Abstract
Rising atmospheric CO2 generally increases yield of indica rice, one of the two main Asian cultivated rice subspecies, more strongly than japonica rice, the other main subspecies. The molecular mechanisms driving this difference remain unclear, limiting the potential of future rice yield increases through breeding efforts. Here, we show that between-species variation in the DNR1 (DULL NITROGEN RESPONSE1) allele, a regulator of nitrate-use efficiency in rice plants, explains the divergent response to elevated atmospheric CO2 (eCO2) conditions. eCO2 increased rice yield by 22.8–32.3% in plants carrying or mimicking the indica DNR1 allele, but only by 3.6–11.1% in plants carrying the japonica DNR1 allele. Rice plants carrying or mimicking the indica DNR1 allele exhibit decreased eCO2-responsive transcription and protein abundance of DNR1, which activates genes involved in nitrate transport and assimilation, driving the increase in plant growth. Our findings identify the indica DNR1 gene as a key breeding resource for sustainably enhancing nitrate uptake and rice yields in japonica varieties, potentially contributing to global food security as atmospheric CO2 levels continue to increase.
Subject terms: Plant ecology, Plant breeding, Agricultural genetics, Genetic variation
Variation in DNR1, a negative regulator of nitrate-use efficiency, contributes to a stronger response to elevated CO2 levels in indica rice than in japonica rice. This finding could inform breeding efforts to enhance rice yields as CO2 levels rise.
Introduction
Atmospheric CO2 concentrations have increased from 315 ppm in 1958 to 423 ppm in 2024 and are expected to further increase to 800 ppm by the end of the 21st century due to human activities such as fossil fuel consumption and deforestation1,2. Elevated CO2 concentration (eCO2) often stimulates photosynthesis and yields of C3 plants3–5, also known as the CO2 fertilization effect. Global meta-analyses of free air CO2 enrichment (FACE) experiments showed that eCO2 increased photosynthesis by 22% and yield by 14% in rice4,6. Rice sustains approximately half of the global population and provides about one-fifth of the world’s dietary energy supply7. Furthermore, global rice yields are predicted to reduce due to global climate warming8,9. In this context, the role of atmospheric changes, particularly the increase in CO2 levels, becomes a pivotal factor in understanding future trends in rice production and global food security.
Around 90% of rice cultivation occurs in Asia, where two major subspecies, indica (Xian) and japonica (Geng), each with distinct developmental and physiological traits, are cultivated10. Japonica rice, domesticated in the Yangtze River basin around 9000 to 6000 years ago, is predominantly grown and consumed in East Asia, comprising about 40% of the rice cultivation area in China, Japan, and Korea11. In contrast, indica rice, first domesticated in the Ganges region between 8500 and 4500 years ago, is more prevalent in most other regions. However, CO2 fertilization effect varies between indica and japonica6,12–14, with indica varieties showing a more pronounced increase in yield (+20.4%) under eCO2 conditions compared to japonica varieties (+12.7%)12. The disparity in the CO2 fertilization between subspecies may strongly affect future global total rice yields; the difference between all rice globally responding to eCO2 like japonica or like indica amounts to ~109 Tg per year, equal to roughly half of China’s annual rice production. However, the underlying mechanisms driving this divergence in response to eCO2 between rice subspecies, especially at the molecular level, remain largely unexplored.
Nitrogen (N) is a critical nutrient for plant growth, and its availability and uptake significantly affect the response of C3 plants, including rice, to eCO2 conditions3,15,16. In rice, nitrate (NO3-) and ammonium (NH4+) serve as the primary inorganic N sources17. Due to nitrification in the rhizosphere, up to 40% of the total N absorbed and utilized by rice is NO3−18. Notably, indica rice varieties generally exhibit higher capacities of both NO3− uptake and assimilation compared to japonica, whereas the two subspecies show similar NH4+ uptake rates19,20. Consistent with these results, we found that japonica varieties exhibited significantly lower NO3− uptake rates that were less sensitive to changes in external CO2 status compared with indica varieties (Supplementary Fig. 1). Furthermore, we observed an interactive effect on NO₃⁻ uptake (P = 0.001), but not on NH₄⁺ absorption (P = 0.373), between eCO₂ and rice species. The NO₃⁻ uptake rates in japonica varieties were less responsive to eCO₂ (+40%) compared to indica varieties (+69%) (Supplementary Fig. 1). Based on these observations, we hypothesize that variations in NO3−-use efficiency might contribute to the divergent yield responses to eCO2 between indica and japonica.
Recent studies have identified five genes-OsNRT1.1B, OsNR2, DULL NITROGEN RESPONSE1 (DNR1), REGULATOR OF N-RESPONSIVE RSA ON CHROMOSOME 10 (RNR10), and MYB61-that are involved in NO3− use efficiency and that exhibit allelic variations between indica and japonica rice subspecies19,21–24 (Supplementary Tables 1, 2). To evaluate the response of genetic variations to eCO2, we performed an RNA-seq analysis, using the flag leaves of a typical indica variety Yangdao 6 (YD6) and a typical japonica variety Zhonghua 11 (ZH11) at the heading stage in a FACE experiment (See supplementary method). We found that while eCO2 (~ +150 ppm) decreased DNR1 abundance in the leaves of YD6, it did not significantly affect its expression in ZH11 (Supplementary Fig. 2a). In contrast, the responses of the other four genes to eCO2 were similar between the two varieties (Supplementary Fig. 2b–e). Furthermore, we found that the DNR1 allele varies between japonica and indica varieties frequently used in FACE experiments (Supplementary Table 3). Notably, rice varieties with the indica DNR1 allele were more responsive to eCO₂ (Supplementary Fig. 3).
DNR1, a negative regulator of auxin biosynthesis, has diverged in sequence between the two rice subspecies23. Plants carrying the indica DNR1 variant confers reduced DNR1 mRNA and protein abundance and subsequently increased auxin accumulation, thereby inducing transcriptional activation of genes coding NO3− uptake and downstream NO3− assimilation enzymes (Supplementary Fig. 4, Supplementary Table 2), leading to high N-use efficiency (NUE) and grain yield23. As expected, eCO2 increased the rice biomass, N uptake and yields more strongly in YD6 than those in ZH11 (Supplementary Fig. 5). These findings suggest that DNR1 variations could play a significant role in the divergent rice responses to eCO2 between rice subspecies.
Results
DNR1 variation drives divergent yield responses to elevated CO2
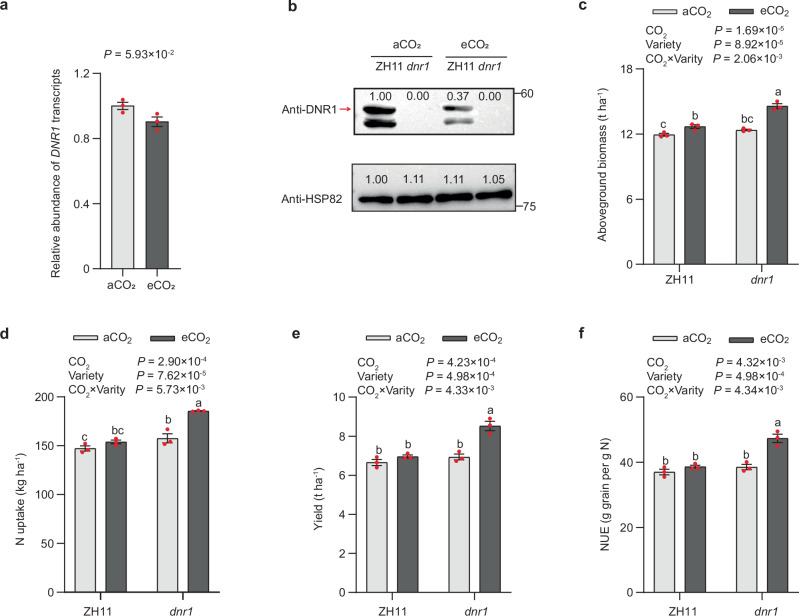
To evaluate whether the DNR1 variation contributes to the divergent responses to eCO2 between rice subspecies, we carried out a field experiment within a FACE system to compare typical japonica variety ZH11 and its dnr1 mutants, which mimics the indica DNR1 allele against a ZH11 background by reducing DNR1 abundance23. eCO2 reduced both DNR1 transcript and protein abundances in ZH11, and DNR1 protein levels were undetectable in the dnr1 mutants under both eCO2 and aCO2 conditions, due to the complete loss-of-function nature of the DNR1 variant (Fig. 1a, b). We found that eCO2 stimulated the growth of the dnr1 mutants more strongly than the ZH11 variety (Fig. 1c). At the heading stage, eCO2 increased the light-saturated net photosynthesis rate more strongly in dnr1 mutants than in ZH11 (Supplementary Fig. 6a). eCO2 significantly reduced stomatal conductance (gs), maximum rate of RuBP carboxylation (Vcmax), maximum rate of electron transport driving RuBP regeneration (J max), and non-photochemical quenching (NPQ) (Supplementary Figs. 6, 7). While dnr1 mutants exhibited higher Vcmax and NPQ compared to ZH11, eCO2 had no effect on the quantum yield of Photosystem II (Y(II)), the photochemical quenching coefficient (qL), or the maximum photochemical quantum yield of PSII (Fv/Fm) (Supplementary Fig. 7). eCO2 increased leaf area by 40.8% in dnr1 mutants, compared to only 6.6% increases in ZH11 (Supplementary Fig. 8a). In agreement with previous studies6, eCO2 tended to reduce the N content of flag leaf due to the dilution effect (Supplementary Fig. 8b). eCO2 increased N uptake more substantially in dnr1 mutants than in ZH11 (Supplementary Fig. 8c).
Fig. 1. Different responses of japonica variety Zhonghua 11 (ZH11) and its dnr1 mutants mimicking the indica DNR1 allele to elevated CO2.
a DNR1 transcript abundance in ZH11 shoots under ambient CO2 (aCO2) and elevated CO2 (eCO2) conditions. Transcript abundance was measured relative to ZH11 under aCO2 (set to 1). Data are mean ± s.e.m. (n = 3 biological replicates). P-value was generated from two-sided Student’s t tests. b DNR1 protein abundance in the shoots of ZH11 and dnr1 under aCO2 and eCO2 conditions. HSP82 serves as a loading control. The red arrow indicates the DNR1 bands. Data are representative of three independent experiments, with similar results. Aboveground biomass (c), N uptake (d), yield (e) and N-use efficiency (f) under aCO2 and eCO2 conditions were measured at maturity. ZH11 and dnr1 indicate japonica variety Zhonghua 11 and its dnr1 mutants, respectively. c–f Data are mean ± s.e.m. (n = 3 biological replicates). P-values were generated from two-way ANOVA. Different letters indicate significant differences among treatments (P < 0.05). Source data are provided as a Source Data file.
At maturity stage, the aboveground biomass, N uptake, and rice yield were all significantly higher in the dnr1 mutants compared to ZH11 (Fig. 1c–e). The impacts of eCO2 on these parameters were clearly dependent on the DNR1 allele. In dnr1 mutants, eCO2 stimulated the aboveground biomass, N uptake, and rice yield by 17.8%, 17.8%, and 22.8%, respectively, while it had no significant effects on ZH11 (Fig. 1c–e). Also, eCO2 stimulated N-use efficiency (NUE) more strongly in dnr1 than in ZH11 (Fig. 1f).
To test whether DNR1 variation also causes differential responses to eCO2 within an indica variety, we compared the effects of eCO2 on the typical indica variety, Hua-Jing-Xian 74 (HJX74), with a near-isogenic line (NIL) carrying the japonica DNR1 allele for two years. The indica HJX74 exhibited lower DNR1 transcripts and protein abundance compared to NIL, and the changes induced by eCO2 were stronger in HJX74 (Supplementary Fig. 9a, b). More importantly, eCO2 stimulated rice plant growth parameters (i.e., photosynthesis rate, leaf area, biomass, N uptake, and NUE) more strongly in HJX74 than in NIL (Supplementary Figs. 9, 10). At maturity, eCO2 significantly increased rice yield by 27.6–32.3% in HJX74, but only by 3.6–11.1% in NIL (Supplementary Figs. 9j, 10h). Taken together, our findings indicate that the variation in the DNR1 allele plays a crucial role in driving the divergent responses to eCO2 between rice subspecies.
DNR1 variation affects response of N metabolism to eCO2
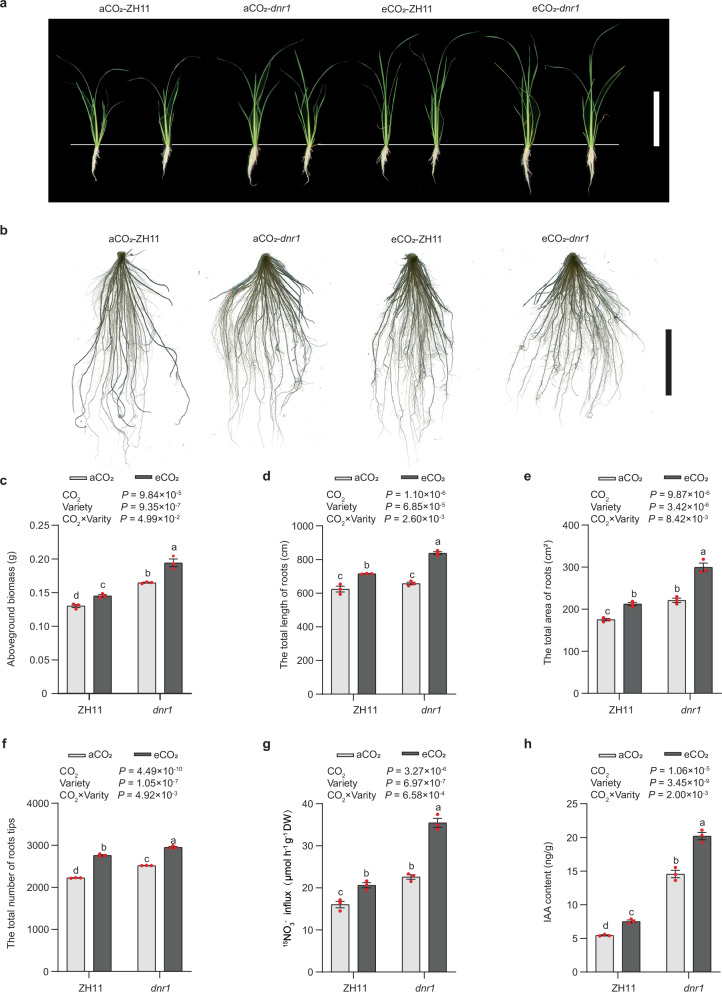
To determine how DNR1 variation affects the response of N metabolism to eCO2, we conducted a 14-day hydroponic experiment in walk-in growth chambers, administering either aCO2 or eCO2 treatments to ZH11 and dnr1 mutants, as well as HJX74 and NIL. Similar with results of FACE experiment, eCO2 enhanced aboveground biomass, especially in dnr1 mutants (Fig. 2a, c). eCO2 also enlarged root system architecture (RSA), including total root length, total area, and the total number of root tips (Fig. 2b, d–f). While eCO2 increased 15NO3− uptake rates in both ZH11 and dnr1 mutants (Fig. 2g), the positive effect of eCO2 was more pronounced in the dnr1 mutants (Fig. 2). Similarly, eCO2 promotes aboveground biomass, RSA and 15NO3− absorption rate more strongly in HJX74 than in NIL (Supplementary Fig. 11a–g). These effects are attributed to increased IAA content under eCO2 treatment, caused by the inhibition of DNR1 expression, particularly in the dnr1 mutant and HJX74 (Fig. 2h; Supplementary Fig. 11h).
Fig. 2. Differences in plant growth and root developmental plasticity in response to elevated CO2 between japonica variety Zhonghua 11 (ZH11) and its dnr1 mutants mimicking the indica DNR1 allele.
a, b 14-day-old japonica variety Zhonghua 11 (ZH11) and its dnr1 mutants (dnr1) rice plants grown under ambient CO2 (aCO2) and elevated CO2 (eCO2) conditions, respectively. Morphology of plants (a) and root systems (b). a Scale bar, 20 cm. b Scale bar, 5 cm. c Aboveground biomass. Root statistics of total length of visible roots (d), total area of visible roots (e), and number of root tips (f). 15NO3- uptake rates (g) and root free IAA content (h) of japonica variety ZH11 and dnr1 under aCO2 and eCO2 conditions. c–h Data are mean ± s.e.m. (n = 3 biological replicates). P-values were generated from two-way ANOVA. Different letters indicate significant differences among treatments (P < 0.05). Source data are provided as a Source Data file.
Additionally, we conducted hydroponic experiments using either NO3− (KNO3) or NH4+ ((NH4)2SO4) as the N source for ZH11 and dnr1 plants. dnr1 plants consistently showed a higher responsiveness to variable CO2 levels in both NO3− and NH4+ conditions compared to ZH11 (Supplementary Fig. 12). However, the responses of aboveground and root biomass in both ZH11 and dnr1 were more pronounced under eCO2 when NO3− was provided, compared to NH4+ (Supplementary Fig. 12). Therefore, it is reasonable to conclude that DNR1 plays a crucial role in mediating plant growth in response to varying CO2 levels, particularly with NO3−.
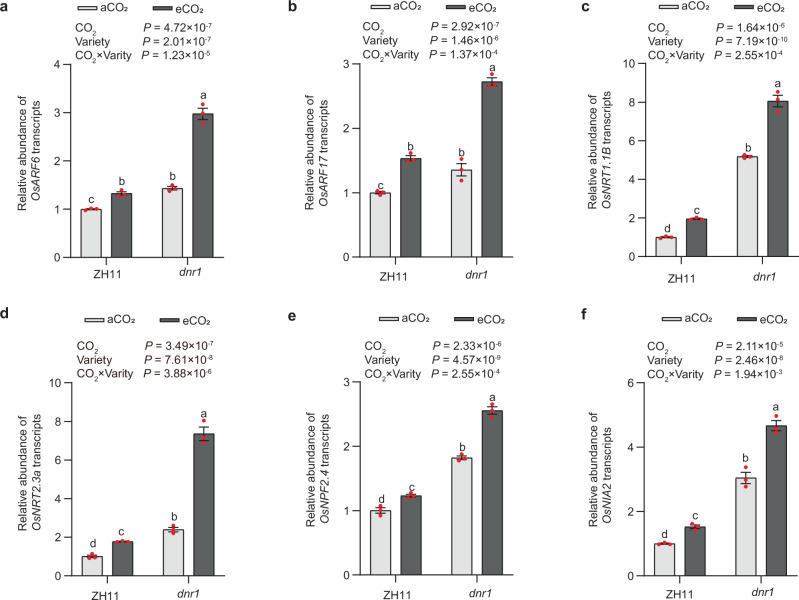
DNR1 acts as an antagonist to auxin biosynthesis, triggering AUXIN RESPONSE FACTORs (OsARF6 and OsARF17)-mediated activation of genes associated with NO3− uptake and metabolism, thereby contributing to enhanced NUE and grain yield23 (Supplementary Fig. 1). To determine whether eCO2 influences NO3− metabolism by modulating DNR1-mediated auxin homeostasis, we conducted RT-qPCR analysis of key genes involved in the DNR1-auxin-N pathway, including OsARF6, OsARF17, OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2. As anticipated, eCO2 significantly increased transcription levels of OsARF6, OsARF17, OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 (Figs. 2h and 3a–f). Correspondingly, nitrate reductase (NR) activity was markedly enhanced under eCO2 conditions (Supplementary Fig. 13a). Notably, eCO2 stimulated the expression of these genes in the dnr1 mutants more strongly than in ZH11, resulting in higher NO3− uptake rate and NR activity (Fig. 3; Supplementary Fig. 13a). Similar trends were observed in HJX74 and NIL, with or without eCO2 treatment (Supplementary Figs. 13b and 14).
Fig. 3. Elevated CO2 enhances NO3- metabolism via DNR1-mediated auxin homeostasis in japonica variety Zhonghua 11 (ZH11) and its dnr1 mutants mimicking the indica DNR1 allele.
Root mRNA abundances of OsARF6 (a) and OsARF17 (b) grown under ambient CO2 (aCO2) and elevated CO2 (eCO2) conditions, relative to ZH11 under aCO2 (set to 1). Root mRNA abundances of OsNRT1.1B (c) and OsNRT2.3a (d) relative to ZH11 under aCO2 (set to 1). Shoot mRNA abundances of OsNPF2.4 (e) and OsNIA2 (f) relative to ZH11 under aCO2 (set to 1). ZH11 and dnr1 indicate japonica variety Zhonghua 11 and its dnr1 mutants, respectively. a–f Data are mean ± s.e.m. (n = 3 biological replicates). P-values were generated from two-way ANOVA. Different letters indicate significant differences among treatments (P < 0.05). Source data are provided as a Source Data file.
Next, we investigated whether eCO2 could stimulate NO3− uptake and assimilation through the involvement of OsARFs, which act as downstream transcription factors of auxin homeostasis mediated by DNR1. Firstly, we conducted a time course assessment of the expression levels of OsARF6, OsARF17, OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 and found that the expression patterns of these four N-related genes and OsARFs are aligned. Specifically, when OsARFs expression is strongly induced by eCO2 (30 min to 2 h), the expression of OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 also significantly increases, suggesting that OsARFs regulates these genes in response to eCO2 (Supplementary Fig. 15). Interestingly, under eCO2 conditions, OsNRT1.1B shows a quicker response, which may be due to its direct response to CO2 concentration changes.
To test this hypothesis, we used HJX74 and a single-segment substitution line (SSSL-064), generated by crossing IRAT261 (donor parent) with HJX74 (recurrent parent), which incorporates a chromosome segment containing OsNRT1.1B from japonica IRAT261 into the HJX74 genetic background to investigate the response of OsNRT1.1B to eCO2. Both HJX74 and SSSL-064 exhibited increased biomass and 15NO3− uptake under eCO2 compared to aCO2, with HJX74 showing a slightly more pronounced increase (Supplementary Fig. 16). These results indicate that OsNRT1.1B itself can influence 15NO3− absorption and thereby affect growth to some extent in response to eCO2.
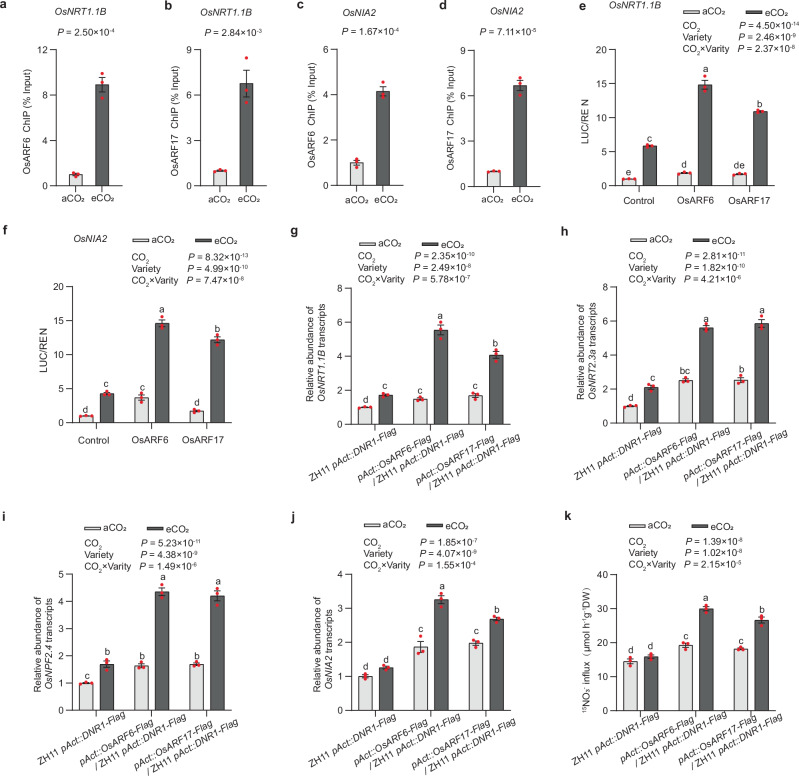
Secondly, to further illustrate the regulatory effects of OsARF6 and OsARF17 under aCO2 and eCO2 conditions, we performed ChIP-qPCR assays. These assays indicated that eCO2 enhanced the enrichment of TGTCTC/GAGACA motif-containing fragments from the promoters of OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 compared to the aCO2 treatment (Fig. 4a–d; Supplementary Fig. 17a–f). The following in vitro transient transactivation assays revealed that eCO2 increased the transcriptional activation capacities of OsARF6 and OsARF17 towards their downstream genes, OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2, when compared to aCO2 treatment (Fig. 4e, f; Supplementary Fig. 17g, h). Accordingly, under both aCO2 and eCO2 conditions, the expression levels of OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 were upregulated in the OsARF6 or OsARF17 overexpression lines within the ZH11/pACT::DNR1-Flag background (Fig. 4g–j). These results further confirm the transactivation activities of OsARFs towards N-related genes in rice protoplasts (Fig. 4e, f; Supplementary Fig. 17g, h).
Fig. 4. The effect of elevated CO2 on the expression of N metabolism-related genes and NO3- uptake is mediated by DNR1-OsARFs module.
Extent of OsARF6 and OsARF17-mediated ChIP-qPCR enrichment (relative to Input) of TGTCTC-containing promoter fragments from OsNRT1.1B (a, b) and OsNIA2 (c, d) under ambient CO2 (aCO2) and elevated CO2 (eCO2) conditions. Data are mean ± s.e.m. (n = 3 biological replicates). P-values were generated from two-sided Student’s t tests. OsARF6 and OsARF17 activate OsNRT1.1B (e) and OsNIA2 (f) promoter-LUC fusion constructs in transient transactivation assays. The LUC/REN activity obtained from a co-transfection with an empty effector construct and indicated reporter constructs under ambient CO2 (aCO2) was set to 1. Root mRNA abundances of OsNRT1.1B (g) and OsNRT2.3a (h) relative to ZH11 pAct::DNR1-Flag under aCO2 (set to 1). i, j Shoot mRNA abundances of OsNPF2.4 (i) and OsNIA2 (g) relative to ZH11 pAct::DNR1-Flag under aCO2 (set to 1). k Root 15NO3- uptake rate of OsARF6 and OsARF17 overexpression lines in the ZH11 pAct::DNR1-Flag background grown under aCO2 and elevated CO2 (eCO2) conditions, respectively. e–k Data are mean ± s.e.m. (n = 3 biological replicates). P-values were generated from two-way ANOVA. Different letters indicate significant differences among treatments (P < 0.05). Source data are provided as a Source Data file.
Finally, we tested whether the altered transcriptional activation capacities of OsARF6 and OsARF17 led to changes in NO3− uptake in response to eCO2. Under eCO2, the over-expression of OsARF6 or OsARF17 in the ZH11/pACT::DNR1-Flag background significantly restored the insensitivities of NO3− absorption caused by auxin depletion (Fig. 4k). This suggests that eCO2 promotes NO3− uptake and assimilation by enhancing the transcriptional activation capacities of OsARFs.
DNR1 variation affects response of photosynthetic genes to eCO2
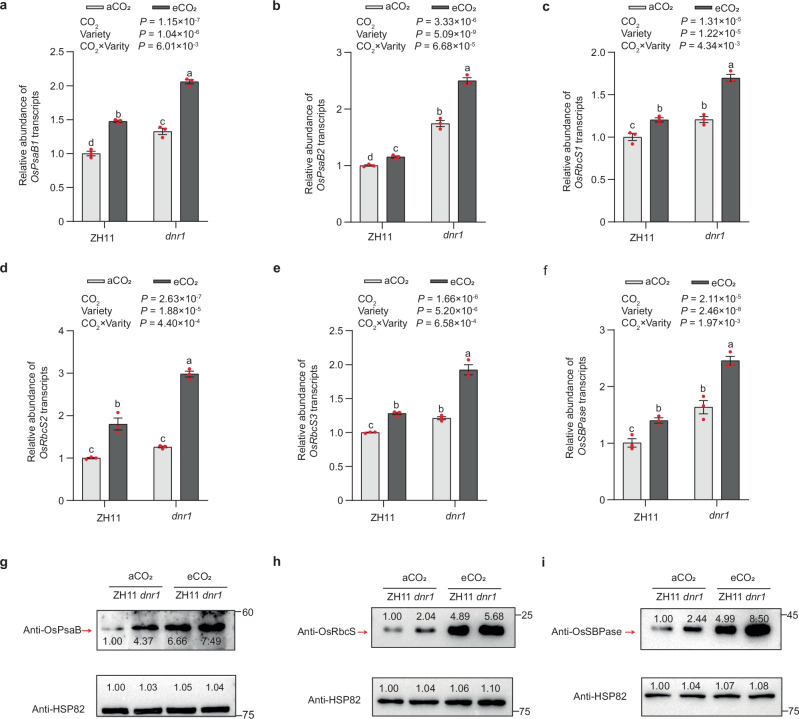
As a crucial substrate for photosynthesis, CO2 actively participates in the photosynthetic processes of plants. Thus, we compared the transcription and protein levels of core elements in photosystems (OsPsbA, OsPsaB), and rate-limiting factors in the Calvin-Benson cycle (OsRbcs, OsRbcL and OsSBPase) (Supplementary Table 4), in both ZH11 and dnr1 mutants with or without CO2 treatment. Both transcription and protein levels of OsPsaB, OsRbcS and OsSBPase were significantly higher in dnr1 mutants compared to ZH11, and the changes induced by eCO2 were more pronounced in dnr1 mutants (Fig. 5). However, although the expression levels of OsPsbA, OsRbcS4 and OsRbcL slightly increased in dnr1, they appear to be unaffected by eCO2 (Supplementary Fig. 18). Similar results were found for HJX74 and NIL (Supplementary Fig. 19), confirming that the abundance patterns of photosynthetic genes induced by eCO2 in ZH11 and dnr1 mutants were shared by these varieties.
Fig. 5. The impact of elevated CO2 on DNR1-regulated abundances of photosynthetic genes in japonica variety Zhonghua 11 (ZH11) and its dnr1 mutants mimicking the indica DNR1 allele.
Shoot mRNA abundances of OsPsaB1 (a), OsPsaB2 (b), OsRbcS1 (c), OsRbcS2 (d), OsRbcS3 (e), and OsSBPase (f) grown ambient CO2 (aCO2) and elevated CO2 (eCO2) conditions, respectively, relative to Zhonghua 11 under aCO2 (set to 1). Data are mean ± s.e.m. (n = 3 biological replicates). P-values were generated from two-way ANOVA. Different letters indicate significant differences among treatments (P < 0.05). OsPsaB (g), OsRbcS (h) and OsSBPase (i) protein abundances in shoots. HSP82 serves as a loading control. The red arrows indicate the OsPsaB, OsRbcS and OsSBPase bands, respectively. Data are representative of three independent experiments, with similar results. Source data are provided as a Source Data file.
We found that OsARF6 and OsARF17 do not possess transcriptional activation abilities for these genes under both aCO2 and eCO2 conditions (Supplementary Fig. 20a), suggesting that eCO2 promotes the accumulation of photosynthesis-related proteins and enhances photosynthesis independent on OsARFs. Additionally, examining the expression levels of these genes in plants overexpressing OsARF6 and OsARF17 within the ZH11/pACT::DNR1-Flag background revealed no changes compared to ZH11/pACT::DNR1-Flag with either aCO2 or eCO2 treatment (Supplementary Fig. 20b–g). This suggests that DNR1 does not regulate photosynthetic efficiency through the transcriptional activation of OsARF6 and OsARF17.
We conducted RNA sequencing analysis on ZH11 and the dnr1 mutant under both aCO2 and eCO2 conditions and identified 397 target genes regulated by both CO2 and DNR1. Among these, 9 transcription factors are upregulated by both eCO2 and null-DNR1 allele, and 4 transcription factors are downregulated by both eCO2 and null-DNR1 allele (Supplementary Table 5). These 13 transcription factors may serve as potential candidates for regulating photosynthetic efficiency in response to eCO2 and DNR1 interactions, offering promising avenues for future research. Overall, eCO2 enhances photosynthetic efficiency by improving the C and N cycles through various mechanisms.
Discussion
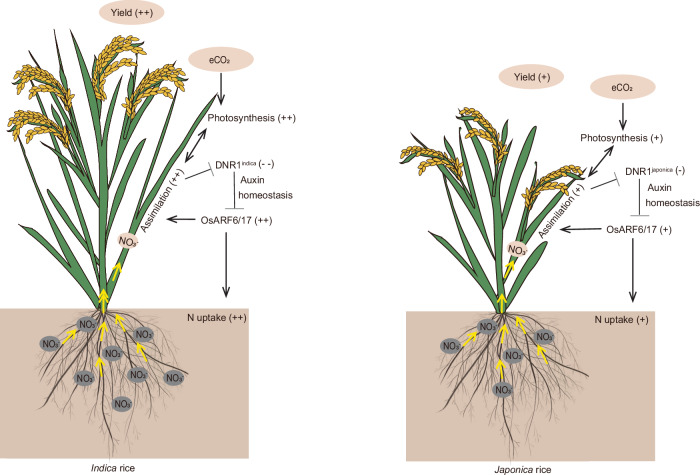
We present evidence of the key role of DNR1 in driving the divergent responses of indica and japonica rice varieties to eCO2 (Fig. 6). eCO2 likely influences DNR1 indirectly through changes in N status. eCO2 can increase photosynthesis and plant growth, thereby raising the demand for nitrogen. This, in turn, could lead to a decrease in DNR1 abundance, stimulating N uptake. Specifically, plants carrying the japonica DNR1 allele, which exhibit higher DNR1 abundance leading to reduced auxin accumulation, key traits such as biomass, nitrogen content, and yield respond relatively weakly to eCO2. Conversely, in dnr1 mutants, eCO2 decreases the expression of DNR1 and the following transcriptional activation of OsARF6 and OsARF17, which in turn upregulates genes associated with NO3− uptake and assimilation (OsNRT1.1B, OsNRT2.3a, OsNIA2, and OsNPF2.4). Consequently, N uptake and assimilation increased, resulting in elevated N content in rice plants, which enhanced the photosynthetic capacity of the plants under eCO2 concentrations. Additionally, previous studies have shown that IAA levels rise under eCO225–27. Our results suggest that the inhibition of DNR1 by eCO2 may also contribute to this increase in IAA content, which further enhances our understanding of the relationship between eCO2 and IAA homeostasis.
Fig. 6. Schematic overview of the role of DNR1 in driving the divergent responses of indica and japonica rice to elevated CO2.
Rising CO2 concentrations increase photosynthesis and rice yields by affecting a range of genes involved in uptake, transport, and assimilation of nitrate, which in turn are regulated by DNR1. +, ++, −, and -- indicate positive, strongly positive, negative, and strongly negative, respectively.
Previous studies have extensively explored the impacts of eCO2 on leaf photosynthesis28–30. A meta-analysis of 20-year rice FACE studies revealed that eCO2 significantly increased the light-saturated photosynthetic rate of leaves but reduced gs, Vcmax, and Jmax 6, consistence with our study. However, the response of photosynthetic genes to elevated CO2 has rarely been investigated. In this study, we showed that eCO2 promotes the abundance of photosynthetic genes (OsPsaB, OsRbcs and OsSBPase) more strongly in plants carrying the indica DNR1 allele, leading to higher yield response to eCO2. Interestingly, DNR1 mediated the regulation of photosynthetic genes expression independently of OsARFs. Further research is needed to elucidate why DNR1 influences C and N cycles through different mechanisms.
Together, our findings demonstrate that DNR1 plays a crucial role in coordinating N metabolism and C fixation to enhance plant growth in response to eCO2. This insight provides a valuable breeding strategy for adapting to increasing atmospheric CO2 levels by modulating DNR1, which acts as a pivotal bridge in this process. Our findings also provide insights into the relationship between N metabolism and the rate of photosynthetic C fixation in plants, particularly in the context of molecular coupling mechanisms. This study identify molecular mechanisms driving CO2 effects among rice subspecies.
Our findings corroborate previous studies showing that in seed crops, sink capacity is a critical factor limiting yield response to eCO₂4,31,32. Specifically, we found that eCO2 increased sink capacity more strongly in HJX74 than in NIL, and more strongly in dnr1 mutants than in ZH11, resulting in higher yields (Supplementary Table 6). Similarly, eCO₂ increases the spikelet numbers per panicle more strongly in indica rice as compared to japonica rice, resulting in a heightened overall yield response12,14. Importantly, this sink capacity is intricately regulated by N availability. Indeed, by increasing NO3− use efficiency, dnr1 stimulates sink capacity, as evidenced by increased panicle size, spikelet number, and seed size23. Furthermore, a previous FACE study also showed that effects of eCO2 on rice yield are affected by N application rate33,34. Collectively, these results indicate that the absorption and utilization of N by rice plants is a pivotal factor influencing yield responses to eCO₂.
Two limitations of our experiment should be noted. First, we only used ZH11 as the typical japonica and YD6 and HJX74 as the typical indica. While more recipient varieties would strengthen the robustness of our results, the layout of the FACE experiment did not allow for testing more varieties. However, YD6 has been often utilized in other FACE experiments14. In our experiments, both YD6 and HJX74 exhibited similar responses to eCO2 levels. Additionally, ZH11’s response aligns with the average response observed in other FACE experiments involving japonica rice varieties14. These results strongly suggest that the chosen cultivars are broadly representative for japonica and indica. Second, although data from the FACE experiment have been used as a basis for hydroponic cultivation, the different environmental conditions between the two experiments may have affected rice growth and eCO2 responses. Unfortunately, this issue is difficult to avoid. On the other hand, the results from our FACE experiment and hydroponic culture experiment consistently support the findings regarding nitrogen absorption and plant growth, suggesting that our findings are robust.
Optimizing yield response to rising atmospheric CO₂ is crucial in mitigating the anticipated supply-demand shortfall this century due to rice yield loss of climatic warming and demand increase4,35. Field experiments have revealed large variation in crop productivity and quality responses to eCO₂ between rice varieties, underscoring the importance of genetic variation in breeding for enhanced productivity and quality under eCO₂ conditions4,36,37. Our study builds on this understanding by showing that DNR1-mediated variations in NO₃− utilization largely explains the difference in yield responses to eCO₂ between rice subspecies.
Importantly, our phylogenetic analysis of ~3000 rice accessions showed that indica and japonica DNR1 alleles belong to two separate clades23,24. Haplotype analysis of the DNR1 gene of these varieties revealed four distinct haplotypes (Hap. I-IV). Notably, 98.1% of the indica subpopulation belongs to Hap. I, while 75.7% and 22.2% of the japonica subpopulation belongs to Hap. II and Hap. III, respectively24, demonstrating consistent differentiation across existing varieties. This divergence may be attributed to high-fertilizer breeding conditions that have led to the effective utilization of the indica-type DNR1, while it remains underutilized in japonica rice. Together, these results suggest that the CO2 fertilization effect for the vast majority japonica varieties can be increased by manipulating DNR1.
As japonica rice accounts for ~15% of global rice production38, breeding efforts focused on NUE increase are key. Other breeding approaches may also show potential. For instance, eCO2 increased yield by up to 30.3% in a low-yielding old japonica variety, suggesting that other loci are also important in determining the rice response to elevated CO2. Together these results underscore the importance of rice varieties and agronomic practices with high NUE to improve food security as atmospheric CO2 concentrations continue to increase.
Methods
Field experiment
FACE system
In 2021, we established a Free Air CO2 Enrichment (FACE) system in Baolin village (31.9°N, 119.5°E), Yanling Town, Danyang City, Jiangsu Province, China. The FACE system comprises six octagonal rings, each 8 m in diameter. Three rings were designated for ambient atmospheric CO2 concentration treatment (aCO2), and three for elevated atmospheric CO2 treatment (eCO2). To prevent crossover effects, a minimum distance of 25 m was maintained between any aCO2 and eCO2 rings. At the center of each ring, a monitoring system was installed to track CO2 concentration, air temperature, and wind speed. Within each eCO2 ring, eight CO2 sensors (VC2008T, SenseAir, Sweden) were placed above the rice canopy, evenly distributed within a 4-m circle at the ring’s center.
In line with IPCC1 predictions, atmospheric CO2 concentrations are expected to reach between 500 (SSP5) and 800 ppm (SSP3) by 2100. Hence, we set eCO2 treatments to mimic end-of-century conditions at ~550 ppm. PVC emission tubes, arranged to form an octagon around each eCO2 ring at 50 cm above the rice canopy, facilitated CO2 distribution. Considering operational costs, pure CO2 gas was released only during the daytime (5:00 a.m. to 7:00 p.m.). This release was regulated automatically based on wind direction and speed and was halted when wind speeds exceeded 5 m s−1. The average daytime CO2 concentrations were maintained at 560 ± 19 ppm in eCO2 treatments and 387 ± 10 ppm in aCO2 treatments. Details of climate condition, soil properties, and FACE system can be found in Qian et al.39.
Experimental design
To determine whether variation in the DNR1 allele drives the divergent responses to eCO2 between rice subspecies, we planted a typical japonica variety, Zhonghua 11 (ZH11), with its dnr1 mutants, which mimics the indica DNR1 allele in ZH11 background by reducing DNR1 abundance in 2023 and 2024. We also planted a typical indica variety, Hua-Jing-Xian 74 (HJX74), with a near-isogenic line (NIL) carrying the japonica DNR1 allele in 2022 and 2023. Details of these plant materials can be found in Zhang et al.23. The plots for each treatment measured 1.5 m × 2 m. Two healthy rice seedlings per hill were transplanted at planting spaced at 15 cm × 25 cm intervals. N fertilizer, in the form of urea, was applied at a total rate of 180 kg N ha−1. The N fertilizers were divided into three applications: 40% at soil tillage, 30% at the tillering stage, and the remaining 30% at the jointing stage. Phosphorus fertilizer, at a rate of 120 kg P2O5 ha−1, was applied at soil tillage, while potassium fertilizer, at 160 kg K2O ha−1, was split equally between the soil tillage and jointing stages. All other agronomic practices were conducted in accordance with local agricultural recommendations.
Sampling and measurements
Light response of net photosynthesis (An) and stomatal conductance (gs) of flag leaves were measured at 11 photosynthetically active radiation (PAR) levels (in decreasing order of 1800, 1500, 1200, 1000, 800, 600, 400, 200, 100, 50 and 0 μmol m−2 s−1) by a portable photosynthesis system (LI-6800, LI-COR, Lincoln, America) between 8:30 and 11:30 a.m. Air temperature and relative humidity in the chamber were set at 35 °C, and 60%, respectively. CO2 concentrations were set at 400 ppm for the aCO2 and 550 ppm for the eCO2 treatment.
We used the rapid A - Ci response (RACiR) technique to obtain the CO2 response of net photosynthesis rate (An) of flag leaves by a portable photosynthesis system (LI-6800, LI-COR, Lincoln, America) between 8:30 and 11:30 a.m according to Joseph et al.40. We set the initial and final CO2 concentration at 50 and 1200 ppm respective, and ramp rates at 300 ppm min−1. Photosynthetic active radiation, air temperature and relative humidity in the chamber were set at 1800 μmol m−2 s−1, 35 °C, and 60%, respectively. Then, the resulting functional relationship (A-Ci curves) was used to estimate the Vcmax and Jmax FvCB model.
Chlorophyll fluorescence parameters photosystem II quantum yield (Y(II)), non-photochemical quenching (NPQ), photochemical quenching coefficient (qL), and maximum photochemical quantum yield of Photosystem II (Fv/Fm) of flag leaves were measured using the FluorPen (FluorPen FP110 - LM/D, Photon Systems Instruments, Czech) after 1-h period of darkness.
For each plot, three hills were selected to measure leaf area using a table leaf area instrument (LI-3100C, LI-COR, America). Rice plants were harvested at the heading stage and the mature stage, oven-dried at 70 °C to obtain a constant weight and then crushed. The nitrogen content was measured using an elemental analyser (vario PYRO, Elemental, German), and N uptake was calculated by multiplying the aboveground biomass by the N content. NUE is defined as grain weight divided by nitrogen supply41.
RNA-seq analysis
Total RNAs were extracted from heading stage rice flag grown in field under the FACE system using the QIAGEN RNeasy plant mini kit (QIAGEN, 74904) following the manufacturer’s instructions. Three replicate RNA-seq libraries were prepared from YD6 and ZH11 plants under aCO2 or eCO2, respectively. A total of the four libraries were sequenced separately using the Illumina Novaseq platform. Raw sequencing reads were cleaned by removing adaptor sequences, reads containing poly-N sequences, and low-quality reads. Approximately 44,226,120 clean reads were mapped to the Nipponbare reference genome using Hisat2 v2.0.542. After data were mapped, normalization was performed and then FPKM (fragments per kilobase per million mapped reads) was calculated using RESM software43. As previously described44, a false discovery rate (FDR) <0.05 and absolute value of log2 ratio≥2 were used to identify differentially expressed genes in YD6 and ZH11 samples under aCO2 or eCO2, respectively.
Hydroponic experiment
Plant materials
The plant materials including a pair of near isogenic materials of NIL-DNR1HJX74 (HJX74) and NIL-DNR1IRAP9 (NIL), as well as pAct::DNR1-Flag and dnr1 under Zhonghua 11 (ZH11) background, and pAct::OsARF6-Flag and pAct::OsARF17-Flag under ZH11/pAct::DNR1-Flag. Details of these plant materials can be found in Zhang et al.21 and Huang et al.22.
Hydroponic culture
Seeds were soaked in 20% sodium hypochlorite solution for 30 min for disinfection and selected with uniform growth for further analyses as described previously45. The 7-day-old seedlings were transferred to PVC pots containing 10 L of nutrient solution (1.25 mM NH4NO3, 0.3 mM NaH2PO4·2H2O, 0.35 mM K2SO4, 1 mM CaCl2, 1 mM MgSO4·7H2O, 20 µM EDTA-Fe, 0.5 mM Na2SiO3, 9 µM MnCl2, 20 µM H3BO3, 0.77 µM ZnSO4, 0.32 µM CuSO4, and 0.39 µM (NH4)6Mo7O24, pH 5.5) and grown at either aCO2 levels or eCO2 levels for 2 weeks. All nutrient solutions were replaced twice per week, pH was adjusted to 5.5 daily. The average daytime (8:00 a.m. to 8:00 p.m.) CO2 concentrations were maintained at 598 ± 32 ppm in eCO2 treatments and 408 ± 28 ppm in aCO2 treatments. The mean day/night air temperature was maintained at 30 °C/25 °C with 60% relative humidity. The LED lamps were positioned 50 cm above the rice, providing a mean photon flux of 500 μmol m−2 s−1 during the daytime.
15N uptake analysis
After growth in hydroponic condition (1.25 mM NH4NO3) for 2 weeks, rice root 15NH4+ and 15NO3- influx measurements were performed as described elsewhere44,46. Specifically, 14-day-old rice plants were transferred first to 0.1 mM CaSO4 for 1 min, then to a complete nutrient solution containing 2.5 mM K15NO3 (Sigma, 335134, 98% atom excess 15N) or 1.25 mM (15NH4)2SO4 (Aladdin, A110168, 99% atom excess 15N) instead of 1.25 mM NH4NO3 as the N source for 5 min. The plants were incubated in 0.1 mM CaSO4 for 1 min before the roots were collected and dried at 80 °C for 72 h. Root dry weight was recorded and the 15 N content was measured using the IsoPrime100 elemental analyser (Elementar, Germany). Finally, influx of 15NH4+ and 15NO3- was calculated as described elsewhere24.
Root system analysis
Roots from rice plants grown in the aCO2 and eCO2 treatments for 14 d were cut off and spread out in water in a transparent dish. Subsequently, the root system was scanned as described previously24.
Measurement of NR activity
Fresh plant material (~1 g) from individual rice plant grown at either aCO2 levels or eCO2 levels for 2 weeks was used to measure NR activity, following the instruction manual of the NR Kit (Solarbio LIFE SCIENCES, BC0080).
Quantitative real time PCR (RT-qPCR) analysis
After growth in hydroponic condition (1.25 mM NH4NO3) for 2 weeks, total RNAs were extracted from different plant tissues using the TRIzol reagent (Ambion), and full-length cDNAs were reverse transcribed using a cDNA synthesis kit (Accurate Biology, AG11728). Subsequent RT-qPCR was performed according to the manufacturer’s instructions (Accurate Biology, AG11718). Each RT-qPCR assay included at least three biological replicates. As for time course experiment, selected eight time points (0, 15 min, 30 min, 2 h, 24 h, 72 h, 7 d, 14 d) for sample preparation and extracted RNA for RT-qPCR. The rice ACTIN1 gene was used as an internal reference. Relevant RT-qPCR primer sequences are listed in Supplementary Table 4.
ChIP-qPCR assays
The ChIP-qPCR protocol has been previously described23. Approximately 2 g of two-week-old pAct::OsARF6-Flag and pAct::OsARF17-Flag overexpression lines in the Wuyunjing 7 (WYJ7) background, grown under aCO2 and eCO2 conditions, were cross-linked with 1% formaldehyde under vacuum for 15 min to stabilize protein-DNA interactions. The samples were then ground to a fine powder in liquid N. Nuclei were isolated and lysed, and chromatin was fragmented by sonication into ~500 bp fragments. The chromatin was incubated overnight at 4 °C with 7 μg of anti-Flag antibody (Sigma, F1804) for immunoprecipitation. The following day, the samples were washed, eluted, and reverse-cross-linked, followed by DNA purification. Enrichment of specific DNA fragments was analyzed by RT-qPCR using three biological replicates. Relevant qPCR primer sequences are provided in Supplementary Table 7.
Western blotting
Total protein was extracted in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40 detergent, 10% Glycerol, 1 mM DTT with added protease inhibitor cocktail (Roche LifeScience) and transferred onto a nitrocellulose membrane. Then, after blocking with 5% milk solution for 1 h, the nitrocellulose membrane was incubated with antibodies. The DNR1 protein was detected by probing the membrane with anti-DNR1 (ABclonal) and proteins involved in photosynthesis were detected by anti-PsbA (Agrisera, AS05084), anti-PsaB (Agrisera, AS10695), anti-RbcS (Agrisera, AS07259), anti-RbcL (Agrisera, AS03037) and anti-SBPase (Agrisera, AS152873), respectively. The result of immunoblotting was visualized on the Tanon-5200 Chemiluminescent Imaging System and grayscale analysis was used Tanon image GIS Semi-quantitative analysis (Tanon Science and Technology).
In vitro transient transactivation assays
As described elsewhere23, the indica variety YD6 was planted under different CO2 treatments for 10 days. Firstly, the free IAA content of YD6 under two conditions was detected and then extracted rice protoplasts, subsequently, used the effector plasmids pRTBD-OsARF6 and pRTBD-OsARF17 to drive the reporters 5×GAL4-OsNRT1.1B, -OsNRT2.3a, -OsNPF2.4, -OsNIA2, -OsPsbA2, -OsPsaB2, -OsRbcs2, OsRbcs4, OsRbcL and OsSBPase, respectively. Transient transactivation assays were performed as described elsewhere47. The Dual-Luciferase Reporter Assay System (Promega, E1960) was used to perform the luciferase activity assays, with the Renilla LUC gene as an internal control. Relevant PCR primer sequences are listed in Supplementary Table 8.
Determination of free IAA content
Root tip samples (~50 mg) were ground into a powder in liquid N and extracted with methanol/water/formic acid (15:4:1, V/V/V). The combined extracts were evaporated to dryness under a N gas stream, reconstituted in 80% methanol (V/V), and filtered through a PTFE membrane (0.22 μm, Anpel). The final solution was then analyzed using an LC-ESI-MS/MS system and an ESI-triple quadrupole-linear ion trap (QTRAP)-MS system (Wuhan Triploid Biotech).
Statistical analysis
We analyzed the data in the field and hydroponic experiments through two-sided Student’s t tests and two-way ANOVA, including CO2 and variety as fixed effects. When the ANOVA indicated interactive effects, multiple comparisons were performed using the least significant difference test at the significance level of P = 0.05. All analyses were performed with the statistical package SPSS 27.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Dr. Chunwu Zhu for valuable comments. This work was supported by the National Natural Science Foundation of China (32122065 to S.L., 32271635 to Y.J., 32372121 and 32441056 to S.L., 32301354 to H.Q. and 32401866 to S.Z.), the Natural Science Foundation of Jiangsu Province (BK20230979 to H.Q. and BK20241559 to S.Z.), Jiangsu Carbon Peak Carbon Neutrality Science and Technology Innovation Fund project (BE2022308 to Y.D.), and the China Postdoctoral Science Foundation (GZB20240320 to S.Z.).
Author contributions
Y.J. and S.L. designed the study. Y.L., H.Q., and Y.W. conducted the field experiment. S.Z., C.S., and Y.L. conducted the hydroponic experiment. Y.J., S.L., Y.D., Y.L., and S.Z. drafted the paper. Shuijun Hu, W.Z., Shan Huang, S.W., Z.L., G.L., X.F., and K.J.v.G. reviewed and commented on the manuscript.
Peer review
Peer review information
Nature Communications thanks Chengcai Chu, Brent Kaiser and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Raw RNA-seq data were deposited at the National Genomics Data Center, Genome Sequence Archive (GSA) (accession number PRJCA024327 [https://ngdc.cncb.ac.cn/gsa/s/Hz4L1U30]). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yunlong Liu, Siyu Zhang.
Contributor Information
Yanfeng Ding, Email: dingyf@njau.edu.cn.
Shan Li, Email: shanli@njau.edu.cn.
Yu Jiang, Email: yujiang@njau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55809-3.
References
- 1.IPCC. Working Group 1 Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Climate Change 2021: The Physical Science Basis (Cambridge University Press, Cambridge, USA, 2021).
- 2.National Aeronautics and Space Administration (NASA), 2024. https://climate.nasa.gov/vital-signs/carbon-dioxide/.
- 3.Wang, S. et al. Recent global decline of CO2 fertilization effects on vegetation photosynthesis. Science370, 1295–1300 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Ainsworth, E. A. & Long, S. P. 30 years of free‐air carbon dioxide enrichment (FACE): What have we learned about future crop productivity and its potential for adaptation? Glob. Chang Biol.27, 27–49 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Rezaei, E. E. et al. Climate change impacts on crop yields. Nat. Rev. Earth Environ.4, 831–846 (2023). [Google Scholar]
- 6.Hu, S. et al. Response of rice growth and leaf physiology to elevated CO2 concentrations: a meta-analysis of 20-year FACE studies. Sci. Total Environ.807, 151017 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Fahad, S. et al. Major constraints for global rice production. in Advances in Rice Research for Abiotic Stress Tolerance, 1–22 (Elsevier, 2019).
- 8.Wang, X. et al. Emergent constraint on crop yield response to warmer temperature from field experiments. Nat. Sustain.3, 908–916 (2020). [Google Scholar]
- 9.Zhu, P. Warming reduces global agricultural production by decreasing cropping frequency and yields. Nat. Clim. Change12, 1–8 (2022). [Google Scholar]
- 10.Jing, C. et al. Multiple domestications of Asian rice. Nat. Plants9, 1221–1235 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Purugganan, M. D. & Fuller, D. Q. The nature of selection during plant domestication. Nature457, 843–848 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Hu, S., Wang, Y. & Yang, L. Response of rice yield traits to elevated atmospheric CO2 concentration and its interaction with cultivar, nitrogen application rate and temperature: a meta-analysis of 20 years FACE studies. Sci. Total Environ.764, 142797 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Wang, W. et al. Elevated CO2-induced changes in cytokinin and nitrogen metabolism are associated with different responses in the panicle architecture of two contrasting rice genotypes. Plant Growth Regul.89, 119–129 (2019). [Google Scholar]
- 14.Lv, C. et al. Response of rice yield and yield components to elevated [CO2]: a synthesis of updated data from FACE experiments. Eur. J. Agron.112, 125961 (2020). [Google Scholar]
- 15.Bloom, A. J., Burger, M., Asensio, J. S. R. & Cousins, A. B. Carbon dioxide enrichment inhibits nitrate assimilation in wheat and arabidopsis. Science328, 899–903 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Terrer, C. et al. Nitrogen and phosphorus constrain the CO2 fertilization of global plant biomass. Nat. Clim. Change9, 684–689 (2019). [Google Scholar]
- 17.Wang, Y. Y., Cheng, Y. H., Chen, K. E. & Tsay, Y. F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol.69, 85–122 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Li, Y., Fan, X. & Shen, Q. The relationship between rhizosphere nitrification and nitrogen‐use efficiency in rice plants. Plant Cell Environ.31, 73–85 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Hu, B. et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet.47, 834–838 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Zhang, Z. & Chu, C. Nitrogen-use divergence between indica and japonica rice: variation at nitrate assimilation. Mol. Plant13, 6–7 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Gao, Z. et al. The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun.10, 5207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, Y. et al. MYB61 is regulated by GRF4 and promotes nitrogen utilization and biomass production in rice. Nat. Commun.11, 5219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, S. et al. Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell33, 566–580 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y. et al. Improving rice nitrogen-use efficiency by modulating a novel monouniquitination machinery for optimal root plasticity response to nitrogen. Nat. Plants9, 1902–1914 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Li, C. R. et al. Responses of carboxylating enzymes, sucrose metabolizing enzymes and plant hormones in a tropical epiphytic CAM orchid to CO2 enrichment. Plant Cell Environ.25, 369–377 (2002). [Google Scholar]
- 26.Teng, N. J. et al. Elevated CO2 induces physiological, biochemical and structural changes in leaves of Arabidopsis thaliana. New Phytol.172, 92–103 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Li, X. M. et al. Influence of elevated CO2 and O3 on IAA, IAA oxidase and peroxidase in the leaves of ginkgo trees. Biol. Plant53, 339–342 (2009). [Google Scholar]
- 28.Bernacchi, C. J. et al. Photosynthesis and stomatal conductance responses of poplars to free‐air CO2 enrichment (PopFACE) during the first growth cycle and immediately following coppice. New Phytol.159, 609–621 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Long, S. P. et al. Rising atmospheric carbon dioxide: plants FACE the future. Annu. Rev. Plant Biol.55, 591–628 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Cai, C. et al. Do all leaf photosynthesis parameters of rice acclimate to elevated CO2, elevated temperature, and their combination, in FACE environments? Glob. Change Biol.24, 1685–1707 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Nakano, H. et al. Quantitative trait loci for large sink capacity enhance rice grain yield under free-air CO2 enrichment conditions. Sci. Rep.7, 1827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takai, T. et al. MORE PANICLES 3, a natural allele of OsTB1/FC1, impacts rice yield in paddy fields at elevated CO2 levels. Plant J.114, 729–742 (2023). [DOI] [PubMed] [Google Scholar]
- 33.Kim, H. Y. et al. Growth and nitrogen uptake of CO2‐enriched rice under field conditions. New Phytol.150, 223–229 (2001). [Google Scholar]
- 34.Kim, H. et al. Seasonal changes in the effects of elevated CO2 on rice at three levels of nitrogen supply: a free air CO2 enrichment (FACE) experiment. Glob. Change Biol.9, 826–837 (2003). [Google Scholar]
- 35.Ray, D. K. et al. Yield trends are insufficient to double global crop production by 2050. PLoS ONE8, e66428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziska, L. H. et al. Food security and climate change: on the potential to adapt global crop production by active selection to rising atmospheric carbon dioxide. Proc. R. Soc. B.279, 4097–4105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, C. et al. Carbon dioxide (CO2) levels this century will alter the protein, micronutrients, and vitamin content of rice grains with potential health consequences for the poorest rice-dependent countries. Sci. Adv.4, eaaq1012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koizumi, T. & Furuhashi, G. Global rice market projections distinguishing japonica and indica rice under climate change. Jpn Agric. Res. Q.50, 63–91 (2020). [Google Scholar]
- 39.Qian, H. et al. Intermittent flooding lowers the impact of elevated atmospheric CO2 on CH4 emissions from rice paddies. Agric. Ecosyst. Environ.329, 107872 (2022). [Google Scholar]
- 40.Stinziano, J. R. et al. The rapid A-Ci response: photosynthesis in the phenomic era. Plant Cell Environ.40, 1256–1262 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Good, A. G., Shrawat, A. K. & Muench, D. G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci.9, 597–605 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Kim, D. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform.12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini, Y. et al. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res.125, 279–284 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Ho, C., Lin, S. H., Hu, H. & Tsay, Y. F. CHL1 functions as a nitrate sensor in plants. Cell138, 1184–1194 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Li, S. et al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature560, 595–600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, S. et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet.47, 949–954 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-seq data were deposited at the National Genomics Data Center, Genome Sequence Archive (GSA) (accession number PRJCA024327 [https://ngdc.cncb.ac.cn/gsa/s/Hz4L1U30]). Source data are provided with this paper.