Abstract
Autophagy is essential for maintaining material balance and energy circulation and plays a critical role as a regulatory mechanism in tissue regeneration. However, current studies primarily describe this phenotype, with limited exploration of its molecular mechanisms. In this study, we provided the first evidence that autophagy is required for intestinal regeneration in Apostichopus japonicus and identified a previously unrecognized regulatory mechanism involved in this process. We observed that autophagy activation was significantly associated with enhanced regeneration, and its upregulation was shown to be regulated by reactive oxygen species (ROS) bursts. Mechanistically, ROS induced the dephosphorylation of Forkhead box protein O (FoxO) through AjAKT dephosphorylation. The dephosphorylated AjFoxO translocated to the nucleus, where it bound to the promoters of AjLC3 and AjATG4, inducing their transcription. This study highlights the ROS-AjAKT-AjFoxO-AjATG4/AjLC3 pathway as a novel regulatory mechanism underlying autophagy-mediated intestinal regeneration in echinoderms, providing a reference for studying regenerative processes and cytological mechanisms in economically important echinoderms.
Graphical Abstract
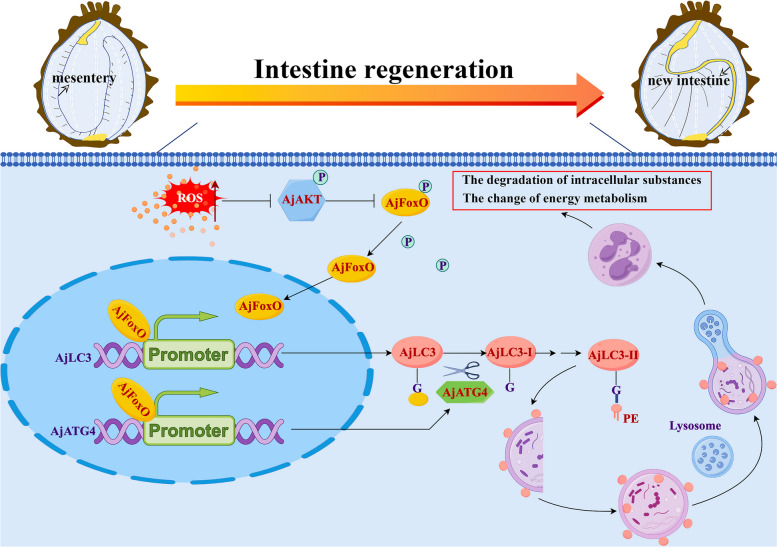
Scheme depicting the necessity and molecular regulatory mechanisms of autophagy during the process of intestinal regeneration in A. japonicus.The activation of autophagy during intestinal regeneration is positively correlated with the accumulation of ROS, which induces the dephosphorylation of AjFoxO by mediating the dephosphorylation of AjAKT. The dephosphorylated AjFoxO then entered the nucleus, interacted with the promoters of AjATG4 and AjLC3, and regulated autophagy levels by mediating the transcription of autophagy-related genes AjATG4 and AjLC3, thereby regulating intestinal regeneration probably via ROS-AjAKT-AjFoxO-AjATG4/AjLC3 pathway. The figure was constructed by FigDraw (www.figdraw.com/ID:rrPwA263e3).
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01993-0.
Keywords: Apostichopus japonicus, Intestine regeneration, Autophagy, Forkhead box protein O
Introduction
Regeneration is an incredible ability to replace lost body parts that has fascinated humans since ancient times. The regenerative capabilities of metazoans vary considerably, ranging from whole-body regeneration in invertebrates to organ and tissue regeneration in vertebrates [1]. Insights gained from animal studies not only enhance our fundamental understanding of biology but also provide valuable information for developing clinical strategies to repair damaged organs and tissues in humans. In recent decades, several model organisms, such as planarians, salamanders, and zebrafish, known for their strong regenerative potential-such as regenerating heads, eyes, limbs, and skin-have been used to explore the mechanisms behind injury-induced cellular plasticity and regeneration [2–4]. However, many fundamental processes remain poorly understood. Given the vast biodiversity on Earth, current model organisms may not be sufficient to uncover the general principles of regeneration and its underlying regulatory mechanisms [5]. Therefore, it is crucial to investigate the regeneration mechanisms in emerging non-standard model organisms, which could pave the way for advancements in human health and regenerative science research.
Organisms in the phylum Echinodermata exhibit remarkable regenerative abilities, making them key subjects in regeneration research [6]. As deuterostomes, echinoderms share approximately 70% of their genes with humans [7]. Therefore, studying echinoderm regeneration could lead to advanced therapeutic options for human diseases, particularly in intestinal regeneration in sea cucumbers. Unlike higher vertebrates and most invertebrates, which can only perform localized intestinal epithelial renewal, sea cucumbers can regenerate their entire intestine. In some species, this regeneration occurs in less than 2–3 weeks after evisceration [8, 9]. Additionally, the evisceration process in sea cucumbers can be induced through a simple KCl injection, minimizing procedural irritation [10]. These characteristics make sea cucumbers an attractive model for studying intestinal regeneration and developing treatments for intestinal diseases. Research has documented various morphological changes in the new intestine following self-evisceration. The prevailing view is that regeneration begins with the thickening of the mesentery tip. This process requires precise coordination of several cellular events, including cell migration, autophagy, dedifferentiation, proliferation, and apoptosis [10]. Among these, cell migration and dedifferentiation are considered the primary sources for early intestinal rudiment formation [11]. Following this, cell proliferation generates additional cells needed for the growing intestine [12]. Moreover, apoptosis likely plays a vital role in managing cell proliferation within the rudiment and in timing muscle cell dedifferentiation [12]. However, research on autophagy’s role in sea cucumber intestinal regeneration has been limited. Although autophagy is a key cellular regulatory mechanism associated with various cellular processes and is crucial for maintaining energy supply and material cycling, its molecular mechanism during intestinal regeneration remain poorly understood. Therefore, there is significant potential to explore autophagy in the context of sea cucumber intestinal regeneration, necessitating further investigations to uncover its mysteries.
Autophagy is an evolutionarily conserved process that enables cells to degrade and recycle their misfolded proteins and damaged organelles. This self-digestion helps maintain macromolecular synthesis and ATP production, supporting cellular homeostasis and remodeling at both cellular and tissue levels [13]. During autophagy, a small structure known as the isolation membrane, or phagophore, elongates to encircle a portion of the cytoplasm, forming a double-membraned structure called the autophagosome [14]. Autophagosomes can fuse with late endosomes to create amphisomes, which subsequently merge with lysosomes, or they can fuse directly with lysosomes. This fusion forms autolysosomes, where the internal contents are digested. In mammalian cells, autophagy depends on multiple autophagy-related (ATG) proteins. Among these, the microtubule-associated protein 1 light chain 3 (MAP1LC3/LC3), an ortholog of the yeast ATG8 protein, is vital for growing isolation membranes and recognizing autophagic cargoes [15]. Newly synthesized LC3 undergoes several processes, including cleavage by the cysteine protease ATG4 at the carboxyl terminal glycine 120 site, resulting in LC3-I. LC3-I then conjugates with phosphatidylethanolamine (PE) through a ubiquitylation-like system to form membrane-bound LC3-II [16]. Both PE-conjugated LC3 (LC3-II) and unconjugated LC3 (LC3-I) can be distinguished by immunoblot analysis. The ratio of LC3-II to LC3-I is commonly used to quantify autophagic activity [17]. Recent studies have highlighted autophagy’s role in tissue repair and organ regeneration in vertebrates. For instance, during mouse muscle fiber regeneration, autophagy aids in maintaining proteostasis and supports survival mechanisms in regenerating fibers [18]. In zebrafish eye muscle regeneration, autophagy facilitates dedifferentiation and accelerates muscle repair by clearing damaged organelles and proteins, as well as regulating oxidative stress signals [19]. Similarly, in lizard limb regeneration, autophagy plays a crucial role in maintaining morphology, generating new tissue, and remodeling existing tissue [20]. Although these studies across various animal models highlight autophagy’s importance in regeneration, the specific regulatory mechanisms remain underexplored, particularly in marine invertebrates. Gaining deeper insights into the processes of autophagy may help identify regenerative targets applicable across species.
In this study, we used Apostichopus japonicus (A. japonicus), a representative echinoderm species, to investigate the mechanisms underlying autophagy during intestinal regeneration. This research provides the first evidence that autophagy is crucial for intestinal regeneration, and there is a positive correlation between autophagy activation and the accumulation of reactive oxygen species (ROS) during this process in A. japonicus. Further analysis revealed that ROS induces dephosphorylation of AjFoxO via dephosphorylation of AjAKT. This process promotes the entry of dephosphorylated AjFoxO into the nucleus, leading to the expression of AjATG4 and AjLC3, which ultimately regulates autophagy levels during intestinal regeneration. This work demonstrates that autophagy is necessary for intestinal regeneration and identifies a previously unknown mechanism involving the ROS-AjAKT-AjFoxO-AjATG4/AjLC3 signaling pathway that regulates autophagy in echinoderms. The insights gained from echinoderms fill a gap in basic biological knowledge and provide foundational data for researching organ regeneration in higher metazoans.
Methods and materials
Ethics statement
The BALB/c mouse and A. japonicus, utilized in this study were commercially bred, and all studies followed the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. The study procedure was authorized by Ningbo University’s Experimental Animal Ethics Committee (No. NBU.ES-2021–11180).
Animals and treatment
Healthy adult A. japonicus (weight 110 ± 10 g) were obtained from the Dalian Pacific Aquaculture Company. They were housed in natural saltwater (salinity 28, temperature 11℃) and fed daily with commercially produced feed for one week before treatment. Following acclimation, A. japonicus were divided into 6 groups, each with three replicates containing 12 individuals. Group 1 served as the control and did not undergo visceral removal; their mesentery was sampled. Groups 2–6 were treated to induce evisceration via intracoelomic injection of 0.35 M KCl (3–5 mL) and were placed in seawater aquaria to allow for intestinal regeneration [9]. At 2, 7, 12, 20, and 28 days post-evisceration (dpe), samples from groups 2–6 were collected sequentially. For analysis, the mesentery and regenerated intestines of 6 A. japonicus (weight 100 mg each) were sampled and placed into centrifuge tubes containing 500 µL of cell lysate buffer (Cat#P0013, Beyotime, Beijing, China) for protein extraction and western blotting (WB). The mesentery and regenerated intestines of 3 additional A. japonicus were wrapped in frozen section compound (OCT, Cat#4583, Sakura, CA, USA) and stored at −80℃ for indirect immunofluorescence assay (IIFA). The remaining 3 A. japonicus had their mesentery and regenerated intestines fixed in a 2.5% glutaraldehyde solution for subsequent transmission electron microscopy (TEM).
To investigate the effect of autophagy inhibition on A. japonicus intestinal regeneration, 243 A. japonicus were eviscerated following an intracoelomic injection of KCl (0.35 M, 3 mL). They were divided into 3 treatment groups: the 3-Methyladenine (3-MA, 5 mM, Cat#5142-23-4, MedChemExpress, New Jersey, USA) group, the Bafilomycin A1 (Baf-A1, 20 nM, Cat#88899-55-2, MedChemExpress, New Jersey, USA) group, and the Dimethyl Sulfoxide (DMSO, Cat#D8371, Solarbio, Beijing, China) group. Each treatment group consisted of 3 replicates, each with 27 A. japonicus. For drug administration, A. japonicus sampled at 2-dpe were individually injected with one of the three reagents 6 h after evisceration. Those sampled at 7-dpe received injections at 6 h post-evisceration, as well as at 2-dpe, 4-dpe, and 6-dpe. Specimens taken at 12-dpe were injected at 6 h post-evisceration and additionally at 2-dpe, 4-dpe, 6-dpe, 8-dpe, and 10-dpe. For sampling, the mesentery and regenerated intestines of 6 A. japonicus (weighing 100 mg each) from the 3 treatment groups at 2-dpe, 7-dpe, and 12-dpe were individually collected. These were washed with phosphate buffered saline (PBS, Sigma-Aldrich, Saint-Louis, USA) and placed into centrifuge tubes for protein extraction and WB. The tissue of an additional 3 A. japonicus was also sampled individually at 2-dpe, 7-dpe, and 12-dpe. This tissue was embedded in a frozen section compound (OCT, Cat#4583, Sakura, USA) for IIFA.
To clarify the interaction between ROS and autophagy during the regeneration of the intestine in A. japonicus, the following experimental steps were carried out. First, 36 A. japonicus were randomly selected and subjected to evisceration. At various time points—0-dpe, 2-dpe, 7-dpe, 12-dpe, 20-dpe, and 28-dpe—samples of the mesentery and regenerating intestine from 3 A. japonicus (each weighing 50 mg) were collected and processed to measure ROS concentrations. To facilitate this, 2’, 7’-dichlorodihydrofluorescein diacetate (H2DCFDA, 10 µM, Cat#HY-D0940, MedChemExpress, New Jersey, USA) and DAPI (3 µM, Cat#C1002, Beyotime, Beijing, China) were administered 2 h and 30 min before each sampling, respectively. The collected tissues from the 3 A. japonicus were then embedded in a frozen section compound (Cat#4583, Sakura, CA, USA) for fluorescence observation. After establishing a significant association between ROS, autophagy, and intestinal regeneration, Apocynin (APO, 0.1 µM, a common ROS inhibitor, Cat#HY-N0088, MedChemExpress, New Jersey, USA) was introduced to further investigate the role of ROS in autophagy and intestinal regeneration. A total of 54 A. japonicus were randomly divided into 2 groups: the APO group and the DMSO (solvent control) group, with 27 A. japonicus in each. After evisceration, starting at 6 h post-procedure, both groups received intraperitoneal injections of either APO or DMSO every 48 h. At critical time points of 2-dpe, 7-dpe, and 12-dpe, mesentery and regenerating intestine samples from 3 A. japonicus (each weighing 50 mg) were taken and immediately immersed in cell lysate buffer (Cat#P0013, Beyotime, Beijing, China) for protein extraction and WB analysis. Additionally, samples from another 3 A. japonicus were embedded in a frozen section compound (Cat#4583, Sakura, CA, USA) for immunofluorescence experiments. Tissue samples from the remaining 3 A. japonicus were used for ROS content detection.
Phos-tag SDS-PAGE and WB
The total protein from mesentery and regenerated intestine samples in each group was ground at −10℃ using a tissue grinder. These samples were then lysed with cell lysis buffer (Cat#P0013, Beyotime Biotechnology, Shanghai, China), which contains 20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, and several inhibitors, including sodium pyrophosphate, β-glycerophosphate, EDTA, Na3VO4, and leupeptin. Nuclear and cytoplasmic proteins were isolated from each group using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Cat#78833, Thermo Scientific, MA, USA) according to the manufacturer’s instructions. The extracted proteins were analyzed for concentration using a BCA Protein Assay Kit (Cat#CW0014S, CWBIO, Beijing, China). A total of 50 µg of protein was separated by 12% or 15% SDS-PAGE in a Tris-Gly buffer system under constant voltage at 120 V. After electrophoresis, wet transfer was performed using a mini-transfer tank (Bio-Rad Laboratories) with NcmBlot Rapid Transfer Buffer (Cat#WB4600, NCM Biotech, Suzhou, China) at 400 mA for 30 min onto PVDF or nitrocellulose membranes (MilliporeSigma). The membranes were then blocked using 5% non-fat milk in TBST buffer (25 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20, pH 7.4) at room temperature for 2 h. Next, the membranes were incubated overnight at 4℃ with relevant primary antibodies (Supplementary Table 1). On the following day, the membranes were washed three times with TBST and incubated at room temperature for 2 h with horseradish peroxidase (HRP)-conjugated secondary antibodies (Supplementary Table 1). The membranes were then washed three additional times with TBST. Finally, the proteins were visualized using a chemiluminescence assay with NcmECL Ultra (Cat#P10200, NCM, Suzhou, China) and imaged with an Aplegen Omega Lum C (Gel Company, San Francisco, CA, USA). The protein band intensities were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The mean values of band intensities from three independent experiments were calculated, and the ratio of target protein intensity to tubulin intensity was determined.
The phosphorylation status of AjFoxO was analyzed using Phosphate-binding tag (Phos-tag™) acrylamide gels, a phospho-affinity SDS-PAGE technique developed by Kinoshita et al. [21]. The separating gel was created through the copolymerization of acrylamide and Phos-tag acrylamide. Phosphorylated proteins migrate more slowly in the gel than their unphosphorylated counterparts due to the reversible trapping of phosphoproteins by Phos-tag during electrophoresis. The Phos-tag SDS-PAGE was performed on gels containing 10% polyacrylamide, incorporating 50 µM Phos-tag Acrylamide AAL-107 (Cat#300–93523, Wako Chemicals, Osaka, Japan) and 100 µM MnCl2, as outlined in reference [22], under a current of 30 mA. This method allows for the differentiation of proteins by observing the mobility shift in phosphoproteins, which appear as distinct bands corresponding to their degree of phosphorylation. After separation, the proteins were blotted onto PVDF membranes using a submerged apparatus. The immunoreaction was carried out according to the protocol described in the previous paragraph.
Tissue immunofluorescence and image analysis
The mesentery and regenerated intestine from both experimental and control groups were embedded in frozen section compound (OCT, Cat#4583, Sakura, CA, USA) and frozen in liquid nitrogen. Slices were made using a Leica CM1900 cryostat at −20℃ and adhered to adhesion microscope slides (Cat#188105, Citoglas, Jiangsu, China). The Sect. (6 μm) were then removed and placed in 4% formaldehyde for 15 min. Permeabilization followed with 0.3% Triton X-100 for 20 min, and incubation with 10% goat serum for 1 h at room temperature. Next, the sections were incubated overnight at 4℃ with rabbit anti-AjLC3 antibody (1:1000 dilution, Cat#EPR18709, Abcam, Cambridge, UK) and/or mouse anti-AjFoxO antibody (1:500 dilution). The following day, the sections were washed three times with PBS containing 0.05% Tween-20 (PBST) and then incubated with Alexa488-conjugated goat anti-rabbit IgG (1:1000 dilution, Cat#A11008, Invitrogen, Carlsbad, CA, USA) at 37℃ for 90 min. Afterward, they underwent three additional washes with PBST and were incubated with Cy3-conjugated goat anti-mouse IgG (1:1000 dilution, Cat#M30010, Invitrogen, Carlsbad, CA, USA) at 37℃ for another 90 min. Following three more washes with PBST, the sections were stained with DAPI (10 mg/ml in PBS, Cat#C1002, Beyotime, Beijing, China) for 10 min at room temperature. Finally, fluorescence microscopy (Olympus, BX51) equipped with DAPI (330–380 nm), Alexa488 (460–490 nm), and Cy3 (510–550 nm) filters, along with objectives offering 5x magnification for broad views of the regenerated intestine and 100x magnification for detailed local structures, was performed for comprehensive examination. This setup enabled precise observation of fluorescence-positive signals and facilitated high-quality imaging of the sections.
Fluorescence signals and the area size of regenerated tissues across different regeneration periods and treatment groups were measured using ImageJ software (National Institutes of Health, Bethesda, MD, USA). To quantify fluorescence signal intensity, fluorescence images were initially converted to an 8-bit grayscale format. Regions of interest (ROIs) were defined using the Rectangle or Ellipse selection tools. Mean grayscale intensity values were measured for each ROI. This procedure was repeated for all ROIs within the image. Consistent settings were maintained during image capture and analysis to ensure accuracy and reliability of the results. The freehand selection tool in ImageJ was used to capture and measure the area of the regenerated intestine. The average area of these intestinal rudiments was calculated from at least three discontinuous tissue sections.
Transmission electron microscopy
TEM is considered the gold-standard method for detecting autophagy by analyzing morphological structures [23]. To directly observe the number, morphology, and structure of autophagosomes in regenerated intestinal tissue cells, the mesentery and regenerated intestine of A. japonicus from the control and 2-dpe groups were sampled. These samples were treated with 1% osmium tetroxide for 2 h at 25 °C and washed three times with 0.1 M PBS. The tissues were then dehydrated through a series of increasing ethanol concentrations and incubated in pure propylene oxide. During dehydration, the tissues were stained with 1% uranyl acetate (Serva, Heidelberg, Germany) in 70% ethanol. The tissues were subsequently embedded in a mixture of propylene oxide (Electron Microscopy Sciences, PA, USA) and Epon resin (Serva, Heidelberg, Germany), followed by pure Epon resin. After polymerization at 60 °C, 70 nm thick sections were cut using a Leica ultramicrotome (Leica UC7, Germany) and collected on TEM copper grids (Ted Pella, CA, USA). Electron micrographs were obtained using a transmission electron microscope (HITACHI HT7800, Japan) at 5000–10,000x magnification. Three optimal fields of view from each control/treatment group were selected for photography, and the number of autophagosomes in the cells of each group was counted for statistical analysis.
ROS probe-based chemiluminescence and fluorescence microscopy assay
We investigated the dynamic changes and tissue distribution of ROS in the mesentery and regenerating intestines of A. japonicus at different time points during the intestinal regeneration process in the Control, DMSO, and MK2206 (a novel allosteric inhibitor of Akt, 2 µM, Cat# SF2712, Beyotime, Shanghai, China) groups. Briefly, the entire length of the mesentery and regenerative primordia (30 mg) from the end of the esophagus to the cloaca of 6 A. japonicus from the control (non-regenerating stage mesentery), 2-dpe, 7-dpe, 12-dpe, 20-dpe, and 28-dpe groups were collected, ground into a homogenate in a −10°C cryogrinder, with 3 replicates per group. The intracellular ROS concentrations in these tissues were measured using the Tissue ROS Test Kit (DHE) (Cat# HR8821, Baiaolaibo, Beijing, China) according to the manufacturer’s instructions. Fluorescence intensity absorbance was detected using a microplate reader (Thermo Varioskan Flash 3001, USA) at 610 nm. Additionally, the distribution of ROS in A. japonicus was assessed in the control, 2-dpe, 7-dpe, 12-dpe, 20-dpe, and 28-dpe groups using 2’, 7’-dichlorodihydrofluorescein diacetate (H2DCFDA, HY-D0940, MedChemExpress, New Jersey, USA). Three A. japonicus were sampled from each group. At each sampling time point of intestinal regeneration, H2DCFDA (10 µM) and DAPI (3 µM) were intraperitoneally injected 30 min and 2 h in advance, respectively. The samples were then embedded in frozen section compounds (OCT, Cat# 4583, Sakura, CA, USA), frozen in liquid nitrogen, and cut into 6 μm sections using a Leica CM1900 low-temperature cryostat. Images were captured using a fluorescence microscope (Olympus BX51). For co-localization of ROS and AjLC3 fluorescence, we first used the free radical-permeable H2DCFDA to visualize ROS localization. Indirect immunofluorescence staining was then performed as described in the section “Tissue Immunofluorescence and Image Analysis” to visualize AjLC3 localization. Images of ROS probe fluorescence and AjLC3 fluorescence were taken sequentially from the same location in the same tissue and merged using ImageJ software.
Recombinant expression and antibody preparation
To obtain a recombinant Escherichia coli Rosetta (DE3) strain capable of expressing A. japonicus FoxO (AjFoxO) protein, the plasmid vector pET28a(+)-AjFoxO was constructed. First, the pET28a(+) plasmid was prepared by digestion with BamHI and XhoI. Next, the AjFoxO sequence (1704 bp, A. japonicus genomic Bioproject: PRJNA354676: MRZV01000610.1:105711|106124, MRZV01000610.1:122773|124062) was amplified from cDNA of A. japonicus using the primers F: 5’-AGCAAATGGGTCGCGGATCCATGGATGAAATAGATCCTGATT-3’ (homology arm underlined) and R: 5’-GGTGGTGGTGGTGCTCGAGTTTAGTGCACCCAATTGGTTC-3’ (homology arm underlined), then purified using a PCR product purification kit (Cat#B610363-0050, Sangon Biotech, Shanghai, China). The PCR products were ligated to the pET28a(+) plasmid with homologous arms at both ends using a pEASY®-Basic Seamless Cloning and Assembly Kit (Cat#CU201, TransGen Biotech, Beijing, China) to generate the recombinant pET28a(+)-AjFoxO vector in E. coli DH5α cells. The structure of pET28a(+)-AjFoxO was confirmed by DNA sequencing (Sangon Biotech). Following successful sequencing confirmation, the recombinant pET28a(+)-AjFoxO vector was transformed into E. coli Rosetta (DE3) and induced with isopropyl-β-D-thiogalactopyranoside at 18 °C overnight. The expressed recombinant AjFoxO protein (rAjFoxO) was purified using Ni-NTA Agarose (QIAGEN, Germany) and validated by SDS-PAGE. The purified rAjFoxO was used as an immunogen to prepare mouse anti-AjFoxO polyclonal antisera. Six BALB/c mice were immunized with rAjFoxO protein in four separate doses. The immunization procedure was as follows: 50 µg of rAjFoxO protein suspension was mixed with Complete Freund’s Adjuvant (Cat#HY-153808, MedChemExpress, New Jersey, USA) in a 1:1 ratio, and 0.2 mL of the mixture was intraperitoneally injected into the mice. Two weeks later, the first booster immunization was performed, using a mixture of rAjFoxO protein suspension and Incomplete Freund’s Adjuvant (Cat#HY-153808 A, MedChemExpress, New Jersey, USA) in a 1:1 ratio, injected intraperitoneally. Subsequently, immunizations were strengthened weekly with 0.2 mL of adjuvant-free rAjFoxO protein suspension injected into the tail vein. On the 7th day after the last booster, blood was collected from the mice by eyeball exsanguination. The collected blood was incubated at room temperature for 2 h, then left overnight at 4 °C. The next day, the serum was separated by centrifugation at 10,000 rpm for 5 min. Mouse IgG was purified using a protein G-agarose rapid flow column (Sigma, USA) and characterized by enzyme-linked immunosorbent assay (ELISA) and WB.
Enzyme-linked immunosorbent assay
ELISA was used to detect the potency titer of the anti-AjFoxO antibody. Specifically, 100 µL of 100 µg/mL rAjFoxO protein was coated onto 96-well flat-bottom microplates (Costar, USA) and incubated overnight at 4 °C. The wells were then washed three times with PBST and blocked with 200 µL of 3% bovine serum albumin (BSA) solution in PBS for 1 h at 37 °C. After washing, the wells were incubated with 100 µL of serially diluted anti-AjFoxO polyclonal antibodies in PBS at concentrations of 1:500, 1:1000, 1:2000, 1:4000, 1:8000, 1:16000, 1:32000, 1:64000, and 1:128000 for 1 h at 37 °C. Following additional washes with PBST, 100 µL of AP-conjugated goat anti-mouse IgG (Cat#ZY2044, Ziyunbao, Shanghai, China) at a 1:1000 dilution in PBS was added and incubated for 1 h at 37 °C. After the final wash, 100 µL of 0.1% (w/v) p-nitrophenyl phosphate (pNPP, Sigma, USA) in pNPP buffer (1% diethanolamine, 0.5 mM MgCl2, pH 9.8) was added to each well and incubated for 30 min at 37 °C in the dark. The reaction was stopped by adding 50 µL of 2 M NaOH per well, and the absorbance was read at 450 nm using an automatic ELISA reader (Molecular Devices, USA). Serum from a non-immunized mouse served as the negative control. Each experimental step was replicated three times to ensure data reliability. The potency of the polyclonal antibody was determined when the ratio (P/N) of the OD 450 value of the polyclonal antibody well to the negative control well exceeded 2.1, with the highest dilution representing the antibody potency.
Cell immunofluorescence staining
Immunofluorescence staining was performed to examine the subcellular distribution and translocation of AjFoxO in the mesentery and regenerating primordium during intestinal regeneration. The mesentery and regenerated intestine from the DMSO, Apocynin, and APO + MK2206 groups were minced and digested in a mixture of PBS containing 0.2% Trypsin (Cat#25200-072, Gibco, Grand Island, NY, USA) and 0.2% Collagenase IV (Cat#17104019, Gibco, Grand Island, NY, USA) at 4 °C for 30 min to form a single-cell suspension. The suspension was washed twice with PBS containing 10% fetal bovine serum (FBS, Cat#10270-106, Gibco, Grand Island, NY, USA) and twice with PBS alone. The suspension was then dropped onto adhesion microscope slides (Cat#188105, Citoglas, Jiangsu, China) and allowed to settle for 1 h. After removing the liquid, the slides were air-dried, and a circle was drawn around the dried surface using a PAP pen to prevent fluid loss. The slides were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.3% Triton-X-100 in PBS for 15 min, and blocked with 5% BSA at room temperature for 1.5 h. The slides were then incubated overnight at 4 °C with anti-AjFoxO mouse antibody (1:500 dilution), followed by incubation with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:1000 dilution, Cat#A0516, Beyotime, Beijing, China) for 1 h at room temperature. The cell membrane was stained with DiL (red) (Cat#C1991S, Byotiome, Beijing, China) and the nuclei were stained with DAPI (Cat#C1002, Byotiome, Beijing, China). Images were captured using a confocal laser scanning microscope (LSM 880; ZEISS, Germany) with a 40× objective lens.
RNA interference (RNAi) and inhibitor assays
The specific siRNA (Supplementary Table 2) targeting AjFoxO (siAjFoxO-1 and siAjFoxO-2) and a control siRNA (Negative control, NC) were designed and synthesized by GenePharma Company. Before the formal experiment, the interference reagents were tested at various concentrations (0 µM, 10 µM, 20 µM, 40 µM) to evaluate their interference ability and off-target effects. After determining the optimal concentration and specificity of the interfering primers, the experimental and control siRNAs were dissolved in RNase-free water to prepare 20 µM (optimal interference concentration) stock solutions of siAjFoxO-1/siAjFoxO-2 or siNC. The interference reagent mixture for each A. japonicus was prepared in 100 µL total volume: 10 µL of Lipo6000 transfection reagent (Cat#C0526, Byotiome, Shanghai, China), 10 µL of siAjFoxO-1/siAjFoxO-2 (20 µM) or siNC (20 µM), and 80 µL of PBS. This mixture was intraperitoneally injected into the coelom of A. japonicus every 48 h throughout the intestinal regeneration process. The first injection was administered 6 h post-evisceration, with the last injection given at 6-dpe. Mesentery and regenerating intestine samples were collected at 2-dpe and 7-dpe and stored at −20 °C for subsequent RT-qPCR (AjFoxO mRNA levels) and WB (AjFoxO, AjLC3-II/I, Ajp62 protein levels).
All chemical inhibition treatments were performed in vivo. Control groups were treated with DMSO dilution. The inhibitors applied in this study are listed in Supplementary Table S1.
Quantitative Real-Time RT-PCR
Total RNA was extracted from 100 mg of intestinal regeneration tissues (6 A. japonicus) at different stages of regeneration (0-, 2-, 7-, 12-dpe) and from the regenerated tissues of the AjFoxO interference and control groups at 2-dpe, following the manufacturer’s instructions using Trizol reagent (Cat#T9108, Takara, Otsu, Japan). RNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific), and RNA integrity was assessed by 1.5% agarose gel electrophoresis. To remove genomic DNA (gDNA), 1 µg of total RNA was treated with 1 unit of DNase I for 15 min at 37 °C. cDNA was synthesized using the PrimeScript™ RT Reagent Kit (Cat#RR047A, Takara, Otsu, Japan). The cDNA from each sample was diluted 10-fold and used as a template for RT-qPCR on an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, USA) with TB Green Premix Ex Taq™ II mix (Cat#CN830A, Takara, Otsu, Japan). Oligonucleotide primers for AjLC3, AjATG4, and AjFoxO were designed using Primer 5 software, based on sequences from the A. japonicus genomic (NCBI accession number: MRZV00000000.1) and transcriptome (NCBI accession number: SAMN29388289) databases [24]. The amplification efficiency and specificity of the primers were evaluated using the standard curve, amplification plot, and melt curve (Supplementary Figure S1). For validation of the housekeeping gene expression stability during intestinal regeneration, the expression stability of 8 candidate housekeeping genes (β-actin, β-Tubulin, Elongation factor-1 (EF1α), 40 S ribosomal protein S9 (RPS9), RPS18, Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), NADH dehydrogenase (NADH) and 60 S ribosomal protein L18a (RPL18A)) was evaluated using four algorithms: geNorm (qbase+, version 3.1) [25], NormFinder (version 0.953) [26], BestKeeper (version 1) [27], and RefFinder (https://blooge.cn/RefFinder/?type=reference). The amplification efficiency and specificity of these 8 housekeeping genes were previously reported [28], and their expression stability is shown in Supplementary Figure S2. Amplification was conducted in a 20 µL reaction volume containing 10 µL of SYBR Green I Master, 2 µL of diluted cDNA, 0.4 µL of each forward and reverse primer (10 µM) (Supplementary Table 2), 0.4 µL of ROXII (Takara), and 6.8 µL of RNase-free water. The reaction was carried out at 95 °C for 30 s, followed by 45 cycles of denaturation at 95 °C for 5 s and extension at 60 °C for 30 s [29]. Each sample was run in triplicate, and the experiments were repeated three times using the Ajβ-tubulin gene as the endogenous control. Data were analyzed relative to the Ajβ-tubulin gene using the 2−△△Ct method [30].
Transcriptomic analysis of intestinal regeneration after DMSO/APO treatment in A. japonicus
The mesentery and regenerated intestines from 9 A. japonicus in the APO group and the corresponding control (DMSO) group were sampled at 2-dpe and stored at −80 °C for transcriptome sequencing. Total mRNA was extracted as described above. RNA integrity was evaluated using the RNA Nano 6000 Assay Kit (Agilent Technologies). RNA-seq libraries were constructed using the NEBNext Ultra RNA Library Prep Kit, following the manufacturer’s guidelines. Libraries were subjected to paired-end sequencing with a read length of 150 bp on the Illumina NovaSeq 6000 platform (Illumina, USA). Adapter sequences and low-quality reads were filtered using Fastp (v0.19.7, parameters: -g -q 5 -u 50 -n 15 -l 150), and the remaining clean reads were aligned to the A. japonicus genome (NCBI accession number: MRZV00000000.1) using HISAT2 software (v2.0.5) [31]. Potential novel transcripts were identified using StringTie and CPC2 (version 1.0.1) [32]. Gene expression levels were quantified using RSEM (v1.2.15) software [33]. Differentially expressed genes (DEGs) between the APO and DMSO groups were identified using the DESeq2 package (Bioconductor, v1.14.1, p < 0.05 and |log2(FoldChange)| > 1) [34]. DEGs were further analyzed by Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (R software 4.2.1) and heatmap analysis (heatmap.2 in R package v3.6.0).
Cell culture
Human embryonic kidney epithelial cells (HEK 293T) were cultured in DMEM medium (Cat#10013102, Corning, NY, USA) supplemented with 10% FBS. The cells were maintained in a humidified incubator at 37 °C with 5% CO2. Petri dishes and culture plates were purchased from Jet Bio-Filtration Co., Ltd. (Guangzhou, China) and NEST Biotechnology Co., Ltd. (Wuxi, China).
Chromatin immunoprecipitation (ChIP) assay
To systematically investigate the downstream genes regulated by AjFoxO and their specific binding sites during the sea cucumber intestine regeneration process, we selected the mesentery and regenerated intestine at 2-dpe and prepared DNA fragments following the protocol of a commercial ChIP kit (Cat# P2078, Beyotime, Beijing, China). Specifically, 200 mg of mesentery and regenerated intestine tissues were chopped into small pieces and incubated in RBI buffer (10 mM KCl, 5 mM MgCl2, 5 mM EGTA pH 8, 5 mM Na pyrophosphate, 1 mM PMSF, 1X Complete™ protease inhibitors from Roche Diagnostics, Basel, Switzerland) on ice for 10 min. The samples were then centrifuged at 1000 g for 5 min and resuspended in ice-cold PBS containing 1X Complete™ protease inhibitors (PBS-C) and 1% formaldehyde (FA, Cat#28906, Thermo Fisher Scientific, Waltham, USA) for 10 min at 37 °C under agitation. Reactions were stopped by adding 1X glycine and incubating for 5 min at room temperature whereas agitating. Tubes were centrifuged for 5 min at 1000 g, and the tissues were washed three times with ice-cold PBS-C. The samples were then ground using an ice-cold mortar and pestle until homogeneous and incubated for 10 min on ice with 1 mL of ice-cold Lysis Buffer (10 mM Tris-HCl pH 8, 5 mM EDTA pH 8, 85 mM KCl, 0.5% NP-40, 1 mM PMSF, 1X Complete™ protease inhibitors). The tissue lysate was centrifuged at 5000 g for 5 min, and the cellular pellets were resuspended in 500 µL of SDS Lysis Buffer (50 mM Tris-HCl pH 8, 10 mM EDTA pH 8, 1% SDS, 10% glycerol, 1X Complete™ protease inhibitors) and incubated for 10 min. The tubes were vortexed for 20 s, incubated on ice for 10 min, and vortexed again for 20 s. Ultrasonic treatment was performed using an Ultrasonic Homogenizer (Cat# JY92-IIN, Scientz, Ningbo, China) set at a frequency of 15 W (10 s on, 10 s off, 8–10 cycles). Then, 0.2 mL of the ultrasonically treated sample was transferred to a centrifuge tube, 8 µL of 5 M NaCl solution was added, and the mixture was thoroughly mixed. The sample was incubated at 65 °C for 4 h. DNA purification was carried out using the Macherey Nagel NucleoSpin® Gel and PCR Clean-up kit (Macherey Nagel, Duren, Germany), and DNA smears were visualized by migration on a 1% agarose gel electrophoresis stained with Gel Red (Biotium, Fremont, USA).
ChIP experiments were conducted on samples with qualified ultrasonic treatment, which achieved the desired fragment size (200–1000 bp). The samples were centrifuged at 12,000–14,000 g for 5 min. A volume of 0.2 mL of the supernatant was transferred into a 2 mL centrifuge tube, and 1.8 mL of ChIP Dilution Buffer containing 1 mM PMSF was added and mixed thoroughly. A 20 µL aliquot was set aside as the “input” for subsequent testing. To the remaining 2 mL of sample, 70 µL of Protein A + G Agarose was added, and the mixture was gently rotated at 4 °C for 30 min. The samples were then centrifuged at 4 °C and 1000 g for 1 min, and the supernatant was carefully transferred to a fresh 2 mL centrifuge tube. Subsequently, 10 µL of AjFoxO antibody was added, and the mixture was incubated overnight at 4 °C with rotation. After incubation, 60 µL of Protein A + G Agarose was added, and the mixture was rotated or shaken at 4 °C for 60 min to precipitate the protein or corresponding complex recognized by the AjFoxO antibody. The solution was then centrifuged at 1000 g for 1 min at 4 °C. The precipitate was washed sequentially with Low Salt Immune Complex Wash Buffer, High Salt Immune Complex Wash Buffer, LiCl Immune Complex Wash Buffer, and TE Buffer. Each wash used 1 mL of solution, with the precipitate gently rotated at 4 °C for 3–5 min after each addition. After the final wash, the liquid was carefully removed to avoid contact with the sediment. The precipitate was sent to Cloud-Seq Biotech (Shanghai, China) for ChIP-seq sequencing. Sequencing libraries were prepared using the NEB Next Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, USA). The process involved 10 ng of immunoprecipitated (IP) DNA and 100 ng of input DNA, with PCR amplification cycles totaling 7 for IP DNA and 4 for input DNA. DNA fragments of approximately 200 bp were selected using Agencourt AMPure XP beads (Beckman Coulter, Brea, USA). Library concentrations were quantified using Qubit fluorometry and stored at − 80 °C prior to sequencing. Sequencing was performed in single-end 50-base format on an Illumina 4000 system by the IGBMC GenomEast Platform (Illkirch, France).
For bioinformatics analysis, sequencing quality was checked with FastQC. Low-quality reads were removed using cutadapt software (v1.9.3) [35], and the remaining clean reads were aligned to the A. japonicus genome using Bowtie2 (v2.2.6.2) [36]. Peak detection was performed using Epic (v0.1.23), a SICER rework, with the following settings: fragment-size 50 -gaps-allowed 2 -false-discovery-rate-cutoff 0.05 [37]. AjFoxO regions were identified per sample. To account for read count differences, 40 million unique reads were randomly selected for analysis. Common peaks were identified by intersecting the results, and peak visualization was done using the Integrative Genomics Viewer. For BigWig files, the mapped reads were determined using flagstat with 40 million reads, and input data was scaled to AjFoxO data using genomeCoverageBed, then converted to bedGraph. AjFoxO files were converted directly to bedGraph without scaling. BigWig files were generated and normalized [log2(AjFoxO/input)] using bigwigCompare (default settings except for –pseudocount 0.1 and –bs 1) [37].
Dual luciferase reporter assay
To verify the regulatory effect of AjFoxO transcription factor on AjLC3 and AjATG4 genes, fluorescent reporter vectors for wild-type and mutant-type AjLC3 and AjATG4 were constructed. The steps are as follows: First, the pGL3 dual enzyme digestion vector was prepared by digestion with XhoI and HindIII. Then, the promoter sequences containing the conserved FoxO protein binding site (TGTTT) on AjLC3 and AjATG4 (Supplementary Fig. 19, underlined sections) were amplified from A. japonicus genomic DNA using the primers listed in Supplementary Table 2 (homology arm underlined) and purified using a PCR product purification kit (Cat# B610363-0050, Sangon Biotech, Shanghai, China). The PCR products were ligated to the pGL3 plasmid using a pEASY®-Basic Seamless Cloning and Assembly Kit (Cat# CU201, Transgene, Beijing, China) to generate the recombinant pGL3-AjLC3-promo-WT and pGL3-AjATG4-promo-WT vectors. The structure of these vectors was confirmed by DNA sequencing (Sangon Biotech). After confirming the sequencing results, the vectors were transformed into E. coli DH5α for large-scale amplification. These amplified plasmids served as templates for further mutation. Using mutant primers detailed in Supplementary Table 2 (homology arm underlined) and the Mut Express® II Fast Mutagenesis Kit (Cat# C214-01, Vazyme, Nanjing, China), PCR amplification was carried out. The mutant primers were designed via a website (https://crm.vazyme.com/cetool/singlepoint.html). Upon completion of amplification, the products were treated with Dpn I enzyme to digest methylated plasmid templates, ensuring that only mutated sequences were retained. The amplified products were then recombined with homologous arms at both ends and transformed into Trans1-T1 cell (Cat# CD501-02, Transgene, Beijing, China). Single colonies were selected and sequenced for validation. After verification, large-scale plasmid amplification was performed to yield pGL3-AjLC3-promo-MUT and pGL3-AjATG4-promo-MUT vectors. Next, the pcDNA3.1-Flag vector was prepared by double enzyme digestion using EcoRI and XhoI. The AjFoxO sequence was amplified from A. japonicus cDNA using primers from Supplementary Table 2 (homology arms underlined), designed to match the homologous arms of the double-digested plasmid. The recombinant pcDNA3.1-AjFoxO-Flag plasmid was obtained following the homologous recombination method.
For the Dual-Luciferase Reporter assay, 20 ng of pRL-TK vector (containing the Renilla luciferase gene as an internal control), 300 ng of pcDNA3.1-AjFoxO-Flag or pcDNA3.1-Flag, and 200 ng of pGL3 or one of the constructed pGL3 vectors were transfected into HEK-293T cells in 24-well plates using Lipofectamine 6000 (Cat# C0526, Beyotime, Beijing, China). After 48 h of transfection, cells were harvested, and both firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay Kit (Cat# E1910, Promega, WI, USA) and a Tecan Spark 10 M multimode microplate reader. Relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity. The results represent the average of three independent assays.
DNA electrophoretic mobility shift assay (EMSA)
Biotin-labeled wild-type and mutant-type AjLC3 and AjATG4 probes were synthesized by Youkang Biotech (Hangzhou, China) (Supplementary Table 2). These probes contained the conserved AjFoxO binding site “CTGTTT” or its mutated sequence, along with 20–30 bases upstream and downstream to ensure the GC content was between 40% and 60%. The sequence information was submitted to Youkang Biotech for probe synthesis, HPLC purification, 3’ biotin labeling, and annealing to obtain biotin-labeled complementary double-stranded probes. DNA EMSA experiments were performed following the manufacturer’s instructions from the LightShift™ Chemiluminescent DNA EMSA Kit (Thermo Fisher Scientific, USA) [38].
Biotin-labeled probes (wild-type and mutant-type) and purified rAjFoxO (500 ng) were combined and incubated in a binding buffer at room temperature for 20 min. For super-shift tests, rAjFoxO was pre-incubated with AjFoxO antibodies for 20 min before incubation with biotin-labeled probes for 20 min. DNA-protein complexes were separated on a 6% non-denaturing polyacrylamide gel at 90 V for 1 h. After electrophoresis, wet transfer was performed using a mini-transfer tank (Bio-Rad Laboratories, Inc.) in pre-cooled 0.5× TBE buffer (Cat# R0223, Beyotime, Beijing, China) at 300 mA for 30 min at 4 °C onto a nylon membrane. The membrane was crosslinked with UV light (254 nm) for 15 min, blocked with 15 mL blocking solution (Cat# GS009B, Beyotime, Beijing, China) for 15 min, and treated with horseradish peroxidase-linked streptavidin (dilution 1:2000) in blocking solution for 15 min at room temperature. After three washes with ddH2O, the membrane was treated with NcmECL Ultra (Cat# P10200, NCM, Suzhou, China) and imaged using an Aplegen Omega Lum C imaging system (Gel Company, San Francisco, CA, USA).
Statistical analysis
All data are expressed as the mean ± SD from three independent replicate studies. The Student’s t-test was used to compare results between two groups, including protein expression levels, ROS content, regenerated intestinal area, and fluorescence intensity between the control group and other drug treatment groups at the same regeneration stage. One-way ANOVA followed by Duncan’s post-hoc test was used to detect statistical significance among three or more groups. For example, the analysis of protein expression grayscale values, regenerated intestinal area, and fluorescence intensity at different stages (control, 2-dpe, 7-dpe, 12-dpe, 20-dpe, 28-dpe) of intestinal regeneration, or protein grayscale analysis, ROS content analysis, tissue fluorescence intensity analysis, and regenerated area analysis of different drug treatment groups (DMSO, APO, APO + MK2206) at the same time point. A p-value < 0.05 was considered statistically significant. Statistical analyses were conducted using Prism version 7.0 (GraphPad Software).
Results
Autophagy is significantly increased in the early phase of intestinal regeneration
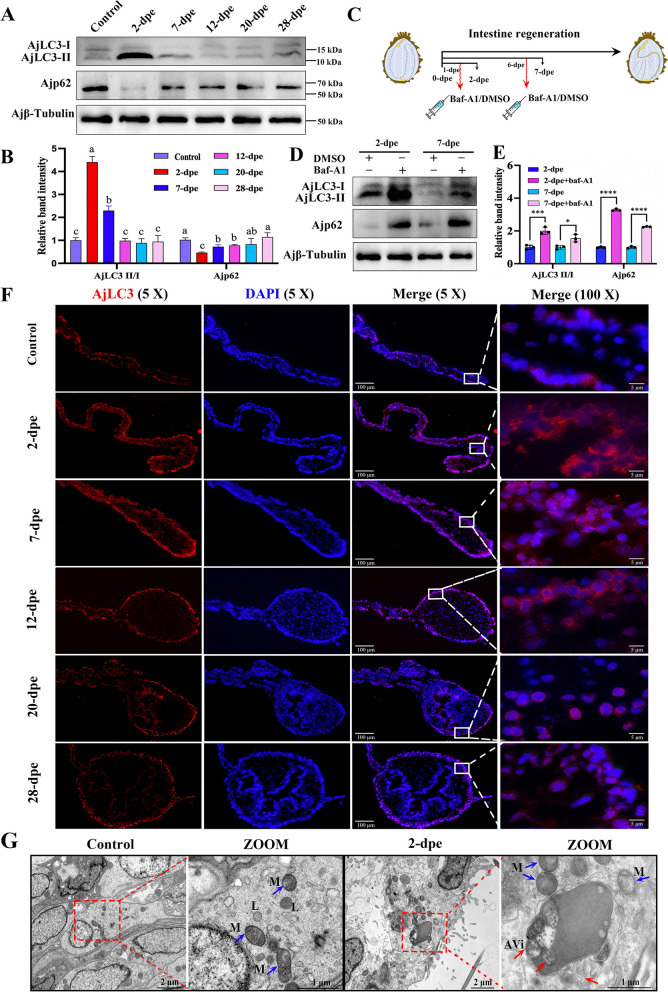
To determine whether autophagy is induced throughout the course of intestine regeneration in A. japonicus, several methodological approaches were employed [39]. These included the monitoring of fluctuations in autophagy-associated proteins, such as LC3 and p62, through WB in conjunction with autophagy inhibitors, the localization of LC3 within cells using immunofluorescence, and the observation of autophagosomes via TEM. It is well-established that during autophagy formation, cytoplasmic LC3 (LC3-I) is enzymatically hydrolyzed to form LC3-II, a membrane-bound form associated with autophagosomes [40]. The ratio of LC3-II to LC3-I can be used to estimate autophagic activity, with a higher ratio indicating increased autophagic activity. Additionally, p62, a linker molecule between ubiquitinated proteins and the autophagy system, plays a critical role in autophagic degradation and reflects autophagic activity [18]. In this study, the WB results showed that the ratio of AjLC3-II/I increased initially, peaking at 2-dpe, and then gradually decreased to baseline levels as regeneration progressed. Notably, the levels remained similar to the control group starting at 12-dpe (Fig. 1A, B). In parallel, the protein expression level of the autophagic substrate Ajp62 was low at baseline in the control group, sharply decreased by 2-dpe, reaching the lowest point during the observation period, suggesting substantial consumption of the autophagic substrate during this phase. As regeneration continued, Ajp62 levels gradually recovered, reflecting its dynamic regulation in intestinal regeneration (Fig. 1A, B). To further investigate autophagy, autophagy-lysosome inhibitors, such as Baf-A1, were used to examine the effects on autophagic activity (Fig. 1C). The results revealed that the ratio of AjLC3-II/I in Baf-A1-treated groups was significantly higher than in the DMSO-treated groups. Moreover, the protein levels of Ajp62 in the Baf-A1 treatment groups were also higher, indicating that Baf-A1 effectively blocked autophagy flow and confirming autophagy activation during intestinal regeneration at 2-dpe and 7-dpe (Fig. 1D, E).
Fig. 2.
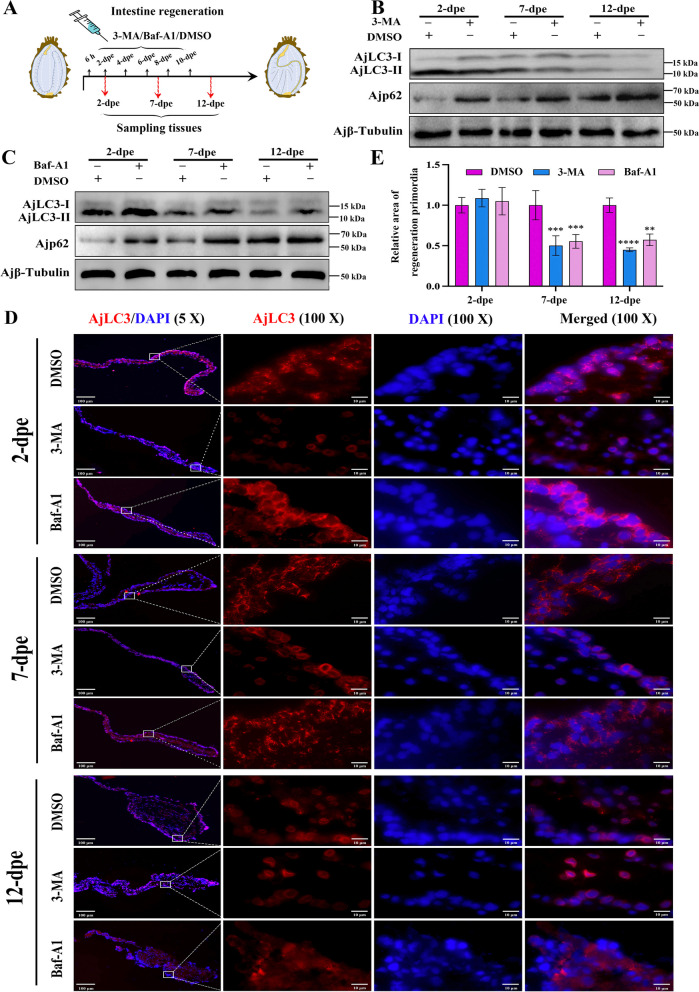
Autophagy is required for regenerative primordia in intestine regeneration. A Schematic diagram of the injection method for DMSO and autophagy inhibitors (3-MA, Baf-A1) during the intestinal regeneration process. B Protein expression levels of AjLC3-II/I and Ajp62 in the regenerative primordia at 2-, 7-, and 12-dpe post 3-MA treatment were determined by WB. C Protein expression levels of AjLC3-II/I and Ajp62 in the regenerative primordia at 2-, 7-, and 12-dpe after Baf-A1 treatment were determined by WB. D Indirect immunofluorescence was used to measure the expression of AjLC3 at 2-, 7-, and 12-dpe after DMSO, 3-MA, or Baf-A1 treatment. All samples were taken from the same location in A. japonicus. Blue indicates DAPI-stained nuclei, and red indicates the expression of AjLC3. E The size of the regeneration primordium in different treatment groups of (D) was detected using the ImageJ program. Data are presented as mean ± SD, n = 3 replicates. Statistical analysis was performed using the Student’s t-test: **p < 0.01; ***p < 0.001; ****p < 0.0001
Fluorescence microscopy was next used to observe autophagosome formation, as indicated by the redistribution of AjLC3 from a diffuse pattern to punctate spots. Compared to the control group (non-regenerating stage mesentery), the 2-dpe group exhibited numerous AjLC3-specific puncta on coelomic epithelial cells (Fig. 1F). These fluorescent spots gradually decreased as the regenerated intestinal area increased (Figure S3), eventually showing no significant difference from the control group at 20-dpe (Figure S4). To observe autophagosomes more accurately, TEM was employed to examine autophagosomes in normal and regenerating intestinal cells at 2-dpe. TEM qualitatively characterizes autophagy; early autophagic compartments (autophagosomes) appear as vacuolar structures with double or multiple membranes containing intact cytosol or organelles, whereas late autophagic structures (autolysosomes) contain partially degraded cytoplasmic and organelle material [41]. As shown in Fig. 1G, the cells in the control group exhibited intact mitochondria and close cell-cell connections, with few autophagosomes visible (blue arrowhead: mitochondria; red arrowhead: autophagosome). In contrast, at 2-dpe, the mitochondria cristae disappeared, and numerous double-membraned autophagosomes containing cytoplasmic components appeared (Figure S5). In summary, the results from these experiments confirm that autophagy occurs in cells of the regeneration primordia during the intestinal regeneration process of A. japonicus. Autophagy reaches its peak in the early phases (2-dpe) and gradually returns to normal levels as regeneration progresses.
Autophagy is required for intestinal regeneration
To investigate the necessity of autophagy in the intestinal regeneration of A. japonicus, we used a dual-pronged approach: early-stage autophagy was inhibited using the 3-MA inhibitor to block the formation and progression of autophagosomes, whereas late-stage autophagy was inhibited using Baf-A1 to prevent the fusion of autophagosomes with lysosomes [42]. Through these two steps, we carefully evaluated the impact of inhibited autophagic processes on intestinal regeneration. For the early-stage autophagy inhibitor 3-MA, the results in Fig. 2B and Figure S6A and B showed that, compared to the DMSO control groups, the AjLC3-II/I ratio decreased and Ajp62 expression increased in the 3-MA treatment groups, indicating successful inhibition of autophagy by 3-MA. Similarly, in Fig. 2D and Figure S7, the AjLC3 fluorescence level was significantly lower in the 3-MA treatment groups compared to the DMSO control groups. Statistical analysis of the regenerated intestinal area at the free end of the mesentery revealed that the regeneration area in the 3-MA group was significantly smaller than in the DMSO control groups at 7-dpe and 12-dpe (Fig. 2E). At 2-dpe, the free end of the mesentery was still in the wound repair phase, so no significant difference in regeneration area was observed following 3-MA treatment. These results indicate that 3-MA inhibits autophagy and blocks the progression of A. japonicus intestinal regeneration. Regarding the late-stage autophagy inhibitor Baf-A1, as shown in Fig. 2C and Figure S6C and D, the AjLC3-II/I ratio and Ajp62 expression increased in the Baf-A1 treatment groups, confirming autophagy inhibition by Baf-A1. Additionally, the AjLC3 fluorescence level in the Baf-A1 treatment groups was significantly higher than in the DMSO control groups (Figure S7), and statistical analysis of the regenerated intestinal area at the free end of the mesentery revealed that the regenerated area in the Baf-A1 group was significantly larger than in the DMSO control groups at 7-dpe and 12-dpe (Fig. 2E). These results suggest that Baf-A1 inhibits autophagy and blocks the development of A. japonicus intestinal regeneration. The experimental results using early-stage autophagy inhibitor (3-MA) and late-stage autophagy inhibitor (Baf-A1) indicate that during A. japonicu intestinal regeneration, inhibition of the autophagy pathway at any stage disrupts the regeneration process.
Fig. 1.
Autophagy is upregulated in the early phases of intestinal regeneration. A WB was performed to evaluate the expression changes of AjLC3-II/I and Ajp62 proteins in the control (non-regenerating stage mesentery) and various regenerative stages (from 2 to 28-dpe). B The band density of AjLC3-II/I and Ajp62 proteins in (A) was quantified using the ImageJ program. Data are presented as mean ± SD, n = 3 replicates. Statistical analysis was performed using one-way ANOVA and Duncan’s post-hoc test, with p < 0.05 considered statistically significant. Different letters above each bar indicate significant differences. C Schematic diagram of the injection method for DMSO and autophagy inhibitors (Baf-A1) during the intestinal regeneration process. D WB was used to detect the protein expression changes of AjLC3-II/I and Ajp62 in regenerating mesentery and intestine at 2 and 7-dpe post Baf-A1 or DMSO treatment. E The band density of AjLC3-II/I and Ajp62 proteins in (D) was quantified using the ImageJ program. Data are presented as mean ± SD, n = 3 replicates. Statistical significance was determined using the Student’s t-test: *p < 0.05; ***p < 0.001; ****p < 0.0001. F Indirect immunofluorescence was used to detect the expression changes of AjLC3 during the course of intestine regeneration (2-28 dpe). All samples were taken from the same location in different individuals. Blue indicates DAPI-stained nuclei, and red indicates the expression of AjLC3. Bar = 100 μm. G TEM was used to observe mesentery and regenerative primordia in control (non-regenerating stage mesentery) and regenerating intestinal cells at 2-dpe. In the regenerative primordia at 2-dpe, autophagic vacuoles accumulated (red arrowheads). The left panel shows low magnification images, whereas the right panels provide high magnification views of specific areas. These images are enlargements of specific portions of the images in the left panel. M: mitochondria. L: lysosome. AVi: initial autophagic vacuoles
Autophagy depends on the induction of ROS during the intestinal regeneration process
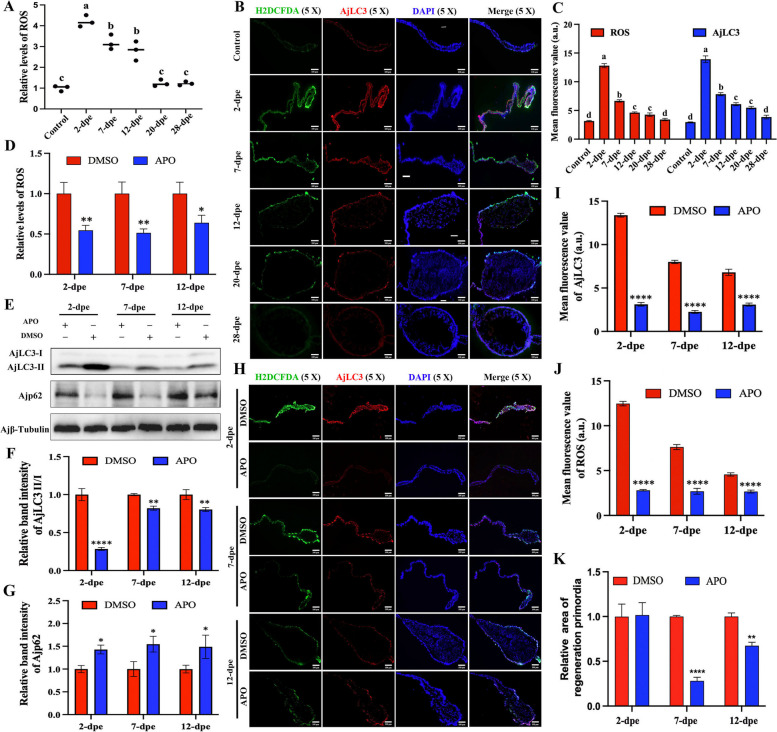
Recent studies have shown that oxidative stress is a key regulator of autophagy, with ROS playing a significant role in the signaling pathways that maintain autophagic processes [9, 43]. In the TEM results of 2-dpe regenerated primordia, we observed a large number of autophagosomes along with mitochondrial cristae disappearance (Fig. 1G). Since mitochondrial damage is widely reported to release large amounts of ROS [44], we hypothesized that the autophagy observed during intestinal regeneration is associated with ROS production. To test this hypothesis, we first examined ROS levels following A. japonicus evisceration and during intestinal regeneration. The results showed that ROS production initially increased, peaking at 2-dpe, and then decreased, with no significant difference at 20-dpe compared to the control group (Fig. 3A). This pattern closely mirrored the changes observed in autophagy (Fig. 1A and B). Next, we investigated the co-localization of AjLC3 protein and ROS in situ tissues. The results showed that ROS and AjLC3 were co-localized in the regenerative tissues (Fig. 3B), and fluorescence-based ROS measurements (Fig. 3C) were consistent with the ROS levels measured by enzyme-linked immunosorbent assay (Fig. 3A). ROS were found not only at the cut edge of the mesentery but also along the entire mesentery from the free end to the wall end. Furthermore, the co-localization of ROS and AjLC3 in tissues indicates an association between ROS and autophagy during intestinal regeneration. To further explore the association between ROS and autophagy, we used APO, a ROS inhibitor, to suppress ROS production during intestinal regeneration. The results showed that APO effectively inhibited ROS production (Fig. 3D, H, J), and the expression of AjLC3 (Fig. 3H, I) as well as the size of the regenerative primordia (Fig. 3K) were reduced following APO treatment compared to the DMSO control. Additionally, the ratio of AjLC3-II/I in the ROS inhibition group was significantly lower (Fig. 3E, F), whereas Ajp62 expression increased (Fig. 3E, G), suggesting that reduced ROS production impaired autophagy during intestinal regeneration. In summary, autophagy levels during intestinal regeneration were positively correlated with ROS levels. Inhibition of ROS production reduced autophagy induction, which in turn hindered intestinal regeneration.
Fig. 3.
Autophagy depends on the induction of ROS during the intestinal regeneration process. A Dynamic changes in ROS levels were detected using the DHE-ROS detection kit in the control group (non-regenerating stage mesentery) and during intestinal regeneration (2–28 dpe). Data are presented as mean ± SD, n = 3 replicates. Statistical analysis was performed using one-way ANOVA, followed by Duncan’s post-hoc test, with p < 0.05 considered statistically significant. Different letters above each bar indicate significant differences. B The localization of ROS and AjLC3 protein during intestinal regeneration was detected using the H2DCFDA probe and anti-AjLC3 antibody, followed by fluorescence imaging. The fluorescent signals were captured in the same field of view and superimposed. Bar = 100 μm. C Mean fluorescence values of ROS and AjLC3 in the regenerated intestine of A. japonicus at different stages (control (normal mesentery), 2-dpe, 7-dpe, 12-dpe, 20-dpe, 28-dpe) from (B). D Measurement of ROS production in regenerated intestine mesentery at 2-, 7-, and 12-dpe after APO or DMSO treatment using the DHE-ROS detection kit. E The ratio of AjLC3-II/I and the expression level of Ajp62 protein were measured by WB in regenerative primordia during intestinal regeneration (2-dpe, 7-dpe, 12-dpe) after continuous administration of APO inhibitors or DMSO. F Band density of AjLC3-II/I proteins in (E) was quantified using the ImageJ program. Data are presented as mean ± SD, n = 3 replicates. Statistical significance was assessed using the Student’s t-test: **p < 0.01, ****p < 0.0001. G Band density of Ajp62 proteins in (E) was quantified using the ImageJ program. Data are presented as mean ± SD, n = 3 replicates. Statistical significance was assessed using the Student’s t-test: *p < 0.05. H The localization of ROS and AjLC3 protein in the regenerative primordia post APO or DMSO treatment at 2-, 7-, and 12-dpe was detected using the H2DCFDA probe and anti-AjLC3 antibody. The fluorescent signals were photographed in the same field of view and superimposed. Bar = 100 μm. I Mean fluorescence value of AjLC3 in the regenerated intestine of A. japonicus at different stages (2-dpe, 7-dpe, 12-dpe) in the APO or DMSO group of (H). Data are presented as mean ± SD, n = 3 replicates. Statistical significance was assessed using the Student’s t-test: ****p < 0.0001. J Mean fluorescence value of ROS in the regenerated intestine of A. japonicus at different stages (2-dpe, 7-dpe, 12-dpe) in the APO or DMSO group of (H). Data are presented as mean ± SD, n = 3 replicates. Statistical significance was assessed using the Student’s t-test: ****p < 0.0001. K The size of the intestine rudiment area corresponding to (H) was determined and summarized. Data are presented as mean ± SD, n = 3 replicates. Statistical significance was assessed using the Student’s t-test: **p < 0.01, ****p < 0.0001
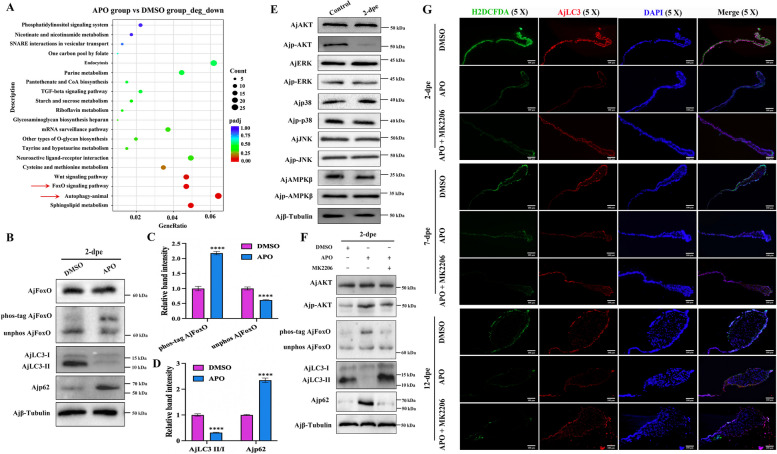
High concentrations of ROS regulate cell autophagy through dephosphorylation AjAKT-mediated AjFoxO dephosphorylation
To investigate how ROS regulate autophagy, we injected ROS inhibitors (APO) into A. japonicus following intestine evisceration and conducted RNA-seq (NCBI accession number: PRJNA1035161) to identify potential molecular pathways in the presence or absence of ROS at the 2-dpe stage. The results showed that, compared to the DMSO group, the “autophagy-animal” pathway was significantly downregulated in the APO treatment group (Fig. 4A). Notably, the “FoxO signaling pathway,” a key regulator of autophagy, was also significantly enriched in the ROS-downregulated group (Fig. 4A), suggesting a possible association between AjFoxO and ROS-mediated autophagy during intestinal regeneration. Subsequent analysis of the differential gene heat map for the AjFoxO pathway (Figure S8) revealed that downstream genes of the AjFoxO pathway were significantly downregulated in the APO group, although no significant differences were observed in the transcript levels of AjFoxO itself. Since AjFoxO plays a pivotal role in autophagy regulation through phosphorylation, we hypothesized that AjFoxO may regulate ROS-mediated autophagy by altering its phosphorylation state during intestinal regeneration. To test this hypothesis, we used APO to inhibit ROS production and assessed changes in autophagic flow and the phosphorylation status of AjFoxO (the specificity and titer (1:64000) of AjFoxO polyclonal antibody are shown in Figures S9 and S10). Phosphorylated AjFoxO migrates more slowly in gels due to the reversible trapping of phosphoproteins by Phos-tag during electrophoresis. By comparing protein bands running more slowly than nonphosphorylated AjFoxO, we could assess differences in phosphorylation levels. The results showed no significant difference in the total protein levels of AjFoxO, but a significant difference in phospho-AjFoxO between the APO and DMSO groups (Fig. 4B, C). This indicates that ROS may induce dephosphorylation of AjFoxO. Moreover, as the phosphorylation level of AjFoxO increased, autophagy levels decreased proportionally (Fig. 4B, D), suggesting a negative regulatory association between AjFoxO phosphorylation and autophagy induction.
Fig. 4.
High concentrations of ROS regulate cell autophagy through dephosphorylation AjAKT-mediated AjFoxO dephosphorylation. A Functional analysis of differentially downregulated DEGs in the regenerative primordia between APO- and DMSO-treated A. japonicus at the 2-dpe stage based on Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment. B Protein expression levels of AjFoxO, AjLC3-II/I, and Ajp62 in the mesentery and newly formed intestinal rudiment at the 2-dpe stage after APO and DMSO treatment were detected by WB. Phosphorylation levels of AjFoxO were assessed using Phos-tag SDS-PAGE followed by WB analysis. C Band density of phospho-AjFoxO and unphosphorylated AjFoxO proteins in (B) was quantified using the ImageJ program. Data are presented as mean ± SD, n = 3 replicates. Statistical significance was assessed using the Student’s t-test: ****p < 0.0001. D Band density of AjLC3-II/I and Ajp62 proteins in (B) was quantified using the ImageJ program. Data are presented as mean ± SD, n = 3 replicates. Statistical significance was assessed using the Student’s t-test: ****p < 0.0001. E The protein expression and phosphorylation levels of AjAKT, AjERK, Ajp38, AjJNK, and AjAMPKβ in the control group (non-regenerating stage mesentery) and regenerative primordia at the 2-dpe stage were measured by WB analysis. F The protein expression levels of AjAKT, Ajp-AKT, AjFoxO, AjLC3 I/II, and Ajp62 in the regenerative primordia at the 2-dpe stage post DMSO, APO, and APO + MK2206 treatment were detected by WB. Phosphorylation levels of AjFoxO were assessed using Phos-tag SDS-PAGE followed by WB. G The distribution of ROS and AjLC3 protein in regenerated intestinal mesentery at 2-, 7-, and 12-dpe post DMSO, APO, and APO + MK2206 treatment was localized using the H2DCFDA probe and anti-AjLC3 antibody, with the fluorescent signals photographed in the same field of view and superimposed on each other. Bar = 100 μm
To further investigate how ROS mediate AjFoxO dephosphorylation during intestinal regeneration, we measured the expression levels of key phosphorylated kinases (AjAMPK, AjAKT, AjJNK, Ajp38, and AjERK) downstream of ROS by WB [45]. The results showed that, compared to the control group, only Ajp-AKT was significantly downregulated at 2-dpe, with no significant changes observed in other proteins such as AjERK, AjP38, AjJNK, or AjAMPK (Fig. 4E and Figure S11). These findings suggest that ROS may mediate AjFoxO dephosphorylation via regulation of p-AKT. To validate this hypothesis, we set up three experimental groups, with DMSO as the control: one group treated with APO to inhibit ROS production, another with MK2206 to inhibit AjAKT phosphorylation following APO treatment. Results showed that, compared to the DMSO group, inhibition of ROS by APO led to upregulation of AjAKT and AjFoxO phosphorylation levels. In contrast, phosphorylation levels of both AjAKT and AjFoxO were significantly downregulated in the APO + MK2206 group (Fig. 4F and Figure S12). These results suggest that ROS regulate AjFoxO phosphorylation through the mediation of AjAKT phosphorylation. Furthermore, autophagy levels were assessed in these groups, revealing that compared to the DMSO group, the AjLC3-II/I ratio was downregulated, and the Ajp62 protein level was upregulated in the APO group (Fig. 4F and Figure S12), indicating inhibited autophagy after ROS inhibition. In contrast, the APO + MK2206 group showed a significant increase in the AjLC3-II/I ratio and a decrease in Ajp62 protein levels, suggesting that autophagy was restored following inhibition of both ROS and AjAKT phosphorylation. These findings were further confirmed by immunofluorescence (Fig. 4G), and the effect of these treatments on intestinal regeneration was assessed. In the APO group, fluorescence signals for both ROS and AjLC3 were diminished, along with reduced intestinal regeneration (Figure S13). In the APO + MK2206 group, fluorescence signals for both ROS and AjLC3 were restored, and the regeneration area of A. japonicus intestine was recovered (Figure S14), likely due to the inhibitory effect of MK2206 on AKT phosphorylation. Taken together, these results confirm that ROS mediate AjAKT-dependent AjFoxO dephosphorylation and finely regulate autophagy during intestinal regeneration.
Dephosphorylated AjFoxO driven by ROS-AKT translocates into the nucleus and regulates the progress of intestinal regeneration
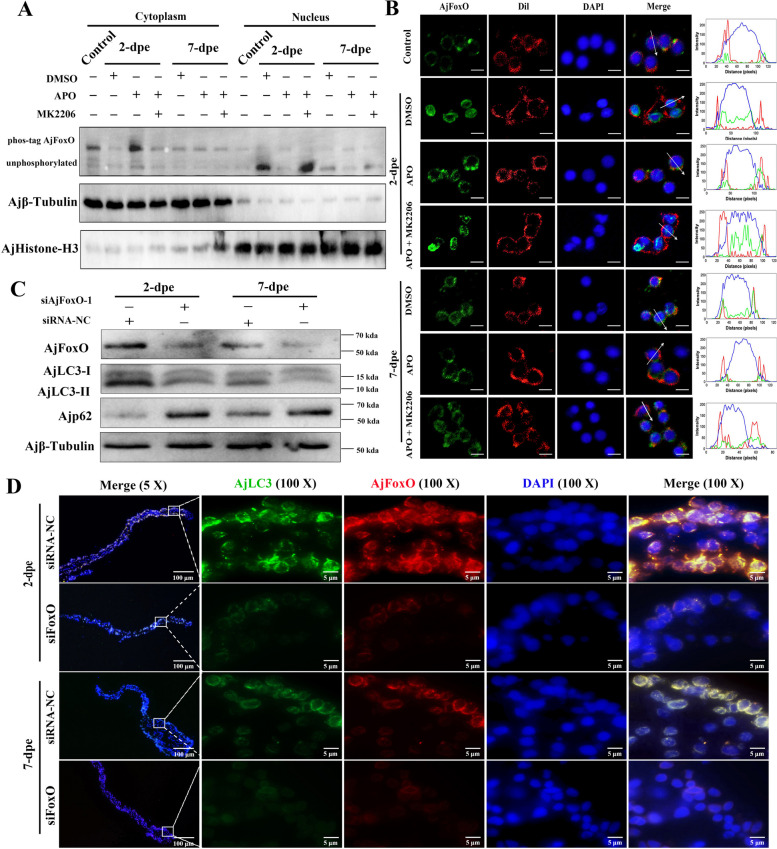
It has been reported that phosphorylation of FoxO transcription factors under growth conditions results in their export from the nucleus and subsequent inactivation of their transcriptional activity [46]. To investigate the localization of dephosphorylated AjFoxO during intestinal regeneration, we purified nuclear and cytoplasmic protein components from samples at different stages of intestinal regeneration. The dynamic distribution of both phosphorylated and non-phosphorylated AjFoxO between the nucleus and cytoplasm was systematically tracked using Phos-tag technology combined with WB. Additionally, the distribution of AjFoxO in the nucleus and cytoplasm was visualized through immunofluorescence analysis. As shown in Fig. 5A, in the control group (non-regenerating stage A. japonicus), phosphorylated AjFoxO was mainly localized in the cytoplasm. As intestinal regeneration progressed, the level of non-phosphorylated AjFoxO in the nucleus rapidly increased, peaking at 2-dpe, and then gradually decreased at 7-dpe. These results indicate that AjFoxO is dephosphorylated and translocated to the nucleus during intestinal regeneration, with the most pronounced effect occurring at 2-dpe.
Fig. 5.
Dephosphorylated AjFoxO driven by ROS-AKT translocated into the nucleus and regulates the progress of intestinal regeneration. A The levels of phosphorylated and non-phosphorylated AjFoxO in the control group (non-regenerating stage mesentery) and different regeneration groups (2- and 7-dpe) in the DMSO, APO, and APO + MK2206 groups were detected by WB. B Immunofluorescence was used to detect the dynamic distribution of AjFoxO at the subcellular level in the single-cell lysate of control and different regeneration groups (2- and 7-dpe) post DMSO, APO, and APO + MK2206 treatment. The distribution of DAPI-stained nuclei (blue), AjFoxO (green), and Dil-stained cell membranes (red) at the white arrowheads in the merged region was measured by ImageJ. Scale bar = 5 μm. C WB was performed to detect the expression changes of AjFoxO, AjLC3-II/I, and Ajp62 proteins in different regeneration groups (2- and 7-dpe) post siRNA-NC and siFoxO-1 treatment. D Immunocytochemistry was used to detect the expression of AjFoxO and AjLC3 proteins in different regeneration groups (2- and 7-dpe) post siRNA-NC and siFoxO treatment. Blue color indicates DAPI-stained nuclei, green color represents FITC-labeled AjLC3, and red color represents Cy3-stained AjFoxO. Scale bar = 5 μm
It has been reported that phosphorylation of FoxO transcription factors under growth conditions leads to nuclear export and subsequent inactivation of their transcriptional activity [46]. To investigate the localization of dephosphorylated AjFoxO during intestinal regeneration, we purified nuclear and cytoplasmic protein components from samples at various stages of intestinal regeneration. The dynamic distribution of both non-phosphorylated and phosphorylated AjFoxO between the nucleus and cytoplasm was systematically analyzed using phosphate tag technology combined with WB method. Additionally, the subcellular distribution of AjFoxO was visualized through immunofluorescence analysis. As shown in Fig. 5A, in the control group (non-regenerating stage A. japonicus), phosphorylated AjFoxO was predominantly located in the cytoplasm. As intestinal regeneration progressed, the level of non-phosphorylated AjFoxO in the nucleus rapidly increased, peaking at 2-dpe, and then gradually decreased by 7-dpe. These results indicate that AjFoxO is dephosphorylated and translocates to the nucleus during intestinal regeneration, with the most pronounced translocation observed at 2-dpe. We also analyzed the dynamic distribution of phosphorylated and non-phosphorylated AjFoxO in the cytoplasm and cell membrane of the regenerating intestine after treatment with ROS inhibitor (APO), AKT phosphorylation inhibitor (MK2206), and control reagent (DMSO) at 2-dpe and 7-dpe. At 2-dpe, most AjFoxO protein in the DMSO group was in a non-phosphorylated state and localized in the nucleus. After treatment with the ROS inhibitor (APO), AjFoxO was phosphorylated and detected in the cytoplasm. However, in the APO + MK2206 group, AjFoxO was dephosphorylated and translocated back to the nucleus. These findings are further illustrated in Fig. 5B, where, at 2-dpe, the DMSO group showed AjFoxO predominantly in the nucleus, while in the APO treatment group, it was primarily located in the cytoplasm. In the APO + MK2206 treatment group, AjFoxO was restored to the nucleus. Combining the results from Fig. 5A and B, we conclude that AjFoxO undergoes dephosphorylation and translocates to the nucleus during intestinal regeneration, particularly at the 2-dpe stage. Inhibiting ROS production during regeneration promotes the phosphorylation and nuclear export of AjFoxO, whereas inhibiting both ROS production and AKT phosphorylation leads to the dephosphorylation and nuclear import of AjFoxO.
To further explore the effects of AjFoxO dephosphorylation on autophagy and intestinal regeneration, we faced the limitation of lacking specific AjFoxO phosphorylation inhibitors or enhancers for A. japonicus. Therefore, we were unable to directly verify the effects of inhibiting or enhancing AjFoxO phosphorylation on autophagy and regeneration. This limitation is a critical issue that needs to be addressed in future studies. As a preliminary approach, we interfered with the expression of total AjFoxO protein and investigated its effects on autophagy and intestinal regeneration. First, we inhibited AjFoxO expression using different concentrations of siAjFoxO (siAjFoxO-1 and siAjFoxO-2). Both siAjFoxO-1 and siAjFoxO-2 specifically inhibited the expression of AjFoxO mRNA, with optimal inhibition achieved at 20 µM (Figure S15). Next, silencing AjFoxO expression at 2-dpe and 7-dpe resulted in a decrease in the ratio of AjLC3-II/I, while Ajp62 protein expression increased in the siAjFoxO group (Fig. 5C, Figure S16, and Figure S17). Additionally, compared to the siNC group, the average fluorescence values of AjLC3 and AjFoxO, as well as the area of regenerated intestines, were significantly reduced in the siAjFoxO group (Fig. 5D, Figure S18, and Figure S19). Since regeneration at 2-dpe is naturally slow [47], differences were more pronounced at 7-dpe. These findings suggest that downregulation of AjFoxO expression during intestinal regeneration may inhibit the activation of autophagy mechanisms, thereby impeding the progression of intestinal regeneration.
Translocation of dephosphorylated AjFoxO into the nucleus initiates the expression of autophagy-related genes
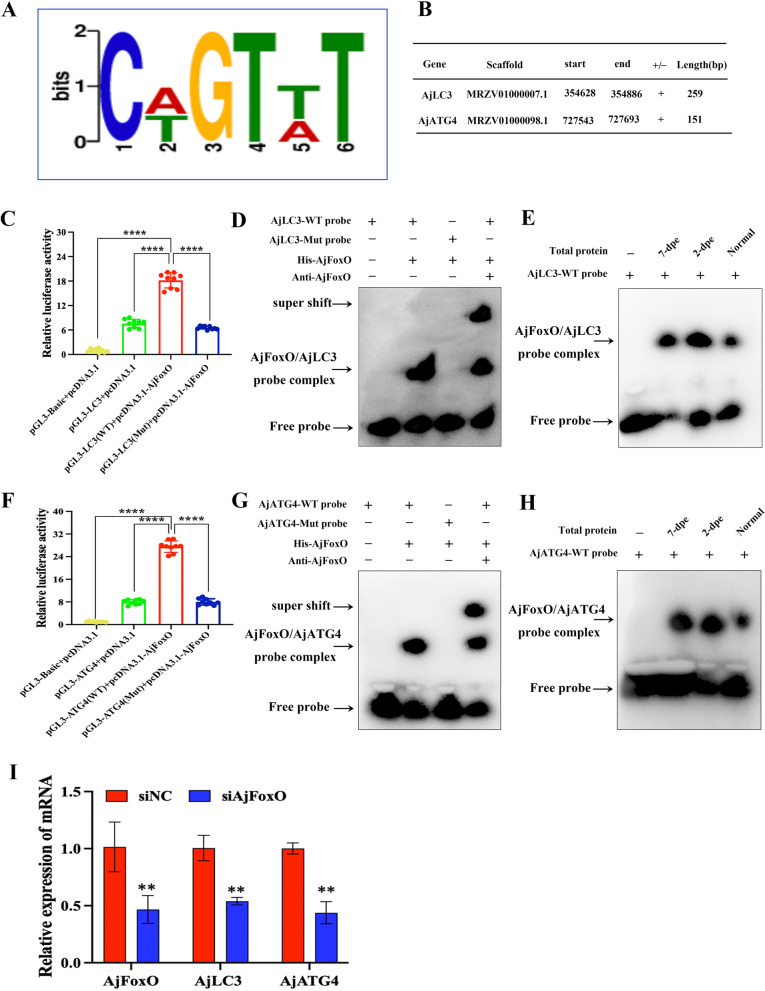
To explore how AjFoxO regulates autophagy during intestinal regeneration, we performed ChIP-seq using chromatin from the regenerated mesentery and primordia at 2-dpe. The raw data has been uploaded to the NCBI Sequence Read Archive (SRA) Public Database under accession number PRJNA1168870. We identified 13,748 unique peaks of AjFoxO binding, which target 2,747 genes in the A. japonicus transcriptome. Using the annotation file linking the reference genome and transcriptome of A. japonicus, we defined the sequence within 2 kb upstream of the transcription initiation site as the promoter region. We then used MEME software (https://meme-suite.org/meme/tools/meme) to search for enriched motifs within the AjFoxO-binding peaks and identified the ten highest-rated motifs (Figure S20). Among these, Fig. 6A shows the motif “TGTTT” a conserved sequence previously reported for FoxO. Among the 2,747 potential target genes, at least 20 are likely involved in autophagy. Further analysis revealed that AjFoxO may bind to the promoters of AjATG4 and AjLC3, as they share the conserved motif at their enrichment peaks. The detailed gene sequences are shown in Fig. 6B and Figure S21. To verify whether AjFoxO specifically binds to the regions of these candidate target genes, we first constructed wild-type and mutant PGL-3 vectors of ATG4 and LC3 near the motif region and conducted dual-luciferase experiments. The results confirmed that AjFoxO proteins could bind to the promoters of AjATG4 and AjLC3 in vitro (Fig. 6C, F). Next, we designed wild-type and mutant probes for AjATG4 and AjLC3 and verified the binding of these probes to both prokaryotically expressed AjFoxO proteins and the native proteins in the regenerating primordia during intestinal regeneration of A. japonicus using EMSA experiments. The results showed that the wild-type AjATG4 and AjLC3 probes bound to the prokaryotic AjFoxO proteins, whereas the mutant probes did not (Fig. 6D, G). Similarly, wild-type probes for AjLC3 and AjATG4 bound to native AjFoxO proteins in the regenerating primordia during intestinal regeneration (Fig. 6E, H). Additionally, the effect of inhibiting AjFoxO on AjATG4 and AjLC3 transcription was assessed by RT-qPCR. As shown in Fig. 6I, compared to the siNC treatment, siAjFoxO significantly suppressed the transcription of AjFoxO, which, in turn, affected the mRNA expression levels of AjATG4 and AjLC3. These results confirm that AjFoxO binds to the promoters of AjLC3 and AjATG4 genes upon nuclear translocation, regulates their transcription, and ultimately controls the activation of autophagy.
Fig. 6.
Identification of autophagy-related genes potentially regulated by AjFoxO. A Highly enriched motif of AjFoxO in A. japonicus. B Annotation of AjFoxO binding to AjATG4 and AjLC3 promoters based on ChIP-seq data. C, F Relative luciferase activity of wild-type and mutant pGL3-AjLC3/pGL3-AjATG4 luciferase reporter constructs in AjFoxO-overexpressing cells. Overexpression of AjFoxO significantly upregulated luciferase activity in the pGL3-AjLC3 (WT) and pGL3-AjATG4 (WT) groups, whereas activity in pGL3-AjLC3 (Mut) and pGL3-AjATG4 (Mut) showed no significant change. Renilla luciferase activity was measured and normalized to firefly luciferase activity. Asterisks indicate significant differences between groups. n = 9 replicates. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. D, G DNA EMSA showing the binding capability of purified His-AjFoxO with biotin-labeled AjLC3 and AjATG4 wild-type and mutant probes. E, H DNA EMSA showing the binding of biotin-labeled AjLC3 and AjATG4 wild-type and mutant probes to AjFoxO protein in different stages of the regenerative mesentery. I mRNA expression of AjATG4 and AjLC3 under AjFoxO interference. Data are presented as mean ± SD, n = 3 replicates. *p < 0.05 and **p < 0.01
Discussion
In recent years, autophagy has been associated with many physiological processes and is considered a crucial pro-survival mechanism to counteract tissue damage and loss in harsh environments by mediating repair and regeneration [48]. However, the regulatory mechanisms of autophagy during regeneration require further verification and research. In this study, we provide the first evidence that autophagy is required for intestinal regeneration in A. japonicus. Our further investigation revealed that the upregulation of autophagy during regeneration is controlled by ROS bursts, and this process involves the dephosphorylation of AjFoxO through AjAKT signaling. The dephosphorylated AjFoxO translocates to the nucleus, binds to the promoters of AjLC3 and AjATG4, and regulates their transcription, ultimately controlling autophagy and influencing intestinal regeneration. This study offers novel insights into the role of autophagy in intestinal regeneration and highlights the ROS-AjAKT-AjFoxO-AjATG4/AjLC3 pathway as an unrecognized regulatory mechanism for autophagy-mediated intestinal regeneration in echinoderms.
Autophagy may play an important role in energy metabolism and protein cycling during the regeneration process
Tissue/organ regeneration is inherently an anabolic process that demands a sufficient supply of energy, nucleic acids, and amino acids to build new cells for tissue formation [49]. For A. japonicus, the prolonged starvation caused by self-evisceration may present a challenge for intestinal regeneration, as this process requires maintaining metabolism and consuming substantial amounts of energy. However, our previous study demonstrated that A. japonicus is capable of generating extensive cell proliferation, migration, and dedifferentiation, along with other cellular events, after evisceration, to support the swelling of the distal mesentery and ultimately form a new intestine [47]. Autophagy is known to be crucial for material balance and energy circulation, and inducing autophagy to generate energy during intestinal regeneration is an advantageous strategy for A. japonicus, which cannot obtain energy through digestion after self-evisceration. In this study, we demonstrated that the intestinal regeneration process is accompanied by significant autophagy activation, including an increased ratio of AjLC3-II/I, decreased Ajp62 expression, and the enrichment of AjLC3-specific autophagosomes and autolysosomes in primordial cells, as observed by fluorescence microscopy and TEM. Additionally, treatment with lysosomal inhibitors (Baf-A1) or specific autophagy inhibitors (3-MA) led to an increase/decrease in the AjLC3-II/I ratio, an increase in Ajp62 expression, and a reduction in the size of the regeneration primordia. All of these results indicate that autophagy is essential for intestinal regeneration in A. japonicus, a finding consistent with other regeneration models, such as adult muscle regeneration, gecko tail regeneration, and zebrafish caudal fin and heart regeneration [20, 50]. In addition to energy metabolism, autophagy is also considered a mechanism for degrading protein aggregates or large organelles during cell dedifferentiation, which supports cell survival and organelle turnover through basal autophagy levels [51]. For example, in a zebrafish model of muscle regeneration, autophagy plays a critical role in cell dedifferentiation by regulating the degradation of sarcomeres and nuclear fragments, facilitating the transition to a less differentiated cell identity [13]. In our study, numerous autophagosomes were observed in mesentery and regenerated intestinal cells via transmission electron microscopy (Fig. 2G). Some autophagic vacuoles appeared to engulf and degrade aggregates of spindle-like structures (SLS) proteins (Fig. 2G, red arrow), leading to the appearance of numerous white vacuoles in the cytoplasm. The formation of SLS is a hallmark of muscle fiber dedifferentiation, and the elimination or degradation of SLS is a critical step in completing cell dedifferentiation [11]. Based on our results, we speculate that large-scale activation of autophagy facilitates the degradation of SLS, thus promoting muscle fiber dedifferentiation. Overall, these findings suggest that autophagy is required for intestinal regeneration, both for energy supply and the degradation of waste proteins to support cell dedifferentiation.
The activation of ROS and it’s downstream pathway is crucial for inducing autophagy
Organ regeneration is a dynamic biological process that requires the coordinated action of various external and intracellular signals. Recently, the physiological role of ROS in cellular signaling has been recognized, with ROS serving as upstream regulators that play a pivotal role in mediating tissue regeneration [52]. In this study, we found that ROS levels significantly increased in the regenerative primordia during intestinal regeneration, especially in the early stages. When the ROS inhibitor APO was used, ROS production was suppressed, and regeneration was severely impaired, indicating that ROS upregulation is necessary for intestinal regeneration. A similar conclusion was observed in Xenopus tadpoles, where tail amputation induced sustained ROS production during regeneration. Decreased ROS levels, particularly early after amputation, resulted in impaired tail regeneration [53]. Similarly, ROS generated by tail fin amputation also induced nerve regeneration, as increased H2O2 produced by DUOX1 at the wound site was crucial for peripheral sensory axon and skin nerve regeneration [54]. These studies indicate that ROS production is a key feature in the regeneration process. Moreover, recent evidence has established a tight correlation between ROS production and the induction of autophagy during regeneration. Previous studies have shown that ROS induced by damage primarily affects regeneration by activating cell proliferation [53], inducing apoptosis, or interfering with early neonatal cell differentiation [55]. In this study, we confirmed a positive regulatory association between ROS and autophagy during intestinal regeneration in A. japonicus. The suppression of ROS production by APO reduced autophagy induction, thereby impairing intestinal regeneration. These results demonstrate that ROS accumulation is critical for autophagy induction. However, the extent of ROS production and its interactions with downstream signaling pathways, including the autophagic machinery and feedback loops, are key to understanding this process. The classic signaling pathways through which ROS promote autophagy include the activation of MAPK, AMPK, and attenuation of AKT. In the early stage of intestinal regeneration, massive ROS production inhibited AjAKT activation, and as regeneration progressed, the decrease in ROS levels relieved the inhibitory effect on AjAKT (Fig. 4). These results suggest that ROS regulate autophagy during intestinal regeneration by selectively inhibiting AKT activity. To test this hypothesis, we assessed the effects of ROS on autophagy in the presence and absence of APO (ROS inhibition) and MK2206 (AKT phosphorylation inhibitor). The results showed that in the presence of APO, AjAKT phosphorylation increased, and autophagy levels decreased. When AjAKT phosphorylation was inhibited, autophagy levels were restored. These findings indicate that increased AjAKT phosphorylation in response to ROS inhibition, likely through its kinase activity, suppresses autophagy. AjAKT appears to be a critical target of endogenous ROS, and the sustained inhibition of its activity by physiological levels of ROS is required for the promotion of autophagy during intestinal regeneration.
FoxO transcription factors regulate autophagy activation by inducing transcription of autophagy related genes
FoxO transcription factors are evolutionarily conserved regulators known to control many physiological processes, including metabolic homeostasis, skeletal muscle homeostasis, stem cell regulation, cardiac remodeling, and tumor suppression [56, 57]. Recently, increasing evidence has linked FoxO transcription factors to autophagy during organ/tissue regeneration by enhancing the transcription of several autophagy-related genes, such as LC3b, Gabarapl1, Pi3kIII, Ulk2, Atg12, Beclin1, Atg7, and Bnip3 [58]. In this study, by analyzing the promoter motifs in the AjFoxO ChIP-seq data, we identified a conserved core binding site “TGTTT” in AjFoxO, similar to that found in other higher organisms [59]. Although direct targets of AjFoxO have been identified genome-wide in organisms such as worms, flies, mice, and human cells using chromatin immunoprecipitation followed by sequencing (ChIP-seq), this is the first identification of such targets in echinoderms. This result highlights the functional conservation of AjFoxO across species, suggesting the potential to extend its regulatory mechanisms to various species. Furthermore, we analyzed potential genes targeted by AjFoxO and identified two autophagy-related genes, AjATG4 and AjLC3. Through a series of experiments, including dual-luciferase and EMSA, we demonstrated that AjFoxO directly binds to the promoters of AjATG4 and AjLC3 and regulates their transcription. Given the important roles of AjLC3 and AjATG4 in autophagosome formation [16], the regulation of their transcription by AjFoxO directly affects autophagy levels during intestinal regeneration in A. japonicus.
The regulation of autophagy by the ROS-AjAKT-AjFoxO-AjATG4/AjLC3 pathway exhibits a certain degree of conservation in different species or life activities
This research provides the first evidence that autophagy is required for intestinal regeneration in A. japonicus and reveals the regulatory mechanism through the ROS-AjAKT-AjFoxO-AjATG4/AjLC3 pathway. On one hand, this molecular pathway offers valuable insights into the regeneration processes and cellular mechanisms of economic echinoderms. On the other hand, from an evolutionary perspective, sea cucumbers are an ideal model for phylogenetic research, as they occupy a unique position in the evolutionary transition from invertebrates to vertebrates. The discovery of this molecular mechanism is not limited to echinoderms and may be extended to other invertebrates and vertebrates, potentially influencing the regulation of various life processes beyond regeneration. For example, in both cancer cells and colon xenograft models, sodium selenite has been shown to induce ROS/AMPK/FoxO3a/GABARAPL1-mediated autophagy and downregulate apoptosis. This finding suggests the potential of sodium selenite as an anti-cancer agent in clinical applications [60]. Similarly, in studies on the use of glucocorticoids in treating human osteoarthritis (OA) and rheumatoid arthritis (RA), dexamethasone (Dex), a synthetic glucocorticoid, upregulates ROS through the ROS/AKT/FoxO3 signaling pathway, leading to autophagy and promoting cartilage degeneration [61]. In research on hypercholesterolemia-induced tendinopathy, it was found that cholesterol lipotoxicity is regulated by the ROS/AKT/FoxO1 signaling pathways, with autophagy having a protective effect on tendon-derived stem cells against apoptosis. This study contributes to developing effective treatment strategies for hypercholesterolemia-induced tendinopathy, such as combining statins and autophagy inhibitors [62]. These studies highlight that although there are minor differences in autophagy regulatory pathways among different species or life-regulatory processes, they generally exhibit strong conservation. Thus, exploring the autophagy regulation mechanism during sea cucumber intestine regeneration not only deepens our understanding of regeneration mechanisms but also provides valuable insights for studying similar processes in other species.
Supplementary Information
Authors’ contributions
Chuili Zeng performed the experiments, interpreted the data, and wrote the manuscript. Chenghua Li participated in the experimental design, interpreted the data, contributed new reagents, analytic tools, and revised the manuscript. Ming Guo revised the manuscript. Ke Xiao performed the partial experiment. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32325050), The State Key Program of Natural Science Foundation of Ningbo (2023J005), The Fundamental Research Funds for the Provincial Universities of Zhejiang (SJLY2024003), The Natural Science Foundation of Zhejiang Province (QN25C190009), The General Projects of Zhejiang Provincial Department of Education (Y202456633) and the K.C.Wong Magna Fund in Ningbo University to Chenghua Li. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Conflict of interest: The authors have no relevant financial or non-financial interests to disclose. Ethics approval All work performed for this study complied with the governing body in each respective country. The sea cucumbers A. japonicus were commercially cultured animals, and all the experiments were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The study protocol was approved by the Experimental Animal Ethics Committee of Ningbo University, China.
Consent for publication
Informed consent for publication was obtained from all participants involved in the study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alvarado AS. Regeneration in the metazoans: why does it happen? BioEssays. 2000;22(6):578–90. [DOI] [PubMed] [Google Scholar]
- 2.Marques IJ, Lupi E, Mercader N. Model systems for regeneration: zebrafish. Development. 2019;146(18):dev167692. [DOI] [PubMed] [Google Scholar]
- 3.Joven A, Elewa A, András S. Model systems for regeneration: salamanders. Development. 2019;146(14):dev167700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivankovic M, Radmila H, Albert T, Markus AV, Miguel VF, Steffen W, Jochen CR. Mode systems for regeneration: planarians. Development. 2019;146(17):dev167684. [DOI] [PubMed] [Google Scholar]
- 5.Zhao L, Gao F, Gao S, Liang Y, Long H, Lv Z, et al. Biodiversity-based development and evolution: the emerging research systems in model and non-model organisms. Sci China Life Sci. 2021;64(8):1236–80. [DOI] [PubMed] [Google Scholar]
- 6.Carnevali MDC, Sugni M, Bonasoro F. Regeneration Potential in Echinoderms: Revisiting the Regeneration Concept. In: Saber S, Sally PL, Robert DR, Iain CW, editors. Frontiers in Invertebrate Physiology: A Collection of Reviews, Volume 3. New York: Apple Academic Press; 2024. p. 185–302.
- 7.Dupont S, Thorndyke M. Bridging the regeneration gap: insights from echinoderm models. Nat Rev Genet. 2007;8(4):320–320. [Google Scholar]
- 8.Dignass AU. Mechanisms and modulation of intestinal epithelial repair. Inflamm Bowel Dis. 2001;7(1):68–77. [DOI] [PubMed] [Google Scholar]
- 9.Su F, Yang H, Sun L. A review of histocytological events and molecular mechanisms involved in intestine regeneration in holothurians. Biology. 2022;11(8):1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quispe-Parra D, Valentín G, García-Arrarás JE. A roadmap for intestinal regeneration. Int J Dev Biol. 2021;65:427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Arrarás JE, Valentín-Tirado G, Flores JE, Rosa RJ, Rivera-Cruz A, Miguel-Ruiz JE, Tossas K. Cell dedifferentiation and epithelial to mesenchymal transitions during intestinal regeneration in H. glaberrima. BMC Dev Biol. 2011;11:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes-Rivera J, Grillo-Alvarado V, Soriano-López AE, García-Arrarás JE. Evidence of interactions among apoptosis, cell proliferation, and dedifferentiation in the rudiment during whole-organ intestinal regeneration in the sea cucumber. Dev Biol. 2024;505:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saera-Vila A, Kish PE, Louie KW, Grzegorski SJ, Klionsky DJ, Kahana A. Autophagy regulates cytoplasmic remodeling during cell reprogramming in a zebrafish model of muscle regeneration. Autophagy. 2016;12(10):1864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura S, Yoshimori T. New insights into autophagosome-lysosome fusion[J]. J Cell Sci. 2017;130(7):1209–16. [DOI] [PubMed] [Google Scholar]
- 15.Park SW, Jeon P, Yamasaki A, Lee HE, Choi H, Mun JY. Development of new tools to study membrane-anchored mammalian Atg8 proteins[J]. Autophagy. 2023;19(5):1424–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou YY, Wang ZK, Huang YJ, Bai CJ, Zhang XL, Fang MF, Ju ZY, Liu B. Membrane dynamics of ATG4B and LC3 in autophagosome formation[J]. J Mol Cell Biol. 2021;13(12):853–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadowaki M, Karim MR. Cytosolic LC3 ratio as a quantitative index of macroautophagy[J]. Methods Enzymol. 2009;452:199–213. [DOI] [PubMed] [Google Scholar]
- 18.Trelford CB, Di Guglielmo GM. Molecular mechanisms of mammalian autophagy[J]. Biochem J. 2021;478(18):3395–421. [DOI] [PubMed] [Google Scholar]
- 19.Saera-Vila A, Kish PE, Kahana A. Autophagy in Zebrafish Extraocular Muscle Regeneration. In: Turksen K, editor. Autophagy in Differentiation and Tissue Maintenance. Methods in Molecular Biology, volume 1854. New York: Humana Press; 2018. p. 105-117. [DOI] [PubMed]
- 20.Zhang Q, Wang Y, Man L, Zhu Z, Bai X, Liu Y, et al. Reactive oxygen species generated from skeletal muscles are required for gecko tail regeneration. Sci Rep. 2016;6(1):20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteom. 2006;5:749–57. [DOI] [PubMed] [Google Scholar]
- 22.Hosokawa T, Saito T, Asada A, Fukunaga K, Hisanaga S. Quantitative measurement of in vivo phosphorylation states of Cdk5 activator p35 by Phos-tag SDS-PAGE. Mol Cell Proteom. 2010;9:1133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YX, Wang Y, Wang H. Recent advances in nanotechnology for autophagy detection[J]. Small. 2017;13(33):1700996. [DOI] [PubMed] [Google Scholar]
- 24.Zhang XJ, Sun LN, Yuan JB, Sun YM, Gao Y, Zhang LB, et al. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 2017;15(10):e2003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for nor-malization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15. [DOI] [PubMed] [Google Scholar]
- 28.Fu XM, Guo M, Liu JQ, Li CH. circRNA432 enhances the coelomocyte phagocytosis via regulating the miR-2008-ELMO1 axis in Vibrio splendidus-challenged Apostichopus japonicus[J]. Commun Biol. 2023;6(1):115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo M, Li X, Tao WJ, Teng F, Li CH. Vibrio splendidus infection promotes circRNA-FGL1-regulated coelomocyte apoptosis via competitive binding to Myc with the deubiquitinase OTUB1 in Apostichopus japonicus[J]. Plos Pathog. 2024;20(8):e1012463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analy sis of relative gene expression data using real-time quantitative PCR and the 2– ∆∆CT method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 31.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–8. [DOI] [PubMed] [Google Scholar]
- 32.Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei LP, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Y, Huo K, Pei X, Liang C, Meng X, Song X, et al. Full-length transcriptome revealed the accumulation of polyunsaturated fatty acids in developing seeds of Plukenetia volubilis. PeerJ. 2022;10:e13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundaram AYM, Hughes T, Biondi S, Bolduc N, Bowman SK, Camilli A, et al. A comparative study of ChIP-seq sequencing library preparation methods[J]. BMC Genomics. 2016;17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David SA, Piégu B, Hennequet-Antier C, Pannetier M, Aguirre-Lavin T, Crochet S, et al. An assessment of fixed and native chromatin preparation methods to study histone post-translational modifications at a whole genome scale in skeletal muscle tissue. Biol Proced Online. 2017;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.HC-Jr BL, Ortego TJ, Payne J, Dienstag JL. by using a LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer’s protocol. Nuclear extract of the cell lines and liver tissue was prepared with an NE-PER nuclear. PNAS. 2007;104(5):1665. [Google Scholar]
- 39.Shao Y, Wang Z, Chen K, Li D, Lv Z, Zhang C et al. Xenophagy of invasive bacteria is differentially activated and modulated via a TLR-TRAF6-Beclin1 axis in echinoderms. J Biol Chem. 2022; 298(3):101667. [DOI] [PMC free article] [PubMed]
- 40.González-Polo RA, Pizarro-Estrella E, Yakhine-Diop SMS, Rodriguez-Arribas M, Gomez-Sanchez R, Casado-Naeanjo I, Bravo-San J, Fuentes JM. The basics of autophagy. In: Maiuri M, De Stefano D, editors. Autophagy networks in inflammation. Progress in Inflammation Research. Springer, Cham; 2016. p. 3-20.
- 41.Nadal M, Gold SE. Assessment of autophagosome formation by transmission electron microscop. Methods Mol Biol. 2012; 835: 481–489. [DOI] [PubMed]
- 42.Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11(8):1437–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J, Zhou L, Yang T, Deng B, Bao Y, Gu L et al. P16INK4a Regulates ROS-Related Autophagy and CDK4/6-Mediated Proliferation: A New Target of Myocardial Regeneration Therapy. Oxid Med Cell Longev. 2023; 2023: 1696190. [DOI] [PMC free article] [PubMed]
- 44.Zhao K, Huang X, Zhao W, Lu B, Yang Z. LONP1-mediated mitochondrial quality control safeguards metabolic shifts in heart development. Development. 2022;149(6):dev200458. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Hu X, Liu Y, Dong S, Wen Z, He W, et al. ROS signaling under metabolistress: cross-talk between AMPK and AKT pathway. Mol Cancer. 2017;16:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klotz LO, Sánchez-Ramos C, Prieto-Arroyo I, Urbanek P, Steinbrenner H, Monsalve M. Redox regulation of FoxO transcription factors. Redox Biol. 2015;6:51–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng CL, Guo M, Xiang YX, Song MS, Xiao K, Li CH. Mesentery AjFGF4–AjFGFR2–ERK pathway modulates intestinal regeneration via targeting cell cycle in echinoderms. Cell Proliferat. 2023;56(2):e13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu X, Wang J, Xia Y, Yin Y, Zhu T, Chen F, et al. Autophagy, a double-edged sword for oral tissue regeneration. J Adv Res. 2023;59:141–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng ZL, Yin BX, Ren RM, Liao YL, Cai H, Wang H. Altered metabolic state impedes limb regeneration in salamanders. Zool Res. 2021;42(6):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee DE, Bareja A, Bartlett DB, White JP. Autophagy as a therapeutic target to enhance aged muscle regeneration. Cells. 2019;8(2):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boya P, Codogno P, Rodriguez-Muela N. Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development. 2018;145(4):dev146506. [DOI] [PubMed] [Google Scholar]
- 52.Pirotte N, Stevens AS, Fraguas S, Plusquin M, Roten AV, Belleghem FV et al. Reactive oxygen species in planarian regeneration: an upstream necessity for correct patterning and brain formation. Oxid Med Cell Longev. 2015; 2015. [DOI] [PMC free article] [PubMed]
- 53.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, et al. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15(2):222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rieger S, Sagasti A. Hydrogen peroxide promotes injury-induced peripheral sensory axon regeneration in the zebrafish skin. Plos Biol. 2011;9(5):e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gauron C, Rampon C, Bouzaffour M, Lpendey E, Teillon J, Volovitch M, et al. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci Rep. 2013;3(1):2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakae J, Oki M, Cao Y. The FoxO transcription factors and metabolic regulation. FEBS Lett. 2008;582(1):54–67. [DOI] [PubMed] [Google Scholar]
- 57.Sanchez AMJ, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci. 2014;71:1657–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39(4):159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webb AE, Kundaje A, Brunet A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell. 2016;15(4):673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu HL, Huang Y, Ge YM, Hong XP, Lin X, Tang KX, Wang Q, Yang Y, Sun WM, Huang YQ, Luo H. Selenite-induced ROS/AMPK/FoxO3a/GABARAPL-1 signaling pathway modulates autophagy that antagonize apoptosis in colorectal cancer cells. Discov Oncol. 2021;12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen C, Cai GQ, Peng JP, Chen XD. Autophagy protects chondrocytes from glucocorticoids-induced apoptosis via ROS/Akt/FOXO3 signaling. Osteoarthr Cartil. 2015;23(12):2279–87. [DOI] [PubMed] [Google Scholar]
- 62.Li KQ, Deng Y, Deng GM, Chen PY, Wang YT, Wu HT, Ji ZG, Yao ZL, Zhang XR, Yu B, Zhang KR. High cholesterol induces apoptosis and autophagy through the ROS-activated AKT/FOXO1 pathway in tendon-derived stem cells. Stem Cell Res Ther. 2020;11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.