Abstract
The Natural Product Research Laboratory (NPRL) of China Medical University Hospital (CMUH) was established in collaboration with CMUH and Professor Kuo-Hsiung Lee from the University of North Carolina at Chapel Hill. The laboratory collection features over 6000 natural products worldwide, including pure compounds and semi-synthetic derivatives. This is the most comprehensive and fully operational natural product database in Taiwan. This review article explores the history and development of the NPRL of CMUH. We then provide an overview of the recent applications and impact of artificial intelligence (AI) in new drug discovery. Finally, we examine advanced powerful AI-tools and related software to explain how these resources can be utilized in research on large-scale drug data libraries. This article presents a drug research and development (R&D) platform that combines AI with the NPRL. We believe that this approach will reduce resource wastage and enhance the research capabilities of Taiwan's academic and industrial sectors in biotechnology and pharmaceuticals.
Keywords: Natural Products Research Laboratories (NPRL), Artificial Intelligence (AI), Natural products, Drug research and development (R&D), Drug discovery
1. Introduction
Living organisms such as plants, invertebrates, and microorganisms produce chemical molecules known as natural products. These compounds exhibit a range of biological and pharmacological activities, including anti-cancer, antioxidant, anti-aging, and anti-inflammatory properties, making them valuable for the research and development (R&D) of new drugs [1–4]. Identifying bioactive compounds typically involves several steps: obtaining natural products from biological sources, testing their efficacy as medicines, isolating bioactive substances, determining their structures, identifying their molecular targets, and utilizing bioinformatics for further analysis [5]. However, these traditional research models are often time-consuming and require significant financial investment.
To keep pace with evolving trends in new drug development, it is essential to establish a large-scale, integrated compound database R&D platform in Taiwan, especially given the rapid advancements in artificial intelligence (AI) and biotechnology [6–8]. The Natural Product Research Laboratory (NPRL) database aims to create a comprehensive resource of compound big data, employ high-speed AI computational tools to develop potential lead compounds, and facilitate clinical applications in translational medicine [9–12]. China Medical University Hospital (CMUH) and Professor Kuo-Hsiung Lee are funding the NPRL-CMUH initiative, which focuses on sourcing compounds from fruits, vegetables, microorganisms, traditional Chinese medicine (TCM), and Chinese herbal materials (Fig. 1). Detailed information on NPRL-CMUH is provided in the following sections.
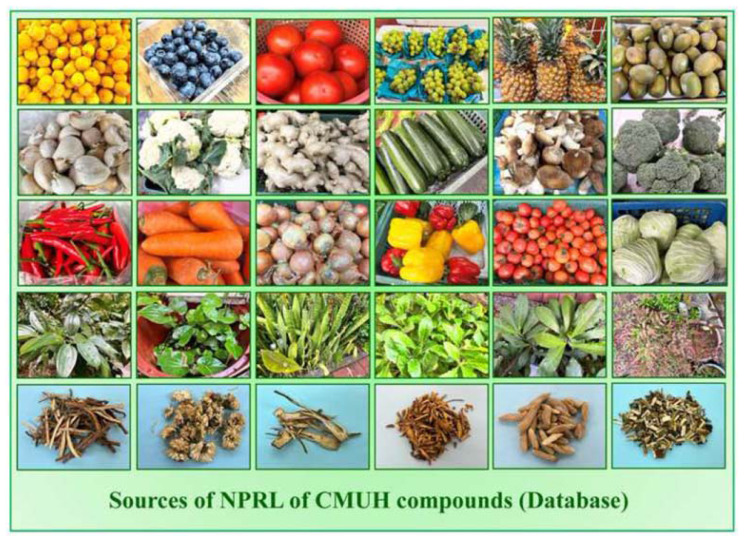
Fig. 1.
Sources of NPRL of CMUH compounds database. The NPRL of CMUH utilizes a variety of natural compounds. This collection includes plant-based materials, such as fruits, vegetables, herbs, roots, and plants used in traditional medicine. The samples ranged from everyday food items such as tomatoes, carrots, garlic, and ginger to rare herbs and medicinal plants being investigated for their potential therapeutic properties. NPRL utilizes these varied resources as fundamental materials for extracting and developing natural compounds, facilitating drug discovery efforts and supporting studies in pharmacological research.
This review gathers relevant literature on the use of artificial intelligence (AI) tools and techniques in drug discovery applied throughout all phases of drug development. These methods aim to expedite the research process while minimizing the risks and costs of clinical trials.
2. Summary of history and process for constructing NPRL of CMUH
2.1. Development and establishment for the NPRL of CMUH: A comprehensive approach to constructing a natural product compound database
The development of the NPRL compound database was initiated with a planning stage in 2017 and finalized in 2019, spanning three years [13–15]. A crucial aspect of establishing NPRL is the comprehensive documentation and cataloging of all the compounds. A thorough action plan was formulated before the commencement of the project. Academics from CMUH collaborated with Professor Lee's research group to examine the various elements of these compounds, including their storage requirements, physical locations, individual scientific documentation, and related publication lists. Our team gathered general project details and identified potential challenges that might arise during the process. For example, we discovered that certain compounds and experimental data were stored in separate containers, necessitating meticulous verification. We also addressed issues related to the storage and preservation of specific compounds, particularly those that require preparation before shipment back to Taiwan. Furthermore, we developed a specialized AI software application to inventory all existing and future compounds within the NPRL.
Establishing this software is crucial before successfully entering the compound data.
2.2. Systematic organization and documentation of NPRL compounds
Our initial approach involved establishing a primary method for systematically organizing and documenting NPRL compounds. The process illustrated in Fig. 2 began with collecting all sample containers. Once gathered, the containers were sorted according to their designated names. Subsequently, we meticulously inventoried the contents of each container and correlated them with existing experimental data, various documents, and digital records. The final step involved assigning unique identification codes to individual samples. The NPRL of CMUH database requires the inclusion of specific details for each entry. These include a unique NPRL Code Number, the Original Code Number, CID Number, PubChem/CAS Number, and Common Name. Additionally, the structure, molecular weight, and quantity must be determined. The Sample State (e.g., solid or liquid) and Sample Location (e.g., freezer or storage box) are also required. Finally, any related publications and other data pertinent to entry should be included.
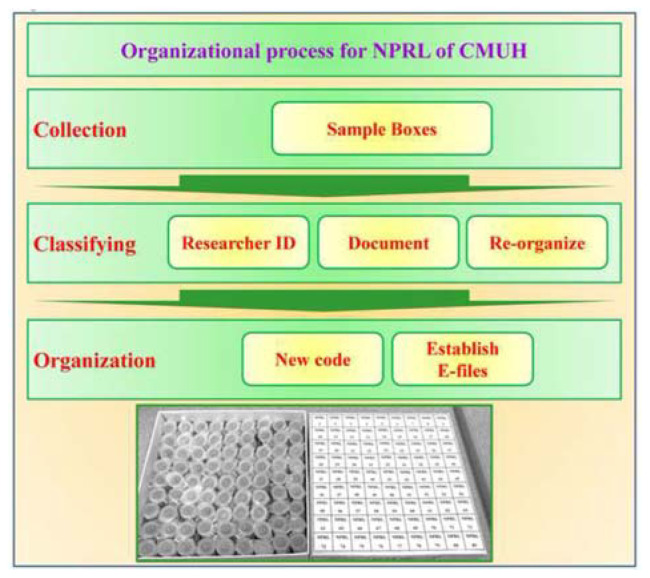
Fig. 2.
Organizational process for the NPRL of CMUH. The structured approach for establishing NPRL of CMUH outlines the steps for acquiring, categorizing, and arranging compound specimens. Acquisition: the initial phase involved obtaining sample containers containing diverse compounds. Categorization: specimens are sorted based on crucial information, such as the investigator's identity, pertinent paperwork, and reorganization specifications. This approach ensures precise and effortless retrieval. Arrangement: each specimen is given a unique identity, and digital records are generated to support computerized data management and streamline future access.
2.3. Development of NPRL: Taiwan's largest natural products database and AI-driven drug R&D
The NPRL of CMUH repository contains 6782 natural products, including both pure and semi-synthetic derivatives, making it Taiwan's most extensive and comprehensive collection of natural products. Through structural refinement, 37,682 compounds with diverse configurations were obtained. The development of NPRL was finalized in 2019, after which work began on creating in silico and in vitro platforms for various medical conditions, thereby enabling the expansion of this substantial database. Fig. 3 illustrates the construction process, research initiatives, and current research directions for NPRL from 2017 to 2024.
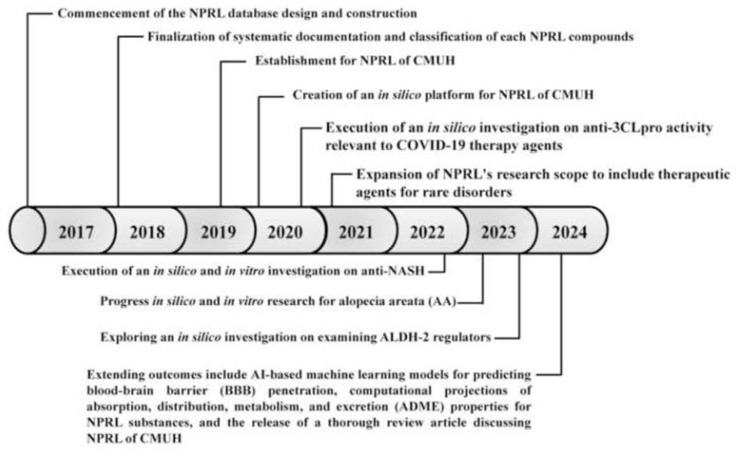
Fig. 3.
Chronology of NPRL evolution and utilization. The timeline of significant events, developmental phases, and scientific applications associated with the NPRL of CMUH from 2017 to 2024.
2017: Commencement of the NPRL database design and construction.
2018: Finalization of systematic documentation and classification of NPRL compounds.
2019: Establishment for NPRL of CMUH.
2020: Creation of an in silico platform for NPRL of CMUH.
2020: Execution of an in silico investigation on anti-3CLpro activity relevant to COVID-19 therapy agents.
2021: Expansion of NPRL's research scope to include therapeutic agents for rare disorders.
2022: Execution of an in silico and in vitro investigation on anti-NASH.
2023: Progress in silico and in vitro research for alopecia areata (AA) therapeutic agents.
2023: Execution of an in silico investigation on examining ALDH-2 regulators with NPRL compounds.
2024: Additional progress and research outcomes include AI-based machine learning (ML) models for predicting blood–brain barrier (BBB) penetration, computational projections of ADME (absorption, distribution, metabolism, and excretion) properties for NPRL-CMUH substances, and the release of a thorough review article discussing NPRL at CMUH.
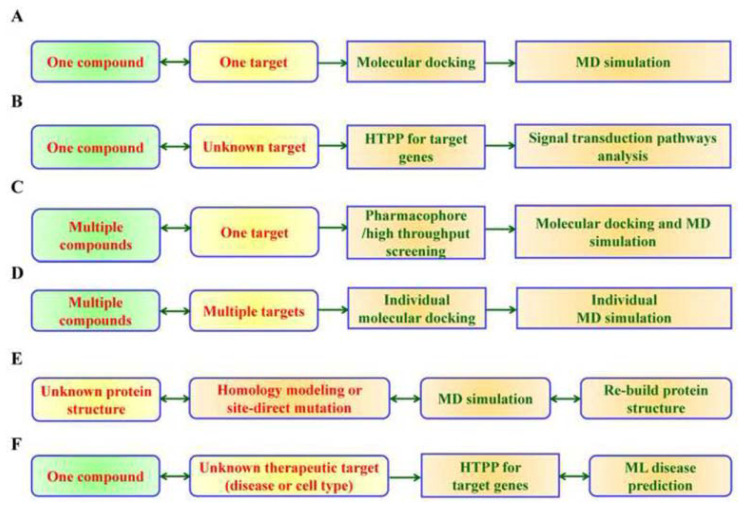
Table 1 presents a compilation of websites and databases containing extensive compound libraries relevant to pharmaceutical R&D [13,16–30]. Fig. 4 presents a comprehension design of AI-driven in silico drug R&D platform that CMUH created to address various medical conditions. The system is composed of different parts: (A) a compound connected to a model for predicting a target, (B) a compound connected to a model for predicting an unidentified target, (C) a library of compounds linked to a model for predicting a target, (D) multiple compounds combined with a model for predicting multiple targets, (E) a model for homology modeling or site-directed mutation prediction, and (F) models for finding combined therapeutic targets. Fig. 4 shows the AI-driven in silico drug R&D platform of CMUH, which was developed to address various diseases.
Table 1.
Websites and databases of large compound libraries for drug research and development (R&D).
| Name | Type | Website URL | References |
|---|---|---|---|
| ClinicalTrials.gov | Clinical trials database | https://www.clinicaltrials.gov/ | [16] |
| Chemical entities of biological Interest (ChEBI) | Small chemical molecule database | https://www.ebi.ac.uk/chebi/ | [17] |
| ChEMBL database | Database of bioactive molecules | https://www.ebi.ac.uk/chembl/ | [18] |
| ChemSpider The cambridge structural | Chemical molecule database | https://www.chemspider.com/ | [19] |
| Database (CSD) | Chemical molecule database | https://www.ccdc.cam.ac.uk/solutions/software/csd/ | [20] |
| DailyMed database | FDA-regulated products | https://dailymed.nlm.nih.gov/dailymed/ | [21] |
| DrugBank | Drug database | https://go.drugbank.com/ | [22] |
| Drugs@FDA | FDA approved drugs database | https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm | [25] |
| FDA online label repository | FDA drug label database | https://labels.fda.gov/ | [23] |
| FooDB IUPHAR/BPS guide to | Nature products database | https://foodb.ca/ | [24] |
| Pharmacology | Pharmacology database | https://www.guidetopharmacology.org/ | [26] |
| PubChem | Chemical molecule database | https://pubchem.ncbi.nlm.nih.gov/ | [27] |
| PKIDB | Kinase inhibitor database | https://www.icoa.fr/pkidb/ | [28] |
| TargetMol | Natural products database | https://www.targetmol.com/search?keyword=home/ | [29] |
| TCMBank | Traditional Chinese medicines | https://tcmbank.cn/ | [30] |
| ZINC | Available compounds | zinc.docking.org/ | [13] |
Fig. 4.
Pipeline of In Silico R&D platforms for various disease models at NPRL. Each component (A–F) showed a distinct computational approach for analyzing compounds and targets. (A) Single-compound single-target model: Molecular docking was used to test a single compound against a specific target, and MD simulation was used to improve the interaction analysis. (B) Single compound unknown target model: HTPP is used to identify possible target genes for compounds with unknown targets. A signal transduction pathway analysis is added to clarify the compound's mode of action. (C) Multiple compound-single target model: pharmacophore modeling or high-throughput screening tests of a group of compounds against a single target. Molecular docking and MD simulations are then used for a more in-depth analysis. (D) The multiple compound-multiple target model tests different compounds against different targets. Molecular docking and MD simulations improve the connection between each compound and its target. (E) Homology Modeling and site-directed mutation prediction model: Homology modeling or site-directed mutagenesis was used for proteins with unknown structures or mutations. This is followed by MD simulations and protein structure reconstruction. (F) Therapeutic Target Identification Model: If the compound's therapeutic target (disease or cell type) is unknown, HTPP is used to guess the target genes. ML techniques are used to treat diseases.
3. AI applications in drug R&D
3.1. Advancing drug development: Utilizing AI for efficient pharmaceutical R&D
Recently, AI has made substantial progress across various social domains with notable advancements in the pharmaceutical sector [31,32]. AI encompasses diverse, sophisticated tools, including reasoning capabilities, knowledge representation systems, solution search algorithms, and networking technologies [33]. The pharmaceutical industry has witnessed the extensive digitization of experimental and clinical data over the past few decades, enabling the analysis and processing of big data [33,34]. By implementing AI modules, the sector can improve the automation of large-scale data management and proactively tackle potential future issues, enabling the earlier identification of solutions [35]. Traditionally, drug development has relied on identifying and creating numerous small molecule compounds. Various compound libraries have been established over the last decade. Conventional approaches to drug development are often expensive and time-intensive, hampering the process's efficiency. Nevertheless, AI can address these challenges. This study examined AI techniques for drug R&D [33–36].
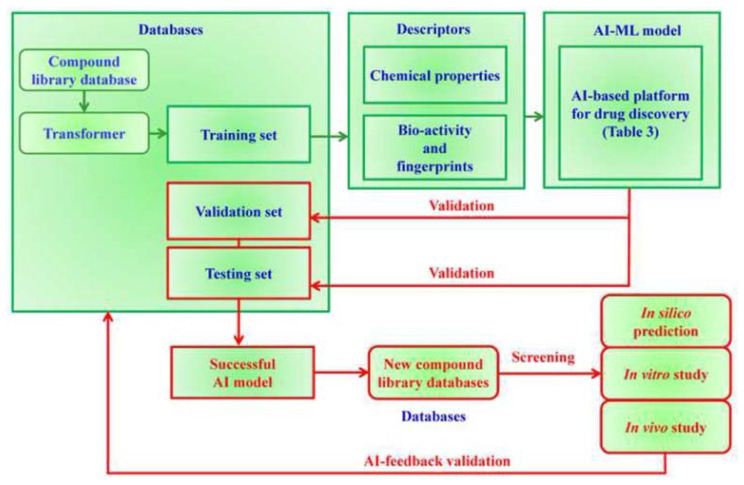
Fig. 5 provides a detailed illustration of the workflow of the AI model. The process began with data extraction from the compound library database, followed by using a transformer to separate the data into training, validation, and testing sets. The training set comprised chemical characteristics, biological activity, and molecular fingerprints subjected to computational analysis and ML [13,37]. Validation and testing sets were used to verify the results. Once the AI model has been successfully validated, it can be used to analyze other compound library database. Commonly used ML methods for developing classification models in AI include linear discriminant analysis (LDA), k-nearest neighbors (kNN), kNN regression (kNNR), artificial neural networks (−), probabilistic neural networks (PNN), support vector machines (SVM), support vector regression (SVR), C4.5 decision trees (C4.5DT), recursive partitioning (RP) classifiers, random forests (RF), naïve Bayes classifiers, multiple linear regression (MLR), partial least squares regression (PLSR), and logistic PLS, among others. Several AI-driven platforms for discovering novel pharmaceuticals are listed in Table 2 [38–47].
Fig. 5.
AI-powered drug discovery leveraging compound library databases. The process consists of three main phases: 1. Database management and data preparation; 2. Model development and verification; and 3. Model application and screening. The drug discovery workflow can be optimized using AI-based modeling and validation techniques, enabling more effective screening and prediction of potential therapeutic agents.
Table 2.
AI-based tools and platform for drug research and development (R&D).
3.2. AI-driven approaches for disease classification and novel drug development
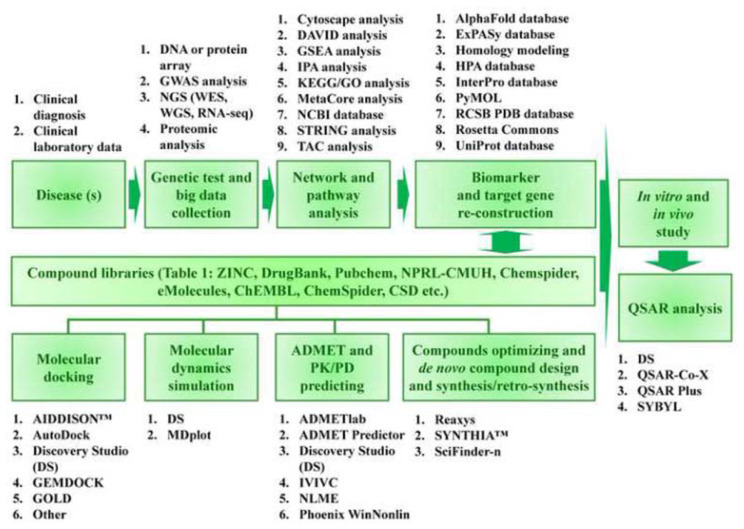
Standard analytical software processes large compound libraries with AI-driven models. Fig. 6 and Table 3 present an overview of the workflow for AI-enhanced drug discovery and development, along with a list of commonly used software programs. Disease classification can be achieved by compiling the clinical diagnoses and laboratory findings. Following Institutional Review Board (IRB) approval for specimen collection, researchers can investigate the relationship between diseases and genes [48–50]. Various techniques have been employed, including DNA or protein arrays, Genome-Wide Association Studies (GWAS), and next-generation sequencing (NGS) methods, such as whole-exome sequencing (WES), whole-genome sequencing (WGS), RNA sequencing, and proteomic analysis [51–55]. Examination of genetic big data, encompassing network and pathway analyses, enables the identification of disease biomarkers and target genes. These analyses utilize multiple approaches, such as Cytoscape analysis, DAVID analysis, Gene Set Enrichment Analysis (GSEA), Ingenuity Pathway Analysis (IPA), KEGG/GO analysis, MetaCore analysis, NCBI database utilization, STRING analysis, and TCA analysis. Various protein structure repositories, including the Alpha-Fold database, Binding MOAD, ExPASy, HPA, InterPro, RCSB PDB, Rosetta Commons, and Uni-Prot, allow access to known crystal structures [49,50,56–72]. AI-based homology modeling techniques can be employed to predict the three-dimensional configurations [14,73].
Fig. 6.
Workflow of AI-enhanced drug discovery and development processes. This diagram showcases a holistic AI-based drug discovery and development strategy, from disease identification to experimental testing utilizing various computational and empirical methodologies. The process is divided into eight crucial phases: 1. Disease identification and data collection; 2. Network and pathway examination; 3. Biomarker and target gene reconstruction; 4. Compound library screening; 5. Molecular docking and dynamics simulation; 6. ADMET and PK/PD property prediction; 7. Compound optimization and novel design; and 8. QSAR analysis and experimental verification. This approach effectively integrates AI and computational techniques with traditional research methods to accelerate drug discovery, improve candidate selection, and enhance the efficacy of experimental validation.
Table 3.
Common analytical methods and software for analyzing big data in compound libraries with AI models.
Various software tools have been employed for molecular docking between molecules and proteins, including AutoDock, AIDDISON™, Discovery Studio (DS), GEMDOCK, GOLD, and PotentialNet. Following docking, programs such as DS and MDplot can be utilized to perform molecular dynamics simulations. Upon confirming the binding affinity of a compound to a protein, additional characteristics, such as pharmacokinetics/pharmacodynamics (PK/PD) and blood–brain barrier (BBB) permeability, can be estimated using platforms such as ADMETlab, ADMET Predictor, DS, IVIVC, NLME, and Phoenix WinNonlin. The creation of innovative molecular structures is crucial in novel drug development. Software such as Reaxys, SYNTHIA ™, and SciFinder-n can be used for compound optimization, de novo compound design, and synthesis/retrosynthesis [14,74–89].
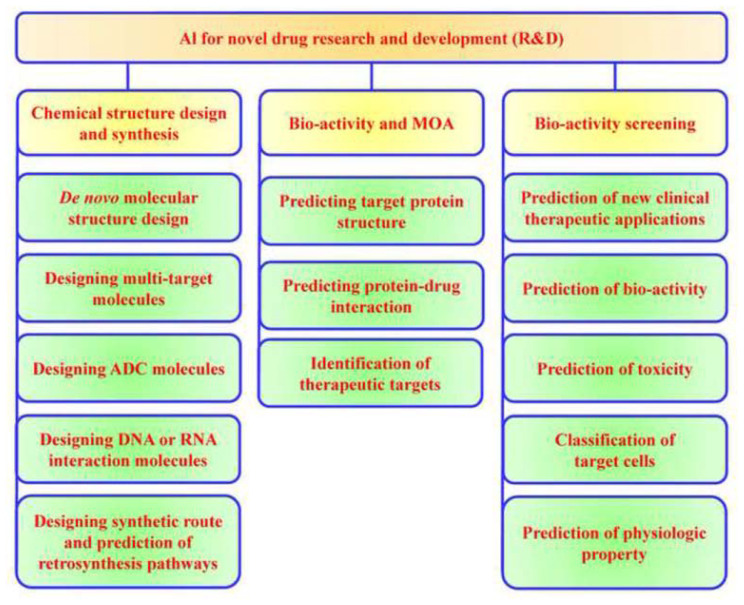
Fig. 7 outlines the roles and benefits of AI in novel drug R&D. AI-driven tools offer diverse capabilities that significantly enhance the new drug discovery process. These tools facilitate the creation of novel molecular structures, the development of multi-target compounds, and the generation of antibody-drug conjugates (ADCs) [90,91]. They also support the design of nucleic acid (DNA or RNA)-interacting compounds, streamline chemical synthesis, and enable the reverse engineering of chemical structures, thus transforming various facets of chemical and molecular design [92,93]. In the field of bioactivity and mechanism of action (MOA) research, AI assists in predicting target protein structures, modeling protein–drug interactions, and identifying potential therapeutic targets [94,95]. Furthermore, AI technologies are instrumental in uncovering new clinical applications through bioactivity screening, assessing bioactivity and toxicity, classifying target cells, and predicting physiological properties [6,8,32].
Fig. 7.
AI applications in drug development: Improving the efficiency of chemical design, bioactivity assessment, and predictive screening. The field of novel medication research and development employs three main categories of AI applications, each with a specific role: (1) Chemical structure design and synthesis, (2) Bioactivity and MOA, and (3) Bioactivity screening and prediction. AI plays a crucial role in various stages of drug development, including molecular design, therapeutic outcome forecasting, and safety profile evaluation. These AI-driven applications shorten the research and development process, enhancing prospects for achieving successful outcomes.
4. Conclusion
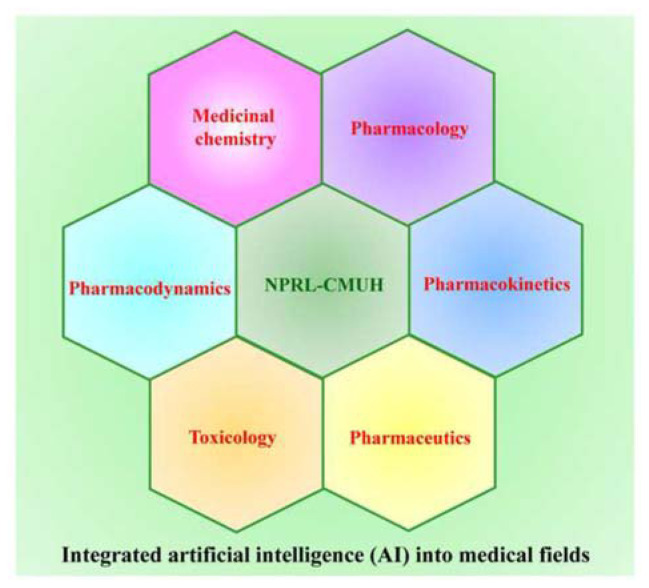
Having the right tools is essential for achieving excellence in any task. Table 4 outlines the benefits and prospects for creating an NPRL of CMUH database. Combining NPRL compounds with AI technology can significantly improve the creation of new small-molecule structures, uncover novel therapeutic targets, and reveal new pharmacological uses of natural product lead compounds. This synergy offers crucial insights for treating various human diseases and developing new drugs, ultimately enhancing patient care and quality of life. Integrating AI into pharmacies within NPRL is anticipated to spur progress and innovations in fields such as medicinal chemistry, pharmacology, pharmacodynamics, pharmacokinetics, toxicology, and pharmaceutics (Fig. 8).
Table 4.
Advantages and opportunities in establishing the NPRL of CMUH database.
| Advantages | The number of samples includes up to 6782 distinct types |
| Cultivate research talents in nature products, phytochemicals, Chinese herbal and traditional Chinese medicine (TCM) | |
| Enrich the diversity of research resources in nature products, phytochemicals, Chinese herbal and TCM | |
| Receive support from leading research centers both domestically and internationally | |
| Advanced research outcomes in the fields of TCM and natural products | |
| Facilitate the integration of Chinese and Western medicine | |
| Accelerate the pace of new drug discovery | |
| Establish a foundation for international collaboration | |
| Opportunities | Establish AI models for TCM diseases and treatments |
| Utilize innovative technologies and methods | |
| Enhance the applications of translational medicine | |
| Advance drug discovery and patent technology transfer | |
| Increase research outcomes and publications in TCM | |
| Create job opportunities for TCM professionals | |
| Expand avenues for international collaboration | |
| Improve patient health and well-being |
Fig. 8.
Impact of AI integration on NPRL of CMUH and related pharmaceutical fields. Integrating AI within the NPRL framework significantly impacts various pharmaceutical disciplines, including medicinal chemistry, pharmacology, pharmacokinetics, pharmacodynamics, toxicology, and pharmaceutics. Incorporating AI fosters a more efficient, data-driven approach to drug discovery and development, ultimately enhancing patient care and medical treatments.
Acknowledgement
We sincerely thank Professor Kuo-Hsiung Lee, a National Academician of Taiwan. Throughout his career, Professor Lee has dedicated himself to advancing natural medicine and pharmacy, making significant contributions through persistent research and innovation. His unwavering commitment to academic progress is evident in his research tenacity and active support for the development and realization of the Liberty project. Professor Lee's forwardthinking leadership and altruistic dedication serve as invaluable models for emerging researchers, inspiring us to pursue excellence. We feel truly privileged to have benefited from his support and encouragement, which have greatly enhanced our research endeavors.
We also sincerely thank NVIDIA AI Technology Center (NVIDIA Corporation, USA), Merck Ltd. Taiwan Company, Mr. Kuan-Wen Chen (GGA Corporation, Molecular Science and Digital Innovation Center, Taiwan), Miss Nian-Gu Chen and Miss Pei-Jen Chung (UNI-ONEARD Corporation, Taiwan) for their assistance on this work.
List of abbreviations
- AA
Alopecia areata
- ADC
Antibody-drug conjugates
- AI
Artificial intelligence
- AlphaFold
Alphafold protein structure database
- CMUH
China Medical University Hospital
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DS
Discovery studio
- E-files
Electronic files
- ExPASy
Expert protein analysis system
- GO
Gene ontology
- GOLD
Genetic Optimisation for Ligand Docking
- GSEA
Gene set enrichment analysis
- GWAS
Genome-Wide Association Study
- HPA
The human protein atlas
- HTPP
High-throughput protein production
- IPA
Ingenuity pathway analysis
- IRB
Institutional Review Board
- IVIVC
In vivo in vitro correlation
- MD
Molecular dynamics
- Metacore
Metacore enrichment analysis
- ML
Machine learning
- MOAD
Mother of all databases
- NASH
Non-alcoholic steatohepatitis
- NCBI
National center for biotechnology information
- NGS
Next generation sequencing
- NPRL
Natural Product Research Laboratory
- NLME
WinNonMix
- PD
Pharmacodynamics
- PK
Pharmacokinetics
- PPB
Plasma protein binding
- QSAR
Quantitative structure-activity relationship
- RCSB_PDB
RCSB protein data bank
- R&D
Research and development
- SMILES
Simplified molecular input line entry system
- STRING
Search tool for the retrieval of interacting genes
- TAC
Transcriptome analysis console
- UniProt
Universal Protein
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
- RNA-seq
RNA sequencing
Funding Statement
This work was supported in part of the project (DMR-113-109) from China Medical University Hospital, Taiwan
Footnotes
Authors' contributions: JSY, SCT, YMH and FJT were involved in the conception of this study. YMH, DTB, CWT, WSC, CCY and YJC were involved in the literature search and critical reviewing of the manuscript. DTB, CWT, WSC, CCY and YJC were involved in the preparation of the draft of the manuscript. JSY, SCT, YMH, DTB and FJT were involved in the revising and editing of the manuscript. All authors have read and approved the final manuscript.
Conflict of interest: The authors declare that they have no competing interests.
Funding: This work was supported in part of the project (DMR-113-109) from China Medical University Hospital, Taiwan.
References
- 1. Gaspar A, Garrido E, Borges F, Garrido J. Biological and medicinal properties of natural chromones and chromanones. ACS Omega. 2024;9:21706–26. doi: 10.1021/acsomega.4c00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahuja A, Singh S, Murti Y. Chemical probes review: choosing the right path towards pharmacological targets in drug discovery, challenges and future perspectives. Comb Chem High Throughput Screen. 2024;27:2544–64. doi: 10.2174/0113862073283304231118155730. [DOI] [PubMed] [Google Scholar]
- 3. Wang Z, Yang L. Natural-product-based, carrier-free, non-covalent nanoparticles for tumor chemo-photodynamic combination therapy. Pharmacol Res. 2024;203:107150. doi: 10.1016/j.phrs.2024.107150. [DOI] [PubMed] [Google Scholar]
- 4. Sun Y, Sun H, Zhang Z, Tan F, Qu Y, Lei X, et al. New insight into oxidative stress and inflammatory responses to kidney stones: potential therapeutic strategies with natural active ingredients. Biomed Pharmacother. 2024;179:117333. doi: 10.1016/j.biopha.2024.117333. [DOI] [PubMed] [Google Scholar]
- 5. Atanasov AG, Zotchev SB, Dirsch VM. International Natural Product Sciences T, Supuran CT. Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov. 2021;20:200–16. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wei D, Peslherbe GH, Selvaraj G, Wang Y. Advances in drug design and development for human therapeutics using artificial intelligence-I. Biomolecules. 2022:12. doi: 10.3390/biom12121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ryan DK, Maclean RH, Balston A, Scourfield A, Shah AD, Ross J. Artificial intelligence and machine learning for clinical pharmacology. Br J Clin Pharmacol. 2024;90:629–39. doi: 10.1111/bcp.15930. [DOI] [PubMed] [Google Scholar]
- 8. Frusciante L, Visibelli A, Geminiani M, Santucci A, Spiga O. Artificial intelligence Approaches in Drug Discovery: owards the laboratory of the future. Curr Top Med Chem. 2022;22:2176–89. doi: 10.2174/1568026622666221006140825. [DOI] [PubMed] [Google Scholar]
- 9. Collaborators GBDSRF. Global, regional, and national burden of stroke and its risk factors, 1990–2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet Neurol. 2024;23:973–1003. doi: 10.1016/S1474-4422(24)00369-7. [DOI] [PubMed] [Google Scholar]
- 10. Xu M, Chen Z, Zheng J, Zhao Q, Yuan Z. Artificial intelligence-aided optical imaging for cancer theranostics. Semin Cancer Biol. 2023;94:62–80. doi: 10.1016/j.semcancer.2023.06.003. [DOI] [PubMed] [Google Scholar]
- 11. Dergaa I, Chamari K, Zmijewski P, Ben Saad H. From human writing to artificial intelligence generated text: examining the prospects and potential threats of ChatGPT in academic writing. Biol Sport. 2023;40:615–22. doi: 10.5114/biolsport.2023.125623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng JY, Abel JT, Balis UGJ, McClintock DS, Pantanowitz L. Challenges in the development, deployment, and regulation of artificial intelligence in anatomic pathology. Am J Pathol. 2021;191:1684–92. doi: 10.1016/j.ajpath.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 13. Huang ETC, Yang JS, Liao KYK, Tseng WCW, Lee CK, Gill M, et al. Predicting blood-brain barrier permeability of molecules with a large language model and machine learning. Sci Rep. 2024;14:15844. doi: 10.1038/s41598-024-66897-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J-S, Chiang J-H, Tsai SC, Hsu Y-M, Bau D-T, Lee K-H, et al. In silico de novo curcuminoid derivatives from the compound library of natural products research laboratories inhibit COVID-19 3CLpro activity. Nat Prod Commun. 2020;15:1934578X20953262. [Google Scholar]
- 15. Wu SY, Pan SL, Xiao ZY, Hsu JL, Chen MC, Lee KH, et al. NPRL-Z-1, as a new topoisomerase II poison, induces cell apoptosis and ROS generation in human renal carcinoma cells. PLoS One. 2014;9:e112220. doi: 10.1371/journal.pone.0112220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen HA, Hsu RH, Fang CY, Desai AK, Lee NC, Hwu WL, et al. Optimizing treatment outcomes: immune tolerance induction in Pompe disease patients undergoing enzyme replacement therapy. Front Immunol. 2024;15:1336599. doi: 10.3389/fimmu.2024.1336599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Degtyarenko K, Hastings J, de Matos P, Ennis M. ChEBI: an open bioinformatics and cheminformatics resource. Curr Protoc Bioinformatics. 2009;Chapter 14:14 9 1–14 9 20. doi: 10.1002/0471250953.bi1409s26. [DOI] [PubMed] [Google Scholar]
- 18. Bazzi-Allahri F, Shiri F, Ahmadi S, Toropova AP, Toropov AA. SMILES-based QSAR virtual screening to identify potential therapeutics for COVID-19 by targeting 3CL(pro) and RdRp viral proteins. BMC Chem. 2024;18:191. doi: 10.1186/s13065-024-01302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu SQ, Shen BB, Li HY, Yao YX, Li B, Yu HH, et al. Integrating UPLC-Q-Exactive Orbitrap/MS, Network pharmacology and experimental validation to reveal the potential mechanism of Kadsuracoccinea roots in Colon Cancer. J Ethnopharmacol. 2024;337:118934. doi: 10.1016/j.jep.2024.118934. [DOI] [PubMed] [Google Scholar]
- 20. Burgi HB. The cambridge structural database and structural dynamics. Struct Dyn. 2024;11:021302. doi: 10.1063/4.0000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ajimura CM, Jagan N, Morrow LE, Malesker MA. Drug interactions with oral inhaled medications. J Pharm Technol. 2018;34:273–80. doi: 10.1177/8755122518788809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weng J, Wu XF, Shao P, Liu XP, Wang CX. Medicine for chronic atrophic gastritis: a systematic review, meta- and network pharmacology analysis. Ann Med. 2023;55:2299352. doi: 10.1080/07853890.2023.2299352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Shi Q. Drugs and diseases interacting with cigarette smoking in US prescription drug labelling. Clin Pharmacokinet. 2015;54:493–501. doi: 10.1007/s40262-015-0246-6. [DOI] [PubMed] [Google Scholar]
- 24. Chavez-Hernandez AL, Sanchez-Cruz N, Medina-Franco JL. Fragment library of natural products and compound databases for drug discovery. Biomolecules. 2020:10. doi: 10.3390/biom10111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vojnits K, Feng Z, Johnson P, Porras D, Manocha E, Vandersluis S, et al. Targeting of human cancer stem cells predicts efficacy and toxicity of FDA-approved oncology drugs. Cancer Lett. 2024;599:217108. doi: 10.1016/j.canlet.2024.217108. [DOI] [PubMed] [Google Scholar]
- 26. Harding SD, Armstrong JF, Faccenda E, Southan C, Alexander SPH, Davenport AP, et al. The IUPHAR/BPS guide to pharmacology in 2024. Nucleic Acids Res. 2024;52:D1438–49. doi: 10.1093/nar/gkad944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bisht A, Tewari D, Kumar S, Chandra S. Network pharmacology, molecular docking, and molecular dynamics simulation to elucidate the mechanism of anti-aging action of Tinospora cordifolia. Mol Divers. 2024;28:1743–63. doi: 10.1007/s11030-023-10684-w. [DOI] [PubMed] [Google Scholar]
- 28. Carles F, Bourg S, Meyer C, Bonnet P. PKIDB: a curated, annotated and updated database of protein kinase inhibitors in clinical trials. Molecules. 2018;23 doi: 10.3390/molecules23040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olmedo DA, Durant-Archibold AA, Lopez-Perez JL, Medina-Franco JL. Design and diversity analysis of chemical libraries in drug discovery. Comb Chem High Throughput Screen. 2024;27:502–15. doi: 10.2174/1386207326666230705150110. [DOI] [PubMed] [Google Scholar]
- 30. Lv Q, Chen G, He H, Yang Z, Zhao L, Zhang K, et al. TCMBank-the largest TCM database provides deep learning-based Chinese-Western medicine exclusion prediction. Signal Transduct Targeted Ther. 2023;8:127. doi: 10.1038/s41392-023-01339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bajorath J. Chemical language models for molecular design. Mol Inform. 2024;43:e202300288. doi: 10.1002/minf.202300288. [DOI] [PubMed] [Google Scholar]
- 32. Hasselgren C, Oprea TI. Artificial intelligence for drug discovery: are we there yet? Annu Rev Pharmacol Toxicol. 2024;64:527–50. doi: 10.1146/annurev-pharmtox-040323-040828. [DOI] [PubMed] [Google Scholar]
- 33. Xu Y, Liu X, Cao X, Huang C, Liu E, Qian S, et al. Artificial intelligence: a powerful paradigm for scientific research. Innovation. 2021;2:100179. doi: 10.1016/j.xinn.2021.100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vora LK, Gholap AD, Jetha K, Thakur RRS, Solanki HK, Chavda VP. Artificial intelligence in pharmaceutical technology and drug delivery design. Pharmaceutics. 2023:15. doi: 10.3390/pharmaceutics15071916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta R, Srivastava D, Sahu M, Tiwari S, Ambasta RK, Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers. 2021;25:1315–60. doi: 10.1007/s11030-021-10217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gholap AD, Uddin MJ, Faiyazuddin M, Omri A, Gowri S, Khalid M. Advances in artificial intelligence for drug delivery and development: a comprehensive review. Comput Biol Med. 2024;178:108702. doi: 10.1016/j.compbiomed.2024.108702. [DOI] [PubMed] [Google Scholar]
- 37. Chen H, Bajorath J. Generative design of compounds with desired potency from target protein sequences using a multi-modal biochemical language model. J Cheminf. 2024;16:55. doi: 10.1186/s13321-024-00852-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tao L, Zhang P, Qin C, Chen SY, Zhang C, Chen Z, et al. Recent progresses in the exploration of machine learning methods as in-silico ADME prediction tools. Adv Drug Deliv Rev. 2015;86:83–100. doi: 10.1016/j.addr.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 39. Swanson K, Walther P, Leitz J, Mukherjee S, Wu JC, Shivnaraine RV, et al. ADMET-AI: a machine learning ADMET platform for evaluation of large-scale chemical libraries. Bioinformatics. 2024:40. doi: 10.1093/bioinformatics/btae416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakiyama H, Fukuda M, Okuno T. Prediction of blood-brain barrier penetration (BBBP) based on molecular descriptors of the free-form and in-blood-form datasets. Molecules. 2021:26. doi: 10.3390/molecules26247428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramsundar B, Liu B, Wu Z, Verras A, Tudor M, Sheridan RP, et al. Is multitask deep learning practical for pharma? J Chem Inf Model. 2017;57:2068–76. doi: 10.1021/acs.jcim.7b00146. [DOI] [PubMed] [Google Scholar]
- 42. Shulga DA, Shaimardanov AR, Ivanov NN, Palyulin VA. Assessing how residual errors of scoring functions correlate to ligand structural features. Int J Mol Sci. 2022:23. doi: 10.3390/ijms232315018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu J, Hou X, Wang C, Zhang Y. Incorporating explicit water molecules and ligand conformation stability in machine-learning scoring functions. J Chem Inf Model. 2019;59:4540–9. doi: 10.1021/acs.jcim.9b00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stork C, Chen Y, Sicho M, Kirchmair J. Hit dexter 2.0: machine-learning models for the prediction of frequent hitters. J Chem Inf Model. 2019;59:1030–43. doi: 10.1021/acs.jcim.8b00677. [DOI] [PubMed] [Google Scholar]
- 45. Stork C, Wagner J, Friedrich NO, de Bruyn Kops C, Sicho M, Kirchmair J. Hit dexter: a machine-learning model for the prediction of frequent hitters. ChemMedChem. 2018;13:564–71. doi: 10.1002/cmdc.201700673. [DOI] [PubMed] [Google Scholar]
- 46. Yang K, Xie Z, Li Z, Qian X, Sun N, He T, et al. MolProphet: a one-stop, general purpose, and AI-based platform for the early stages of drug discovery. J Chem Inf Model. 2024;64:2941–7. doi: 10.1021/acs.jcim.3c01979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hetmann M, Parigger L, Sirelkhatim H, Stern A, Krassnigg A, Gruber K, et al. Folding the human proteome using BioNeMo: a fused dataset of structural models for machine learning purposes. Sci Data. 2024;11:591. doi: 10.1038/s41597-024-03403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bau DT, Liu TY, Yang JS, Chen WT, Tsai CW, Chang WS, et al. Characterizing genetic susceptibility to colorectal cancer in Taiwan through genome-wide association study. Mol Carcinog. 2024 Oct;:1–8. doi: 10.1002/mc.23823. [DOI] [PubMed] [Google Scholar]
- 49. Yang JS, Liu TY, Lu HF, Tsai SC, Liao WL, Chiu YJ, et al. Genome-wide association study and polygenic risk scores predict psoriasis and its shared phenotypes in Taiwan. Mol Med Rep. 2024:30. doi: 10.3892/mmr.2024.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang JS, Liu TY, Chen YC, Tsai SC, Chiu YJ, Liao CC, et al. Genome-Wide association study of alopecia areata in Taiwan: the conflict between individuals and hair follicles. Clin Cosmet Invest Dermatol. 2023;16:2597–612. doi: 10.2147/CCID.S428788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang LC, Hsieh MT, Yang JS, Lu CC, Tsai FJ, Tsao JW, et al. Effect of bis(hydroxymethyl) alkanoate curcuminoid derivative MTH-3 on cell cycle arrest, apoptotic and auto-phagic pathway in triple-negative breast adenocarcinoma MDA-MB-231 cells: an in vitro study. Int J Oncol. 2018;52:67–76. doi: 10.3892/ijo.2017.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu CY, Yang JS, Huang SM, Chiang JH, Chen MH, Huang LJ, et al. Smh-3 induces G(2)/M arrest and apoptosis through calcium-mediated endoplasmic reticulum stress and mitochondrial signaling in human hepatocellular carcinoma Hep3B cells. Oncol Rep. 2013;29:751–62. doi: 10.3892/or.2012.2166. [DOI] [PubMed] [Google Scholar]
- 53. Chung JG, Yang JS, Huang LJ, Lee FY, Teng CM, Tsai SC, et al. Proteomic approach to studying the cytotoxicity of YC-1 on U937 leukemia cells and antileukemia activity in orthotopic model of leukemia mice. Proteomics. 2007;7:3305–17. doi: 10.1002/pmic.200700200. [DOI] [PubMed] [Google Scholar]
- 54. Wu RS, Liu KC, Tang NY, Chung HK, Ip SW, Yang JS, et al. cDNA microarray analysis of the gene expression of murine leukemia RAW 264.7 cells after exposure to propofol. Environ Toxicol. 2013;28:471–8. doi: 10.1002/tox.20742. [DOI] [PubMed] [Google Scholar]
- 55. Cheng YD, Lu CC, Hsu YM, Tsai FJ, Bau DT, Tsai SC, et al. In Silico and in Vitro studies of taiwan chingguan yihau (NRICM101) on TNF-alpha/IL-1beta-induced human lung cells. Biomedicine. 2022;12:56–71. doi: 10.37796/2211-8039.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Song M, Yang X, Zhang X, Li J, Xu Y, Shi J. The Masquelet technique triggers the formation of a network involving LncRNA, circRNA, miRNA, and mRNA during bone repair. Ann Med. 2024;56:2395591. doi: 10.1080/07853890.2024.2395591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sherman BT, Panzade G, Imamichi T, Chang W. DAVID ortholog: an integrative tool to enhance functional analysis through orthologs. Bioinformatics. 2024 Oct 1;40(10):btae615. doi: 10.1093/bioinformatics/btae615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tiong KL, Yeang CH. MGSEA - a multivariate Gene set enrichment analysis. BMC Bioinf. 2019;20:145. doi: 10.1186/s12859-019-2716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shan C, Wu Z, Xia Y, Ji X, Zhang W, Peng X, et al. Network pharmacological study and in vitro studies validation-Molecular dynamics simulation of Cistanche deserticola in promoting periodontitis and bone remodeling. Int Immunopharm. 2024;135:112299. doi: 10.1016/j.intimp.2024.112299. [DOI] [PubMed] [Google Scholar]
- 60. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–50. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 62. Ptak RG, Fu W, Sanders-Beer BE, Dickerson JE, Pinney JW, Robertson DL, et al. Cataloguing the HIV type 1 human protein interaction network. AIDS Res Hum Retrovir. 2008;24:1497–502. doi: 10.1089/aid.2008.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hedgpeth B, Missall R, Bambaci A, Smolen M, Yavuz S, Cottrell J, et al. Medicines (Basel) 2019. A review of bioinformatics tools to understand acetaminophen-alcohol interaction; p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. David A, Islam S, Tankhilevich E, Sternberg MJE. The AlphaFold database of protein structures: a Biologist's guide. J Mol Biol. 2022;434:167336. doi: 10.1016/j.jmb.2021.167336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schneider M, Tognolli M, Bairoch A. The Swiss-Prot protein knowledgebase and ExPASy: providing the plant community with high quality proteomic data and tools. Plant Physiol Biochem. 2004;42:1013–21. doi: 10.1016/j.plaphy.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 67. Colwill K. Renewable Protein Binder Working G, Graslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods. 2011;8:551–8. doi: 10.1038/nmeth.1607. [DOI] [PubMed] [Google Scholar]
- 68. Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, et al. InterPro in 2022. Nucleic Acids Res. 2023;51:D418–27. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martinez X, Krone M, Alharbi N, Rose AS, Laramee RS, O'Donoghue S, et al. Molecular graphics: bridging structural biologists and computer scientists. Structure. 2019;27:1617–23. doi: 10.1016/j.str.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 70. Choudhary P, Feng Z, Berrisford J, Chao H, Ikegawa Y, Peisach E, et al. PDB NextGen Archive: centralizing access to integrated annotations and enriched structural information by the Worldwide Protein Data Bank. Database. 2024:2024. doi: 10.1093/database/baae041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cheng J, Li Z, Liu Y, Li C, Huang X, Tian Y, et al. [Bioinformatics analysis and validation of the interaction between PML protein and TAB1 protein]. Nan Fang Yi Ke Da Xue Xue Bao. 2024;44:179–86. doi: 10.12122/j.issn.1673-4254.2024.01.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–69. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen L, Li Q, Nasif KFA, Xie Y, Deng B, Niu S, et al. AI-driven deep learning techniques in protein structure prediction. Int J Mol Sci. 2024:25. doi: 10.3390/ijms25158426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rusinko A, Rezaei M, Friedrich L, Buchstaller HP, Kuhn D, Ghogare A. AIDDISON: empowering drug discovery with AI/ML and CADD tools in a secure, web-based SaaS platform. J Chem Inf Model. 2024;64:3–8. doi: 10.1021/acs.jcim.3c01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Solis-Vasquez L, Tillack AF, Santos-Martins D, Koch A, LeGrand S, Forli S. Benchmarking the performance of irregular computations in AutoDock-GPU molecular docking. Parallel Comput. 2022:109. doi: 10.1016/j.parco.2021.102861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang JM, Chen CC. GEMDOCK: a generic evolutionary method for molecular docking. Proteins. 2004;55:288–304. doi: 10.1002/prot.20035. [DOI] [PubMed] [Google Scholar]
- 77. Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–23. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 78. Sedaghatkish A, Gossen BD, McDonald MR. Characterization of a virulence factor in Plasmodiophora brassicae, with molecular markers for identification. PLoS One. 2023;18:e0289842. doi: 10.1371/journal.pone.0289842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017;9:91–102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Margreitter C, Oostenbrink C. MDplot: visualise molecular dynamics. R J. 2017;9:164–86. [PMC free article] [PubMed] [Google Scholar]
- 81. Fu L, Shi S, Yi J, Wang N, He Y, Wu Z, et al. ADMETlab 3.0: an updated comprehensive online ADMET prediction platform enhanced with broader coverage, improved performance, API functionality and decision support. Nucleic Acids Res. 2024;52:W422–31. doi: 10.1093/nar/gkae236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Du BX, Xu Y, Yiu SM, Yu H, Shi JY. ADMET property prediction via multi-task graph learning under adaptive auxiliary task selection. iScience. 2023;26:108285. doi: 10.1016/j.isci.2023.108285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jiang Y, Ray A, Junaid MSA, Bhattaccharjee SA, Kelley K, Banga AK, et al. The pharmacokinetics of 3-fluoroamphetamine following delivery using clinically relevant routes of administration. Drug Deliv Transl Res. 2020;10:271–81. doi: 10.1007/s13346-019-00685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Trumbull EJ, Papich MG, Peters M, Whitmer ER, Rivard M, Field CL. Comparative pharmacokinetics of a single oral dose of meloxicam in the California sea lion (Zalophus californianus) and Pacific harbor seal (Phoca vitulina richardii) J Vet Pharmacol Therapeut. 2024;47:485–91. doi: 10.1111/jvp.13469. [DOI] [PubMed] [Google Scholar]
- 85. Ariano RE, Zelenitsky SA, Davis C, Sathianathan C, Wolowich WR. Comparative simulation of intraperitoneal aminoglycoside regimens for patients with peritonitis on automated peritoneal dialysis. Perit Dial Int. 2024;44:438–44. doi: 10.1177/08968608231221062. [DOI] [PubMed] [Google Scholar]
- 86. Kumar S, Chang YC, Lai KH, Hwang TL. Resveratrol, a molecule with anti-inflammatory and anti-cancer activities: natural product to chemical synthesis. Curr Med Chem. 2021;28:3773–86. doi: 10.2174/0929867327999200918100746. [DOI] [PubMed] [Google Scholar]
- 87. Chiu YJ, Chiang JH, Fu CW, Hour MJ, Ha HA, Kuo SC, et al. Analysis of COVID-19 prevention and treatment in Taiwan. Biomedicine. 2021;11:1–18. doi: 10.37796/2211-8039.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Halder AK, Dias Soeiro Cordeiro MN. QSAR-Co-X: an open source toolkit for multitarget QSAR modelling. J Cheminf. 2021;13:29. doi: 10.1186/s13321-021-00508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hao M, Li Y, Wang Y, Yan Y, Zhang S, Li G, et al. Combined 3D-QSAR, molecular docking, and molecular dynamics study on piperazinyl-glutamate-pyridines/pyrimidines as potent P2Y12 antagonists for inhibition of platelet aggregation. J Chem Inf Model. 2011;51:2560–72. doi: 10.1021/ci2002878. [DOI] [PubMed] [Google Scholar]
- 90. Sobhani N, D'Angelo A, Pittacolo M, Mondani G, Generali D. Future AI will most likely predict antibody-drug conjugate response in oncology: a review and expert opinion. Cancers. 2024:16. doi: 10.3390/cancers16173089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang Z, Wang G, Lu H, Li H, Tang M, Tong A. Development of therapeutic antibodies for the treatment of diseases. Mol Biomed. 2022;3:35. doi: 10.1186/s43556-022-00100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chakraborty C, Bhattacharya M, Lee SS, Wen ZH, Lo YH. The changing scenario of drug discovery using AI to deep learning: recent advancement, success stories, collaborations, and challenges. Mol Ther Nucleic Acids. 2024;35:102295. doi: 10.1016/j.omtn.2024.102295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen Z, Hu L, Zhang BT, Lu A, Wang Y, Yu Y, et al. Artificial intelligence in aptamer-target binding prediction. Int J Mol Sci. 2021:22. doi: 10.3390/ijms22073605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wenteler A, Cabrera CP, Wei W, Neduva V, Barnes MR. AI approaches for the discovery and validation of drug targets. Camb Prism Precis Med. 2024;2:e7. doi: 10.1017/pcm.2024.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Song Z, Chen G, Chen CY. AI empowering traditional Chinese medicine? Chem Sci. 2024;15:16844–86. doi: 10.1039/d4sc04107k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gupta M, Madan AK. Diverse models for the prediction of HIV integrase inhibitory activity of substituted quinolone carboxylic acids. Arch Pharm (Weinheim) 2012;345:989–1000. doi: 10.1002/ardp.201100316. [DOI] [PubMed] [Google Scholar]