Abstract
Seamounts are biodiversity hotspots that face increasing threats from anthropogenic activities. Seamounts host diverse sessile suspension-feeding organisms such as sponges and anthozoans, which are crucial for seamount ecosystems as they construct three-dimensional habitats utilized by numerous other animals. Therefore, accurate identification of seamount fauna, in particular of sessile suspension-feeding organisms, is of paramount importance for robust conservation efforts. This study focused on Zoantharia, a sessile anthozoan group, and specifically the family Parazoanthidae, known for associations with many different host taxa, prominently including octocorals and sponges. We collected Parazoanthidae specimens from northwestern Pacific seamounts and formally describe a new species, Vitrumanthusflosculus Kise & Reimer, sp. nov., based on morphological and molecular analyses. We also report the complete mitochondrial genomes of this new species and the related species Churabanakuroshioae. Our results reconfirm the phylogenetic positions of these two species within Parazoanthidae, while demonstrating much remains to be learned about the benthic diversity of northwestern Pacific seamounts.
Key words: Baseline data, Hexasterophora, mitochondrial genome, mitogenome, MPA, phylogeny, seamount, taxonomy, zoantharian
Introduction
Seamounts are diversity hotspots for deep-sea organisms (Worm et al. 2003; Samadi et al. 2006; Clark et al. 2010; Morato et al. 2010; Rowden et al. 2010), harboring diverse assemblages of sessile suspension-feeding organisms due to turbulent and hydrodynamic water flowing around their peaks, which delivers planktonic food and nutrients to the benthos in the immediate area (Clark et al. 2010; Watling and Auster 2017). Seamount habitats and their fauna face threats from anthropogenic activities, such as bottom trawling that may damage or destroy these diverse marine animal forests (Worm et al. 2006; Clark et al. 2007; Althaus et al. 2009; Rossi et al. 2022). Sessile suspension-feeding organisms, especially sponges and anthozoans, play important roles in seamount communities, as they construct three-dimensional habitats utilized by numerous other animals such as crustaceans, ophiuroids, and polychaetes (Glasby and Watson 2001; Buhl-Mortensen and Mortensen 2004; Mosher and Watling 2009; Watling et al. 2011; Bracken-Grissom et al. 2018; Okanishi and Mah 2020; Komai et al. 2022). Sponges and anthozoans are vulnerable to damage from anthropogenic activities, as they are often large, fragile, long-lived, and extremely slow-growing (Probert et al. 1997; Clark et al. 2016; Molodtsova and Opresko 2017). It has been estimated that the recovery of these organisms from such anthropogenic damage will take decades to centuries (Clark et al. 2016). Therefore, accurate identification and documentation of seamount fauna, in particular of sessile suspension-feeding organisms, is important to generate robust baseline datasets that can be utilized to better protect the biological communities of seamounts.
The order Zoantharia is a group of sessile cnidarians consisting of > 300 species (Reimer and Sinniger 2024). In the deep sea, species of zoantharians within the family Parazoanthidae are known to associate with many different host taxa, prominently including octocorals and sponges (e.g., Carlgren 1923; Sinniger et al. 2005, 2013; Reimer et al. 2008, 2019; Carreiro-Silva et al. 2017; Kise et al. 2022; Montenegro et al. 2024). Four zoantharian genera have been reported to be associated with Hexasterophora sponges; Churabana Kise, Montenegro & Reimer, 2021, Parachurabana Kise, 2023, Thoracactis Gravier, 1918, and Vitrumanthus Kise, Montenegro & Reimer, 2022. Churabana, Parachurabana, and Thoracactis are monotypic genera while Vitrumanthus includes three species from the Pacific and Atlantic oceans. Although the Hexasterophora-zoantharian association thus has a wide distribution across the global oceans (Reiswig and Wheeler 2002; Dohrmann et al. 2011; Reiswig and Dohrmann 2014; Van Soest et al. 2014; Montenegro et al. 2020; Kise et al. 2022, 2023), the diversity of these associations is still poorly known. In this study, we collected specimens of Churabana and Vitrumanthus from the Shoho and An’ei seamounts along the Nishi-Shichito Ridge in the northwestern Pacific Ocean, and formally describe one species, Vitrumanthusflosculus sp. nov., utilizing a combination of morphological observations and molecular phylogenetic analyses. In addition, we report the complete mitochondrial genomes of two Hexasterophora-associated species, Churabanakuroshioae and Vitrumanthusflosculus sp. nov., which further reinforce the phylogenetic position of these species within Parazoanthidae.
Materials and methods
Specimen collection
Hexasterophora-associated zoantharians were collected from Shoho and An’ei seamounts on 29 November 2020 and 17 October 2021 by the remotely operated vehicle (KM-ROV) aboard the R/V Kaimei at depths of 400 and 770 m, respectively (Fig. 1). Photographs of the specimens were taken in situ for gross external morphological observation before collection using a camera mounted on the KM-ROV. Upon specimen retrieval, each specimen was anesthetized with magnesium chloride and subsequently fixed in 10% seawater formalin with subsamples preserved in 99.5% ethanol. The specimens examined in this study have been deposited in the National Museum of Nature and Science, Tsukuba, Japan (NSMT).
Figure 1.
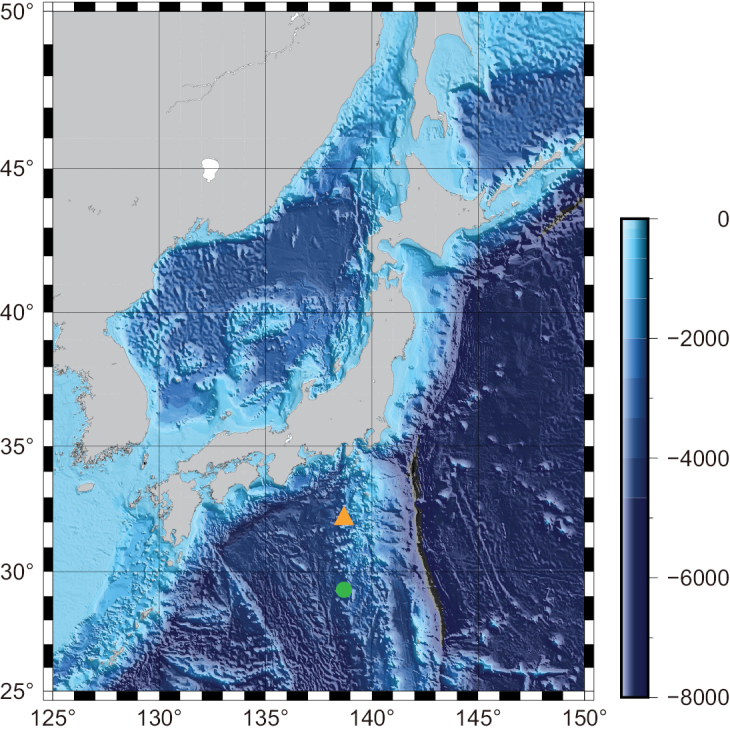
Research area and location of sampling sites. Enclosed symbols indicate sampling sites of two species examined in this study. Vitrumanthusflosculus sp. nov. (orange triangle) and Churabanakuroshioae (green circle).
DNA extraction, sequencing, and assembly
Tissues preserved in 99.5% ethanol were used for DNA extraction with a spin-column DNeasy Blood and Tissue Extraction kit following the manufacturer’s instructions (Qiagen, Hilden, Germany). Extracted DNA was quantified using a Qubit dsDNA BR assay kit (ThermoFisher Scientific, Waltham, USA). Whole-genome shotgun sequencing was performed by Bioengineering Lab. Co., Ltd. (Sagamihara, Japan) with DNBSEQ-G400 platforms (MGI Tech, Shenzhen, China) to produce pair-end 200 bp reads. The raw reads were filtered using Trimmomatic v. 0.39 (Bolger et al. 2014) with default parameters. Filtered reads were de novo assembled with GetOrganelle v.1.7.5 (Jin et al. 2020), which used implemented SPAdes v.3.6.2 genome assembler (Bankevich et al. 2012) with K-mer = 115. The mitochondrial genome annotation was performed with MITOS webserver (Bernt et al. 2013), and manually inspected and adjusted using Geneious Prime 2022.1.1 (https://www.geneious.com). Transfer RNA genes were identified using the tRNAscan-SE v2.0 (Chan et al. 2021). The annotated mitochondrial genomes were deposited in GenBank with the accession numbers PQ554681 and PQ554682. Sequences of Cox1 (mitochondrial cytochrome c oxidase subunit I), 12S rDNA (mitochondrial 12S ribosomal DNA), and 16S rDNA (mitochondrial 16S ribosomal DNA) were extracted from newly obtained mitochondria genomes. Three nuclear sequences, 18S rDNA (nuclear 18S ribosomal DNA), ITS rDNA (nuclear internal transcribed spacer region of ribosomal DNA), and 28S rDNA (nuclear 28S ribosomal DNA) were recovered from filtered and trimmed reads according to reference fragment sequences of Churabanakuroshioae (Accession numbers: MK377416, MZ329753, and MZ329743) and Vitrumanthusschrieri (Accession numbers: MZ329701, MZ329735, and MZ329712) using the Geneious Read Mapper (https://www.geneious.com).
Molecular phylogenetic analyses
Partial fragments of mitochondrial genes (Cox1, 12S rDNA, and 16S rDNA) and of the nuclear genes (18S rDNA, ITSrDNA, and 28S rDNA) were added to the alignment dataset used in Kise et al. (2023). In addition, previously reported sequences of Thoracactistopsenti (Kise et al. 2024) were also added to the alignment dataset. GenBank accession numbers used for phylogenetic analyses in this study are listed in Suppl. material 1. Subsequently, these sequences were manually trimmed and realigned using MAFFT (Katoh and Standley 2013) with the auto algorithm under default parameters for all genetic markers, and finally these alignments for each genetic marker were concatenated (hereafter six-gene dataset). Phylogenetic analyses were performed on the concatenated dataset using maximum likelihood (ML) and Bayesian inference (BI). ModelTest-NG v.0.1.6 (Darriba et al. 2020) under the Akaike information criterion was used to select the best-fitting model for each molecular marker independently for both ML and BI analyses. The best-selected models for ML and BI analyses are listed in Suppl. material 2. The final dataset consisted of 5148 bp and was used for ML and BI analyses. ML analyses were performed by RAxML-NG (Kozlov et al. 2019) with 1000 bootstrap replicates. BI analyses were performed with MrBayes; four Markov chain Monte Carlo (MCMC) heated chains were run for 5,000,000 generations with the temperature of the heated chain set to 0.2. Chains were sampled every 200 generations. Burn-in was set to 1,250,000 generations, at which point the average standard deviation of split frequency was consistently below 0.01. Tracer v.1.7.1 (Rambaut et al. 2018) was used to inspect the convergence of MCMC.
In addition, 13 protein-coding genes were extracted from newly sequenced mitochondrial genomes and other zoantharian mitochondrial genomes listed in Poliseno et al. (2020) and Fourreau et al. (2023) (Suppl. material 3). These protein-coding genes were individually aligned using MAFFT with the auto algorithm under default parameters. The concatenated dataset consisted of 35 zoantharian species and 13025 sites. For this mitochondrial genome dataset, ML reconstruction was performed using IQ-TREE2 (Minh et al. 2020) with best-fitting models for each protein-coding gene selected using ModelFinder (Kalyaanamoorthy et al. 2017) implemented in the IQ-TREE2 under Bayesian information criterion (Suppl. material 4). Support for each node was evaluated using 10,000 ultrafast bootstrap (UFBoot2) approximations (Hoang et al. 2018). BI was performed with MrBayes v.3.2.7 (Ronquist et al. 2012); four Markov chain Monte Carlo (MCMC) heated chains were run for 5,000,000 generations with the temperature of the heated chain set to 0.2. Chains were sampled every 200 generations. Best-fitting models for BI analyses were selected from models available in MrBayes using IQ-TREE2 (-mset mrbayes) (Suppl. material 4). Burn-in was set to 1,250,000 generations, at which point the average standard deviation of split frequency was consistently below 0.01. Tracer v.1.7.1 (Rambaut et al. 2018) was used to inspect the convergence of MCMC. The mitochondrial genomes of two antipatharian species, Stichopathesluetkeni Brook, 1889 and Myriopathesjaponica (Brook, 1889), were used as outgroups according to Poliseno et al. (2020).
Morphological observations
External morphological characteristics were observed and dissected under a Stemi 305 microscope (Carl Zeiss, Oberkochen, Germany), and photographs were taken using a Zeiss Axiocam 208 color camera (Carl Zeiss, Oberkochen, Germany). In addition, in-situ photographs were used for morphological observations. Internal morphological characters were examined by histological sections; 10–15-mm thickness serial sections were made with a microtome (Leica RM2145, Leica Biosystems, Wetzlar, Germany) and stained with haematoxylin and eosin after desilication with 20% hydrofluoric acid for 18–24 h. Classification of marginal muscle shapes followed Swain et al. (2015). Cnidae analyses were conducted using undischarged nematocysts and spirocysts from tentacles, column, actinopharynx, and mesenterial filaments using a Nikon Eclipse80i stereomicroscope (Nikon, Tokyo, Japan), and photographs were taken by a Nikon DS-Qi2 (Nikon, Tokyo, Japan). Cnidae sizes were measured using ImageJ v.1.45s (Rasband 2012). The reported frequencies were the relative amounts based on numbers from all slides in the cnidae analyses. Cnidae classification generally followed England (1991) and Ryland and Lancaster (2004) except for the treatment of basitrichs and microbasic b-mastigophores as in Kise et al. (2019).
Results
Taxonomic account
Order Zoantharia Rafinesque, 1815
Suborder Macrocnemina Haddon & Shackleton, 1891
Family Parazoanthidae Delage & Hérouard, 1901
Genus. Vitrumanthus
Kise, Montenegro & Reimer, 2022
06E67587-080B-56D6-8189-5AEABE104B5B
Type species.
Vitrumanthusschrieri Kise, Montenegro & Reimer, 2022.
Diagnosis.
Parazoanthidae characterized by obligate symbiotic relationship with massive hexasterophoran and Demospongiae sponges. Preserved polyps 0.3–3.1 mm in length, 0.8–3.4 mm in diameter. Azooxanthellate. Cyclically transitional marginal musculature.
. Vitrumanthus flosculus
Kise & Reimer sp. nov.
79CE76E9-E591-5693-91B4-28BAD1978DAE
https://zoobank.org/BD579CA0-C245-4CBD-85DA-389A18CBAD7E
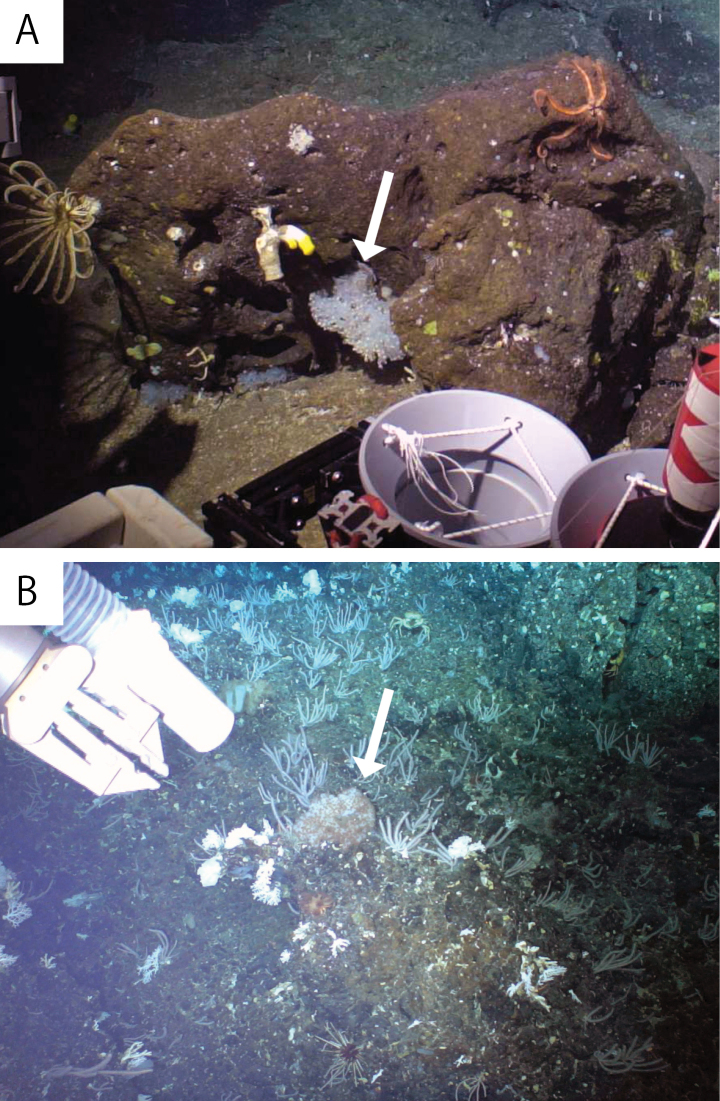
Figure 2.
In-situ image of AVitrumanthusflosculus sp. nov. and BChurabanakuroshioae. White arrows indicate each species associated with Hexasterophora sponges.
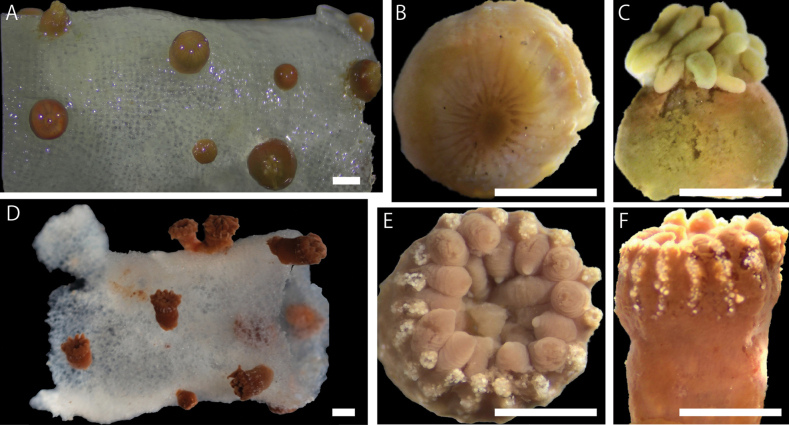
Figure 3.
Images of external morphology of (A–C) Vitrumanthusflosculus sp. nov. and (D–F) ChurabanakuroshioaeA preserved polyps attached to Farrea sp. B and C close-up image of a preserved polyp D preserved polyps attached to Pararete sp. E and F close-up image of a preserved polyp. Scale bars: 1.0 mm (A–C); 2.0 mm (D–F).
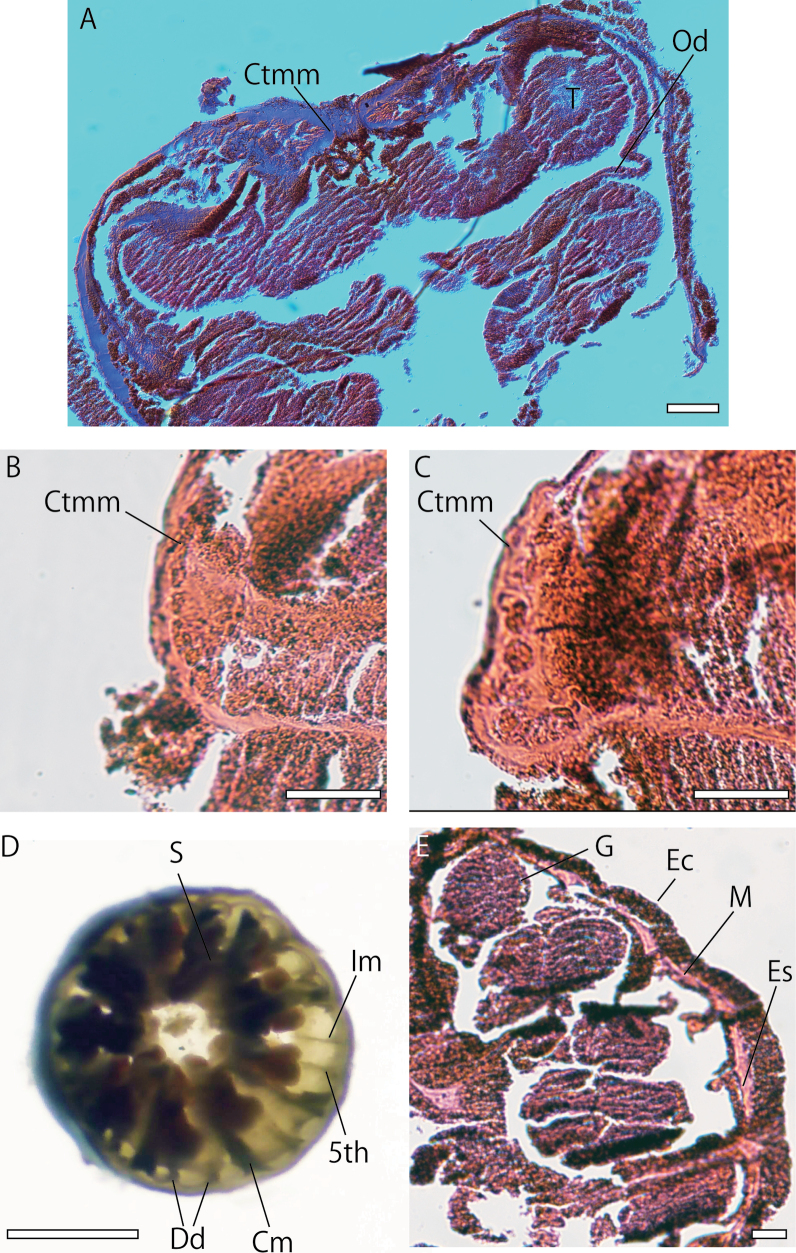
Figure 4.
Images of internal morphology of Vitrumanthusflosculus sp. nov. A longitudinal section of a polyp B and C closed-up image of cyclically transitional marginal musculature D transverse-section of polyp at level of actinopharynx by hand-cutting E transverse-section of polyp. Ctmm cyclically transitional marginal musculature, CM complete mesentery, Dd dorsal directives, Ec ectoderm, Es encircling sinus, IM incomplete mesentery, G gonads, M mesoglea, Od oral disk, T tentacles, S siphonoglyph, 5th 5th mesentery from dorsal directives. Scale bars: 100 µm (A); 50 µm (B, C, E); 1 mm (D).
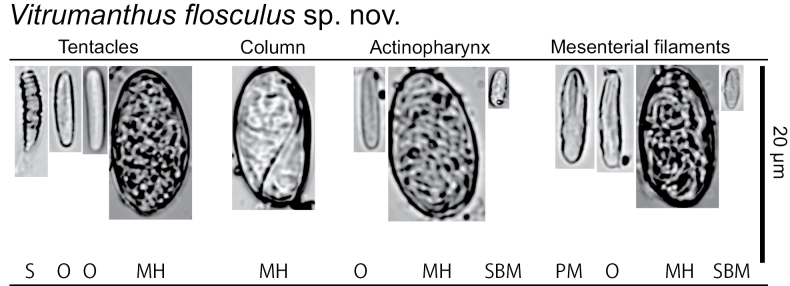
Figure 5.
Cnidae in the tentacles, column, actinopharynx and mesenterial filaments of Vitrumanthusflosculus sp. nov. HM holotrich medium O basitrichs and microbasic b-mastigophores PM microbasic p-mastigophores S spriocysts SBM special microbasic b-mastigophores.
Material examined.
Holotype • NSMT-Co 1898, Shoho Seamount, Nishi-Shichito Ridge, Japan (32°19.73'N, 138°44.28'E), 400 m depth, November 29, 2020.
Type locality.
Shoho Seamount, Nishi-Shichito Ridge, Japan.
Etymology.
“flosculus” meaning “small flower” or “floweret” in Latin.
Description.
External morphology. Colonial macrocnemic zoantharians associated with host hexasterophoran sponge Farrea Bowerbank, 1862 (Fig. 2A). Solitary or colonial polyps rise irregularly from all over the three-dimensional structure of host hexasterophoran sponge with base of polyps embedded in tissue of sponge (Fig. 3A). Preserved specimens consist of cylindrical polyps (Fig. 3B, C), dark brown in coloration. The living polyps and tentacles transparent yellowish in coloration. Surface of column smooth and ectoderm continuous (Fig. 3C). No encrustations of sand and silica particles in ectoderm of capitulum but ectoderm of scapus encrusted with small-sized sand and silica particles. Contracted preserved polyps 1.5–2.5 mm in height, 1.0–2.5 mm in diameter. Capitulary ridges indiscernible when contracted. Tentacles 22–26 in number.
Internal morphology. Zooxanthellae absent. Cyclically transitional marginal musculature (Fig. 4A–C). Encircling sinus or mesogleal canal present and basal canals of mesenteries absent (Fig. 4E). Mesenteries 22–26 in number, in brachycnemic arrangement (Fig. 4D). Mesoglea thickness 20–30 µm. Siphonoglyph distinct and V-shaped. Mesenterial filaments present. Complete mesenteries fertile (Fig. 4E).
Cnidae. Basitrichs and microbasic b-mastigophores, microbasic p-mastigophores, holotrichs, special b-mastigophores, and spirocysts (see Fig. 5, Table 1 for sizes and distributions).
Table 1.
Cnidae types and sizes observed in this study. Frequency: relative abundance of cnidae type in decreasing order; numerous, common, occasional, rare. N = number of cnidae measured.
| Vitrumanthusflosculus sp. nov. | |||||
|---|---|---|---|---|---|
| Tissue | Type of cnidae | Length (min-max, mean) | Width (min-max, mean) | Frequency | n |
| Tentacle | Spirocysts | 18.5–8.5, 12.7 | 3.3–1.3, 2.0 | Numerous | 215 |
| Basitrichs and microbasic b-mastigophores | 12.4–8.5, 10.4 | 3.2–1.4, 2.1 | Numerous | 73 | |
| Holotrichs (M) | 20.5 | 9.7 | Rare | 1 | |
| Column | Holotrich (M) | 20.9–17.0, 18.7 | 11.9–8.7, 10.0 | Common | 25 |
| Actinopharynx | Basitrichs and microbasic b-mastigophores | 18.2–8.7, 12.0 | 3.5–1.6, 2.3 | Common | 38 |
| Special microbasic b-mastigophores | 8.6–7.0, 7.8 | 3.3–1.6, 2.3 | Common | 13 | |
| Holotrichs (L) | 19.7–13.0, 17.5 | 11.3–8.5, 10.2 | Common | 18 | |
| Mesenterial filaments | Bastrichs and microbasic b-mastigophores | 14.7–7.9, 11.8 | 3.1–1.4, 1.9 | Common | 21 |
| Microbasic p-mastigophores | 16.9–10.7, 13.2 | 3.7–2.5, 3.3 | Common | 32 | |
| Special microbasic b-mastigophores | 9.3–7.2, 8.1 | 3.0–2.1, 2.5 | Occasional | 5 | |
| Holotrichs (M) | 22.2–15.4, 18.6 | 11.3–7.6, 9.4 | Common | 25 | |
Habitat and distribution.
Northwestern Pacific Ocean: known from the Shoho Seamount, Nishi-Shichito Ridge, Japan at a depth of 400 m. The new species was found on a glass sponge, Farrea sp., attached to rocks on the summit of the Shoho Seamount.
Associated host.
Farrea sp.
Remarks.
Regarding host sponges, Vitrumanthusflosculus sp. nov. is associated with Farrea, while other Vitrumanthus species are associated with other, different host sponges (Vitrumanthusschrieri: Verrucocoeloidea, Parahigginsia and Cyrtaulon, Vitrumanthusvanderlandi: Aphrocallistes, and Vitrumanthusoligomyarius: Tretochone). Vitrumanthusflosculus sp. nov. has holotrich nematocysts in all tissues we examined, while V.vanderlandi does not have holotrichs in any tissues. The surface of the column is smooth in Vitrumanthusflosculus sp. nov. with no encrustation of sand and silica particles in the ectoderm of capitulum, while the surface of the column is rough in V.schrieri with encrustation in the ectoderm of capitulum. The mesenteric arrangement of both Vitrumanthusflosculus sp. nov. and V.oligomyarius is brachycnemic, an exceptional characteristic for species within the suborder Macrocnemina. However, these two species can be distinguished by their numbers of tentacles and the sizes of the polyps; Vitrumanthusflosculus sp. nov. has 22–26 tentacles, while V.oligomyarius has 32–36 tentacles. Vitrumanthusflosculus sp. nov. has relatively smaller polyps than those of V.oligomyarius (1.5–2.5 mm in height and 1.0–2.5 mm in diameter vs. 0.5–3.1 mm in height and 1.2–3.4 mm in diameter). Furthermore, the host sponges of Vitrumanthusflosculus sp. nov. and V.oligomyarius are different (Farrea vs. Tretochone).
Parachurabana is a monotypic genus, and the diagnostic feature of this genus is described as having an association with Farreidae sponges. Although Vitrumanthusflosculus sp. nov. is associated with Farrea sp., Vitrumanthusflosculus sp. nov. can be easily distinguished from Parachurbana by the different shape of its sphincter muscle (cyclically transitional vs. cteniform endodermal marginal musculature) and different mesenterial arrangement (brachycnemic vs macrocnemic arrangement). The diagnosis of Parachurabana may need to be updated based on examinations of additional specimens.
Genus. Churabana
Kise, Montenegro & Reimer, 2021
A851B0FE-9CD0-5A73-8438-05E7F06730FF
Type species.
Churabanakuroshioae Kise, Montenegro & Reimer, 2021.
Diagnosis.
(modified from the diagnosis given by Kise et al. 2022). Parazoanthidae with obligate symbiotic relationship with Pararete sponges. Preserved polyps 3.0–10.0 mm in height, 2.8–5.0 mm in diameter. Azooxanthellate. Cteniform endodermal marginal musculature.
Remarks.
We modified the generic diagnosis based on a newly collected specimen of Churabanakuroshioae. This species seems to have host specificity to Pararete species based on this study and Kise et al. (2022), although further investigations are required to confirm this.
. Churabana kuroshioae
Kise, Montenegro & Reimer, 2021
A29E3662-4B6C-5808-8A85-CE5C747CAB78
Material examined.
• NSMT-Co 1899, An’ei Seamount, Nishi-Shichito Ridge, Japan (29°17.03'N, 138°37.85'E), 770 m depth, October 17, 2021.
Type locality.
Near Iejima Island, Motobu, Okinawa, Japan.
Description.
External morphology. Parazoanthidae associated with host hexasterophoran sponge Pararete Ijima, 1927. Approximately 100 truncated cone-shaped cylindrical polyps in preserved specimen. Solitary or colonial polyps rise irregularly from host Pararete sponges (Figs 2B, 3D). The living and preserved polyps dark brown and tentacles brown in coloration. Ectoderm and mesoglea of capitulum encrusted with numerous and comparatively large sizes of sand and silica particles (approximately < 100 µm). No encrustations of sand and silica particles in the ectoderm or mesoglea of scapus (Fig. 3F). Contracted preserved polyps 3.0–10.0 mm in height, 2.8–5.0 mm in diameter. Capitulary ridges discernible when contracted, 15–16 in number, and 30–32 tentacles (Fig. 3E).
Habitat and distribution.
Northwestern Pacific Ocean: Churabanakuroshioae was originally reported from the Ryukyu Archipelago, Japan at depths of 520–650 m (Kise et al. 2022). The findings in this study reveal that this species is also distributed at the An’ei Seamount, Nishi-Shichito Ridge, Japan at a depth of 770 m. Churabanakuroshioae was found on the summit of An’ei Seamount on glass sponge Pararete sp. attached to rocky substrate.
Associated host.
Pararete sp.
Remarks.
The polyp coloration of Churabanakuroshioae is cream-pink or beige with cream or whitish transparent tentacles in the original description, while the specimen of C.kuroshioae collected from An’ei Seamount has dark brown polyps with brown tentacles. As well, the polyp sizes of the specimen examined in this study were relatively larger than that of the original description (3.0–4.0 mm in height, 2.8–4.0 mm in diameter) by Kise et al. (2022). Based on the results of molecular phylogenetic analyses, the differences in coloration and polyp sizes found in this study are considered intraspecific variation, although detailed molecular analyses in the future may warrant reconsideration of this.
Mitochondrial genome
The complete mitochondrial genome sizes of Churabanakuroshioae and Vitrumanthusflosculus sp. nov. were 22,738 and 20,556 bp, respectively. The mitochondrial gene order and content of these two species were the same, including 13 protein-coding genes, two rRNA genes, and two transfer RNA genes. The sequences of the protein-coding region covered 54.0% (Churabanakuroshioae) and 58.8% (Vitrumanthusflosculus sp. nov.) of the mitochondrial genomes, while GC contents of Churabanakuroshioae and Vitrumanthusflosculus sp. nov. were 49.8% and 50.0%, respectively. Regarding stop codons, both C.kuroshioae and V.flosculus sp. nov. have either TAA and TAG for all protein-coding genes, with the start codon being ATG. The mitochondrial base composition was A: 22.6%, T: 27.6%, G: 26.1%, C: 23.7% in C.kuroshioae, and A: 22.2%, T: 27.8%, G: 26.3%, C: 23.6% in V.flosculus sp. nov.
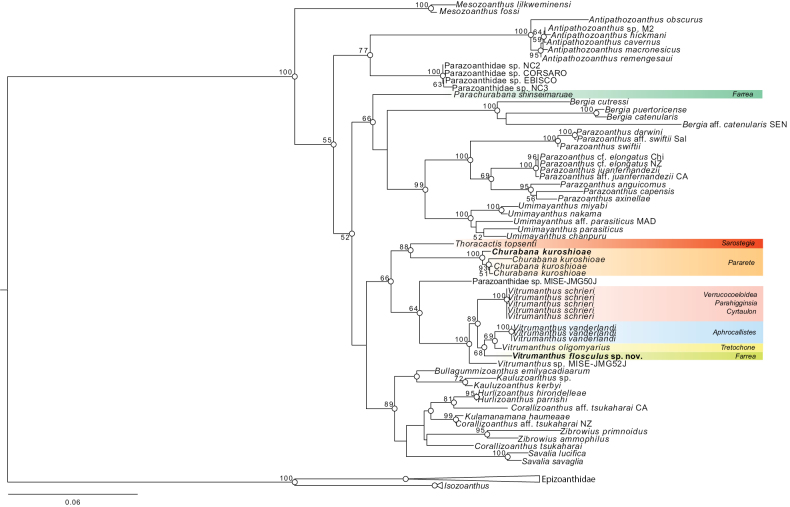
Molecular phylogeny
ML and BI phylogenetic analyses based on the six-gene dataset indicated that Churabana and Vitrumanthus were both monophyletic clades with complete support (ML = 100%, BI = 1). Churabana was sister to Thoracactis (ML = 88%, BI = 0.96). ML and BI phylogenetic topologies were congruent (Fig. 6). Vitrumanthusflosculus sp. nov. was sister to Vitrumanthusoligomyarius and Vitrumanthusvanderlandi (ML = 68%, BI = 0.99).
Figure 6.
Maximum-likelihood tree based on combined dataset of CoxI, 12S rDNA, 16S rDNA, 18S rDNA, 28S rDNA, and ITS rDNA sequences. Numbers at nodes represent ML bootstrap values (>50% are shown). White circles on nodes indicate high support of Bayesian posterior probabilities (PP) (>0.95).
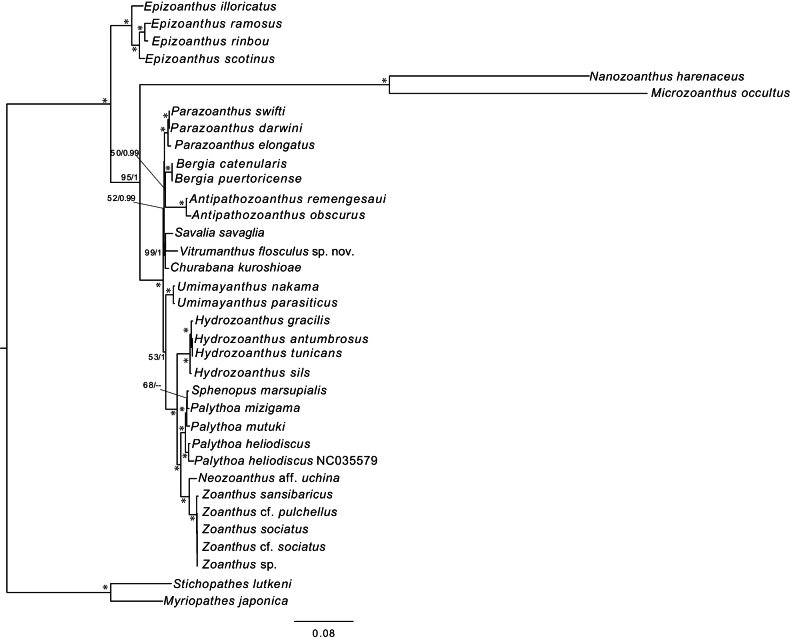
ML and BI phylogenetic topologies based on the complete mitochondrial genome dataset were also congruent (Fig. 7). Churabana and Vitrumanthus formed a monophyletic clade with Savaliasavaglia (Bertoloni, 1819) with strong support (ML = 99%, BI = 1).
Figure 7.
Maximum-likelihood tree based on complete mitochondrial genome dataset. Numbers at each node indicates ML bootstrap value and Bayesian posterior probabilities. Asterisks indicate ML/BI = 100/1.00.
Discussion
Thoracactistopsenti was the first zoantharian species to be described as Hexasterophora-associated (Gravier 1918). Subsequent studies have more recently described three Parazoanthidae genera associated with hexasterophorans from the Indo-Pacific and the Atlantic (Kise et al. 2022, 2023). Although Thoracactis was originally placed in the family Epizoanthidae, Kise et al. (2024) have recently transferred T.topsenti to Parazoanthidae based on molecular phylogenetic and morphological results, indicating that the association with Hexasterophora is unique to the family Parazoanthidae. Kise et al. (2023) found that Hexasterophora-associated species were not monophyletic, but instead that Parachurabana was recovered as basal to Demospongiae-associated species (Bergia, Parazoanthus, and Umimayanthus), indicating that Parazoanthidae species may have switched its host from Hexasterophora to Demospongiae. However, the phylogenetic tree based on a six-gene dataset of this study and previous studies have shown weak support at some nodes in Parazoanthidae. Therefore, further studies using phylogenetically informative loci, as mentioned below, are needed to better understand the evolutionary history of host switching in Parazoanthidae.
This study sequenced the complete mitochondrial genomes of Churabana and Vitrumanthus for the first time. The mitochondrial gene arrangements of these two genera were in the same order as those of other zoantharians (Poliseno et al. 2020), further reinforcing the conservative nature of zoantharian mitochondrial gene orders.
Based on both our six-gene and complete mitochondrial genome analyses, it is apparent that much of the diversity of Parazoanthidae has comparatively recently evolved, resulting in weak support at many generic–level nodes, with short genetic distances as reported in Poliseno et al. (2020). Perhaps more robust genomic analytical methods (e.g., ultra-conserved elements; Cowman et al. 2020; Quattrini et al. 2020) may help resolve the weak phylogenetic structure of Parazoanthidae, which would then help in taxonomic reconsideration of the family. Most of the genera contained within Parazoanthidae have been erected since 2008 (12/17 genera), with each genus erected based on its uniqueness from other genera, and little consideration has yet been given to the phylogeny and taxonomy of the family. It may be time to reconsider the framework of Parazoanthidae, and it is hoped that the current study provides the impetus to begin this future work.
Shoho and An’ei seamounts are on the Nishi-Shichito Ridge, which has been designated as a marine protected area (MPA) (Ministry of the Environment of Japan 2020), and recent studies have described a number of previously unknown species including sea pens, sea stars, ribbon worms, and parasitic crustacean from the Shoho and An’ei seamounts (Hookabe et al. 2021, 2023; Kobayashi et al. 2022; Jimi et al. 2023; Kushida et al. 2024). Our results echo these recent studies, highlighting the overall lack of diversity studies in this MPA. Documentation of local faunal biodiversity is one important key for effective monitoring of MPAs, and further taxonomic studies of many taxa are needed to better understand the true marine diversity of this MPA.
Supplementary Material
Acknowledgements
We thank the captain and crew of the R/V Kaimei, the operation team of KM-ROV, and all the research members during the KM20-10C and KM21-E04 cruises. This research was performed by the Environment Research and Technology Development Fund (JPMEERF20S20700) of the Environmental Restoration and Conservation Agency Provided by the Ministry of Environment of Japan.
Citation
Kise H, Reimer JD, Iguchi A, Ise Y, Tsuchida S, Fujiwara Y (2024) Parazoanthidae (Cnidaria, Zoantharia) associated with glass sponges on the Nishi-Shichito Ridge, northwestern Pacific Ocean, with the description of a new species. ZooKeys 1221: 343–362. https://doi.org/10.3897/zookeys.1221.131258
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This study was supported by the Environment Research and Technology Development Fund (JPMEERF20S20700) of the Environmental Restoration and Conservation Agency Provided by the Ministry of Environment of Japan. This study was also partially supported by the Research Laboratory on Environmentally Conscious Developments and Technologies (E-code) at the National Institute of Advanced Industrial Science and Technology. The first author was supported in part by JSPS KAKENHI grant number 23KJ2206 from the Japan Society for the Promotion of Science. We thank an anonymous reviewer and the journal editor for their valuable comments that improved this work.
Author contributions
Data curation: HK. Formal analysis: HK. Funding acquisition: YF, AI. Investigation: HK, YI. Project administration: YF, ST. Resources: YI, JDR, YF. Validation: HK. Visualization: HK. Writing - original draft: HK. Writing - review and editing: ST, YI, JDR, AI, YF.
Author ORCIDs
Hiroki Kise https://orcid.org/0000-0002-4099-6469
James Davis Reimer https://orcid.org/0000-0003-0453-8804
Akira Iguchi https://orcid.org/0000-0002-8894-1977
Yuji Ise https://orcid.org/0009-0009-4887-611X
Shinji Tsuchida https://orcid.org/0000-0002-8600-2386
Yoshihiro Fujiwara https://orcid.org/0000-0002-1833-1866
Data availability
All of the data that support the findings of this study are available in the main text or Supplementary Information. Obtained sequences have been deposited in NCBI GenBank (accession number PQ308072–PQ308077 and PQ554681–PQ554682).
Supplementary materials
GenBank accession numbers used for phylogenetic analyses in this study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
Best fitting models for ML and BI phylogenetic analyses based on six-gene dataset
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
Information of Zoantharian species used for phylogenomic analyses of mitochondrial genomes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
Best fitting models for ML and BI phylogenetic analyses based on complete mitochondrial genome dataset
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
References
- Althaus F, Williams A, Schlacher TA, Kloser RJ, Green MA, Barker BA, Bax NJ, Brodie P, Schlacher-Hoenlinger MA. (2009) Impacts of bottom trawling on deep-coral ecosystems of seamounts are long-lasting. Marine Ecology Progress Series 397: 279–294. 10.3354/meps08248 [DOI] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski Ad, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler Gl, Alekseyev MA, Pevzner PA. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19(5): 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution 69(2): 313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15): 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerbank JS. (1862) On the anatomy and physiology of the Spongiadae. Part III On the generic characters, the specific characters, and on the method of examination. Philosophical Transactions of the Royal Society 152(2): 1087–1135. 10.1098/rstl.1862.0044 [DOI] [Google Scholar]
- Bracken-Grissom H, Widder E, Johnsen S, Messing C, Frank T. (2018) Decapod diversity associated with deep-sea octocorals in the Gulf of Mexico. Crustaceana 91(10): 1267–1275. 10.1163/15685403-00003829 [DOI] [Google Scholar]
- Brook G. (1889) Report on the Antipatharia collected by H.M.S. Challenger during the years 1873–76. Report on the Scientific Results of the Voyage of H.M.S. Challenger during the years 1873–76. Zoology 32[part 80]: [i–iii,]1–222 [pl. 1–15]. http://www.19thcenturyscience.org/HMSC/HMSC-Reports/Zool-80/README.html
- Buhl-Mortensen L, Mortensen PB. (2004) Symbiosis in deep-water corals. Symbiosis.
- Carlgren O. (1923) Ceriantharia and Zoantharia. Wissensch Ergebn Deutsch Tiefsee-Exp Dampfer ‘Valdivia’ 1898–99 19(7): 242–337.
- Carreiro-Silva M, Ocaña O, Stanković D, Sampaio I, Porteiro FM, Fabri MC, Stefanni S. (2017) Zoantharians (Hexacorallia: Zoantharia) associated with cold-water corals in the Azores region: new species and associations in the deep sea. Frontiers in Marine Science 4: 88. 10.3389/fmars.2017.00088 [DOI]
- Chan PP, Lin BY, Mak AJ, Lowe TM. (2021) tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Research 49(16): 9077–9096. 10.1093/nar/gkab688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MR, Vinnichenko VI, Gordon JD, Beck-Bulat GZ, Kukharev NN, Kakora AF. (2007) Large-scale distant-water trawl fisheries on seamounts. Seamounts: Ecology, Fisheries, and Conservation 12: 361–399. 10.1002/9780470691953.ch17 [DOI] [Google Scholar]
- Clark MR, Rowden AA, Schlacher T, Williams A, Consalvey M, Stocks KI, Rogers AD, O’Hara TD, White M, Shank TM, Hall-Spencer JM. (2010) The ecology of seamounts: structure, function, and human impacts. Annual Review of Marine Science 2: 253–278. 10.1146/annurev-marine-120308-081109 [DOI] [PubMed] [Google Scholar]
- Clark MR, Althaus F, Schlacher TA, Williams A, Bowden DA, Rowden AA. (2016) The impacts of deep-sea fisheries on benthic communities: a review. ICES Journal of Marine Science 73(suppl_1): i51–i69. 10.1093/icesjms/fsv123 [DOI]
- Cowman PF, Quattrini AM, Bridge TC, Watkins-Colwell GJ, Fadli N, Grinblat M, Roberts TE, McFadden CS, Miller DJ, Baird AH. (2020) An enhanced target-enrichment bait set for Hexacorallia provides phylogenomic resolution of the staghorn corals (Acroporidae) and close relatives. Molecular Phylogenetics and Evolution 153: 106944. 10.1016/j.ympev.2020.106944 [DOI] [PubMed]
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. (2020) ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution 37(1): 291–294. 10.1093/molbev/msz189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage Y, Hérouard E. (1901) Zoanthidés. – Zoanthidae. In Traité de Zoologie concrète, 2eme partie. Les Cœlentérés. C. Reinwald, Paris, 654–667.
- Dohrmann M, Haen KM, Lavrov DV, Wörheide G. (2011) Molecular phylogeny of glass sponges (Porifera, Hexactinellida): increased taxon sampling and inclusion of the mitochondrial protein-coding gene, cytochrome oxidase subunit I. In Ancient Animals, New Challenges (pp. 11–20). Springer, Dordrecht. 10.1007/s10750-011-0727-z [DOI]
- England KW. (1991) Nematocysts of sea anemones (Actiniaria, Ceriantharia and Corallimorpharia: Cnidaria): nomenclature. Hydrobiologia 216: 691–697. 10.1007/BF00026532 [DOI] [Google Scholar]
- Fourreau CJL, Kise H, Santander MD, Pirro S, Maronna MM, Poliseno A, Santos MEA, Reimer JD. (2023) Genome sizes and repeatome evolution in zoantharians (Cnidaria: Hexacorallia: Zoantharia). PeerJ 11: e16188. 10.7717/peerj.16188 [DOI] [PMC free article] [PubMed]
- Glasby CJ, Watson C. (2001) A new genus and species of Syllidae (Annelida: Polychaeta) commensal with octocorals. Beagle: Records of the Museums and Art Galleries of the Northern Territory, The Beagle 17: 43–51. 10.5962/p.286289 [DOI] [Google Scholar]
- Gravier C. (1918) Note sur une actinie (*Thoracactis* n. g., *topsenti* n. sp.) et un annélide polychète (*Hermadion Fauveli* n. sp.), commensaux d’une Éponge siliceuse (*Sarostegiaoculata* Topsent). Bulletin de l’Institut océanographique de Monaco 344: 1–20. 10.5962/bhl.part.8664 [DOI] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35(2): 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookabe N, Jimi N, Yokooka H, Tsuchida S, Fujiwara Y. (2021) Lacydoniashohoensis (Annelida, Lacydoniidae) sp. nov.–a new lacydonid species from deep-sea sunken wood discovered at the Nishi-Shichito Ridge, north-western Pacific Ocean. Journal of the Marine Biological Association of the United Kingdom 101(6): 927–933. 10.1017/S0025315421000862 [DOI] [Google Scholar]
- Hookabe N, Kohtsuka H, Fujiwara Y, Tsuchida S, Ueshima R. (2023) Three new species in Tetrastemma Ehrenberg, 1828 (Nemertea, Monostilifera) from sublittoral to upper bathyal zones of the northwestern Pacific. ZooKeys 1146: 135–146. 10.3897/zookeys.1146.95004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimi N, Kobayashi I, Moritaki T, Woo SP, Tsuchida S, Fujiwara Y. (2023) Insights into the diversification of deep-sea endoparasites: Phylogenetic relationships within Dendrogaster (Crustacea: Ascothoracida) and a new species description from a western Pacific seamount. Deep Sea Research Part I: Oceanographic Research Papers 196: 104025. 10.1016/j.dsr.2023.104025 [DOI]
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. (2020) GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology 21(1): 1–31. 10.1186/s13059-020-02154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kise H, Montenegro J, Ekins M, Moritaki T, Reimer JD. (2019) A molecular phylogeny of carcinoecium-forming Epizoanthus (Hexacorallia: Zoantharia) from the Western Pacific Ocean with descriptions of three new species. Systematics and Biodiversity 17(8): 773–786. 10.1080/14772000.2019.1693439 [DOI] [Google Scholar]
- Kise H, Montenegro J, Santos MEA, Hoeksema BW, Ekins M, Ise Y, Higashiji T, Fernandez-Silva I, Reimer JD. (2022) Evolution and phylogeny of glass-sponge-associated zoantharians, with a description of two new genera and three new species. Zoological Journal of the Linnean Society 194(1): 323–347. 10.1093/zoolinnean/zlab068 [DOI] [Google Scholar]
- Kise H, Nishijima M, Iguchi A, Minatoya J, Yokooka H, Ise Y, Suzuki A. (2023) A new hexactinellid-sponge-associated zoantharian (Porifera, Hexasterophora) from the northwestern Pacific Ocean. ZooKeys 1156: 71–85. 10.3897/zookeys.1156.96698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kise H, Montenegro J, Corrêa PV, Clemente MV, Sumida PY, Hoeksema BW, Reimer JD. (2024) A taxonomic revision of the sponge-associated genus Thoracactis Gravier, 1918 (Anthozoa: Zoantharia) based on an integrated approach. Contributions to Zoology 93(3): 229–251. 10.1163/18759866-bja10059 [DOI] [Google Scholar]
- Kobayashi I, Yamamoto M, Fujiwara Y, Tsuchida S, Fujita T. (2022) First record of the family Myxasteridae (Asteroidea: Velatida) from western north Pacific with description of a new species of Asthenactis. Species Diversity 27(2): 251–258. 10.12782/specdiv.27.251 [DOI] [Google Scholar]
- Komai T, Tsuchida S, Fujiwara Y. (2022) New record of a rarely collected caridean shrimp Bathypalaemonellapandaloides (Rathbun, 1906) (Decapoda: Bathypalaemonellidae) from the West Mariana Ridge, northwestern Pacific. Zootaxa 5129(2): 272–284. 10.11646/zootaxa.5129.2.7 [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35(21): 4453–4455. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushida Y, Kise H, Iguchi A, Fujiwara Y, Tsuchida S. (2024) Description of the fifth sea pen species that attaches to hard substrates by modifying its peduncle. Deep Sea Research Part I: Oceanographic Research Papers 203: 104212. 10.1016/j.dsr.2023.104212 [DOI]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37(5): 1530–1534. 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of the Environment of Japan (2020) Ministry of the Environment of Japan. https://www.env.go.jp/nature/naturebiodic/kaiyo-hogoku.html [Accessed 20th Mar 2024]
- Molodtsova TN, Opresko DM. (2017) Black corals (Anthozoa: Antipatharia) of the Clarion-Clipperton fracture zone. Marine Biodiversity 47(2): 349–365. 10.1007/s12526-017-0659-6 [DOI] [Google Scholar]
- Montenegro J, Hoeksema BW, Santos MEA, Kise H, Reimer JD. (2020) Zoantharia (Cnidaria: Hexacorallia) of the Dutch Caribbean and one new species of Parazoanthus. Diversity 12(5): 190. 10.3390/d12050190 [DOI] [Google Scholar]
- Montenegro J, Fromont J, Richards Z, Kise H, Gomez O, Hoeksema BW, Reimer JD. (2024) Museum collections as untapped sources of undescribed diversity of sponge-zoantharian associations with the description of six new species of Umimayanthus (Zoantharia: Parazoanthidae) from Western Australia and eastern Indonesia. Contributions to Zoology 1: 1–57. 10.1163/18759866-bja10069 [DOI] [Google Scholar]
- Morato T, Hoyle SD, Allain V, Nicol SJ. (2010) Seamounts are hotspots of pelagic biodiversity in the open ocean. Proceedings of the National Academy of Sciences 107(21): 9707–9711. 10.1073/pnas.0910290107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher CV, Watling L. (2009) Partners for life: a brittle star and its octocoral host. Marine Ecology Progress Series 397: 81–88. 10.3354/meps08113 [DOI] [Google Scholar]
- Okanishi M, Mah CL. (2020) Overlooked biodiversity from museum collections: four new species and one new genus of Ophiuroidea (Echinodermata) from Antarctica and adjacent regions with notes on multi-armed ophiuroids. Marine Biodiversity 50(5): 1–26. 10.1007/s12526-020-01080-w [DOI] [Google Scholar]
- Poliseno A, Santos MEA, Kise H, Macdonald B, Quattrini AM, McFadden CS, Reimer JD. (2020) Evolutionary implications of analyses of complete mitochondrial genomes across order Zoantharia (Cnidaria: Hexacorallia). Journal of Zoological Systematics and Evolutionary Research 58(4): 858–868. 10.1111/jzs.12380 [DOI] [Google Scholar]
- Probert PK, Mcknight DG, Grove SL. (1997) Benthic invertebrate bycatch from a deep‐water trawl fishery, Chatham Rise, New Zealand. Aquatic Conservation: Marine and Freshwater Ecosystems 7(1): 27–40. [DOI] [Google Scholar]
- Quattrini AM, Rodríguez E, Faircloth BC, Cowman PF, Brugler MR, Farfan GA, Hellberg ME, Kitahara MV, Morrison CL, Paz-García DA, Reimer JD, McFadden CS. (2020) Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nature Ecology & Evolution 4(11): 1531–1538. 10.1038/s41559-020-01291-1 [DOI] [PubMed] [Google Scholar]
- Rafinesque CS. (1815) Analyse de la nature: or, Tableau de l'univers et des corps organisés. Palermo, Italy, 224 pp. 10.5962/bhl.title.106607 [DOI] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67(5): 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. (2012) ImageJ: Image processing and analysis in Java. Astrophysics Source Code Library 1: e06013.
- Reimer JD, Sinniger F. (2024) World List of Zoantharia. Zoantharia. [World Register of Marine Species] https://www.marinespecies.org/aphia.php?p=taxdetails&id=607338 [on 2024-04-27]
- Reimer JD, Nonaka M, Sinniger F, Iwase F. (2008) Morphological and molecular characterization of a new genus and new species of parazoanthid (Anthozoa: Hexacorallia: Zoantharia) associated with Japanese Red Coral. Coral Reefs 27: 935–949. 10.1007/s00338-008-0389-0 [DOI] [Google Scholar]
- Reimer JD, Kise H, Santos MEA, Lindsay DJ, Pyle RL, Copus JM, Bowen BW, Nonaka M, Higashiji T, Benayahu Y. (2019) Exploring the biodiversity of understudied benthic taxa at mesophotic and deeper depths: examples from the order Zoantharia (Anthozoa: Hexacorallia). Frontiers in Marine Science 6: 305. 10.3389/fmars.2019.00305 [DOI]
- Reiswig HM, Dohrmann M. (2014) Three new species of glass sponges (Porifera: Hexactinellida) from the West Indies, and molecular phylogenetics of Euretidae and Auloplacidae (Sceptrulophora). Zoological Journal of the Linnean Society 171: 233–253. 10.1111/zoj.12138 [DOI] [Google Scholar]
- Reiswig HM, Wheeler B. (2002) Family Euretidae Zittel, 1877. In: Hooper JNA, Van Soest RWM, Willenz P. (Eds) Systema Porifera.Springer, Boston, MA, 1301–1331. 10.1007/978-1-4615-0747-5_135 [DOI]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematics Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Bramanti L, Horta P, Allcock L, Carreiro-Silva M, Coppari M, Denis V, Hadjioannou L, Isla E, Jimenez C, Johnson M, Mohn C, Orejas C, Ramšak A, Reimer J, Rinkevich B, Rizzo L, Salomidi M, Samaai T, Schubert N, Soares M, Thurstan R, Vassallo P, Ziveri P, Zorrilla-Pujana J. (2022) Protecting global marine animal forests. Science 376(6596): 929–929. 10.1126/science.abq7583 [DOI] [PubMed] [Google Scholar]
- Rowden AA, Dower JF, Schlacher TA, Consalvey M, Clark MR. (2010) Paradigms in seamount ecology: fact, fiction and future. Marine Ecology 31: 226–241. 10.1111/j.1439-0485.2010.00400.x [DOI] [Google Scholar]
- Ryland JS, Lancaster JE. (2004) A review of zoanthid nematocyst types and their population structure. Hydrobiologia 530: 179–187. 10.1007/s10750-004-2685-1 [DOI] [Google Scholar]
- Samadi S, Bottan L, Macpherson E, De Forges BR, Boisselier MC. (2006) Seamount endemism questioned by the geographic distribution and population genetic structure of marine invertebrates. Marine Biology 149(6): 1463–1475. 10.1007/s00227-006-0306-4 [DOI] [Google Scholar]
- Sinniger F, Montoya-Burgos JI, Chevaldonne P, Pawlowski J. (2005) Phylogeny of the order Zoantharia (Anthozoa, Hexacorallia) based on the mitochondrial ribosomal genes. Marine Biology 147: 1121–1128. 10.1007/s00227-005-0016-3 [DOI] [Google Scholar]
- Sinniger F, Ocaña OV, Baco AR. (2013) Diversity of zoanthids (Anthozoa: Hexacorallia) on Hawaiian seamounts: description of the Hawaiian gold coral and additional zoanthids. PLoS ONE 8: e52607. 10.1371/journal.pone.0052607 [DOI] [PMC free article] [PubMed]
- Swain TD, Schellinger JL, Strimaitis AM, Reuter KE. (2015) Evolution of anthozoan polyp retraction mechanisms: convergent functional morphology and evolutionary allometry of the marginal musculature in order Zoanthidea (Cnidaria: Anthozoa: Hexacorallia). BioMed Central Evolutionary Biology 15: 1–19. 10.1186/s12862-015-0406-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest RW, Meesters EH, Becking LE. (2014) Deep-water sponges (Porifera) from Bonaire and Klein Curaçao, Southern Caribbean. Zootaxa 387: 401–443. 10.11646/zootaxa.3878.5.1 [DOI] [PubMed] [Google Scholar]
- Watling L, Auster PJ. (2017) Seamounts on the high seas should be managed as vulnerable marine ecosystems. Frontiers in Marine Science 4: 14. 10.3389/fmars.2017.00014 [DOI]
- Watling L, France SC, Pante E, Simpson A. (2011) Biology of deep-water octocorals. Advances in Marine Biology 60: 41–122. 10.1016/B978-0-12-385529-9.00002-0 [DOI] [PubMed] [Google Scholar]
- Worm B, Lotze HK, Myers RA. (2003) Predator diversity hotspots in the blue ocean. Proceedings of the National Academy of Sciences 100(17): 9884–9888. 10.1073/pnas.1333941100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm B, Barbier EB, Beaumont N, Duffy JE, Folke C, Halpern BS, Jackson JBC, Lotze HK, Micheli F, Palumbi SR, Sala E, Selkoe KA, Stachowicz JJ, Watson R. (2006) Impacts of biodiversity loss on ocean ecosystem services. Science 314(5800): 787–790. 10.1126/science.1132294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenBank accession numbers used for phylogenetic analyses in this study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
Best fitting models for ML and BI phylogenetic analyses based on six-gene dataset
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
Information of Zoantharian species used for phylogenomic analyses of mitochondrial genomes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
Best fitting models for ML and BI phylogenetic analyses based on complete mitochondrial genome dataset
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Hiroki Kise, James Davis Reimer, Akira Iguchi, Yuji Ise, Shinji Tsuchida, Yoshihiro Fujiwara
Data type
xlsx
Data Availability Statement
All of the data that support the findings of this study are available in the main text or Supplementary Information. Obtained sequences have been deposited in NCBI GenBank (accession number PQ308072–PQ308077 and PQ554681–PQ554682).