Abstract
Tigecycline is a last-resort antibiotic to treat complicated infections caused by multidrug-resistant pathogens, while the emergence of plasmid-mediated tet(X) family severely compromises its clinical efficacy. Novel antimicrobial strategies not limited to new antibiotics in pharmaceutical pipeline are urgently needed. Herein, we reveal the metabolic disparities between tet(X)-negative and -positive E. coli, including distinct energy demand patterns under tigecycline exposure. In particular, the cysteine and methionine metabolism pathway is remarkably downregulated in tet(X)-positive bacteria. More importantly, we find that the addition of exogenous L-methionine (Met) effectively resensitizes tet(X)-positive pathogens to tigecycline. Our mechanistic analysis demonstrates that exogenous Met promotes intracellular tigecycline accumulation by upregulating bacterial proton motive force. Moreover, Met accelerates the conversion to S-adenosyl-L-methionine, an essential methyl donor, thereby enhancing 5mC methylation modification in the promoter region of tet(X4) gene and reducing its expression. Consistently, the potentiation of Met to tigecycline is abolished in tet(X4)-carrying E. coli Δdcm but restored in dcm-complementary bacteria, which encodes DNA-cytosine methyltransferase. In multiple animal models of infection, Met markedly potentiates the effectiveness of tigecycline against pathogenic E. coli and K. pneumoniae. Overall, this work highlights the therapeutic potential of Met in overcoming plasmid-mediated high-level tigecycline resistance, and provides a new paradigm to enhance antibiotic efficacy by harnessing cellular metabolic networks as well as epigenetic modifications.
Subject terms: Microbiology, Drug discovery, Antimicrobial resistance
The dissemination of plasmid-encoded tigecycline resistance gene tet(X) poses a global threat to public health, necessitating the development of innovative strategies. Here, the authors reveal that the supplementation of L-methionine not only facilitates intracellular accumulation of tigecycline but also reduces tet(X) expression by augmenting the 5mC methylation modification in its promoter region, thereby overcoming plasmid-mediated high-level tigecycline resistance.
Introduction
Antimicrobial resistance (AMR) has emerged as a public health threat of the 21st century, due to the indiscriminate use of antibiotics, the growing prevalence of antibiotic-resistance genes (ARGs) and the deceleration of antibiotic discovery1. In 2016, there were an estimated 0.7 million deaths caused by AMR, but in 2019, this burden increased exponentially to 1.27 million around the world2,3. If left uncontrolled, AMR would incur 10 million people deaths per year by 20502. Nowadays, tigecycline (Tig) is one of the last-resort antibiotics approved by the Food and Drugs Administration (FDA) in 20054, for its therapeutic benefits in complicated skin and skin structure infections, community-acquired bacterial pneumonia, and intra-abdominal infections5. However, plasmid-mediated tet(X) determinants have been numerously detected in human samples, food animals, and ecological environments in China since 2015. Even worse, the prevalence of tet(X3/4) genes has conferred high level of Tig resistance (minimal inhibitory concentration (MIC) values increased by 64-128-fold), potentially creating a barrier for treating serious infections, especially those caused by Enterobacteriaceae and Acinetobacter species6–8.
The molecular basis regarding different tet(X) genes has been extensively investigated. Briefly, tet(X) encodes a flavin-dependent monooxygenase to compromise not only Tig but also almost all tetracycline antibiotics’ efficacy in the ways of substrate hydroxylation. Recent studies indicated that Tet(X4) displayed the highest enzymatic activity among Tet(X1-X7), and tet(X4) gene was commonly located on different types of plasmids such as IncQ1, IncX1, IncHI1 etc.9. These tet(X4)-carrying plasmids share the same replicon type, facilitating horizontal transfer and widespread dissemination of the tet(X4) gene across various reservoirs in the “One Health” environment. It is reported that China has the highest prevalence of tet(X) among six continents, with tet(X4) mediating the highest level of Tig resistance among tet(X) variants, thereby exacerbating public health harm10. Therefore, it is imperative to exploit efficient strategies for overcoming tet(X)-conferred Tig resistance.
Previous studies have shown a strong association between antibiotic efficacy and bacterial metabolism11. Antibiotic lethality should be considered as a complex physiological process, involving the primary target inhibition elicited by antibiotics, and downstream events followed by antibiotics exposure. For example, the two-component system, iron homeostasis, and nucleotide metabolism all indispensably contributed to reactive oxygen species (ROS)-mediated antibiotic lethality in Escherichia coli12–14. Conversely, the alternation of bacterial metabolism can also manipulate antibiotic efficacy. Our previous studies suggested that exogenous indole-3-acetic acid enhanced antibiotic efficacy in eliminating persister cells, a sub-population of bacterial cells that can escape transient antibiotic killing15, by a mechanism that involving activated bacterial metabolic pathways16, and deoxycholic acid strikingly potentiated antibiotic efficacy by promoting antibiotic accumulation and destroying oxidant-antioxidant system17. Moreover, the treatment regimen of glutamine or inosine plus ampicillin (AMP) restored antibiotic activity against AMP-resistant strains, by upregulating ompF expression and intracellular AMP uptake18. These pioneering studies suggest that novel antimicrobial strategies can be designed from the perspective of bacterial metabolism.
In this study, we comprehensively delineated the differential metabolic profiles and fluxes between Tig-susceptible (Tig-S) and Tig-resistant bacteria (Tig-R), as well as their energy response patterns upon Tig exposure. On the basis of differential network biology between Tig-S and -R, we deployed a metabolite-facilitated strategy to modulate bacterial biochemistry and metabolic homeostasis, thereby reverting the phenotype of antibiotic resistance. Regarding tet(X4)-dominated resistance mechanism, the survival or death of bacteria can be determined by the action of Tet(X4) enzymes and Tig accumulation, which are two counteractive forces in bacterial cells upon antibiotic exposure. From these perspectives, we elucidated the underlying mechanisms of action. Additionally, we explored the relationship between antibiotic-resistance genes and reprogrammed pathways. Finally, we assessed the therapeutic prospects of the potent metabolite to restore Tig efficacy in multiple animal modes of infection.
Results
The global transcriptomic and metabolomic profiling between Tig-R and -S
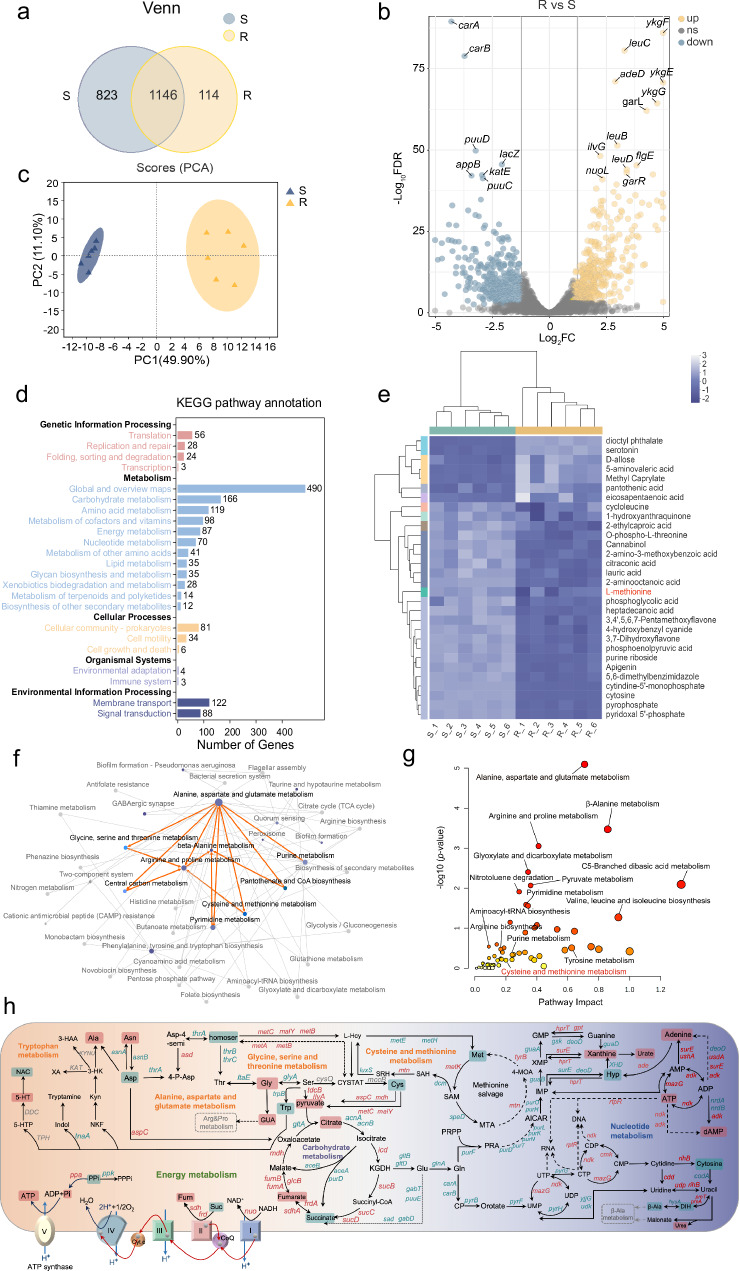
Firstly, we applied an integrated multi-omics analysis of E. coli DH5α (pUC19) (Tig-S) and E. coli DH5α (pUC19-tet(X4)) (Tig-R). In transcriptomics, the shared and independent genes numbers in Tig-R and -S were shown by the Venn diagram (Fig. 1a). Among these genes, differently expressed genes (DEGs) were distinguished with log2FC ≥ 1 or log2FC ≤ −1 (Fig. 1b). Gene Ontology (GO) enrichment highlighted the cellular process and metabolic process as the major altered pathways (Supplementary Fig. 1a, b), and in Kyoto Encyclopedia and Genomes (KEGG) enrichment, carbohydrate metabolism and amino acid metabolism were the most affected pathways (Fig. 1d). These enrichment analyses further highlighted a significant perturbation of pathways associated with amino acid metabolism, energy metabolism, and nucleotide metabolism in Tig-R, of which detailed differences have been pictured in Fig. 2a, Supplementary Figs. 2 and 3. Directional connectivity analysis further demonstrated the intrinsic connections of these prominent pathways, of which alanine, aspartate and glutamate metabolism, glycine, serine and threonine metabolism, cysteine and methionine metabolism were the most affected pathways in amino acid metabolism when compared Tig-R and -S (Fig. 1f).
Fig. 1. Differential metabolomic flux and profiling between tigecycline-resistant and -susceptible bacteria.
a Venn diagrams of annotated genes, 1146 represents the shared genes between two bacteria (Tig-R and -S). b Differentially expressed genes (DEGs) in Tig-R vs Tig-S. Tig-S, E. coli DH5α (pUC19); Tig-R, E. coli DH5α (pUC19-tet(X4)). One subset of significant DEGs was annotated. Yellow dots, up-regulated; gray dots, not-significant; blue dots, down-regulated. Log2FC were calculated to take the direction of the expression difference into account. −Log10FDR value ≥ 1 and log2FC ≥ 1 or log2FC ≤ −1 were identified as significant different DEGs. c PCA analysis of metabolomics, an unsupervised PCA plot was applied to examine the clustering characteristics of different metabolites. Colored dots representing individual samples, n = 6 per group. d KEGG pathway classification based on the significantly altered genes, including the first and secondary pathways. The pathways involved in genetic information processing (pink), metabolism (blue), cellular processes (yellow), organismal systems (purple) and environmental information processing (dark blue). e Clustering heatmap showing an abundance of top 30 differential metabolites in Tig-R compared with Tig-S. The light blue region on the left (S_1 ~ S_6) contains metabolites that were downregulated in Tig-R, while the corresponding dark blue region contains upregulated metabolites. L-Met was highlighted with red. f A network graph visualizing the relationships between various pathways based on KEGG enrichment analysis. The node size represents the number of genes enriched in KEGG pathway, while the gradient color of the node represents the P-value in KEGG enrichment analysis. The connections between certain amino acid pathways were specifically highlighted with orange lines. g A functional multi-omics integration and covariate adjustment of genomics and metabolomics, 61 associations at −log10 (P-value) > 0 identified mainly discrepant pathways between Tig-R and -S. Pathway impact displays an integration of the centrality and pathway enrichment pathway. Cys & Met metabolism (Pathway Impact 0.2) was labeled red. Adjusted P values were determined using two-way ANOVA with Sidak’s multiple comparison test, the Fisher’s method (−2*∑Log(P)) was used for information integration. h The global metabolic difference network when compared Tig-R with Tig-S, of focus on amino acids metabolism, energy metabolism, nucleotide metabolism and carbohydrate metabolism. Red represents up-regulated genes or metabolites, and green represents down-regulated genes or metabolites in Tig-R. GUA Guanidoacetic acid, CYSTAT Cystathionine, Xob 2-Oxobutanoate, Asp-4-semi L-Aspartate-4-semialdehyde, Homoser L-homoserine, 4-P-Asp L-4-Aspartyl-phosphate, SRH S-D-Ribosyl-L-homocysteine, SAH S-Adenosyl-L-methionine, KGDH α-Ketoglutaric acid, 4-MOA 4-Methylthio-2-oxobutanoic acid, CP Carbamoyl phosphate, PRA 5-Phosphoribosylamine, Xyp Xypoxanthine, Hyp Hypoxanthine, NAC N-Acetylserotonin. SRH S-D-Ribosyl-L-homocysteine.
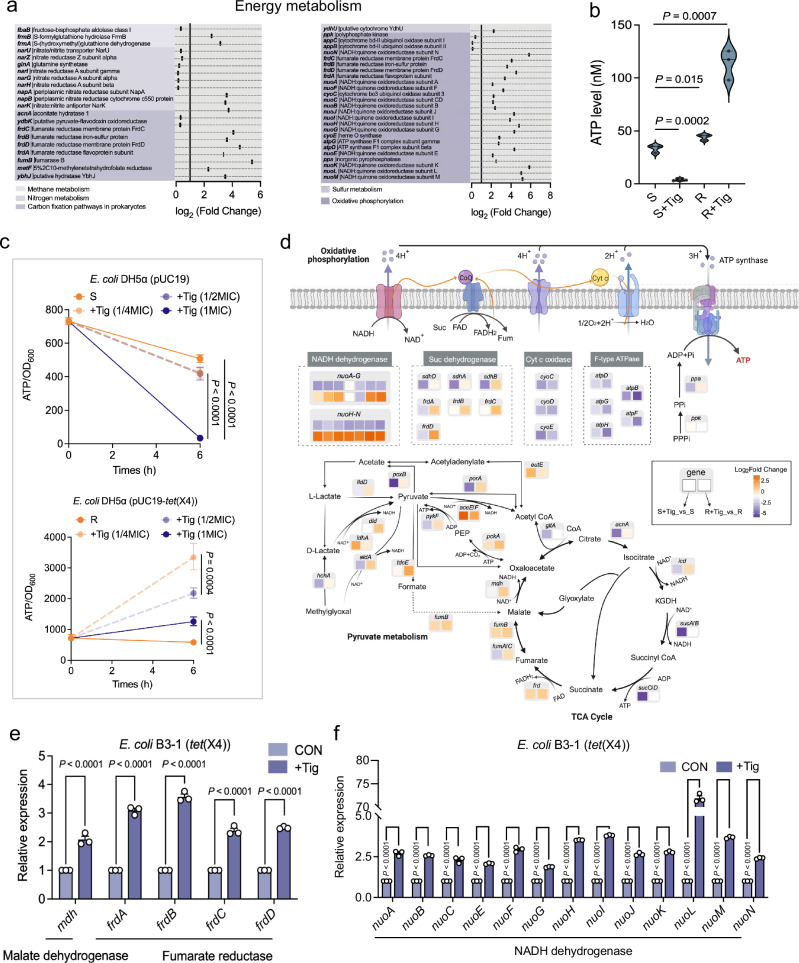
Fig. 2. Distinct energy demand patterns between tigecycline-resistant and -susceptible bacteria under antibiotic exposure.
a Systematic analysis of DEGs and encoded enzymes in energy metabolism. The fold changes were calculated for taking the direction of the expression difference into account. FC reflects the fold change of gene expression between Tig-R and -S. b Intracellular ATP dynamic in E. coli cells, with or without 1-fold MIC Tig treatment. 1-fold MICs of Tig-S and Tig-R were 0.25 and 16 μg/mL, respectively. c Ratio of the intracellular ATP and OD600 during growth of E. coli DH5α (pUC19) (S) and E. coli DH5α (pUC19-tet(X4)) (R) under different Tig concentrations. d Schematic representation of bacterial ATP generation, including pyruvate metabolism, TCA cycle, and oxidative phosphorylation. Box indicates changes in mRNA when compared S + Tig with S (left), R + Tig with R (right), respectively, except for NADH dehydrogenase, the color blocks on the top and bottom represent the two sets of comparisons. Color coding is based on the level of log2-fold change as indicated. e mRNA levels of malate dehydrogenase (mdh) and succinate dehydrogenase (frdA-D) genes in E. coli B3-1 (tet(X4)) cells with Tig treatment for 6 h. f mRNA levels of NADH dehydrogenase (nuo) genes in E. coli B3-1 (tet(X4)) cells with Tig treatment for 6 h. Experiments were performed with three biological replicates and data are means ± SEM. In b and c, the P values were determined using an unpaired two-tailed Student’s t-test. In e and f, adjusted P values were determined using two-way ANOVA with Sidak’s multiple comparison test.
In metabolomics, PCA analysis and volcano map showed an apparent separation in metabolite profiles between Tig-R and -S (Fig. 1c and Supplementary Fig. 4a). Among 74 metabolites, organic acids and derivatives accounted for the largest proportion in the Human Metabolome Database (HMDB), while diverse bacterial physiology processes were assigned to the KEGG pathway (Supplementary Fig. 4b, c). Hierarchical clustering analysis identified L-methionine (Met) as one of the critically down-regulated metabolites among the top 30 differential metabolites in Tig-R, suggesting a potential reprogramming property for Met (Fig. 1e). Notably, Met linked to cysteine and methionine metabolism, which was also a different pathway signature between Tig-S and -R as described above (Supplementary Fig. 2). In line with this, cysteine and methionine metabolism were also marked as a salient differential pathway based on a multi-omics data-driven network analysis (Fig. 1g). Together, we charted the critical pathways that differed strikingly between Tig-R versus -S, which can be characterized by downregulated amino acid metabolism (alanine, aspartate, and glutamate metabolism; glycine, serine and threonine metabolism; cysteine and methionine metabolism), nucleotide metabolism (pyrimidine metabolism), as well as upregulated energy metabolism, carbohydrate metabolism and nucleotide metabolism (purine metabolism) (Fig. 1h and Supplementary Fig. 5). Meanwhile, Met, cysteine and aspartic acid were positively correlated, which was indicative of the concordance between altered metabolites abundance and altered pathways when compared Tig-R with S, implying that there was a high accordance between transcriptome and metabolome data (Supplementary Fig. 2 and Fig. 4d, e).
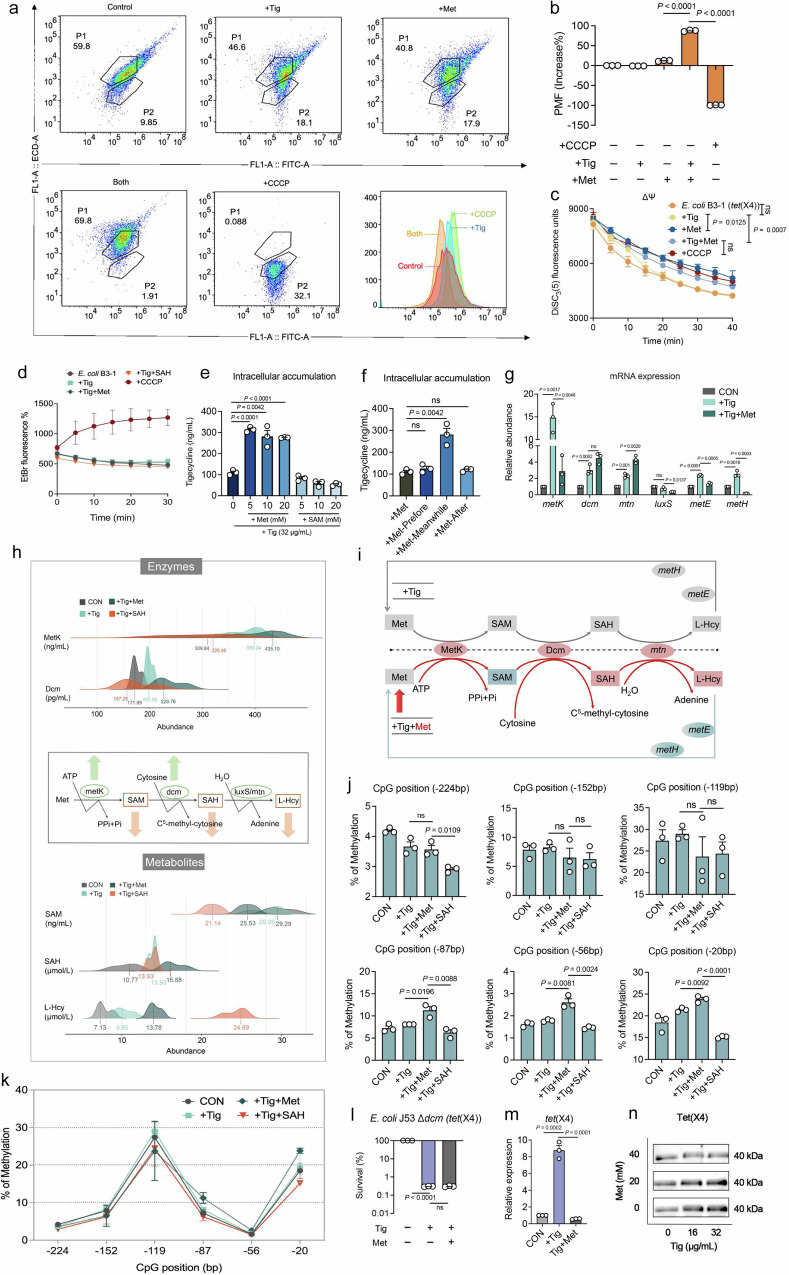
Fig. 4. Exogenous Met promotes intracellular Tig accumulation and 5mC methylation in the promoter region of tet(X4) gene.
a Flow cytometry analysis of the PMF of E. coli B3-1 (tet(X4)) in the presence of Tig (16 μg/mL) with or without Met (20 mM) for 6 h. Cells were incubated with DiOC2(3) (30 μM) for 30 min and then analyzed using 488 nm excitation and 530 nm emission filter. CCCP (10 μM) was recognized as the negative control to blunt the original PMF in cells. b The changes of bacterial PMF under different treatments, are calculated as follows: PMF= Log(103/2*(red fluorescence/green fluorescence)). c Dynamic monitoring of membrane potential (ΔΨ) changes in bacteria was performed using the fluorescent probe DiSC3(5) (0.5 μM), in the presence of Tig, Met, or both. The fluorescence units were monitored during 40 min. d The activity of efflux pump in bacteria after exposure Tig, Met or both, assessed using a fluorescent probe Ethidium Bromide (EtBr, 5 μM). CCCP was used as a known efflux pump inhibitor. EtBr efflux from cells was detected with the excitation wavelength at 530 nm and emission wavelength at 600 nm within 30 min. e The accumulation of intracellular antibiotics in E. coli B3-1 (tet(X4)) after being treated with Tig (1-fold MIC) with or without metabolites supplement, including Met and SAM. f The accumulation of intracellular antibiotic in E. coli B3-1 (tet(X4)) treated by Tig (1-fold MIC), and Met (20 mM) at different time intervals (the first 2 h before Tig addition or the subsequent 2 h). g mRNA expression of Met degradation-related enzymes in E. coli B3-1 (tet(X4)) under the treatment of Tig (32 μg/mL) with or without Met (20 mM) at 6 h. h Metabolic networks surveillance on Met degradation pathway when exogenous Met was added to Tig-treated E. coli B3-1 (tet(X4)), including 2 enzymes (MetK, S-adenosylmethionine synthetase; Dcm, DNA-cytosine methyltransferase) and 3 metabolites (SAM, S-Adenosyl-L-methionine; SAH, S-Adenosyl-L-homocysteine; L-Hcy, L-homocysteine). SAH was recognized as a methyltransferase inhibitor. i Metabolic networks summarized reprogramming process for exogenous Met in Tig-treated cells. The upper part represents the case of Tig treatment, with the normalized processing shown in gray. The lower part represents the changes under Tig plus Met treatment, with upregulated metabolites or enzymes or genes marked in red, and downregulated metabolites or enzymes or genes marked in green. Metabolites (SAM, SAH, L-Hcy); Enzymes (MetK, Dcm); Genes (mtn, metE, metH). j, k Changes in cytosine methylation rates at the six CG sites in the tet(X) promoter region under different treatments. Bisulfite methylation sequencing was performed on E. coli B3-1 (tet(X4)) after exposure to Tig (32 μg/mL) or Tig-Met combination or Tig-SAH combination for 6 h. l Percent survival of 5mC methylation knockout strain (Δdcm) after 6 h treatment of Tig (8 μg/mL) with or without Met (20 mM). m Relative expression of tet(X4) gene of E. coli B3-1 (tet(X4)) by RT-qPCR analysis. Cells were grown in LB broth and suspended in M9 medium in the treatment of Tig (1-fold MIC) with or without Met (20 mM). n Tet(X4) production in E. coli B3-1 (tet(X4)) cells following coculture with different concentrations of Tig (0, 16, and 32 μg/mL) and Met (0, 20, and 40 mM) by western blotting assays. The expression of internal reference protein GroEL and integrated density were shown in Supplementary Fig. 17. Data were displayed as mean ± SEM. Three biological repeats were carried out. In b, c, e, f, j, l, m, the P values were determined using an unpaired two-tailed Student’s t-test. In g, adjusted P values were determined using two-way ANOVA with Sidak’s multiple comparison test. ns, not significant.
Tig-R and -S show distinct energy demand patterns under antibiotic exposure
Having shown the differential features between Tig-R and -S without antibiotic exposure, we next sought to investigate their metabolic difference in antibiotic-induced cellular death physiology (Fig. 2a). Interestingly, we observed that ATP levels differed not only between Tig-R and -S but also in their response patterns upon Tig exposure (Fig. 2b). As shown in Fig. 2c, the ATP demand rapidly increased in Tig-R but decreased in Tig-S under subinhibitory concentrations of Tig. Next, we further elucidated the mechanistic basis of energy metabolism in antibiotic-treated Tig-S and -R. As shown in Fig. 2d and Supplementary Fig. 6a, detailed landscape of metabolic states corresponding to energy network perturbations in Tig-R and -S under antibiotic exposure was presented. The most characteristic difference was that nuo-encoded NADH dehydrogenases exhibited opposed expression at the transcriptional level, which largely hindered electron transport and the subsequent ATP synthesis in Tig-S but drove more ATP generation in Tig-R upon Tig exposure. Consistently, mRNA expression of a set of nuo genes in clinical-derived E. coli B3-1 (tet(X4)) further confirmed this finding (Fig. 2f)19. In addition, we observed consistent major shifts in the central carbon metabolism (TCA cycle and pyruvate metabolism). FrdA-D, encoding fumarate reductase proteins for presenting sufficient redox substrate (succinate) to the respiratory chain, were accordingly upregulated under Tig stress to satisfy the increased ATP demand in Tig-R (Fig. 2e). Likewise, genes involved in producing reducing-equivalents (NADH) for respiratory chain (dld, aldA, mdh, sucB) or catalyzing oxidative phosphorylation at the substrate level (pykF, sucC, sucD) were all elevated in Tig-R-manipulated antibiotic response progresses when compared with that in Tig-S (Fig. 2d). Collectively, these results indicated that Tig-R and -S performed two different modes of energy demand, and the underlying mechanism was predominantly due to nuo-mediated NADH dehydrogenases, accompanied by reducing-equivalent generation and oxidative phosphorylation at the substrate level.
Methionine restores the susceptibility of tet(X)-positive bacteria to tigecycline
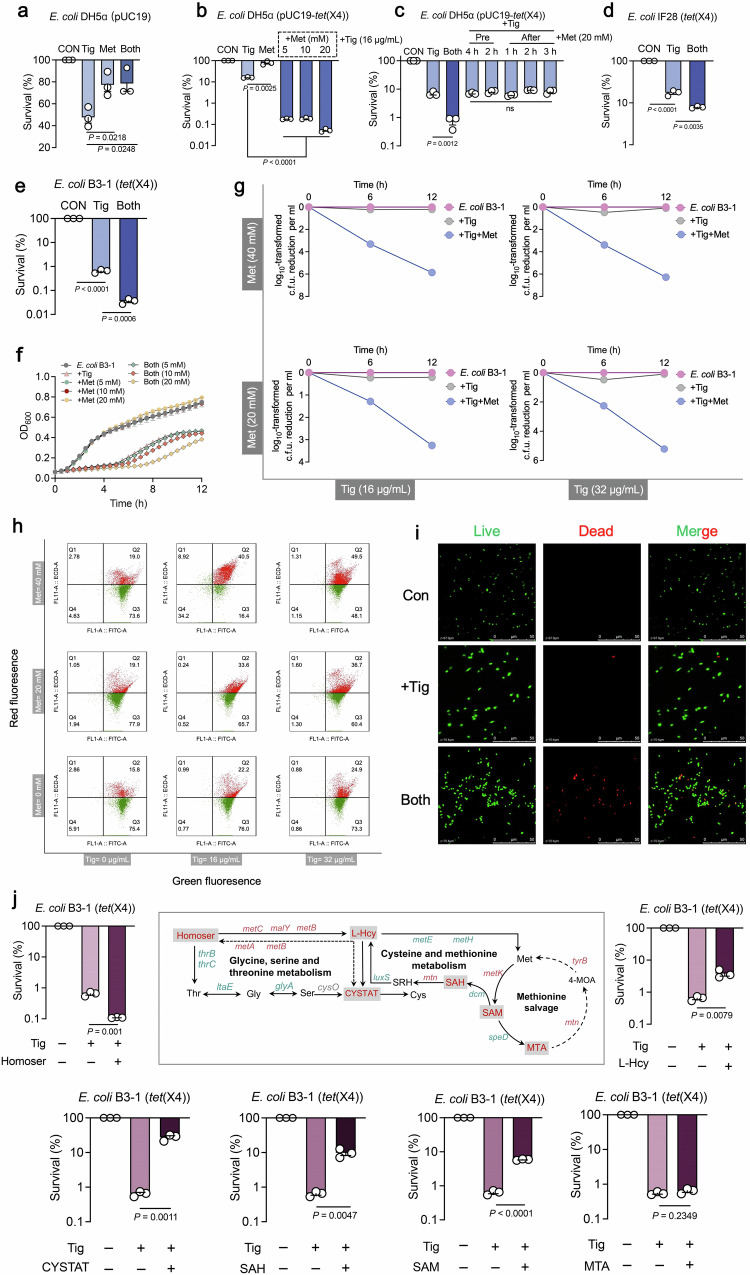
Reasoning that Met was differentially depleted, and cysteine and methionine metabolism strikingly differed in tet(X)-positive bacteria, we hypothesized that exogenous Met supplement might compulsively reprogram this pathway and renewably contribute to high susceptibility to antibiotics. To test this hypothesis, the antibacterial capability of Met plus Tig against tet(X)-positive or -negative bacteria were evaluated. Interestingly, exogenous Met failed to potentiate the antibacterial activity of Tig against tet(X)-negative strains, regardless of engineered or from clinic sources (Fig. 3a and Supplementary Fig. 7), whereas it succeeds in restoring Tig sensitivity to a set of tet(X)-positive bacteria, including engineered E. coli DH5α (pUC19- tet(X4)) (Fig. 3b), and clinically isolated tet(X4)-harboring E. coli IF28 (tet(X4)) and E. coli B3-1 (tet(X4)) (Fig. 3d, e). The growth of resistant strain was unaffected by Met addition, and the combination treatment restrained the growth of E. coli B3-1 (tet(X4)) (Fig. 3f). In line with this, flow cytometry and confocal scanning microscope analysis revealed the dead cells visibility in combination treatment via fluorescent dyes SYTO 9 and propidium iodide (PI) (Fig. 3h, i). Furthermore, the efficacy of Met potentiating Tig enhanced over time, with 1.06 ~ 5.66 orders of magnitude higher than Tig-treated group (Fig. 3g). Interestingly, this action was only observed when Met and Tig were added simultaneously or pre-treated within 20 min, not applied for the condition that Met was added before or after Tig exposure over 1 h (Fig. 3c and Supplementary Fig. 8a, b), which was consistent with the notable Tig accumulation in simultaneous supplement (Supplementary Fig. 8c). Moreover, continuous supplementation of Met, Tig or both at multiple time points after combined therapy had a slight sensitization effect (Supplementary Fig. 9). In addition, Met potentiated Tig against other tet(X)-positive bacteria (Supplementary Fig. 10a), whereas failed to restore other classes of antibiotics [colistin (Col), ciprofloxacin (Cip), gentamicin (Gen) and meropenem (Mem)] activity against multidrug-resistant bacteria, including Gram-negative E. coli B2 (blaNDM5 + mcr-1) and Gram-positive MRSA T144 (Supplementary Fig. 10b, c), suggesting that this reversion utilized a specific manner to disturb tet(X)-mediated resistance pattern.
Fig. 3. Methionine supplementation potentiates tigecycline activity against tet(X)-carrying bacteria.
a Percent survival of E. coli DH5α (pUC19) in the presence of Tig, Met, or both. Tig, tigecycline, 1-fold MIC; Met, L-methionine, 20 mM. b Percent survival of E. coli DH5α (pUC19-tet(X4)) in the presence of Tig (1-fold MIC) or Tig plus Met (5, 10, 20 mM) for 6 h. c The impact of adding Met (20 mM) before or after the addition of antibiotic (Tig, 1-fold MIC) on bacterial survival at 6 h. d Percent survival of clinic-sourced E. coli IF28 (tet(X4)) in the presence of Tig (1-fold MIC), Met (20 mM), or both. e Percent survival of clinic-sourced E. coli B3-1 (tet(X4)) in the presence of Tig (1-fold MIC), or Tig plus Met (20 mM) for 6 h. f Growth curves of E. coli B3-1 (tet(X4)) within 12 h in the presence of Tig (32 μg/mL), Met (5, 10, 20 mM) or both. The mean of three biological replicates is shown and error bars represent the SEM. g Time-dependent killing curves of E. coli B3-1 (tet(X4)) treated by Tig (16 or 32 μg/mL) and Met (20 or 40 mM). h Bacterial LIVE/DEAD BacLight viability in the presence of Tig (16 or 32 μg/mL) or Met (20 or 40 mM) or both via flow cytometry analysis. Viable cells were demonstrated using green fluorescence (SYTO 9 staining, 1.67 mM) with an excitation/emission wavelength of 485 nm/498 nm, while dead cells were shown using red fluorescence (Propidium Iodide staining, 10 mM) with an excitation/emission wavelength of 535 nm/615 nm. i Confocal scanning microscope of the living and dead states of E. coli B3-1 (tet(X4)) when Tig was added alone or in combination with Met. Red was used to label dead bacteria and green was used to dye living bacteria. j Percent survival of E. coli B3-1 (tet(X4)) in the addition of other upstream or downstream metabolites of Met, covering cystathionine (CYSTAT), L-homoserine (Homoser), L-homocysteine (L-Hcy), 5’-Methylthioadenosine (MTA), S-Adenosyl-L-homocysteine (SAH), and S-Adenosyl-L-methionine (SAM). An overview of the changes of Met-related pathways were presented, including methionine salvage, cysteine and methionine metabolism, and a part of glycine, serine, and threonine metabolism. Red, up-regulated genes in E. coli DH5α (pUC19-tet(X4)) compared with E. coli DH5α (pUC19); Green, down-regulated genes in E. coli DH5α (pUC19-tet(X4)) compared with E. coli DH5α (pUC19). Data in a––g and j were displayed as mean ± SEM. Three biological replicates were carried out. Statistical significance in a–e & j were assessed by an unpaired two-tailed Student’s t-test. ns, not significant.
Given that cysteine and methionine metabolism was one of the most repressed pathways, we next fully assessed the sensitization activity of these participated metabolites. As shown in Fig. 3j, homoserine (Homoser) modestly potentiated Tig against E. coli B3-1 (tet(X4)), while other metabolites (Cystathionine, CYSTAT; L-homocysteine, L-Hcy; 5’-methylthioadenosine, MTA; S-adenosyl-L-homocysteine, SAH; S-adenosyl-L-methionine, SAM) exhibited no or negative effect on Tig efficacy. Furthermore, checkerboard assays indicated that Met and related metabolites did not exert direct drug-drug interaction with Tig (Supplementary Fig. 11a–g), implying that Met or Homoser did not function as conventional antibiotic potentiators which can be mechanistically explained as resistance enzyme inhibitors or active site competitors. Conversely, these results demonstrate that Met-manipulated remodeling involves the interplay between these biological networks and drug-triggered resistance processes.
Methionine activates 5mC methylation modification to repress tet(X) expression
Current understanding of how plasmid-mediated tet(X4) defense bacterial cells against Tig is centered on degradative enzyme mechanism, for tet(X4) encoding a flavin-dependent monooxygenase Tet(X4) in the presence of FAD, Mg2+, O2 and NADPH, thus rendering Tig ineffective10. Therefore, drug availability and degradative processes are considered as two antagonistic forces. To evaluate Tig uptake in the presence of Met, we determined bacterial proton motive force (PMF), with functional features in transporting molecules on the membrane20. As shown in Fig. 4a, b, such a combination substantially upregulated bacterial PMF, which was consistent with the visible measurement of PMF through the swimming motility test (Supplementary Fig. 12a, b). Specifically, we found that bacterial electric potential (ΔΨ), one of the components of PMF, dissipated in the presence of Tig and Met (Fig. 4c), leading to a compensatory increase of transmembrane proton gradient (ΔpH) and thus an augmented influx of Tig (Supplementary Fig. 12c). Besides, intracellular antibiotic accumulation is mainly dependent on drug influx and efflux. The EtBr analysis showed that the efflux function of tigecycline-resistant bacteria was not affected under different experimental settings (Fig. 4d). Consistently, an increase of Tig in E. coli B3-1 (tet(X4)) was found, and such an accumulation was limited to the prerequisite condition that bacteria challenged with Tig and Met at the same time (Fig. 4e, f), which supports our previous hypothesis that metabolic reprogramming happened immediately to disturb the resistance procedures in which bacteria respond to antibiotic stress, rather than modulating those procedures that pre- or post- antibiotic exposure.
Next, we investigated which pathway was the critically altered flux after Met supplement. RT-qPCR analysis showed that the expression levels of dcm and mtn increased under Met plus Tig treatment compared with Tig alone, but metK, luxS, metE and metH were decreased (Fig. 4g). Therefore, we hypothesized that a cycled pathway characterized as “Met, SAM, SAH, L-Hcy, and finally convert to Met” was largely reprogrammed. Through quantifying enzymes production and metabolites abundance of this pathway (Fig. 4h), we found that the transform of Met to SAM, SAM to SAH, SAH to L-Hcy was primarily activated in combination treatment, accompanied by increased protein production of MetK, Dcm, and metabolites abundance of SAH and L-Hcy (Fig. 4i). Notably, as the most critical methyl donor for nearly all methylation processes, intracellular SAM was reduced by Tig in the presence of Met. We proposed that SAM was highly consumed due to the over-activation of methylation modification, which raised the possibility of dcm-mediated DNA modifications 5-methylcytosine (5mC) in regulating tet(X4) gene expression. To test this, we utilized bisulfite-sequencing to analyze the 5mC modification of the promoter region of tet(X4) gene in the presence of Tig alone, Tig plus Met, Tig plus SAH, for SAH is a known methylation inhibitor21 (Supplementary Fig. 13a, b). There were six CG sites in the promoter region of tet(X4), where the methyl group of SAM was transferred to the 5-position of the cytosine base with the help of dcm-encoded methyltransferase (Supplementary Fig. 14a). Interestingly, Tig plus Met stimulated methylation modification by 13.82%, 45.98% and 23.99% more than Tig alone on the last three CG sites, while SAH supplement restored it remarkably (Fig. 4j, k), which is consistent with the phenotypic features shown in Fig. 3j. Namely, less methylation modification in tet(X4) promoter region by SAH addition contributed to more bacterial survival of E. coli B3-1 (tet(X4)).
Furthermore, E. coli J53 knockout strain defective in cytosine methyl-transfer was constructed to explore whether 5mC methylation modification is indispensable in Met-mediated sensitization (Supplementary Fig. 14b, c). The results indicated that no growth deficiency was observed in E. coli J53 as well as E. coli J53 Δdcm, E. coli J53 (tet(X4)) as well as E. coli J53 (Δdcm tet(X4)) (Supplementary Fig. 14e). Notably, we found that the potentiation of Met to Tig in E. coli J53 Δdcm tet(X4) was abolished (Fig. 4l and Supplementary Fig. 15a), but restored in E. coli J53 Δdcm tet(X4) pBAD-dcm (Supplementary Fig. 14d), implying the importance of dcm in this process. Consistently, the total cytosine methylation rates and 5mC methylation modification at 6 CG sites in the tet(X4) promoter region were reduced in E. coli J53 Δdcm tet(X4) despite of the addition of Met (Supplementary Figs. 15b, c and S16). Subsquently, we investigated the effect of Met-driven 5mC methylation modification on the expression of tet(X4) genes. Interestingly, RT-qPCR and western blotting analysis indicated that the addition of Met downregulated the transcription and translation of tet(X4) gene in E. coli B3-1 (tet(X4)) (Fig. 4m, n and Supplementary Fig. 17). Additionally, considering that ROS has been identified as a comprehensive factor participating in cellular death pathway22, we assessed the impact of Met supplementation on the ROS and ATP production in E. coli B3-1 (tet(X4)). Consequently, we found that the addition of Met reduced both ROS and ATP levels in tet(X4)-positive bacteria, suggesting that Met functions via a ROS-independent sensitization mechanism (Supplementary Fig. 18). Together, these results demonstrate that exogenous Met reprograms methionine metabolism and accelerates SAM converting to SAH, which activates 5mC methylation modification, and leads to the repression of tet(X4) expression in resistant bacteria upon Tig exposure. Meanwhile, exogenous Met triggers the intracellular accumulation of Tig by activating PMF. Combining these two aspects, Met supplementation resensitizes tet(X4)-carrying multidrug-resistant bacteria to Tig.
tet(X) expression correlates with the adjustment of methionine pathway
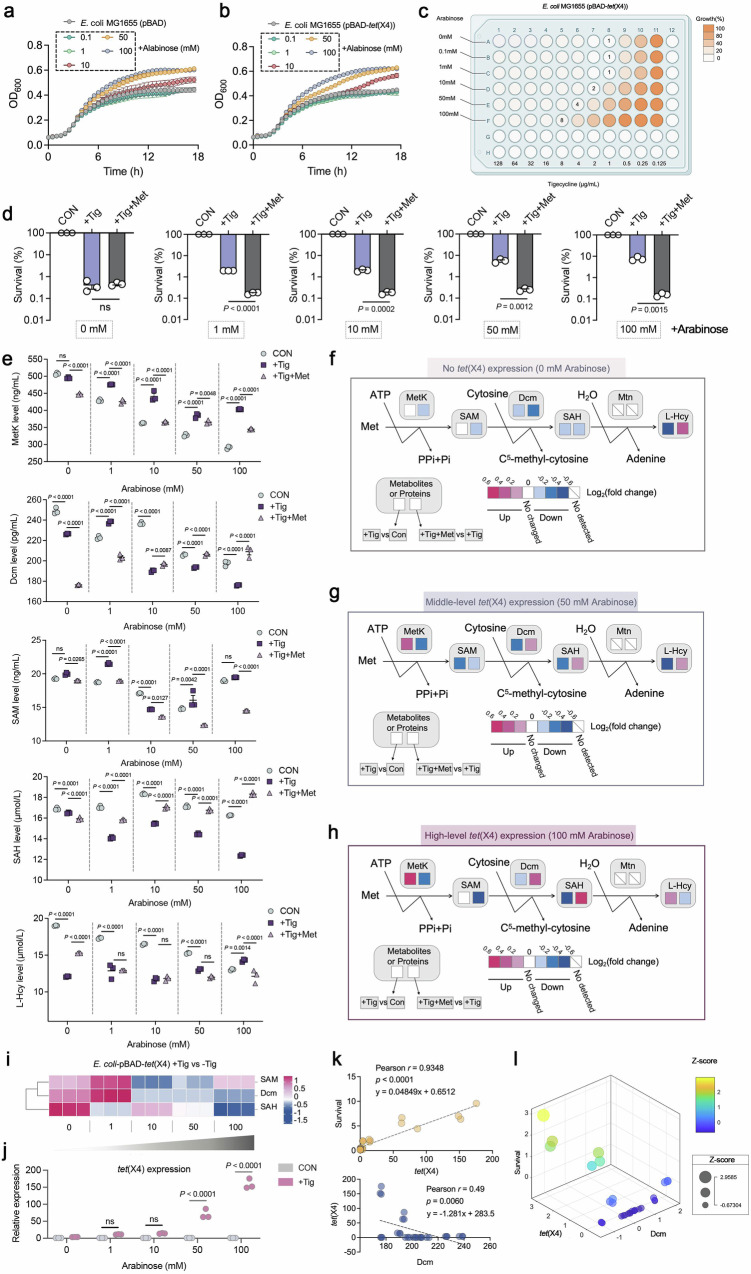
Further investigation of the correlation between tet(X4) expression and methionine pathway was performed to fully assess the co-evolutionary association of both. Firstly, we constructed the tet(X4)-carried E. coli MG1655 with pBAD plasmid, which featured as a tet(X4) gene inducible-expressing system in the presence of L-arabinose. No growth deficiency was observed between E. coli MG1655 pBAD and E. coli MG1655 pBAD-tet(X4), which even gained a slight growth advantage by 50 mM or 100 mM L-arabinose supplement (Fig. 5a, b). Next, we determined the MIC values of E. coli MG1655 pBAD-tet(X4) against Tig with varied concentrations of L-arabinose. As expected, higher L-arabinose concentrations conferred more robust Tig-resistant phenotype of E. coli MG1655 pBAD-tet(X4) (Fig. 5c). The direct connections between L-arabinose induction and tet(X4) transcription were also confirmed via bacterial enumeration and RT-qPCR assay (Fig. 5d and j).
Fig. 5. 5mC methylation modification negatively correlates with tet(X) expression.
a, b Growth curves of a reference strain E. coli MG1655 pBAD or pBAD-tet(X4), cultured with varied concentrations of L-arabinose ranging from 0.1 to 100 mM. Data are presented as mean values ± SEM. c MIC values of engineered E. coli MG1655 (pBAD-tet(X4)) in the presence of varied concentrations of L-arabinose (0.1 to 100 mM). Created with Biorender.com. d Percent survival of E. coli MG1655 (pBAD-tet(X4)) in the presence of Tig or Tig + Met. Tig, 1-fold MIC; Met, 20 mM. e Metabolic networks surveillance on Met degradation pathway in E. coli MG1655 (pBAD-tet(X4)) after exposure to exogenous L-arabinose (1, 10, 50 and 100 mM), corresponding to different expression levels of tet(X4), respectively. The group without Tig was served as a blank control, with the supplement of Tig or Tig + Met was the experimental setting. In 5mC methylation-related pathway, two enzymes (MetK, S-adenosylmethionine synthetase; Dcm, DNA-cytosine methyltransferase) and three metabolites (SAM, S-Adenosyl-L-methionine; SAH, S-Adenosyl-L-homocysteine; L-Hcy, L-homocysteine) were investigated. SAH was recognized as a methyltransferase inhibitor. f–h Effect of different levels of tet(X4) expression on the Met degradation pathway. Box indicates changes in metabolites or proteins when compared E. coli MG1655 (pBAD-tet(X4)) plus Tig with E. coli MG1655 (pBAD-tet(X4)) alone (left), and the right section indicates the comparison between Tig alone and combination treatment. Color coding is based on the level of log2(fold change) as indicated. i Heatmap of SAM (5mC methylated donors), Dcm (5mC methyltransferases), and SAH (downstream metabolite) level in E. coli MG1655 (pBAD-tet(X4)), in response to the Tig treatment with the increasing concentration of L-arabinose. j mRNA expression of tet(X4) gene of E. coli MG1655 (pBAD-tet(X4)) in the presence of varied concentration of L-arabinose. k Pearson correlation analyses between the survival of E. coli MG1655 (pBAD-tet(X4)) and the mRNA expression of tet(X4) (yellow, Pearson r = 0.9348), the mRNA expression of tet(X4) and the protein expression of Dcm (blue, Pearson r = 0.49). The correlation was fitted alongside linear curves, shown by the grey dashed lines. Colored dots represent the actual values. l A three-dimension bubble chart for summarizing the fitting relationship among the bacterial survival, tet(X4) expression, and Dcm expression. The value of these three were shown by the three axes, and the size of a bubble in was proportional to the survival data that has been normalized according to Z-score. The color intensity from blue to yellow represented the span between death to survival. Data in d, e, and j were displayed as mean ± SEM. Three biological repeats were performed. In d, the P values were determined using an unpaired two-tailed Student’s t-test. In e and j, adjusted P values were determined using two-way ANOVA with Sidak’s multiple comparison test.
To explore the correlation between resistance degree and Met pathway, particularly between the tet(X4)-mediated target inhibition and dcm-mediated 5mC methylation modification, we measured the changes of 5mC methylation related-enzymes and metabolites under increasing concentrations of L-arabinose. Interestingly, “Met, SAM, SAH, L-Hcy” pathway displayed significant differences under different L-arabinose concentrations (Fig. 5e). Notably, changes of this pathway did not directly follow an absolute linear trend along with the change of tet(X4) expression (Fig. 5i and j), possibly can be explained as the metabolic process of bacterial population under different induction conditions are more likely to follow the Gaussian distribution, which generally considering variability and uncertainty in population behavior and permitting error distribution23. Next, we compared the +Tig versus control (left in the box), +Tig+Met versus +Tig groups (right in the box) under no-, middle-, or high-level tet(X4) expression respectively, and found that Tig treatment only slightly inhibited Dcm and SAH production, but had no effect on the production of key metabolites (SAM) and enzymes (MetK) under no-tet(X4) expression (Fig. 5f, left box). However, when tet(X4) expression reached a moderately high level, Dcm was significantly inhibited, the accumulation of SAM increased or remained unchanged, and the accumulation of its downstream SAH was also downregulated (Fig. 5g, h, left box). These results suggested that middle- or high-level tet(X4) expression inhibited the conversion from SAM to SAH, and thus reducing modification of 5mC DNA methylation. Consistent with the above findings in E. coli B3-1 (tet(X4)) (Fig. 4i), exogenous Met supplementation likewise reversed such a metabolic flux in E. coli MG1655 pBAD-tet(X4) (Fig. 5f–h). Furthermore, Pearson correlation analysis and multivariate linear regression analysis displayed that these two parameters, tet(X4) and Dcm, significantly co-determined the survival of tet(X4)-positive Tig-resistant bacteria (Fig. 5k, l). We reasoned that both of them realized a compensatory regulation in the co-evolutionary trajectory without growth fitness, which opens avenues for Met-mediated reprogramming intervention strategy.
Methionine effectively potentiates tigecycline efficacy in vivo
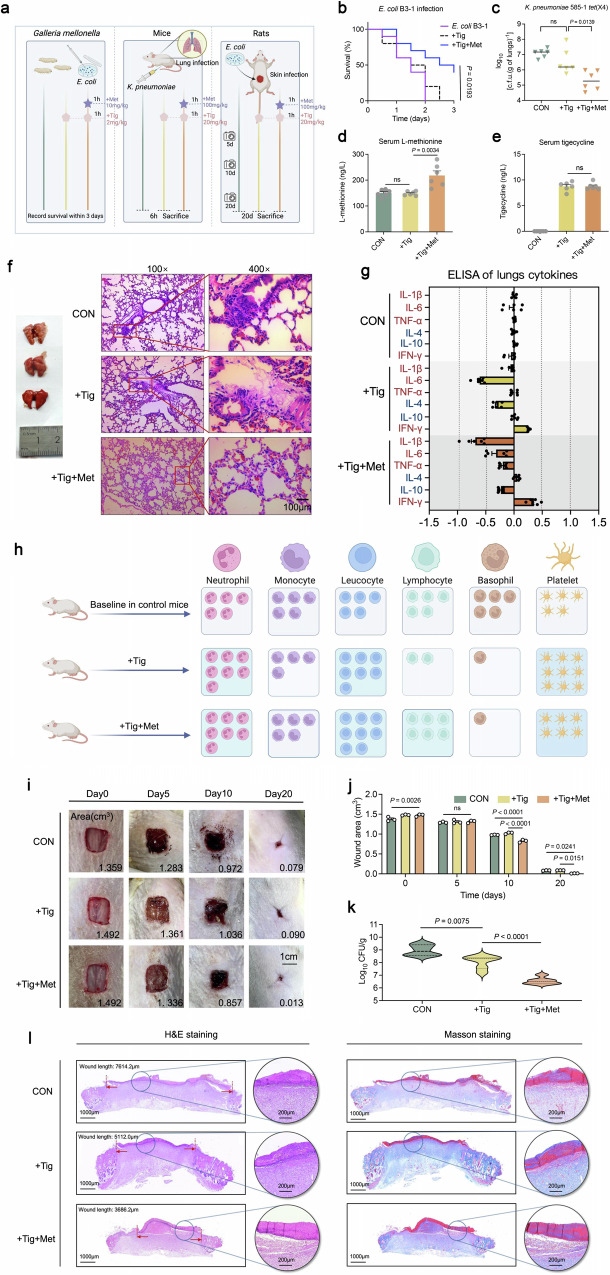
To fully demonstrate Met-enabled Tig killing of tet(X4)-positive pathogens, we next assessed the in vivo efficacy of Tig plus Met in various infection models (Fig. 6a). In E. coli B3-1 (tet(X4))-infected macrophages RAW264.7 cells, we found that the combination of Met and Tig significantly reduced the loads of intracellular bacteria, and this process was accompanied by an increased intracellular accumulation of Met (Supplementary Fig. 19). Furthermore, we constructed a Galleria mellonella infection model, which was infected with E. coli B3-1 (tet(X4)) and then treated by Tig alone or Met plus Tig. Consequently, the larvae in the control and Tig-treated group all died within three days, while the survival rate was enhanced to 40% when the combination treatment was applied (Fig. 6b).
Fig. 6. Met plus Tig promotes pathogens elimination and alleviates inflammatory responses in animal models of infection.
a The experimental protocols for assessing the potentiation of Met to Tig in multiple animal infection models. Created with Biorender.com. b Survival rate in the G. mellonella larvae infected with a lethal dose of E. coli B3-1 (tet(X4)) (1*106 CFU/mL). n = 10 biologically independent animals per group. c Bacterial loads of infected mice lung after Tig treatment, or in combination with Met. The mice were randomly divided into three groups (n = 6 per group), including K. pneumoniae 585-1 (tet(X4)), Tig, Tig + Met groups. Mice were infected with K. pneumoniae 585-1 (tet(X4)) (1*109 CFU/mL, 200 μL per mice, i.p.) and administrated with Met (100 mg/kg) at 1 h after bacterial infection, as well as Tig (20 mg/kg) at 1 h after Met administration, namely at 2 h after bacterial infection. d, e Met and Tig concentrations of mice serum at 6 h post mouse peritonitis infection. Samples were collected from 6 biologically independent mice per group. f Hematoxylin and Eosin (H&E) staining of lung from mice at 6 h post-infection (Scar bar, 100 μm). g Fold changes of inflammatory cytokines between three groups were compared. K. pneumoniae 585-1 (tet(X4)) group was used as the control allowed normalization of the inflammatory data. Pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, INF-γ) were marked with red and anti-inflammatory cytokines (IL-4, IL-10) were marked with blue. Abscissa represents the value after the logarithm of FC (Log2FC). Lungs samples were collected from 6 biologically independent mice per group. h Relative shifts of major leukocyte populations and platelets compared with the Control, Tig, Tig + Met groups. Baseline cell content in the control group was defined as “5 cells”. Results in Tig, Tig + Met groups were calculated in relative relation to the control group. Detailed results for each group were depicted in Supplementary Fig. 21. Created with Biorender.com. i Photograph the healing process of rat skin wound infection model and the wound areas were calculated on days 0, 5, 10, and 20 among the three groups. n = 6 biologically independent animals per group. Scale bars, 1 cm. The representative wound areas for each group were labeled in the bottom right corner. j Statistical comparison of wound areas in rats of each group on days 0, 5, 10, and 20. Samples were collected from three biologically independent rats per group. k Effects of Tig treatment or Tig plus Met treatment against E. coli B3-1 (tet(X4)) in wound infection model, manifested by the scabs bacterial colonies at 10th day. l Representative histopathological changes of the wound by H&E staining (left) and Masson staining (right). For Masson staining, the collagen deposition was dyed blue, muscle fibers and cellulose were dyed red. Three biological replicates were performed per group. In d, e, g and j, data are presented as mean values ± SEM. In k, the solid line represents the median, the dotted line represents the quartiles. In b, the P value was determined using a two-sided log-rank (Mantel-Cox) test. In c–e and k, the P values were determined using an unpaired two-tailed Student’s t-test. In j, adjusted P values were determined using two-way ANOVA with Sidak’s multiple comparison test.
Next, we established a mouse peritonitis infection model infected by tet(X4)-carrying Klebsiella pneumoniae 585-1. To exclude the possibility that Met might influence the antibiotic resorption kinetics or penetration, we measured Met and Tig concentrations in serum. As shown in Fig. 6d, e, Met supplementation efficiently enhanced Met concentration in serum but had no effect on Tig levels. As expected, lungs collected from Tig plus Met-treated mice showed a significant decrease in K. pneumoniae loads compared with Tig-treated mice (Fig. 6c). Consistently, we observed lung atrophy and congestion, and excessive infiltration of lymphocytes in the control and Tig-treated mice, whereas it was alleviated in the Tig plus Met-treated group (Fig. 6f). ELISA analysis of inflammatory cytokines in serum samples suggested that combination treatment downregulated the pro-inflammatory cytokines (IL-1β and TNF-α) and upregulated the anti-inflammatory cytokine (IL-4) compared with that in Tig-treated mice. Meanwhile, IFN-γ, one of the robust protective effector molecules that govern host immunity against various pathogens24, was strongly boosted in Tig plus Met group, which is indicative of the sensitized host immune in combating pathogens invasion in the early stage of infection (Fig. 6g and Supplementary Fig. 20). The shift for major leucocyte classes (neutrophils, monocytes, lymphocytes, and basophils) in mice serum counted respectively in Supplementary Fig. 21 and represented graphically in Fig. 6h. Notably, leucocyte cells experienced an increase in Tig + Met-treated mice, which mainly attributed to the increase of neutrophils and lymphocytes.
In addition, we constructed a model of acute lung infection with K. pneumoniae 585-1 (tet(X4)) via nasal drip infection route (Supplementary Fig. 22a). As shown in Supplementary Fig. 22c, Met plus Tig effectively reduced bacterial loads both in lungs and serum of mice, and alleviated inflammatory damage (Supplementary Fig. 22b, c). Lastly, we performed a rat skin wound infection model and continuously monitored wound healing within 20 days. As shown in Fig. 6i–k, Met plus Tig-treated rats displayed better wound healing and lower pathogen loads at 10th days post infection. Furthermore, Hematoxylin and Eosin (H&E) and Masson staining suggested that combination therapy strikingly ameliorated wound inflammatory and breakage, which can be manifested by the lightest lymphocytes infiltration and interweaved collagen tissue in Tig plus Met group (Fig. 6l and Supplementary Fig. 23).
Discussion
Recent ongoing efforts have yielded insights into overcoming Tig resistance spanning multiple scales. Our previous study showed that bismuth nitrate, commonly used in the clinical treatment of stomach-associated diseases, effectively boosted the antibacterial activity of Tig by competitively antagonizing the enzymatic activity of Tet(X4)25. Another example was the anti-HIV agent azidothymidine, which was identified as a potent Tig adjuvant against tet(X4)-positive E. coli and the underlying mechanism involved the suppression of both DNA synthesis and resistance enzyme activity26. Plumbagin, a natural compound targeting Tet(X) monooxygenase activity, also displayed a synergistic effect with Tig against tet(X4)-positive E. coli27. These advances established a inhibitors-oriented sensitization strategy by inhibiting monooxygenase activity. However, little evidence highlighted knowledge gaps in metabolic networks that can be optimally exploited to potentiate antibiotic efficacy. In fact, there is a close relationship between bacterial metabolic homeostasis and cell fate determination. For example, fumarate functioned as a tobramycin potentiator against metabolically tolerant bacteria, which resulted from activating cellular respiration and enhancing PMF28. A recent study uncovered that clinically relevant mutations in core metabolic genes conferred antibiotic resistance29, which tremendously broadens our current understanding of the interaction between bacterial metabolism and population destiny.
Antibiotic exposure has been shown to influence a broader range of biological functions including amino acids, nucleotides, and lipids metabolism, which lays the foundation for reprogramming-based sensitization strategy11,30. Recent studies have indicated that both exogenous nitrogen sources (eg. pyruvate) and carbon sources (eg. glucose) could potentiate aminoglycoside antibiotics against pathogenic Edwardsiella tarda by increasing TCA cycle flux and PMF-mediated drug uptake31,32. However, the metabolic profiles between tet(X4)-negative and -positive bacteria remain unclear, and it is uncertain whether manipulating these pathways with exogenous metabolites could reverse antibiotic resistance. Our study revealed the differential metabolic characteristics and fluxes between tet(X)-negative and -positive E. coli, including distinct energy demand patterns under tigecycline exposure. More investigations are warranted to validate whether this patterns also apply to other antibiotics or under different conditions. On the basis of these findings, we showed that the supplementation of Met drastically potentiated Tig efficacy against tet(X4)-positive bacteria, and elucidated the underlying mechanism. Notably, a delicate correlation between antibiotic resistance genes and crucial metabolic pathways was deciphered.
Nowadays, the interactions between metabolic reprogramming and epigenetic modifications in cancer have made great progression in clinical setting33. However, metabolic reprogramming, as well as epigenome investigation in prokaryotes, especially in the bacteria population, remain poorly understood. DNA methylation is a common epigenetic modification after replication in prokaryotic and eukaryotic genomes and is also a critical regulation of gene expression in mammals, playing a crucial role in gene transcription inhibition. There are three major forms of DNA methylation in bacterial genomes, including 5-methylcytosine (5mC), N4-methylcytosine (4mC), and N6-methyladenine (6 mA)34. However, it remains largely unknown whether there is a correlation between DNA methylation and drug-resistant gene expression in bacteria. In this study, we first revealed that dcm-mediated 5mC methylation modification could precisely modulate the expression of tet(X) resistance genes in E. coli. In the future, a global methylation profiling of the tet(X4)-positive E. coli genome will be required to gain a more comprehensive understanding of the effects of epigenetic modification on bacterial biochemistry and metabolic homeostasis.
Met, a sulfur-containing proteinogenic amino acid, plays a critical role in various metabolic processes, including immune function, mammalian nutrition, and digestive function35,36. Clinically, Met supplementation within the range of 236-705 mg/kg/day is recommended37, and the highest dose used in this study, 100 mg/kg, is considered safe. Our findings of increased lymphocyte count in Met-supplemented mice serum suggest its role in immune modulation (Fig. 6h). Met is particularly involved in T cell activation and differentiation, such as manipulating epigenetic reprogramming in CD4(+) T helper (Th) cells. Sufficient Met is critical for suppressing Th17 cell proliferation and the subsequent cytokines production, with a mechanism involving histone H3K4 methylation38. Therefore, we speculated that alleviated inflammatory response in Tig plus Met-treated mice can be attributed not only to the therapeutic efficacy of this combination but also to Met-mediated immunotherapy.
In the future, how Met supplementation accelerated the elimination of pathogens and its roles in immune regulation within the host necessitate further research. Indeed, host-directed therapy (HDT) is an emerging strategy in the field of anti-infective fields. For example, an all-in-one strategy that combined antibacterial effects with anti-inflammatory actions in the host by neutralizing and detoxifying serum LPS was proposed39. Additionally, it has been reported that the increased ROS in macrophages by enrofloxacin treatment significantly reduced internalized pathogens and alleviated inflammatory injury40. Identifying metabolic hallmarks from both bacterial and host perspectives is essential for optimizing current antibiotics and understanding pathogens-immunity and host-pathogen interactions.
In summary, we comprehensively elaborate the metabolic profiles between tet(X)-negative and -positive bacteria, uncovering that exogenous supplement of Met can effectively restore the susceptibility of tet(X)-carrying pathogens to Tig. As a proof of concept, the in vivo efficacy of this combination therapy was evidenced in multiple animal infection models. Certainly, more investigation of Met-assisted strategy in treating bacterial infections will be required to fully assess its clinical prospects. Mechanistically, the combination of Met and Tig enhances bacterial PMF, which subsequently promotes intracellular accumulation of Tig. Meanwhile, exogenous Met enhances the 5mC methylation modification in the promoter of tet(X4) gene. The increase in drug accumulation and the decrease in tet(X) gene expression collectively contribute to cell death (Fig. 7). Overall, our work indicates that Met is a promising antibiotic enhancer and has the potential to address life-threatening infections by tet(X)-carrying multidrug-resistant bacteria.
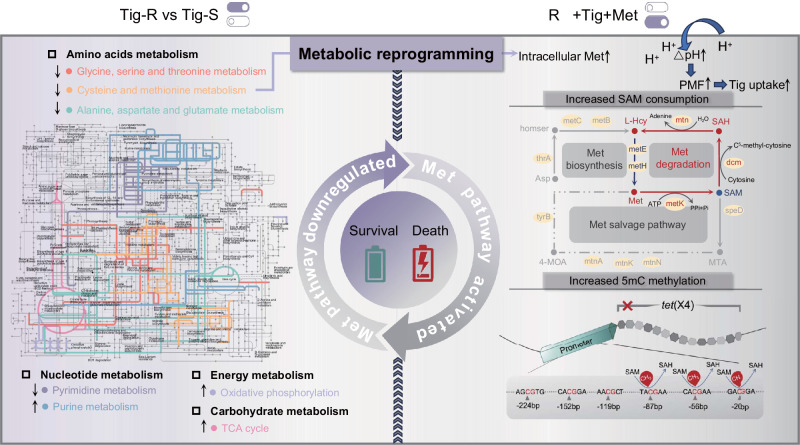
Fig. 7. Schematic illustration of the proposed mechanisms of met-driven metabolic reprogramming against tet(X)-carrying bacteria.
The left shows the comparative metabolomics network between Tig-R and -S. Colored lines represent the major altered pathways, including several amino acid metabolism pathways, nucleotide metabolism, energy metabolism, and carbohydrate metabolism. The gray sections are derived from Interactive Pathways Explorer 3 (iPath3) annotated pathway backgrounds. The right section depicts the specific mechanism of action of Met-mediated metabolic reprogramming. Briefly, exogenous Met induced the high-level depletion of SAM, which activated 5mC methylation modification at the promoter region of tet(X4), therefore down-regulating the transcription of resistance progress. Meanwhile, exogenous Met promoted the uptake of Tig by upregulating the ΔpH component of bacterial PMF. These actions collectively resensitize tet(X)-carrying multidrug-resistant bacteria to Tig treatment.
Methods
Ethical statement and animal studies
This study was conducted according to the relevant guidelines of Jiangsu Laboratory Animal Welfare and Ethical of Jiangsu Administrative Committee of Laboratory Animals (SYXK-2022-0044). All animal experiments were approved by the Animal Care Committee of Yangzhou University.
Bacterial strains and reagents
All the strains used in this study were listed in Supplementary Table 1. Briefly, a bacterial single colony was incubated in 1 mL fresh Luria-Bertani broth (LB, Qingdao Hope Bio-technology) overnight at 37 °C and 200 g in 2 mL flasks. Then cultures were diluted to 1:100 using fresh 30 mL LB broth in 50 mL flasks. After 4 ~ 5 h of cultivation to exponential phase, bacterial cells were centrifuged at 4000 g for 7 min and washed with sterile saline solution to remove the external nutrients. Then cells were resuspended in M9 minimal medium (Na2HPO4 6.78 g/L, KH2PO4 3.0 g/L, NaCl 0.5 g/L, NH4Cl 1.0 g/L, MgSO4 0.241 g/L, CaCl2 0.011 g/L, glucose 4 g/L, pH = 6.8) to arrive at OD600 of 0.1, and antibiotics or metabolites were added at given concentrations. Collected cells were used for following plate counting or biochemical analysis. Antibiotics were obtained from the China Institute of Veterinary Drug Control, and the other chemical reagents were purchased from YuanYe or Solarbio or Aladdin company. M9 minimal medium was purchased from Coolaber company (SL0060). Methionine refers to L-methionine. Arabinose refers to L-arabinose. To culture Vero and RAW264.7 cells, we used Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco) with 10% heat-inactivated fetal bovine serum, 1% (w/v) penicillin-streptomycin, and 1% (w/v) sodium pyruvate (Sigma Aldrich, Oakville, Ontario).
Growth curves measurement
Overnight cultures were diluted 1:1000 in 1 mL fresh LB broth and challenged different treatments, then bacterial suspensions were cultured in a clear UV-sterilized 96-well microliter plate with total volume of 200 μL, and then 50 μL mineral oil was added to prevent volatilization. OD600 was continuously determined by the Infinite E Plex Microplate reader (Tecan).
Transcriptomics sequencing and analysis
E. coli DH5α (pUC19) (Tig-S) and E. coli DH5α (pUC19-tet(X4)) (Tig-R) were cultured in 1.5 mL fresh LB broth without antibiotics for 6 h until the exponential phase. Meanwhile, another experimental setting was Tig-R and -S treated with Tig (1MIC) for 6 h, respectively, and then the supernatant was discarded after centrifugation at 3000 g for 7 min. Subsequently, bacterial cells were collected for following RNA extraction and sequencing according to our previous study41.
Metabolomics profiling
Samples pre-treatment were described as above. After the LC-MS raw data were calibrated, the mass spectrometry information was simultaneously matched with the metabolism public database HMDB and majorbio (https://metlin.scripps.edu/) to obtain metabolite information. The pathways involved by differential metabolites were obtained by metabolic pathway annotation in KEGG database (https://www.kegg.jp/kegg/pathway.htmL), GO database, and Interactive Pathways Explorer3 (iPath3) (https://pathways.embl.de). The python software package scipy.stats was used for pathway enrichment analysis, and the most relevant biological pathways were obtained by Fisher exact test.
Minimum inhibitory concentration (MIC) determination
MIC values of Tig with or without Met were determined by the standard broth microdilution method, according to the CLSI 2021 guideline42. In brief, Tig (2560 μg/mL) was 2-fold diluted in MHB with a total volume of 100 μL and mixed with an equal volume of bacterial suspensions (approximately 1.0×105 ~ 106 CFU per mL) in a clear UV-sterilized 96-well microliter plate. After 18 h incubation at 37 °C, the MIC values were defined as the lowest concentration of antibiotics without any visible growth of bacteria. The detailed MIC values were shown in Supplementary Table 1.
Live/Dead cells staining
The resuspended E. coli B3-1 (tet(X4)) in M9 medium were co-incubated with different treatments as described above for 5 h at 37 °C and 300 g. SYTO 9 (1.67 mM) and PI (10 mM) dyes were added to cultures for 15 min co-incubation. Flow cytometry assay and confocal scanning microscope analysis were performed to characterize the transient state of bacterial population. 10,000 ungated events were measured using CytExpert Flow Cytometer (Beckman, USA) and the results were analyzed using FlowJo software 10.8.1. In confocal laser scanning microscopy analysis, a CLSM microscope (Leica TCS SP2, Heidelberg, Germany) were used.
PMF determination
Briefly, 1 mL of bacterial suspension in M9 minimal media (OD600 = 0.1) was collected and diluted to 106 CFU per mL, then co-incubated with Tig or Met or both for 4 h. Meanwhile, Carbonyl Cyanide 3-ChloroPhenylhydrazone (CCCP), the inhibitor of oxidative phosphorylation, were chosen as the positive control with the final concentration of 5 μM. Next, bacterial cultures immediately mixed with 10 μL 3 mM DiOC2(3) (3, 3’- diethyloxy carbonyl iodide cyanine). The mixture was incubated for 30 min at 37 °C with 300 g shaking. The membrane potential was evaluated by FACScalibur flow cytometer (Becton Dickinson, San Jose, CA, USA). Green fluorescence was identified by a filter that had a bandwidth of 488 to 530 nm, and red fluorescence was identified by a filter that had a bandwidth of 488 to 610 nm. The membrane potential was determined and normalized to the intensity ratio of red/green fluorescence. PMF was calculated with the following formula: Log(103/2*(red fluorescence/green fluorescence)).
ΔΨ detection
Bacteria were cultured overnight and diluted 1:100 to fresh LB broth for 4 h, and then resuspended with M9 media until OD600 of 0.1. Then, cells were co-incubated with a fluorescence dye DiSC3(5) (final concentration, 0.5 μM) for 30 min43. Fluorescence intensity of E. coli B3-1 (tet(X4)) in the treatment of Tig with or without Met within 40 min was continuously monitored, using the Infinite E Plex Microplate reader (Tecan). The excitation wavelength is 622 nm, and the emission wavelength is 670 nm.
ΔpH detection
Bacteria were cultured overnight and diluted 1:100 for 4 h, and then resuspended with M9 media until OD600 of 0.1. Then, cells were co-incubated with a fluorescence dye BCECF-AM (final concentration, 0.5 μM) for 30 min43. Fluorescence intensity of E. coli B3-1 (tet(X4)) in the treatment of Tig with or without Met within 40 min was continuously monitored, using the Infinite E Plex Microplate reader (Tecan). The excitation wavelength is 488 nm, and the emission wavelength is 535 nm.
Efflux pump function assessment
Cells were co-incubated with EtBr (final concentration, 5 μM) and Tig, or Met, or combination, or known efflux pump inhibitor CCCP (10 μM) at 37 °C to an OD600 of 0.5. After centrifuging at 4000 g for 7 min, the cells were collected and resuspended in fresh M9 minimal media. Subsequently, fluorescence intensity was monitored with the excitation wavelength at 530 nm and emission wavelength at 600 nm during 40 min.
Total ROS measurement
We used DCFH-DA (10 μM) to determine the intracellular ROS production in E. coli B3-1 (tet(X4)). After harvesting and washing the labeled bacteria three times with PBS, they were then co-cultured with Tig, Met, or their combination. Fluorescence intensity was monitored with the excitation wavelength at 525 nm and emission wavelength at 488 nm.
Intracellular ATP measurement
The ATP levels of E. coli B3-1 (tet(X4)) were quantified utilizing an Enhanced ATP Assay Kit (Beyotime, S0027). In brief, following the pretreatment described above, the bacteria were centrifuged, washed, and resuspended in lysis buffer. Centrifuged at a speed of 12,000 g for 5 min at 4 °C, the bacterial debris was removed after lysis, and the supernatant was taken as a sample for later use. Subsequently, 100 μL ATP detection working solution was put into a 96-well black plate and placed for 5 min at room temperature. Afterwards, 20 μL sample was added and mixed them evenly. The relative light unit was measured using an Infinite E Plex Microplate reader (TECAN) immediately.
Intracellular Tig assessment
Culturing E. coli B3-1 (tet(X4)) overnight and 1:100 diluted into fresh LB broth for a 4 h incubation, with a setting of 37 °C, 200 g. Three biological replicates were performed. The bacteria were centrifuged at 12000 g for 10 min at 4 °C and the supernatant was discarded. Then bacteria cells were resuspended in PBS, and this step was repeated once. Subsequently, bacteria were diluted to 105 CFU/mL with M9 and aliquoted into a 1.5 mL centrifuge tube. A final concentration of 32 μg/mL Tig was added, then cultured at 37 °C and 200 g for 1 h. Then cells were centrifuged at 12000 g for 3 min. The precipitates were dissolved in 200 μL water. Freeze thaw was repeated in liquid nitrogen for three times (3 min each), and then treated in a water bath at 65 °C for another three minutes to fully lyse the samples. The supernatant was collected by centrifugation at 12,000 g for 3 min. The remaining pellet was resuspended in 200 μL methanol, vortexed, centrifuged, and the supernatant was taken and combined with the supernatant collected previously. 600 μL acetonitrile (acidified by glacial acetic acid) was added, and then cells were vortexed and centrifuged. After centrifuging at 12000 g for 3 min to remove the residual debris, supernatant was taken, and filtered by 0.22 μM filter for following LC-MS/MS analysis (QTRAP 6500, Applied Biosystem). HPLC separation conditions: mobile phase A (0.1% formic acid aqueous solution) and mobile phase B (acetonitrile solution); Chromatographic column: C18 column (4.6 mm*250 mm). The flow rate was 0.5 mL/min. The gradient elution ratio was as follows: 0.1–1.0 min, 85%A; 1.0–1.5 min 85%-60%A; 1.5–3.5 min, 60%A; 3.5–4.0 min, 60%-85%A; 4.0–4.5 min, 85%A; The injection volume was 10 μL. The positive ion mode of multiple reaction monitoring (MRM) was used to quantify the intracellular antibiotic concentration.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA extraction was carried out as previous description44. Total RNA of E. coli B3-1 (tet(X4)) treated with Tig, Met or their combination was isolated and quantified by the ratio of absorbance (260 nm/280 nm) using a Nanodrop spectrophotometer (Thermo Scientific, MA, USA). The first-strand cDNA from all bacterial cells was synthesized using the PrimeScript RTreagent Kit with gDNA Eraser (Vazyme, Nanjing, China) following the manufacturer’s protocols. Thermal cycling was performed by a two-step PCR amplification standard procedure with 95 °C for 30 s and 40 cycles of 95 °C for 5 s, 60 °C for 30 s. RT-qPCR test was performed using a 7500 Fast Real-Time PCR System (Applied Biosystem, CA, USA). The fold changes of gene expression were determined using the 2−ΔΔCt method. Primer sequences used in this study were listed in Supplementary Table 2.
Enzyme-linked immunosorbent assay (ELISA)
ELISA kits (MLbio, Shanghai, China) were used to quantified metabolites (SAM, SAH and L-Hcy), enzymes (MetK, Dcm), cytokines (IL-1β, mL098416; IL-6, mL098430; TNF-α, mL002095; INF-γ, mL002277; IL-4, mL064310; IL-10, mL037873), serum Tig and serum Met (YJ954470, YJ983622) levels. Briefly, a 50 μL volume of standard reagent with different concentrations was added to the standard wells, while 40 μL of sample dilution reagent and 10 μL of sample solution were added to the enzyme-coated plate. Horseradish peroxidase (HRP) conjugate reagent (100 μL) was added to both the standard and sample wells. The reaction well was sealed with a plate sealing membrane and incubated at 37 °C for 60 min. The sealing film was removed, the liquid was discarded, and the plate is spun dry. Each well was filled with washing liquid, allowed to stand for 30 sec, then discarded, and this process was repeated five times. Color developer A (50 μL) was added to each well, followed by color developer B (50 μL). Incubation from light at 37 °C for 10 min occurs, after which 50 μL of stop solution was added to each well to terminate the reaction. The absorbance of each well is measured at a wavelength of 450 nm using a microplate reader. The corresponding concentration is calculated from the standard curve according to the OD value of the sample wells.
Bisulfite methylation sequencing
This method can analyze methylated cytosines in the genome from the individual base level. Firstly, the target sequence is treated with bisulfate to deamination cytosine without methylation to uracil. The cytosine that has been methylated is not deamination, so methylation sites can be found by comparing bisulfite-treated and untreated sequencing samples. E. coli B3-1 (tet(X4)) were treated with Tig (1-fold MIC), Tig + Met (20 mM), Tig + SAH (20 mM) for 6 h, and samples DNA were isolated through FastPure Bacteria DNA Isolation Mini Kit (DC103-01, Vazyme, Nanjing, China). Bisulfite methylation sequencing was performed by Weiaobio company (Shanghai, China).
Western blot analysis
An overnight culture of E. coli B3-1 (tet(X4)) cells in LB broth was diluted 1:100 and grown to mid-exponential phase (5 h) at 37 °C with 200 g, then the culture was back-diluted to OD600 of 0.1 with M9 medium and treated with Tig (16 μg/mL, 32 μg/mL), Tig + Met (20 mM, 40 mM) for 6 h. Next, 10 mL bacterial culture was collected and resuspended with 1 mL NaCl, then the bacterial ultrasonic crusher was used to lyse intracellular proteins. Using BCA Protein Quantification Kit (E112-01, Vazyme), the final protein concentrations of samples were detected and adjusted to the same level. 200 μL adjusted samples were mixed with 5×loading buffer and then boiled at 100 °C for 10 min. After SDS-PAGE separation, all samples were transferred onto a polyvinylidene difluoride (PVDF) membrane. Then, the membrane was immersed with NcmBlot blocking buffer (P30500) for 10 min at the room temperature and further incubated with the primary antibodies against Tet(X4) (supported by Prof. Wang laboratory27, 1:500 dulited) and HPR-conjugated goat anit-mouse secondary antibodies (1:10000 dulited). Recombinant E. coli GroEL protein (ab53107) were purchased from Abcam.
Dcm-knockout strain construction
Firstly, the reverse enzyme recognition site FRT + resistance gene expression element + reverse enzyme recognition site FRT were amplified with universal primers using pKD3 (P1:5’-ATATGAATATCCTCCTTAG-3’; P2:5’-TGTAGGCTGGAGCTGCTTCG-3 ‘). Primers with homologous arms were used to amplify the resistance fragment, and the corresponding PCR gel was cut for electric conversion. Secondly, pKD46 was transformed into E. coli J53 by electric conversion. Thirdly, the knockout fragment was mixed with the competent state of strain carrying pKD46 plasmid and placed in an electric rotating cup with a voltage of 2.1 kv for electric shock. After the electric shock, 1 mL LB liquid medium containing 0.2% L-arabinose was quickly added. After mixing, it was transferred to a 15 mL tube, placed in a 30 °C incubator for overnight culture, then coated on a resistant plate containing chloromycetin (Cm), and cultured at 37 °C to obtain a single colony (E. coli J53: Cm + ). pCP20 were transferred into E. coli J53: Cm+ and inoculated in LB plate containing Amp and Cm. Primers were designed at 100 bp upstream and downstream of the homologous exchange site. The PCR products of the knockout strains were compared with those of the non-knockout strains, and then the PCR products of the knockout strains were sent to sequencing.
Checkerboard assays
Synergistic activity was evaluated by checkerboard assays with two-fold serially dilution of drugs and metabolites (8*8 matrix)45. After 18 h co-incubation with bacterial suspension (1.0*106 CFUs/well), the absorbance of bacterial culture at 600 nm was measured by Microplate reader. Three biological replicates were performed for each combination and the means were used for FIC index (FICI) calculation according to the formula as follows: FIC index = FICIa + FICIb = MICab/MICa + MICba/MICb.
Swimming motility test
0.3% (w/v) agar media containing trypticase peptone (10 g/L), NaCl (10 g/L), and yeast extract (5 g/L) was used to evaluate bacterial swimming motility46. Wait until the temperature drops to 50 °C, Tig (8, 16, 32 μg/mL) or Met (20, 40 mM) or the combination were added. In the center of each plate, a volume of 2 μL of bacterial suspensions (106 CFUs per mL) was deposited. Swimming motility of the bacteria was evaluated by measuring the microsphere diameter after incubation at 37 °C for 48 h.
tet(X4) inducible expression system in E. coli
Firstly, tet(X4) gene fragment containing SacI and HindIII endonuclease sites were amplified with tet(X4)-F and tet(X4)-R as primers, and the PCR products were purified to obtain DNA. Secondly, the target gene and plasmid pBAD were double digested by SacI and HindIII, respectively. After PCR verification, the gel was recovered and purified for use. Thirdly, the target gene was linked to the pBAD vector with T4-DNA ligase, and then transformed into E. coli MG1655 competent cells treated with Calcium chloride. The pBAD vector was also transferred into E. coli MG1655 competent cells as well for blank control. Both were coated in LB plate containing AMP for 37 °C overnight. Single bacterial colony was randomly selected and inoculated in LB broth containing AMP. Finally, the extracted plasmids from amplified cultures were sequenced for verification. The validated strains were used for further experiments.
Animal infection models
Galleria mellonella infection model, mouse lung infection model and rat skin wound infection model were performed, respectively. In Galleria mellonella infection model, the larvae were randomly divided into three groups (n = 10 per group, one control group and two experimental groups) and infected with 10 μL of E. coli B3-1 (tet(X4)) suspension (1*106 CFU/mL) at the right posterior gastropoda. After 1 h post-infection, one group of larvae was treated with Met (10 mg/kg) at the left posterior gastropoda. Another 1 h later, the Tig+Met and Tig groups were injected with a single dose of Tig at 2 mg/kg, then the survival rate was monitored for 3 days.
In the mouse peritonitis infection model, the mice were housed in individually ventilated cages in a room maintained at 22 ± 2°C, relative moisture of 50 ± 10% and with a regular 14-h light.: 10-h dark cycle. Mice were randomly divided into three groups (n = 6 per group), including K. pneumoniae 585-1 (tet(X4)), Tig, Tig+Met groups. A week of adaptation later, mice were intraperitoneal infected with K. pneumoniae 585-1 (tet(X4)) (1*109 CFU per mL, 200 μL per mice, i.p.). After 1 h post-infection, Tig+Met group of mice were supplied with Met administration (100 mg/kg). Another 1 h later, Tig group and Tig + Met group mice were subjected to Tig administration (20 mg/kg). After 6 h of infection, mice were sacrificed, and serum and lung tissue were collected for further examination.
In the mouse acute lung injury model, mice were randomly divided into three groups (n = 10 per group), including K. pneumoniae 585-1 (tet(X4)), Tig, Tig + Met groups. Mice were infected with K. pneumoniae 585-1 (tet(X4)) (5.2*109 CFU/mL, 50 μL per mice) via nasal drip infection route. After 30 min post-infection, treated group of mice were supplied with Met administration (100 mg/kg) or Tig (20 mg/kg) administration or both of them (i.p.). After 3 and 6 h of infection, mice were sacrificed, and serum and lung tissue were collected for further examination.
In rat skin wound infection, rats were divided into three groups, namely CON, Tig, Tig + Met groups. Rats were anesthetized with sedative, 1*1 cm wound was cut on the skin under anesthesia. Then wounds were administered with 200 μL E. coli B3-1 (tet(X4)) bacterial solution (1.0 × 109). After 1 h of infection, Met (100 mg/kg) was administered locally to the wounds in the Tig + Met group. After another 1 h, Tig (20 mg/kg) was administered topically to the wounds of rats in the Tig and Tig + Met groups. After 10 days of infection, half of the number of rats were sacrificed for wound load counting, H&E and Masson straining. The remaining rat wounds were photographed on days 5, 10, and 20, and the wound area was calculated using ImageJ software.
H&E staining
Samples from the lung or wound were embedded in O.C.T. (Sakura), snap-frozen in liquid nitrogen, and stored at −80 °C. Sections (5 μm) were mounted on glass slides, air-dried at room temperature for 2 h, and stained with hematoxylin and eosin (H&E). H&E pictures were taken using an optical microscope (Leica 1100). An automatic best fit was applied for the best contrast.
Masson staining
The rat was euthanized, then the skin wound site and surrounding healthy tissue were removed, fixed on a clear paper piece to fully spread the wound, and then loaded with 4% tissue fixative solution (P1110, Solarbio). The corresponding wound samples were prepared according to experimental Standard Operation Procedure (SOP) of Servicebio (Wuhan, China).
Blood routine test
In the mice lung infection model, blood from mice was collected from sampling tubes containing anticoagulant for routine blood testing.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.0, Matlab version R2021a and FlowJo version 10.8.1 software. Data were presented as mean ± SEM. For the in vitro studies, unpaired two-tailed Student’s t-test (normally distributed data) between two groups were used to calculate P-values, and two-way ANOVA with Sidak’s multiple comparison among multiple groups were used to calculate adjusted P-values. For the in vivo studies, data between two groups were analyzed by unpaired t-test if the data exhibited a Gaussian distribution and equal variance, or by unpaired t-test with Welch’s correction. Differences with P < 0.05 were considered significant. The exact P values were annotated in Figures. The data of metabolite profiles and microbiota profiles were analyzed on the online platform of Majorbio Cloud Platform (https://cloud.majorbio.com/), metabioanalyst5.0 (https://www.metaboanalyst.ca/) and Omic Share platform (https://www.omicshare.com/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32222084 and 32172907), National Key Research and Development Program of China (2021YFD1801000), Key R&D Program of Jiangsu Province (Modern Agriculture) (BE2023332), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX24_3814), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and 111 Project D18007.
Author contributions
Y.L. and Z.W. designed and conceived the project. D.F., T.X., F.L., Y.S., J.S., Y.Y. and H.Z. performed all experiments. Y.L. and D.F. analyzed the data and prepared all figures. Y.L. and D.F. wrote the manuscript. All authors reviewed the manuscript.
Peer review
Peer review information
Nature Communications thanks Javier Ignacio Sanchez-Villamil, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Metabolomics data have been deposited in figshare database (10.6084/m9.figshare.28039505.v1). RNA-sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (PRJNA759745). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhiqiang Wang, Email: zqwang@yzu.edu.cn.
Yuan Liu, Email: liuyuan2018@yzu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55791-w.
References
- 1.Cassini, A. et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis.19, 56–66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes, A. H. et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet387, 176–187 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Stein, G. E. & Babinchak, T. Tigecycline: an update. Diagn. Microbiol. Infect. Dis.75, 331–336 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Yaghoubi, S. et al. Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: narrative review. Eur. J. Clin. Microbiol. Infect. Dis.41, 1003–1022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He, T. et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol.4, 1450–1456 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Sun, J. et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol.4, 1457–1464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu, Y., Liu, L., Sun, J. & Feng, Y. Limited distribution and mechanism of the TetX4 tetracycline resistance enzyme. Sci. Bull.64, 1478–1481 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Fang, L. X. et al. Emerging high-level tigecycline resistance: novel tetracycline destructases spread via the mobile Tet(X). Bioessays42, e2000014 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Zhang, S. et al. Dissemination and prevalence of plasmid-mediated high-level tigecycline resistance gene tet(X4). Front. Microbiol.13, 969769 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stokes, J. M., Lopatkin, A. J., Lobritz, M. A. & Collins, J. J. Bacterial metabolism and antibiotic efficacy. Cell Metab.30, 251–259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohanski, M. A., Dwyer, D. J., Wierzbowski, J., Cottarel, G. & Collins, J. J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell135, 679–690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehi, O. et al. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol.31, 2793–2804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foti, J. J., Devadoss, B., Winkler, J. A., Collins, J. J. & Walker, G. C. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science336, 315–319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, R. A., Gollan, B. & Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol.15, 453–464 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y. et al. Gut microbiome alterations in high-fat-diet-fed mice are associated with antibiotic tolerance. Nat. Microbiol6, 874–884 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Fang, D. et al. Nicotinamide mononucleotide ameliorates sleep deprivation-induced gut microbiota dysbiosis and restores colonization resistance against intestinal infections. Adv. Sci.10, e2207170 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao, X. L. et al. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med.13, eabj0716 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Peng, K. et al. QitanTech nanopore long-read sequencing enables rapid resolution of complete genomes of multi-drug resistant pathogens. Front. Microbiol.13, 778659 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang, B., Tong, Z., Shi, J., Wang, Z. & Liu, Y. Bacterial proton motive force as an unprecedented target to control antimicrobial resistance. Med. Res. Rev.43, 1068–1090 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Milvae, R. A., Alila, H. W. & Hansel, W. Methylation in bovine luteal cells as a regulator of luteinizing hormone action. Biol. Reprod.29, 849–855 (1983). [DOI] [PubMed] [Google Scholar]
- 22.Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A. & Collins, J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell130, 797–810 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Hiura, S., Abe, H., Koyama, K. & Koseki, S. Bayesian generalized linear model for simulating bacterial inactivation/growth considering variability and uncertainty. Front. Microbiol.12, 674364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kak, G., Raza, M. & Tiwari, B. K. Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol. Concepts9, 64–79 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Deng, T. et al. Bismuth Drugs Reverse Tet(X)-conferred tigecycline resistance in gram-negative bacteria. Microbiol. Spectr.10, e0157821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Y. et al. Anti-HIV agent azidothymidine decreases Tet(X)-mediated bacterial resistance to tigecycline in Escherichia coli. Commun. Biol.3, 162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, L. et al. A novel inhibitor of monooxygenase reversed the activity of tetracyclines against tet(X3)/tet(X4)-positive bacteria. EBioMedicine78, 103943 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meylan, S. et al. Carbon sources tune antibiotic susceptibility in pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem. Biol.24, 195–206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopatkin, A. J. et al. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science371, eaba0862 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobritz, M. A. et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl Acad. Sci. USA112, 8173–8180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng, B. et al. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab.21, 249–262 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Su, Y. B. et al. Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc. Natl Acad. Sci. USA115, E1578–E1587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, L., Zhang, H. & Gao, P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell13, 877–919 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blow, M. J. et al. The epigenomic landscape of prokaryotes. PLoS Genet.12, e1005854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blachier, F., Mariotti, F., Huneau, J. F. & Tomé, D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids33, 547–562 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Martínez, Y. et al. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids49, 2091–2098 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Navik, U. et al. Methionine as a double-edged sword in health and disease: Current perspective and future challenges. Ageing Res. Rev.72, 101500 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Roy, D. G. et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab.31, 250–266.e259 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Li, X. et al. In situ neutralization and detoxification of LPS to attenuate hyperinflammation. Adv. Sci.10, e2302950 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, Y., Gong, X., Shen, J. & Zhu, K. Postantibiotic leukocyte enhancement-mediated reduction of intracellular bacteria by macrophages. J. Adv. Res.58, 117–128 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, T., Fang, D., Li, F., Wang, Z. & Liu, Y. A dietary source of high level of fluoroquinolone tolerance in mcr-carrying Gram-negative bacteria. Research6, 0245 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphries, R., Bobenchik, A. M., Hindler, J. A. & Schuetz, A. N. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st Edition. J. Clin. Microbiol.59, e0021321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, J., Chen, C., Wang, D., Wang, Z. & Liu, Y. The antimicrobial peptide LI14 combats multidrug-resistant bacterial infections. Commun. Biol.5, 926 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, Y. et al. Metformin restores tetracyclines susceptibility against multidrug resistant bacteria. Adv. Sci.7, 1902227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, C., Cai, J., Shi, J., Wang, Z. & Liu, Y. Resensitizing multidrug-resistant Gram-negative bacteria to carbapenems and colistin using disulfiram. Commun. Biol.6, 810 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai, J. et al. Structural-activity relationship-inspired the discovery of saturated fatty acids as novel colistin enhancers. Adv. Sci.10, e2302182 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metabolomics data have been deposited in figshare database (10.6084/m9.figshare.28039505.v1). RNA-sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (PRJNA759745). Source data are provided with this paper.