Abstract
The antimicrobial agent pentamidine inhibits the self-splicing of the group I intron Ca.LSU from the transcripts of the 26S rRNA gene of Candida albicans, but the mechanism of pentamidine inhibition is not clear. We show that preincubation of the ribozyme with pentamidine enhances the inhibitory effect of the drug and alters the folding of the ribozyme in a pattern varying with drug concentration. Pentamidine at 25 µM prevents formation of the catalytically active F band conformation of the precursor RNA and alters the ribonuclease T1 cleavage pattern of Ca.LSU RNA. The effects on cleavage suggest that pentamidine mainly binds to specific sites in or near asymmetric loops of helices P2 and P2.1 on the ribozyme, as well as to the tetraloop of P9.2 and the loosely paired helix P9, resulting in an altered structure of helix P7, which contains the active site. Positively charged molecules antagonize pentamidine inhibition of catalysis and relieve the drug effect on ribozyme folding, suggesting that pentamidine binds to a magnesium binding site(s) of the ribozyme to exert its inhibitory effect.
INTRODUCTION
Pentamidine, 1,5-bis(4-amidinophenoxy)pentane, is used clinically to treat the severe infections caused by Pneumocystis carinii in AIDS and other immunodeficient states; it is also used to treat African trypanosomiasis and leishmaniasis (1,2). The mechanism of pentamidine action is not understood, although several mechanisms have been suggested for its action against different microorganisms (3–6). Pentamidine has long been known to bind to DNA, RNA and nucleotides (7). Pentamidine has been shown to be selectively bound in the minor groove of AT-rich DNA duplexes, with the amidinium groups hydrogen bonded to N3 atoms of adenine and the drug molecule occupying a groove site spanning 4–5 bp (8–10). However, the antimicrobial effects of pentamidine have not been proven to be due to DNA binding (11).
Recently, pentamidine has been demonstrated to inhibit self-splicing of various group I introns in vitro, including the nuclear group I introns present in the rRNA genes of P.carinii (12,13), the Ca.LSU nuclear group I intron of the 26S rRNA gene of Candida albicans (14) and the mitochondrial introns of Saccharomyces cerevisiae (15). Pentamidine is also a potent inhibitor of mitochondrial translation in S.cerevisiae (15). The splicing of Ca.LSU in living C.albicans cells has also been shown to be sensitive to pentamidine, and strains lacking this intron are relatively pentamidine resistant (14).
Another RNA-binding class of antibiotics, the aminoglycosides, displace metal ions to bind to specific sites on RNA, and thus inhibit the activity of ribosomes and various ribozymes (reviewed in 16). Aminoglycosides appear to bind to the phage sunY small group I intron in precursor RNA at specific bases in bulges or regions where double helices are distorted, as indicated by protection of these sites from chemical attack. Although the protected bases include G96 in the catalytically active P7 helix, several bases near this site are sensitized to cleavage by aminoglycosides (17). Another small phage group I intron, td, has been suggested to interact with neomycin B at two sites, one at the catalytic center and one distant, based on chemical protection, analysis of drug-resistant mutations and use of a computer docking program (18). Neither of these phage introns are as large or complex as the nuclear group I introns of eukaryotic cells, such as the well-characterized group I intron of Tetrahymena (19,20).
We have previously shown that group I intron Ca.LSU can partially fold in the absence of divalent cations, and this folding results in a conformation with increased catalytic activity. In the presence of divalent cations, Ca.LSU folds into a more ordered, stable but misfolded conformation, which is less able to convert into the catalytically active form into which the ribozyme folded in the absence of cations (21). We demonstrate here that pentamidine interacts with Ca.LSU RNA, preventing both the correct folding and the self-splicing activity of the intron. We suggest that, as neomycin B binds to various RNA targets, pentamidine appears to bind to specific regions of ribozyme Ca.LSU. This binding appears to alter both RNA folding and inhibit catalysis by the ribozyme. Like neomycin B, pentamidine appears to compete with positively charged molecules for binding to its RNA target. In this work, for simplicity we refer to the interaction of pentamidine with the ribozyme RNA as ‘binding’. We have not directly measured the binding of pentamidine to the RNA; rather, we have measured the effect of the drug on ribonuclease sensitivity, electrophoretic mobility and catalytic activity of the RNA.
MATERIALS AND METHODS
Labeling and self-splicing of Ca.LSU precursor
The PCR amplified DNA fragment containing the SP6 promoter at the 5′ end of the sense strand of a fragment of the rRNA gene containing Ca.LSU was transcribed in vitro in the presence of 500 µM rATP, rCTP and rGTP, 200 µM rUTP (Promega) and 20 µCi [α-32P]UTP (3000 Ci/mmol; NEN-DuPont), as previously described (21). This 717 nt precursor RNA consists of a 129 nt fragment of the 5′ exon (E1) and a 209 nt fragment of the 3′ exon (E2) flanking the 379 nt intron (I). The transcribed precursor RNA was purified by PAGE on a 5% polyacrylamide–8 M urea gel (12,15). The standard self-splicing assays and precincubations were performed as previously described (21), unless otherwise indicated. The standard self-splicing assay was performed in 10 µl reactions containing 50 mM Tris–HCl (pH 7.5), 1.25 mM MgCl2, 0.4 mM spermidine, 10 µM GTP and 10 U RNasin (Promega) at 37°C for 20 min. Where indicated, precursor RNA was preincubated in 8 µl of 62.5 mM Tris–HCl (pH 7.5) plus 10 U RNasin with the indicated components at 37°C for the indicated times prior to initiating the self-splicing reaction by addition of 2 µl of reaction mix containing all reaction components at concentrations to bring each to the standard assay conditions. Splicing products were analyzed by PAGE on 5% polyacrylamide–8 M urea gels that were exposed to X-ray film to visualize the bands or exposed to a phosphorimager screen (GS-525 Molecular Imager; Bio-Rad) to quantify band intensity (21).
Native gel analysis of Ca.LSU precursor RNA
Homogeneous radiolabeled precursor RNA (0.3 nM), purified as described above, was incubated in 8 µl of 62.5 mM Tris–HCl (pH 7.5) containing 10 U RNasin and the indicated concentrations of cations and/or pentamidine at 37°C for 20 min. The control sample was incubated on ice for 20 min. The samples were then analyzed by native PAGE, as described previously (21).
Ribonuclease T1 protection assay
Ca.LSU intron RNA was labeled at its 5′ end by reaction of [α-32P]GTP with non-radioactive in vitro transcribed Ca.LSU precursor RNA by the self-splicing reaction (21). The radioactive 5′-guanylylated intron RNA (3 nM) was preincubated with the indicated concentrations of positively charged molecules and/or pentamidine in 8 µl of 62.5 mM Tris–HCl (pH 7.5) and 10 U RNasin (Promega) at 37°C for 20 min, and then 2 µl of ribonuclease T1 (0.006 U; Pharmacia) was added to each sample and the digestion was continued for another 20 min. RNA ladders were generated by cleaving the 5′ end labeled Ca.LSU intron using 0.01 U ribonuclease T1 for G, 0.2 U ribonuclease U2 for A and 0.2 U ribonuclease PhyM for A+U in 2.8 M urea buffers at 50°C. All cleavage reactions were stopped by chilling on ice and analyzed by PAGE as previously described (21). Several gels were run with the same samples to obtain optimal resolution of the products resulting from cleavage at each individual nucleotide.
RESULTS
Preincubation of Ca.LSU precursor with pentamidine increases drug efficacy
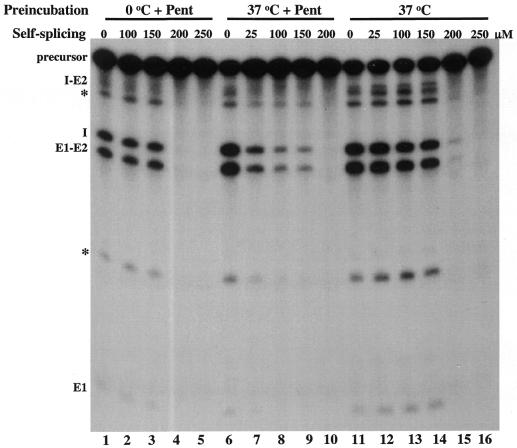
The self-splicing activity of group I introns is measured by the rate of conversion of radioactive precursor (E1–I–E2) to the splicing intermediates (I–E2 and E1) and products (I and E1–E2) of the splicing reaction (Fig. 1). Pentamidine completely inhibits the self-splicing activity of Ca.LSU at 200 µM concentration, while no inhibition was observed at 150 µM (Fig. 1, lanes 1–5) (14). Similar inhibition by pentamidine was observed of the self-splicing of Pc1.LSU, a nuclear group I intron of P.carinii (13). As shown in Figure 2A, the complete inhibition of activity of Ca.LSU by 200 µM pentamidine was evident at 2 min, the shortest time examined, indicating the lack of an obvious delay in the interaction of pentamidine with the RNA. We speculated that some splicing components, such as MgCl2, spermidine and GTP, might compete with the pentamidine for binding to the ribozyme RNA. This speculation is consistent with our finding that preincubation of a mitochondrial intron-containing precursor from yeast with pentamidine prior to the addition of other splicing components increases the inhibitory potency of the drug by up to 10-fold (15,22). Therefore, Ca.LSU precursor RNA was preincubated with pentamidine at 37°C before the splicing reaction was started to examine whether pentamidine can effectively interact with the RNA molecule at lower drug concentrations in the absence of MgCl2, spermidine and GTP. Figures 1 (lanes 6–10) and 2B show that when added to the precursor RNA during preincubation prior to addition of the other reaction components, 25 µM pentamidine substantially inhibited both the rate and efficiency of the Ca.LSU self-splicing reaction; greater inhibition was observed at 100–150 µM and complete inhibition was achieved at 200 µM. Therefore, pentamidine at concentrations of 25–150 µM can interact with Ca.LSU precursor RNA effectively if added during preincubation, exerting its inhibitory effect on the subsequent splicing reaction. These observations are consistent with the hypothesis that at least one of the three splicing components may compete with pentamidine for interaction with or binding to the ribozyme RNA. Since we had previously shown that divalent cations or spermidine present during preincubation altered the folding of Ca.LSU (21), it seemed likely that these positively charged molecules might compete with positively charged pentamidine for binding to the ribozyme RNA, or they might alter ribozyme conformation in a manner that reduced its availability for interaction with pentamidine.
Figure 1.
Preincubation of Ca.LSU precursor with pentamidine enhances its inhibitory effect on splicing. Equal amounts of radiolabeled Ca.LSU precursor RNA were added to each reaction. The non-preincubated reactions were run under standard splicing conditions with the indicated concentrations of pentamidine (lanes 1–5). The other samples were preincubated in Tris buffer at 37°C for 20 min before addition of MgCl2, spermidine and GTP. The indicated concentrations of pentamidine were added either during preincubation (lanes 6–10) or after preincubation together with the other splicing components (lanes 11–16) and reaction products were analyzed by PAGE as described in Materials and Methods. The asterisks indicate products possibly generated by Mg2+-induced hydrolysis of the Ca.LSU precursor RNA.
Figure 2.
Kinetics of pentamidine inhibition of splicing by Ca.LSU. (A) Indicated concentrations of pentamidine were added to the splicing reaction together with all splicing components without preincubation of the precursor RNA (Materials and Methods). (B) The indicated concentrations of pentamidine were present during a 20 min preincubation of precursor RNA and splicing was initiated by addition of all other splicing components. In both panels the time course of the splicing reaction was analyzed as in Figure 1. All experiments were performed four times and the average splicing fraction [the ratio of spliced intron (I) and ligated exons (E1–E2) to total RNA] is presented (standard deviations indicated).
Pentamidine alters the electophoretic mobility of native Ca.LSU precursor RNA
Native PAGE analysis was used to examine the possible conformational changes of the precursor RNA during preincubation with pentamidine. Ca.LSU precursor RNA was preincubated with the indicated concentrations of pentamidine under the same conditions used for preincubation prior to the splicing reaction assayed above; the preincubated samples were chilled on ice and analyzed by native PAGE at 4°C (Fig. 3). As described previously (21), two distinct RNA conformations (the S and F bands) existed for the homogeneous Ca.LSU precursor prior to preincubation. In addition, another precursor RNA population migrated more slowly as a polydisperse smear, probably reflecting RNA aggregation in the absence of any cations. Such a polydisperse distribution of RNA conformations has also been reported for the Tetrahymena intron precursor (23). Preincubation of the Ca.LSU precursor RNA at 37°C converted the precursor RNA predominantly into the catalytically active F band conformation, as previously reported (21). However, preincubation in the presence of 25 µM pentamidine resulted in most of the RNA migrating as did the S band conformation. This effect of 25 µM pentamidine during preincubation is similar to that of MgCl2, CaCl2 or spermidine, which similarly convert some precursor RNA into the S band form, resulting in decreased self-splicing activity (21). However, the possibility remains that the RNA conformations migrating at the S band position may not be the same for RNA preincubated with MgCl2, CaCl2 or spermidine as opposed to pentamidine. The altered form of precursor RNA, induced by preincubation in the presence of 25 µM pentamidine, is also apparently much less active in catalysis, as indicated by the reduced self-splicing activity shown in Figures 1 and 2B. This result can be readily explained by pentamidine binding to precursor RNA and preventing the RNA from folding into the F band conformation. Apparently, the drug keeps the RNA in the S band or similar conformation, which shows reduced catalytic activity. Preincubation of precursor RNA in the presence of 100 µM pentamidine caused the disappearance of the two distinct precursor RNA conformations with a corresponding increase in a very broad polydisperse smear with mobility ranging from approximately zero to a position above the S band. This pentamidine-induced RNA smear is not identical to that present in the original precursor RNA (Fig. 3, Exposure 2). The appearance of this slow moving polydisperse RNA at high concentrations of pentamidine (100 µM) may indicate that pentamidine causes intermolecular binding between RNA molecules by its two symmetric amidinium groups, causing RNA aggregation. The sum of the intensities of these aggregated RNAs is substantially less than that of the RNAs in the S and F bands analyzed in parallel samples containing the same amount of homogeneous RNA, which is consistent with our observation that some radioactivity is retained in the reaction tubes, resulting in loss of some radiolabeled RNA in the presence of 100 µM pentamidine. However, pentamidine-induced RNA retention in the tube was not observed when loading buffer containing 7 M urea was used, as in the splicing reaction analysis and ribonuclease T1 sensitivity assay, described below. Radiolabeled Ca.LSU intron RNA was also assayed for its gel mobility change in the presence of pentamidine, and similar results were obtained (data not shown).
Figure 3.
Pentamidine alters the folding of Ca.LSU precursor RNA. The same amount of radiolabeled Ca.LSU precursor in Tris buffer was incubated at 0 or 37°C for 20 min in the presence of the indicated concentration of pentamidine and was then analyzed by native PAGE (Materials and Methods). The two different exposures were to allow clear visualization of the S and F bands (Exposure 1) and smear RNA species (Exposure 2).
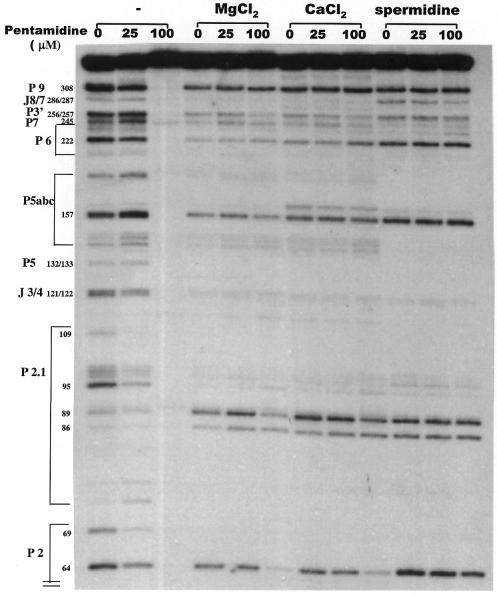
Interaction of pentamidine with Ca.LSU intron alters the ribonuclease T1 cleavage pattern
Susceptibility to ribonuclease T1 cleavage was used to probe possible pentamidine binding sites on 5′ end labeled Ca.LSU intron RNA (Materials and Methods). The radiolabeled intron RNA was preincubated with the indicated concentrations of pentamidine at 37°C for 20 min, ribonuclease T1 was then added to partially digest the intron RNA and the resultant RNA fragments were fractionated by PAGE on an 8% polyacrylamide–7 M urea gel (Fig. 4A). In the absence of pentamidine during preincubation, many predicted helical structures of Ca.LSU intron formed in Tris buffer, as indicated by resistance to ribonuclease T1 cleavage at the predicted paired G sites (21). However, the presence of 25 µM pentamidine altered the ribonuclease T1 cleavage pattern at many G sites that are accessible to cleavage by ribonuclease T1 in the absence of pentamidine (Fig. 4A and B) and most of the inaccessible G sites remained inaccessible. Since ribonuclease T1 is unable to cleave at paired G residues, we cannot draw conclusions about the ability of pentamidine to bind to RNA double helices. We cannot determine whether the failure of the paired G residues to be cleaved by ribonuclease T1 in the presence of pentamidine indicates that pentamidine binds to these sites or that the double helical structure of these sites persists despite presence of the drug. In the catalytic domain P4–P6, 25 µM pentamidine increased ribonuclease T1 accessibility at all of the otherwise cleavable G sites in domain P5abc, but not at most of the G residues in the other helices in the P4–P6 structural domain. In the other catalytic domain, P3–P9, the drug increased ribonuclease T1 cleavage at the ribozyme active site G247 and the adjacent G245 in the P7 helix, as well as at G residues in P3, but decreased ribonuclease T1 cleavage at G residues in P9 and P9.2. The cleavability by ribonuclease T1 of accessible G residues in P9.0 was unchanged by the presence of 25 µM pentamidine. Helix P8 was completely resistant to ribonuclease T1 cleavage in the presence or absence of the drug, while a number of G sites of the otherwise resistant P9.1 were sensitized by the drug. In the peripheral helices P2 and P2.1, pentamidine decreased ribonuclease T1 cleavage at most G sites. Based on its effects on the ribonuclease T1 cleavage pattern of Ca.LSU intron RNA, pentamidine at 25 µM had the least effect on cleavable sites in helices P5, P6 and P9.0, while the folding of the other ribonuclease-accessible helices was altered by the drug with P2, P2.1, P9 and P9.2 being protected and the other helices being sensitized. The lack of pentamidine effect on ribonclease T1 accessibility of P5, P6 and P9.0 might suggest that these helices are folded inside the intron molecule, making them less accessible to the planar pentamidine molecule. This hypothesis is consistent with the crystal structure of the Tetrahymena ribozyme (19,20,25), which is in the same subclass (IC) of group I introns as Ca.LSU.
Figure 4.
Pentamidine alters the folding of Ca.LSU precursor RNA. The same amount of radiolabeled Ca.LSU precursor in Tris buffer was incubated at 0 or 37°C for 20 min in the presence of the indicated concentration of pentamidine and was then analyzed by native PAGE (Materials and Methods). The two different exposures were to allow clear visualization of the S and F bands (Exposure 1) and smear RNA species (Exposure 2).
In contrast, the presence of 100 µM pentamidine resulted in blocking of ribonuclease T1 cleavage at all G residues in the intron RNA and the appearance of cleavage at several U residues (circled in lane 6 in Fig. 4A and in red in Fig. 4B). Cleavage of U243, but not G242, of the folded intron RNA in the presence of ribonuclease T1 has been confirmed by alignment with RNA ladders generated by ribonucleases PhyM (cleaving A and U), U2 (cleaving A), and T1 (cleaving G) under RNA-denaturing conditions. We found that pentamidine at 100 µM greatly increased cleavage at U243 and induced substantial cleavage at U258. The increased cleavage at these U sites observed in the presence of 100 µM pentamidine but not at 25 µM is consistent with pentamidine interacting with the RNA molecule differently at these two concentrations. We cannot exclude the possibility that contaminating nucleases are responsible for cleavage at U residues, but it is hard to explain the limitation of this cleavage to only a few U sites of the RNA. However, we have not determined whether this cleavage represents a reaction catalyzed by cationic molecules or by a contaminating ribonuclease.
Blockage of ribonuclease T1 cleavage at all intron G residues by 100 µM pentamidine might be explained by the drug binding to all of the G residues and blocking accessibility by ribonuclease T1. Because the molar ratio of pentamidine (100 µM) to intron RNA nucleotide (∼5 µM) in these experiments is about 20, such blockage of enzyme accessibility to all of the RNA nucleotides by this positively charged drug is possible. Taken together with the smear forms of precursor RNA observed with native PAGE analysis, these results suggest that pentamidine binds to ribozyme RNA in both an intra- and intermolecular manner, by means of its two flexible symmetric amidinium groups. The difference between the effects of 25 and 100 µM pentamidine, demonstrated by both native gel analysis and ribonuclease T1 protection assays, suggests that the binding of multiple pentamidine molecules to ribozyme RNA might be cooperative. This hypothesis is consistent with the previous observations that pentamidine inhibits the self-splicing of group I introns in a threshold pattern, i.e. the inhibition occurred in a very narrow dynamic concentration range, with a very steep dose-dependent inhibition curve (Fig. 1) (13,14).
Magnesium, calcium and spermidine reduce the inhibitory effect of pentamidine on ribozyme splicing
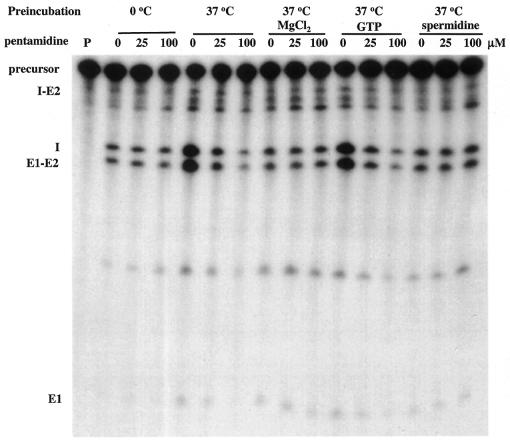
We have noted that components in the ribozyme assay buffer may compete with pentamidine for interaction with ribozyme RNA and reduce its inhibitory effect. Therefore, each of these components was individually included with pentamidine in the preincubation of ribozyme precursor RNA, and the remaining splicing components were then added to initiate the splicing reaction after preincubation (Fig. 5). In the absence of preincubation, 100 µM pentamidine failed to inhibit splicing of ribozyme Ca.LSU. In contrast, the addition of 25 µM pentamidine during preincubation substantially inhibited splicing, with 100 µM pentamidine reducing splicing even further. Addition of either 1.25 mM MgCl2 or 0.4 mM spermidine (the same concentrations present in the standard ribozyme assay) prevented both the preincubation-induced stimulation of ribozyme activity (21) and the enhanced sensitivity to pentamidine present during preincubation. In contrast to the effect of positively charged Mg2+ and spermidine, the presence during preincubation of 10 µM GTP failed to alter the effects of preincubation on both ribozyme activity (Fig. 5) (21) and sensitivity to pentamidine (Fig. 5). This result is consistent with kinetic data indicating that pentamidine inhibition is non-competitive with respect to the GTP substrate of the splicing reaction of this and other ribozymes (13,14). Addition of MgCl2 to the preincubation of three mitochondrial group I intron ribozymes (aI3α, aI5α and bI5) similarly abolishes their preincubation-enhanced sensitivity to pentamidine (22).
Figure 5.
Mg2+ and spermidine reduce pentamidine inhibition of Ca.LSU catalysis. Equal amounts of radiolabeled Ca.LSU precursor were either assayed for ribozyme activity without preincubation in the presence of varying levels of pentamidine or preincubated before ribozyme assays in buffer alone or in the presence of 1.25 mM MgCl2, 10 µM GTP or 0.4 mM spermidine. All assays with and without preincubation were performed under standard splicing conditions. Lane P is a sample preincubated in buffer alone without adding splicing components (magnesium, spermidine, GTP).
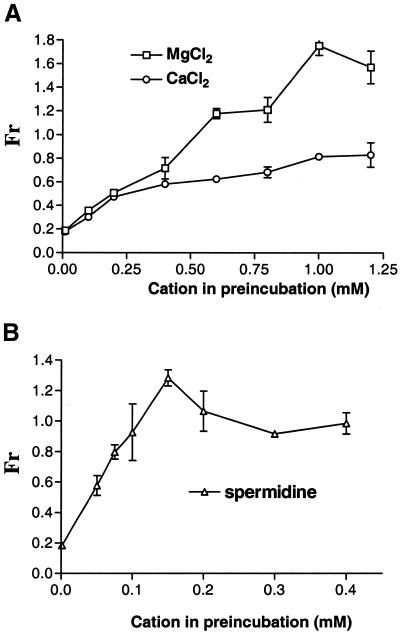
The ability of MgCl2 and spermidine, as well as another divalent metal ion CaCl2, to prevent pentamidine from interacting with ribozyme RNA during preincubation was determined by varying the concentrations of these molecules in the presence of 100 µM pentamidine. As can be seen in Figure 6, all three cations caused a dose-dependent reduction in ribozyme sensitivity to pentamidine during preincubation. Spermidine was more potent in protecting Ca.LSU from pentamidine inhibition, with complete protection being achieved by 0.1 mM spermidine, while complete protection was seen at 0.6 mM MgCl2; CaCl2 did not afford complete protection even at 1.2 mM. When added to preincubation mixes, chloride salts of the monovalent cations NH4+ and Na+ did not alter pentamidine sensitivity at concentrations as high as 50 mM (data not shown).
Figure 6.
Dose-dependent effect of positively charged molecules on ribozyme sensitivity to pentamidine. Equal amounts of radiolabeled Ca.LSU precursor RNA were preincubated in the presence or absence of 100 µM pentamidine with the indicated concentrations of CaCl2, MgCl2 or spermidine at 37°C for 20 min. The missing splicing reaction components (and varying concentrations of CaCl2, MgCl2 or spermidine) were then added. However, in the subsequent splicing reactions, divalent cation concentrations were 1.25 mM MgCl2 and 1.25 mM CaCl2 for experiments in which CaCl2 was present during preincubation and 2.5 mM MgCl2 when MgCl2 was present in the preincubation (A), and 2.5 mM MgCl2 when spermidine (B) was present in the preincubation. Spermidine concentration was 0.4 mM in all splicing reactions. All data were obtained from triplicate experiments. Each gel was scanned and analyzed by PhosphorImager and the splicing percentage was obtained for each reaction. The fraction pentamidine resistance (Fr) represents the ratio of Ca.LSU splicing activity of the reaction including pentamidine to that of the pentamidine-free reaction at each cation concentration. These data were plotted using the program PRISM v.2.01 (GraphPad Software, San Diego, CA).
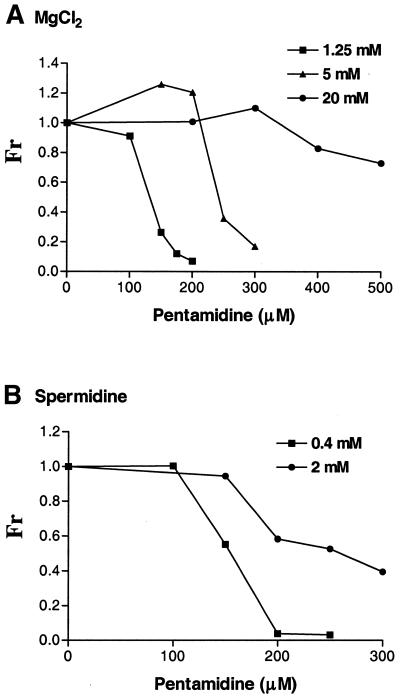
During preincubation in the presence of at least 0.5 mM Mg2+, the presence of 100 µM pentamidine enhanced the subsequent splicing activity of Ca.LSU (Fig. 6A). This apparent ‘stimulation’ of ribozyme activity by pentamidine may reflect pentamidine competition with Mg2+ for similar binding sites, relieving kinetically trapped folding intermediates induced by Mg2+, as previously described for ribozyme Tt.LSU (26,27) and postulated to be present in Ca.LSU (21). In ribozyme reactions run without preincubation, MgCl2 and spermidine at concentrations higher than the standard ribozyme assay conditions dramatically increased the pentamidine concentrations required to completely inhibit the self-splicing activity of the ribozyme (Fig. 7). These results indicate that both during preincubation and in the ribozyme reaction, divalent cations and spermidine can diminish the inhibitory effect of pentamidine on splicing by Ca.LSU. However, higher concentrations of pentamidine added during preincubation or to the ribozyme reaction can inhibit the ribozyme even in the presence of these added positively charged molecules (Fig. 7 and data not shown). Thus, these positively charged molecules appear to compete with pentamidine for binding to the ribozyme RNA.
Figure 7.
Positively charged molecules antagonize pentamidine inhibition directly in the splicing reaction. Equal amounts of radiolabeled Ca.LSU precursor RNA were spliced in the presence of the indicated concentrations of pentamidine and MgCl2 (A) or spermidine (B) at 37°C for 20 min. Only the concentrations of the indicated splicing component varied, while the other splicing components remained as in the standard splicing reaction. The fraction pentamidine resistance (Fr) is defined as in Figure 6.
Positively charged molecules oppose pentamidine effects on ribozyme folding
The ribonuclease T1 cleavage assay was used to study the effects of these positively charged molecules on the folding of Ca.LSU RNA in the presence of pentamidine. Consistent with the elimination of inhibition by 25 and 100 µM pentamidine of Ca.LSU self-splicing in the presence of 1.25 mM MgCl2 or 0.4 mM spermidine, addition of these concentrations of MgCl2 or spermidine or 1.25 mM CaCl2 during preincubation resulted in reversal of the effects of pentamidine on sensitivity of all of the G residues to ribonuclease T1 in the presence of pentamidine compared with the control samples receiving no drug (Fig. 8). This result strongly suggests that oligovalent cations compete with pentamidine for binding to the intron RNA molecule and thereby protect the self-splicing of such RNA from pentamidine inhibition. As demonstrated previously for the effect of the different positively charged molecules on folding of Ca.LSU (21), folding of the intron RNA is modified somewhat differently in the presence of these three molecules, with more difference observed between the divalent metal ions and spermidine than between the two different divalent cations. Moreover, native PAGE of the precursor RNA preincubated under the same conditions revealed that all three positively charged molecules, at the same concentrations that were effective in reducing sensitivity to preincubation with pentamidine, were also able to eliminate the smear RNA resulting from the presence of 100 µM pentamidine during preincubation. Instead, in the presence of any of these positively charged molecules and 100 µM pentamidine, a band of RNA migrating near the F band position appeared, and the intensity of the newly formed band was about the same as the reduction in the smear RNA (Fig. 9). Thus, positively charged molecules can antagonize the effects of pentamidine on the folding of both precursor and intron RNA, which is consistent with the ability of these molecules to reduce the inhibitory effect of pentamidine on ribozyme activity.
Figure 8.
Positively charged molecules eliminate the pentamidine-induced alteration in ribonuclease T1 sensitivity. Equal amounts of 5′ end radiolabeled intron RNA was preincubated in Tris–HCl in the presence of 0, 25 or 100 µM pentamidine and 1.25 mM MgCl2, 1.25 mM CaCl2 or 0.4 mM spermidine at 37°C for 20 min. Then ribonuclease T1 cleavage and PAGE were performed as described in Materials and Methods. The major cleaved positions are indicated at the left of the gel (nucleotide numbers as in Figure 4).
Figure 9.
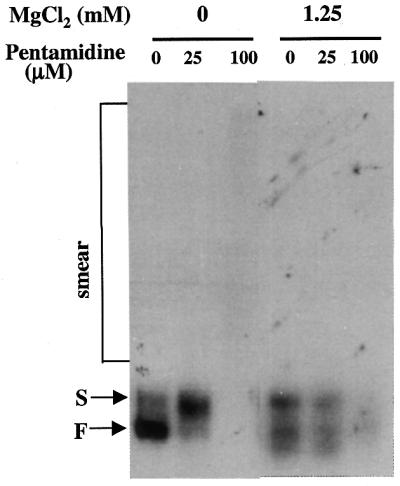
MgCl2 antagonizes the alteration of folding of Ca.LSU precursor RNA induced by pentamidine. Equal amounts of radiolabeled precursor RNA were preincubated in Tris–HCl in the presence of 0, 25 or 100 µM pentamidine and in the absence or presence of 1.25 mM MgCl2, 1.25 mM CaCl2 or 0.4 mM spermidine at 37°C for 20 min, then analyzed by native PAGE as described in Materials and Methods.
DISCUSSION
Pentamidine interacts with Ca.LSU RNA, altering ribozyme folding and inhibiting catalysis
The anti-infective utility and toxicity of pentamidine in prophylaxis and treatment of P.carinii pneumonia and of protozoan infections have stimulated the study of the molecular target of this drug in living organisms for several decades. Although pentamidine has been known to bind to the minor groove of DNA (9), the DNA affinity of different pentamidine analogs does not correlate with their antimicrobial activity (11), so identification of DNA as the sole target of pentamidine action remained uncertain (1). It has long been known that pentamidine interacts with both RNA and DNA (7), although the interaction with DNA has been much more extensively studied. Pentamidine has been shown to inhibit splicing in vitro of various group I introns (13–15). In addition, the splicing of Ca.LSU in C.albicans (14) and mitochondrial translation in S.cerevisiae (15) are inhibited by pentamidine, as is protein synthesis in a cell-free rat liver extract (28), suggesting that pentamidine is able to bind to RNA in vitro and in vivo to inhibit RNA biological functions. In this report, we demonstrate that pentamidine interacts with Ca.LSU intron and precursor RNAs, altering the folding and the catalytic activity of this ribozyme. These results indicate a possible mechanism for the antimicrobial activity of pentamidine based on its interaction with specific RNA targets. While we have not directly measured pentamidine binding to RNA, the effects of pentamidine on ribonuclease sensitivity, gel mobility and catalytic activity of the Ca.LSU ribozyme are consistent with it binding to that RNA molecule.
The ribonuclease T1 protection pattern of 25 µM pentamidine on the free intron (Fig. 4) indicates several specific sites at which pentamidine alters cleavage of RNA. Note that most of the G residues that are protected by this concentration of pentamidine are located in domains P2 and P2.1, generally in asymmetric internal loops or base paired residues predicted to be near loops. Protection at ribonuclease T1-sensitive G residues in loosely paired helix P9 and the tetraloop of P9.2 is also substantial. On the other hand, this same concentration of pentamidine resulted in increased cleavability in or near the internal loops of domains P5a and P5b, in domain P3 and in the catalytically active domain P7. These results suggest that pentamidine might bind preferentially to some sites in domains P2, P2.1, P9 and P9.2 and that this binding might secondarily alter the folding in P3, P5a, P5b and P7 to make them more accessible to ribonuclease T1; this altered folding might explain the altered gel mobility and reduced catalytic activity seen after preincubation of precursor RNA in the presence of 25 µM pentamidine. We propose that pentamidine binds to specific asymmetric loops and other structures of Ca.LSU and that this binding secondarily affects the structure of the active site and the catalytic activity of the ribozyme. Our finding that pentamidine inhibits self-splicing of group I introns non-competitively with respect to guanosine cofactor is consistent with the nuclease protection data indicating that the drug does not directly bind to the guanosine binding site. Note that for the smaller td (18) and sunY group I introns (17), neomycin B shows similar specific protection of sites distant from the catalytic site at drug concentrations inhibiting catalysis. Although the drugs and introns are distinct, it is noteworthy that asymmetric loops are among the strongest sites at which neomycin protection is seen. Selective pentamidine binding to asymmetric loops and other RNA structures in the ribozyme helices P2, P2.1, P9 and P9.2, but not those in the other ribozyme domains, suggests that these helices may fold in the outer surface of the ribozyme tertiary structure, making them accessible to the drug. This speculation is consistent with the crystal structure resolved for the catalytic core of the Tetrahymena intron (19,20,25) and group I intron structures revealed by other methods (29–31).
Because one pentamidine molecule has two symmetric amidinium groups, it may cause intermolecular conjugation of different RNA strands, as well as intramolecular conjugation. The broad smear on native gels of precursor RNA treated with 100 µM pentamidine may represent such randomly conjugated RNA molecules (Fig. 3). However, since inhibition is seen at concentrations of 25 µM pentamidine added during preincubation (Fig. 5), drug interaction with specific sites seen at that concentration indicates functionally important targets at which the drug binds to the ribozyme.
Because folding of the catalytically active F band form of Ca.LSU precursor RNA is completed within 1 min at 37°C in the absence of pentamidine (21), competition between Ca.LSU intron RNA folding and pentamidine binding to the RNA should occur when the ribozyme is incubated with pentamidine. The altered native gel mobility and ribonuclease T1 cleavage pattern of the RNA presumably represents the outcome of this competition. Figures 3 and 4 show that the binding of pentamidine (100 µM) to Ca.LSU precursor RNA competes effectively with correct Ca.LSU folding, as indicated by the disappearance of the F band conformation of native RNA and loss of accessibility of all intron G residues to ribonuclease T1 after preincubation with pentamidine.
Antagonism between positively charged molecules and pentamidine suggests that drug amidinium groups interact with RNA
The effects of pentamidine on both the folding and catalysis of Ca.LSU ribozyme RNA are opposed by oligovalent cations. It has been demonstrated that Mg2+, Ca2+ and spermidine each potentiates folding of Ca.LSU to a stable (but relatively inactive) structure with the predicted local and long distance base pairing (21). Therefore, it is likely that these positively charged species compete with pentamidine for binding to the RNA molecule, and the positively charged molecule-induced ribozyme structure may be more resistant to pentamidine binding because of the stable and extensive base pairing of the ribozyme RNA. Moreover, we found that less spermidine is required to compactly fold the ribozyme RNA and to counter the effect of pentamidine on the RNA, compared with the divalent metal ions. Spermidine at 0.1 mM added during preincubation of the precursor RNA in the presence of 0.1 mM pentamidine renders the ribozyme RNA completely resistant to pentamidine inhibition, indicating a dominant effect of spermidine relative to pentamidine on Ca.LSU folding. Another way of viewing this competitive interaction is that pentamidine binds to the sites of the ribozyme that function as magnesium binding sites, as appears to be the case for aminoglycoside binding to other group I introns (18) and other RNA molecules (reviewed in 16). If this is the case, the magnesium binding sites of Ca.LSU should be located in domains P2 and P2.1 and/or P9 and P9.2, which is different from magnesium binding sites of the Tetrahymena intron and the phage td intron, located in the catalytic core of the ribozyme (18–20). However, previous findings have suggested a different response of ribozyme Ca.LSU to magnesium from that of the Tetrahymena intron (21). Further evidence is required to verify the magnesium binding sites of the Ca.LSU intron.
Note that pentamidine and its analogs interact with nucleic acids by virtue of their positive charge, aromaticity and structure. The variation between these analogs in their effect on activity of Ca.LSU (14,22) and of similar ribozymes from P.carinii (13) indicates that features in addition to positive charge determine the relative potency as ribozyme inhibitors. Thus, the analog 1,5-di[4-(2-imidazolinyl)-2-methoxyphenoxyl]pentane, which has the same charge as pentamidine, is more inhibitory than is pentamidine for ribozyme Pcl.LSU (13). On the other hand, propamidine, which only differs from pentamidine by having two fewer methylene groups, is inactive against both ribozymes Ca.LSU and Pc1.LSU (13,14). Further study of the structures of these analogs and the pentamidine binding sites of Ca.LSU could lead to the design of more potent and specific inhibitors of microbial ribozymes.
RNA functions may be important physiological targets of pentamidine
This work demonstrates that pentamidine inhibits catalytic activity of group I intron ribozyme Ca.LSU by binding to the RNA and disturbing the folding of this ribozyme. This is also the first demonstration that pentamidine, a known minor groove binder of DNA, can interact with RNA, apparently at sites quite distinct from the binding sites on DNA, resulting in altered RNA conformation and inhibition of the biological functions of that RNA. Pentamidine appears to bind to magnesium binding sites at specific asymmetric loops and regions of distorted double helix conformation on RNA molecules. Unlike DNA molecules, RNA molecules have a variety of important catalytic functions in living cells in addition to their function as coding (mRNA) molecules. These RNA-catalyzed reactions include self-splicing of ribozyme introns (32,33), such as the Ca.LSU intron that we have studied, and protein synthesis by ribosomes, in which rRNA is catalytic (34–36). Therefore, binding of pentamidine to these RNA molecules in living cells may be cytotoxic, resulting in the antimicrobial activity and toxicity of the drug. Further elucidation of the structural requirements for the binding of pentamidine and related congeners to specific microbial RNA targets should provide a pathway for development of more potent and less toxic antimicrobial agents.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Marc Gartenberg, Terri Goss Kinzy and Jonathan Dinman for critical reading of this manuscript. This study was supported by NIH grant 1R01 GM53815, a grant from the Foundation for UMDNJ (M.J.L.), NSFC grant 30040033 and Wuhan University grant 0000028 (Y.Z.).
REFERENCES
- 1.Sands M., Kron,M.A. and Brown,R.B. (1985) Pentamidine: a review. Rev. Infect. Dis., 7, 625–634. [DOI] [PubMed] [Google Scholar]
- 2.Van Voorhis W.C. (1990) Therapy and prophylaxis of systemic protozoan infections. Drugs, 40, 176–202. [DOI] [PubMed] [Google Scholar]
- 3.Grady R.W., Blobstein,S.H., Meshnick,S.R., Ulrich,P.C., Cerami,A., Amirmoazzami,J. and Hodnett,E.M. (1984) The in vitro trypanocidal activity of N-substituted p-benzoquinone imines: assessment of biochemical structure-activity relationships using the Hansch approach. J. Cell. Biochem., 25, 15–29. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro T.A. and Englund,P.T. (1990) Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc. Natl Acad. Sci. USA, 87, 950–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dykstra C.C. and Tidwell,R.R. (1991) Inhibition of topoisomerases from Pneumocystis carinii by aromatic dicationic molecules. J. Protozool., 38, 78S–81S. [PubMed] [Google Scholar]
- 6.Shapiro T.A. (1993) Inhibition of topoisomerases in African trypanosomes. Acta Trop., 54, 251–260. [DOI] [PubMed] [Google Scholar]
- 7.Makulu D.R. and Waalkes,T.P. (1975) Interaction between aromatic diamines and nucleic acids: possible implications for chemotherapy. J. Natl Cancer Inst., 54, 305–309. [PubMed] [Google Scholar]
- 8.Nunn C.M., Jenkins,T.C. and Neidle,S. (1993) Crystal structure of d(CGCGAATTCGCG) complexed with propamidine, a short-chain homologue of the drug pentamidine. Biochemistry, 32, 13838–13843. [DOI] [PubMed] [Google Scholar]
- 9.Edwards K.J., Jenkins,T.C. and Neidle,S. (1992) Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry, 31, 7104–7109. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins T.C. and Lane,A.N. (1997) AT selectivity and DNA minor groove binding: modelling, NMR and structural studies of the interactions of propamidine and pentamidine with d(CGCGAATTCGCG)2. Biochim. Biophys. Acta, 1350, 189–204. [DOI] [PubMed] [Google Scholar]
- 11.Tidwell R.R., Jones,S.K., Geratz,J.D., Ohemeng,K.A., Cory,M. and Hall,J.E. (1990) Analogues of 1,5-bis(4-amidiniumphenoxy)pentane (pentamidine) in the treatment of experimental Pneumocystis carinii pneumonia. J. Med. Chem., 33, 1252–1257. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y. and Leibowitz,M.J. (1993) Variation and in vitro splicing of group I introns in rRNA genes of Pneumocystis carinii. Nucleic Acids Res., 21, 2415–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Tidwell,R.R. and Leibowitz,M.J. (1994) Inhibition of in vitro splicing of a group I intron of Pneumocystis carinii. J. Eukaryot. Microbiol., 41, 31–38. [DOI] [PubMed] [Google Scholar]
- 14.Miletti K.E. and Leibowitz,M.J. (2000) Pentamidine inhibition of group I intron splicing in Candida albicans correlates with growth inhibition. Antimicrob. Agents Chemother., 44, 958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Bell,A., Perlman,P.S. and Leibowitz,M.J. (2000) Pentamidine inhibits mitochondrial intron splicing and translation in Saccharomyces cerevisiae. RNA, 6, 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter F., Vicens,Q. and Westhof,E. (1999) Aminoglycoside-RNA interactions. Curr. Opin. Chem. Biol., 3, 694–704. [DOI] [PubMed] [Google Scholar]
- 17.von Ahsen U. and Noller,H.F. (1993) Footprinting the sites of interaction of antibiotics with catalytic group I intron RNA. Science, 260, 1500–1503. [DOI] [PubMed] [Google Scholar]
- 18.Hoch I., Berens,C., Westhof,E. and Schroeder,R. (1998) Antibiotic inhibition of RNA catalysis: neomycin B binds to the catalytic core of the td group I intron displacing essential metal ions. J. Mol. Biol., 282, 557–569. [DOI] [PubMed] [Google Scholar]
- 19.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a Group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 20.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Szewczak,A.A., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) RNA tertiary structure mediation by adenosine platforms. Science, 273, 1696–1699. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y. and Leibowitz,M.J. (2001) Folding of the group I intron ribozyme from the 26S rRNA gene of Candida albicans. Nucleic Acids Res., 29, 2644–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y. (1999) Group I introns: potential antimicrobial targets. PhD dissertation, UMDNJ Graduate School of Biomedical Sciences, Piscataway, NJ.
- 23.Emerick V.L. and Woodson,S.A. (1994) Fingerprinting the folding of a group I precursor RNA. Proc. Natl Acad. Sci. USA, 91, 9675–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mercure S., Montplaisir,S. and Lemay,G. (1993) Correlation between the presence of a self-splicing intron in the 25S rDNA of C.albicans and strains susceptibility to 5-fluorocytosine. Nucleic Acids Res., 21, 6020–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golden B.L., Gooding,A.R., Podell,E.R. and Cech,T.R. (1998) A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science, 282, 259–264. [DOI] [PubMed] [Google Scholar]
- 26.Pan J., Thirumalai,D. and Woodson,S.A. (1999) Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc. Natl Acad. Sci. USA, 96, 6149–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rook M.S., Treiber,D.K. and Williamson,J.R. (1999) An optimal Mg2+ concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl Acad. Sci. USA, 96, 12471–12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bielawski K., Galicka,A., Bielawska,A. and Sredzinska,K. (2000) Inhibitory effects of pentamidine analogues on protein biosynthesis in vitro. Acta Biochim. Pol., 47, 113–120. [PubMed] [Google Scholar]
- 29.Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 30.Murphy F.L. and Cech,T.R. (1993) An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry, 20, 5291–5300. [DOI] [PubMed] [Google Scholar]
- 31.Cech T.R., Damberger,S.H. and Gutell,R.R. (1994) Representation of the secondary and tertiary structure of group I introns. Nature Struct. Biol., 1, 273–280. [DOI] [PubMed] [Google Scholar]
- 32.Cech T.R. (1990) Self-splicing of group I introns. Annu. Rev. Biochem., 59, 543–568. [DOI] [PubMed] [Google Scholar]
- 33.Michel F. and Ferat,J.-C. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 34.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 35.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 36.Cech T.R. (2000) The ribosome is a ribozyme. Science, 289, 878. [DOI] [PubMed] [Google Scholar]