Abstract
Coscinium fenestratum, a medicinal plant traditionally used in Southeast Asia, exerts protective effects against various inflammatory diseases, primarily due to its rich alkaloid content. Despite substantial evidence supporting its anti-inflammatory properties, the biological activities of C. fenestratum are unclear. This study aimed to elucidate anticolitis mechanisms of C. fenestratum alkaloids (CFAs) using an integrative approach of network pharmacology and molecular docking analyses. Key active alkaloids and core target genes were identified through pharmacological and protein-protein interaction networks. The core targets were enriched in the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways to determine the functional properties of active CFA. Finally, the binding affinity of the key compounds with the core targets was determined using molecular docking. The results showed that 11 active CFAs interactively interfered with 10 hub genes related to ulcerative colitis, including prostaglandin-endoperoxide synthase 2 (PTGS2), selectin E (SELE), kinase insert domain receptor (KDR), fms-related receptor tyrosine kinase 1 (FLT1), intracellular adhesion molecule 1 (ICAM1), C-X-C motif chemokine receptor 4 (CXCR4), hypoxia-inducible factor-1 (HIF1A), matrix metalloproteinase (MMP)-2, MMP3, and MMP9, which were functionally involved in the immunological response, tumor necrosis factor signaling pathway, and interleukin-17 signaling pathway. The molecular docking results indicated that CFA compounds had a strong binding affinity for the hub genes. The findings reveal, for the first time, a therapeutic role of CFA in alleviating ulcerative colitis through its predicted interactions with relevant biological targets.
Keywords: alkaloids, colitis, molecular docking simulation, network pharmacology
INTRODUCTION
Ulcerative colitis (UC) is a chronic gastrointestinal inflammatory disease that affects the colon and rectum (Gupta et al., 2022; Li et al., 2022). Its pathogenesis has been linked to lesions in the colonic mucosa and submucosa, resulting in intestinal inflammation, injury, and fibrosis (Li et al., 2022; Yang et al., 2022). Although the etiology of UC is unclear, many factors are believed to induce disease onset, including genetics, oxidative stress, immune function dysregulation, and environmental factors (Gupta et al., 2022; Yang et al., 2022). Abdominal pain, hematuria, diarrhea, tenesmus, and weight loss are symptoms of UC (Gupta et al., 2022; Li et al., 2022; Yang et al., 2022). Effective treatment strategies have not been developed because of the multifactorial nature of UC and the lack of understanding of its causes. Therapeutic strategies involve administering aminosalicylate, immunomodulators, corticosteroids, and biological agents to inhibit the inflammatory response and maintain remission (Peng et al., 2019; Gupta et al., 2022; Yang et al., 2022). However, prolonged use of therapeutic drugs causes many side effects, including nausea, fever, headache, kidney failure, and congestive heart failure (Gupta et al., 2022; Yang et al., 2022). Therefore, researchers have become more interested in investigating natural alternatives because they have therapeutic potential in various ailments.
Native to South Asia and Southeast Asia, Coscinium fenestratum is a flowering woody climber that belongs to the family Menispermaceae (Sudharshan et al., 2010; Nayak et al., 2012; Okechukwu et al., 2013; Vite et al., 2016). C. fenestratum is traditionally used in herbal remedies in Southeast Asia because all parts of the plant contain medicinal properties-from the root to the fruit. This plant has been used as a decoction and tincture to treat several inflammatory diseases, metabolic disorders, and oxidative stress-related diseases, such as diabetes mellitus, arthritis, cancer, ulcers, and skin diseases (Deevanhxay et al., 2009; Rojsanga et al., 2010; Okechukwu et al., 2013; Rai et al., 2013; Vite et al., 2016). Many studies have demonstrated the health benefits of C. fenestratum, such as antifungal, hypotensive, antioxidant, antihepatotoxic, antiseptic, and antidiabetic properties (Deevanhxay et al., 2009; Nayak et al., 2012; Okechukwu et al., 2013; Rai et al., 2013). In vivo and in vitro studies have shown that C. fenestratum extracts exhibit anti-inflammatory activity (Sudharshan et al., 2010), antigastric ulcer activity (Vite et al., 2016), and antiproliferative properties on colorectal cancer cells (Rojsanga et al., 2010). Although the mechanisms involved are unknown, the biological activities of C. fenestratum are attributed to its high content of alkaloids, notably isoquinoline alkaloids, such as berberine, palmatine, and jatrorrhizine (Rojsanga et al., 2010; Nayak et al., 2012; Rai et al., 2013). Isoquinoline alkaloids, such as berberine, are active compounds with various biological properties, such as antitumor, anti-inflammatory, antioxidant, and antiulcer effects, especially for treating UC ( Li et al., 2019; Peng et al., 2019; Liao et al., 2020; Li et al., 2022).
The investigation of the pharmacological properties of drugs or medicinal herbals can be time-consuming, expensive, and error prone. Thus, in silico studies have become a valuable alternative to investigate the therapeutic potentials of synthetic and natural compounds (Agu et al., 2023). Network pharmacology is one of the in silico methods used to analyze biological systems. It is based on analyzing the interactions between drugs and target genes and target genes and biological functions and predicting potential signaling pathways (Hopkins, 2008). Several studies have demonstrated the effectiveness of this methodology in predicting drug targets, drug function to specific diseases, and the interaction between protein targets that help in enhancing the effectiveness of the developed compounds and improving clinical trial success rate and lowering development costs (Zhao et al., 2023). Studies have shown that network pharmacology is effective in deciphering how plant extracts or decoctions treat UC (Yang et al., 2022; Qu et al., 2023). Molecular docking is an effective virtual screening method that assesses the interaction between biologically active compounds and the target of interest (Jiao et al., 2021). This method provides insight into the accuracy of the target network through the binding affinity between the compounds and proteins (Yang et al., 2022). Therefore, this study aimed to evaluate the anti-UC effects of C. fenestratum alkaloids (CFAs) using an integrative approach of network pharmacology and molecular docking validation.
MATERIALS AND METHODS
Research overview of C. fenestratum alkaloids for treating ulcerative colitis
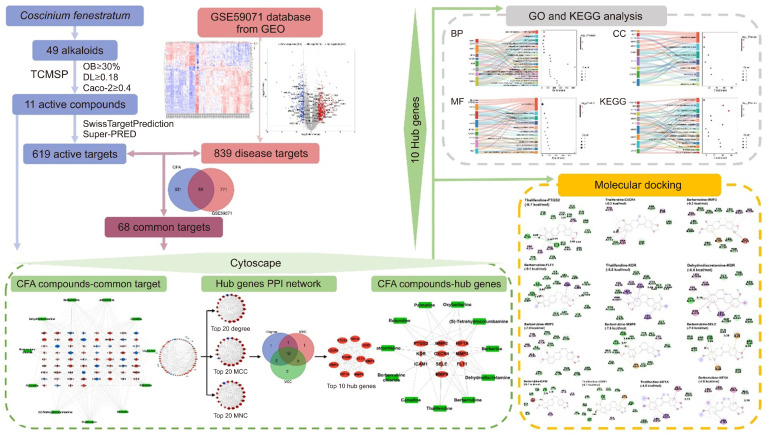
Alkaloid compounds from C. fenestratum obtained from the Indian Medicinal Plants, Phytochemistry And Therapeutics (IMPPAT) database and literature searches were filtered in Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) following the absorption, distribution, metabolism, and excretion (ADME) criteria. The target genes associated with CFA were retrieved from the SwissTargetPrediction and SuperPred databases, and those associated with UC were obtained from the Gene Expression Omnibus (GEO) database. The common targets between CFA and UC were determined to clarify the potential interaction of CFA with UC-related genes. The Limma method was used to identify genes from the GEO dataset to determine those differentially expressed between samples of UC and normal mucosal biopsies. CFA compound target and protein-protein interaction (PPI) networks were constructed. The top 10 hub genes were identified according to the PPI network. Then, the hub genes were enriched following Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Finally, molecular docking analysis was performed to determine the binding affinity between the active alkaloids and key targets. The analysis procedure used is shown as a flowchart (Fig. 1).
Fig. 1.
Flowchart showing the pharmacological mechanisms of CFA against UC. CFA, Coscinium fenestratum alkaloid; OB, oral bioavailability; DL, drug-likeness; UC, ulcerative colitis; TCMSP, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform; BP, biological process; CC, cellular component; MF, molecular function; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction. GEO, Gene Expression Omnibus; MNC, maximum neighborhood component; MCC, maximal clique centrality.
Collection and screening of alkaloid compounds from C. fenestratum and their targets
CFA were obtained from the IMPPAT database (Mohanraj et al., 2018) (https://cb.imsc.res.in/imppat/) and literature research (Malhotra et al., 1989; Pinho et al., 1992; Deevanhxay et al., 2009; Nayak et al., 2012). The ADME properties of CFA were screened from the TCMSP (https://tcmsp-e.com/) (Ru et al., 2014). Compounds that met the criteria of oral bioavailability (OB ≥30%) and drug-likeness (DL ≥0.18) were selected for subsequent analysis (Duan et al., 2023).
The putative human targets of the active compounds in CFA were retrieved from the SuperPred (https://prediction.charite.de/) and the SwissTargetPrediction (http://www.swisstargetprediction.ch/). These are online tools that provide insight into the most probable small molecular protein targets according to the principles of similarity of a given compound (Gfeller et al., 2014; Nickel et al., 2014). The relationship between the CFA active compounds and target genes was analyzed through the construction of the CFA compound-target network using Cytoscape 3.9.1 (National Human Genome Research Institute).
Collection of ulcerative colitis targets and identification of differentially expressed genes
The microarray of the gene expression of “ulcerative colitis” in “Homo sapiens” was acquired from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The gene symbols were obtained by following the annotation information using the Affymetrix GPL6244 platform [GPL6244 (HuGene-1_0-st) Affymetrix Human Gene 1.0 Array {transcript (gene) version}]. The transcriptional profiles of mucosal biopsy from 97 patients with UC and 11 healthy controls were obtained from the GSE59071 dataset. The differentially expressed genes (DEGs) between UC and healthy samples were investigated based on the Limma R package (R Foundation for Statistical Computing) using an online analysis tool, GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r). The missing values were removed to obtain a standardized gene expression matrix. Significant DEGs were defined following a P-value of <0.05 and |logFC|>1 (logFC=log2 fold-change). Subsequently, the DEGs were visualized in a volcano plot.
Determination of ulcerative colitis-specific genes related to C. fenestratum alkaloids
The UC-related genes and CFA compound target genes overlapped in the Venn diagram (https://bioinformatics.psb.ugent.be/webtools/Venn/). The expression of UC-related genes in the dataset was analyzed through a clustering heatmap using online bioinformatics tools (https://www.bioinformatics.com.cn/). The interaction between CFA active compounds and UC-related genes was analyzed by constructing a compound-disease-target (C-D-T) network using Cytoscape 3.9.1.
Determination of protein-protein interaction and screening for the core targets
The common genes between UC genes and CFA targets were input into the STRING database (https://string-db.org/), which contains extensively known human PPIs (Szklarczyk et al., 2021). With a required interaction at 0.400 and other default settings, PPI analysis was set to “Homo sapiens” type of protein. The obtained PPI network was exported to Cytoscape for analysis. The core targets were determined by analyzing the intersection of the top 20 ranked genes following degree, maximum clique centrality, and maximum neighborhood component methods using the CytoHubba plugin (Chin et al., 2014). A network of the core target and CFA active compounds was generated using Cytoscape 3.9.1 to determine the interaction of the key active compounds with the key genes of UC.
Gene Ontology and KEGG enrichment analysis
The functional properties of CFA were investigated through GO and KEGG enrichment analysis using the DAVID database (https://david.ncifcrf.gov/), with a cut-off P-value of 0.05 (Sherman et al., 2022). In brief, the common genes, core genes, and top 10 hub genes were input into the DAVID database to obtain the functional annotation following GO, including biological process (BP), cellular component (CC), and molecular function (MF), and the enriched pathways according to the KEGG database. Using a bioinformatic tool, the top terms of great significance were visualized through Sankey and dot plot (https://www.bioinformatics.com.cn/).
Molecular docking analysis
The structures of the active compounds were obtained from the PubChem database and were converted into the protein data bank, partial charge (Q) and atom type (T) (PDBQT) format using OpenBabel 2.3.1 (Open Babel Development Team). The crystal structures of the hub genes were retrieved from RCSB Protein Data Bank (PDB, https://www.rcsb.org/). Water and ligand molecules were removed from the structure, and the resulting PDB format was saved using the Discovery Studio 2021 software (BIOVIA). The protein was saved in the PDBQT format after adding polar hydrogen and Kollman charges in AutoDock Tools 1.5.7 (Scripps Research Institute). AutoDock Vina-1.1.2 (Scripps Research Institute) was used to perform molecular docking simulations, and binding free energy was calculated using AutoDock Tools version 1.5.7. The resulting binding interactions were visualized using Discovery Studio 2021 software.
RESULTS
Screening C. fenestratum alkaloids and predicting their targets
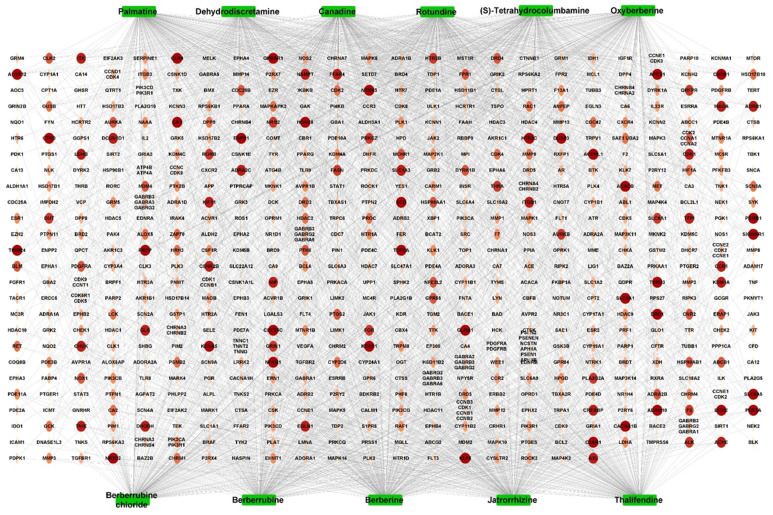
Based on the screening on online databases, including IMPPAT and literature investigation (Malhotra et al., 1989; Pinho et al., 1992; Deevanhxay et al., 2009; Nayak et al., 2012; Rai et al., 2013), 67 active compounds in C. fenestratum were obtained after removing duplicates, of which 49 active compounds were alkaloids. These were further screened on TCMSP based on the ADME screening criteria, OB ≥30%, DL ≥0.18, and Caco-2 permeability ≥0 (Table 1). Finally, 11 CFAs were selected for subsequent analysis. According to SwissTargetPrediction and SuperPred, after deleting duplicates, 619 human genes were obtained. The interaction between CFA and the target genes was analyzed using the compound-target network (Fig. 2). The network contained 2,382 edges and 630 nodes, with 11 green square nodes denoting active compounds and 619 circular nodes indicating target genes. Among the alkaloids, berberine had the strongest interactions with 240 genes, followed by canadine with 234 genes and palmatine with 231 genes. In addition, all 11 alkaloids shared 36 genes as targets, including NFKB1 and KEAP1. This indicates that the active compounds in the plant can synergistically interact with the same target gene, whereas one active compound may interact with several targets. Therefore, CFA may have strong pharmacological effects on inflammatory diseases, such as UC.
Table 1.
ADME characteristics of the active isoquinoline alkaloids in Coscinium fenestratum
| Name | MW | Formula | OB (%) | DL | Caco-2 |
|---|---|---|---|---|---|
| Oxyberberine | 351.38 | C20H17NO5 | 36.68 | 0.82 | 0.97 |
| Thalifendine | 322.36 | C19H16NO4+ | 44.41 | 0.73 | 1.12 |
| Palmatine | 352.44 | C21H22NO4+ | 64.60 | 0.65 | 1.33 |
| Berberine | 336.39 | C20H18NO4+ | 36.86 | 0.78 | 1.24 |
| Berberrubine chloride | 357.80 | C19H16ClNO4 | 35.74 | 0.73 | 1.07 |
| Jatrorrhizine | 338.40 | C20H20NO4+ | 30.44 | 0.75 | 1.38 |
| Canadine | 339.42 | C20H21NO4 | 55.37 | 0.77 | 1.04 |
| Berberrubine | 322.36 | C19H16NO4+ | 35.74 | 0.73 | 1.07 |
| (S)-Tetrahydrocolumbamine | 341.44 | C20H23NO4 | 35.77 | 0.59 | 0.85 |
| Rotundine | 355.47 | C21H25NO4 | 73.94 | 0.64 | 1.00 |
| Dehydrodiscretamine | 324.38 | C19H18NO4+ | 71.29 | 0.54 | 1.10 |
ADME, absorption, distribution, metabolism, and excretion; MW, molecular weight; OB, oral bioavailability; DL, drug-likeness.
Fig. 2.
The active compound-target network of Coscinium fenestratum alkaloid. Green square nodes indicate the 11 active compounds. The ellipse nodes in the orange-red shades indicate the 619 target genes.
C. fenestratum alkaloids target genes related to ulcerative colitis-specific genes
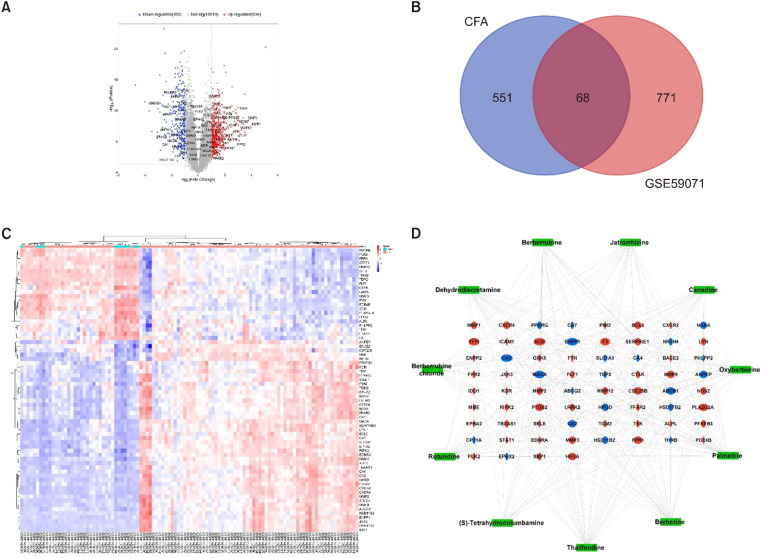
This study evaluated the expression of genes obtained from 108 mucosal biopsy samples, including 97 UC and 11 healthy tissue samples, in the GSE59071 dataset. The DEGs were then identified using GEO2R. Based on the cut-off parameters, 534 upregulated genes and 305 downregulated genes were identified. The volcano map indicated that the DEGs had a normal distribution in the disease samples. Significantly expressed genes were highlighted in the map (Fig. 3A). The intersection between 619 CFA-related genes and 839 UC-related genes revealed 68 common genes that were designated as UC-specific CFA target genes (Fig. 3B). Accordingly, CFA could modulate 47 upregulated genes related to UC, such as prostaglandin-endoperoxide synthase 2 (PTGS2), matrix metalloproteinase (MMP)-1, MMP2, MMP3, MMP9, C-X-C chemokine receptor (CXCR)2, and CXCR4, and 21 downregulated genes related to UC, such as peroxisome proliferator activated receptor gamma (PPARG), alanyl aminopeptidase, membrane (ANPEP), ATP binding cassette subfamily B member 1 (ABCB1), nuclear receptor subfamily 1 group H member 4 (NR1H4), 15-hydroxyprostaglandin dehydrogenase (HPGD), and ATP binding cassette subfamily G member 2 (ABCG2) (Supplementary Table 1 and Fig. 3C).
Fig. 3.
Screening of CFA-UC common targets. (A) Volcano plot of gene expression data (GSE59071). Red and blue indicate upregulated genes (logFC ≥1) and downregulated genes (logFC ≤—1), respectively, whereas those in gray show no significant difference. (B) Venn diagram of the intersection of potential targets of CFA and DEGs in UC-related genes in the GSE59071 dataset. (C) The heatmap represents the 68 genes in UC-related to dataset GSE59071. Red and blue indicate up- and downregulated genes, respectively. Among them, UC tissues expressed 41 up- and 21 downregulated genes. (D) CFA active compound-disease-target network. The 68 common targets are represented by red ellipses for upregulated genes and blue ellipses for downregulated genes. The green square nodes indicate the 11 alkaloid compounds. CFA, Coscinium fenestratum alkaloid; UC, ulcerative colitis; DEGs, differentially expressed genes.
Construction of the compound-disease-target and protein-protein interaction networks and screening of core targets
To evaluate the involvement of CFA active compounds in modulating UC, a C-D-T network was constructed using Cytoscape 3.9.1 (Fig. 3D). The network contained 79 nodes, including 11 green square nodes of active compounds, 47 red circular nodes of upregulated genes, and 21 blue circular nodes of downregulated genes, which could interact through 271 edges. Five compounds in this network, namely thalifendine, berberine, palmitine, canadine, and oxyberberine, had >24.63 degrees (average degree of the compound-nodes) and can be considered key compounds in CFA.
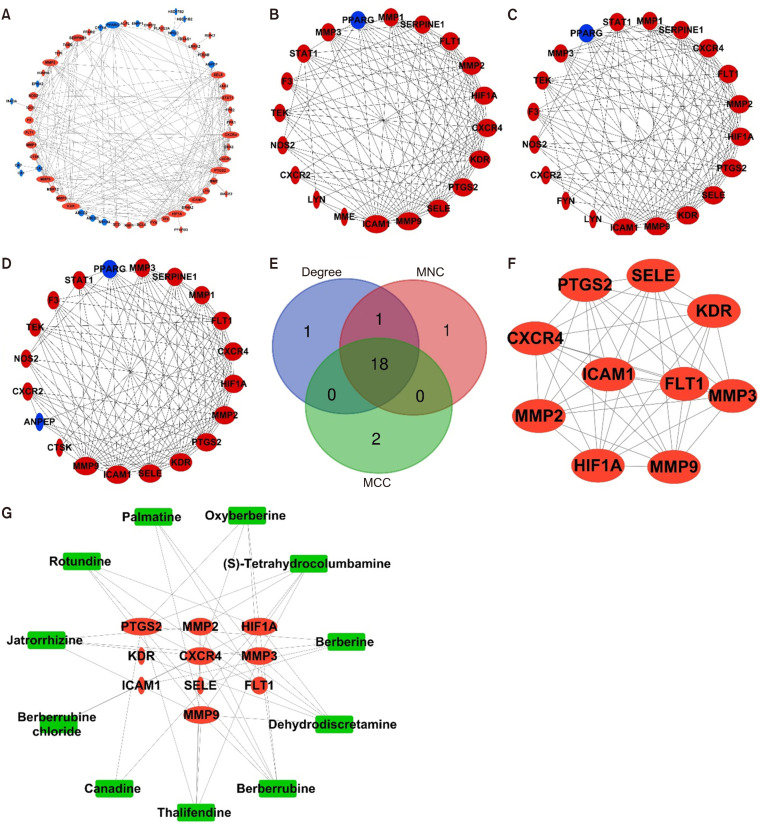
The common genes were uploaded into the STRING database to obtain the PPI to understand the mechanisms by which CFA modulate UC, and the PPI network was constructed and analyzed using Cytoscape. After the disconnected nodes were hidden, the PPI network exhibited interactions between 59 proteins through 220 edges (Fig. 4A). The PPI network was then analyzed using a network analyzer, where node sizes varied according to the degree value. In total, 18 nodes had degree values higher than the average of 7.458 (Supplementary Table 2). Therefore, CFA could modulate UC by targeting these genes. Using the CytoHubba plugin through degree, maximum neighborhood component, and maximum clique centrality methods helped identify the top 20 hub genes in the network (Fig. 4B-4D). By accounting for the intersection of these genes from the three methods, 18 genes were detected and the top 10 highly connected nodes between these genes were identified as hub genes, comprising PTGS2, E-selectin (SELE), kinase insert domain receptor (KDR), fms-related tyrosine kinase 1 (FLT1), intercellular adhesion molecule-1 (ICAM-1), CXCR4, hypoxia inducible factor 1 subunit alpha (HIF1A), MMP2, MMP3, and MMP9 (Fig. 4E and 4F). The 10 hub genes were connected through 44 edges, with an average node degree of 8.80. After identifying the active compounds related to the hub genes, a compound-hub gene-disease network was constructed (Supplementary Table 3, Fig. 4G). Therefore, out of the 11 active compounds in CFA, dehydrodiscretamine, berberrubine, thalifendine, and (S)-tetrahydrocolumbamine were identified as key compounds that modulate UC, because their degree values surpassed the network mean of 4.286. PTGS2 was highly connected to the compounds, suggesting that it can be a key target of CFA in modulating UC.
Fig. 4.
Protein-protein interaction (PPI) networks. (A) A PPI network between the 59 genes after hiding the disconnected genes. Larger size represents a higher degree of confidence. The top 20 gene networks of Coscinium fenestratum alkaloid (CFA) against ulcerative colitis (UC) are evaluated by degree of nodes (B), maximum neighborhood component (MNC) (C), and maximal clique centrality (MCC), where up- and downregulated genes (D) are represented by red and blue, respectively. Identification of the hub genes modulated by CFA. (E) Venn diagram illustrating the intersections of the three methods and (F) PPI network of the overlapping genes identified by the degree method. (G) CFA active compound-hub gene network. The red ellipses indicate the top hub genes, which all belong to the upregulated genes, and the green square nodes indicate the active compounds. PTGS2, prostaglandin-endoperoxide synthase 2; CXCR4, C-X-C motif chemokine receptor 4; MMP, matrix metallopeptidase; FLT1, fms-related receptor tyrosine kinase 1; KDR, kinase insert domain receptor; SELE, selectin E; ICAM1, intercellular adhesion molecule 1; HIF1A, hypoxia-inducible factor 1.
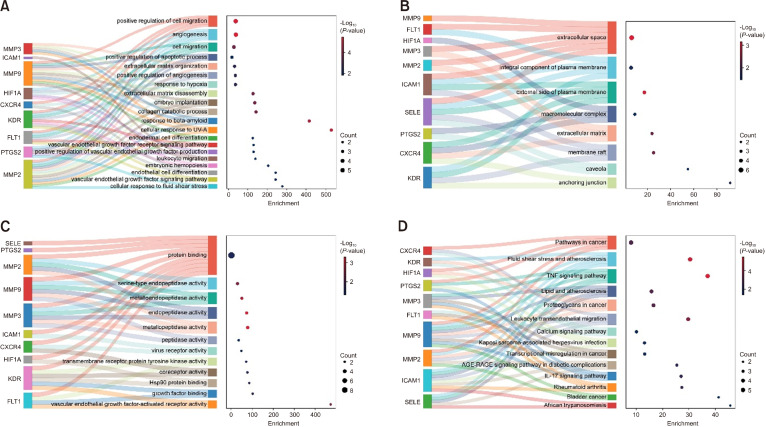
Functional properties of C. fenestratum alkaloid identified through Gene Ontology and KEGG enrichment analysis
To investigate the functional properties according to GO functions and KEGG pathways, the 10 hub genes of CFA were enriched using the DAVID database. A total of 66 GO terms, including 46 BP, 8 CC, and 12 MF, were significantly enriched (P<0.05). The Sankey and dot plot showed the top 20 BP, 8 CC, and 12 MF terms (Fig. 5A-5C). According to the enrichment analysis, the hub genes were functionally associated with the immunological response, cell migration, and cell response to various stimuli, which included the positive regulation of cell migration, angiogenesis, extracellular matrix organization and disassembly, and collagen catabolism (Fig. 5A). According to the CC terms, these processes may primarily take place in the extracellular space and matrix as well as in both the integral component and the external side of the plasma membrane (Fig. 5B). MFs were mostly related to the binding of proteins and growth factors as well as the activities of serine-type endopeptidase, metalloendopeptidase, endopeptidase, metallopeptidase, and peptidase (Fig. 5C).
Fig. 5.
Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways significantly enriched by the hub genes. (A) The top 20 GO-biological process terms. (B) The eight significant GO-cellular component terms. (C) The 12 noteworthy GO-molecular function terms. (D) The most important 14-KEGG signaling pathways. MMP, matrix metallopeptidase; FLT1, fms-related receptor tyrosine kinase 1; HIF1A, hypoxia-inducible factor 1; ICAM1, intercellular adhesion molecule 1; SELE, selectin E; PTGS2, prostaglandin-endoperoxide synthase 2; CXCR4, C-X-C motif chemokine receptor 4; KDR, kinase insert domain receptor.
The 10 key targets were significantly enriched in 14 KEGG pathways, which were primarily associated with the inflammatory response and onset of diseases (Fig. 5D). The KEGG enrichment pathways included the tumor necrosis factor (TNF) signaling pathway, interleukin (IL)-17 signaling pathway, leukocyte transendothelial migration, fluid shear stress and atherosclerosis, lipid and atherosclerosis, advanced glycation end-products (AGE)-receptor for AGE (RAGE) signaling pathway in diabetic complications, and rheumatoid arthritis. The analysis revealed that PTCGS2, MMP3, MMP9, MMP2, ICAM1, and CXCR4 were enriched in the TNF signaling pathway, IL-17 signaling pathway, and leukocyte transendothelial migration, which are crucial in UC pathogenesis.
Molecular docking analysis
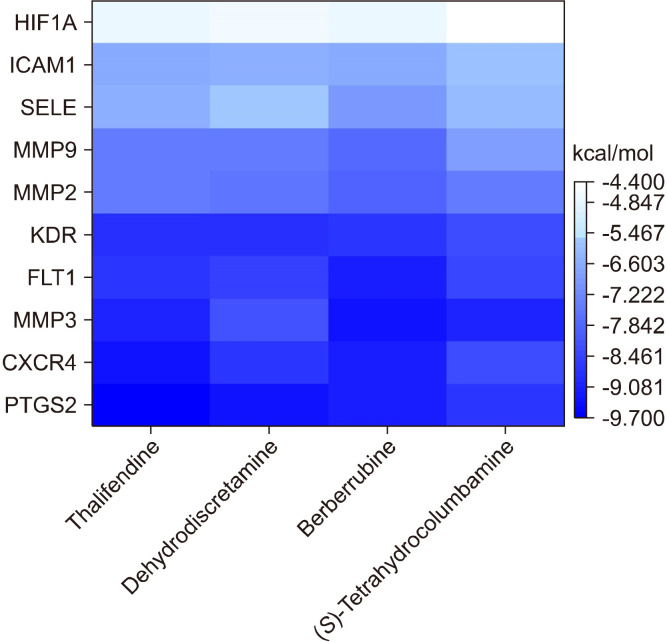
The hub gene-compound network showed that dehydrodiscretamine, berberrubine, thalifendine, and (S)-tetrahydrocolumbamine were highly connected with the hub genes (Fig. 4G). Therefore, the binding potential of these key active compounds with the hub genes was evaluated using molecular docking simulation. The heatmap shows the negative binding energies between the key active compounds in CFA and the hub genes (Fig. 6). Most of the active compounds had a goof affinity with hub genes, as evidenced by binding energies of <−5 kcal/mol. In total, 28/40 docking results exhibited binding energies of −7 kcal/mol. The active compounds, including dehydrodiscretamine, berberrubine, thalifendine, and (S)-tetrahydrocolumbamine, exhibited strong binding activities and stable conformations with PTGS2, CXCR4, MMP3, FLT1, and KDR, with energies of <−8 kcal/mol. The binding poses of the representative docking results are illustrated in Fig. 7.
Fig. 6.
Heatmap representing the binding energies (kcal/mol) between the key active alkaloids of Coscinium fenestratum alkaloid and the top 10 hub genes. Light to dark blue indicates high to small values of the binding energy according to the AutoDock Vina scores, respectively. HIF1A, hypoxia-inducible factor 1; ICAM1, intercellular adhesion molecule 1; SELE, selectin E; MMP, matrix metallopeptidase; KDR, kinase insert domain receptor; FLT1, fms-related receptor tyrosine kinase 1;
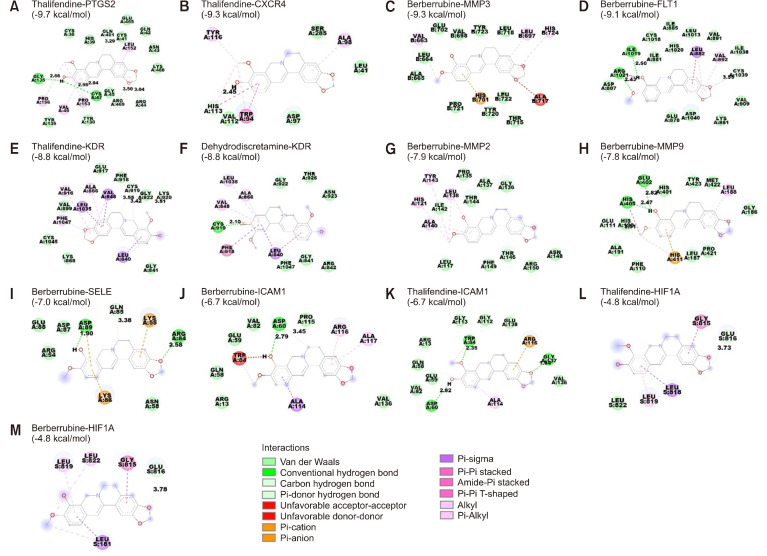
Fig. 7.
Representation of the docking poses between the active compounds and hub genes. The dashed lines in green indicate the interaction through hydrogen bonding, orange indicates Pi-cation or Pi-anion interactions, purple indicates Pi-sigma interactions, and pink indicates hydrophobic interactions, including alkyl, Pi-alkyl, and amide-Pi-stacked. Interactions between (A) thalifendine and PTGS2, (B) thalifendine and CXCR4, (C) berberrubine and MMP3, (D) berberrubine and FLT1, (E) thalifendine and KDR, (F) dehydrodiscretamine and KDR, (G) berberrubine and MMP2, (H) berberrubine and MMP9, (I) berberrubine and SELE, (J) berberrubine and ICAM1, (K) thalifendine and ICAM1, (L) thalifendine and HIF1A, and (M) berberrubine and HIF1A. PTGS2, prostaglandin-endoperoxide synthase 2; CXCR4, C-X-C motif chemokine receptor 4; MMP, matrix metallopeptidase; FLT1, fms-related receptor tyrosine kinase 1; KDR, kinase insert domain receptor; SELE, selectin E; ICAM1, intercellular adhesion molecule 1; HIF1A, hypoxia-inducible factor 1.
Thalifendine exhibited the strongest binding affinity with PTGS2 (−9.7 kcal/mol) through five hydrogen bonds with Cys47, Arg44, Gly135, Gln461, and Arg469, in addition to alkyl, Pi-alkyl, and Van der Waals interactions with other residues (Fig. 7A). Thalifendine had the strongest affinity with CXCR4 (−9.3 kcal/mol) resulting from the hydrogen bond, Pi-Pi stacked, and Pi-alkyl interactions with Trp94, Ala98, Val112, His113, and Tyr116 as well as Van der Waals forces with other residues (Fig. 7B). Thalifendine had the highest binding affinity with KDR (−8.8 kcal/mol), as seen from the hydrogen bond, Pi-sigma, alkyl, and Pi-alkyl interactions formed with the residues at the ligand-binding pocket of KRD, along with Van der Waals interaction formed with other residues (Fig. 7E). Thalifendine demonstrated a relatively strong binding conformation with ICAM1 (−6.7 kcal/mol) but showed weaker binding affinity with HIF1A (−4.8 kcal/mol) (Fig. 7K and 7L). Similarly, dehydrodiscretamine bound strongly to KDR (−8.8 kcal/mol) through one hydrogen bond with Cys919; two Pi-Pi stacked bonds with Leu840 and Phe918; three Pi-alkyl interactions with Val848, Ala866, and Leu1035; and Van der Waals forces with the other residues (Fig. 7F). These findings suggest that thalifendine is a prominent modulator of PTGS2, CXCR4, and KDR, and dehydrodiscretamine could be a potential regulator of KDR.
Berberrubine exhibited the most stable binding to MMP3 (−9.3 kcal/mol) and FLT1 (−9.1 kcal/mol), and the interactions between berberrubine and the residues at the ligand-binding pockets included hydrogen bond, Pi-sigma, Pi-alkyl, alkyl, Pi-Pi stacked, Pi-cation, and Van der Waals interactions (Fig. 7C and 7D). The strongest interactions were between berberrubine and MMP2 (−7.9 kcal/mol), MMP9 (−7.8 kcal/mol), SELE (−7.0 kcal/mol), ICAM1 (−6.7 kcal/mol), and HIF1A (−4.8 kcal/mol) (Fig. 7G-7J and 7M). These interactions may be caused by different bonds, such as hydrogen bond, Pi-cation, Pi-anion, Pi-sigma, Pi-Pi T-shaped, Pi-alkyl, and Van der Waals. (S)-tetrahydrocolumbamine displayed good binding with PTGS2, CXCR4, MMP3, FLT1, and KDR, with binding energies of <−8.0 kcal/mol (Supplementary Table 4).
Taken together, the results suggest that CFA can alleviate UC by targeting PTGS2, CXCR4, FLT1, KDR, MMP2, MMP3, and MMP9 through the synergistic mechanisms of thalifendine, berberrubine, dehydrodiscretamine, and (S)-tetrahydrocolumbamine.
DISCUSSION
UC is chronic inflammation of the mucosa and submucosa in the lower part of the gastrointestinal tract, particularly the colon and rectum. Although many factors, including genetics, diet, and environment, contribute to the development of UC, its etiology is unclear. Therefore, treating UC is challenging due to its occurrence of remission and recurrence. Various drugs are used to treat UC, including corticosteroids, aminosalicylate, immunosuppressors, and biologic agents. However, such drugs are ineffective for radical treatment and when used for long term, trigger side effects (Gupta et al., 2022; Li et al., 2022; Yang et al., 2022). Therefore, the interest in finding natural alternatives to treat UC has increased (Gupta et al., 2022). A growing body of research has shown that medicinal plants, which contain various secondary metabolites, such as polyphenols, flavonoids, and alkaloids, with antioxidant and anti-inflammatory properties, might exert protective effects against UC (Peng et al., 2019; Gupta et al., 2022; Li et al., 2022). Alkaloids, a large class of nitrogen-containing organic compounds, are considered prominent regulators of UC due to their anticancer, anti-inflammatory, and immunoregulatory potential (Peng et al., 2019; Li et al., 2022). In addition, alkaloids are the characteristic phytochemicals of C. fenestratum, a medicinal plant used to treat diarrhea, inflammation, and ulcers in Southeast Asia (Deevanhxay et al., 2009; Rai et al., 2013). This study investigated the protective mechanisms of CFA against UC using an integrative approach of network pharmacology and molecular docking validation.
Of the 49 alkaloids gathered from the IMPPAT database and the literature (Malhotra et al., 1989; Pinho et al., 1992; Deevanhxay et al., 2009; Karthika et al., 2019), 11 belonging to the isoquinoline alkaloid group were identified as active compounds with regard to the OB, DL, and Caco-2 permeability in the TCMSP filter. Thalifendine, berberrubine, dehydrodiscretamine, and (S)-tetrahydrocolumbamine were significant in the network due to the interactions between these alkaloids and the 10 UC-related hub genes. Good binding affinity of these compounds with key targets was demonstrated by molecular docking analysis. These compounds belong to the isoquinoline alkaloid group, a group of active alkaloid derivatives of berberine, which have a broad spectrum of biological activities, such as anti-inflammatory, antioxidant, antitumor, hepatoprotective, and antimicrobial properties (Peng et al., 2019; Zhang et al., 2019; Gaba et al., 2021). Accumulating evidence has shown that berberine and its derivative alkaloids exert anti-inflammatory effects by inhibiting nuclear factor kappa B (NF-κB) signaling pathway and inflammatory mediators, such as cyclooxygenase-2 (COX-2), TNF-α, and ILs (Gaba et al., 2021). Among these berberine alkaloids, berberrubine has been extensively studied for its anti-inflammatory properties in a mouse model of colitis (Li et al., 2022). Berberrubine exerts a curative effect on UC in dextran sulfate sodium-induced mice model, as evidenced by its inhibitory effect on inflammatory cytokines, including TNF-α, IL-1β, IL-6, and interferon-gamma, as well as on myeloperoxidase activity, and by its enhancement of tight junction proteins and mucins (Peng et al., 2019; Li et al., 2022). Although thalifendine, dehydrodiscretamine, and (S)-tetrahydrocolumbamine are characteristic berberine alkaloids in C. fenestratum, there is little information on their biological activities, particularly in modulating inflammatory-associated diseases, such as UC. Through the biological properties ascertained for these alkaloids, CFA could potentially modulate UC.
According to network pharmacology analysis, CFA modulated 68 UC-related target genes. The most interactive genes among targets were evaluated through the PPI network, followed by degree, maximum clique centrality, and maximum neighborhood component method calculation in CytoHubba. Therefore, the top 10 UC-related hub genes were identified. These included PTGS2, SELE, KDR, FLT1, ICAM1, CXCR4, HIF1A, MMP2, MMP3, and MMP9. In the network analysis, PTGS2, also known as COX-2, was targeted by eight alkaloids. COX-2 is an inducible form of COX that converts arachidonic acid into prostaglandins, prostacyclin, and thromboxane, which act as mediators of inflammation (Li et al., 2018; Agoff et al., 2000). Several studies have established that the overexpression of COX-2 in the colonic mucosa is linked to the early onset of inflammation in UC (Agoff et al., 2000). Therefore, inhibiting COX-2 is an effective strategy for managing UC (Spisni et al., 2015). COX-2 was shown to be involved in angiogenesis during UC by activating the epidermal growth factor receptor and stimulating the expression of FLT1 and KDR through the action of prostaglandin E2 (Li et al., 2021). Both FLT1 and KDR are receptors involved in the growth of endothelial cells by vascular endothelial growth factor in UC. Although both KDR and FLT1 are expressed during the active phase of UC, FLT1 is responsible for the formation of new vessels in inflamed tissues in UC, which results in disease severity (Frysz-Naglak et al., 2011; Li et al., 2021). FLT1 has been implicated in the activation of MMPs (Konno et al., 2004). MMPs are a family of zinc-dependent endopeptidases that cause damage to the colonic mucosa matrix and inflammation when overexpressed and activated (Konno et al., 2004; Yang et al., 2022). MMP2 and MMP9, which are gelatinase MMPs, have been associated with the acute phase of UC and the stimulation of neutrophil migration (O’Sullivan et al., 2015; He et al., 2023). MMP3, which belongs to stromelysin MMP, plays an important role in the onset of UC because it is an effector activator of MMP9 and is involved in TNF-α-induced intestinal damage (Yang et al., 2022; He et al., 2023). MMPs exert a wide range of biological functions through the regulation and alteration of several cytokines, chemokines, receptors, proteases, and adhesion molecules (O’Sullivan et al., 2015). SELE, also known as endothelial leukocyte adhesion molecule-1, and ICAM1 are cell surface adhesion molecules (CAMs) that are induced by inflammatory cytokines, including IL-1β, TNF, and interferon-gamma, as well as bacterial endotoxins (e.g., lipopolysaccharide) (Lazaris et al., 1999). On the one hand, SELE, an inducible CAM, is markedly expressed during the initial stage of acute UC by recruiting neutrophils to the site of inflammation (Lazaris et al., 1999; Murahashi et al., 2022). On the other hand, ICAM1 is highly expressed in chronic inflammation as a result of its infiltration into lymphoplasmacytic areas, which may contribute to the persistence of inflammatory disorders (Lazaris et al., 1999). CXCR4 functions as a receptor for the C-X-C motif chemokine ligand (CXCL12). Growing data suggests that CXCR4/CXCL12 plays a pathogenic role in UC by attracting lymphocytes and mononuclear cells to inflamed colon tissue (Xia et al., 2010). The blockade of CXRC4 in mice delayed the progression of dextran sulfate sodium-induced colitis (Lin et al., 2017; Xia et al., 2010). Hypoxia-inducible factor 1α (HIF-1α) is a hypoxia-regulated transcriptional factor that mainly controls the gene expression responsible for angiogenesis, an integrated mechanism in the tissue regeneration of UC (Giatromanolaki et al., 2003). HIF-1α has several functions in UC development, depending on the site of action. In T cells, HIF-1α stimulates the differentiation of Th17 cells, which results in the release of IL-17. HIF-1α can directly induce the transcription of proinflammatory cytokines, such as IL-1β, in activated macrophages via toll-like receptor 4 (TLR4) signaling (Giatromanolaki et al., 2003; Yin et al., 2022). Our study indicated that CFA might exhibit protective effects against colitis by targeting these genes. In addition, molecular docking analysis demonstrated strong binding affinities of the active compounds in CFA with the hub genes, which supported CFA activity.
KEGG analysis highlighted that CFA compounds can interfere with the immune response, inflammatory reaction, and disease processes. The two most important pathways of UC pathogenesis-IL-17 and TNF signaling pathways-may in fact be regulated by CFA (Shou et al., 2022). IL-17 is a proinflammatory cytokine that is widely distributed and mainly released by differentiated Th17 cells (Monin and Gaffen, 2018; Shou et al., 2022). Its overexpression in the mucosa and serum has been associated with UC progression (Monin and Gaffen, 2018; Yang et al., 2022). IL-17 amplifies the inflammatory response in UC by triggering chemokines, such as CXCL2, and maintaining the release of other inflammatory mediators, including IL-6 and TNF-α, by triggering mitogen-activated protein kinases (MAPKs) and NF-κB signaling pathways (Noviello et al., 2021). TNF is a proinflammatory cytokine that orchestrates inflammatory pathways in chronic intestinal inflammation (Yang et al., 2022). TNF-α is involved in the pathogenesis of UC by stimulating the production of several cytokines and chemokines, prostaglandin E2, and reactive oxygen species (Oliveira et al., 2021). By binding to its receptors, comprising tumor necrosis factor receptor (TNFR)1 and TNFR2, TNF-α stimulated the activation of key immunoregulatory pathways, such as NF-κB and MAPKs, which resulted in the persistence of the inflammatory conditions, such as those observed in UC (Liu, 2005; Sabio and Davis, 2014). NF-κB is a transcription factor that controls innate immunity and inflammation (Liu et al., 2017), whereas MAPKs are a group of serine/threonine protein kinases that play a role in intracellular signaling pathways that regulate inflammation (Kaminska, 2005). Taken together, CFA may effectively inhibit UC by modulating inflammation-related signaling.
In conclusion, this study is the first to investigate the molecular mechanisms of CFA in preventing colitis using an integrative approach of network pharmacology and molecular docking analysis. Our study showed that the key active alkaloids in C. fenestratum, consisting of thalifendine, berberrubine, dehydrodiscretamine, and (S)-tetrahydrocolumbamine, can alleviate UC. The potential therapeutic targets of CFA in UC are PTGS2, SELE, KDR, FLT1, ICAM1, CXCR4, HIF1A, MMP2, MMP3, and MMP9. The anti-UC effects of CFA may be brought about by modulating pathways related to inflammation and the immune system. Insight into a predictive mechanism is provided by the network pharmacology approach, but further experimental verification is needed to confirm the biological activity of CFA.
SUPPLEMENTARY MATERIALS
Supplementary materials can be found via https://doi.org/10.3746/pnf.2024.29.4.441
Footnotes
FUNDING
This work was support in part by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2021R1A2C2006745).
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Concept and design: VLT. Methodology: VLT. Supervision: VLT. Data collection: JWP, RHGR. Data analysis: JWP, RHGR. Investigation: JWP, RHGR. Formal analysis: JWP, RHGR. Writing the article: JWP, RHGR. Critical revision of the article: VLT. Project administration: WSJ. Obtained funding: WSJ. Final approval of the article: all authors. Overall responsibility: WSJ.
REFERENCES
- Agoff SN, Brentnall TA, Crispin DA, Taylor SL, Raaka S, Haggitt RC, et al. The role of cyclooxygenase 2 in ulcerative colitis-associated neoplasia. Am J Pathol. 2000;157:737–745. doi: 10.1016/S0002-9440(10)64587-7. https://doi.org/10.1016/s0002-9440(10)64587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agu PC, Afiukwa CA, Orji OU, Ezeh EM, Ofoke IH, Ogbu CO, et al. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci Rep. 2023;13:13398. doi: 10.1038/s41598-023-40160-2. https://doi.org/10.1038/s41598-023-40160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. doi: 10.1186/1752-0509-8-S4-S11. https://doi.org/10.1186/1752-0509-8-s4-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deevanhxay P, Suzuki M, Maeshibu N, Li H, Tanaka K, Hirose S. Simultaneous characterization of quaternary alkaloids, 8-oxoprotoberberine alkaloids, and a steroid compound in Coscinium fenestratum by liquid chromatography hybrid ion trap time-of-flight mass spectrometry. J Pharm Biomed Anal. 2009;50:413–425. doi: 10.1016/j.jpba.2009.05.023. https://doi.org/10.1016/j.jpba.2009.05.023. [DOI] [PubMed] [Google Scholar]
- Duan ZL, Wang YJ, Lu ZH, Tian L, Xia ZQ, Wang KL, et al. Wumei Wan attenuates angiogenesis and inflammation by modulating RAGE signaling pathway in IBD: Network pharmacology analysis and experimental evidence. Phytomedicine. 2023;111:154658. doi: 10.1016/j.phymed.2023.154658. https://doi.org/10.1016/j.phymed.2023.154658. [DOI] [PubMed] [Google Scholar]
- Frysz-Naglak D, Fryc B, Klimacka-Nawrot E, Mazurek U, Suchecka W, Kajor M, et al. Expression, localization and systemic concentration of vascular endothelial growth factor (VEGF) and its receptors in patients with ulcerative colitis. Int Immunopharmacol. 2011;11:220–225. doi: 10.1016/j.intimp.2010.11.023. https://doi.org/10.1016/j.intimp.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Gaba S, Saini A, Singh G, Monga V. An insight into the medicinal attributes of berberine derivatives: A review. Bioorg Med Chem. 2021;38:116143. doi: 10.1016/j.bmc.2021.116143. https://doi.org/10.1016/j.bmc.2021.116143. [DOI] [PubMed] [Google Scholar]
- Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–W38. doi: 10.1093/nar/gku293. https://doi.org/10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giatromanolaki A, Sivridis E, Maltezos E, Papazoglou D, Simopoulos C, Gatter KC, et al. Hypoxia inducible factor 1α and 2α overexpression in inflammatory bowel disease. J Clin Pathol. 2003;56:209–213. doi: 10.1136/jcp.56.3.209. https://doi.org/10.1136/jcp.56.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Mishra V, Gulati M, Kapoor B, Kaur A, Gupta R, et al. Natural compounds as safe therapeutic options for ulcerative colitis. Inflammopharmacology. 2022;30:397–434. doi: 10.1007/s10787-022-00931-1. https://doi.org/10.1007/s10787-022-00931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Kang Q, Chan KI, Zhang Y, Zhong Z, Tan W. The immunomodulatory role of matrix metalloproteinases in colitis-associated cancer. Front Immunol. 2023;13:1093990. doi: 10.3389/fimmu.2022.1093990. https://doi.org/10.3389/fimmu.2022.1093990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. https://doi.org/10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Jiao X, Jin X, Ma Y, Yang Y, Li J, Liang L, et al. A comprehensive application: Molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Comput Biol Chem. 2021;90:107402. doi: 10.1016/j.compbiolchem.2020.107402. https://doi.org/10.1016/j.compbiolchem.2020.107402. [DOI] [PubMed] [Google Scholar]
- Kaminska B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy-from molecular mechanisms to therapeutic benefits. Biochim Biophys Acta. 2005;1754:253–262. doi: 10.1016/j.bbapap.2005.08.017. https://doi.org/10.1016/j.bbapap.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Karthika K, Gargi G, Jamuna S, Paulsamy S, Ajmal Ali M, Al-Hemaid F, et al. The potential of antioxidant activity of methanolic extract of Coscinium fenestratum (Goetgh.) Colebr (Menispermaceae) Saudi J Biol Sci. 2019;26:1037–1042. doi: 10.1016/j.sjbs.2018.08.010. https://doi.org/10.1016/j.sjbs.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno S, Iizuka M, Yukawa M, Sasaki K, Sato A, Horie Y, et al. Altered expression of angiogenic factors in the VEGF-Ets-1 cascades in inflammatory bowel disease. J Gastroenterol. 2004;39:931–939. doi: 10.1007/s00535-004-1423-9. https://doi.org/10.1007/s00535-004-1423-9. [DOI] [PubMed] [Google Scholar]
- Lazaris AC, Dicoglou C, Tseleni-Balafouta S, Paraskevakou H, Davaris PS. In situ expression of E-selectin and intercellular adhesion molecule-1 in chronic inflammatory diseases of the gastrointestinal tract. APMIS. 1999;107:819–827. doi: 10.1111/j.1699-0463.1999.tb01477.x. https://doi.org/10.1111/j.1699-0463.1999.tb01477.x. [DOI] [PubMed] [Google Scholar]
- Li C, Wang J, Ma R, Li L, Wu W, Cai D, et al. Natural-derived alkaloids exhibit great potential in the treatment of ulcerative colitis. Pharmacol Res. 2022;175:105972. doi: 10.1016/j.phrs.2021.105972. https://doi.org/10.1016/j.phrs.2021.105972. [DOI] [PubMed] [Google Scholar]
- Li MY, Li MX, Xu N, Li ZH, Zhang YM, Gan YX, et al. Effects of Huangqin Decoction on ulcerative colitis by targeting estrogen receptor alpha and ameliorating endothelial dysfunction based on system pharmacology. J Ethnopharmacol. 2021;271:113886. doi: 10.1016/j.jep.2021.113886. https://doi.org/10.1016/j.jep.2021.113886. [DOI] [PubMed] [Google Scholar]
- Li Y, Soendergaard C, Bergenheim FH, Aronoff DM, Milne G, Riis LB, et al. COX-2-PGE2 signaling impairs intestinal epithelial regeneration and associates with TNF inhibitor responsiveness in ulcerative colitis. EBioMedicine. 2018;36:497–507. doi: 10.1016/j.ebiom.2018.08.040. https://doi.org/10.1016/j.ebiom.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Sun W, Zhou BJ, Rosenstein A, Zhao J, Wang J, et al. iTRAQ-based pharmacoproteomics reveals potential targets of berberine, a promising therapy for ulcerative colitis. Eur J Pharmacol. 2019;850:167–179. doi: 10.1016/j.ejphar.2019.02.021. https://doi.org/10.1016/j.ejphar.2019.02.021. [DOI] [PubMed] [Google Scholar]
- Liao Z, Xie Y, Zhou B, Zou B, Xiao D, Liu W, et al. Berberine ameliorates colonic damage accompanied with the modulation of dysfunctional bacteria and functions in ulcerative colitis rats. Appl Microbiol Biotechnol. 2020;104:1737–1749. doi: 10.1007/s00253-019-10307-1. https://doi.org/10.1007/s00253-019-10307-1. [DOI] [PubMed] [Google Scholar]
- Lin X, Wang H, Li Y, Yang J, Yang R, Wei D, et al. Functional characterization of CXCR4 in mediating the expression of protein C system in experimental ulcerative colitis. Am J Transl Res. 2017;9:4821–4835. [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Sig Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. https://doi.org/10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZG. Molecular mechanism of TNF signaling and beyond. Cell Res. 2005;15:24–27. doi: 10.1038/sj.cr.7290259. https://doi.org/10.1038/sj.cr.7290259. [DOI] [PubMed] [Google Scholar]
- Malhotra S, Taneja SC, Dhar KL. Minor alkaloid from Coscinium fenestratum. Phytochemistry. 1989;28:1998–1999. doi: 10.1016/S0031-9422(00)97910-X. https://doi.org/10.1016/S0031-9422(00)97910-X. [DOI] [Google Scholar]
- Mohanraj K, Karthikeyan BS, Vivek-Ananth RP, Chand RPB, Aparna SR, Mangalapandi P, et al. IMPPAT: A curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci Rep. 2018;8:4329. doi: 10.1038/s41598-018-22631-z. https://doi.org/10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monin L, Gaffen SL. Interleukin 17 family cytokines: signaling mechanisms, biological activities, and therapeutic implications. Cold Spring Harb Perspect Biol. 2018;10:a028522. doi: 10.1101/cshperspect.a028522. https://doi.org/10.1101/cshperspect.a028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murahashi M, Kogami A, Muramoto A, Hoshino H, Akama TO, Mitoma J, et al. Vascular E-selectin expression detected in formalin-fixed, paraffin-embedded sections with an E-selectin monoclonal antibody correlates with ulcerative colitis activity. J Histochem Cytochem. 2022;70:299–310. doi: 10.1369/00221554221085336. https://doi.org/10.1369/00221554221085336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Sajini RJ, Padmavati D, Karthik R. Chemistry and medicinal properties of Coscinium fenestratum (Gaertn.) colebr (tree turmeric) Res J Pharm Technol. 2012;5:198–202. [Google Scholar]
- Nickel J, Gohlke BO, Erehman J, Banerjee P, Rong WW, Goede A, et al. SuperPred: update on drug classification and target prediction. Nucleic Acids Res. 2014;42(Web Server issue):W26–W31. doi: 10.1093/nar/gku477. https://doi.org/10.1093/nar/gku477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noviello D, Mager R, Roda G, Borroni RG, Fiorino G, Vetrano S. The IL23-IL17 immune axis in the treatment of ulcerative colitis: Successes, defeats, and ongoing challenges. Front Immunol. 2021;12:611256. doi: 10.3389/fimmu.2021.611256. https://doi.org/10.3389/fimmu.2021.611256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okechukwu PN, Ndyeabura AW, Chiang CN, Akowuah GA. Effect of standardized extract of Cosinium fenestratum stem bark on liver and kidney function parameters in streptozotocin-induced diabetic rats. J Acute Dis. 2013;2:201–206. doi: 10.1016/S2221-6189(13)60127-4. https://doi.org/10.1016/S2221-6189(13)60127-4. [DOI] [Google Scholar]
- Oliveira RG, Damazo AS, Antonielli LF, Miyajima F, Pavan E, Duckworth CA, et al. Dilodendron bipinnatum Radlk. extract alleviates ulcerative colitis induced by TNBS in rats by reducing inflammatory cell infiltration, TNF-α and IL-1β concentrations, IL-17 and COX-2 expressions, supporting mucus production and promotes an antioxidant effect. J Ethnopharmacol. 2021;269:113735. doi: 10.1016/j.jep.2020.113735. https://doi.org/10.1016/j.jep.2020.113735. [DOI] [PubMed] [Google Scholar]
- O'Sullivan S, Gilmer JF, Medina C. Matrix metalloproteinases in inflammatory bowel disease: an update. Mediators Inflamm. 2015;2015:964131. doi: 10.1155/2015/964131. https://doi.org/10.1155/2015/964131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Zheng TT, Li X, Liang Y, Wang LJ, Huang YC, et al. Plant-derived alkaloids: The promising disease-modifying agents for inflammatory bowel disease. Front Pharmacol. 2019;10:351. doi: 10.3389/fphar.2019.00351. https://doi.org/10.3389/fphar.2019.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho PMM, Pinto MMM, Kijjoa A, Pharadai K, Díaz JG, Herz W. Protoberberine alkaloids from Coscinium fenestratum. Phytochemistry. 1992;31:1403–1407. doi: 10.1016/0031-9422(92)80301-T. https://doi.org/10.1016/0031-9422(92)80301-T. [DOI] [Google Scholar]
- Qu F, Li D, Zhang S, Zhang C, Shen A. The potential mechanism of qinghua quyu jianpi decoction in the treatment of ulcerative colitis based on network pharmacology and experimental validation. J Ethnopharmacol. 2023;310:116396. doi: 10.1016/j.jep.2023.116396. https://doi.org/10.1016/j.jep.2023.116396. [DOI] [PubMed] [Google Scholar]
- Rai RV, Rajesh PS, Kim HM. Medicinal use of Coscinium fenestratum (Gaertn.) Colebr.: an short review. Orient Pharm Exp Med. 2013;13:1–9. doi: 10.1007/s13596-012-0094-y. https://doi.org/10.1007/s13596-012-0094-y. [DOI] [Google Scholar]
- Rojsanga P, Sukhthankar M, Krisanapun C, Gritsanapan W, Lawson DB, Baek SJ. In vitro anti-proliferative activity of alcoholic stem extract of Coscinium fenestratum in human colorectal cancer cells. Exp Ther Med. 2010;1:181–186. doi: 10.3892/etm_00000029. https://doi.org/10.3892/etm_00000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. https://doi.org/10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio G, Davis RJ. TNF and MAP kinase signalling pathways. Semin Immunol. 2014;26:237–245. doi: 10.1016/j.smim.2014.02.009. https://doi.org/10.1016/j.smim.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50(W1):W216–W221. doi: 10.1093/nar/gkac194. https://doi.org/10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou X, Wang Y, Zhang X, Zhang Y, Yang Y, Duan C, et al. Network pharmacology and molecular docking analysis on molecular mechanism of Qingzi Zhitong decoction in the treatment of ulcerative colitis. Front Pharmacol. 2022;13:727608. doi: 10.3389/fphar.2022.727608. https://doi.org/10.3389/fphar.2022.727608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spisni E, Valerii MC, De Fazio L, Cavazza E, Borsetti F, Sgromo A, et al. Cyclooxygenase-2 silencing for the treatment of colitis: a combined in vivo strategy based on RNA interference and engineered Escherichia coli. Mol Ther. 2015;23:278–289. doi: 10.1038/mt.2014.222. https://doi.org/10.1038/mt.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudharshan SJ, Kekuda TRP, Sujatha ML. Antiinflammatory activity of Curcuma aromatica Salisb and Coscinium fenestratum Colebr: a comparative study. J Pharm Res. 2010;3:24–25. [Google Scholar]
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. https://doi.org/10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vite MH, Nangude SL, Patil JS. Pharmacognostical and phytochemical studies and evaluation of anti-ulcer activity of Coscinium fenestratum (gaertn) colebr. SPER J Adv Nov Drug Deliv. 2016;1:22–27. [Google Scholar]
- Xia XM, Wang FY, Xu WA, Wang ZK, Liu J, Lu YK, et al. CXCR4 antagonist AMD3100 attenuates colonic damage in mice with experimental colitis. World J Gastroenterol. 2010;16:2873–2880. doi: 10.3748/wjg.v16.i23.2873. https://doi.org/10.3748/wjg.v16.i23.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tang C, Jin R, Liu B, Wang P, Chen Y, et al. Molecular mechanisms of Huanglian jiedu decoction on ulcerative colitis based on network pharmacology and molecular docking. Sci Rep. 2022;12:5526. doi: 10.1038/s41598-022-09559-1. https://doi.org/10.1038/s41598-022-09559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Ren Y, Yang K, Wang W, Wang T, Xiao W, et al. The role of hypoxia-inducible factor 1-alpha in inflammatory bowel disease. Cell Biol Int. 2022;46:46–51. doi: 10.1002/cbin.11712. https://doi.org/10.1002/cbin.11712. [DOI] [PubMed] [Google Scholar]
- Zhang YT, Yu YQ, Yan XX, Wang WJ, Tian XT, Wang L, et al. Different structures of berberine and five other protoberberine alkaloids that affect P-glycoprotein-mediated efflux capacity. Acta Pharmacol Sin. 2019;40:133–142. doi: 10.1038/s41401-018-0183-7. https://doi.org/10.1038/s41401-018-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zhang H, Li N, Chen J, Xu H, Wang Y, et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J Ethnopharmacol. 2023;309:116306. doi: 10.1016/j.jep.2023.116306. https://doi.org/10.1016/j.jep.2023.116306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.