Abstract
Although telomeres are not recognized as double-strand breaks (DSBs), some DSB repair proteins are present at telomeres and are required for telomere maintenance. To learn more about the telomeric function of proteins from the homologous recombination (HR) and non-homologous end joining pathways (NHEJ), we have screened a panel of chicken DT40 knockout cell lines for changes in telomere structure. In contrast to what has been observed in Ku-deficient mice, we found that Ku70 disruption did not result in telomere–telomere fusions and had no effect on telomere length or the structure of the telomeric G-strand overhang. G-overhang length was increased by Rad51 disruption but unchanged by disruption of DNA-PKcs, Mre11, Rad52, Rad54, XRCC2 or XRCC3. The effect of Rad51 depletion was unexpected because gross alterations in telomere structure have not been detected in yeast HR mutants. Thus, our results indicate that Rad51 has a previously undiscovered function at vertebrate telomeres. They also indicate that Mre11 is not required to generate G-overhangs. Although Mre11 has been implicated in overhang generation, overhang structure had not previously been examined in Mre11-deficient cells. Overall our findings indicate that there are significant species-specific differences in the telomeric function of DSB repair proteins.
INTRODUCTION
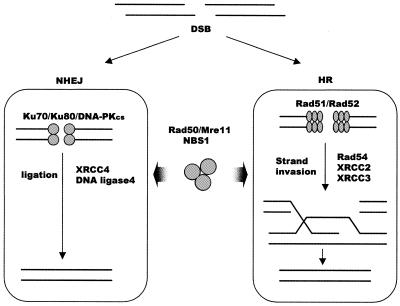
Although a telomere and a double-strand break (DSB) both correspond to the end of a DNA molecule, eukaryotic cells respond very differently to these two types of DNA terminus. DSBs are quickly recognized as a threat to genome integrity and cause cell cycle arrest and/or DNA repair. Because repair of DSBs is such a fundamental mechanism of genome protection, cells have two well-conserved repair pathways (see Fig 1): homologous recombination (HR) and non-homologous end joining (NHEJ) (1,2). Unlike DSBs, telomeres are stable structures that appear to be invisible to the DNA repair machinery (3). They exist as specialized DNA–protein complexes that protect the DNA terminus from unwanted DNA processing activities such as nucleases, but allow access to enzymes such as telomerase that are required for telomere maintenance.
Figure 1.
Proteins involved in DSB repair in vertebrate cells. During NHEJ the DNA-PK complex, which consists of the Ku70/Ku80 heterodimer and the DNA-PK catalytic subunit, binds the broken ends and facilitates rejoining by DNA ligase IV and XRCC4. HR is carried out by proteins in the Rad52 epistasis group. These include Rad51, Rad52, Rad54 and the Rad51 paralogs Rad51B, Rad51C, Rad51D, XRCC2, XRCC3, Rad50, Mre11 and Nbs1. During the repair process, Rad51 aided by other accessory factors catalyzes strand exchange with a homologous DNA molecule. The Rad50/Mre11/Nbs1 complex, which has nuclease and DNA-unwinding activities, participates in both HR and NHEJ in ways that are still not fully understood.
In vertebrates, telomeric DNA consists of tandem repeats of the sequence T2AG3·C3TA2 and the length of the telomeric tract is maintained around a fixed average value (e.g. ∼5–15 kb in humans) (4). The G-rich strand extends beyond the C-rich strand to generate a 3′ overhang of several hundred nucleotides. This overhang may be protected by single-strand telomere-binding proteins or incorporated into a t-loop, a lariat-like structure formed by the overhang invading the duplex portion of the telomeric tract (5,6). As telomeric DNA cannot be completely replicated by conventional DNA polymerases, telomere length is normally maintained by telomerase, an unusual reverse transcriptase that uses its RNA subunit to template addition of new telomeric repeats (7). However, in the absence of telomerase, telomeres can sometimes be maintained by a recombination-based pathway (8). In yeast this pathway utilizes proteins in the Rad52 epistasis group (9).
The difference in cellular response to a telomere versus a DSB initially suggested that there would be little overlap between the proteins that package telomeric DNA and those used to process DSBs. While many unique telomere proteins have been identified, it is now apparent that at least a subset of the DSB repair proteins also associate with telomeres in both vertebrates and yeast (3,10). Deletion or mutation of these repair proteins causes a variety of telomere defects, including telomere shortening, alterations to the terminal DNA structure and end-to-end fusions, indicating that they are required for telomere maintenance. However, we do not know why DSB repair proteins are needed at telomeres or which of the many HR and NHEJ proteins associate with telomeres rather than functioning only in repair. To learn more about this aspect of telomere biology in vertebrate cells, we have used a panel of chicken DT40 knockout cell lines to examine how telomeric DNA structure is affected by the removal of specific HR or NHEJ proteins.
Chicken DT40 cells are useful for dissecting vertebrate gene function via gene disruption because this B cell line exhibits an unusually high efficiency of gene conversion and targeted integration (11). The panel of knockout cell lines used for this study was generated to determine the role of specific HR and NHEJ proteins in DSB repair (12). While all the cell lines exhibit deficiencies in DNA repair (Table 1), the effect of the gene disruptions on telomere maintenance had not been examined. For some of the repair proteins (Rad51, Rad54 and the Rad51 paralogs XRCC2 and XRCC3) this was the first time that a role in vertebrate telomere biology had been sought, whereas for other proteins (Ku, DNA-PKcs and Mre11) there was good reason to suspect that a gene disruption might affect telomere structure because their homologs were known to associate with mammalian telomeres and/or affect telomere structure in yeast. For example, loss of Ku and DNA-PKcs from mammalian cells results in end-to-end fusion of chromosomes (13–16), while loss of Saccharomyces cerevisiae Ku results in telomere shortening and resection of the C-strand to create long G-strand overhangs (17–19). Mre11 is present at vertebrate telomeres (20) and in yeast has been shown to function in the telomerase-mediated pathway of telomere replication (21). As Mre11 has endo and exonuclease activity, and the yeast Mre11/Rad50/Xrs1 complex increases 5′→3′ resection of DSBs, this protein has been proposed to participate in formation of telomeric G-strand overhangs (21–24).
Table 1. Phenotypes of DT40 cell lines deficient for DSB repair proteins.
| Gene | Viability | Chromosome breaks | DNA damage sensitivity | Rad51 focus formation | DSB repair | Reference | |
|---|---|---|---|---|---|---|---|
| IR | X-linking | ||||||
| Ku70 | Viable | No | Yes | No | Yes | NHEJ defective | (31) |
| DNA-PKcs | Viable | NHEJ defective | (33) | ||||
| Mre11 | Lethal | Yes +++a | Yes | Yes | HR defective | (32) | |
| Rad51 | Lethal | Yes ++++ | No | HR defective | (28) | ||
| Rad52 | Viable | No | No | No | Yes | Almost wild-type | (29) |
| Rad54 | Viable | Yes + | Yes | Yes | Enhanced | HR defective | (31,48) |
| XRCC2 | Viable | Yes ++ | Yes | Yes | Defective | HR defective | (47) |
| XRCC3 | Viable | Yes ++ | Yes | Yes | Defective | HR defective | (47) |
a++++ denotes most and + fewest breaks.
Chicken telomeres are structurally very similar to mammalian telomeres. Like human telomeres they consist of 5–15 kb of T2AG3 repeats, end with a 3′ G-strand overhang and are maintained by telomerase (25). Moreover, telomere proteins such as TRF1, TRF2, Pot1, RAP1 and tankyrase are well conserved between chickens and humans (25–27; C.Wei, M.De Rycker, M.Tan and C.Price, unpublished results). To determine whether the DSB repair proteins Ku70, DNA-PKcs, Mre11, Rad51, Rad52, Rad54, XRCC2 and XRCC3 play a role in telomere maintenance in chicken cells, we analyzed G-overhang structure and/or telomere length in the corresponding DT40 knockout cell lines. Our findings indicate that several of these proteins play a different role at vertebrate telomeres from what had been predicted based on discoveries made in yeast.
MATERIALS AND METHODS
Cell culture and cell lines
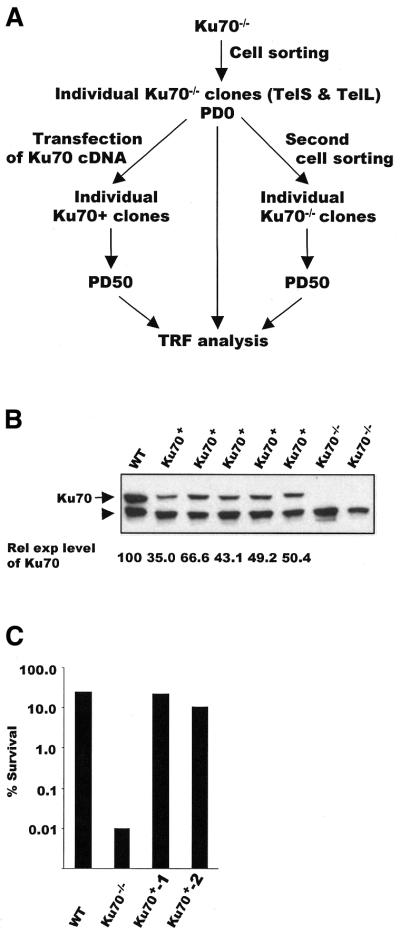
Wild-type DT40 and the various knockout cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, 10 µM β-mercaptoethanol, glutamine, penicillin and streptomycin. The Rad51–/–, Rad52–/–, Rad54–/–, Ku70–/–, DNA-PKcs–/–, Mre11–/–, XRCC2–/– and XRCC3–/– cell lines were as described previously (28–33). The Rad54–/–, Rad51–/– and Mre11–/– conditional cell lines contained human Rad54 or chicken Rad51 and Mre11 under control of a tetracycline (tet)-repressible promoter (28,30,32). Expression of the rescuing allele was repressed by growth in 100 ng/ml doxycycline. Ku70–/– clones with short (TelS) and long (TelL) telomeres were established by using cell sorting to isolate single cells from the parental Ku70–/– cell line (Fig. 2A). Ku70+ clones were generated by subcloning flag-tagged full-length chicken Ku70 cDNA (FcKu70) into pcDNA3 (Invitrogen) and transfecting the construct into Ku70–/– TelL or TelS clones. Expression of FcKu70 was verified by western blot analysis using an antibody to chicken Ku70. The relative level of FcKu70 expression was determined by quantifying the amount of Ku70 signal from wild-type and Ku70+ cells. The corresponding signal from a cross-reacting band was used to normalize for the amount of protein loaded in each lane. The tTA77 cell line was established by transfecting wild-type DT40 with a plasmid containing the VP16TetRepressor transgene (34) and the PuroR gene and selecting with puromycin (T.de Lange and B.Li, personal communication).
Figure 2.
Isolation of Ku70+ and Ku70–/– clones. (A) Scheme for isolating individual Ku70 clones. (B) Western blot showing expression of FcKu70 in Ku–/– cells transfected with FcKu70 cDNA. Lane 1, wild-type DT40; lanes 2–6, Ku+ TelL clones; lanes 7 and 8, Ku70–/– cells. The arrow marks the position of Ku70 and FcKu70; the arrowhead marks a cross-reacting band that was used to normalize for the amount of protein loaded in each lane. The level of FcKu70 expression relative to wild-type cKu70 expression is given below each lane. (C) Rescue of γ-radiation sensitivity by FcKu70 expression. Cells were treated with 2 Gy γ-radiation at the indicated times following release from nocodazole arrest. The fraction of colonies surviving compared with non-irradiated controls of the same genotype are shown for the 5 h time point. Ku70+-1 and Ku70+-2 represent two different Ku+ TelL clones.
Measurement of γ-radiation sensitivity
Cells were enriched in prometaphase by growth in 0.5 µg/ml nocodazole for 7 h. The cells were washed three times with pre-warmed medium and 75, 750 or 7500 cells were plated into 6-well microtiter dishes containing 5 ml/well 1.5% methylcellulose in DMEM/F-12. The cells were γ-irradiated with 2 Gy 3–7 h later using a 137Co source (Gammacel 40; Atomic Energy of Canada). Colonies were counted 7 days later. Percent survival was calculated based on the number of colonies in untreated samples.
Karyotype analysis
Cells were grown in medium containing 1 mg/ml colcemid for 3 h then harvested and swelled in 0.9% sodium citrate for 15 min at room temperature. Following fixation in 5 ml freshly prepared methanol/acetic acid (3:1), the cell suspension was dropped onto glass slides, flame dried and stained with 4% Giemsa solution at pH 6.4 for 10 min (28).
Measurement of telomere length
Genomic DNA was isolated from wild-type and knockout cell lines using the Amersham-Pharmacia genomic-prep cell and tissue DNA isolation kit and digested with MspI, HinfI and HaeIII. The DNA (10 µg/lane) was separated in 0.7% agarose gels, the gels were dried and telomeric restriction fragments were detected by non-denaturing in-gel hybridization with 32P-labeled (C3TA2)4 probe (25). The signal in each lane was measured by PhosphorImager analysis; each lane was divided into 50 separate boxes and the signal in each box was determined along with its position in base pairs. The weighted mean telomere length was determined using the formula LWM = [∑(Li × Sigi)]/∑Sig, where LWM is the mean length (in bp) of the telomere signal, Li is the length in bp of an individual box and Sigi is the signal intensity of that box (35,36).
G-overhang analysis
Following digestion with MspI, HaeIII and HinfI, duplicate samples of genomic DNA were separated in 0.7% gels until the Orange-G loading dye reached the bottom of the gel. The bottom portion of the gel containing DNA fragments <800 bp was cut away from the rest of the gel, denatured, the DNA transferred to nylon membrane and the membrane hybridized with 32P-labeled DT40 genomic DNA. The upper portion of the gel (which contained the telomeric restriction fragments) was subjected to in-gel hybridization with a (C3TA2)4 probe as described above. The signal resulting from hybridization to the G-overhang was determined for each lane by PhosphorImager analysis. The corresponding signal from a defined segment of the genomic DNA blot was used to normalize for the amount of DNA loaded in each lane. The relative G-overhang signal was then calculated by comparing the level of signal obtained with DNA from each knockout cell line to the level obtained with DNA from cells expressing a rescuing allele or wild-type cells run on the same gel. A mean relative overhang signal intensity was calculated for each cell line by using the values obtained from several DNA samples run on multiple gels.
RESULTS
Effect of Ku70 deletion on telomere length and telomere fusions
Although loss of Ku causes telomere shortening in yeast, the effect of Ku deficiency on mammalian telomere length is still controversial (14,16,17). Consequently, we decided to investigate whether Ku regulates telomere length in chicken cells. We were unable to generate a conditional Ku70-deficient cell line so we instead re-introduced flag-tagged chicken Ku70 cDNA (FcKu70) into Ku70–/– cells under control of the CMV promotor and compared the length of the telomeres from the resulting Ku70+ and the parental Ku70–/– cells. DT40 cells resemble certain human cell lines in that they show considerable cell-to-cell variation in telomere length (C.Price and C.Wei, unpublished results; 37), so a culture that has been propagated for several years can contain some cells with telomeres that are only 3–4 kb and others with telomeres of 10–15 kb. To address the issue of clonal variation, we established a series of single cell clones from the parental Ku70–/– cell line by cell sorting (Fig. 2A). We then introduced the FcKu70 expression construct into one clone with short ∼3 kb telomeres (TelS) and one with longer 5–6 kb telomeres (TelL).
Following transfection of the expression construct, five of the TelL and three of the TelS Ku70-expressing clones were cultured for 50 population doublings (PD), DNA was then isolated for telomere length analysis. To ensure that each clone expressed FcKu70, the level of expression was examined by western blotting at the time of DNA isolation. As shown in Figure 2B, the FcKu70 was generally expressed at lower levels than wild-type Ku70. We therefore tested for functional complementation in the Ku+ clones by measuring the percentage of cells that survived relative to wild-type DT40 following γ-irradiation. Since Ku–/– cells show greatly increased sensitivity to γ-irradiation in G1/early S phase (31), cells were partially synchronized in prometaphase by treatment with nocodazole, released from the nocodazole block and irradiated 3–7 h later (31). Despite the lower level of FcKu70 expression, the Ku+ clones were almost as radiation resistant as the wild-type DT40 and were 100–1000-fold more resistant than the Ku70–/– cells (Fig. 2C). Thus, expression of the FcKu70 was sufficient to rescue DNA repair by NHEJ.
To monitor the extent to which telomere length varied in the Ku70–/– cells during prolonged cell culture, single cell clones were established from the parental Ku70–/– TelL and TelS clones by another round of cell sorting (Fig. 2A). These were also cultured for 50 PD prior to DNA isolation. Finally, DNA was isolated from the parental Ku70–/– TelL or TelS cultures at the time of transfection (PD0). The telomeric restriction fragments were identified by non-denaturing in-gel hybridization using a probe to the G-strand overhang. This approach prevents hybridization to the abundant interstitial T2AG3 sequence that is present on many chicken chromosomes (25). The weighted mean telomere length was calculated for each sample following PhosphorImager analysis of the hybridization signal.
When the telomere length was determined for each set of Ku70+ or Ku70–/– clones, a ≤10% difference in length was found between the individual clones within a set (Fig. 3A–C). For example, the telomere length was 2970 bp for the Ku70–/– TelS clone with the longest telomeres and 2880 bp for the Ku70–/– TelS clone with the shortest telomeres (Fig. 3C). Thus, 50 PD is not sufficient for clonal variation in telomere length to become apparent. Comparison of the average telomere length of the Ku70–/– and Ku70-expressing clones revealed that even the clones with the highest levels of Ku70 expression showed no significant difference in telomere length from the Ku70–/– cells at PD50 or from the parental Ku70–/– cells at PD0. The telomeres of the TelS Ku70–/– and Ku70+ clones were essentially identical to those from the TelS parental cells. The telomeres of the TelL Ku70–/– clones were slightly longer than those of the parental TelL cells, but this difference was not statistically significant. Although the Ku70+ clones expressed lower levels of Ku70 than wild-type DT40, 50 PD should have been sufficient for any Ku-mediated changes in telomere length to become apparent. Since no such changes were observed, our results indicate that Ku70 does not regulate telomere length in chicken cells.
Figure 3.
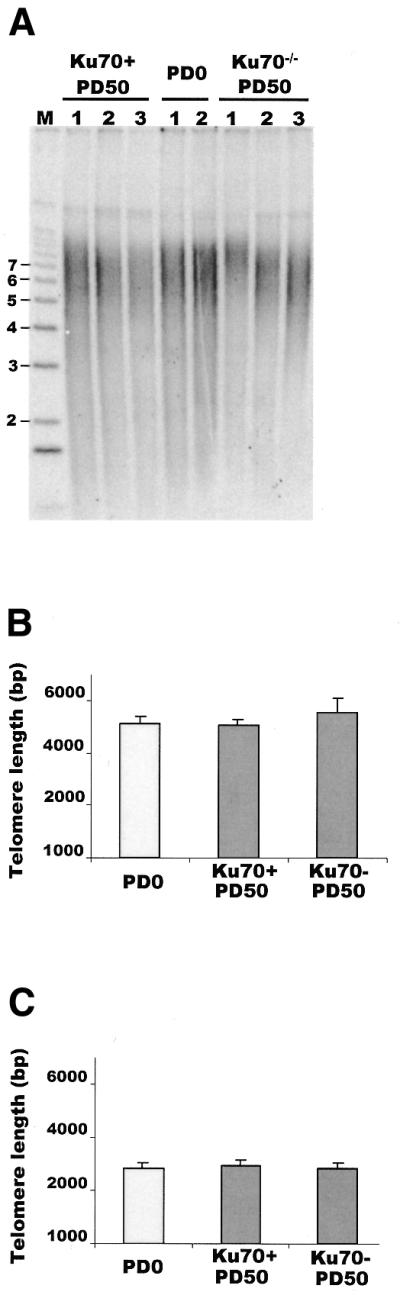
Effect of Ku70 deficiency on telomere length. (A) Telomere length analysis of Ku70–/– and Ku70+ TelL clones. Telomeric restriction fragments were identified by in-gel hybridization to 32P-labeled (C3TA2)4 probe and the weighted mean telomere length was calculated following quantification of the hybridization signal by PhosphorImager. Size (in kb) of DNA markers is shown to the left. (B) Histogram showing telomere lengths in Ku70–/– and Ku70+ TelL clones. Column 1, telomere length of parental Ku70–/– TelL clone at PD0; column 2, average telomere length at PD50 of five Ku70+ TelL clones; column 3, average telomere length at PD50 of five Ku70–/– TelL clones. (C) Histogram showing telomere lengths in Ku70–/– and Ku70+ TelS clones. Column 1, average telomere length of parental Ku70–/– TelS clone at PD0; column 2, average telomere length at PD50 of three Ku70+ TelS clones; column 3, average telomere length at PD50 of three Ku70–/– TelS clones.
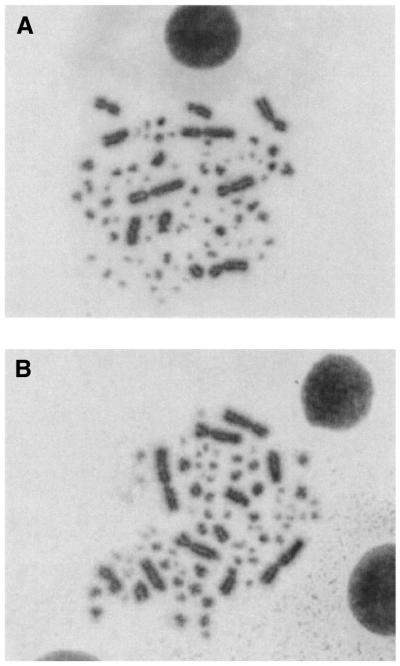
Cytogenetic analysis of chromosomes from Ku70–/– DT40 cells had previously revealed that Ku deficiency does not cause an increase in spontaneous chromosome breaks (31). Further examination of metaphase spreads from Ku70–/– cells revealed that end-to-end fusions are also absent and the karyotype is completely normal (Fig. 4). This contrasts with the situation in mouse cells where Ku deficiency causes end-to-end fusion of chromosomes (14,16).
Figure 4.
Representative metaphase spreads from wild-type and Ku70–/– cells. Chromosomes were Giemsa stained. Note the lack of end-to-end fusions in the Ku–/– cells. (A) Wild-type cells. (B) Ku70–/–.
Effect of Ku70 deficiency on G-overhang length
Since deletion of yeast Ku70 causes loss of telomere end protection and a dramatic change in G-strand overhang structure (18,19), we wished to determine whether a deficiency in Ku70 causes a similar effect in vertebrate cells. To detect changes in overhang structure we made use of a ‘G-overhang’ assay that compares how much 32P-labeled (C3TA2)4 oligonucleotide can hybridize to the G-strand overhang on telomeric restriction fragments from different cell lines. The amount of hybridization is proportional to the length of the overhang. This assay has been used extensively in yeast and mammalian cells to identify telomere-binding and replication proteins that affect G-overhang structure (14,15,18,38,39), and will detect gross changes in overhang length (e.g. a 50% increase in length), such as those observed in a yeast Ku70 knockout or human TRF1 mutant cell lines (18,38).
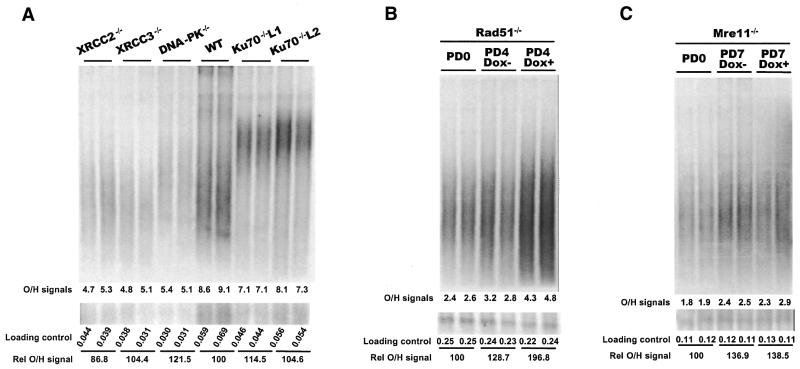
Genomic DNA from wild-type DT40, two separate clones with short telomeres (Ku70–/– TelS) and two with longer telomeres (Ku70–/– TelL), was separated on agarose gels and the telomeric restriction fragments were identified by non-denaturing in-gel hybridization to the (C3TA2)4 probe (Fig. 5A and Table 2) (25,40). The relative amount of overhang available for hybridization was then determined for each cell line by using a PhosphorImager to quantify the total hybridization signal in a lane and normalizing this signal to the amount of genomic DNA loaded in that lane. The normalized hybridization signal from each knockout cell line (Fig. 5A, lanes 9–12) was then compared with the signal obtained with a sample of wild-type DT40 DNA run on the same gel (Fig. 5A, lanes 7–8). To increase the accuracy of the assay, two separate DNA samples were isolated from each knockout cell line and each sample was run in duplicate on two gels. To ensure that the probe was only hybridizing to G-strand overhangs and not regions of duplex telomeric DNA, control samples were digested with the 3′→5′ nucleases exonuclease 1 or T4 DNA polymerase prior to the hybridization step. This treatment abolished all hybridization (25) (data not shown).
Figure 5.
Comparison of G-strand overhang length by in-gel hybridization. The upper panels show in-gel hybridization of 32P-labeled (C3AT2)4 probe to genomic DNA from DT40 cells deficient for various DSB repair proteins. The lower panels show loading controls. The bottom portion of each gel was removed before the in-gel hybridization, denatured, blotted to membrane and hybridized with a probe for total genomic DNA. (A) Ku70-, XRCC2-, XRCC3- and DNA-PKcs-deficient cell lines. As none of these cell lines contained a conditional allele, the hybridization signal for each mutant cell line was compared with the signal from wild-type samples run on the same gel (lanes 7 and 8). Lanes 9 and 10, and 11 and 12, contain samples from two different Ku70–/– clones with long telomeres. The total amount of G-overhang signal (×10–6) from the PhosphorImager analysis is given at the bottom of the upper panel while the total signal from the loading control is given at the bottom of the lower panel. The relative G-overhang signal for each lane is given as a percentage of the wild-type signal. (B) Rad51 conditional cells. Lanes 1 and 2, cells isolated at PD0; lanes 3 and 4, cells grown without doxycycline isolated at PD4; lanes 5 and 6, cells grown with doxycycline isolated at PD4. (C) Mre11 conditional cells. Lanes 1 and 2, cells isolated at PD0; lanes 3 and 4, cells grown without doxycycline isolated at PD7; lanes 5 and 6, cells grown with doxycycline isolated at PD7.
Table 2. Relative G-strand overhang signals from DT40 knockout cell lines Ku70–/–, Rad52–/–, XRCC2–/–, XRCC3–/– and DNA-PKcs–/–
| Cell line | G-overhang signal intensity (%) |
|---|---|
| Wild-type | 100 |
| Ku70–/– (TelL) | 109 ± 12 |
| Ku70–/– (TelS) | 93 ± 12 |
| Rad52–/– | 96 ± 24 |
| XRCC2–/– | 106 ± 21 |
| XRCC3–/– | 89 ± 15 |
| DNA-PK–/– | 97 ± 22 |
The G-overhang signal was measured for each lane and normalized to the amount of genomic DNA loaded in that lane. The amount of overhang signal from each knockout cell line was compared with the signal from wild-type DT40 DNA run in duplicate on the same gel. Ku70–/– (TelS), Ku70-deficient clones with short (∼3 kb) telomeres; Ku70–/– (TelL), Ku70-deficient clones with longer (5–6 kb) telomeres. The standard deviations refer to the variation in relative G-overhang signal obtained from at least two DNA preparations run in duplicate on two different gels.
Analysis of each Ku70–/– clone revealed that loss of Ku70 has no significant effect on overhang length (Table 2). This lack of change in overhang structure was unexpected because the specificity of Ku for DNA termini, together with its physical presence at mammalian telomeres, had suggested a role for Ku in vertebrate telomere maintenance. Moreover, the result with the Ku70–/– DT40 cells is in direct contrast to what has been observed in S.cerevisiae (18,19), where Ku plays an important role in maintaining proper telomere structure.
Effect of other DSB repair proteins on G-overhang length
As Mre11 and other DSB repair proteins also have activities that could affect overhang structure, we next decided to screen a selection of cell lines that had disruptions in the Mre11, Rad51, Rad52, Rad54, XRCC2 , XRCC3 or DNA-PKcs genes for changes in G-overhang length. As before, we made use of the G-overhang hybridization assay to compare the amount of 32P-labeled (C3TA2)4 oligonucleotide that could hybridize to the G-strand overhang on telomeric restriction fragments from different cell lines.
For the Rad51, Rad54 and Mre11 gene knockouts, conditional cell lines were available that had a rescuing allele under control of a tet-repressible promoter and the tet-responsive tTA transcriptional activator (28,30,32). Thus, we were able to compare the relative amount of hybridization with the G-overhang from cells expressing the rescuing allele versus cells where rescuing allele expression was repressed by doxycycline (Fig. 5B and C and Table 3). Northern blots were used to check that repression of the rescuing allele was complete (data not shown). As the Rad54–/– cells continue to divide indefinitely after rescuing allele repression, DNA was isolated from these cells prior to addition of doxycycline (PD0) and after 90 PD with and without doxycycline. Rad51 and Mre11 are essential genes, so the conditional cell lines die in the presence of doxycycline (Table 1). For both these cell lines, DNA was isolated approximately one cell division prior to cell death to ensure telomere replication had taken place when the levels of Rad51 and Mre11 were greatly depleted or absent. Thus, DNA was isolated from the Rad51-deficient cells at PD0 and PD4 and from the Mre11-deficient cells at PD0 and PD7. For each cell line the normalized G-overhang signal from the PD0 sample was set at 100%. As a control, DNA from the tTA transactivator-expressing cell line tTA77 was also examined at PD0 and after 90 PD in the presence or absence of doxycycline. Conditional cell lines were not available for the Rad52, XRCC2, XRCC3 or DNA-PKcs knockout cell lines so we compared the normalized hybridization signal from each knockout cell line with the signal obtained with a sample of wild-type DT40 DNA run on the same gel (Fig. 5A and Table 2).
Table 3. Overhang signals from conditional Rad54–/–, Rad51–/– and Mre11–/– cells.
| Cell line | G-overhang signal intensity (%) | ||
|---|---|---|---|
| Dox– | Dox+ | ||
| tTA77 | PD0 100 | PD90 115 ± 24 | PD90 121 ± 41 |
| Rad54–/– | PD0 100 | PD90 82 ± 14 | PD90 124 ± 35 |
| Rad51–/– | PD0 100 | PD4 115 ± 9 | PD4 179 ± 20 |
| Mre11–/– | PD0 100 | PD7 95 ± 27 | PD7 102 ± 27 |
The amount of overhang signal at PD0 was set at 100% and the overhang signal for subsequent PD was calculated relative to this. Dox– and Dox+, growth in doxycycline for indicated number of population doublings; tTA77, cell line expressing the tTA transactivator. The values for the Rad51–/– cells were derived from three separate DNA samples while the values for the Mre11–/– and Rad54–/– cells were from two DNA samples.
Although most of the gene knockouts had no significant effect on overhang length, Rad51 depletion caused a >1.5-fold increase in the relative G-overhang signal intensity. These results indicate that while the majority of genes tested probably do not play a role in generating or maintaining G-strand overhangs at vertebrate telomeres, Rad51 is somehow involved in this process. The effect of Rad51 depletion on overhang structure is both interesting and surprising because proteins in the HR pathway have not previously been shown to be involved in telomere maintenance in any telomerase-expressing cells. In yeast, deletion or mutation of genes in the Rad52 epistasis group prevents telomeres from being maintained by the alternative non-telomerase-based pathway but gross alterations in telomere structure have not been found (9). The lack of change in overhang structure in the Mre11–/– cells was also notable because this protein is physically present at telomeres and has a nuclease activity that has been implicated in the generation of G-overhangs. Our results indicate that Mre11 is not required to generate structurally normal G-overhangs.
DISCUSSION
We have used a panel of chicken DT40 cell lines to screen an array of DSB repair genes from several different repair pathways for effects on telomere structure. Our results provide new information about the role of DSB repair proteins in vertebrate telomere maintenance. Surprisingly, a number of these proteins play a different role at vertebrate telomeres from what might be predicted based on their in vitro activities or their known function at either yeast telomeres or DSBs. The advantage of using DT40 cell lines for this type of screen is that the only genetic difference between the cell lines lies in the genes that have been intentionally disrupted. This contrasts with the situation in knockout mice where genetic variability is introduced by the back-crosses needed to establish and maintain a mouse colony. Such genetic variability can lead to conflicting results when different laboratories study the effect of a gene disruption using mice that originated from the same founder colony (14,16). An additional advantage of DT40 cells is that the telomeres are similar in length to human telomeres (25), so it is much easier to detect telomere length changes in DT40 cells than in mouse cells, which have much longer telomeres (41).
NHEJ pathway; Ku70 and DNA-PKcs
Studies with knockout mice and mouse embryonic fibroblasts have established that Ku, and to a lesser extent DNA-PKcs, are important for telomere capping because chromosomes from Ku- and DNA-PKcs-deficient cells display an elevated level of end-to-end fusions (13–16). However, the precise role of Ku in end capping is unclear because different studies with the same Ku80 knockout mouse strain (42) have given conflicting results (14,16). In one study, telomere length and G-overhang structure was unchanged and most of the chromosome fusions had telomeric DNA at the fusion site (14). This result is consistent with the loss of telomere capping being caused by disruption of the telomeric protein complex rather than loss of telomeric DNA. However, a second study showed a clear decrease in telomere length in both a Ku80 knockout and a Ku70 knockout, and most of the fused chromosomes were found to lack telomeric DNA at the fusion junction (16). Heterozygous Ku80+/– mice showed a smaller decrease in telomere length and a level of end-to-end fusions that was between that observed in the homozygous Ku80–/– and wild-type mice. The results of this study suggested that a lack of Ku causes loss of telomere capping by causing the telomeres to shorten beyond a critical point. The reason for the discrepancy between the two studies remains unclear. In some ways our results resemble those of the first study, as we found no correlation between telomere length and the level of Ku70 expression. Even those clones expressing the highest levels of Ku70 had the same length telomeres and G-overhang structure as the Ku70–/– cell line. However, it is striking that loss of Ku70 does not cause end-to-end fusion of chromosomes in chicken cells. A similar lack of chromosome fusions has been observed in Ku70–/– Arabidopsis thaliana, although in this case the telomeres undergo a gradual elongation (K.Riha and D.Shippen, personal communication). Thus, it appears that there is significant interspecies variation in the precise manner in which Ku functions at the telomeres of higher eukaryotes. The data with the Ku80–/– mice suggest that Ku function may also be influenced by genetic background.
Mre11 complex
Although the Mre11 complex promotes the formation of 3′ overhangs at DSBs, its role at telomeres is unclear because the Mre11 3′→5′ exonuclease activity is the wrong polarity to generate 3′ overhangs. Thus, the Mre11 complex has been suggested to promote C-strand resection, either via the Mre11 endonuclease activity or by recruitment of a separate 5′→3′ exonuclease (22,23). However, recent studies suggest that the Mre11 complex may instead be important for recruiting telomerase to the telomere (43). Despite the controversy surrounding the role of Mre11 at telomeres, our examination of telomere structure in Mre11-deficient chicken cells provides the first direct analysis of the effect of Mre11 depletion on G-overhang structure.
If Mre11 played a major role in generating the G-overhangs on vertebrate telomeres, changes in overhang structure would occur in a knockout cell line as soon as DNA replication proceeded in the presence of limiting Mre11 levels. Thus, we should have seen a decrease in the amount of G-strand overhang signal from the conditional Mre11–/– DT40 cell line when DNA was isolated 7 PD after repression of the rescuing allele. Our observation that Mre11 depletion has no detectable effect on G-overhang length indicates that Mre11 is not required to generate normal G-strand overhangs. However, our results do not indicate that Mre11 has no function in overhang generation because another exonuclease (e.g. exonuclease 1) might be able to substitute for Mre11 in the knockout cell line (44).
Rad51 and the HR pathway
Rad51, the vertebrate homolog of Escherichia coli RecA, is a key protein in HR as it forms the filament on single-stranded DNA that promotes strand invasion (1). Our finding that loss of Rad51 causes G-overhang elongation is surprising as there is currently no evidence that the protein normally binds to the G-overhang or is part of the telomeric DNA–protein complex. However, yeast that lack telomerase can maintain their telomeres by two alternative recombination-based pathways, one of which is dependent on Rad51 (45,46). Thus, under some circumstances Rad51 clearly can gain access to the telomeric DNA. Our results indicate that in vertebrate cells, Rad51 acts at the telomere during normal telomere maintenance in telomerase-positive cells and not just when telomeres are maintained by a recombination-based pathway. Intriguingly, we have found that Rad54, a protein which acts in the same recombination pathway as Rad51, may also influence telomere structure. When DT40 cells become depleted for Rad54, the telomeres show a small but consistent increase in telomere length (∼16 bp/PD; C.Wei and C.Price, unpublished data). This change in telomere length is too small to say conclusively that Rad54 plays a role in telomere length regulation, but the data fit with our observation that Rad51, another protein in the HR pathway, has a telomeric function.
It is noteworthy that the XRCC2 and XRCC3 knockouts have no effect on G-overhang structure because these Rad51 paralogs facilitate the action of Rad51 during DSB repair (47) and hence might have been expected to also affect Rad51 function at the telomere. However, our results suggest that at least a subset of the Rad51 paralogs are unnecessary for this aspect of Rad51 activity. This reinforces our finding that while some DSB repair proteins clearly have a telomeric function, both their role and mechanism of action seem to differ significantly at telomeres and DSBs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Titia de Lange and Bibo Li for their kind gift of the tTA77 cell line, Ranganathan Venkatesan for performing preliminary experiments, K. Arumaganathan for help with cell sorting, Michelle Zeleny for assistance with the γ-irradiation experiments and Chad Price and members of the Price and Shippen laboratories for helpful comments. This work was supported by grant RO1 AG17212 to C.M.P. from the National Institutes of Health.
REFERENCES
- 1.Karran P. (2000) DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev., 10, 144–150. [DOI] [PubMed] [Google Scholar]
- 2.Khanna K.K. and Jackson,S.P. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genet., 27, 247–254. [DOI] [PubMed] [Google Scholar]
- 3.Lundblad V. (2000) DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res., 451, 227–240. [DOI] [PubMed] [Google Scholar]
- 4.Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- 5.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 6.Baumann P. and Cech,T.R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science, 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- 7.Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- 8.Dunham M.A., Neumann,A.A., Fasching,C.L. and Reddel,R.R. (2000) Telomere maintenance by recombination in human cells. Nature Genet., 26, 447–450. [DOI] [PubMed] [Google Scholar]
- 9.Kass-Eisler A. and Greider,C.W. (2000) Recombination in telomere-length maintenance. Trends Biochem. Sci., 25, 200–204. [DOI] [PubMed] [Google Scholar]
- 10.Shore D. (2001) Telomeric chromatin: replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev., 11, 189–198. [DOI] [PubMed] [Google Scholar]
- 11.Lahti J.M. (1999) Use of gene knockouts in cultured cells to study apoptosis. Methods, 17, 305–312. [DOI] [PubMed] [Google Scholar]
- 12.Sonoda E., Takata,M., Yamashita,Y.M., Morrison,C. and Takeda,S. (2001) Homologous DNA recombination in vertebrate cells. Proc. Natl Acad. Sci. USA, 98, 8388–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilley D., Tanaka,H., Hande,M.P., Kurimasa,A., Li,G.C., Oshimura,M. and Chen,D.J. (2001) DNA-PKcs is critical for telomere capping. Proc. Natl Acad. Sci. USA, 98, 15084–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samper E., Goytisolo,F.A., Slijepcevic,P., van Buul,P.P. and Blasco,M.A. (2000) Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep., 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goytisolo F.A., Samper,E., Edmonson,S., Taccioli,G.E. and Blasco,M.A. (2001) The absence of the DNA-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol., 21, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.d’Adda di Fagagna F., Hande,M.P., Tong,W., Roth,D., Lansdorp,P.M., Wang,Z. and Jackson,S.P. (2001) Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol., 11, 1192–1196. [DOI] [PubMed] [Google Scholar]
- 17.Boulton S.J. and Jackson,S.P. (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double-strand break rejoining and in telomeric maintenance. Nucleic Acids Res., 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 19.Polotnianka R.M., Li,J. and Lustig,A.J. (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X.D., Kuster,B., Mann,M., Petrini,J.H. and Lange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nature Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]
- 21.Nugent C.I., Bosco,G., Ross,L.O., Evans,S.K., Salinger,A.P., Moore,J.K., Haber,J.E. and Lundblad,V. (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol., 8, 657–660. [DOI] [PubMed] [Google Scholar]
- 22.Diede S.J. and Gottschling,D.E. (2001) Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol., 11, 1336–1340. [DOI] [PubMed] [Google Scholar]
- 23.Haber J.E. (1998) The many interfaces of Mre11. Cell, 95, 583–586. [DOI] [PubMed] [Google Scholar]
- 24.Hopfner K.P., Putnam,C.D. and Tainer,J.A. (2002) DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol., 12, 115–122. [DOI] [PubMed] [Google Scholar]
- 25.Venkatesan R.N. and Price,C. (1998) Telomerase expression in chickens: constitutive activity in somatic tissues and down-regulation in culture. Proc. Natl Acad. Sci. USA, 95, 14763–14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konrad J.P., Mills,W., Easty,D.J. and Farr,C.J. (1999) Cloning and characterization of the chicken gene encoding the telomeric protein TRF2. Gene, 239, 81–90. [DOI] [PubMed] [Google Scholar]
- 27.Price C.M. (2001) How many proteins does it take to make a telomere? Trends Genet., 17, 437–438. [Google Scholar]
- 28.Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi-Iwai Y., Sonoda,E., Buerstedde,J.M., Bezzubova,O., Morrison,C., Takata,M., Shinohara,A. and Takeda,S. (1998) Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol., 18, 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison C., Sonoda,E., Takao,N., Shinohara,A., Yamamoto,K. and Takeda,S. (2000) The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J., 19, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takata M., Sasaki,M.S., Sonoda,E., Morrison,C., Hashimoto,M., Utsumi,H., Yamaguchi-Iwai,Y., Shinohara,A. and Takeda,S. (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J., 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi-Iwai Y., Sonoda,E., Sasaki,M.S., Morrison,C., Haraguchi,T., Hiraoka,Y., Yamashita,Y.M., Yagi,T., Takata,M., Price,C. et al. (1999) Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J., 18, 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukushima T., Takata,M., Morrison,C., Araki,R., Fujimori,A., Abe,M., Tatsumi,K., Jasin,M., Dhar,P.K., Sonoda,E. et al. (2001) Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem., 276, 44413–44418. [DOI] [PubMed] [Google Scholar]
- 34.Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy M.Z., Allsopp,R.C., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere end-replication problem and cell aging. J. Mol. Biol., 225, 951–960. [DOI] [PubMed] [Google Scholar]
- 36.Ouellette M.M., Liao,M., Herbert,B.S., Johnson,M., Holt,S.E., Liss,H.S., Shay,J.W. and Wright,W.E. (2000) Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem., 275, 10072–10076. [DOI] [PubMed] [Google Scholar]
- 37.Bryan T.M., Englezou,A., Dunham,M.A. and Reddel,R.R. (1998) Telomere length dynamics in telomerase-positive immortal human cell populations. Exp. Cell Res., 239, 370–378. [DOI] [PubMed] [Google Scholar]
- 38.van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 39.Parenteau J. and Wellinger,R.J. (1999) Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol., 19, 4143–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wellinger R.J., Ethier,K., Labrecque,P. and Zakian,V.A. (1996) Evidence for a new step in telomere maintenance. Cell, 85, 423–433. [DOI] [PubMed] [Google Scholar]
- 41.Blasco M.A., Lee,H.W., Hande,M.P., Samper,E., Lansdorp,P.M., DePinho,R.A. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- 42.Zhu C., Bogue,M.A., Lim,D.S., Hasty,P. and Roth,D.B. (1996) Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell, 86, 379–389. [DOI] [PubMed] [Google Scholar]
- 43.Tsukamoto Y., Taggart,A.K. and Zakian,V.A. (2001) The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol., 11, 1328–1335. [DOI] [PubMed] [Google Scholar]
- 44.Lewis L.K., Karthikeyan,G., Westmoreland,J.W. and Resnick,M.A. (2002) Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics, 160, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teng S.C., Chang,J., McCowan,B. and Zakian,V.A. (2000) Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell, 6, 947–952. [DOI] [PubMed] [Google Scholar]
- 46.Chen Q., Ijpma,A. and Greider,C.W. (2001) Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol., 21, 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takata M., Sasaki,M.S., Tachiiri,S., Fukushima,T., Sonoda,E., Schild,D., Thompson,L.H. and Takeda,S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol., 21, 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bezzubova O., Silbergleit,A., Yamaguchi-Iwai,Y., Takeda,S. and Buerstedde,J.M. (1997) Reduced X-ray resistance and homologous recombination frequencies in a RAD54–/– mutant of the chicken DT40 cell line. Cell, 89, 185–193. [DOI] [PubMed] [Google Scholar]