Abstract
The relative contributions of mutation rate variation, selection, and recombination in shaping genomic variation in bacterial populations remain poorly understood. Here we analyze 3318 Yersinia pestis genomes, spanning nearly a century and including 2336 newly sequenced strains, to shed light on the patterns of genetic diversity and variation distribution at the population level. We identify 45 genomic regions (“hot regions”, HRs) that, although comprising a minor fraction of the genome, are hotbeds of genetic variation. These HRs are distributed non-randomly across Y. pestis phylogenetic lineages and are primarily linked to regulatory genes, underscoring their potential functional significance. We explore various factors contributing to the shaping and maintenance of HRs, including genomic context, homologous recombination, mutation rate variation and natural selection. Our findings suggest that positive selection is likely the primary driver behind the emergence of HRs, but not the sole force, as evidenced by the pronounced trend of variation purging within these regions.
Subject terms: Bacterial evolution, Evolutionary theory, Bacterial genomics, Phylogenetics, Pathogens
The relative contributions of mutation rate variation, selection, and recombination in shaping genomic variation in bacterial populations remain poorly understood. Here, the authors analyze over 3,300 Yersinia pestis genomes, spanning nearly a century, to shed light on the patterns of genetic diversity and variation distribution at the population level.
Introduction
The evolutionary trajectory of bacteria is profoundly shaped by spontaneous mutations1–3, which are not uniformly scattered across the bacterial chromosome4. This disparity leads to the emergence of genomic hot spots and cold spots for mutations, a phenomenon well-illustrated by contingency loci in pathogenic bacteria5. These regions, marked by increased mutability, are assumed to be a result of evolutionary adaptations that facilitate rapid phenotypic changes in response to the unpredictable challenges posed by environmental pressures.
The proliferation of next-generation sequencing (NGS) technology has confirmed the existence of mutation biases within the genomes of a wide array of bacterial species6–9. However, the distribution patterns and functional units of non-random mutation in large-scale natural populations are not fully understood, and no single explanatory model has gained universal acceptance. This gap in understanding has led to ongoing debates, highlighted by the conflicting conclusions from studies on Escherichia coli6,10. Martincorena et al. examined the genomes of 34 E. coli natural isolates and discovered distinct non-random patterns in synonymous substitutions, with certain regions exhibiting more than a 20-fold difference in synonymous diversity (θs)6. They proposed that these variations might reflect evolutionarily shaped local mutation rates. Conversely, Maddamsetti et al. who analyzed data from the long-term evolution experiment (LTEE) with E. coli, observed an uneven distribution of point substitutions as well but linked it to the gene length rather than to variations in mutation rates across genes10. They suggested that observed differences in θs within natural E. coli populations could be due to variations in effective population size (Ne), influenced by selective pressures, gene flow, or population structure.
Yersinia pestis, the infamous agent behind the historical plague pandemics, persists in natural reservoirs, continuing to pose a significant public health risk11,12. A detailed examination of 133 Y. pestis genomes revealed a significantly high density of nonsynonymous SNPs in seven out of 3450 core genes13. In a separate study focusing on 78 Y. pestis strains collected from a localized natural plague focus more than four decades, the rpoZ gene stood out as a genetic variation hotspot, displaying a variation density that exceeded other genomic regions by over 2000-fold14. These observations accentuate the non-random distribution of substitution within the Y. pestis genome. Building upon these findings, we have scrutinized 3318 Y. pestis genomes that span a historical period of 97 years and encompass samples from the primary natural plague foci globally. Our comprehensive study seeks to shed light on the genetic diversity and substitution distribution patterns at the natural population level. By using Y. pestis as a model organism, we provide a detailed view of the hot regions of genetic variations and hypothesize about the mechanisms that dictate non-random variation distribution.
Results
Global diversity of the current largest dataset of Y. pestis
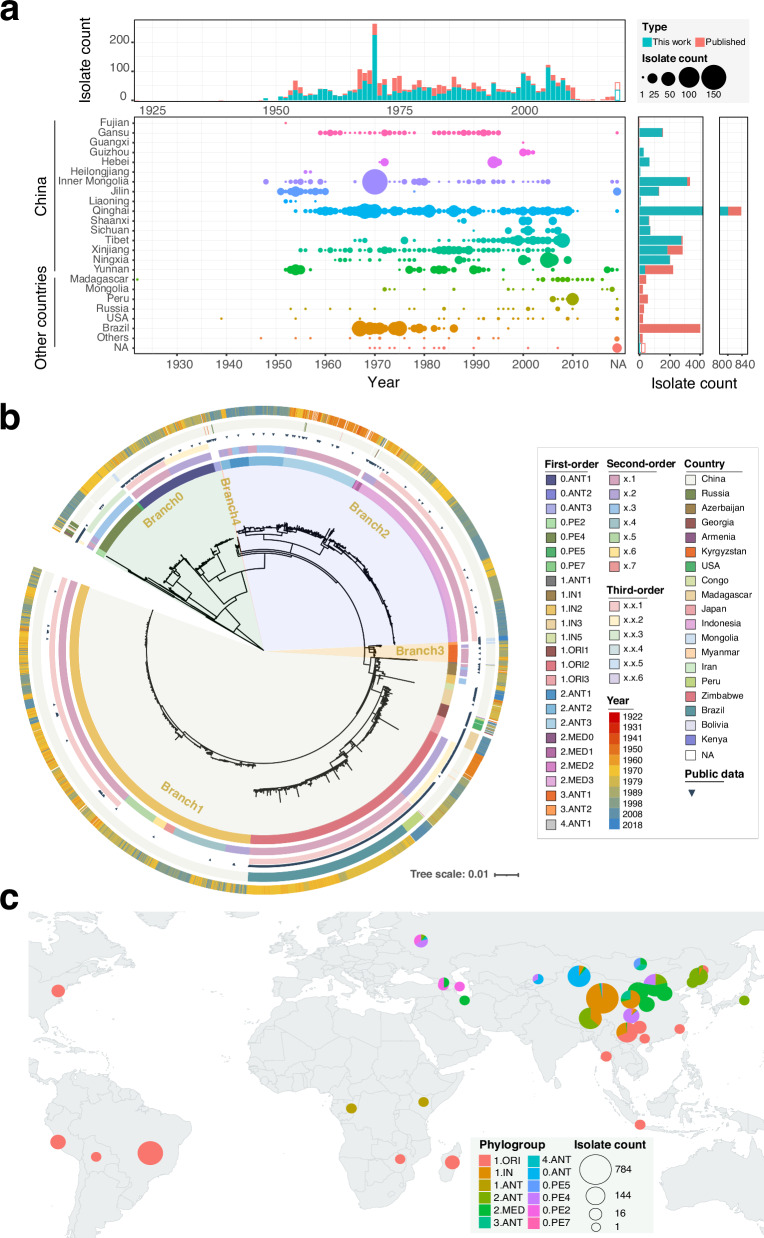
Our comprehensive collection of 3318 Y. pestis genomic sequences, including 2336 newly sequenced samples (Supplementary Data 1) and 982 from the NCBI database (Supplementary Data 2), spans 19 countries across 5 continents from 1922 to 2018 (Fig. 1a). This dataset represents the most extensive Y. pestis genome collection to date. Utilizing 6552 high-quality SNPs from the core genome, we constructed a maximum-likelihood (ML) phylogenetic tree (Fig. 1b), which affirmed the geographical distribution of Y. pestis populations and supported previously described constraints (Fig. 1c)13,15. The median pairwise genetic distance among strains is 118 SNPs, reinforcing the genetically monomorphic nature of the Y. pestis core genome11,16 (Supplementary Fig. 1).
Fig. 1. Phylogenetic analysis and global distribution of 3318 Y. pestis genomes.
a Bubble chart illustrating the distribution of sampling year and location for public and newly sequenced data. The x-axis represents years, while the y-axis shows locations globally and provinces within China. Bubble size corresponds to strain counts, with additional bar charts at the top and right displaying counts by time and location. Color variations differentiate between public and new data. Source data are provided as a Source Data file. b Maximum likelihood tree based on 6552 SNP sites from the core genomes of 3318 Y. pestis strains. Circles from inner to outer as follows: first-order clades, second-order clades, third-order clades, public data, isolation country, and year. c Global geographical distribution of major Y. pestis populations. Pie chart size indicates strain count, with different colors representing unique phylogroups. The world map was created using the ggplot2 and maps packages in R. Source data are provided as a Source Data file.
While no major new branches were identified beyond the established designations (Branches 0–4)13,17, our data refined the phylogenetic structure, revealing multiple novel tip clades (Fig. 1b). We proposed a three-level hierarchical nomenclature system for Y. pestis clade assignment, inspired by the dynamic nomenclature used for SARS-CoV-2 (see Methods)18, resulting in 31 first-order clades, 34 second-order clades, and 23 third-order clades (Supplementary Fig. 2). Five clades contained genomes exclusively sequenced in this study (n = 196, Supplementary Data 1), filling previous gaps in our understanding of Y. pestis diversity.
Detection of non-randomly distributed variations
In a genome where genetic variations are fixed by chance, their distribution should conform to a binomial model. Contrary to this expectation, our analysis of 9370 polymorphic sites—comprising 6552 SNPs and 2818 indels—uncovered 45 variation hot regions (HRs) scattered across the core genome of Y. pestis (Fig. 2a and Supplementary Data 3). These HRs displayed a level of nucleotide diversity (θ) that significantly exceeded the binomial distribution’s theoretical forecasts (Padjusted < 0.05), as determined by a binomial-based sliding window and random sampling procedures (refer to Methods).
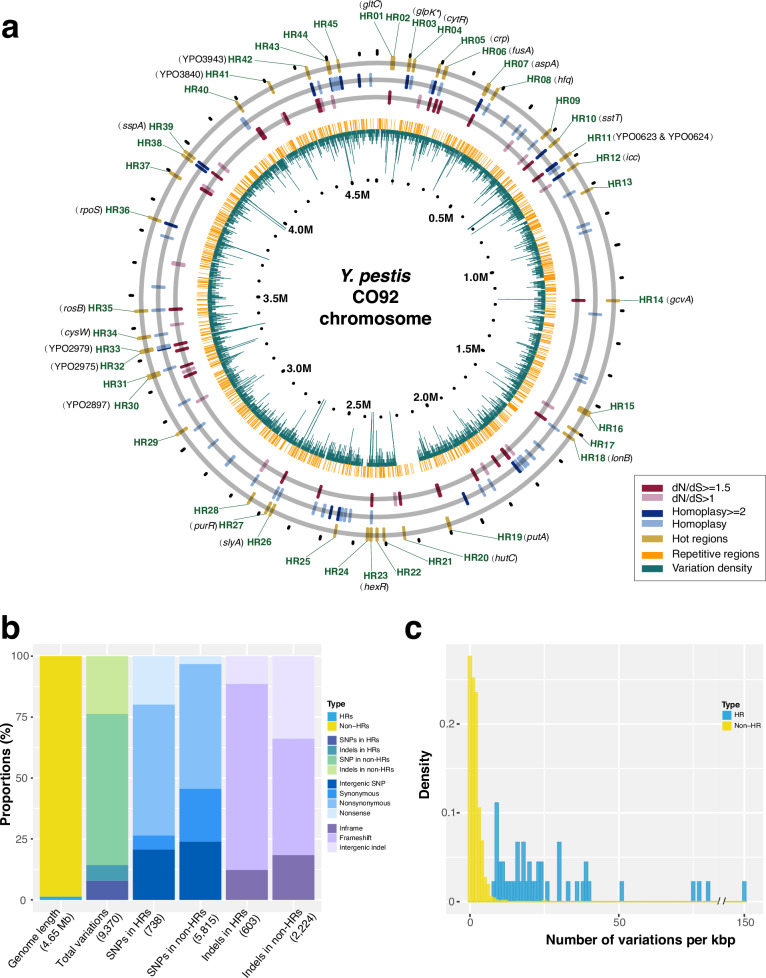
Fig. 2. Genomic features of variation hot regions (HRs).
a Distribution of 45 HRs on the CO92 chromosome. The outermost circle displays the identified HRs (HR01-HR45). Two middle circles represent genes with homoplasy sites or dN/dS ratios above 1, with genes meeting these criteria labeled in black. The two inner circles, shown in orange and green lines, correspond to repeat regions and variation density (window size = 500 bp), respectively. b Statistical comparison of sequence length, variation count, and variation types for HRs and non-HRs. Numbers in parentheses represent the total genome length, variation sites, or mutation types per bar. Some SNPs and indels are multiallelic, so the number of variation sites may be fewer than the number of mutation types. SNPs in HRs: 737 sites (738 types); indels in HRs: 599 sites (603 types); indels in non-HRs: 2219 sites (2224 types). Source data are provided as a Source Data file. c Vatiation density comparison between HRs and randomly selected non-HRs of equal length (averaged over 10,000 repetitions). Source data are provided as a Source Data file.
These HRs, labeled HR01-HR45, span 40–4436 bp and contain 6–226 variations (Supplementary Data 3). While they represent only 1.35% of the reference genome’s chromosome length, they include 14.26% of all genome-wide variations (n = 1336/9370) (Fig. 2b and Supplementary Data 4). Variation density in HRs ranges from 8 to 150 per kbp, far exceeding the genome-wide average of about 2 per kbp (Fig. 2c). HRs also show significantly higher ratios of nonsense (6.10-fold increase) and frameshift variations (1.60-fold increase) compared to other genomic regions (Fig. 2b and Supplementary Data 4, Fisher’s exact test, P < 2.2 × 10−16 and P = 1.19 × 10−10). Moreover, the nonsynonymous-to-synonymous substitution ratio per site in HRs is 1.541 × 10−4, substantially higher than the 5.165 × 10−7 ratio for non-HR regions.
Chromosome and population distribution of hot regions
Among the 45 identified HRs, only five were confined to specific genes (HR01, HR18, HR20, HR41, and HR42, ranging from 505 to 1382 bp) (Supplementary Fig. 3a), and six were located within intergenic regions (HR13, HR21, HR25, HR43, HR44, and HR45, ranging from 40 to 429 bp) (Supplementary Fig. 3b). The preponderance of HRs (75.56%, n = 34) spanned multigenic domains, covering 1 to 4 genes and extending from 527 to 4436 bp in length (Supplementary Fig. 3c).
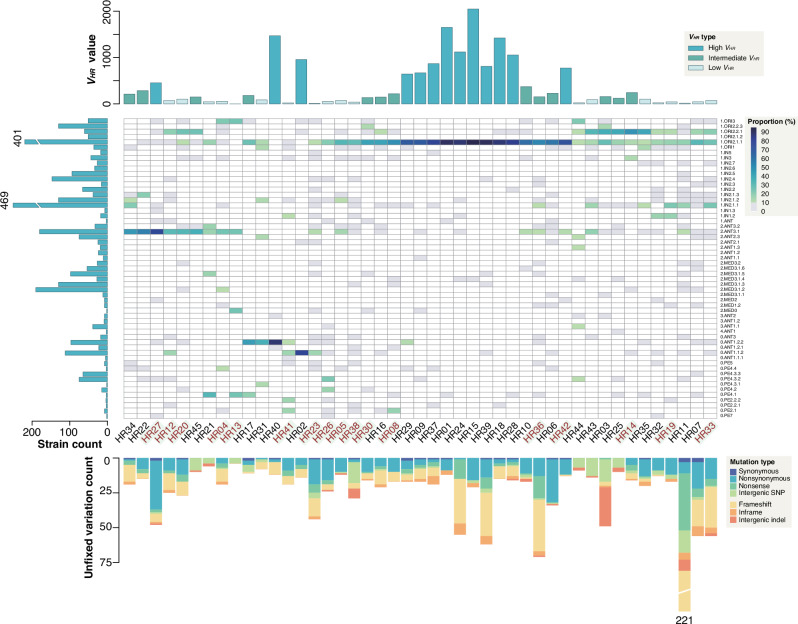
The distribution of HRs across the Y. pestis phylogeny is strikingly uneven, with a marked preference for certain phylogroups (Fig. 3 and Supplementary Fig. 4). For example, 68.75% of variations within HR02 were exclusive to the 0.ANT1 population (Supplementary Fig. 4 and Supplementary Data 5). We quantified the dispersion of variation across phylogroups per HR by calculating the variance of relative abundance of variation sites, denoted as VHR (see Methods). This metric revealed that 28.89% of HRs (n = 13/45) had high VHR values (over 400), indicating a concentration within specific phylogroups (Fig. 3 and Supplementary Data 6). Conversely, 42.22% of HRs (n = 19/45) showed low VHR (below 101), suggesting variations affecting the species at large, as they are distributed across 4–44 top-level phylogroups. HRs with intermediate VHR values (101–400) present a mixed distribution pattern; with variations predominantly found in one or two phylogroups, yet also sporadically present in several others.
Fig. 3. Mutation distribution and composition in 45 HRs across populations.
The heatmap, with columns clustered, illustrates the proportions of variation sites across different populations for each HR, with dark shades representing higher values, gray for values between 0 and 10, and white for no variation. HRs labeled in red on the x-axis highlight regions containing regulator-related genes. The upper bar chart displays the variance of the proportions; the left bar chart depicts the strain count for each phylogroup; and the lower stacked bar chart presents the number of unfixed variations within each HR, with different colors representing distinct genetic variation types. Source data are provided as a Source Data file.
To explore potential epistatic interactions among HRs that could shape the evolutionary path of Y. pestis, we analyzed the phylogroup proportions of HR variations and discerned a non-random pattern of associations, grouping the 45 HRs into four distinct clusters (Supplementary Fig. 5). Group 1 HR variations are chiefly found in the 1.ORI2.1.1 phylogroup, while Group 3’s are mainly in 2.ANT3.1. Group 2, characterized by HRs with low to intermediate variances, show a broader phylogroup presence, particularly within 1.ORI2.2.1, 1.ORI2.1.1, and 1.IN2.1.1. Group 4, although more scattered, revealed closely related HRs, such as HR17, HR30, and HR41, as well as HR02 and HR41, with denser distributions in the 0.ANT1.2.2 and 0.ANT1.1.2 phylogroups, respectively. These patterns hint at the possibility of positive epistasis among these clustered regions.
Functional categorization of genes in HRs
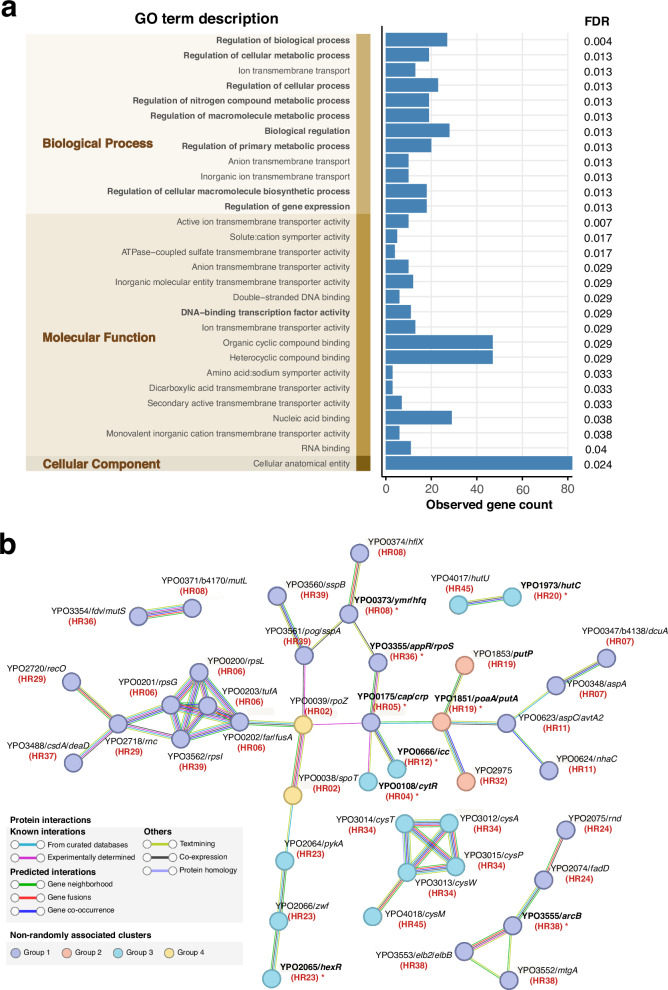
Gene Ontology (GO) term enrichment analysis was conducted on 96 genes associated with identified HRs, including genes within HRs and those potentially influenced by adjacent intergenic regions. We identified 12 significantly enriched GO terms in Biological Process, 16 in Molecular Function, and one in Cellular Component (FDR < 0.05, Fig. 4a).
Fig. 4. Functional enrichment analysis and protein-protein interaction analysis of HR-associated genes.
a Enrichment analysis of Gene Ontology (GO) terms for 96 HR-related genes against the whole genome background. GO terms are categorized into three distinct domains, represented as vertical stacked bars and highlighted in the shaded area. Regulatory function-related GO terms are highlighted in bold text. The blue bar chart represents the observed count of HR-related genes in corresponding GO terms (noting that each gene can correspond to multiple GO terms). The false discovery rate (FDR) for significantly enriched GO terms (FDR < 0.05) are labeled in the rightmost. Source data are provided as a Source Data file. b STRING association analysis for 96 HR-related genes. Only node interactions with a confidence score greater than 0.7 are displayed, while disconnected nodes in the network are hidden and nodes connected solely through physical interactions were removed manually. Nodes are color-coded according to four clustered groups with possible epistatic signals, while connecting lines are colored based on the evidence supporting the interaction within the STRING database. Bold gene names with asterisks indicate regulatory proteins, and the red text in parentheses below the gene name indicates the HR associated with the corresponding gene.
A notable finding was the prevalence of regulatory proteins among the HR-associated genes. This was evidenced by 9 of the 12 significantly enriched GO terms in Biological Process and one enriched GO term (DNA-binding transcription factor activity) in Molecular Function being related to regulatory functions (Fig. 4a). To further support this observation, we utilized the P2RP platform for regulatory protein prediction, augmented by CO92 annotations19. A pronounced overrepresentation of 18 regulatory proteins, involving 17 HRs (Fig. 3 and Supplementary Data 6), was found within the 96 HR-associated genes compared to 358 potential regulators identified across the entire CO92 chromosome (Hypergeometric test, P = 0.0017). HRs linked to regulatory proteins showed a significantly lower VHR than those not associated with regulators (Wilcoxon rank sum test, P = 0.0069), suggesting that regulator-related HRs may exert an influence on the regulatory networks at the species level, potentially affecting the evolutionary trajectory of Y. pestis as a whole.
To examine protein-protein interactions (PPI) among HR-related genes, we constructed a PPI network using STRING database information20. This network revealed five gene clusters with interaction confidence scores exceeding 0.7 (Fig. 4b and Supplementary Data 7). The predominant cluster comprised 28 genes, including seven regulatory proteins, and featured three high-confidence sub-clusters (score ≥0.9). Correlating gene functions with the COG database, we observed strong connections between genes from different HRs sharing similar functions. For instance, genes rpsG, rpsL, tufA, and fusA from HR06, along with rpsI from HR39, fall under the same COG category and exhibit close connectivity. These genes, which are integral to information storage and processing, also associate with recO and rnc from HR29 and deaD from HR37 (Fig. 4b). Intriguingly, HRs comprising interacting genes share similar patterns in population distribution (Fig. 3 and Supplementary Fig. 5). For example, HR06, HR29, HR37, and HR39 all show a high frequency of variations within the 1.ORI2.1.1 phylogroup. These observations lend support to the notion of positive epistasis among these HRs, underscoring the need for further study to unravel their functional interdependencies and their collective influence on the evolution of Y. pestis.
Diverse factors that may contribute to the observed patterns of HRs
Four potential mechanisms are hypothesized to influence the emergence of HRs. Firstly, the distinct genomic context of these regions may inherently predispose them to greater variability than other genomic segments, such as known contingency loci5,21. Secondly, homologous recombination may introduce new genetic material with higher density of variations than typically seen throughout the genome22,23. Thirdly, variation in mutation rates could precipitate non-random distributions of genetic variations across the genome, a pattern that emerges even without a specific genomic context and is described as an evolutionary risk management strategy6,24. The final mechanism involves selection pressure; genome regions under intense positive selection are likely to show a surge in variation density25,26. Finally, our research scrutinizes each of these potential influences to infer the most credible explanations for the patterns observed in our dataset.
Genomic context
We assess the impact of local sequence context on genomic diversity by comparing the GC content and the complexity of genomic regions between HRs and non-HRs. Our analysis did not reveal a significant difference in GC content (two-tailed Welch’s t test, P = 0.17) (Supplementary Fig. 6a). Although regions of lower genomic complexity, such as homopolymers, are generally associated with higher mutation rates, our comparison of genomic complexity—measured by linguistic complexity (LC) score and Shannon’s entropy (H) score27—showed no statistically significant differences between HRs and non-HRs (two-tailed Welch’s t test, P = 0.21 and P = 0.24) (Supplementary Fig. 6a). These results suggest that the complexity of the genome does not predominantly influence the formation of HRs.
Homologous recombination
Y. pestis is typically characterized by a clonally structure, with scant evidence of recombination events. Our analysis of homologous recombination in the core genome of 3318 Y. pestis strains revealed that, although the recombination to mutation ratio (r/m) ranged from 0.17 to 5.09 across different methods and datasets, the recombination coverage was exceedingly low (<1.6% overall, median <0.3% per sample). In comparison, its ancestor Y. pseudotuberculosis exhibited recombination coverage exceeding 64.9% overall, with a median over 16.27% per sample (see Methods, Supplementary Fig. 7a–d, and Supplementary Data 8). These findings underscore the minimal contribution of recombination to the genetic diversity of this species. This finding aligns with previous observations and reinforces the notion that genetic variations in Y. pestis are predominantly the result of spontaneous mutations rather than recombination events.
We also examined the overlap between recombination regions and the HRs, as well as the number of variations in relation to the ancestral state within each HR for every isolate. Our analysis revealed that only HR06 partially overlapped with a single recombination region identified by two methods, covering approximately 5% of HR06’s length. Additionally, the number of variations within each genome was constrained at any given HR (≤4, Supplementary Fig. 7e). After excluding potential non-single-introduction events (refer to Methods for details), we identified only 31 clusters of physically linked variations (0.07%, n = 31/42,963) that could be attributed to recombination (Supplementary Data 9). Of these, 29 clusters involved the concurrent introduction of two variations, with five clusters where linked variations became fixed in the population; only two clusters potentially involved three linked variations introduced simultaneously in two separate isolates (Supplementary Fig. 7f). These data indicate that recombination has a negligible impact on the development of HRs.
Mutation rate variation
In alignment with the neutral theory of molecular evolution (θ = 2Neμ), we anticipate that population diversity (θ) would be a reflection of both the mutation rate (μ) and the effective population size (Ne)28. Given the low recombination coverage in Y. pestis, each genomic site’s history is congruent with the history of the organism itself, suggesting that the effective population size’s demographic influence on individual sites might be minimal. Our analysis revealed no substantial codon usage bias impacting synonymous diversity within the population (paired Wilcoxon signed-rank test, with P-values ranging from 0.57 to 1, see Methods), thereby supporting the use of synonymous diversity (θs) as an indicative measure of mutation rates.
We observed that the θs within HRs was 2.38-fold higher than in the remaining genomic landscape, indicating a potential acceleration of mutation rates within these regions (binomial test, P < 1.19 × 10−6). While most HRs exhibited synonymous mutation counts and θs values close to the genome-wide average, five HRs (HR11, HR17, HR27, HR37, and HR42) displayed significantly elevated θs, ranging from 4.65 to 9.12 times higher, suggesting a potential mutation rate bias in these regions. Moreover, we identified a distinct mutation spectrum between HRs and non-HRs, as well as between HRs with high θs and non-HRs (Chi-squared test, P = 0.011 and P = 0.0031, respectively; Supplementary Fig. 6b). HRs with high θs showed a greater proportion of G:C- > T:A (transversion) and a lower proportion of G:C- > A:T (transition) compared to other HRs and non-HRs. These findings suggest that we cannot exclude the possibility of elevated mutation rates occurring in several HRs, potentially driven by complex evolutionary processes, which requires further validation.
Selection pressure
Previous research has shown limited evidence of natural selection across the whole genome of Y. pestis during its evolution13. Our study supports this observation, identifying only 47 genes with high dN/dS ratios (dN/dS > 1) and 17 genes that, despite lacking synonymous SNPs, demonstrated a significantly higher occurrence of nonsynonymous SNPs (P < 0.05). Importantly, a substantial proportion of genes with selection signatures are located within HRs. We identified 18 genes with high dN/dS ratios and 11 with an excess of nonsynonymous SNPs, suggesting adaptive evolution within these regions (Supplementary Data 10). This is further supported by the prevalence of homoplasies, with 62.32% occurring within HRs (Fig. 2a and Supplementary Data 11). Additionally, the fixation of variations within the Y. pestis population in 37 out of the 45 HRs points to the influence of positive selection (Supplementary Data 5). For instance, in HR31, seven variations, including six within the ail gene and one upstream of the ail gene by 50 base pairs, were independently fixed in the main branch and across multiple phylogroups (Supplementary Fig. 8). Collectively, these findings suggest that positive selection may significantly influence the genetic landscape of HRs.
However, the majority of HR variations (93.11%) had low allele frequencies, with many observed in only one or two genomes out of the 3318 analyzed (Supplementary Data 5). For example, in HR36, a mere 1.4% were fixed across the species. This suggests that while positive selection appears to be the main driver in shaping HRs, most variations within HRs are purged over time, contrary to expectation if positive selection was the sole force at play.
To further investigate whether variations within HRs could confer a fitness advantage, we focused on HR06, which encompasses the rpsL gene associated with antibiotic resistance. The K43R mutation in rpsL gene has been documented to confer resistance to streptomycin, the first-line antibiotic for Y. pestis infection29,30. Within HR06, we identified 36 variations: nine within rpsL, 25 across the adjacent rpsG, fusA and tuf genes, and two in the intergenic regions. Despite this, only the previously reported K43R mutation in rpsL29,30 demonstrated a resistance phenotype; the other seven variations we tested did not result in drug resistance (Supplementary Fig. 9 and Supplementary Data 12). Notably, a strain from the same region as the streptomycin-resistant variant, carrying the R86S mutation in rpsL, the mutation frequently observed in E. coli streptomycin resistance study31, remained as sensitive to the antibiotic as wild-type strains. This suggest that many of the variations in HR06 do not offer a selective advantage under antibiotic pressure.
Discussion
The surge in high throughput genome sequencing has exponentially increased the availability of pathogen genomes, offering unprecedented opportunities into the evolutionary intricacies of genetically monomorphic species like Y. pestis. The previously hidden evolutionary patterns can now be exposed due to whole-genome level resolution in determining diversity. Nevertheless, this abundance of genomic information also brings to the fore the challenge of developing a robust and coherent nomenclature system to classify emerging clades. Drawing inspiration from the comprehensive naming system employed for SARS-CoV-2, which effectively manages over 10 million genomes18, we propose a similar structured three-tiered hierarchical naming framework for Y. pestis clades. This proposed system harmonizes with existing nomenclature practices13,15,32, ensuring continuity while also being scalable enough to accommodate the anticipated influx of genomic data. It promises to streamline communication within the scientific and public health sectors regarding evolution, ecology, and epidemiology of Y. pestis.
Echoing the seminal work by Tenaillon et al. on E. coli7, our findings corroborate the notion that selection operates at multiple biological scales, from genes to operons and beyond. In Y. pestis, we observed that the distribution of clustered genetic variations, or HRs, reflect this multi-level selection. While a minority of HRs are confined to single genes or intergenic spaces, the majority traverse both, suggesting that evolutionary mechanisms of Y. pestis may act on expansive genomic segments. This observation expands our understanding of the units of adaptive evolution in this pathogen.
Our analysis also reveals that HR variations are stratified across the Y. pestis phylogeny. Some HRs, like the one encompassing the ail gene linked to phage resistance33–36 are widespread, indicating their pivotal role in the species’ survival across diverse environmental challenges. Conversely, other HRs appear more temporally and phylogenetically localized, such as the one involving the rpoZ gene, hinting at adaptations to unique environmental pressures faced by certain phylogroups14.
We observed a pronounced overrepresentation of regulatory proteins within the HRs, with over one-third of HRs implicated. The widespread distribution of these HRs across different phylogroups aligns with their potential role in bolstering Y. pestis’ adaptability in response to environmental fluctuations. Interaction network analysis via STRING revealed significant protein-protein interactions among these HRs, suggesting the possibility that hotspots may coalesce into functional complexes. Furthermore, HRs displaying protein interactions were frequently found to cluster within particular phylogroups, suggesting a linkage between STRING-defined functional complexes and HR groupings. This observed pattern may reflect positive epistasis among HRs, potentially underpinning their synergistic action. Although correlating HR variations with specific phenotypes remains a challenging task due to the pleiotropic nature of the associated genes, our findings identified novel avenues for probing the physiology and pathogenic mechanisms of Y. pestis.
Despite the increased number of variations observed within hotspots, many exhibit low allele frequencies and do not appear to be adaptive. One hypothesis rationalizes this pattern is transient Darwinian selection, which echoes findings from a population genomics study on Salmonella enterica serovar Paratyphi A8. These regions may have undergone diversifying selection over short timescale. Initially, certain variants were favored and increased in prevalence, but they were subsequently subject to negative selection, leading to their gradual purging. This may be due to fitness reductions caused by changing environmental selection pressures or the absence of compensatory epistatic mutations. Another hypothesis involves the combined effects of selection and genetic drift. An estimate of the Ne for Y. pestis, based on the Bayesian Coalescent Skyline model, was 1 × 103-1 × 104 (95% CI: 1 × 102-1 × 105)37, indicating a small Ne for Y. pestis. This finding is supported by a comparison of Ne across 152 bacterial species and one archaeon using different evaluation methods38. Thus, such a small Ne for Y. pestis could “purge” alleles in HRs and prevent them from reaching fixation. Realistic mathematical models for bacterial genome evolution are needed in future work to quantitatively clarify the impacts of these driver forces.
We observed higher θs in a few HRs compared to other genomic regions, therefore it cannot fully exclude the potentially accelerated mutation rate in these regions. An elevated mutation rate in localized genome regions could provide a greater probability for the emergence of advantageous mutations, conferring fitness benefits while maintaining a reduced evolutionary risk in contrast to global mutator phenotypes6,24. Although several explanations had been inferred related with the upsurge of localized mutation rates, including transcription-induced mutagenesis, replication-repair system bottlenecks, the chromosomal spatial organization, epigenetic variation, and targeted error-prone polymerases4,39, the mechanisms need to be further investigated, possibly through long-term evolutionary experiment and mutation accumulation studies.
Our findings are based on the comprehensive sampling of genetically monomorphic population with negligible homologous recombination. Limited sampling could overlook some HRs, particular since many HR mutations are likely lost over evolutionary timescales. With the extensive accumulation of bacterial genome sequences in current age, previously undetectable evolutionary details, such as HR characters identified in this study, are likely discovered in other bacterial species. This will provide a more robust foundation for developing quantitative models for bacterial evolution. Such efforts will not only shed light on the fate of mutations within populations, including their emergence, fixation, and potential extinction, but also enrich our comprehension of bacterial adaptive evolution, with practical implications for understanding phenomena like antibiotic resistance from a novel perspective.
Methods
Data collection
In this study, we analyzed 3318 Y. pestis genome sequences, including 2336 newly sequenced strains (Supplementary Data 1), 982 publicly available genomes from NCBI databases (Supplementary Data 2). The new strains were collected from natural plague foci in 14 provinces across China over the past 60 years (1948–2011), except for two strains from Myanmar and 13 of unknown origin. Among them, 1661 strains were isolated from rodents, 424 from vectors, 175 from humans, one from the environment, 53 from other hosts, and 22 from unknown hosts. The 982 public NCBI genomes consist of short-read sequencing data from the SRA database, excluding 78 Guertu strains (Project ID: PRJNA412676)14 with locally available raw data and the CO92 reference genome (Accession: GCA_000009065.1). We filtered out lab-passaged strains, genomes with less than 20X sequencing depth, and strains with duplicate BioSample IDs.
Additionally, we included 164 GenBank assemblies without available SRA short-read data, and 93 ancient genomes (aDNAs) from the SRA database (Supplementary Data 2). Together with the 3318 genomes, we analyzed a total of 3575 Y. pestis strains to construct the most comprehensive phylogenetic tree.
Whole-genome sequencing and assembly
We extracted genomic DNA from 2336 Yersinia pestis strains using the Qiagen DNeasy Blood Tissue Kit (No. 69506) and performed whole-genome sequencing on the Illumina Novaseq 6000 platform, generating paired-end reads with an average length of 150 bp and greater than 300X sequencing depth.
After filtering out adapters and low-quality reads (<Q20) using Trimmomatic software (v0.38)40, we downsized the data to 100X for downstream analysis. We employed Shovill software (v1.0.1) for de novo assembly of newly sequenced strains and SPAdes software (v3.13.0)41 for NCBI publicly available SRA data (modern genomes), which includes both paired-end and single-end sequencing data.
Genome annotation and pan-genome analysis
Genome annotation was performed using Prokka (v1.14.6)42 for all 3318 genomes. Genome sizes, including both chromosomes and plasmids, ranged from 4.15 to 4.93 Mb, with a median of 4.63 Mb. The number of annotated genes per genome varied from 3622 to 4420 coding sequences (CDS), with a median of 4075 CDS. Pan-genome analysis was performed using Panaroo (v1.2.8)43 in strict mode with a sequence identity threshold set to 90%, and Roary (v3.13.0)44 with a minimum blastp identity threshold of 90%. The Panaroo analysis identified a total of 5402 genes, including 3850 core and soft-core genes present in more than 95% of the strains, 270 shell genes present in 15–95% of the strains, and 1282 cloud genes present in less than 15% of the strains. The Roary analysis identified a comparable number of 3667 core and soft-core genes, 240 shell genes, and a higher number of 3060 cloud genes, resulting in a total of 6967 genes.
SNP and indel calling
MUMmer software (v3.23)45 was used to align the assemblies with the CO92 chromosome and extract SNPs from core genomes shared by at least 95% of strains. Short-read sequencing data was aligned to the reference using BWA software (v0.7.17)46, and GATK’s HaplotypeCaller module (v4.2.4.0)47 identified mutation sites for each strain. Joint mutation detection was performed using GATK’s CombineGVCFs and GenotypeGVCFs modules, and SelectVariants module was used to extract SNP variations. Only high-quality SNPs supported by both assemblies and sequencing reads (with ≥10 reads coverage and ≥90% base proportion) and corresponding genomic sites present in over 95% of all strains were retained. SNPs located in repeat regions, identified using Tandem Repeat Finder (v4.07b, with a minimum alignment score of 50) and BLAST software (v2.12.0+, for nucleotide identities ≥95%) were removed from the final dataset. Pairwise SNP differences among 3318 strains were computed using snp-dists software (v0.8.2) and visualized through violin plots in R software (v3.6.2). For the 3575 genomes, due to the low sequencing quality of aDNA, we reduced the SNP calling threshold for aDNAs to ≥3 reads and ≥90% base proportion without assembly verification, and manually corrected some sites as needed
Indels were extracted from the combined GVCF file of 3318 strains in SNP calling using GATK’s SelectVariants module. In every strain, indels supported by ≥10 reads coverage and a base proportion of ≥80% were retained. We focused on indels within the soft-core genome shared by at least 95% of genomes and with lengths under 30 bp, while removing indels located in repetitive regions identified during the SNP calling step.
Naming rules of the updated hierarchical nomenclature system
Maximum likelihood (ML) tree was constructed based on concatenated SNPs using IQ-TREE software (v1.6.5)48 with the GTR + G model and ultrafast bootstrapping (bootstrap = 1000), and visualized using FigTree (v1.4.3) and online tool iTOL (v6.8)49.
The Y. pestis nomenclature system was established in 2004 and has undergone refinements in subsequent studies. However, the use of different naming rules in various studies, especially those with multiple genomes sequenced within a specific phylogroup, has led to difficulties in referring to or designating new classifications. To overcome this challenge, we utilized the largest dataset available to date and proposed an updated three-level hierarchical nomenclature system that provides a more detailed phylogenetic topology of Y. pestis. Our first-order clade designation references published literature and EnteroBase13,15,32,50,51, while the second and third orders follow dynamic naming rules of influenza viruses and SARS-CoV-218,52–54.
Descendant lineages must meet four criteria to be named at a higher level: (1) it exhibits clade-specific SNP differences that distinguish it from other clades; (2) it comprises at least three non-redundant genomic sequences that differ either in SNP variation or have distinct origins, such as geographical regions or isolation years; (3) the clade-defining node has a bootstrap value >50%; (4) it forms a geographically-clustered branch or parallel branches that have diverged from the same ancestral node. If all criteria are met, a higher-level clade is determined; if not, it is classified into the lowest level that meets inclusion criteria.
For example, we divided 1.ORI2 into two main lineages: 1.ORI2.1 (South American strains) and 1.ORI2.2 (Chinese and Southeast Asian strains). Within 1.ORI2.1, Brazilian and Peruvian isolates form sub-lineages 1.ORI2.1.1 and 1.ORI2.1.2, respectively. In 1.ORI2.2, isolates from China and Southeast Asia form three sub-lineages: 1.ORI2.2.1, 1.ORI2.2.2, and 1.ORI2.2.3. Scattered strains were excluded from second and third-level classifications due to limited sampling. To standardize naming, we assigned 0.PE4A ~ 0.PE4D to the second-level classification as 0.PE4.1-0.PE4.4, while all other first-level naming systems remain consistent with existing nomenclature.
In our study, we newly identified five clades, including 3 second-order clades (1.IN2.5, 1.IN2.7, and 3.ANT1.2), along with an additional 2 third-order clades (1.IN2.1.3 and 2.MED3.1.4). These clades are delineated by genomes exclusively sequenced for this study (n = 196, Supplementary Data 1).
Hot regions identification and verification
To identify variation hotspots (HRs), we investigated the distribution of mutations within specific genomic lengths and compared our observations to theoretical expectations. Under the assumption of neutral evolution, we posited that mutations are uniformly distributed throughout the chromosome, following a binomial distribution B ~ (n, v), where n represents the genomic length and v denotes the mutation rate. The mutation rate is computed as: v = N/L, with N denoting the total number of mutations and L signifying the chromosome length.
Employing a 500 bp sliding window approach, we selected regions that exceeded the 95% confidence interval and contained at least two mutations. Preliminary HRs were established by merging overlapping or adjacent regions within a 1000 bp distance, with their start and end points corresponding to the nearest variant positions. We then randomly selected an equivalent number of mutations from the reference genome using sampling without replacement and repeated this procedure 100,000 times (T). Instances (M) were recorded when evaluated hotspots had a higher mutation count within length ranges, allowing us to calculate the occurrence probability P = M/T for each hotspot. After applying the Benjamini-Hochberg (BH) method for multiple hypothesis testing correction, we derived corrected P-values and identified regions with P-values < 0.05 as final HRs.
To access the robustness of our method, we performed simulations under a model of neutral evolution using fastSimBac software55. These simulations mirrored our empirical data in terms of genome count, genomic dimensions, and mutation rate, without the influence of selective forces. We conducted 100 simulations, each featuring a mutation rate of 0.0002 per generation per site, a genome length of 5 Mb, and 3300 genomes, without recombination. Identifying HRs in these simulations utilized the same pipeline, but we reduced the random sampling to 10,000 times for increased efficiency. Remarkably, a mere two HRs emerged in only two of the 100 simulation iterations, underscoring the exceptional nature of such clusters in a neutrally evolving landscape (Supplementary Data 13).
Ancestral state inference and fixed/unfixed mutations identification
We identified ancestral mutation states using two Y. pseudotuberculosis strains (IP32953 and IP31758, Accession: GCF_000047365.1 and GCF_000016945.1), 28 Y. pestis prehistoric aDNAs, and two 0.PE7 Y. pestis strains. Ancestral states were determined by consistency among prehistoric genomes. In cases of inconsistent alleles or inadequate sequencing quality in prehistoric genomes, we inferred the ancestral state from Y. pseudotuberculosis and 0.PE7 strains. Based on the reconstructed ancestral states, we determined the mutation base state at corresponding genomic positions. If a mutation base appears in over 80% of the genomes within a specific phylogroup, considering only top-level phylogroups containing more than three genomes, it is considered a fixed state; otherwise, it is a non-fixed state.
HR mutation distribution across phylogroups
To quantify the dispersion of phylogroups within a given HR, we first compute the weight (wi) of the phylogroup at each mutation site where it appears as: wi=ci/ti, where ci is the count of mutated strains (relative to the ancestral state) for the phylogroup at site i and ti is the total mutated strain count across all phylogroups at that site. Then, we sum the weights (wi) and calculate the proportion of a specific phylogroup (R) within a given HR using the following formula:
| 1 |
where n is the total number of mutation sites within the HR.
We calculated R for each phylogroup within a given HR and then determined the variance of the relative abundance of variation sites, denoted as VHR, as the variance of R for phylogroups where R > 0 (i.e., those containing strains exhibiting variations within the HR). The variance was calculated using the following formula:
| 2 |
where Rj represents the R values for phylogroups with R > 0, is the mean of the Rj values, and m is the number of phylogroups with R > 0.
To avoid biases caused by population structures, only unfixed mutation sites were considered for the analysis. The numpy.var function from Python’s NumPy library was utilized to calculate variance of phylogroup proportions within each HR. The pheatmap (v1.0.12) and ggplot2 (v3.3.5) packages in R software (v4.3.2) was used to visualize the results.
Functional analysis of HR-related genes
We utilized the STRING database (v11.0)20 to perform GO term enrichment analysis and protein-protein interaction analysis on 96 HR-related genes identified in our study, which included genes within HRs and those potentially regulated by downstream HR intergenic regions. Of these genes, 93 were successfully mapped in the STRING database. For the GO term enrichment analysis, we used the whole genome as a statistical background and considered terms with an FDR < 0.05 as significantly enriched across the three GO domains: Biological Process, Cellular Component, and Molecular Function. We also randomly selecting 96 genes from genome-wide reference genes and organizing them into 45 clusters with 1–4 adjacent genes to mirror the 45 identified HRs yielded no significant GO term enrichment in the STRING analysis. In the protein-protein interaction analysis, we displayed node interactions that exhibited a confidence score greater than 0.7, with disconnected nodes hidden.
We used the P2RP website19 to identify regulatory proteins in the CO92 chromosome, resulting in 284 unique proteins after removing duplicates and missing annotations. Additionally, from the CO92 annotation file, we extracted 74 new genes related to regulation, resulting in a total of 358 potential regulators in CO92. We conducted a hypergeometric test to assess the significance of HR-related regulators, using the whole genome as background.
Genomic sequence complexity and GC content analysis
We utilized the FindingInfo tool (v1.0.0)27 to calculate the genomic sequence complexity, including the linguistic complexity (LC) score and the Shannon’s entropy (H) score. This analysis was conducted using a 21 bp window encompassing 10 bases upstream and downstream of each position in the reference genome. The LC and H scores for each position within a specific region were averaged to obtain the region’s LC and H score. The GC content was determined by calculating the ratio of guanine (G) and cytosine (C) nucleotides to the total count of nucleotides within a specific region of the reference genome. Values closer to 1 indicate higher sequence complexity and GC content. Additionally, we randomly selected 45 regions from non-hotspot regions in the reference genome with the same length to match the 45 identified HRs and calculate corresponding GC content, LC and H scores. This process was repeated 1000 times. Subsequently, we employed a two-tailed Welch’s t-test to compare the differences in GC content, LC and H scores between the 45 HRs and the randomly selected regions.
Detection of homologous recombination
We employed three methods, ClonalFrameML (v1.12)56, Gubbins (v3.3.1)57, and mcorr (v20180102)58, to evaluate homologous recombination in the Y. pestis and its ancestor Y. pseudotuberculosis across different dataset sizes. For Y. pestis, we randomly selected 25 strains (one from each first-order phylogroup), 64 strains (one from each top-level phylogroup), 186 strains (three from each first-order or top-level phylogroup), 1167 strains (excluding those with fewer than 2 SNP differences), and the full set of 3318 strains. For Y. pseudotuberculosis, we expanded the dataset from 23 genomes by Torrance et al.59 to 559 by adding 536 genomes from the NCBI SRA database.
In ClonalFrameML analysis, we used the non-repetitive core genome aligned to CO92 and the ML tree from IQ-TREE as inputs. The tree was made bifurcating using ape’s “multi2di” command in R. We applied the “-ignore_user_sites” parameter to exclude non-core and duplicate region sites and set “-emsim” to 100 for reliable confidence interval estimations. The recombination/mutation ratio (r/m) was computed as r/m = R/θ*δ*ν. Recombination events were visualized using ClonalFrameML’s built-in R script.
For Gubbins, we used the same core genome alignments. The “--tree-builder iqtree” option was used for Y. pseudotuberculosis, while the default “raxml” was utilized for Y. pestis to avoid missing bipartition errors. Recombination events were visualized with Phandango (v1.3.1)60. The overall r/m and R/θ values were calculated as per the Gubbins manual. For mcorr, the core genome alignments were converted to XMFA format and analyzed using default parameters.
In ClonalFrameML and Gubbins, total recombination coverage was calculated as the ratio of the total reference-mapped recombination region length (with overlapping regions merged) to the non-repetitive core genome length. Sample-related recombination coverage was determined as the median ratio of total recombination region length per strain (including internal branches and strain-specific events) to the non-repetitive core genome length.
Identification of selection signals
KaKs_Calculator software (v2.0)61 with five distinct models (GLWL, GMYN, GNG, GLPB, and GYN) was used to calculate dN/dS values of coding sequences with SNP mutations in the reference genome. Genes showing dN/dS values greater than 1 in all models were considered under positive selection.
We employed HomoplasyFinder software62 to identify homoplastic sites, with connected SNPs and the IQ-TREE-generated ML tree as inputs. The detected sites were mapped to their original positions in the reference genome, and we excluded homoplastic sites shared by phylogroups with a common ancestor and identical alleles. Due to the higher occurrence of homplasies in indels, our study concentrated on SNPs. Among the 6552 single nucleotide polymorphisms (SNPs) analyzed, 138 homoplasies were identified in 76 genes or intergenic regions, with 62.32% of them (n = 86/138) found within 25 HRs. Thirteen genes and seven intergenic regions were discovered to have more than two homoplastic sites, with 80% of these regions (including 10 in genes and 6 in intergenic region) located within 14 HRs.
Condon usage bias analysis
To analyze codon usage patterns from the Y. pestis population perspective, we employed CodonW software (v1.4.4)63 to calculate Relative Synonymous Codon Usage (RSCU) values for three distinct group datasets: (1) the original coding sequences (CDS) obtained from the reference genome, and the corresponding synonymous mutation-replaced CDSs; (2) a concatenated codon sequence that includes all synonymous mutations, and a matching sequence containing the corresponding codons derived from the reference genome; and (3) a concatenated codon sequence encompassing all synonymous mutations, and a connected sequence of all CDSs extracted from the original reference genome. Afterward, we used an in-house Python script to process the generated output files containing RSCU values. We then employed a paired Wilcoxon signed-rank test in R software to evaluate the differences in RSCU values across each group dataset.
Streptomycin susceptibility testing
In light of the reported resistance to streptomycin in Y. pestis strains with a K43R mutation in the rpsL gene29,30, we assessed streptomycin susceptibility in seven Y. pestis strains that have different mutations in the HR06 hot region (including the rpsL gene) and five strains from neighboring regions without HR06 mutations (Supplementary Data 12). We utilized both the disk diffusion and agar dilution methods (Streptomycin 0.5–32 µg/mL) for antimicrobial testing, adhering to established procedures while identifying streptomycin-resistant Y. pestis S1996012730, which served as a positive control in our study, along with the use of E. coli ATCC 25922 as a negative control. We performed disk diffusion tests on five strains with HR06 mutations and five strains without such mutations, while using the agar dilution method to examine all seven HR06-mutated strains and the five non-mutated strains (Supplementary Data 12 and Supplementary Fig. 9). The study was performed in the Bio-safety Level 3 (BSL-3) laboratory at the Qinghai Institute for Endemic Disease Control and Prevention to ensure proper safety precautions.
World map visualization with pie charts
The world map was generated using the ggplot2 and maps (v3.3.0) packages in R, with country boundaries delineated by the borders() function in ggplot2. Pie charts were plotted on the map using the scatterpie package (v0.1.5).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2022YFC2305304 to Y.C.), National Natural Science Foundation of China (No. 31970002 to Y.C.), Key Scientific and Technology Project of Inner Mongolia Autonomous Region (No. 2021ZD0006 to Y.C.), and the State Key Laboratory of Pathogen and Biosecurity (No. SKLPBS2215 to Y.W.).
Author contributions
Y.C., R.Y., and H.T. conceived and designed the study. Z.Q., Y. X., and X.Y. collected the Y. pestis strains. C.L., Y.X., X. Y., and Y.J. were responsible for strain revival and cultivation. X.Y., H.Z., and Y.X. performed DNA extraction. K.S., Y.G., and Y.T. carried out DNA library construction and sequencing. Y.W., Y.C., and Y.S. conducted bioinformatic analyses. Y.X. and Q.Z. performed streptomycin resistance experiments. Y.C. and Y.W. wrote the manuscript. Y.C. and R.Y. supervised the project.
Peer review
Peer review information
Nature Communications thanks Rohan Maddamsetti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The newly sequenced genomic data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) GenBank database under BioProject PRJNA1177432 and the China National Microbiology Data Center (NMDC, https://nmdc.cn/en) under BioProject NMDC10018536. The publicly available data used in this study are available in the NCBI GenBank and Sequence Read Archive (SRA) databases, with accession numbers provided in Supplementary Data 2. Source data are provided with this paper.
Code availability
The custom scripts associated with this study have been deposited on GitHub (https://github.com/WuYarong/YP_Hotspots), and through 10.5281/zenodo.14219512.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yarong Wu, Youquan Xin, Xiaoyan Yang.
Contributor Information
Zhizhen Qi, Email: qzz7777@163.com.
Ruifu Yang, Email: ruifuyang@gmail.com.
Yujun Cui, Email: cuiyujun.new@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55581-4.
References
- 1.Luria, S. E. & Delbruck, M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics28, 491–511 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederberg, J. & Lederberg, E. M. Replica plating and indirect selection of bacterial mutants. J. Bacteriol.63, 399–406 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershberg, R. Mutation–the engine of evolution: studying mutation and its role in the evolution of bacteria. Cold Spring Harb. Perspect. Biol.7, a018077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch, M. et al. Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet17, 704–714 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Moxon, E. R., Rainey, P. B., Nowak, M. A. & Lenski, R. E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol.4, 24–33 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Martincorena, I., Seshasayee, A. S. & Luscombe, N. M. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature485, 95–98 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Tenaillon, O. et al. The molecular diversity of adaptive convergence. Science335, 457–461 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zhou, Z. et al. Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc. Natl Acad. Sci. USA111, 12199–12204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jee, J. et al. Rates and mechanisms of bacterial mutagenesis from maximum-depth sequencing. Nature534, 693–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maddamsetti, R. et al. Synonymous genetic variation in natural isolates of Escherichia coli does not predict where synonymous substitutions occur in a long-term experiment. Mol. Biol. Evol.32, 2897–2904 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Achtman, M. et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA96, 14043–14048 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry, R. D. & Fetherston, J. D. Yersinia pestis–etiologic agent of plague. Clin. Microbiol Rev.10, 35–66 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, Y. et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc. Natl Acad. Sci. USA110, 577–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, Y. et al. Evolutionary selection of biofilm-mediated extended phenotypes in Yersinia pestis in response to a fluctuating environment. Nat. Commun.11, 281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morelli, G. et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet42, 1140–1143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNally, A., Thomson, N. R., Reuter, S. & Wren, B. W. ‘Add, stir and reduce’: Yersinia spp. as model bacteria for pathogen evolution. Nat. Rev. Microbiol.14, 177–190 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Yang, R. et al. Yersinia pestis and plague: some knowns and unknowns. Zoonoses3, 5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambaut, A. et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol.5, 1403–1407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barakat, M., Ortet, P. & Whitworth, D. E. P2RP: a Web-based framework for the identification and analysis of regulatory proteins in prokaryotic genomes. BMC Genomics14, 269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res.49, D605–D612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moxon, R., Bayliss, C. & Hood, D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev. Genet40, 307–333 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Yahara, K. et al. The landscape of realized homologous recombination in pathogenic bacteria. Mol. Biol. Evol.33, 456–471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold, B. J., Huang, I. T. & Hanage, W. P. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol.20, 206–218 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Wagner, A. Risk management in biological evolution. J. Theor. Biol.225, 45–57 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Liu, Q. et al. Local adaptation of Mycobacterium tuberculosis on the Tibetan Plateau. Proc. Natl Acad. Sci. USA118, e2017831118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiner-Oms, A. et al. Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex. Sci. Adv.5, eaaw3307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta, A. & Alland, D. Reversible gene silencing through frameshift indels and frameshift scars provide adaptive plasticity for Mycobacterium tuberculosis. Nat. Commun.12, 4702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei, M., Suzuki, Y. & Nozawa, M. The neutral theory of molecular evolution in the genomic era. Annu Rev. Genomics Hum. Genet11, 265–289 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Andrianaivoarimanana, V. et al. Transmission of antimicrobial resistant Yersinia pestis during a pneumonic plague outbreak. Clin. Infect. Dis.74, 695–702 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai, R. et al. A novel mechanism of streptomycin resistance in Yersinia pestis: mutation in the rpsL gene. PLoS Negl. Trop. Dis.15, e0009324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar, R., Friedman, N., Molshanski-Mor, S. & Qimron, U. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl Environ. Microbiol.78, 744–751 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achtman, M. et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl Acad. Sci. USA101, 17837–17842 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartra, S. S. et al. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect. Immun.76, 612–622 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolodziejek, A. M., Hovde, C. J. & Minnich, S. A. Yersinia pestis Ail: multiple roles of a single protein. Front. Cell Infect. Microbiol.2, 103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsang, T. M., Wiese, J. S., Felek, S., Kronshage, M. & Krukonis, E. S. Ail proteins of Yersinia pestis and Y. pseudotuberculosis have different cell binding and invasion activities. PLoS One8, e83621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao, L. et al. Interplays of mutations in waaA, cmk, and ail contribute to phage resistance in Yersinia pestis. Front. Cell Infect. Microbiol.13, 1174510 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spyrou, M. A. et al. Analysis of 3800-year-old Yersinia pestis genomes suggests Bronze Age origin for bubonic plague. Nat. Commun.9, 2234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bobay, L. M. & Ochman, H. Factors driving effective population size and pan-genome evolution in bacteria. BMC Evol. Biol.18, 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, X., Xing, X. H. & Zhang, C. Targeted mutagenesis: a sniper-like diversity generator in microbial engineering. Synth. Syst. Biotechnol.2, 75–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput Biol.19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Tonkin-Hill, G. et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol.21, 180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol.5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at 10.48550/arXiv.1303.3997 (2013).
- 47.Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinforma.43, 11 10 11–11 10 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi, L. et al. New genotype of Yersinia pestis found in live rodents in Yunnan Province, China. Front. Microbiol.12, 628335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, Z. et al. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res.30, 138–152 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Group, W. O. F. H. N. E. W. Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza Other Respir. Viruses6, 1–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Group, W. O. F. H. N. E. W. Continuing progress towards a unified nomenclature for the highly pathogenic H5N1 avian influenza viruses: divergence of clade 2.2 viruses. Influenza Other Respir. Viruses3, 59–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Group, W. O. F. H. N. E. W. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis.14, e1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Maio, N. & Wilson, D. J. The bacterial sequential Markov coalescent. Genetics206, 333–343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Didelot, X. & Wilson, D. J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol.11, e1004041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res.43, e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin, M. & Kussell, E. Inferring bacterial recombination rates from large-scale sequencing datasets. Nat. Methods16, 199–204 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Torrance, E. L., Burton, C., Diop, A. & Bobay, L. M. Evolution of homologous recombination rates across bacteria. Proc. Natl Acad. Sci. USA121, e2316302121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hadfield, J. et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics34, 292–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, D., Zhang, Y., Zhang, Z., Zhu, J. & Yu, J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteom. Bioinforma.8, 77–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crispell, J., Balaz, D. & Gordon, S. V. HomoplasyFinder: a simple tool to identify homoplasies on a phylogeny. Micro. Genom.5, e000245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peden, J. F. Analysis of codon usage, PhD Thesis (University of Nottingham, UK., 1999).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The newly sequenced genomic data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) GenBank database under BioProject PRJNA1177432 and the China National Microbiology Data Center (NMDC, https://nmdc.cn/en) under BioProject NMDC10018536. The publicly available data used in this study are available in the NCBI GenBank and Sequence Read Archive (SRA) databases, with accession numbers provided in Supplementary Data 2. Source data are provided with this paper.
The custom scripts associated with this study have been deposited on GitHub (https://github.com/WuYarong/YP_Hotspots), and through 10.5281/zenodo.14219512.