong GPD promoter (pGPD) and employed the B42 (weak) and VP64 (strong) activation domains. They were labelled as pGPD-B42 and pGPD-VP64. The other two receptor parts combined VP64 with two promoters: the complete CMV(pCMVc-VP64) and the synthetic DEG1t_pCYC1noTATA (DEG1t_pCYC1noTATA-VP64) promoter36. The dose-response curves of these four biosensors are shown in Fig. 1A. Similar to what was reported in10, pGPD-VP64 presented high toxicity effects (accompanied by a dramatic drop in fluorescence expression) at 62.50 nM β-estradiol, whereas the ON/OFF ratio of pCMVc-VP64 (1.54) was far below 2. Thus, only DEG1t_pCYC1noTATA-VP64 and pGPD-B42 were kept for the assembly of further circuits. DEG1t_pCYC1noTATA-VP64 was confirmed to be the best receptor part for biosensor construction, whereas pGPD-B42 permitted to implement sharp switches. For this reason, all results involving pGPD-B42 are shown and discussed in Fig. S3 and Tables S10-S13.
Fig. 1.
A new library of β-estradiol biosensors in S. cerevisiae (all the structures of the inducible promoters are shown in Fig. S2). (A) DEG1t_pCYC1noTATA-VP64 and pGPD-B42 are the only receptors that do not show toxicity effects and have an ON/OFF ratio higher than two—the reporter is lexOpR(40-TATA)_pCYC1min (see Table S16 for the statistical analysis). In the next panels (B-F), the receptor is always DEG1t_pCYC1noTATA-VP64. (B) No difference in the biosensor performance is due to the presence of lexOpR or lexOpL in the reporter—except for 0.98 nM β-estradiol (see Table S17). (C) The two strains carrying 2lexOpL express the highest fluorescence levels at every concentration of β-estradiol. Above 500 nM β-estradiol, they clearly overcome 2lexOpR(20,40-TATA) (see Tables S18-S19). (D) The optimal distance between lexOpL and TATA−52 corresponds to 60 nt. The fluorescence intensity of lexOpL(60-TATA)_pCYC1min is significantly higher than that of the other two promoters over the whole concentration range of β-estradiol except for 0.98 and 7.81 nM (see Table S20). (E) The highest fluorescence expression from 2lexOpL is achieved when they are spaced by either 1 or 20 nt—no significant difference was detected between these two configurations except for 15.63 nM (see Table S21). (F) The biosensor hosting, in the reporter part, 8lexOpL(1,60-TATA)_pCYC1min expresses a maximal fluorescence intensity 3.02-fold higher than that measured on 4lexOpL(1,60)_pCYC1min (see Table S22—ns: p-value > 0.05; *: p-value ≤ 0.05; **: p-value ≤ 0.01; two-sided Welch’s t test (black line) or ANOVA (pink line)).
To enhance fluorescence expression and improve the biosensor performance, the reporter part was modified with various combinations of short lex operators upstream of pCYC1min. A single lexOpR was compared to one lexOpL: their dose-response curves were substantially equivalent (see Fig. 1B). We then constructed four cassettes made of two short lex operators of the same kind. They were all placed 40 nt upstream of TATA−52. Moreover, the distance between the two operators was set to either a single (2lexOpR(1,40), 2lexOpL(1,40)) or 20 nt (2lexOpR(20,40), 2lexOpL(20,40))—see Fig. 1C. 2lexOpR(1,40) returned a fluorescence intensity much lower than the other three reporter configurations over the whole range of β-estradiol concentration (0 up to 2000 nM). The other three cassettes showed comparable fluorescence levels up to 250 nM β-estradiol, after which the two biosensors hosting 2lexOpL reached significantly higher fluorescence levels than that carrying 2lexOpR(20,40). According to these results, we decided to consider only lexOpL for the construction of longer operator cassettes. Since, as shown in10, the distance between the operator cassette and TATA−52 and that between two adjacent operators both have a significant impact on the efficiency of a biosensor, we considered a separation of 20, 40, and 60 nt between the operator region and TATA−52 and a spacer sequence of 1, 20, and 40 nt between two consecutive operators. From Fig. 1D, it is apparent that 60 nt between a single lexOpL and TATA−52 assured higher fluorescence at every concentration of β-estradiol. The fluorescence intensity corresponding to 2lexOpL(1,40-TATA) and 2lexOpL(20,40-TATA) appeared to be comparable, whereas the fluorescence levels due to 2lexOpL(40,40-TATA) are significantly lower than the other two in the interval between 3.91 and 1000 nM β-estradiol (see Fig. 1E). Although 1 and 20 nt are equivalent, we chose 1 nt as the most appropriate distance between the operators to reduce the overall promoter length and, therefore, the DNA synthesis cost. With these experiments, we figured out that lexOpL appeared more appropriate than lexOpR to construct multiple-operator-containing promoters for the reporter part. The best distance between the lexOpL cassette and TATA−52 was 60 nt, and two adjacent lexOpL should be separated by 1 nt. Finally, we increased the number of lexOpL to four and eight. As shown in Fig. 1F, the 8lexOpL(1,60-TATA)_pCYC1min synthetic promoter in the reporter part returned the best performance with an average maximal fluorescence 1.38-fold higher than that reached by the strong GPD promoter and 46.02-fold higher than its basal fluorescence. The biosensor detection range spanned a large interval between 1.9 and 2000 nM β-estradiol, whereas the switch-like behavior was not particularly pronounced (n = 1.37), especially if compared with that of the circuit having pGPD-B42 in the receptor part (n= 2.57). Overall, these results are consistent with those in10 and our new most performant biosensor, engineered in the strain byMM1984, slightly overcame the one in byMM38110 for the maximal output, ON/OFF ratio, and lower basal fluorescence.
We made a further test to investigate whether the order of the short operators had an impact on the biosensors’ performance. We engineered two reversed-order promoters: lexOpR(1)lexOpL(40-TATA)_pCYC1min and lexOpR(20)lexOpL(40-TATA)_pCYC1min—where (1) and (20) indicate the distance between the two short operators—and compared them with lex2Op(40-TATA)_pCYC1min (see Fig. S4 and Tables S14-S15). The reversed-order promoter with 20 nt between the two short operators showed, overall, modest performance. In contrast, the reversed-order promoter with 1 nt between the short operators returned lower fluorescence than the original promoter up to 7.81 nM β-estradiol but reached a much higher level from 500 to 2000 nM β-estradiol.
Determination of the β-estradiol biosensor transfer functions and real sample recovery
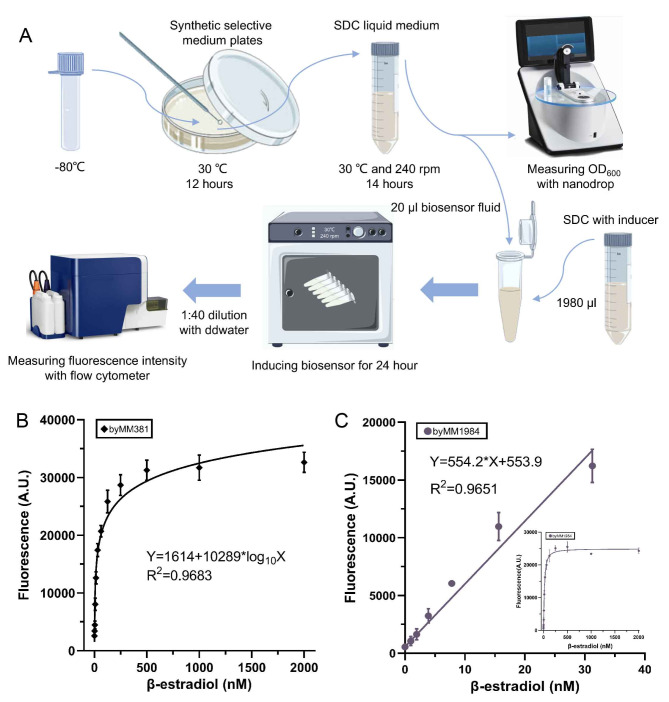
Synthetic gene circuits are characterized by the transfer function that expresses the circuit output as a function of the input. As for the above-described biosensors, the transfer function, which is obtained by fitting the data from the corresponding titration curve, links β-estradiol concentrations to fluorescence levels (see Fig. S5 and Table S23-S24) measured with high precision via flow cytometry (see Fig. 2A).
Fig. 2.
Flow cytometry and β-estradiol biosensors transfer functions. (A) Flow cytometry assay for S. cerevisiae (see “Materials and Methods” section. The drawing, based on Fig. 1 in19, was realized with Procreate software, Savage Interactive, Hobart, Australia). (B) Transfer function of biosensor byMM381. The curve was calculated by fitting the data over the whole range of β-estradiol concentrations considered in this work. (C) Transfer function of biosensor byMM1984. It approximates to a line the relation at low concentrations of β-estradiol. The inset shows the whole curve up to 2000 nM β-estradiol.
Among the β-estradiol biosensors engineered in our lab, byMM38110 did not reach the steady state before 500 nM β-estradiol. Therefore, the corresponding transfer function could determine precisely fluorescence at high concentrations of β-estradiol (see Fig. 2B). In contrast, byMM1984 titration curve is fitted by a line at low concentrations of β-estradiol. Thus, it is particularly appropriate to represent fluorescence in the range 0–30 nM β-estradiol (see Fig. 2C).
By culturing any of these two biosensors in a yeast growth medium supplied with an unknown concentration of β-estradiol, the fluorescence readout from the circuit together with its transfer function can be used to determine the concentration of β-estradiol in the solution. As a test, we prepared two sets of SDC solutions containing a variable concentration of β-estradiol: from 5 to 500 nM (analyzed via byMM381) and from 5 to 30 nM (byMM1984). Both byMM381 and byMM1984 were grown in these solutions (real samples). The efficacy of determining the β-estradiol concentration was calculated—according to the real sample recovery method37—via the ratio between the estimated and the real concentration value (see Table S25). byMM1984 showed a shorter recovery-concentration range (97.13–104.69%) and smaller relative standard deviations (RSDs < 5.0%) compared to byMM381, whose recovery-concentration range went from 91.74 to 110.98%, and RSDs were included between 4.03% and 11.91%. Taken together, byMM1984 and byMM381 appeared to give β-estradiol-concentration estimations reasonably close to their real value. Moreover, the RSD was always lower than 15%37 proving the reproducibility and reliability of the assays developed in this study.
Activity measurement of human CYP enzymes via enzyme bags in S. cerevisiae
The human CYP19A1 (aromatase) represents the focus of our work. Initially, we measured its activity (together with that of the human CYP1B1) by treating S. cerevisiae as enzyme bags where the CYP enzymes reacted with their substrates directly.
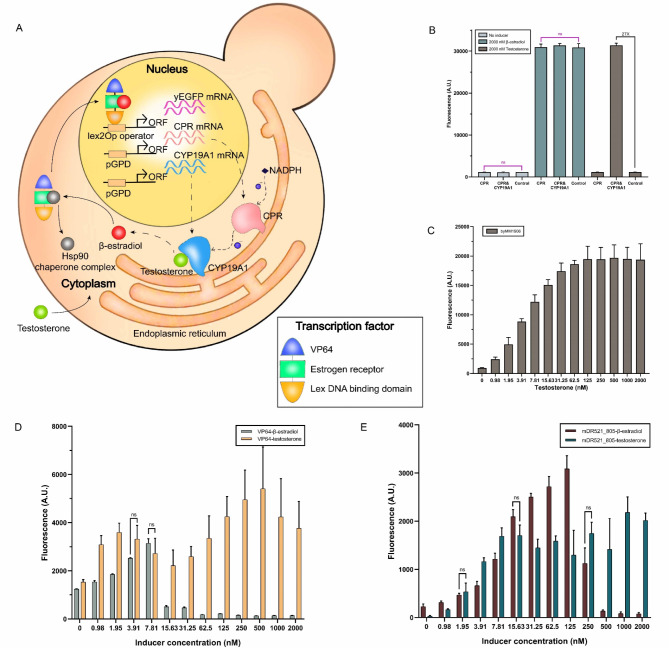
First, we determined the cell culture time needed to maximize the production of the human CYP enzymes. byMM1712 (which expresses CYP19A1 and CPR) and byMM1709 (CPR only, the control) were grown over different time intervals: 7, 14, 21, and 28 h. At the end of each growth period, the cell solutions were diluted to OD600 = 0.5 and distributed to four replicas (1 ml volume each). One ml SDC supplied with 4000 nM testosterone was added to each replica (such that the final concentration of testosterone became 2000 nM). Aromatase and testosterone reacted for 24 h giving rise to β-estradiol. One ml from each reaction tube was used to grow the β-estradiol biosensor byMM381 for 24 more hours. Finally, fluorescence was measured via flow cytometry experiments. As shown in Fig. 3A, fluorescence reached its maximum value when byMM1712 had been grown for 14 h (i.e., it was still in the exponential phase—see Fig. 3B). Therefore, we set to 14 h the culture time of byMM1712 in any further experiment.
Fig. 3.
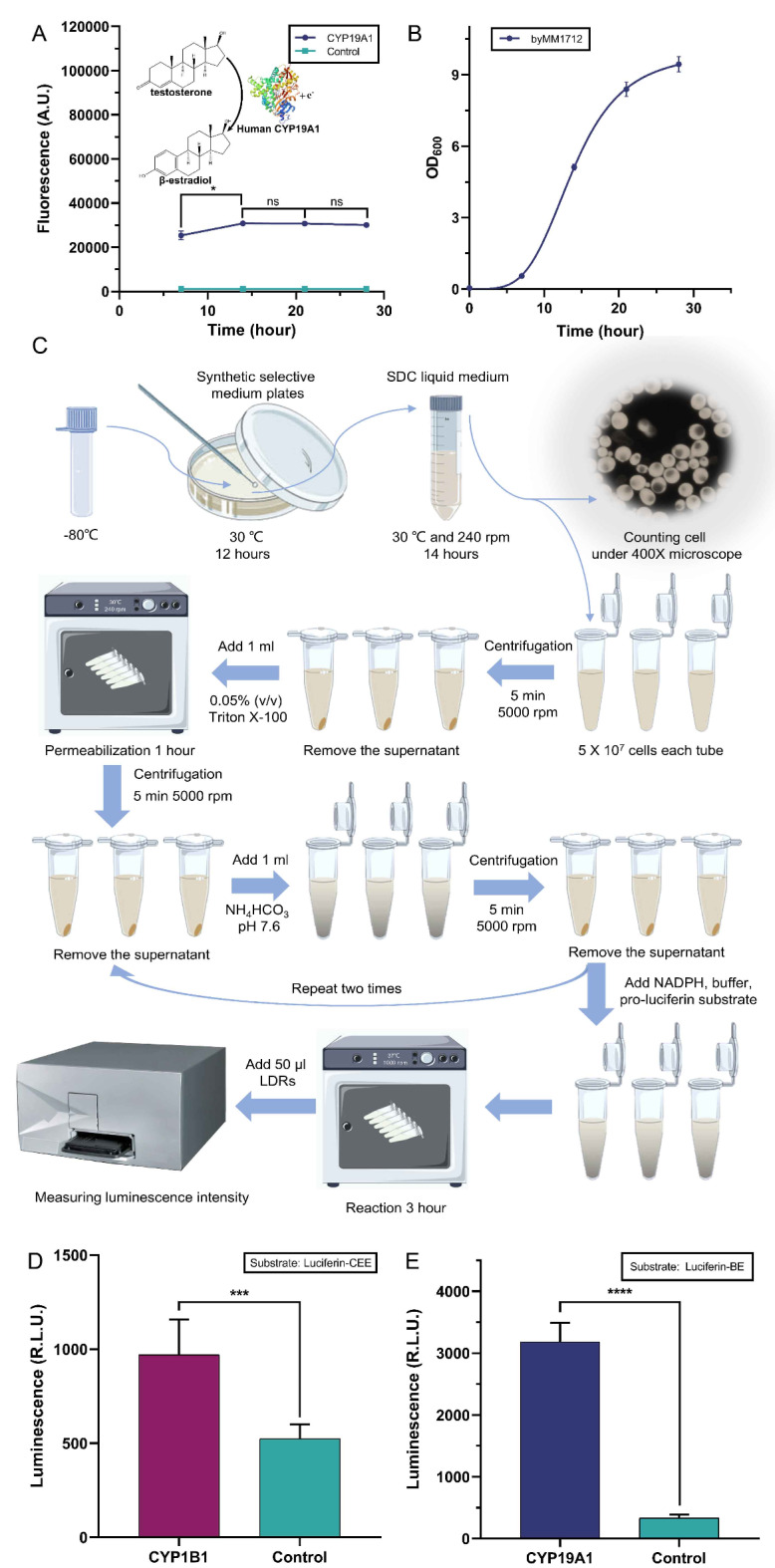
Determining the activity of human CYP enzymes in S. cerevisiae via enzyme bags. (A) Fluorescence due to the β-estradiol produced by strain byMM1712 through the testosterone oxidation by the CPR-CYP19A1 system. The optimal culture time for byMM1712 is 14 h. Fluorescence intensity at 14, 21, and 28 h are statistically equivalent (the structure of human CYP19A1 was obtained from PDB, DOI: 10.2210/pdb4KQ8/pdb). (B) Growth curve of the yeast strain byMM1712 that express both CPR and CYP19A1. (C) Luminescence-based assay (enzyme bags) with S. cerevisiae cells (see “Materials and Methods” section. The drawing, based on Fig. 1 in19, was realized with Procreate software, Savage Interactive, Hobart, Australia). (D-E) Activity of the CYP1B1 and CYP19A1 in S. cerevisiae measured via the enzyme bags technique. Bioluminescence is expressed in relative light unit (R.L.U.) (ns: p-value > 0.05; *: p-value ≤ 0.05; ***: p-value ≤ 0.001; ****: p-value ≤ 0.0001; two-sided Welch’s t test, see Table S26).
We then measured the activity, in S. cerevisiae, of both human CYP19A1 (byMM1712) and CYP1B1 (byMM1713)—on the substrates Luc-BE and Luc-CEE, respectively—via the “luminescence-based assay (enzyme bags)” described in “Materials and Methods” section and illustrated in Fig. 3C. Both enzymes returned an activity significantly higher than that of the control strain (see Fig. 3D-E).
Construction and characterization of a whole-cell testosterone biosensor
The yeast strain byMM381(a β-estradiol biosensor) was the starting point for the construction of a whole-cell testosterone biosensor. byMM381 was transformed with the integrative plasmid carrying the CPR expression cassette. Then, the resultant strain, byMM1894, underwent a transformation with the plasmid hosting the aromatase-containing TU to finalize the assembly of a testosterone biosensor (strain byMM1906). After each transformation, integration was checked via genomic DNA extraction, PCR, and Sanger sequencing.
The mechanism of our testosterone biosensor is illustrated in Fig. 4A. Strain byMM1906 expresses CPR and CYP19A1 that anchor to the endoplasmic reticulum. CPR works as a coenzyme to transfer electrons from NADPH—produced by the yeast strain itself—to CYP19A1. CYP19A1 oxidizes testosterone into β-estradiol that binds LexA-ER-AD at the human estrogen receptor and displaces, in this way, the Hsp90 chaperone complex. Thus, the chimeric transcription factor enters the cell nucleus and enhances the expression of yEGFP after binding the lex operators on the DNA.
Fig. 4.
Whole-cell testosterone biosensor. (A) Cartoon representation of the working of our testosterone biosensor in S. cerevisiae (the drawing was realized with Procreate software, Savage Interactive, Hobart, Australia). (B) Only byMM1906 returns high fluorescence in the presence of 2000 nM testosterone (27-fold higher than that of the control strain byMM381—see Table S27). (C) Testosterone titration curve. (D-E) Titration curves of two original β-estradiol biosensors engineered in our lab (byMM109 and byMM125) and the testosterone biosensors built on top of them (byMM1979 and byMM1987). The reporter part in these biosensors contains the synthetic weak promoter 7lex2Op(2, 37)_truncated_pCYC1min10. The conversion of testosterone into β-estradiol through the action of the CPR-CYP19A1 system appears to reduce toxicity effects drastically (see Table S28-S29) (ns: p-value > 0.05; two-sided Welch’s t test (black line) or ANOVA (pink line)).
byMM1906 was compared to byMM1894 and byMM381 through flow cytometry experiments. Each strain was grown for 14 h in SDC. Then, 1 ml of cell solution was mixed with 1 ml of either pure SDC or SDC supplied with 4000 nM of inducer (β-estradiol or testosterone; OD600 ~ 2.5). After growing for 24 more hours, the three yeast strains expressed comparable fluorescence upon induction with 2000 nM β-estradiol, whereas only byMM1906 returned high fluorescence in the presence of 2000 nM testosterone (see Fig. 4B), which indicated that testosterone was converted into β-estradiol by CPR-CYP19A1 inside byMM1906. We further carried out a full testosterone titration curve on byMM1906 (see Fig. 4C). Here, byMM1906 was first grown for 14 h. Then, only 20 µl cell solution was mixed to SDC plus a variable concentration of testosterone in a total volume of 2 ml (OD600 ~ 0.05). The induced cells grew for 24 h. The titration curve showed a considerably high ON/OFF ratio (21.12), whereas the Hill coefficient resulted in a modest value (n = 1.31). The detection range spanned all chosen testosterone concentrations (from 0.98 to 2000 nM). The maximum fluorescence intensity, detected at 500 nM, corresponded to only 62.7% of that previously measured (and reported in Fig. 4B). This seems to point out that the reduced amount of cells used for the titration curve did not guarantee a complete conversion of testosterone into β-estradiol.
Exploring the cause of cytotoxicity in the presence of transcription factors induced by β-estradiol
Previous works on synthetic systems induced by β-estradiol have reported cytotoxicity effects. The overexpression of chimeric activators carrying a rather strong AD has been indicated as a possible cause9. However, other synthetic activators containing the very strong VPR did not provoke S. cerevisiaecell death38,39. Hence, the real cause of cytotoxicity in β-estradiol-induced systems still appears unclear10. Our testosterone biosensor, which generates β-estradiol upon the interaction between aromatase and testosterone, can help figure out the relationship between β-estradiol and cytotoxicity. The β-estradiol biosensors in strain byMM109 (pGPD-VP64 in the receptor part) and byMM125 (pGPD-mDR521_805) differ only for the AD (with VP64 being stronger than mDR521_805) and show strong toxicity above 7.81 and 125 nM β-estradiol, respectively (see Fig. 4D-E). They were turned into testosterone biosensors upon integration of the TUs expressing CPR and CYP19A1—strains byMM1979 (VP64) and byMM1987 (mDR521_805). Both new strains were induced with crescent concentrations of testosterone (from 0.98 up to 2000 nM) and their titration curves (Fig. 4D-E) did not show any sudden drop in fluorescence expression, which typically signals strong cytotoxicity. Under the hypothesis that testosterone is completely converted into β-estradiol, we think that the slow release of β-estradiol—through the testosterone oxidation by CYP19A1—is what reduced toxicity effects considerably. Therefore, it would be a fast “activation” of relatively strong ADs the cause of cell death in previously published β-estradiol-based systems. It should be noted, though, that the basal fluorescence from byMM1987 is lower than that from byMM125 (see Fig. 4E), possibly because the presence of the TUs expressing CPR and CYP19A1 limits the production of the chimeric activator (which, at very high concentration, diffuses into the nucleus also in the absence of β-estradiol). However, the same effect is absent from byMM1979 (Fig. 4D), which seems to negate the hypothesis that the loss of toxicity in testosterone biosensors is due to a possible lower amount of the activators induced by β-estradiol.
Enzyme activity evaluation of yeast strain as ‘off the shelf’ reagents
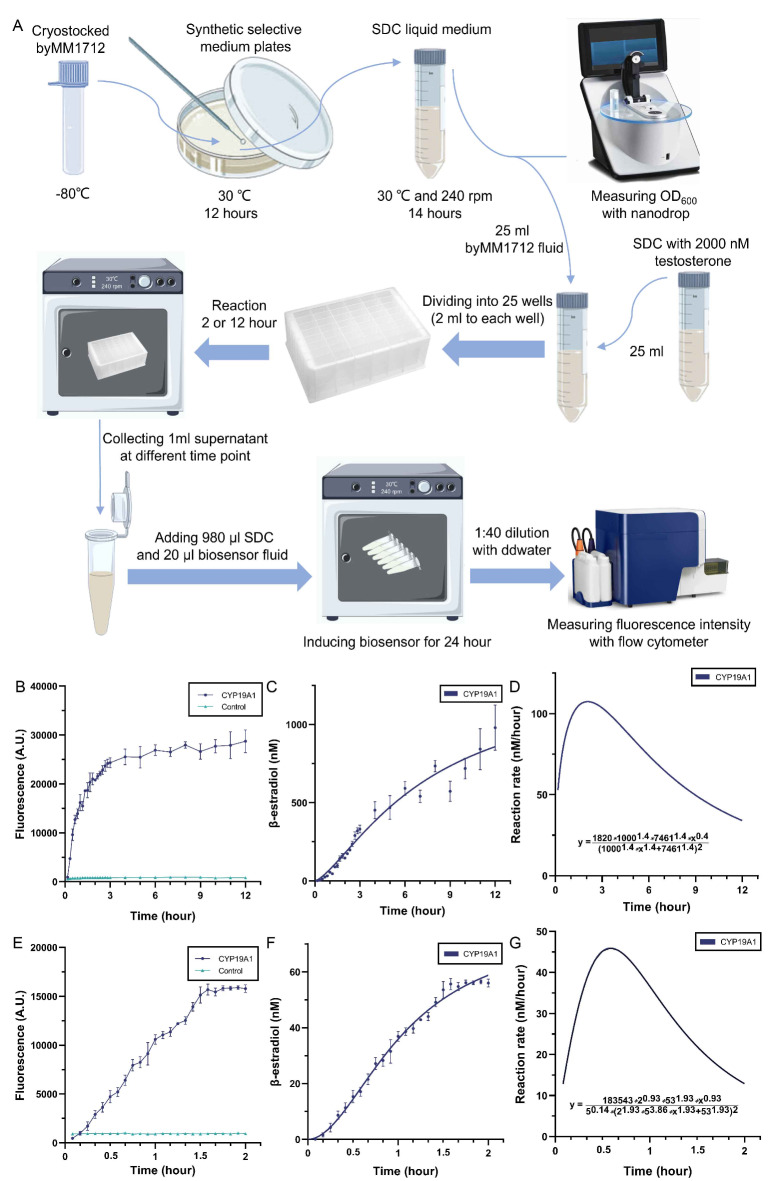
It is necessary to determine the activity of intracellular enzymes if we want yeast strains to work as ‘off the shelf’ reagents40. We used the “aromatase-activity evaluation assay” (see “Materials and Methods” section and Fig. 5A) to study the reaction between CYP19A1 and testosterone inside byMM1712 over a time interval of 12 h. With this assay, we could describe both the concentration of the reaction product (β-estradiol) and the reaction speed as time functions. We measured fluorescence from the β-estradiol biosensors byMM381 and byMM1984 at different time points (Fig. 5B and E) upon exposure to the supernatant from the reaction between aromatase and testosterone inside byMM1712. The concentration of β-estradiol due to byMM1712 was derived by combining the fluorescence-over-time results with the transfer functions of the two β-estradiol biosensors shown in Fig. 2B-C above. We fitted these computed results to express β-estradiol concentration as a function of time (see Fig. 5C and F). The time derivative of these two curves corresponds to the reaction rate over time (see Fig. 5D and G), which shows how the speed of the testosterone conversion into β-estradiol changed over a given time interval. The transformation of testosterone into β-estradiol reached its maximum speed (107 nM/hour) after two hours when byMM1712 was induced with 1000 nM testosterone. In contrast, it was equal to 46 nM/hour at 35 min when byMM1712 was induced with 60 nM testosterone. The same trend was observed in both cases: when yeast strains are used as ‘off the shelf’ reagents, the reaction substrate needs to be transported into the cytoplasm before it can react with the enzymes. In contrast, in vitro reactions with purified enzymes show the maximum reaction rate at the very beginning because the initial substrate concentration is the highest one41.
Fig. 5.
Aromatase-activity evaluation-assay and its result. (A) Aromatase-activity evaluation assay (see “Materials and Methods” section. The drawing, based on Fig. 1 in19, was realized with Procreate software, Savage Interactive, Hobart, Australia). (B) The fluorescence intensities from biosensor byMM381 upon exposure to the supernatant—obtained at different times—of the reaction between aromatase and testosterone in byMM1712 (OD600 = 4). The initial concentration of testosterone was set to 1000 nM. (C) β-estradiol concentrations at different times are derived from byMM381 transfer function. These concentrations are then fitted to the function y = 1300*x^1.4/(7.46^1.4 + x^1.4) (R2 = 0.9648), where y is β-estradiol (nM) and x is time (hour). (D) The reaction rate of the conversion of testosterone into β-estradiol when byMM1712 is exposed to 1000 nM testosterone. (E) The fluorescence intensities from biosensor byMM1984 in supernatants—collected at different time points—of the reaction between aromatase and testosterone inside byMM1712 (OD600 = 1). The initial concentration of testosterone was set to 60 nM. (F) β-estradiol concentrations at different times are derived from byMM1984 transfer function. These concentrations are fitted to the function y = 76.08*x^1.93/(1.06^1.93 + x^1.93) (R2 = 0.9936), where y is β-estradiol (nM) and x is time (hour). (G) The reaction rate of the conversion of testosterone into β-estradiol when byMM1712 is exposed to 60 nM testosterone.
Moreover, according to the result in Figs. 5F and 60 nM testosterone were almost completely consumed after two hours (the initial OD600 of yeast cell solution was 1). Hence, in the “aromatase- inhibitors evaluation assay” (see “Materials and Methods” section) we induced with 60 nM testosterone a solution of byMM1712 at OD600 = 1 and let the enzymatic reaction run for 3 h to ensure that all testosterone could be consumed.
Letrozole IC50 determination through aromatase inhibitors evaluation assay
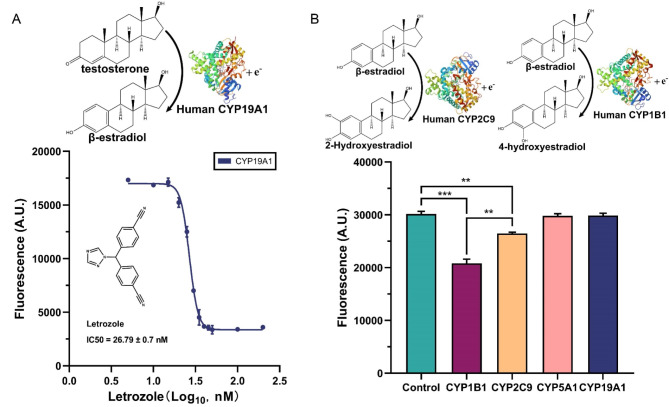
Most of breast cancers shows β-estradiol dependence and aromatase plays a rate-limiting role in β-estradiol biosynthesis. Aromatase inhibitors can specifically repress the activity of aromatase and reduce the level of β-estradiol in blood, which is a necessary condition to treat breast cancer20. Although many drugs based on aromatase inhibitors have been approved by the FDA for clinical treatment42, serious side effects have been reported43. Moreover, tumor resistance to known aromatase inhibitors demands the development of new aromatase repressors44—which should cause fewer side effects. Pharmacophore simulations and computational screening have been widely used to identify new possible inhibitors45–47. However, so far, no wet-lab method has been proposed that can avoid the costly high-throughput evaluation of candidate compounds. We developed an “aromatase-inhibitors evaluation assay” based on β-estradiol biosensor byMM1984 (see “Materials and Methods” section). Beside our engineered yeast strain, it requires only SDC solution, testosterone, and the candidate aromatase inhibitors. The method was tested on letrozole (see Fig. 6A). The IC50 of letrozole (MCE CGS 20267) on aromatase expressed by yeast strain byMM1712 (the ‘off the shelf’ reagent) is equal to 26.79 ± 0.70 nM (mean ± SEM). The small relative error, 2.61%, indicates an accurate measurement. Furthermore, our IC50estimation is reasonably close to the value of 20 nM obtained by using hamsters’ ovarian tissue48. Overall, our aromatase-inhibitors evaluation assay makes it possible to screen and assess a large number of candidate aromatase inhibitors in a cheap way.
Fig. 6.
Results from both the “aromatase-inhibitors evaluation assay” and the “screening for enzymes that metabolize β-estradiol”. (A) Fluorescence from byMM1984 at different concentrations of letrozole. Testing the “aromatase-inhibitors evaluation assay” on letrozole demands to expose byMM1712 to 60 nM testosterone and various concentrations of letrozole. After three hours of enzymatic reactions, the supernatant is used to induce byMM1984 β-estradiol biosensor. Fluorescence data are fitted to the empirical Hill function y = 3363 + (16988 − 3363)/(1 + 10^((1.428-x)*(−8.462))), where y is fluorescence (A.U.), and x is proportional to the letrozole concentration (log10, nM). The IC50 of letrozole corresponds to 26.79 ± 0.70 nM (the structure of human CYP19A1 was obtained from PDB, DOI: 10.2210/pdb4KQ8/pdb). (B) “Screening assay for enzymes that metabolize β-estradiol”. Four CYP proteins (CYP1B1, CYP2C9, CYP5A1, and CYP19A1) were tested to see if our methods could discriminate between CYPs that metabolize β-estradiol and those that are inert on this hormone. Correctly, CYP1B1 and CYP2C9 showed an activity significantly different from the control strain expressing CPR only (**: p-value ≤ 0.01; ***: p-value ≤ 0.001; two-sided Welch’s t test—see Table S30; the structure of human CYP2A9 is available at PDB, DOI: 10.2210/pdb1OG2/pdb. The structure of human CYP1B1 was also obtained from PDB, DOI: 10.2210/pdb1OG2/pdb).
Assay for screening enzymes capable of metabolizing β-estradiol
Enzymes that metabolize β-estradiol are expressed in numerous human tissues and play a crucial role in several physiological processes49. Moreover, metabolites produced during β-estradiol metabolism participate in many different biological reactions49. Understanding which enzymes in the human body can process β-estradiol is of great significance in treating β-estradiol-related diseases. Studies on β-estradiol metabolism have been limited to a few enzymes due to the high cost of enzyme isolation and purification50–52. The “screening assay for enzymes that metabolize β-estradiol”, which we have described in “Materials and Methods” section, utilizes ad-hoc-engineered yeast cells as enzyme factories and the β-estradiol biosensor byMM381 as a detection tool to find out which enzymes are capable of reacting with and transform β-estradiol into new molecules. We tested four CYP enzymes for their possible action on this hormone (see Fig. 6B). In agreement with published data53, only CYP1B1 and CYP2C9 exhibited a significant metabolic activity on β-estradiol (with the former being much stronger than the latter). These results proved the reliability of our assay, which can be easily applied to any other CYP enzyme upon expressing it in S. cerevisiae.
Conclusions
We engineered a library of S. cerevisiae β-estradiol biosensors hosting short lex operators. The best performance was achieved with eight copies of lexOpL. In general, the repetitive usage of the left half of the lex operator guaranteed higher fluorescence expression than the right part (lexOpR).
We calculated the transfer functions of the β-estradiol biosensors in our library in order to determine β-estradiol concentrations from fluorescence measurements. Biosensor strain byMM381 could detect concentrations in a wide range, from 0 to 500 nM. In contrast, biosensor byMM1984 appeared more reliable when responding to lower concentrations, from 0 to 30 nM. The two biosensors were tested via the real sample recovery method: both showed good reliability.
Four kinds of human CYP proteins were engineered, expressed, and analyzed in S. cerevisiae. First, the enzyme bag method was adapted from S. pombe to S. cerevisiae to quantify CYP enzyme activity. The whole process takes only one day instead of the week required by the original procedure for S. pombe. We put in evidence that the human CYP19A1 showed significantly high activity on the luciferin-BE substrate. S. cerevisiae testosterone biosensors were built on the S. cerevisiae β-estradiol biosensors by expressing the human CYP19A1-CPR system, which is able to convert testosterone into β-estradiol. The new biosensor strain byMM1906 returned a remarkably high ON/OFF ratio (21.12) and a detection range from 0.98 nM to 2000 nM, which covered all concentrations we used. The joint usage of strain byMM1712, which allows the conversion of testosterone into β-estradiol, and a β-estradiol biosensors (strain byMM381 or byMM1894) permitted us to quantify aromatase activity with a novel assay, alternative to enzyme bags, that only demands fluorescence measurements. We also developed an “aromatase-inhibitors evaluation assay” that was tested on letrozole giving an IC50 equal to 26.79 ± 0.7 nM (Mean ± SEM). Besides, we described and showed the validity of a further assay to detect “enzymes that metabolize β-estradiol”. We are confident that these two assays will permit to carry out high throughput screening of a large number of candidate aromatase inhibitors and β-estradiol metabolic factors, respectively, rapidly and at extremely low cost.
Materials and methods
Materials and reagents
The nucleotide sequences were synthesized by GENEWIZ (Soochow, China). Acc651 (R0599S), BstXI (R0113S), EcoRV (R0195S), Sacl (R0156S), Stul (R0187S) restriction enzymes, O5 Hot-start polymerase (M0491S), T4 DNA ligase (M0202S), T5 exonuclease (M0663S), and Taq DNA ligase (M0208S) were purchased from New England Biolabs (NEB, USA). DNA ladder (M130-01) was provided by GeneStar (Beijing, China). Histidine (H-25675) and uracil (U-3810) were purchased from Heowns Biochem Technologies. Llc. (HEOWNS, USA), tryptophan (DH357-4) was provided by Ding Guo biolab (Beijing, China), leucine (L0011), glycerol (G8190), and ampicillin (A8180) were purchased from Solarbio (Beijing, China). Phusion DNA polymerase (F530L) and Yeast Nitrogen Base (Q30009) were provided by Thermo Fisher Scientific (TFS, USA). Agarose (92008) was purchased from Invitrogen (TFS, USA). Glucose (HA1967-500) and DNA elution kit (AP-GX-250) were purchased from Axygen (USA). DMSO (D2650) and PEG 3350 (P4338) were provided by Millipore Sigma (MERCK, Germany).
Plasmid construction
All plasmids were constructed via Gibson (isothermal assembly) method54. The yeast-shuttle vectors pRSII40X (obtained from Addgene, a gift from Steven Haase55) were used as backbones (see Fig. S1). A list of all plasmids used/built in this work is given in Table S1.
Construction of plasmids expressing the yeast enhanced green fluorescent protein (yEGFP).Plasmids expressing yEGFP are the reporter parts of our biosensors. Synthetic promoters were designed by adding lex operators to a pCYC1min/pCYC1core56 promoter sequence. The scheme of all synthetic promoters used in this work are shown in Fig. S2, whereas all DNA part sequences are listed in Table S2-S5. Promoter, yEGFP coding region, and terminator were assembled into a cut-open pRSII406 backbone—i.e., digested with Acc65I (NEB-R0599S) and SacI-HF (NEB-R3156S)—via Gibson method.
Construction of plasmids expressing the chimeric protein LexA-ER-AD.Plasmids expressing LexA-ER-AD host the receptor part of our biosensors. Three chimeric proteins with different activation domains (VP6457, B4258, and mDR521_80559) were assembled together with a promoter and a terminator into the cut-open pRSII405 backbone via Gibson method.
Construction of plasmids expressing CPR and CYP protein.The DNA sequences of CPR and CYP proteins had been codon-optimized for expression in fission yeast cells18. The sequence encoding CPR was assembled into either pRSII403 or pRSII406 backbone, whereas the genes coding for the four CYP proteins were placed into either pRSII404 or pRSII405. All backbones were digested with Acc65I and SacI-HF. Every plasmid was assembled via Gibson method.
Touchdown PCR was carried out to amplify DNA fragments. Q5 High-Fidelity DNA Polymerase (NEB-M0491S) was employed. AxyPrep DNA extraction kit (Axigen-AP-GX-250) was adopted to purify PCR products from a gel. 5 µl of solution containing DNA fragments and cut-open backbone in equimolar amount were mixed with 15 µl Gibson mixture inside a PCR tube that was kept at 50 ℃ for 1 h in a thermal cycler. The isothermal assembly products (new plasmids) were finally inserted into E. coli competent cells (DH5α, Life Technology––18263–012). The bacterial cell transformation required a short heatshock (30 s at 42˚C).
Yeast transformation
Strain CEN.PK2-1 C (MATa; his3 1; leu2-3 112; ura3-52; trp1-289; MAL2-8c; SUC2), Euroscarf-30,000 A (Johann Wolfgang Goethe University, Frankfurt, Germany) named as byMM584 was used as the chassis for our circuits. All plasmids constructed in this study were linearized with StuI (NEB-R0187V) or BstXI (NEB-R0113V) and then integrated into the genome of byMM584. The standard lithium acetate-thermal shock method60 was adopted to transform yeast cells. Plates (2% agar, 2% glucose) containing a synthetic selective growth medium were used to incubate transformed yeast cells for 2 days at 30 ℃. All yeast strains engineered in this work are listed in Table S6.
Flow cytometry
Strains hosting a biosensor were taken from plates containing a synthetic selective medium and dissolved in 3 ml SDC (synthetic defined complete) solution (the initial OD600 was adjusted, via dilution, to ~ 0.04). Cells were grown at 30 ℃ and 240 RPM for 14 h. Then, 20 µl of cell solution was added to SDC supplied with the inducer to reach an overall volume of 2 ml (OD600~ 0.05). Cells were incubated for 24 h at 30 ℃ and 240 RPM10. Finally, 5 µl of induced-cell solution was added to 195 µl ddH2O (1:40 dilution) before measuring fluorescence with a flow cytometer (BD FACSVerse machine, laser 488 nm-FITC, filter 527/32 nm). Ten thousand cells (events) were collected at each measurement. Performance Quality Control (PQC) should be passed once a month. Fluorescent beads (BD FACSuite CS&T Research beads 650621) were used to adjust the FITC voltage in order to assure that the flow cytometry results were reliable and reproducible—the relative difference between the bead peaks at the beginning of two consecutive measurements should be less than 5%. The flowCore R-Bioconductor package61 was used to analyze flow cytometry (fcs) files. Fluorescence mean values were calculated on three independent experiments, i.e., performed on different days.
Luminescence-based assay (Enzyme Bags)
The luminescence-based assay developed in19 was adapted to S. cerevisiae. S. cerevisiae strains are first streaked on synthetic selective medium plates and incubated for 12 h at 30˚C. Then, they are transferred into SDC solution and further incubated for 14 h (the initial OD600 should be ~ 0.04) at 30˚C and 240 RPM. Cell number is counted under a microscope (Bresser 5201000) using a hemocytometer (Shanghai Qiu Jing YX-JSB52). 5·107 yeast cells are added into each reaction tube. After centrifugation, cells are resuspended in 1 ml solution—containing 0.05% (v/v) Triton X-100 and 0.1 M Tris-HCl buffer (pH 7. 6)—to be permeabilized. After 1 h incubation at 30˚C and 240 RPM, cells are washed three times with ice-cold NH4HCO3 (pH 7.6). Then, 25 µl 2X NADPH regeneration system—consisting of 22 µl deionized H2O, 2.5 µl NADPH mix A (Promega V952A), and 0.5 µl NADPH mix B (Promega V953A)—are added. At this point, 25 µl 0.1 M Phosphate Buffered Saline (PBS) buffer (pH 7.4) containing pro-luciferin substrate (P450-Glo™ Assays Technical Bulletin TB325) is mixed with the yeast cells62. The reaction takes place at 37˚C and 1000 RPM and lasts 3 h. This mixture is then centrifuged at 16,000 RPM for 1 min to collect 50 µl supernatant to which are added 50 µl Luciferin Detection Reagents (LDRs, Promega V8921). After 20 min, luminescence is measured with a luminometer (TECAN INFINITE M200 Pro) with 1 s integration time and 150 ms settle time. Each mean value is calculated on at least three independent experiments.
Real sample recovery
Real sample recovery experiments were carried out to test if our biosensors could measure β-estradiol concentration (in SDC) precisely37. Six samples with different concentrations of β-estradiol were chosen depending on the biosensor detection range. Recovery (%) and relative standard deviation (%) were calculated on three independent flow cytometry experiments.
Growth curve
Yeast strains, taken from synthetic selective medium plates, are cultured in 3 ml SDC (initial OD600 ~ 0.04). Each strain is grown for 28 h at 30 ℃ and 240 RPM. The OD600 value is measured with an Eppendorf Bio Photometer apparatus every 7 h. OD600 mean values are obtained from three independent experiments (see Table S7).
Aromatase-activity evaluation assay
Biosensor byMM381 (plus a high concentration of testosterone). Yeast strain byMM1712 that expresses both CPR and CYP19A1 is first streaked on a synthetic selective medium plate and then incubated for 14 h in 30 ml SDC at 30 ℃ and 240 RPM. The initial OD600 is ~ 0.2. Yeast strain byMM1709 that expresses only CPR is used as a control group. After the growth time, 25 ml yeast cells solution are added to 25 ml SDC solution that contains 2000 nM testosterone (OD600 ~ 4). The overall 50 ml solution is distributed among 25 wells (2 ml to each well) of a 48-deep-well cell-culture plate (Axygen P-5ML-48-C) and cultured for 12 h at 30 ℃ and 240 RPM. During the first 3 h, every 10 min 1.5 ml solution from one well is moved into a 2 ml tube that is centrifuged at 16,000 RPM for 2 min to collect all the yeast cells. 1.2 ml supernatant is then moved into a new 1.5 ml tube and marked with the time at which it was prepared. During the next 9 h, 1.2 ml supernatant is obtained in the same way, though only every hour. Tubes containing 1.2 ml supernatant are stored at 4 ℃. One ml supernatant (sample solution) from each tube is added to a different well in a 48-deep-well cell-culture plate. Then, 980 µl SDC solution and 20 µl biosensor byMM381 cell solution (previously cultured for 14 h, OD600 ~ 0.05) are added. Cells are finally grown for 24 h at 30 ℃ and 240 RPM before measuring fluorescence via a flow cytometry experiment. Three independent experiments reproducing the whole assay should be performed to calculate mean values and standard deviations.
Biosensor byMM1984 (plus a low concentration of testosterone). Yeast strain byMM1712 that expresses both CPR and CYP19A1 is dissolved into 30 ml SDC and incubated at 30 ℃ and 240 RPM for 14 h. The initial OD600 shall be ~ 0.04. Yeast strain byMM1709 that expresses only CPR is used as a control. After the whole incubation time, 25 ml yeast cell solution is added to 25 ml SDC solution that contains 120 nM testosterone (OD600 ~ 1). The overall solution is divided into 25 wells (2 ml in each well) of a 48-deep-well cell-culture plate (Axygen P-5ML-48-C) and incubated at 30 ℃ and 240 RPM for 2 h. Every 5 min, 1.5 ml solution from one well is taken out and poured into a new 2 ml tube that is centrifuged at 16,000 RPM for 2 min to remove all yeast cells. Afterwards, 1.2 ml supernatant is poured into another 1.5 ml tube that is marked with the time at which it was made and stored at 4 ℃. One ml supernatant (sample solution), from each tube, is transferred to a different well of a 48-deep-well cell-culture plate. Here, 980 µl SDC solution and 20 µl byMM1984 cells, which contain a biosensor and were cultured for 14 h, are added (OD600 ~ 0.05). Cells are then grown for 24 h at 30 ℃ and 240 RPM. Finally, fluorescence is measured. Three independent experiments of the whole assay are carried out to calculate mean fluorescence values and standard deviations.
Aromatase-inhibitors evaluation assay
Yeast strain byMM1712 that expresses both CPR and CYP19A1 is incubated for 14 h in 10 ml SDC solution (the initial OD600 shall be equal to ~ 0.04) at 30 ℃ and 240 RPM. Then, 1 ml of yeast solution is added to 12 different wells of a 48-deep-well cell-culture plate (Axygen P-5ML-48-C). Afterwards, 1 ml SDC solution with 120 nM testosterone and different inhibitor concentrations is added to each well (OD600 ~ 1). The solution is incubated for 3 h at 30 ℃ and 240 RPM. Then, 1.5 ml solution is taken out from each well and poured into a 2 ml tube that is centrifuged at 16,000 RPM to remove all the yeast cells. 1.2 ml supernatant is transferred into a 1.5 ml tube and marked with the inhibitor concentration. 1 ml supernatant (sample solution) from each tube is added to a different well of a 48-deep-well cell culture plate (Axygen P-5ML-48-C), to which both 980 µl SDC solution and 20 µl byMM1984 biosensor cell solution—previously cultured for 14 h—are added (OD600 ~ 0.05). Cells are then grown for 24 h at 30 ℃ and 240 RPM. Fluorescence is finally measured. The whole assay is repeated three times to calculate fluorescence mean values and standard deviations.
Screening assay for enzymes that metabolize β-estradiol
Yeast strains that express both CPR and CYP proteins are dissolved in 3 ml SDC solution (OD600 ~ 0.2) and incubated for 14 h at 30 ℃ and 240 RPM. Yeast strain byMM1709, which expresses only CPR, is used as a control. One ml yeast solution and 1 ml SDC solution with 2000 nM β-estradiol (OD600 ~ 4) are added to the wells of a 48-deep-well cell-culture plate (Axygen P-5ML-48-C), which is incubated for 12 h at 30 ℃ and 240 RPM. Afterwards, 1.5 ml solution is taken out of each well and poured into a 2 ml tube that is centrifuged at 16,000 RPM to remove all yeast cells. 1.2 ml supernatant is then poured into a new 1.5 ml tube and marked with the enzyme name. One ml supernatant (sample solution) from each tube is added to a different well of a 48-deep-well cell culture plate (Axygen P-5ML-48-C) together with 980 µl SDC solution and 20 µl byMM381 biosensor cell solution, which was previously cultured for 14 h (OD600 ~ 0.05). Cells are grown for 24 h at 30 ℃ and 240 RPM before fluorescence measurement via a flow cytometry experiment. The assay is repeated three times to calculate fluorescence mean values and standard deviations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank all students of the Synthetic Biology Laboratory at the School of Pharmaceutical Science and Technology—Tianjin University for their kind help. In particular, we are grateful to Ruihan Bai and Yue Li for their preliminary tests on the β-estradiol biosensors. We also want to thank Zhi Li and Xiangyang Zhang for their assistance in FACS experiments.
Author contributions
J.W.: conceptualization, experiments, data analysis, and drafting the manuscript. M.B.: conceptualization, supervision, and writing the manuscript. M.A.M.: conceptualization, supervision, and writing the manuscript.
Funding
No funding.
Data availability
Flow cytometry data (fcs files) are available at FlowRepository: http://flowrepository.org/id/RvFrHOBKzTD9F0jhiIlpoTokqYCce9LAGCOK5fjoMwLrgmnU1DLEhkz3TZWpuTi9.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Matthias Bureik, Email: matthias@tju.edu.cn.
Mario Andrea Marchisio, Email: mamarchisio@yahoo.com, Email: marchisio@mail.neu.edu.cn.
References
- 1.Brooks, S. M. & Alper, H. S. Applications, challenges, and needs for employing synthetic biology beyond the lab. Nat. Commun.12, 1390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding, N., Zhou, S. & Deng, Y. Transcription-factor-based biosensor engineering for applications in synthetic biology. ACS Synth. Biol.10, 911–922 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Dossani, Z. Y. et al. A combinatorial approach to synthetic transcription factor‐promoter combinations for yeast strain engineering. Yeast35, 273–280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu, W. et al. Molecularly imprinted polymers on graphene oxide surface for EIS sensing of testosterone. Biosens. Bioelectron.92, 305–312 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Grazon, C. et al. A progesterone biosensor derived from microbial screening. Nat. Commun.11, 1276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta, N., Renugopalakrishnan, V., Liepmann, D., Paulmurugan, R. & Malhotra, B. D. Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron.141, 111435 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Ruden, D. M., Ma, J., Li, Y., Wood, K. & Ptashne, M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature350, 250–252 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Kumar, V., Green, S., Staub, A. & Chambon, P. Localisation of the oestradiol-binding and putative DNA‐binding domains of the human oestrogen receptor. EMBO J.5, 2231–2236 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ottoz, D. S., Rudolf, F. & Stelling, J. Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae. Nucleic Acids Res.42, e130–e130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou, T., Liang, Z. & Marchisio, M. A. Engineering a two-gene system to operate as a highly sensitive biosensor or a sharp switch upon induction with β-estradiol. Sci. Rep.12, 21791 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rantasalo, A., Kuivanen, J., Penttilä, M., Jäntti, J. & Mojzita, D. Synthetic toolkit for complex genetic circuit engineering in Saccharomyces cerevisiae. ACS Synth. Biol.7, 1573–1587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb, D. C. & Waterman, M. R. Unusual properties of the cytochrome P450 superfamily. Philos. Trans. R. Soc. B: Biol. Sci.368, 20120434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Nardo, G. & Gilardi, G. Natural compounds as pharmaceuticals: The key role of cytochromes P450 reactivity. Trends Biochem. Sci.45, 511–525 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Munro, A. W., McLean, K. J., Grant, J. L. & Makris, T. M. Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem. Soc. Trans.46, 183–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellor, S. B. et al. Fusion of ferredoxin and cytochrome P450 enables direct light-driven biosynthesis. ACS Chem. Biol.11, 1862–1869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fantuzzi, A., Fairhead, M. & Gilardi, G. Direct electrochemistry of immobilized human cytochrome P450 2E1. J. Am. Chem. Soc.126, 5040–5041 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Imaoka, S. et al. Multiple forms of human P450 expressed in Saccharomyces cerevisiae: Systematic characterization and comparison with those of the rat. Biochem. Pharmacol.51, 1041–1050 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Durairaj, P. et al. Functional expression and activity screening of all human cytochrome P450 enzymes in fission yeast. FEBS Lett.593, 1372–1380 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Sharma, S., Durairaj, P. & Bureik, M. Rapid and convenient biotransformation procedure for human drug metabolizing enzymes using permeabilized fission yeast cells. Anal. Biochem.607, 113704 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Smith, I. E. & Dowsett, M. Aromatase inhibitors in breast cancer. N. Engl. J. Med.348, 2431–2442 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Guengerich, F. P. Cytochrome P450 enzymes in the generation of commercial products. Nat. Rev. Drug Discovery. 1, 359–366 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Thompson, E. A. & Siiteri, P. K. Utilization of oxygen and reduced nicotinamide adenine dinucleotide phosphate by human placental microsomes during aromatization of androstenedione. J. Biol. Chem.249, 5364–5372 (1974). [PubMed] [Google Scholar]
- 23.Simpson, E. R. et al. Aromatase—A brief overview. Annu. Rev. Physiol.64, 93–127 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Recanatini, M. et al. A new class of nonsteroidal aromatase inhibitors: Design and synthesis of chromone and xanthone derivatives and inhibition of the P450 enzymes aromatase and 17α-hydroxylase/C17, 20-lyase. J. Med. Chem.44, 672–680 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Cabrera, E. et al. Cryopreservation and the freeze–thaw stress response in yeast. Genes11, 835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gou, L., Bloom, J. S. & Kruglyak, L. The genetic basis of mutation rate variation in yeast. Genetics211, 731–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikkhah, M. et al. Review of electrochemical and optical biosensors for testosterone measurement. Biotechnol. Appl. Chem.70, 318–329 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Herold, D. A. & Fitzgerald, R. L. Immunoassays for testosterone in women: Better than a guess? Clin. Chem.49, 1250–1251 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Ankarberg-Lindgren, C. & Norjavaara, E. Sensitive RIA measures testosterone concentrations in prepubertal and pubertal children comparable to tandem mass spectrometry. Scand. J. Clin. Lab. Investig.75, 341–344 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Yockell-Lelievre, H. et al. Plasmonic sensors for the competitive detection of testosterone. Analyst140, 5105–5111 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Eguílaz, M. et al. An electrochemical immunosensor for testosterone using functionalized magnetic beads and screen-printed carbon electrodes. Biosens. Bioelectron.26, 517–522 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Huang, Y., Shi, M., Zhao, S. & Liang, H. A sensitive and rapid immunoassay for quantification of testosterone by microchip electrophoresis with enhanced chemiluminescence detection. Electrophoresis32, 3196–3200 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Mak, P., Cruz, F. D. & Chen, S. A yeast screen system for aromatase inhibitors and ligands for androgen receptor: Yeast cells transformed with aromatase and androgen receptor. Environ. Health Perspect.107, 855–860 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazumder, M. & McMillen, D. R. Design and characterization of a dual-mode promoter with activation and repression capability for tuning gene expression in yeast. Nucleic Acids Res.42, 9514–9522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cormack, B. P. et al. Yeast-enhanced green fluorescent protein (yEGFP): A reporter of gene expression in Candida albicans. Microbiology143, 303–311 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Song, W., Li, J., Liang, Q. & Marchisio, M. A. Can terminators be used as insulators into yeast synthetic gene circuits? J. Biol. Eng.10, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali, M. R. et al. Development of an advanced DNA biosensor for pathogenic Vibrio cholerae detection in real sample. Biosens. Bioelectron.188, 113338 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Yu, L. & Marchisio, M. A. Saccharomyces cerevisiae synthetic transcriptional networks harnessing dCas12a and type VA anti-CRISPR proteins. ACS Synth. Biol.10, 870–883 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y. & Marchisio, M. A. Interaction of bare dSpCas9, scaffold gRNA, and type II anti-CRISPR proteins highly favors the control of gene expression in the yeast S. Cerevisiae. ACS Synth. Biol.11, 176–190 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Baruch, A., Jeffery, D. A. & Bogyo, M. Enzyme activity—it’s all about image. Trends Cell Biol.14, 29–35 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Härdin, H. M., Zagaris, A., Krab, K. & Westerhoff, H. V. Simplified yet highly accurate enzyme kinetics for cases of low substrate concentrations. FEBS J.276, 5491–5506 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Chumsri, S., Howes, T., Bao, T., Sabnis, G. & Brodie, A. Aromatase, aromatase inhibitors, and breast cancer. J. Steroid Biochem. Mol. Biol.125, 13–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gnant, M. et al. Duration of adjuvant aromatase-inhibitor therapy in postmenopausal breast cancer. N. Engl. J. Med.385, 395–405 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Ma, C. X., Reinert, T., Chmielewska, I. & Ellis, M. J. Mechanisms of aromatase inhibitor resistance. Nat. Rev. Cancer15, 261–275 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Schuster, D. et al. Pharmacophore modeling and in silico screening for new P450 19 (aromatase) inhibitors. J. Chem. Inf. Model.46, 1301–1311 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Awasthi, M., Singh, S., Pandey, V. P. & Dwivedi, U. N. Molecular docking and 3D-QSAR-based virtual screening of flavonoids as potential aromatase inhibitors against estrogen-dependent breast cancer. J. Biomol. Struct. Dyn.33, 804–819 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Rampogu, S. et al. Sulfonanilide derivatives in identifying novel aromatase inhibitors by applying docking, virtual screening, and MD simulations studies. Biomed. Res. Int.2017, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhatnagar, A. S., Brodie, A. M. H., Long, B. J., Evans, D. B. & Miller, W. R. Intracellular aromatase and its relevance to the pharmacological efficacy of aromatase inhibitors. J. Steroid Biochem. Mol. Biol.76, 199–202 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Zhu, B. T. & Conney, A. H. Functional role of estrogen metabolism in target cells: Review and perspectives. Carcinogenesis19, 1–27 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Breuer, H., Knuppen, R. & Haupt, M. Metabolism of oestrone and oestradiol-17β in human liver in vitro. Nature212, 76–76 (1966). [DOI] [PubMed] [Google Scholar]
- 51.Tsuchiya, Y., Nakajima, M. & Yokoi, T. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett.227, 115–124 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Badawi, A. F., Cavalieri, E. L. & Rogan, E. G. Role of human cytochrome P450 1A1, 1A2, 1B1, and 3A4 in the 2-, 4-, and 16 [alpha]-hydroxylation of 17 [beta]-estradiol. Metab. Clin. Exp.50, 1001–1003 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Lee, A. J., Cai, M. X., Thomas, P. E., Conney, A. H. & Zhu, B. T. Characterization of the oxidative metabolites of 17β-estradiol and estrone formed by 15 selectively expressed human cytochrome P450 isoforms. Endocrinology144, 3382–3398 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Chee, M. K. & Haase, S. B. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3: Genes Genomes Genet.2, 515–526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hahn, S., Hoar, E. T. & Guarente, L. Each of three TATA elements specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci.82, 8562–8566 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. GAL4-VP16 is an unusually potent transcriptional activator. Nature335, 563–564 (1988). [DOI] [PubMed] [Google Scholar]
- 58.Hussey, S. L., Muddana, S. S. & Peterson, B. R. Synthesis of a β-estradiol-biotin chimera that potently heterodimerizes estrogen receptor and streptavidin proteins in a yeast three-hybrid system. J. Am. Chem. Soc.125, 3692–3693 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Whitelaw, M. L., McGuire, J., Picard, D., Gustafsson, J. A. & Poellinger, L. Heat shock protein hsp90 regulates dioxin receptor function in vivo. Proc. Natl. Acad. Sci.92, 4437–4441 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gietz, R. D. & Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol.350, 87–96 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Hahne, F. et al. flowCore: A Bioconductor package for high throughput flow cytometry. BMC Bioinform.10, 1–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashraf, R. A., Bureik, M. & Marchisio, M. A. Design and engineering of logic genetic-enzymatic gates based on the activity of the human CYP2C9 enzyme in permeabilized Saccharomyces cerevisiae cells. Synth. Syst. Biotechnol.9, 406–415 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Flow cytometry data (fcs files) are available at FlowRepository: http://flowrepository.org/id/RvFrHOBKzTD9F0jhiIlpoTokqYCce9LAGCOK5fjoMwLrgmnU1DLEhkz3TZWpuTi9.