Abstract
Abstract
Bacteria-based tumor therapy, which releases therapeutic payloads or remodels the tumor’s immune-suppressive microenvironment and directly kills tumor cells or initiates an anti-tumor immune response, is recently recognized as a promising strategy. Bacteria could be endowed with the capacities of tumor targeting, tumor cell killing, and anti-tumor immune activating by established gene engineering. Furthermore, the integration of synthetic biology and nanomedicine into these engineered bacteria could further enhance their efficacy and controllability. This comprehensive review systematically elucidates the classification and mechanisms of bacterial gene expression induction systems, as well as strategies for constructing bacterial-nanomaterial nanobiohybrids. The review concludes by highlighting the challenges associated with quality control and regulation of bacteria-based tumor therapy while also providing insights into the future prospects of this therapeutic technology.
Key points
• A comprehensive overview of the current status of research on bacteria-based tumor therapy.
• The classification and mechanisms of bacterial gene expression induction systems are summarized.
• The challenges and perspectives in clinical translation.
Keywords: Bacteria-based tumor therapy, Synthetic biology, Gene circuits, Nanomedicine
Introduction
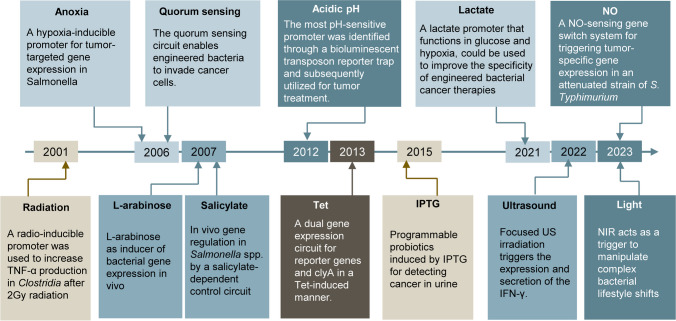
Bacteria-based tumor therapy, with the unique properties of bacteria and their excellent potential for modification, has emerged as a promising strategy for tumor treatment (Feng et al. 2023; Gurbatri et al. 2022; Kwon et al. 2024). The tumor’s disordered vascular system, along with its hypoxic, acidic, and nutrient-rich microenvironment, facilitates the selective colonization of bacteria (Van Mellaert et al. 2006; Xie et al. 2022; Yu et al. 2012). Once inside the tumor, the immunosuppressive microenvironment enables bacteria to accumulate and proliferate (Chandra et al. 2017; Pawelek et al. 1997; Sznol et al. 2000). The inward-to-outward anti-tumor effects induced by bacteria-based tumor therapy can effectively address the limitations of traditional tumor treatments such as non-targeted distribution and limited tumor penetration (Jain 1998; Minchinton and Tannock 2006; Zinger et al. 2019). To enhance the efficacy of local tumor treatment while minimizing the risk of systemic toxicity, bacteria-based tumor therapy must be controlled. Precise regulation of the timing and location of bacterial therapeutic effects is critical for optimizing drug accumulation, resource utilization, and minimizing damage to normal tissues (Zhou et al. 2018).
To date, several techniques have been developed to induce bacterial gene expression (Fig. 1). Various prokaryotic expression regulatory elements have been integrated with exogenous genes to enhance the capacity of engineered bacteria to respond to both the internal and external signals (Cubillos-Ruiz et al. 2021; Omer et al. 2022). Furthermore, the combination of bacteria with nanomedicine offers complementary advantages, as the remarkable optical, magnetic, and acoustic properties of nanomaterials significantly enhance the effectiveness and controllability of bacteria-based tumor therapy (Lou et al. 2021; Sun et al. 2023; Zhou et al. 2024). These technologies can be utilized to manipulate bacterial targeting and colonization, interact with the dynamically changing environment, enable drug delivery within the appropriate therapeutic window, and facilitate post-treatment self-clearance, thereby achieving a balance of specificity, efficacy, and safety in the precision treatment of tumors.
Fig. 1.
The history of these techniques for regulating bacterial gene expression in bacteria-based tumor therapy
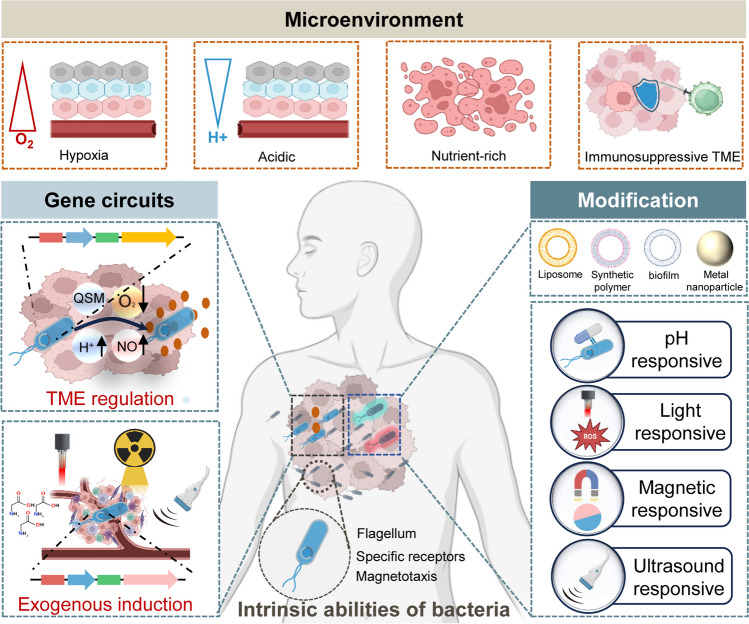
This review is to summarize the engineering and modification of bacteria based on their tumor-targeting and colonization properties to enhance their anti-tumor effects through advanced genetic engineering and nanomedicine. Application of genetically engineered bacteria or bacteria-nanomaterial nanobiohybrids in precise tumor therapy are discussed. Additionally, the challenges related to quality control and regulation of bacterial therapy are summarized to provide insights for clinical translation (Fig. 2).
Fig. 2.
Overview of bacteria-based tumor therapy. The tumor microenvironment (TME) is suitable for targeting and colonization by certain bacteria. On the other hand, some bacteria intrinsically can target the tumor region. Moreover, bacteria could be engineered with gene circuits and other chemical or nano-modifications to eradicate tumors with more accuracy and efficiency (created with BioRender.com)
Bacteria target the TME and colonize
Tumors possess a microenvironment that is markedly different from that of most normal tissues (Jin and Jin 2020), characterized by hypoxia, acidity, and nutrient-rich conditions, which makes them a natural target for bacteria (Van Mellaert et al. 2006; Xie et al. 2022; Yu et al. 2012). The targeting of tumor tissue by bacteria is guided by specific receptors capable of identifying molecules released by dying tumor cells (Kasinskas and Forbes 2007). Bacteria in the bloodstream infiltrate tumor tissue through the disordered vascular system of the tumor (Leschner et al. 2009), where the lower oxygen levels provide an environment conducive to the proliferation of both obligate and facultative anaerobic bacteria (Duong et al. 2019; Lambin et al. 1998). Notably, a specific species of Salmonella has been shown to accumulate preferentially in tumors at ratios exceeding 10,000:1 (Lee et al. 2005). Their robust flagella and small size allow them to penetrate deeply into tumors (Chen et al. 2021a; Dang et al. 2001; Minamino and Imada 2015), while the necrotic areas provide ample nutrients for bacterial growth. Importantly, the immunosuppressive TME facilitates bacterial proliferation within the tumor while protecting it from clearance by the host immune system (Forbes et al. 2003; Zhang and Forbes 2015). Bacteria, due to their inherent immunogenicity, induce a significant influx of neutrophils into the tumor, which subsequently releases neutrophil extracellular traps composed of chromatin fibers and antimicrobial proteins, thereby confining the bacteria to the tumor area (Branzk et al. 2014; Leschner et al. 2009; Westphal et al. 2008).
Furthermore, a special type of bacteria known as magnetotactic bacteria can swim in a directed manner along the magnetic field lines (Faivre and Schuler 2008). This ability is facilitated by their remarkable flagellar propulsion, and a unique cellular organelle called the magnetosome, which contains nanoscale magnetic iron mineral crystals that impart magnetic properties to the bacteria and function as a “biological compass” guiding their movement (Uebe and Schuler 2016). Driven by external magnetic fields, magnetotactic bacteria can navigate toward tumor sites (Kotakadi et al. 2022). Various magnetic field driving approaches have been developed, including rotating magnetic fields (Wu et al. 2022b), gradient magnetic fields, and oscillating magnetic fields. Due to the flexibility of magnetic fields, appropriate magnetic driving schemes can enhance bacterial infiltration within tumors (Gwisai et al. 2022; Mirkhani et al. 2024).
Internal signals induce bacterial gene expression
Based on the characteristics of bacteria that target and colonize the TME, the features of TME can be used to induce bacterial gene expression, thereby achieving drug enrichment and minimizing off-target effects. Figure 3 illustrates two strategies for bacterial expression of exogenous target genes (Boe 1996; Ganusov and Brilkov 2002). Internal signals inducing bacterial gene expression are divided into expression induced by TME characteristics and bacterial self-triggering (Table 1).
Fig. 3.
Comparison of two strategies for bacteria expressing exogenous genes (created with BioRender.com)
Table 1.
Internal signals induce bacterial gene expression
| Induction signals | Species | Cancer | Working mechanism | Reference |
|---|---|---|---|---|
| Anoxia | Attenuated S. typhimurium strain (JRG4401) | 4T1 breast tumors | Bacterial expression of the cytotoxic protein (HlyE) gene in hypoxic regions of mouse mammary tumors under the regulation of a highly hypoxia-inducible promoter (FF + 20) | Ryan et al. 2009 |
| VNP20009 | U87mg glioma | Hypoxia-inducible promoter pflE regulates the expression of wild-type tumor suppressor proteins p53 and Azurin in glioma-hypoxic regions | Mehta et al. 2017 | |
| Salmonella Typhimurium strain YB1 |
MDA-MB-231 breast tumors |
Placing the bacterial essential gene asd under the regulation of the hypoxia-inducible initiator PpepT creates a "specialized" anaerobic Salmonella typhimurium strain to target tumor suppression | Yu et al. 2012 | |
| E. coli Nissle 1917 | B16F10 melanoma | Diadenylate cyclase (DacA), which generates the STING activator cyclic di-nucleotides, was placed under the regulation of the hypoxia-inducible promoter PfnrS | Leventhal et al. 2020 | |
| E. coli BW25133 | spontaneous mouse mammary tumor | A synthetic hypoxia-inducible promoter, nirB, preferentially and selectively induces cardiac peptide expression under hypoxic conditions for tumor suppression | Samadi et al. 2021 | |
| Acidic pH | Salmonella Typhimurium strain SB300A1 | B16F10 melanoma, HCT116 colon cancer | Regulation of Shiga toxin 2 expression within the tumor microenvironment using the screened acidic pH-responsive promoter STM1787 | Flentie et al. 2012 |
| E. coli MG1655 | CT26 colon cancer | Cytolysin A (ClyA) protein-coupled to a green fluorescent protein (GFP) placed under the regulation of an acid-sensitive promoter (adiA) for therapeutic visualization | Qin et al. 2022 | |
| Lactate | E. coli Nissle 1917 | SW480 colorectal cancer cell | A lactate promoter that functions in glucose and hypoxia could be used to improve the specificity of engineered bacterial cancer therapies | Zúñiga et al. 2021 |
| CT26 colon cancer | Integrated bacterial growth with genetic circuits that sense lactate can enhance the tropism of engineered bacteria, thereby regulating the expression of essential genes | Chien et al. 2022 | ||
| NO | ΔppGpp S. typhimurium | / | Elevated NO at the tumor site binds NorR and recruits σ54, causing the expression of the DNA recombinase FimE. This catalyzes the permanent inversion of the DNA switch fimS, leading to sustainable expression of GOI | Qin et al. 2023 |
| Self-triggering | VNP200010 | 4T1 breast tumors | Salmonella carrying the QS trigger system successfully initiates protein production inside tumors but not in healthy tissues | Swofford et al. 2015 |
| E. coli Nissle 1917 |
A20 lymphoma CT26 colon cancer |
Bacteria are utilized as a delivery system for nano-antibodies and as time increases and the bacterial count reaches a lysis threshold, bacterial lysis releases the therapeutic proteins | Gurbatri et al. 2020 | |
| VNP20009 with an additional deletion (Δasd) | 4T1 breast tumors | Salmonella carrying gene circuits autonomously triggers lysis after invading cancer cells, thereby releasing protein drugs | Raman et al. 2021 |
TME regulates bacterial gene expression
Hypoxia-inducible bacterial expression systems primarily rely on the oxygen transcription factor fumarate and nitrate reduction regulatory protein (FNR) naturally present in Salmonella (Arrach et al. 2008; Leschner et al. 2012). This system functions through the binding and dissociation of FNR with [4Fe-4S]2+. In hypoxic environments, FNR binds [4Fe-4S]2+ to form a transcriptionally active homodimer that binds to specific DNA sequences in promoters, recruiting RNA polymerase to transcribe downstream genes (Chien et al. 2022; Ryan et al. 2009). Commonly used hypoxia-inducible promoters include pflE, ansB (Arrach et al. 2008), pepT (Strauch et al. 1985), as well as synthetic promoters like FF + 20 (Ryan et al. 2009), HIP-1 (Mengesha et al. 2006), and nirB (Nasr and Akbari Eidgahi 2014). An engineered hypoxia-inducible Salmonella strain placing the cytotoxic protein hemolysin E (HlyE) gene under the regulation of the hypoxia-inducible promoter FF + 20, can secrete HlyE in the hypoxic TME to kill tumor cells (Ryan et al. 2009). This strategy can also confine bacterial survival to the tumor, reducing systemic toxicity. The asd gene, which encodes a crucial enzyme involved in the synthesis of the bacterial cell wall, is regulated by a hypoxic promoter. Additionally, the asd gene is knocked out of the bacterium, ensuring it cannot survive in normal tissues (Yu et al. 2012).
The acidic microenvironment of tumors is another important feature. Indeed, due to the Warburg effect, even in an aerobic environment, the uptake of glucose by tumor cells is increased. High levels of aerobic glycolysis and glutaminolysis release large amounts of H+ and lactate into the extracellular space, subsequently releasing them into the TME, maintaining a pH of 5.8–6.6 (Huber et al. 2017; Zhang et al. 2023). Exploiting this, Flentie et al. identified a tumor-specific acidic microenvironment inducible promoter STM1787 in Salmonella using a bioluminescent transposon reporter gene; 8 h after exposure to the tumor acidic microenvironment, gene expression increased 90-fold (Flentie et al. 2012). Additionally, acid-induced promoters adiA and Pasr from E. coli responsible for expressing arginine decarboxylase and acid shock RNA, respectively, were identified (Mallick and Das 2023; Qin et al. 2022; Stirling et al. 2020). By linking the cytolysin A (ClyA) with a green fluorescent protein (GFP) under the regulation of the acid-sensitive promoter adiA, visual therapeutics can be effectively achieved (Qin et al. 2022). Lactate in the TME is also used in gene expression systems based on the native lldPRD operon, in which the LldR repressor dimer further inhibits gene expression unless bound to lactate (Aguilera et al. 2008). The tropism of engineered bacteria can be enhanced by integrating bacterial growth with genetic circuits that sense oxygen, pH, or lactate, thereby regulating the expression of essential genes. Bacteria engineered with pH or oxygen sensors demonstrated preferential growth under physiologically relevant acidic or hypoxic conditions (Chien et al. 2022).
Salmonella enterica subsp. enterica serovar (S. typhimurium) colonizing tumor induces upregulation of inducible nitric oxide synthase (iNOS), leading to a 1000-fold increase in NO production at micromolar levels within the tumor site (Barak et al. 2010; Cinelli et al. 2020; Zheng et al. 2017). This enables the use of NO as an internal signal to induce bacterial gene expression. The NO-inducible system is based on NorR, a σ54-dependent regulatory protein in E. coli, which senses and binds NO in the presence of NO, inducing σ54 binding and activating the Pnorv promoter (Tucker et al. 2008, 2010). To achieve permanent activation of NO-induced gene expression, the Pnorv promoter was utilized to regulate the expression of the DNA recombinase FimE, leading to permanent inversion of fimS and, thus, sustained expression of the target gene (Qin et al. 2023).
Self-triggering
Bacterial gene expression can also be regulated by communication between bacteria, which relies on quorum-sensing molecules (QSMs). QSMs are secreted externally, accumulate with increasing community density, and, after reaching a certain threshold, bind to receptors to regulate gene expression (Mukherjee and Bassler 2019). Bacterial QSMs include acyl homoserine lactone (AHL or AI-1), auto-inducible peptide (AIP), and autoinducer-2 (AI-2) (Zeng et al. 2023). The AHL is mainly secreted by Gram-negative bacteria, and the LuxI promoter regulates AHL production, followed by AHL binding to LuxR to turn on downstream gene expression (Din et al. 2016). The quorum-sensing phenomenon of bacteria can be utilized to design many interesting genetic circuits, such as the quorum-sensing lysis circuit, which provides new ideas for drug delivery (Chowdhury et al. 2019; Gurbatri et al. 2020; Raman et al. 2021). By expressing the lytic enzyme gene E of phage FX174 under the control of the LuxI promoter, when the bacteria are enriched in the tumor, and the AHL reaches a certain threshold, bacterial lysis is initiated, and therapeutic proteins pre-expressed constitutively in vivo are released (Din et al. 2016). This strategy enables the delivery of more nanobodies within the tumor, including anti-CD47 nanobodies (Chowdhury et al. 2019), anti-PD-L1/PD-1, and anti-CTLA4 nanobodies (Gurbatri et al. 2020).
External signals induce bacterial gene expression
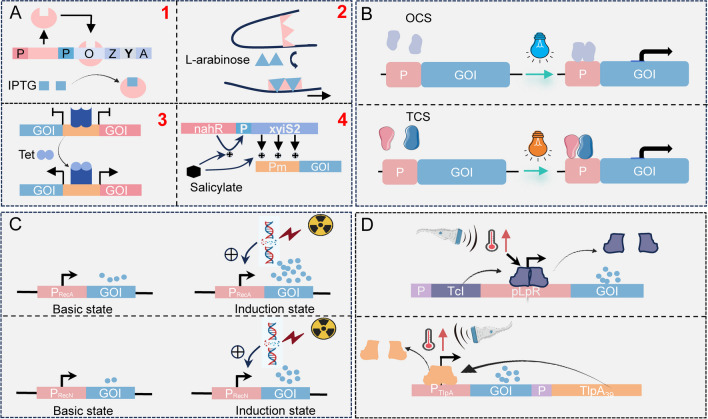
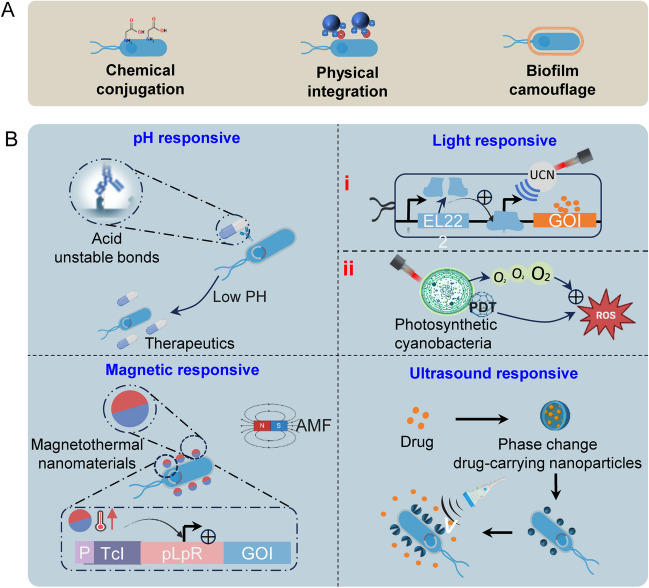
In addition to TME-induced triggering, synthetic biology can also be used to design sophisticated, intelligent gene circuits in response to external signals. Compared with TME-induced triggering, external signal triggering has better initiative and controllability. Depending on the external signals, they are categorized as follows (Fig. 4). The application of external signals to control bacterial expression of exogenous genes to achieve anti-tumor effects is summarized and displayed in Table 2.
Fig. 4.
External signals induce bacterial gene expression. A Four most commonly used chemical inducers to induce bacterial gene expression. B Optogenetic systems for inducing gene expression in bacteria. OCS, one-component systems; TCS, two-component systems. C Radiation-induced bacterial gene expression based on promoters RecN or RecA. D Induction of bacterial gene expression based on the thermal effect of ultrasound
Table 2.
External signals induce bacterial gene expression
| External signals | Species | Cancer | Working mechanism | Reference | |
|---|---|---|---|---|---|
| Chemical inducers | IPTG | E. coli Nissle 1917 | MC26 colon cancer | Integrate the LuxCDABE gene cassette into the IPTG-induced expression vector and induce fluorescent gene expression by intravenous injection of IPTG | Danino et al. 2015 |
| CT26 colon cancer | IPTG induced the expression of NDH-2 enzyme, which could increase the production of H2O2 at tumor sites. On this basis, magnetic Fe3O4 nanoparticles were covalently attached to bacteria to act as catalysts for Fenton-like reactions to convert H2O2 into toxic hydroxyl radicals (-OH) for tumor therapy | Fan et al. 2019 | |||
|
4T1 breast tumors, CT26 colon cancer |
IPTG induction enhances the synthesis of bacterial capsular polysaccharide (CAP) to improve in vivo microbial delivery, increasing the maximum tolerated dose tenfold and enhancing the anti-tumor efficacy in a mouse cancer model | Harimoto et al. 2022 | |||
| L-arabinose | E. coli Nissle 1917 | 4T1 breast tumors | The target gene was placed downstream of the pBAD promoter, and gene expression was selectively induced in vivo using non-toxic L-arabinose in mouse tumor tissues | Stritzker et al. 2007 | |
| VNP20009 |
Human LS174T colon adenocarcinoma |
The combination of the L-arabinose induction system and the bacterial communication system overcame the drawbacks of the induction system alone, prolonged the protein release time, and increased the protein yield | Dai et al. 2013 | ||
| ΔppGpp S. typhimurium | MC26 colon cancer | Under the induction of L-arabinose, the attenuated Salmonella typhimurium strain can express and secrete Vibrio vulnificus flagellin B (FlaB) within tumor tissues. This process leads to the phenotypic and functional activation of intratumoral macrophages, promoting an M1 phenotype while concurrently reducing the M2-like macrophages | Zheng et al. 2017 | ||
| CT26 colon cancer | A strain of S. typhimurium carrying cytotoxic proteins (cytolysin A) and expressing reporter genes that integrate monitoring and therapeutic functions successfully visualized the therapeutic process of these engineered bacteria in mice | Nguyen et al. 2010 | |||
| Glioma | L-arabinose-induced release of tissue inhibitor of matrix metalloproteinase-2 (TIMP2) | Wen et al. 2018 | |||
| SL7207 S. typhimurium | CT26 colon cancer | L-arabinose-induced bioluminescence enables noninvasive in vivo imaging, and the lysate gene E of phage FX174, placed under the control of PBAD for bacterial lysis induced by arabinose, can be engineered to release therapeutic loads | Loessner et al. 2007 | ||
| Tet | ΔppGpp S. typhimurium | CT26 colon cancer | Construction of a dual gene expression circuit for bilateral expression of bioluminescent reporter genes and clya in a tetracycline-induced manner | Jiang et al. 2013 | |
| Salicylate | Salmonella enterica aroA | Fibrosarcoma | Acetylsalicylic acid (ASA) regulates cytosine deaminase (CD) cascade expression, and ASA induction prior to 5-fluorocytosine administration significantly reduces tumor growth | Royo et al. 2007 | |
| Light | NIR | P. aeruginosa strain ExoST | A549 | NIR triggers complex bacterial lifestyle shifts through the optogenetic tool BphS for bacterial adhesion, colonization, and drug release | Fu et al. 2023 |
| Radiation | 2gy γ radiation | VNP20009 | 4T1 breast tumors | Radiation-induced RecA promoter controls secretion of mouse TNF-related apoptosis-inducing ligand (TRAIL) | Ganai et al. 2009 |
| Attenuated Salmonella strain (KST0650) | CT26 colon cancer | Expression of the intracellular pro-apoptotic protein sATF6 is controlled by the radiation-induced recN promoter, and sATF6 is expressed and secreted under γ radiation | Gao et al. 2020 | ||
| Ultrasound | Focused ultrasound | E. coli Nissle 1917 | A20 lymphoma | Alteration of temperature-sensitive repressor protein conformation by the thermal effect of focused ultrasound activates the PLPR promoter to express immune checkpoint inhibitors against CTLA-4 and PDL1 | Abedi et al. 2022 |
| E. coli MG1655 | 4T1 breast tumors | The PL and PR promoters tandemly regulate the IFN-γ gene, and focused ultrasound irradiation and heating trigger the expression and secretion of the IFN-γ gene | Chen et al. 2022 | ||
| 4T1 breast tumors | Introducing an acoustic reporter gene (ARG1) into bacteria that produce gas vesicles (GVs), the IFN-γ gene is under the tandem regulation of the PL and PR promoters, and during ultrasound irradiation, the GVs are used as an ultrasound contrast agent for imaging, while IFN-γ is secreted for tumor treatment | Yang et al. 2024 | |||
| VNP20009 | B16F10 melanoma | The temperature-sensitive transcriptional repressor TlpA39-based gene expression system (SINGER) expresses and secretes PD-L1nb and azurin upon focused ultrasound irradiation and heating | Gao et al. 2024 | ||
Chemical inducers
Chemical induction was first applied to regulate bacterial gene expression, with common chemical inducers including isopropyl β-D-thiogalactoside (IPTG) (Danino et al. 2015; Fan et al. 2019; Harimoto et al. 2022), L-arabinose (Dai et al. 2013; Loessner et al. 2007; Nguyen et al. 2010; Stritzker et al. 2007; Wen et al. 2018), tetracycline (Jiang et al. 2013), and salicylic acid salts (Royo et al. 2007). The regulation of bacterial expression by chemical inducers is mostly based on the operon model, which is a directed DNA sequence that acts as a gene regulator to regulate bacterial gene expression (English et al. 2021).
IPTG is used as an inducer to bind the lac repressor protein in the lactose operon (Fig. 4A1), causing it to dissociate from the DNA-binding domain, thus turning on downstream gene expression (Lewis et al. 1996; Romanuka et al. 2023). Integration of the LuxCDABE gene cassette into an IPTG-induced expression vector and induction of fluorescent gene expression by intravenous injection of IPTG resulted in the detection of bacterial fluorescent signals in approximately 88% of metastases in a mouse colorectal cancer metastasis model (Danino et al. 2015). The expression of downstream target genes of the operon is closely related to the concentration of chemical inducers. Therefore, a tunable microbial surface engineering strategy based on IPTG is utilized to dynamically control the synthesis of bacterial capsular polysaccharides (CAP) to enhance bacterial interactions with their surrounding environment (Harimoto et al. 2022).
The L-arabinose-inducible pBAD promoter is derived from the arabinose operon. When L-arabinose is present, the AraC protein opens the gyratory structure. It acts as an activator to activate target gene transcription (Fig. 4A2). The induction of instantaneous bacterial bioluminescence by L-arabinose was sufficient to localize the bacteria within the tumor but freed the bacteria from the burden of continuous expression of luciferase or fluorescent proteins (Loessner et al. 2007). The induction/repression ratio using the L-arabinose control system can be up to 1200-fold (Guzman et al. 1995). However, recombinant proteins based on the pBAD vector system are expressed at lower levels, and some strains can catabolize L-arabinose.
The Tet-off and Tet-on, bacterial expression regulation systems, induced by tetracycline work in opposite ways. In the Tet-off system, the tTA transcription factor cannot bind to the DNA sequence of the promoter region in the presence of tetracycline, resulting in gene expression inhibition. Conversely, in the Tet-on system, the rtTA transcription factor binds to the DNA sequence and activates downstream gene expression (Fig. 4A3) (Gossen and Bujard 1992; Gossen et al. 1995). Compared to L-arabinose and IPTG, the Tet-on system induced by tetracycline has a lower baseline expression level of target genes while maintaining a higher induction efficiency. By constructing a bidirectional gene circuit, co-expression of the therapeutic gene and reporter gene can be achieved under the induction of doxycycline (or tetracycline) to meet therapeutic needs (Jiang et al. 2013).
Salicylate-induced systems are usually cascade regulatory circuits that achieve amplified expression of specific genes under the control of salicylic acid (Cebolla et al. 2001). The Pm promoter in the TOL operon from Pseudomonas putida catabolism of toluene and xylene can be activated by the xyIS protein, and the transcription of the xyIS gene is controlled by another regulator, XyIR, and the strength of Pm increases as more xyIS protein is expressed. The mutant of xyIS, xyIS2, can be activated by salicylic acid, thus placing the expression of xyIS2 under the control of the Psal promoter and its cognate regulator NahR, which is activated in the presence of salicylic acid to initiate the expression and activation of the xyIS2 protein. These two effects act together on the Pm promoter. This cascade reaction causes a massive amplification of the target gene (Fig. 4A). Placing cytosine deaminase (CD) in a regulatory circuit activated by acetylsalicylic acid (ASA) led to a 20- to 150-fold ex vivo induction and induction with ASA before 5-fluorocytosine administration significantly reduced tumor growth (Royo et al. 2007).
Light
Optogenetics integrates optics and genetics to control biological processes with a high degree of spatiotemporal precision using genetically encoded light-responsive proteins in response to light stimulus (Emiliani et al. 2022). In recent years, driven by synthetic biology, optogenetics has been introduced into bacteria to enable rapid, dynamic, and reversible regulation of bacterial gene expression (Lindner and Diepold 2022). Introducing genetically encoded light-responsive proteins into bacteria to precisely regulate bacterial expression in tumors by using light as an external trigger, the advantages brought by light as a signal are incomparable to chemical induction, even though changes in photoreceptor intensity induced by altering light intensity are similar to those by altering the concentration of a chemical inducer. The fact that light is not only controllable but also reversible and non-invasively transmits signals and that it is possible to restrict the activation region to a specific place independent of diffusion greatly improves spatiotemporal controllability (Deisseroth 2011).
Optogenetic systems used to regulate gene expression in engineered bacteria can usually be categorized into the one-component system (OCS) and two-component system (TCS) (Lindner and Diepold 2022). OCS and TCS are distinguished based on the different mediators responding to light and inducing transcription. The OCS contains only a single protein, which assumes the role of a photoreceptor and is an inducer of transcription. The EL222 protein, first discovered in the marine bacterium Erythrobacter litoralis, is by far the most commonly used blue light-responsive transcription factor in the OCS (Motta-Mena et al. 2014). The TCS consists of a light-responsive kinase and a homologous transcriptional response regulator, and the YF1/FixJ-based pDawn system is one of the most common TCS in use today. OCS is simpler because it uses a single light-responsive protein to induce transcription and does not rely on the fully balanced expression of a second component. Its smaller size saves space for therapeutic genes, allowing for more personalized therapeutic delivery (Hoffman et al. 2022). OCS also have limitations; almost all of them rely on poorly penetrating blue light, but a recently discovered OCS that responds to near-infrared light, called iLight, is very promising (Kaberniuk et al. 2021).
TCS has a wide spectrum of responses, with the optogenetic tools CcaS/CcaR based on CBCRs activating downstream gene expression under 535 nm green light stimulation and inhibiting gene expression under 672 nm red light (Tabor et al. 2011); UirS/ UirR, enabling gene expression initiation in response to ultraviolet light and gene expression inhibition under 535 nm green light (Ramakrishnan and Tabor 2016). BphPs-based optogenetic tools have a faster response rate because BphP1 is activated under near-infrared light and activates the transcription of downstream genes by combining with the transcriptional repressor PpsR2, which does not require the signaling process of the phosphoryl group (Ong et al. 2018). BphS, a mutant of the bacterial photosensitive pigment BphG, is triggered by NIR light, leading to an elevation of c-di-GMP, thereby enhancing biofilm formation, preventing bacterial elimination by neutrophil-mediated killing effects and increasing the illumination power density of NIR triggering bacterial lysis and release of the antitumor toxin HlyE to induce tumor necrosis (Fu et al. 2023).
Since TCS is based on the interaction of two proteins, this has to take into account the sensitivity and balance of the host system to the expression of these two proteins. In addition, in non-blue light-activated systems, the hosts often do not endogenously express the chromophores required by the systems, such as biliverdin, phycocyanin, etc.. So they have to be artificially added or synthesized by the hosts, which will undoubtedly complicate the application of TCS. Nonetheless, TCS has developed into a relatively mature application system, and due to its robustness in regulating proteins, flexibility in module selection, and wide range of response spectra, TCS has unique advantages in regulating bacterial protein expression.
However, it must be recognized that the poor penetration of light into deep tissues has hindered the development of light-induced expression systems. For tumors located deep in the tissue, how to ensure that the light activates enough bacteria to produce effective therapeutic effects is an issue that has to be considered for light-induced expression systems. For specific tumors, such as those in the digestive tract, light-induced expression systems may be able to unite with gastrointestinal endoscope to increase the percentage of activated bacteria to enhance therapeutic efficacy.
Radiation
The current systems for controlling bacterial expression by radiation are all based on the RecA or RecN promoters in the SOS repair system of Clostridium difficile. Various physical, chemical, and biological effects caused by radiation result in DNA breaks and activation of RecA and RecN proteins, which play an important role in repairing damaged DNA, and it is in this process that the RecA and RecN promoters respond to radiation and initiate genes expression (Nuyts et al. 2001c). This induced expression is reproducible and does not differ between aerobic and anaerobic conditions, which is important for using radiation-induced expression promoters in anaerobic bacteria.
Radiation can efficiently penetrate tissues with little diffusion effect, and the release of the therapeutic load from the radiation-responsive promoters RecA or RecN can be precisely controlled in the tumor and avoid systemic toxicity. The reporter gene LacZ was placed under a radiation-responsive promoter, and after 2 Gy of irradiation, β-galactosidase activity was significantly increased by 20–30%, and expression could be induced repeatedly by repeated administration of radiation (Nuyts et al. 2001c). To improve the induction ratio of the radial expression system, after the addition of Cheo cassettes, the RecA promoter induction ratio can be increased tenfold (Nuyts et al. 2001a). Engineering of Clostridium difficile to express TNF-α gene under the regulation of the radiation-responsive promoter RecA successfully expressed TNF-α in situ under 2 Gy of radiation and prolonged the expression of TNF-α at the tumor site by repeated induction, thus enhancing the anti-tumor effect (Nuyts et al. 2001b).
Another radiation-responsive promoter RecN has a much lower basal activity than RecA although the induction ratio is not as high as RecA, which means that RecN is more suitable for expressing proteins with toxicity because it hardly causes leakage (Fu et al. 2023). For example, the intracellular pro-apoptotic protein sATF6, under the regulation of RecN, can control the killing effect at the radiation-exposed site of the tumor while avoiding damage to healthy tissues (Gao et al. 2020).
In conclusion, bacterial expression systems based on the radiation-responsive promoters RecA and RecN show great potential for precise in situ delivery of drugs to tumors. However, radiation is potentially lethal to tissues, so it is important to make an informed choice between the appropriate dose of radiation and the best therapeutic effect that can be elicited and to select bacteria that are best suited to carry the radiation-responsive expression systems to achieve greater effects, including sensitization to radiation therapy, synergistic therapeutic effects, and protection of normal tissues from radiation damage.
Ultrasound
Ultrasound has a natural advantage as an external signal with its non-invasiveness, high penetrability, low attenuation, non-ionizing radiation, and excellent spatial accuracy (Peek et al. 2015; Qian et al. 2017). Currently, ultrasound regulation of expression in engineered bacteria relies mainly on thermal effect and temperature-responsive components, such as RNA thermometers (RNATs) (Neupert and Bock 2009), the PL&PR promoters, and various temperature-sensitive mutants of phage lambda protein “cI” (Valdez-Cruz et al. 2010). Temperature-sensitive mutants of phage lambda protein “cI” exist as monomers at low temperatures and block the PL and PR promoters, then dimerize when they reach their response temperature and release the promoter region, restoring transcriptional activity. In contrast, the temperature-controlled expression mediated by RNATs is at the translational level, where RNATs form stem-loops at body temperature that clamp the ribosome-binding sequences or translation initiation codons tightly by base pairing and only unclamp them when their response temperature is reached to allow ribosome binding, thus facilitating translation (Neupert and Bock 2009; Righetti and Narberhaus 2014).
The system for controlling bacterial expression by the thermal effect of ultrasound benefits from the remote tunability of ultrasound, and the selection of appropriate ultrasound parameters allows the temperature of the target zone to be maintained in the optimal range for initiating gene expression. A temperature-controlled gene expression circuit carrying interferon-γ (IFN-γ) constructed from the pR-pL promoter and cI protein was transferred into an ultrasound-responsive bacterium (URB), and IFN-γ was linked to the N-terminal end of the OmpA secretion signal peptide, and the temperature of the target zone was maintained at 45 ℃ under pulsed ultrasound irradiation to successfully secrete IFN-γ, the percentage of M1-type macrophages, CD4+ T cells, CD8+ T cells, and memory T cells increased significantly, and the immune response of the mice was successfully activated to inhibit tumor growth (Chen et al. 2022). In the clinical setting, where tumors are often treated over several weeks, a switch that is activated only upon ultrasound irradiation is not feasible, necessitating the introduction of a sustained expression function. Serine integrase Bxb1 is a class of enzymes that target DNA sequences at sites known as attP (attachment site of phage) and attB (attachment site of bacterial) and utilize these sites to mediate DNA sequence inversion (Xu et al. 2013). By placing Bxb1 under the regulation of the pR-pL promoter and the cI protein, Bxb1 was expressed when the tissue was heated by focused ultrasound and mediated the inversion of the otherwise inverted constitutive promoter, P7, resulting in sustained expression even when ultrasound was withdrawn (Abedi et al. 2022). Considering that 42–45 °C may cause thermal damage to normal tissues, an ideal system should be activatable in the range of 39–40 °C, with less leakage and a higher induction ratio. Recently, an ultrasound-responsive gene expression system based on the temperature-sensitive transcriptional repressor TlpA39 has been developed, and VNP20009 carrying this system can precisely initiate the efficient expression of the therapeutic gene at 39 °C during ultrasound-mediated local warming. Additionally, the combination of the anticancer protein Azurin protein with the immune checkpoint inhibitor PD-L1nb has been shown to induce apoptosis in tumor cells while activating the host's immune system, leading to a dual anticancer effect (Gao et al. 2024).
Bacteria and nanomedicine unite in cancer therapy
Relying solely on genetically engineered bacteria has limitations, such as rapid clearance of bacteria by the immune system, inadequate drug loading, and difficulty in drug release, which may contribute to poor tumor treatment effects (Liu et al. 2022). With the development of synthetic biology and nanomedicine, it is possible to use a variety of materials such as synthetic polymers (Jackson et al. 2017), liposomes (Gao et al. 2014), and biofilms (Aryal et al. 2013) to modify bacteria, to realize the complementary advantages, and to further improve the safety, precision, and effectiveness of engineered bacteria as well as to improve the shortcomings of traditional nanomaterials, such as poor targeting and high systemic toxicity (Zhou et al. 2024).
Materials camouflage bacteria for tumor targeting and proliferation
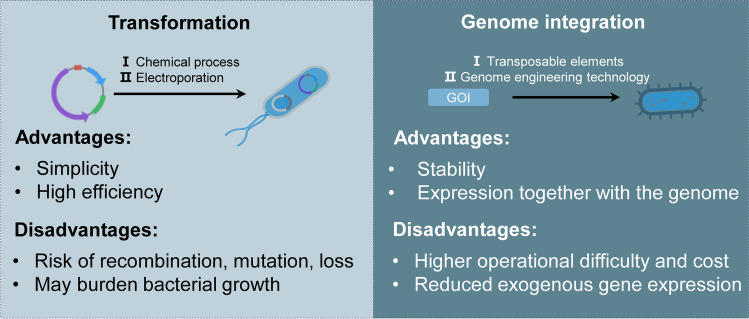
Various biological, chemical, and physical strategies have been developed to modify bacteria to avoid phagocytosis and clearance by the reticuloendothelial system (Fig. 5A) (Liu et al. 2022), thus prolonging the duration in the host and enabling tumor targeting and proliferation.
Fig. 5.
Bacteria and nanomedicine unite in cancer therapy. A Integration strategies for bacteria and nanomedicine. B Nanomedicine-enhanced bacteria exert synergistic anti-tumor effects (created with BioRender.com)
Several biofilms, including macrophages (Thamphiwatana et al. 2017), erythrocytes (Liu et al. 2022), platelets (Xu et al. 2018), neutrophils (Zhang et al. 2018), and cancer cells (Wang et al. 2018) have been used for camouflage. A type of cell membrane encapsulated bacteria formed by encapsulating Porphyromonas gingivalis through erythrocyte membranes had virtually no effect on the intrinsic bioactivities of the bacteria. It reduced bacterial-induced inflammatory responses and side effects, prolonging the persistence of the bacteria in the body (Chen et al. 2021b). Biofilm camouflage of ECN (Cao et al. 2019) and Listeria monocytogenes (Liu et al. 2022) has also been accomplished for tumor therapy. VNP20009 was encapsulated in macrophages by biomodification to form a “Trojan horse,” macrophages were rapidly enriched at the tumor site due to tumor chemotaxis. Then, intracellular bacterial proliferation led to macrophage rupture. This delayed-release strategy avoided premature bacterial clearance and completed bacterial enrichment at the tumor site (Wu et al. 2022a).
Chemical modifications typically utilize chemical reactions, including dopamine co-deposition, chemical bonding covalent linkage, Michael addition, and intermolecular disulfide exchange to couple materials to the bacterial surface (Zhou et al. 2024). Simple co-deposition of EcN with dopamine and simultaneous coupling of a variety of functional small molecules and polymers under biocompatible conditions results in multifunctional coatings that increase the bioavailability of the bacteria by more than 30-fold (Pan et al. 2021a). To further enhance the tumor-targeting property, the tumor-targeting peptide RGD was coupled to the carboxyl group on the surface of Magnetospirillum magneticum AMB-1 by chemical modification to construct the tumor-targeting strain AMB-RGD (Wang et al. 2022).
Physical modifications included electrostatic interactions (Zhao et al. 2022), electroporation (Zoaby et al. 2017), and membrane coating (Wu et al. 2019). Therapeutic agents can be transported by bacteria, which may either be affixed to the external surface of engineered bacteria or loaded into the bacterial body. Electrostatic interactions play a crucial role in enhancing the versatility of nanobiohybrid construction. This is primarily due to the fact that the surface charges of both bacterial cells and various nanomaterials can be readily adjusted. A straightforward electrostatic adsorption method combines black phosphorus quantum dots (BPQD) with E. coli to create a novel hybrid system, referred to as BPQD/E coli (BE), designed to enhance photothermal therapy (PTT) for hypoxic tumors. The BE system effectively targets hypoxic tumors and facilitates the mediation of PTT achieving significant radiotherapy efficacy in response to lower doses of radiation, effectively and specifically damaging hypoxic tumor tissues to suppress tumor growth (Ji et al. 2023). Electroporation is a technique that involves the application of short bursts of electrical pulses to bacterial membranes, leading to the formation of temporary pores. This process effectively enhances the permeability of the membrane, allowing substances to enter the bacterial cells more easily. Through the use of electroporation, the presence of liposomes in 62% of the bacterial samples examined was identified (Zoaby et al. 2017). This demonstrates the effectiveness of electroporation in facilitating the introduction of liposomes into bacterial cells.
Bacterial loading nanomedicines to improve effectiveness and controllability
Bacteria can be greatly enhanced by loading nanomedicines for therapeutic effects; in fact, the ease of modification of nanomaterials opens up great possibilities for material-based bacterial therapies. The utilization of materials to encapsulate bacteria as drug factories for the production of therapeutic agents, along with the integration of nano-photosensitizers or acoustic sensitizers to achieve synergistic therapies, is a promising area of research. Furthermore, nanomaterials, characterized by their exceptional optical, magnetic, and acoustic properties, can precisely control the timing and location of the anti-tumor effects exerted by the material-modified bacteria. (Fig. 5B).
Immobilization of the chemotherapeutic drug Doxorubicin (DOX) onto ECN through the acid-unstable bond of cis-aconitic anhydride can respond to the acidic microenvironment of tumors (Xie et al. 2017). Upconversion nanoparticles (UCN) can respond to penetrating near-infrared (NIR) light and emit blue light (Cui et al. 2021), which can help to solve the limitation of OCS relying on poorly penetrating blue light. OCS-carrying EcN successfully delivered TNF-α locally to tumor using UCN to convert near-infrared light to blue light and activate the blue light-sensitive EL222 protein in tumor, thereby inhibiting tumor growth (Fig. 5B) (Pan et al. 2021b).
In treating tumors with PDT, the intratumoral hypoxic microenvironment inevitably leads to a significant reduction in the efficacy of type II photosensitizers, which are highly dependent on oxygen. Cyanobacteria can both target the tumor site and continuously produce oxygen by laser irradiation (Fig. 5B). Hence, a novel cyanobacteria-based bioreactor, Cyan@BPNSs, uses inorganic two-dimensional black phosphorus nanosheets (BPNSs) to modify cyanobacteria, significantly enhancing the killing effect of photodynamic therapy on tumor cells (Qi et al. 2021). There are many similar strategies, including the formation of photothermophilic bacteria PTB@ZIF-90/MB by surface biomineralization of palladium nanoparticles in combination with methylene blue (MB)-loaded ZIF-90 (Chen et al. 2020), Salmonella typhimurium YB1 in combination with photosensitizing indocyanine green (INPs) nanoparticles (Chen et al. 2019), and Bb@QDs formed by the electrostatic interaction between Bifidobacterium and Ag2S quantum dots (QDs) (Zhao et al. 2022).
Certain magnetic nanomaterials can convert magnetism into heat. Biological hybrid microrobots EcN@MNP with magnetic field, thermal, and hypoxic environment sensing are constructed by coupling nanomaterials with magnetic nanoparticles (MNP) prepared by zinc-iron hybridization with EcN (Ma et al. 2023). Moreover, the expression of mCherry was controlled using a temperature-controlled expression circuit as a dual reporter gene for thermal and localization signals for tumor treatment. According to the fluorescent protein imaging feedback, the microrobot combined with the effects of magnetothermal ablation and thermally expressed NDH-2 enzyme (respiratory streptokinase II) efficiently triggered apoptosis of cancer cells under the remote triggering of an alternating magnetic field (AMF).
Nanodroplet-transition microbubbles can be used as contrast agents to provide echo signals for ultrasound (Wang et al. 2012), and the microbubbles swell and collapse under continuous intensity ultrasound radiation. Based on this principle, phase-change drug-carrying nanoparticles were attached to bacteria to achieve precise drug release under ultrasound imaging via sonomechanical effects. An ultrasound-responsive biohybrid robot, SonoBacteriaBot (Du et al. 2023), encapsulated perfluoropentane (PFP) and the chemotherapeutic drug DOX in polylactic acid-glycolic acid (PLGA) to form DOX-PFP-PLGA nanodroplets, which were attached to the surface of Escherichia coli MG1655 called DOX-PFP-PLGA@EcM. The SonoBacteriaBot was tracked in real-time by the nanodroplet microbubbles, and under the continuous intensity of ultrasound radiation, the microbubbles expanded and collapsed to release the drug.
Summary and outlook
To date, plenty of intelligent and elegant gene circuits have been developed to precisely trigger anti-tumor effects in response to induced signals. In synergy with nanomedicine, bacteria-nanomaterial nanobiohybrids exert robust and effective multimodal synergistic tumor therapies (Zhou et al. 2024). Numerous experimental results have demonstrated that bacterial therapies under precise control systems successfully inhibited tumor growth and prevented systemic toxicity (Abedi et al. 2022; Chen et al. 2022; Fu et al. 2023). Although recent findings have shown the great potential of bacterial therapies for oncology, there are still some challenges that need to be solved to improve their therapeutic efficacy and safety.
It is important to note that the immune function of patients with advanced cancers is low. Although Clostridium novyi-NT (Janku et al. 2021), Listeria monocytogenes (Hassan et al. 2019; Le et al. 2019, 2015), and Salmonella monocytogenes (Toso et al. 2002) have been proven to be safe in healthy people, the residual presence of bacteria in the body as well as the retention of virulence of bacteria may be a potential threat to cancer patients. Maintaining the stability and orthogonality of intelligent gene circuits and nano-biomaterials is very crucial and should be tumor specific regarding the varied TME. It is thus necessary to emphasize the temporal and spatial controllability of bacterial therapeutics by selecting the most appropriate control signals to trigger precise drug synthesis or release.
Anyway, bacterial therapy is a promising strategy in tumor treatment. The therapeutic potential of bacteria has been further amplified by synthetic biology and nanomedicine, which have endowed bacteria with more diversified functions in the complex TME. It is believed that with the steady development of synthetic biology and the deeper and deeper exploration of cancer, bacterial therapy will eventually become a powerful weapon for human beings to fight against tumors.
Author contribution
ZL: writing—review and editing. LW: writing—review and editing. PW: writing—review and editing, supervision, and funding acquisition. LY: writing—review and editing, supervision, and funding acquisition. All authors read and approved the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (82272010) and the Key R&D Project in Shaanxi Province (2023-ZDLSF-22) to L. Yuan.
Data availability
Data will be made available on request.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pengying Wu, Email: wutdyy@163.com.
Lijun Yuan, Email: yuanlj@fmmu.edu.cn.
References
- Abedi MH, Yao MS, Mittelstein DR, Bar-Zion A, Swift MB, Lee-Gosselin A, Barturen-Larrea P, Buss MT, Shapiro MG (2022) Ultrasound-controllable engineered bacteria for cancer immunotherapy. Nat Commun 13(1):1585. 10.1038/s41467-022-29065-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera L, Campos E, Giménez R, Badía J, Aguilar J, Baldoma L (2008) Dual role of LldR in regulation of the lldPRD operon, involved in L-lactate metabolism in Escherichia coli. J Bacteriol 190(8):2997–3005. 10.1128/jb.02013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrach N, Zhao M, Porwollik S, Hoffman RM, McClelland M (2008) Salmonella promoters preferentially activated inside tumors. Can Res 68(12):4827–4832. 10.1158/0008-5472.CAN-08-0552 [DOI] [PubMed] [Google Scholar]
- Aryal S, Hu CM, Fang RH, Dehaini D, Carpenter C, Zhang DE, Zhang L (2013) Erythrocyte membrane-cloaked polymeric nanoparticles for controlled drug loading and release. Nanomedicine (Lond) 8(8):1271–1280. 10.2217/nnm.12.153 [DOI] [PubMed] [Google Scholar]
- Barak Y, Schreiber F, Thorne SH, Contag CH, Debeer D, Matin A (2010) Role of nitric oxide in Salmonella typhimurium-mediated cancer cell killing. BMC Cancer 10:146. 10.1186/1471-2407-10-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L (1996) Estimation of plasmid loss rates in bacterial populations with a reference to the reproducibility of stability experiments. Plasmid 36(3):161–167. 10.1006/plas.1996.0043 [DOI] [PubMed] [Google Scholar]
- Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, Papayannopoulos V (2014) Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol 15(11):1017–1025. 10.1038/ni.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Cheng S, Wang X, Pang Y, Liu J (2019) Camouflaging bacteria by wrapping with cell membranes. Nat Commun 10(1):3452. 10.1038/s41467-019-11390-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, Sousa C, de Lorenzo V (2001) Rational design of a bacterial transcriptional cascade for amplifying gene expression capacity. Nucleic Acids Res 29(3):759–766. 10.1093/nar/29.3.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra D, Selvanesan BC, Yuan Z, Libutti SK, Koba W, Beck A, Zhu K, Casadevall A, Dadachova E, Gravekamp C (2017) 32-Phosphorus selectively delivered by listeria to pancreatic cancer demonstrates a strong therapeutic effect. Oncotarget 8(13):20729–20740. 10.18632/oncotarget.15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zang Z, Chen Z, Cui L, Chang Z, Ma A, Yin T, Liang R, Han Y, Wu Z, Zheng M, Liu C, Cai L (2019) Nanophotosensitizer-engineered Salmonella bacteria with hypoxia targeting and photothermal-assisted mutual bioaccumulation for solid tumor therapy. Biomaterials 214:119226. 10.1016/j.biomaterials.2019.119226 [DOI] [PubMed] [Google Scholar]
- Chen Q-W, Liu X-H, Fan J-X, Peng S-Y, Wang J-W, Wang X-N, Zhang C, Liu C-J, Zhang X-Z (2020) Self-mineralized photothermal bacteria hybridizing with mitochondria-targeted metal–organic frameworks for augmenting photothermal tumor therapy. Adv Func Mater 30(14):1909806. 10.1002/adfm.201909806 [Google Scholar]
- Chen J, Qiao Y, Chen G, Chang C, Dong H, Tang B, Cheng X, Liu X, Hua Z (2021a) Salmonella flagella confer anti-tumor immunological effect via activating Flagellin/TLR5 signalling within tumor microenvironment. Acta Pharm Sin B 11(10):3165–3177. 10.1016/j.apsb.2021.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Liu CY, Liu C, Zhong DN, Hua SY, He J, Wang K, Zhou M (2021) Wrapping Porphyromonas gingivalis for tumor microenvironment immunomodulation and photothermal immunotherapy. Nano Today 41:101311. 10.1016/j.nantod.2021.101311 [Google Scholar]
- Chen Y, Du M, Yuan Z, Chen Z, Yan F (2022) Spatiotemporal control of engineered bacteria to express interferon-gamma by focused ultrasound for tumor immunotherapy. Nat Commun 13(1):4468. 10.1038/s41467-022-31932-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien T, Harimoto T, Kepecs B, Gray K, Coker C, Hou N, Pu K, Azad T, Nolasco A, Pavlicova M, Danino T (2022) Enhancing the tropism of bacteria via genetically programmed biosensors. Nat Biomed Eng 6(1):94–104. 10.1038/s41551-021-00772-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Castro S, Coker C, Hinchliffe TE, Arpaia N, Danino T (2019) Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med 25(7):1057–1063. 10.1038/s41591-019-0498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinelli MA, Do HT, Miley GP, Silverman RB (2020) Inducible nitric oxide synthase: Regulation, structure, and inhibition. Med Res Rev 40(1):158–189. 10.1002/med.21599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz A, Guo T, Sokolovska A, Miller PF, Collins JJ, Lu TK, Lora JM (2021) Engineering living therapeutics with synthetic biology. Nat Rev Drug Discov 20(12):941–960. 10.1038/s41573-021-00285-3 [DOI] [PubMed] [Google Scholar]
- Cui M, Pang G, Zhang T, Sun T, Zhang L, Kang R, Xue X, Pan H, Yang C, Zhang X, Chang J, Liu J, Zhang S, Wang H (2021) Optotheranostic nanosystem with phone visual diagnosis and optogenetic microbial therapy for ulcerative colitis at-home care. ACS Nano 15(4):7040–7052. 10.1021/acsnano.1c00135 [DOI] [PubMed] [Google Scholar]
- Dai Y, Toley BJ, Swofford CA, Forbes NS (2013) Construction of an inducible cell-communication system that amplifies Salmonella gene expression in tumor tissue. Biotechnol Bioeng 110(6):1769–1781. 10.1002/bit.24816 [DOI] [PubMed] [Google Scholar]
- Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B (2001) Combination bacteriolytic therapy for the treatment of experimental tumors. Proc Natl Acad Sci USA 98(26):15155–15160. 10.1073/pnas.251543698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, Hasty J, Bhatia SN (2015) Programmable probiotics for detection of cancer in urine. Sci Transl Med 7(289):289ra84. 10.1126/scitranslmed.aaa3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K (2011) Optogenetics. Nat Methods 8(1):26–29. 10.1038/nmeth.f.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din MO, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN, Hasty J (2016) Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536(7614):81–85. 10.1038/nature18930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Wang T, Feng R, Zeng P, Chen Z (2023) Establishment of ultrasound-responsive SonoBacteriaBot for targeted drug delivery and controlled release. Front Bioeng Biotechnol 11:1144963. 10.3389/fbioe.2023.1144963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong MT, Qin Y, You SH, Min JJ (2019) Bacteria-cancer interactions: bacteria-based cancer therapy. Exp Mol Med 51(12):1–15. 10.1038/s12276-019-0297-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani V, Entcheva E, Hedrich R, Hegemann P, Konrad KR, Luscher C, Mahn M, Pan ZH, Sims RR, Vierock J, Yizhar O (2022) Optogenetics for light control of biological systems. Nat Rev Methods Primers 2. 10.1038/s43586-022-00136-4 [DOI] [PMC free article] [PubMed]
- English MA, Gayet RV, Collins JJ (2021) Designing biological circuits: synthetic biology within the operon model and beyond. Annu Rev Biochem 90:221–244. 10.1146/annurev-biochem-013118-111914 [DOI] [PubMed] [Google Scholar]
- Faivre D, Schuler D (2008) Magnetotactic bacteria and magnetosomes. Chem Rev 108(11):4875–4898. 10.1021/cr078258w [DOI] [PubMed] [Google Scholar]
- Fan JX, Peng MY, Wang H, Zheng HR, Liu ZL, Li CX, Wang XN, Liu XH, Cheng SX, Zhang XZ (2019) Engineered bacterial bioreactor for tumor therapy via fenton-like reaction with localized H(2) O(2) generation. Adv Mater 31(16):e1808278. 10.1002/adma.201808278 [DOI] [PubMed] [Google Scholar]
- Feng Z, Wang Y, Xu H, Guo Y, Xia W, Zhao C, Zhao X, Wu J (2023) Recent advances in bacterial therapeutics based on sense and response. Acta Pharm Sin B 13(3):1014–1027. 10.1016/j.apsb.2022.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentie K, Kocher B, Gammon ST, Novack DV, McKinney JS, Piwnica-Worms D (2012) A bioluminescent transposon reporter-trap identifies tumor-specific microenvironment-induced promoters in Salmonella for conditional bacterial-based tumor therapy. Cancer Discov 2(7):624–637. 10.1158/2159-8290.CD-11-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes NS, Munn LL, Fukumura D, Jain RK (2003) Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Can Res 63(17):5188–5193 [PubMed] [Google Scholar]
- Fu S, Zhang R, Gao Y, Xiong J, Li Y, Pu L, Xia A, Jin F (2023) Programming the lifestyles of engineered bacteria for cancer therapy. Natl Sci Rev 10(5):nwad031. 10.1093/nsr/nwad031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai S, Arenas RB, Forbes NS (2009) Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br J Cancer 101(10):1683–1691. 10.1038/sj.bjc.6605403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusov VV, Brilkov AV (2002) Estimating the instability parameters of plasmid-bearing cells. I. Chemostat culture. J Theor Biol 219(2):193–205. 10.1006/jtbi.2002.3101 [DOI] [PubMed] [Google Scholar]
- Gao W, Vecchio D, Li J, Zhu J, Zhang Q, Fu V, Li J, Thamphiwatana S, Lu D, Zhang L (2014) Hydrogel containing nanoparticle-stabilized liposomes for topical antimicrobial delivery. ACS Nano 8(3):2900–2907. 10.1021/nn500110a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Jung JH, Lin SM, Jang AY, Zhi Y, Bum Ahn K, Ji HJ, Hyang Lim J, Guo H, Choy HE, Lim S, Seo HS (2020) Development of oxytolerant Salmonella typhimurium using radiation mutation technology (RMT) for Cancer Therapy. Sci Rep 10(1):3764. 10.1038/s41598-020-60396-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Niu L, Wu X, Dai D, Zhou Y, Liu M, Wu K, Yu Y, Guan N, Ye H (2024) Sonogenetics-controlled synthetic designer cells for cancer therapy in tumor mouse models. Cell Rep Med 5(5):101513. 10.1016/j.xcrm.2024.101513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 89(12):5547–5551. 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H (1995) Transcriptional activation by tetracyclines in mammalian cells. Science 268(5218):1766–1769. 10.1126/science.7792603 [DOI] [PubMed] [Google Scholar]
- Gurbatri CR, Arpaia N, Danino T (2022) Engineering bacteria as interactive cancer therapies. Science 378(6622):858–864. 10.1126/science.add9667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurbatri CR, Lia I, Vincent R, Coker C, Castro S, Treuting PM, Hinchliffe TE, Arpaia N, Danino T (2020) Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci Transl Med 12(530). 10.1126/scitranslmed.aax0876 [DOI] [PMC free article] [PubMed]
- Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177(14):4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwisai T, Mirkhani N, Christiansen MG, Nguyen TT, Ling V, Schuerle S (2022) Magnetic torque-driven living microrobots for increased tumor infiltration. Sci Robot 7(71):eabo0665. 10.1126/scirobotics.abo0665 [DOI] [PubMed] [Google Scholar]
- Harimoto T, Hahn J, Chen YY, Im J, Zhang J, Hou N, Li F, Coker C, Gray K, Harr N, Chowdhury S, Pu K, Nimura C, Arpaia N, Leong KW, Danino T (2022) A programmable encapsulation system improves delivery of therapeutic bacteria in mice. Nat Biotechnol 40(8):1259–1269. 10.1038/s41587-022-01244-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan R, Alley E, Kindler H, Antonia S, Jahan T, Honarmand S, Nair N, Whiting CC, Enstrom A, Lemmens E, Tsujikawa T, Kumar S, Choe G, Thomas A, McDougall K, Murphy AL, Jaffee E, Coussens LM, Brockstedt DG (2019) Clinical response of live-attenuated, Listeria monocytogenes expressing mesothelin (CRS-207) with chemotherapy in patients with malignant pleural mesothelioma. Clin Cancer Res 25(19):5787–5798. 10.1158/1078-0432.CCR-19-0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman SM, Tang AY, Avalos JL (2022) Optogenetics illuminates applications in microbial engineering. Annu Rev Chem Biomol Eng 13:373–403. 10.1146/annurev-chembioeng-092120-092340 [DOI] [PubMed] [Google Scholar]
- Huber V, Camisaschi C, Berzi A, Ferro S, Lugini L, Triulzi T, Tuccitto A, Tagliabue E, Castelli C, Rivoltini L (2017) Cancer acidity: an ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin Cancer Biol 43:74–89. 10.1016/j.semcancer.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Jackson MA, Werfel TA, Curvino EJ, Yu F, Kavanaugh TE, Sarett SM, Dockery MD, Kilchrist KV, Jackson AN, Giorgio TD, Duvall CL (2017) Zwitterionic nanocarrier surface chemistry improves siRNA tumor delivery and silencing activity relative to polyethylene glycol. ACS Nano 11(6):5680–5696. 10.1021/acsnano.7b01110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK (1998) The next frontier of molecular medicine: delivery of therapeutics. Nat Med 4(6):655–657. 10.1038/nm0698-655 [DOI] [PubMed] [Google Scholar]
- Janku F, Zhang HH, Pezeshki A, Goel S, Murthy R, Wang-Gillam A, Shepard DR, Helgason T, Masters T, Hong DS, Piha-Paul SA, Karp DD, Klang M, Huang SY, Sakamuri D, Raina A, Torrisi J, Solomon SB, Weissfeld A, Trevino E, DeCrescenzo G, Collins A, Miller M, Salstrom JL, Korn RL, Zhang L, Saha S, Leontovich AA, Tung D, Kreider B, Varterasian M, Khazaie K, Gounder MM (2021) Intratumoral injection of Clostridium novyi-NT spores in patients with treatment-refractory advanced solid tumors. Clin Cancer Res 27(1):96–106. 10.1158/1078-0432.CCR-20-2065 [DOI] [PubMed] [Google Scholar]
- Ji P, Chen J, Wang H, Shi L, Tang X, Duo Y (2023) Bacteria-targeted delivery of black phosphorus quantum dots facilitates photothermal therapy against hypoxic tumors and complementary low-dose radiotherapy. Biomater Sci 11(13):4727–4740. 10.1039/d3bm00206c [DOI] [PubMed] [Google Scholar]
- Jiang SN, Park SH, Lee HJ, Zheng JH, Kim HS, Bom HS, Hong Y, Szardenings M, Shin MG, Kim SC, Ntziachristos V, Choy HE, Min JJ (2013) Engineering of bacteria for the visualization of targeted delivery of a cytolytic anticancer agent. Mol Ther 21(11):1985–1995. 10.1038/mt.2013.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MZ, Jin WL (2020) The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther 5(1):166. 10.1038/s41392-020-00280-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberniuk AA, Baloban M, Monakhov MV, Shcherbakova DM, Verkhusha VV (2021) Single-component near-infrared optogenetic systems for gene transcription regulation. Nat Commun 12(1):3859. 10.1038/s41467-021-24212-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinskas RW, Forbes NS (2007) Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Can Res 67(7):3201–3209. 10.1158/0008-5472.CAN-06-2618 [DOI] [PubMed] [Google Scholar]
- Kotakadi SM, Borelli DPR, Nannepaga JS (2022) Therapeutic applications of magnetotactic bacteria and magnetosomes: a review emphasizing on the Cancer Treatment. Front Bioeng Biotechnol 10:789016. 10.3389/fbioe.2022.789016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SY, Thi-Thu Ngo H, Son J, Hong Y, Min JJ (2024) Exploiting bacteria for cancer immunotherapy. Nat Rev Clin Oncol. 10.1038/s41571-024-00908-9 [DOI] [PubMed] [Google Scholar]
- Lambin P, Theys J, Landuyt W, Rijken P, van der Kogel A, van der Schueren E, Hodgkiss R, Fowler J, Nuyts S, de Bruijn E, Van Mellaert L, Anne J (1998) Colonisation of clostridium in the body is restricted to hypoxic and necrotic areas of tumours. Anaerobe 4(4):183–188. 10.1006/anae.1998.0161 [DOI] [PubMed] [Google Scholar]
- Le DT, Wang-Gillam A, Picozzi V, Greten TF, Crocenzi T, Springett G, Morse M, Zeh H, Cohen D, Fine RL, Onners B, Uram JN, Laheru DA, Lutz ER, Solt S, Murphy AL, Skoble J, Lemmens E, Grous J, Dubensky T Jr, Brockstedt DG, Jaffee EM (2015) Safety and survival with GVAX pancreas prime and Listeria monocytogenes-expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J Clin Oncol 33(12):1325–1333. 10.1200/JCO.2014.57.4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Picozzi VJ, Ko AH, Wainberg ZA, Kindler H, Wang-Gillam A, Oberstein P, Morse MA, Zeh HJ 3rd, Weekes C, Reid T, Borazanci E, Crocenzi T, LoConte NK, Musher B, Laheru D, Murphy A, Whiting C, Nair N, Enstrom A, Ferber S, Brockstedt DG, Jaffee EM (2019) Results from a phase IIb, randomized, multicenter study of GVAX pancreas and CRS-207 compared with chemotherapy in adults with previously treated metastatic pancreatic adenocarcinoma (ECLIPSE Study). Clin Cancer Res 25(18):5493–5502. 10.1158/1078-0432.CCR-18-2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Wu CL, Shiau AL (2005) Systemic administration of attenuated Salmonella choleraesuis carrying thrombospondin-1 gene leads to tumor-specific transgene expression, delayed tumor growth and prolonged survival in the murine melanoma model. Cancer Gene Ther 12(2):175–184. 10.1038/sj.cgt.7700777 [DOI] [PubMed] [Google Scholar]
- Leschner S, Westphal K, Dietrich N, Viegas N, Jablonska J, Lyszkiewicz M, Lienenklaus S, Falk W, Gekara N, Loessner H, Weiss S (2009) Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-alpha. PLoS ONE 4(8):e6692. 10.1371/journal.pone.0006692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschner S, Deyneko IV, Lienenklaus S, Wolf K, Bloecker H, Bumann D, Loessner H, Weiss S (2012) Identification of tumor-specific Salmonella typhimurium promoters and their regulatory logic. Nucleic Acids Res 40(7):2984–2994. 10.1093/nar/gkr1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal DS, Sokolovska A, Li N, Plescia C, Kolodziej SA, Gallant CW, Christmas R, Gao JR, James MJ, Abin-Fuentes A, Momin M, Bergeron C, Fisher A, Miller PF, West KA, Lora JM (2020) Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat Commun 11(1):2739. 10.1038/s41467-020-16602-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P (1996) Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271(5253):1247–1254. 10.1126/science.271.5253.1247 [DOI] [PubMed] [Google Scholar]
- Lindner F, Diepold A (2022) Optogenetics in bacteria—applications and opportunities. FEMS Microbiol Rev 46(2). 10.1093/femsre/fuab055 [DOI] [PMC free article] [PubMed]
- Liu Y, Lu Y, Ning B, Su X, Yang B, Dong H, Yin B, Pang Z, Shen S (2022) Intravenous delivery of living Listeria monocytogenes elicits gasdmermin-dependent tumor pyroptosis and motivates anti-tumor immune response. ACS Nano 16(3):4102–4115. 10.1021/acsnano.1c09818 [DOI] [PubMed] [Google Scholar]
- Loessner H, Endmann A, Leschner S, Westphal K, Rohde M, Miloud T, Hammerling G, Neuhaus K, Weiss S (2007) Remote control of tumour-targeted Salmonella enterica serovar Typhimurium by the use of L-arabinose as inducer of bacterial gene expression in vivo. Cell Microbiol 9(6):1529–1537. 10.1111/j.1462-5822.2007.00890.x [DOI] [PubMed] [Google Scholar]
- Lou X, Chen Z, He Z, Sun M, Sun J (2021) Bacteria-mediated synergistic cancer therapy: small microbiome has a Big Hope. Nanomicro Lett 13(1):37. 10.1007/s40820-020-00560-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Liang X, Li Y, Feng Q, Cheng K, Ma N, Zhu F, Guo X, Yue Y, Liu G, Zhang T, Liang J, Ren L, Zhao X, Nie G (2023) Modular-designed engineered bacteria for precision tumor immunotherapy via spatiotemporal manipulation by magnetic field. Nat Commun 14(1):1606. 10.1038/s41467-023-37225-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Das S (2023) Acid-tolerant bacteria and prospects in industrial and environmental applications. Appl Microbiol Biotechnol 107(11):3355–3374. 10.1007/s00253-023-12529-w [DOI] [PubMed] [Google Scholar]
- Mehta N, Lyon JG, Patil K, Mokarram N, Kim C, Bellamkonda RV (2017) Bacterial carriers for glioblastoma yherapy. Mol Ther Oncolytics 4:1–17. 10.1016/j.omto.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengesha A, Dubois L, Lambin P, Landuyt W, Chiu RK, Wouters BG, Theys J (2006) Development of a flexible and potent hypoxia-inducible promoter for tumor-targeted gene expression in attenuated Salmonella. Cancer Biol Ther 5(9):1120–1128. 10.4161/cbt.5.9.2951 [DOI] [PubMed] [Google Scholar]
- Minamino T, Imada K (2015) The bacterial flagellar motor and its structural diversity. Trends Microbiol 23(5):267–274. 10.1016/j.tim.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Minchinton AI, Tannock IF (2006) Drug penetration in solid tumours. Nat Rev Cancer 6(8):583–592. 10.1038/nrc1893 [DOI] [PubMed] [Google Scholar]
- Mirkhani N, Christiansen MG, Gwisai T, Menghini S, Schuerle S (2024) Spatially selective delivery of living magnetic microrobots through torque-focusing. Nat Commun 15(1):2160. 10.1038/s41467-024-46407-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH (2014) An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol 10(3):196–202. 10.1038/nchembio.1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Bassler BL (2019) Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol 17(6):371–382. 10.1038/s41579-019-0186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr R, Akbari Eidgahi MR (2014) Construction of a synthetically engineered nirB promoter for expression of recombinant protein in Escherichia coli. Jundishapur J Microbiol 7(7):e15942. 10.5812/jjm.15942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J, Bock R (2009) Designing and using synthetic RNA thermometers for temperature-controlled gene expression in bacteria. Nat Protoc 4(9):1262–1273. 10.1038/nprot.2009.112 [DOI] [PubMed] [Google Scholar]
- Nguyen VH, Kim HS, Ha JM, Hong Y, Choy HE, Min JJ (2010) Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Can Res 70(1):18–23. 10.1158/0008-5472.CAN-09-3453 [DOI] [PubMed] [Google Scholar]
- Nuyts S, Van Mellaert L, Barbe S, Lammertyn E, Theys J, Landuyt W, Bosmans E, Lambin P, Anne J (2001a) Insertion or deletion of the cheo box modifies radiation inducibility of Clostridium promoters. Appl Environ Microbiol 67(10):4464–4470. 10.1128/AEM.67.10.4464-4470.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuyts S, Van Mellaert L, Theys J, Landuyt W, Bosmans E, Anne J, Lambin P (2001b) Radio-responsive recA promoter significantly increases TNFalpha production in recombinant clostridia after 2 Gy irradiation. Gene Ther 8(15):1197–1201. 10.1038/sj.gt.3301499 [DOI] [PubMed] [Google Scholar]
- Nuyts S, Van Mellaert L, Theys J, Landuyt W, Lambin P, Anne J (2001c) The use of radiation-induced bacterial promoters in anaerobic conditions: a means to control gene expression in clostridium-mediated therapy for cancer. Radiat Res 155(5):716–723. 10.1667/0033-7587(2001)155[0716:tuorib]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- Omer R, Mohsin MZ, Mohsin A, Mushtaq BS, Huang X, Guo M, Zhuang Y, Huang J (2022) Engineered bacteria-based living materials for biotherapeutic applications. Front Bioeng Biotechnol 10:870675. 10.3389/fbioe.2022.870675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong NT, Olson EJ, Tabor JJ (2018) Engineering an E. coli near-infrared light sensor. ACS Synth Biol 7(1):240–248. 10.1021/acssynbio.7b00289 [DOI] [PubMed] [Google Scholar]
- Pan C, Li J, Hou W, Lin S, Wang L, Pang Y, Wang Y, Liu J (2021a) Polymerization-mediated multifunctionalization of living cells for enhanced cell-based therapy. Adv Mater 33(13):e2007379. 10.1002/adma.202007379 [DOI] [PubMed] [Google Scholar]
- Pan H, Li L, Pang G, Han C, Liu B, Zhang Y, Shen Y, Sun T, Liu J, Chang J, Wang H (2021b) Engineered NIR light-responsive bacteria as anti-tumor agent for targeted and precise cancer therapy. Chem Eng J 426:130842. 10.1016/j.cej.2021.130842 [Google Scholar]
- Pawelek JM, Low KB, Bermudes D (1997) Tumor-targeted Salmonella as a novel anticancer vector. Can Res 57(20):4537–4544 [PubMed] [Google Scholar]
- Peek MC, Ahmed M, Napoli A, ten Haken B, McWilliams S, Usiskin SI, Pinder SE, van Hemelrijck M, Douek M (2015) Systematic review of high-intensity focused ultrasound ablation in the treatment of breast cancer. Br J Surg 102(8):873–82. 10.1002/bjs.9793 [DOI] [PubMed] [Google Scholar]
- Qi F, Ji P, Chen Z, Wang L, Yao H, Huo M, Shi J (2021) Photosynthetic cyanobacteria-hybridized lack phosphorus nanosheets for enhanced tumor photodynamic therapy. Small 17(42):2102113. 10.1002/smll.202102113 [DOI] [PubMed] [Google Scholar]
- Qian X, Han X, Chen Y (2017) Insights into the unique functionality of inorganic micro/nanoparticles for versatile ultrasound theranostics. Biomaterials 142:13–30. 10.1016/j.biomaterials.2017.07.016 [DOI] [PubMed] [Google Scholar]
- Qin W, Xu W, Wang L, Ren D, Cheng Y, Song W, Jiang T, Ma L, Zhang C (2022) Bacteria-elicited specific thrombosis utilizing acid-induced cytolysin A expression to enable potent tumor therapy. Adv Sci (Weinh) 9(15):e2105086. 10.1002/advs.202105086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, You SH, Zhang Y, Venu A, Hong Y, Min JJ (2023) Genetic programming by nitric oxide-sensing gene switch system in tumor-targeting bacteria. Biosensors 13(2). 10.3390/bios13020266 [DOI] [PMC free article] [PubMed]
- Ramakrishnan P, Tabor JJ (2016) Repurposing synechocystis PCC6803 UirS-UirR as a UV-violet/green photoreversible transcriptional regulatory tool in E. coli. ACS Synth Biol 5(7):733–40. 10.1021/acssynbio.6b00068 [DOI] [PubMed] [Google Scholar]
- Raman V, Van Dessel N, Hall CL, Wetherby VE, Whitney SA, Kolewe EL, Bloom SMK, Sharma A, Hardy JA, Bollen M, Van Eynde A, Forbes NS (2021) Intracellular delivery of protein drugs with an autonomously lysing bacterial system reduces tumor growth and metastases. Nat Commun 12(1):6116. 10.1038/s41467-021-26367-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righetti F, Narberhaus F (2014) How to find RNA thermometers. Front Cell Infect Microbiol 4:132. 10.3389/fcimb.2014.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanuka J, Folkers GE, Gnida M, Kovacic L, Wienk H, Kaptein R, Boelens R (2023) Genetic switching by the Lac repressor is based on two-state Monod–Wyman–Changeux allostery. Proc Natl Acad Sci USA 120(49):e2311240120. 10.1073/pnas.2311240120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo JL, Becker PD, Camacho EM, Cebolla A, Link C, Santero E, Guzman CA (2007) In vivo gene regulation in Salmonella spp. by a salicylate-dependent control circuit. Nat Methods 4(11):937–42. 10.1038/nmeth1107 [DOI] [PubMed] [Google Scholar]
- Ryan RM, Green J, Williams PJ, Tazzyman S, Hunt S, Harmey JH, Kehoe SC, Lewis CE (2009) Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther 16(3):329–339. 10.1038/gt.2008.188 [DOI] [PubMed] [Google Scholar]
- Samadi M, Majidzadeh AK, Salehi M, Jalili N, Noorinejad Z, Mosayebzadeh M, Muhammadnejad A, Sharif Khatibi A, Moradi-Kalbolandi S, Farahmand L (2021) Engineered hypoxia-responding Escherichia coli carrying cardiac peptide genes, suppresses tumor growth, angiogenesis and metastasis in vivo. J Biol Eng 15(1):20. 10.1186/s13036-021-00269-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling F, Naydich A, Bramante J, Barocio R, Certo M, Wellington H, Redfield E, O’Keefe S, Gao S, Cusolito A, Way J, Silver P (2020) Synthetic cassettes for pH-mediated sensing, counting, and containment. Cell Rep 30(9):3139–3148. 10.1016/j.celrep.2020.02.033 [DOI] [PubMed] [Google Scholar]
- Strauch KL, Lenk JB, Gamble BL, Miller CG (1985) Oxygen regulation in Salmonella typhimurium. J Bacteriol 161(2):673–680. 10.1128/jb.161.2.673-680.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritzker J, Weibel S, Hill PJ, Oelschlaeger TA, Goebel W, Szalay AA (2007) Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol 297(3):151–162. 10.1016/j.ijmm.2007.01.008 [DOI] [PubMed] [Google Scholar]
- Sun L, Liu H, Ye Y, Lei Y, Islam R, Tan S, Tong R, Miao YB, Cai L (2023) Smart nanoparticles for cancer therapy. Signal Transduct Target Ther 8(1):418. 10.1038/s41392-023-01642-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford CA, Van Dessel N, Forbes NS (2015) Quorum-sensing Salmonella selectively trigger protein expression within tumors. Proc Natl Acad Sci USA 112(11):3457–3462. 10.1073/pnas.1414558112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznol M, Lin SL, Bermudes D, Zheng LM, King I (2000) Use of preferentially replicating bacteria for the treatment of cancer. J Clin Invest 105(8):1027–1030. 10.1172/JCI9818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor JJ, Levskaya A, Voigt CA (2011) Multichromatic control of gene expression in Escherichia coli. J Mol Biol 405(2):315–324. 10.1016/j.jmb.2010.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamphiwatana S, Angsantikul P, Escajadillo T, Zhang Q, Olson J, Luk BT, Zhang S, Fang RH, Gao W, Nizet V, Zhang L (2017) Macrophage-like nanoparticles concurrently absorbing endotoxins and proinflammatory cytokines for sepsis management. Proc Natl Acad Sci USA 114(43):11488–11493. 10.1073/pnas.1714267114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, Freezer LJ, Morton KE, Seipp C, Haworth L, Mavroukakis S, White D, MacDonald S, Mao J, Sznol M, Rosenberg SA (2002) Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol 20(1):142–152. 10.1200/JCO.2002.20.1.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker NP, D’Autreaux B, Yousafzai FK, Fairhurst SA, Spiro S, Dixon R (2008) Analysis of the nitric oxide-sensing non-heme iron center in the NorR regulatory protein. J Biol Chem 283(2):908–918. 10.1074/jbc.M705850200 [DOI] [PubMed] [Google Scholar]
- Tucker NP, Ghosh T, Bush M, Zhang X, Dixon R (2010) Essential roles of three enhancer sites in sigma54-dependent transcription by the nitric oxide sensing regulatory protein NorR. Nucleic Acids Res 38(4):1182–1194. 10.1093/nar/gkp1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebe R, Schuler D (2016) Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol 14(10):621–637. 10.1038/nrmicro.2016.99 [DOI] [PubMed] [Google Scholar]
- Valdez-Cruz NA, Caspeta L, Perez NO, Ramirez OT, Trujillo-Roldan MA (2010) Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters. Microb Cell Fact 9:18. 10.1186/1475-2859-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mellaert L, Barbe S, Anne J (2006) Clostridium spores as anti-tumour agents. Trends Microbiol 14(4):190–196. 10.1016/j.tim.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Wang X, Chen H, Chen Y, Ma M, Zhang K, Li F, Zheng Y, Zeng D, Wang Q, Shi J (2012) Perfluorohexane-encapsulated mesoporous silica nanocapsules as enhancement agents for highly efficient high intensity focused ultrasound (HIFU). Adv Mater 24(6):785–791. 10.1002/adma.201104033 [DOI] [PubMed] [Google Scholar]
- Wang D, Dong H, Li M, Cao Y, Yang F, Zhang K, Dai W, Wang C, Zhang X (2018) Erythrocyte-Cancer hybrid membrane camouflaged hollow copper sulfide nanoparticles for prolonged circulation life and homotypic-targeting photothermal/chemotherapy of melanoma. ACS Nano 12(6):5241–5252. 10.1021/acsnano.7b08355 [DOI] [PubMed] [Google Scholar]
- Wang P, Chen C, Wang Q, Chen H, Chen C, Xu J, Wang X, Song T (2022) Tumor inhibition via magneto-mechanical oscillation by magnetotactic bacteria under a swing MF. J Control Release 351:941–953. 10.1016/j.jconrel.2022.09.059 [DOI] [PubMed] [Google Scholar]
- Wen M, Zheng JH, Choi JM, Pei J, Li CH, Li SY, Kim IY, Lim SH, Jung TY, Moon KS, Min JJ, Jung S (2018) Genetically-engineered Salmonella typhimurium expressing TIMP-2 as a therapeutic intervention in an orthotopic glioma mouse model. Cancer Lett 433:140–146. 10.1016/j.canlet.2018.06.031 [DOI] [PubMed] [Google Scholar]
- Westphal K, Leschner S, Jablonska J, Loessner H, Weiss S (2008) Containment of tumor-colonizing bacteria by host neutrophils. Can Res 68(8):2952–2960. 10.1158/0008-5472.CAN-07-2984 [DOI] [PubMed] [Google Scholar]
- Wu M, Wu WB, Duan YK, Li XQ, Qi GB, Liu B (2019) Photosensitizer-bacteria biohybrids promote photodynamic cancer cell ablation and intracellular protein delivery. Chem Mater 31(18):7212–7220. 10.1021/acs.chemmater.9b01518 [Google Scholar]
- Wu L, Li L, Li S, Liu L, Xin W, Li C, Yin X, Xu X, Bao F, Hua Z (2022a) Macrophage-mediated tumor-targeted delivery of engineered Salmonella typhi murium VNP20009 in anti-PD1 therapy against melanoma. Acta Pharm Sin B 12(10):3952–3971. 10.1016/j.apsb.2022.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xu Z, Xu Q (2022b) Design and optimization of a new alternating electromagnetic-field–generation system for an inverted microscope. Micromachines (Basel) 13(4). 10.3390/mi13040542 [DOI] [PMC free article] [PubMed]
- Xie S, Zhao L, Song X, Tang M, Mo C, Li X (2017) Doxorubicin-conjugated Escherichia coli Nissle 1917 swimmers to achieve tumor targeting and responsive drug release. J Control Release 268:390–399. 10.1016/j.jconrel.2017.10.041 [DOI] [PubMed] [Google Scholar]
- Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, Wang F, Yan H, Zeng L, Zhang L, Zhou F (2022) Microbiota in tumors: from understanding to application. Adv Sci (Weinh) 9(21):e2200470. 10.1002/advs.202200470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Thomas L, Davies B, Chalmers R, Smith M, Brown W (2013) Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol 13:87. 10.1186/1472-6750-13-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Gao F, Fan F, Yang L (2018) Platelet membrane coating coupled with solar irradiation endows a photodynamic nanosystem with both improved antitumor efficacy and undetectable skin damage. Biomaterials 159:59–67. 10.1016/j.biomaterials.2017.12.028 [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Zeng F, Chen Y, Chen Z, Yan F (2024) Ultrasound-visible engineered bacteria for tumor chemo-immunotherapy. Cell Rep Med 5(5):101512. 10.1016/j.xcrm.2024.101512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang M, Shi L, Yao Y, Jiang Q, Li X, Tang LH, Zheng BJ, Yuen KY, Smith DK, Song E, Huang JD (2012) Explicit hypoxia targeting with tumor suppression by creating an “obligate” anaerobic Salmonella typhimurium strain. Sci Rep 2:436. 10.1038/srep00436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Zou Y, Zheng J, Qiu S, Liu L, Wei C (2023) Quorum sensing-mediated microbial interactions: mechanisms, applications, challenges and perspectives. Microbiol Res 273:127414. 10.1016/j.micres.2023.127414 [DOI] [PubMed] [Google Scholar]
- Zhang M, Forbes NS (2015) Trg-deficient Salmonella colonize quiescent tumor regions by exclusively penetrating or proliferating. J Control Release 199:180–189. 10.1016/j.jconrel.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Dehaini D, Zhang Y, Zhou J, Chen X, Zhang L, Fang RH, Gao W, Zhang L (2018) Neutrophil membrane-coated nanoparticles inhibit synovial inflammation and alleviate joint damage in inflammatory arthritis. Nat Nanotechnol 13(12):1182–1190. 10.1038/s41565-018-0254-4 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Peng Q, Zheng J, Yang Y, Zhang X, Ma A, Qin Y, Qin Z, Zheng X (2023) The function and mechanism of lactate and lactylation in tumor metabolism and microenvironment. Genes Dis 10(5):2029–2037. 10.1016/j.gendis.2022.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huang H, Zhao J, Xiong X, Zheng S, Wei X, Zhou S (2022) A hybrid bacterium with tumor-associated macrophage polarization for enhanced photothermal-immunotherapy. Acta Pharm Sin B 12(6):2683–2694. 10.1016/j.apsb.2021.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JH, Nguyen VH, Jiang SN, Park SH, Tan W, Hong SH, Shin MG, Chung IJ, Hong Y, Bom HS, Choy HE, Lee SE, Rhee JH, Min JJ (2017) Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci Transl Med 9(376). 10.1126/scitranslmed.aak9537 [DOI] [PubMed]
- Zhou S, Gravekamp C, Bermudes D, Liu K (2018) Tumour-targeting bacteria engineered to fight cancer. Nat Rev Cancer 18(12):727–743. 10.1038/s41568-018-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li Q, Wu Y, Zhang W, Ding L, Ji C, Li P, Chen T, Feng L, Tang BZ, Huang X (2024) Synergistic brilliance: engineered bacteria and nanomedicine unite in cancer therapy. Adv Mater 36(21):e2313953. 10.1002/adma.202313953 [DOI] [PubMed] [Google Scholar]
- Zinger A, Koren L, Adir O, Poley M, Alyan M, Yaari Z, Noor N, Krinsky N, Simon A, Gibori H, Krayem M, Mumblat Y, Kasten S, Ofir S, Fridman E, Milman N, Lubtow MM, Liba L, Shklover J, Shainsky-Roitman J, Binenbaum Y, Hershkovitz D, Gil Z, Dvir T, Luxenhofer R, Satchi-Fainaro R, Schroeder A (2019) Collagenase nanoparticles enhance the penetration of drugs into pancreatic tumors. ACS Nano 13(10):11008–11021. 10.1021/acsnano.9b02395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoaby N, Shainsky-Roitman J, Badarneh S, Abumanhal H, Leshansky A, Yaron S, Schroeder A (2017) Autonomous bacterial nanoswimmers target cancer. J Control Release 257:68–75. 10.1016/j.jconrel.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga A, Camacho M, Chang H-J, Fristot E, Mayonove P, Hani E-H, Bonnet J (2021) Engineered l-lactate responding promoter system operating in glucose-rich and anoxic environments. ACS Synth Biol 10(12):3527–3536. 10.1021/acssynbio.1c00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.