Abstract
Background
Inflammatory diseases impair the reparative properties of endothelial progenitor cells (EPC); however, the involvement of diabetes in EPC dysfunction associated with myocardial infarction (MI) remains unknown.
Methods
A model was established combining high-fat diet (HFD)/streptozotocin (STZ)-induced diabetic mice with myocardial infarction. The therapeutic effects of transplanted wild-type EPC, Nlrp3 knockout EPC, and Nlrp3 overexpression EPC were evaluated. Chip and Luciferase assay revealed CEBPB regulated the transcriptional expression of Nlrp3 as a transcription factor in EPC stimulated by high glucose (HG) or advanced glycation end products (AGEs). CO-IP results suggested that USP14 selectively suppressed NLRP3 degradation. KEGG enrichment revealed PI3K/ Akt/mTOR signaling showed striking significance in the entire pathway.
Results
In our study, wild-type, Nlrp3 knockout and Nlrp3 overexpressed EPC, intracardiac injections effectively improved cardiac function, increased angiogenesis, and reduced infarct size in mice with myocardial infarction. However, in the HFD/STZ-induced diabetic mice model combined with myocardial infarction, Nlrp3 knockout EPC significantly restored angiogenic capacity. Mechanically, CEBPB regulated the transcriptional level of Nlrp3 as a transcription factor in EPC. Meanwhile, we found that USP14 selectively suppressed NLRP3 protein degradation through the USP motif on the NACHT domain in mediating inflammasome activation. Cardiac functional outcomes in recipient mice after intramyocardial injection of shNlrp3 EPC overexpressing CEBPB or USP14 validated the modulation of EPC function by regulating Nlrp3 transcription or post-translational modification. Furthermore, KEGG enrichment and validation at the protein levels revealed PI3K/ Akt/mTOR cascade might be a downstream signal for NLRP3 inflammasome.
Conclusion
Our study provides a new understanding of how diabetes affected progenitor cell-mediated cardiac repair and identifies NLRP3 as a new therapeutic target for improving myocardial infarction repair in inflammatory diseases.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12933-024-02541-3.
Keywords: Myocardial infarction, Diabetes, Endothelial progenitor cell, NLRP3 inflammasome, Angiogenesis
Introduction
Cardiovascular disease (CVD) remains the leading cause of morbidity and mortality globally, new therapeutic targets and complementary prevention that contribute to CVD development and progression need to be identified [1, 2]. Acute myocardial infarction (AMI) is a potentially devastating cardiovascular complication for millions of individuals each year and is strongly associated with morbidity, decreased survival time, and mortality [3–6]. CVD in individuals with diabetes is a complex condition influenced by multiple factors, including metabolic, genetic, and lifestyle elements. Evidence indicates that a comprehensive approach to controlling these risk factors can lead to substantial reductions in the occurrence of cardiovascular events [7]. Evidence suggests that Type 2 diabetes mellitus (T2DM) enhances the risk for CVD by 2- to 6-fold and exacerbates myocardial ischemia/reperfusion injury [8]. Figuring out how T2DM exacerbates myocardial infarction may provide potential therapeutic strategies for myocardial infarction in diabetes.
Stem cell therapy shows potential for repairing the heart after ischemic injury in recent years. Various stem cell types that may repair the injured heart have shown small improvements in cardiac structure and function and a few have shown evidence for myogenesis [9, 10]. Endothelial progenitor cells (EPC) have been isolated from peripheral blood and bone marrow which can enhance angiogenesis after infusion into host animals [11]. However, in clinical trials, EPC transplantation in myocardial infarction shows encouraging but still relatively modest results [12, 13]. The underlying mechanisms of EPC-mediated repair are largely unknown and controversial. In addition, the adverse effects of inflammation on the survival and function of transplanted EPC during angiogenesis remain a challenge.
The NLR Family Pyrin Domain Containing 3 (NLRP3) inflammasome is a multimolecular complex that includes an aminoterminal PYRIN (PYD) domain, a nucleotidebinding NACHT domain, and a carboxyterminal leucinerich repeat (LRR) domain [14–16]. NLRP3 is specifically involved in inflammasome activation under a variety of conditions, as it can respond to a diverse range of stimuli that cause loss of autoinhibition, including ATP, nigericin (Nig), alum, crystals, amyloid-β, lysosomal damage [17]. NLRP3 inflammasome activation causes autotomy and activation of the cysteine protease caspase1, which promotes maturation and secretion of IL-1β and IL-18, and induces pyroptosis. NLRP3 has been implicated in a wide range of pathological sterile inflammation associated with several autoinflammatory, neurodegenerative, and metabolic diseases including Alzheimer’s disease, rheumatoid arthritis, Parkinson’s disease, gout, atherosclerosis, and diabetes [18–21]. However, studies on the relationship between NLRP3 inflammasome and EPC are still insufficient and thus specific research on them is imperative and meaningful.
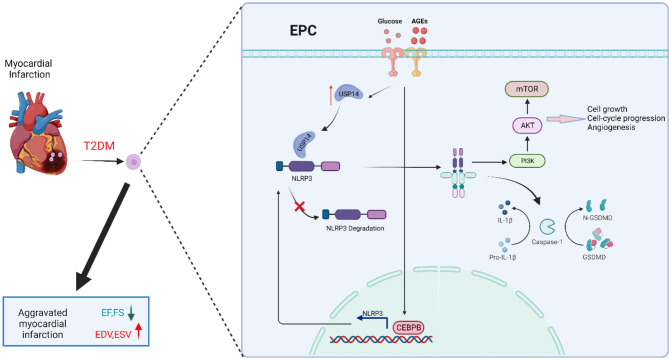
Taken together, our studies demonstrate that knockout of Nlrp3 inhibits the secretion of Caspase1 p20 and IL-1β, and improves cardiac function, restrains left ventricular remodeling after permanent myocardial ischemia in vivo. Here, we show that CEBPB initiates the transcription of Nlrp3 by directly binding to the promoter region of Nlrp3 in EPC. USP14 could interact with NLRP3 to restrain autophagic degradation of NLRP3 at post-transcriptional regulation. Our study thus uncovers mechanisms regulating NLRP3 inflammasome activation in EPC and suggests a promising approach for modulating represent therapeutic targets in diabetic myocardial infarction.
Materials and methods
Animals
Eight-week-old wild-type (C57BL/6J, WT) and Nlrp3 knockout (C57BL/6J, Nlrp3 KO) male mice were procured from Nanjing GemPharmatech company. All experimental protocols were approved by the ethic committee of China Pharmaceutical University, following the Guidelines of Animal Experiment set by the Bureau of Sciences and Techniques of Jiangsu Province, China [NO. SYXK2007-0025].
Preparation of the HFD/STZ-induced diabetic mice model
Adult C57BL/6J male mice were divided into two groups: control, and untreated diabetic group. During the first 6 weeks, mice in the untreated diabetic group were given a high-fat diet (HFD, 10% sucrose, 10% lard, 10% sugar, 5% egg yolk powder, 0.5% cholesterol, 64.5% basic feed) purchased from Qinglong Mountain Laboratory Animals Ltd. Control mice received only a regular diet. At the beginning of the 7th week, HFD-feeding mice were intraperitoneally injected three times with streptozotocin (STZ, 35 mg/kg body weight, dissolved in pH 4.5 citrate buffer, Sigma, N572201) to induce partial loss of pancreatic β-cells as described previously. Control mice only received an equivalent volume of citrate buffer. After 1 week of STZ injection, mice were identified as diabetic mice with fasting blood glucose (FBG) levels reaching 16.7 mM (Supplementary Fig. 1A). We established the HFD/STZ-induced diabetic mice model by assessing multiple indicators including insulin levels (Supplementary Fig. 1B), HbA1c levels (Supplementary Fig. 1C), Oral Glucose Tolerance Test (OGTT) (Supplementary Fig. 1D), and Insulin Tolerance Test (ITT) (Supplementary Fig. 1E).
Myocardial infarction and cell therapy
Myocardial infarction was induced by ligation of the left coronary artery as previously described [22, 23]. Animals are induced with 5% isoflurane in the induction chamber, then intubated and mechanically ventilated with 1–3% isoflurane to maintain the depth of surgical anesthesia during the procedure. Surgical pain was reduced by preincisional subcutaneous injection of buprenorphine 0.05 mg/kg. A left open-heart procedure is performed in the fourth intercostal space of the mouse, followed by a pericardiotomy. The left coronary artery is ligated with a 6 − 0 circular suture, and ST-band elevation is observed on the ECG. EPC were transplanted to the border area of myocardial infarction by injecting at three different but adjacent sites using a syringe. The number of cells injected at each site was 4 × 105, and the PBS group was injected with the same volume of PBS. The heart is rapidly returned to the thorax, which is closed and the mice are allowed to recover using assisted mode ventilation. Pulmonary atelectasis was prevented by generating positive end-expiratory pressure at the end of the procedure. Sham control animals underwent the same surgical procedure in the absence of coronary occlusion. The surgical mortality rate in HFD/STZ-induced diabetic mice after myocardial infarction is approximately 22%, which suggests that our surgical approach is feasible. For euthanasia, mice were placed in a sealed chamber and were euthanized by CO2 asphyxiation, followed by cervical dislocation.
Echocardiography
Echocardiography was performed using a 2% isoflurane anesthesia delivered through a Viking Medical system (Medford, NJ) in mice. Transthoracic two-dimensional M-mode echocardiograms were obtained with the Vevo 3100 equipped with a 30 MHz transducer (Visual Sonics, Toronto, Canada). Echocardiographic studies were conducted before myocardial infarction (baseline) and at 1, 7, and 21 days post-myocardial infarction under anesthesia using a mixture of 1.5% isoflurane and oxygen (1 L/min). The left ventricular (LV) internal diameter was measured in the short-axis view recorded in M-mode; ejection fraction (EF) and fractional shortening (FS) were calculated using the formulas previously described [24].
Bone marrow cell isolation and EPC culture
Briefly, bone marrow mononuclear cells were isolated from mice by density gradient centrifugation with Histopaque-1083 at room temperature and depleted of macrophages by allowing attachment to uncoated plates for 1 h. The unattached cells were removed and plated on 6-well type I collagen-coated tissue culture plates and cultured in phenol red-free endothelial cell basal medium-2 (EBM-2, Clonetics) supplemented with 10% FBS, vascular endothelial growth factor (VEGF)-A, fibroblast growth factor-2, epidermal growth factor, insulin-like growth factor-1, ascorbic acid, and antibiotics. Cells were cultured at 37 °C with 5% CO2 for 48 h before changing media to remove non-adhered cells by washing with PBS. Media was changed every 2 days until colonies appeared after 3 weeks. These EPCs were utilized in subsequent in vivo and in vitro experiments. Similarly, Nlpr3 knockout EPC were isolated from bone marrow mononuclear cells of Nlrp3 knockout mouse according to the above experimental procedure.
FACS sorting
To investigate whether diabetic condition affects MI-induced mobilization of BM-EPC into the circulation, we performed myocardial infarction (MI) in control and HFD/STZ induced diabetic mice and assessed EPC mobilization (Sca-1+ Flk1+ cells) by FACS analysis on peripheral blood mononuclear cells. Freshly isolated mononuclear cells from peripheral blood by histopaque-1083 were stained with Phycoerythrin(PE)-conjugated Rat anti-mouse stem cell antigen 1 (Sca-1) and Allophycocyanin (APC)-Rat conjugated anti-mouse fetal-liver kinase 1 (Flk1) antibodies (BD Pharmingen Inc.) in 1% BSA. Isotype-matched IgG antibodies were used as negative controls. Quantitative fluorescence analyses were performed with a CytoFLEX cytometer (Beckman Coulter, Inc.) and Flow-Jo Software (Tree Star, Inc.); 50,000 events were counted for each sample.
Plasmid construction and transfection
MmNlrp3-coding (Gene ID:216799), MmCaspase1-coding (Gene ID:12362), and MmPycard-coding (Gene ID:66824) sequence were amplified from EPC by touch-down PCR and cloned into pCMV-Myc-N (635689, Clontech). MmUsp14-coding sequence (Gene ID:59025) was cloned into p3XFLAG-CMV-14 (Sigma). The deletion and truncation sequence of Nlrp3 were amplified from EPC by touch-down PCR and cloned into pCMV-Myc-N. pCDH-CMV-MCS-EF1-copGFP-T2A-Puro-Nlrp3 was generated by subcloning the MmNlrp3-coding sequence into the pCDH-CMV-MCS-EF1-copGFP-T2A-Puro (CD513B-1, SBI), and the empty pCDH-CMV-MCS-EF1-copGFP-T2A-Puro was used as a control. Plvx-mcherry-Usp14 was generated by subcloning the MmUsp14-coding sequence into the plvx-mcherry-N1 (632562, Clontech), and the empty plvx-mcherry-N1 was used as a control. In addition, the shRNA targeting Usp14 and Nlrp3 mRNA sequence was introduced into pSIH1-H1-copGFP-T2A-Puro vector (System Biosciences) under the control of the H1 promoter. The Nlrp3 shRNA targeting sequence was 5’- CCGGCCTTACTTCAATCTGTT-3’. The Usp14 shRNA targeting sequence was 5’-CCTCCGAAAGAGATTAAGTAT-3’. Sh-NC was constructed (5’-GAAGCAGCACGACTTCTTC-3’) with no significant homology to any mammalian gene sequence. All constructed plasmids are verified by DNA sequencing. Plasmids were transiently transfected into EPC with Lipofectamine 2000 Transfection Reagent (Invitrogen, USA) according to the manufacturer’s protocol.
Lentivirus production
To make lentiviral particles, HEK 293T cells were grown in a 100 mm cell culture dish in DMEM media without antibiotics. When cells were 70 ~ 80% confluent, 10 µg lentiviral transfer plasmid, 5 µg psPAX2, and 2.5ug pMD2.G were co-transfected into HEK-293T cells with Lipofectamine 2000 Transfection Reagent (Invitrogen, USA) according to the manufacturer’s protocol. Lentiviral particles were harvested at 48 and 72 h post-transfection and concentrated with L-80XP Super Speed Centrifuge (BECKMAN). The lentiviruses were subsequently used to infect EPC.
Luciferase assay
Cells were plated in 96-well plates 24 h before transfection. Plasmids containing different fragments of the Nlrp3 promoter were generated by subcloning the Nlrp3 promoter sequence into the pGL4.17 (Promega) vector. All plasmids containing different fragments of the Nlrp3 promoter were co-transfected with a control Renilla luciferase plasmid (pRL-TK). The ratio of the experimental plasmid to the control plasmid was set at 50:1. Luciferase activity was measured using the Duo-Lite Luciferase Assay System according to the manufacturer’s instructions (DD1205, Vazyme). Briefly, at 24–48 h after transfection, cell lysates were prepared by incubating the cells with the same volume of Duo-Lite Luciferase reagent for 10 min at room temperature. The firefly luminescence signal was detected with Promega GloMax 20 according to the manufacturer’s instructions. After this, duo-Lite Stop reagents are added for the detection Renilla luminescence signal. Firefly luminescence signal in transfected cells was normalized to Renilla luminescence signal.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assays were performed according to the standard ChIP protocol (Millipore). Briefly, EPC was harvested and fixed with 1% formaldehyde for 10 min at room temperature, then crosslinking was quenched with 2-M glycine for 5 min. The chromatin was sonicated until the DNA fragments were 500 bp in size after nuclei isolation. After sonication, 2% percent of the sonicated chromatins were used as input control, and the rest sonicated chromatins were incubated with specific antibodies. Chromatin fragments containing DNA-protein were precipitated by using 5 µl of anti-C/EBPβ (Proteintech) or 5 µl of anti-IgG (Beyotime Biotechnology). The immunoprecipitated complex was washed with low salt immune complex wash buffer, high salt immune complex wash buffer, LiCl immune complex wash buffer, and TE buffer. The DNA was extracted and purified by DNA Extraction Mini Kit (Vazyme).
The enrichment of specific DNA sequences was examined by PCR using specific primer pairs of the different sites of Nlrp3 promoters. Primers were used as follows:
Site1-F 5′-TCCATCCAGATGAGTAACTGCCAATCC-3′.
Site1-R 5′-GGGTCACTGTGACACTGGAGAC-3′.
Site2-F 5′-CCAGATTTCTGGAGACCAACCTAGT-3′.
Site2-R 5′-AGCCAGACTCAGGAAGACAGGAG-3′.
Site3-F 5′-GCTCATCCTCCTATGATGGAGTTG-3′.
Site3-R 5′-GGCTCAAAGAAGCCACTAATGACC-3′.
Nuclear protein extraction
The nuclear protein extraction was performed using the Nuclear and Cytoplasmic Protein Extraction Kit (P0027, Beyotime, China) according to the manufacturer’s instructions. Cells were collected in PBS by scraping from culture flasks and washed twice with cold PBS. The supernatant was removed and the cell pellet was collected into a prechilled microcentrifuge tube. The cell pellet was resuspended with 200 µL Buffer A containing 1 mM PMSF and incubated on ice for 15 min. Then 10 µL buffer B was added and the mixture was vortexed for 5 s for 1 min, followed by centrifugation at 12,000 g, 4 °C for 5 min. The supernatant was removed and the precipitation was resuspended in a 50 µL nuclear protein extraction reagent containing 1 mM PMSF. The mixture was vortexed for 30 min and centrifugated at 12,000 g, 4 °C for 5 min. The supernatant was collected as nuclear protein and analyzed by Western blotting assay.
Co-immunoprecipitation
Cells were washed twice with cold PBS and collected by scraping from culture flasks. Collected cells were lysed in IP lysis buffer containing 1 mM PMSF and centrifuged at 14,000 g, 4 °C for 10 min. The supernatant was transferred into a microcentrifuge tube and incubated with 2 µg primary antibody (Beyotime Biotechnology) overnight at 4 °C. Then the mixture was incubated with 20 µl fully resuspended Protein A + G Agarose for 3 h. Protein A + G Agarose was washed five times with IP buffer. The immunoprecipitates were eluted by boiling with 1% (wt/vol) SDS buffer and analyzed by Western blots assay.
Protein preparation and western blots
Supernatant protein: The precipitation of supernatant proteins was performed using the methanol/chloroform method. Initially, culture media were centrifuged at 2000 × g for 10 min to pellet the cells and cell debris. Subsequently, 600 µL of the supernatant was transferred to a new tube, followed by the addition of 600 µL of methanol and 150 µL of chloroform. The samples were thoroughly mixed and centrifuged at 12,000 × g for 10 min. The upper phase was discarded, and 600 µL of methanol was added to each sample. After mixing well, the samples were centrifuged again at 12,000 × g for 10 min. The protein contents were deter-mined using Bradford reagent (Bio-R ad protein assay kit) and aliquots normalized to equal quantities before loading. The supernatants were removed, and the remaining protein pellets were dried at 55 °C, then resuspended in 2× SDS loading buffer. Finally, the samples were boiled at 98 °C for 10 min until fully dissolved.
Matrigel plug: Tissue samples from mice, embedded in Matrigel, were homogenized using a homogenizer and subsequently lysed in RIPA buffer. Protein concentrations were determined using the Bradford assay, with 60 µg of protein loaded into each well for analysis.
Cell lysates: EPC was lysed in SDS-RIPA lysis buffer and protease inhibitor cocktail. After 15 min centrifugation at 12,000 × g at 4 °C, protein concentrations in the lysates were collected and eluted by boiling with 1% (w/v) SDS sample buffer. Equal amounts of proteins were separated by SDS–PAGE and then were transferred onto PVDF membranes for immunoblot analysis. After membranes were blocked with 5% skim milk for 1.5 h at room temperature, and the blocking buffer was changed to 5% BSA for phosphorylation, they were probed with various primary antibodies overnight at 4 °C. After washing for three times, the membranes were incubated with secondary antibodies for 1.5 h at room temperature and visualized with chemiluminescent systems with LumiGlo and Peroxide. The intensity of protein bands was measured using ImageJ image analysis software (NIH, Littleton, CO, USA).
Immunofluorescence microscopy
EPC was plated on glass coverslips in 24-well plates and fixed with 4% paraformaldehyde for 15 min. Cells were permeabilized with 0.3% Triton-X 100 in phosphate buffer saline (PBS) and blocked with 5% goat serum for 1 h. Then the cells were incubated with anti-USP14 and anti-Nlrp3 diluted in Immunol Staining Primary Antibody Dilution Buffer (Beyotime) at 4 °C overnight. Next, the cells were stained with secondary Antibodies, and nuclei were stained with DAPI (Solarbio, Beijing, China). The fluorescent images were photographed by a Laser scanning confocal microscope (ZEISS Scope. A1, Germany) and the images were processed by ZEN blue 2.3 software (Carl Zeiss, Germany). Pearson’s coefficient which represents colocalization was calculated by Image J software (NIH, Littleton, CO, USA).
ELISA
The concentrations of mouse IL-1β (cat. EK201B), and mouse IL-18 (cat. EK218) were measured using ELISA kits (lianke Biotech Company Ltd., Hangzhou, China) according to the manufacturer’s instructions.
Angiogenesis assay
The angiogenic capability of EPC was determined by matrigel tube formation assay. Briefly, 96-well plates were coated with matrigel (50 µL/well; BD Biosciences) at 37 °C for 30 min. Then, 3 × 104 EPC were plated on matrigel per well and incubated at 37 °C with 5% CO2 for 6 h. Six hours post-treatment, the digital images of the tubes were photographed with a phase-contrast microscope. The total tube length was counted in each well by Image J software.
Matrigel plug assay
The angiogenesis model was established using Matrigel (BD) implants in mice. Specifically, 500µL of Matrigel, with 5 × 105 EPC, was injected subcutaneously into the mid-dorsal region of 8-week-old male mice. The injected Matrigel quickly formed a single, solid plug. After 10 days, the Matrigel plugs were surgically removed from the mice, ensuring the exclusion of connective tissues. The neovascularization of Matrigel plugs was quantified by adding 4 ml of Drabkin’s reagent to 20 ml of well-homogenized neovascularized Matrigel. Following thorough mixing, the absorbance was measured at a wavelength of 540 nm using a spectrophotometer to estimate hemoglobin levels. Hemoglobin concentration was calculated using the formula: Hb (g/dl) = (absorbance of sample / absorbance of standard) × concentration of standard. The excised Matrigel was then embedded in paraffin and sectioned for HE analyses. The infiltrated vessels were counted in three Matrigel H&E-stained sections and expressed as the mean number.
Bioinformatics analyses
RNA sequencing was performed on an Illumina HiSeq 4000 platform of aptbiotech Co., Ltd. (Shanghai, China) to evaluate the differential gene expression profiles in EPC. The gene expression level was normalized by using the EdgeR method. The criterion for significant DEGs (differentially expressed genes) was set to at least a 2-fold change in gene expression, with an adjusted P-value < 0.01. The DEGs were enriched by KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways with the GSEA (gene set enrichment analysis) method in the ClusterProfiler package.
Statistical analysis
Data were presented as mean ± standard error mean (S.E.M.). Statistical analysis was performed with IBM SPSS Statistics 20 software. The Independent-Samples t-test was used to detect significant differences between the two groups. Significant differences between and within multiple groups were examined using ANOVA for repeated measures, followed by Duncan’s multiple-range test. In addition, for the sample groups where n = 3, Mann-Whitney test was employed. Kaplan–Meier survival curve was used for survival analysis, and the log-rank test was used to determine the statistical significance. P < 0.05 was considered statistically significant.
Results
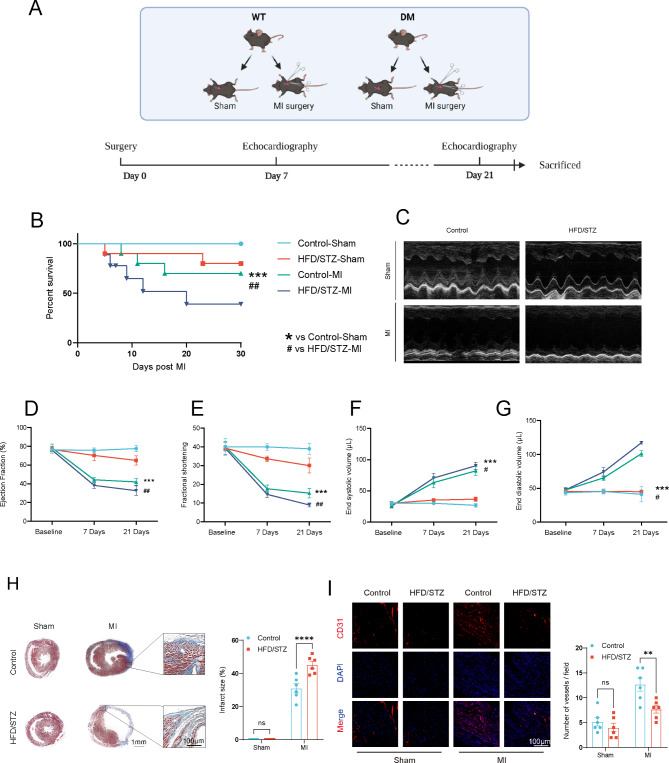
Impaired cardiac function and promoted left ventricular remodeling after permanent myocardial ischemia in HFD/STZ-induced diabetic mice
Diabetes mellitus constitutes a significant risk factor for cardiovascular diseases. The overall schedule of experimental procedures is presented in Fig. 1A. To assess cardiac function impairment in diabetic condition, permanent myocardial ischemia was performed on control and HFD/STZ-induced diabetic mice. Left ventricular cardiac function was measured by echocardiography on day 7 and day 21. Masson trichrome staining was employed on day 21 post-myocardial infarction to analyze cardiac remodeling in heart samples. Our data indicated that the 30-day survival rate for the HFD/STZ-induced diabetic mice with myocardial infarction model was 45%, which was lower than that of the myocardial infarction mice and significantly lower than that of the sham-operated mice (Fig. 1B). The HFD/STZ-induced diabetic mice exhibited significantly impaired left ventricular cardiac function (Fig. 1C), as demonstrated by the reduced ejection fraction and fractional shortening percentages on day 21 post-myocardial infarction (Fig. 1D and E) and a significant difference in end diastolic volume and end systolic volume (Fig. 1F and G). Histological analysis of heart samples on day 21 post-myocardial infarction indicated that the HFD/STZ-induced diabetic mice exhibited a significant increase in myocardial infarction scar size. (Fig. 1H). Immunofluorescence analysis revealed a substantial reduction in neovascularization in the HFD/STZ-induced diabetic mice. (Fig. 1I). Therefore, these findings demonstrated that, compared to control mice, the HFD/STZ-induced diabetic mice exhibited exacerbated myocardial infarction, which adversely affected their prognosis.
Fig. 1.
Impaired cardiac function and promoted left ventricular remodeling after permanent myocardial ischemia in HFD/STZ-induced diabetic mice. (A) a diagram of the experimental protocol. (B) Short-term survival curve after myocardial infarction and the indicated treatments (n = 10–12 mice/group). (C) Representative short-axis M-mode echocardiograms of the left ventricle in the control-Sham, HFD/STZ-Sham, control-MI, and HFD/STZ-MI groups. Left ventricular function was assessed by measurements of ejection fraction (D), LV fractional shortening (E), end diastolic volume (F), end systolic volume (G), and at day 7 and day 21 after MI (n = 6 mice/group). (H) Representative images and quantitative infarct size in Masson’s trichrome stained mice hearts on day 21 after MI (n = 6 mice/group). (I) Representative endothelial CD31 staining at the infarction border zone sections. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Control-sham, # p < 0.05, ## p < 0.01, ### p < 0.001 vs. Control-MI group. Similar results were obtained from three independent experiments
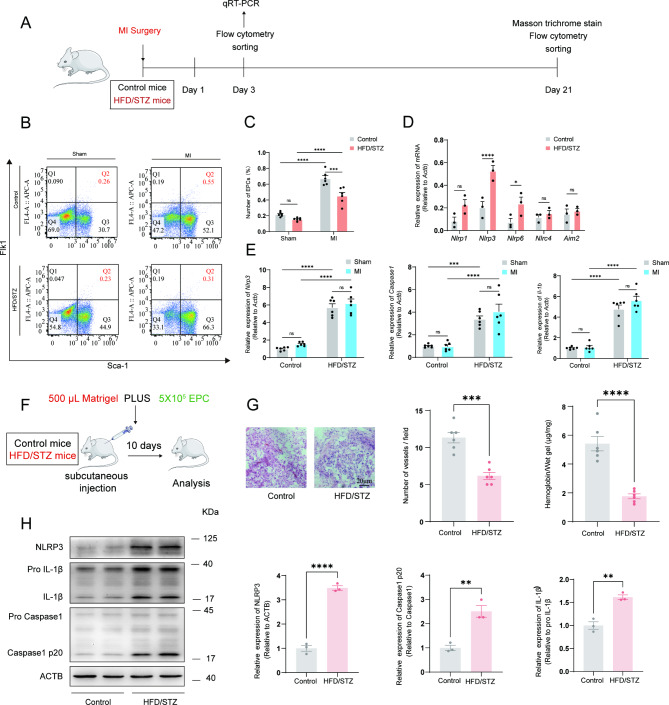
Reduced EPC mobilization and activated NLRP3 inflammasome in HFD/STZ-induced diabetic mice
The schedule of experimental procedures is presented in Fig. 2A. To determine whether diabetic conditions affected MI-induced mobilization EPC in the circulation, we performed MI and assessed relative EPC number in the circulation by FACS analysis on day 3 after MI (Fig. 2B). HFD/STZ-induced diabetic mice showed a decreased number of Flk1+/Sca-1+(double+) cells on day 3 after MI compared to control mice in MI group (Fig. 2C). Furthermore, within the selected inflammasome-related gene family, the expression of Nlrp3 showed significant differences between the control group and the HFD/STZ-induced diabetic mice group in EPC (Fig. 2D). In addition, sorted EPC were analyzed for expression of NLRP3 inflammasome-related mRNA abundance. However, there was no significant difference between sham and MI mice in sham groups or HFD/STZ-induced diabetic mice (Fig. 2E). The expression levels of Nlrp3, Caspase1 and Il1b mRNA in sorted EPC were significantly greater in HFD/STZ-induced diabetic mice than in the control groups with sham and MI groups, suggesting that diabetic condition influenced mobilization of EPC and activated the NLRP3 inflammasome. To further investigate the effect of diabetic condition on EPC in vivo, EPC were obtained from mouse bone marrow mononuclear cells by density gradient centrifugation (Supplementary Fig. 2A and 2B) and a Matrigel plug (Fig. 2F). Angiogenesis assay was performed to study the modulation of in vivo angiogenesis. Hemoglobin content and blood vessel infiltration in implants from HFD/STZ-induced diabetic mice were significantly less than in control mice (Fig. 2G). Similarly, the NLRP3 inflammasome was also activated in EPC from implants of HFD/STZ-induced diabetic mice (Fig. 2H). These findings are consistent with the subsequent NLRP3 inflammasome results observed in the diabetic sorted EPC. Collectively, these results indicated that diabetic condition decreased EPC mobilization, angiogenic which was be relevant to the activation of the NLRP3 inflammasome.
Fig. 2.
Reduced EPC mobilization and activated NLRP3 inflammasome in HFD/STZ-induced diabetic mice. (A) a diagram of the experimental protocol. (B) FACS analysis on peripheral blood mononuclear cells for EPC mobilization in groups. (C) The bar graph showed that EPC mobilization was impaired in HFD/STZ-induced diabetic mice as compared with control mice (n = 6 mice/group). (D) Nlrp1, Nlrp3, Nlrp6, Nlrc4 and Aim2 mRNA expression (RT-PCR) in flow cytometry-sorted EPC were quantified at 3 days after MI. (E) Nlrp3, Caspase1, and Il-1b mRNA expression (RT-PCR) in flow cytometry-sorted EPC were quantified at 3 days after MI. mRNA expression normalized to Actb and depicted as fold change versus control. (F) a diagram of the matrigel plug protocol. (G) Representative HE photographs of matrigel plugs removed from mice 10 days after injection (n = 6 mice/group). The relative amounts of hemoglobin extracted from the excised matrigel plugs and the number of blood vessels in matrigel plugs (n = 6 mice/group). (H) Relative NLRP3, Caspase1 p20, and IL-1β protein expression levels in EPC extracted from the excised matrigel plugs (n = 3 mice/group). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. These experiments were repeated independently at least three times with similar results
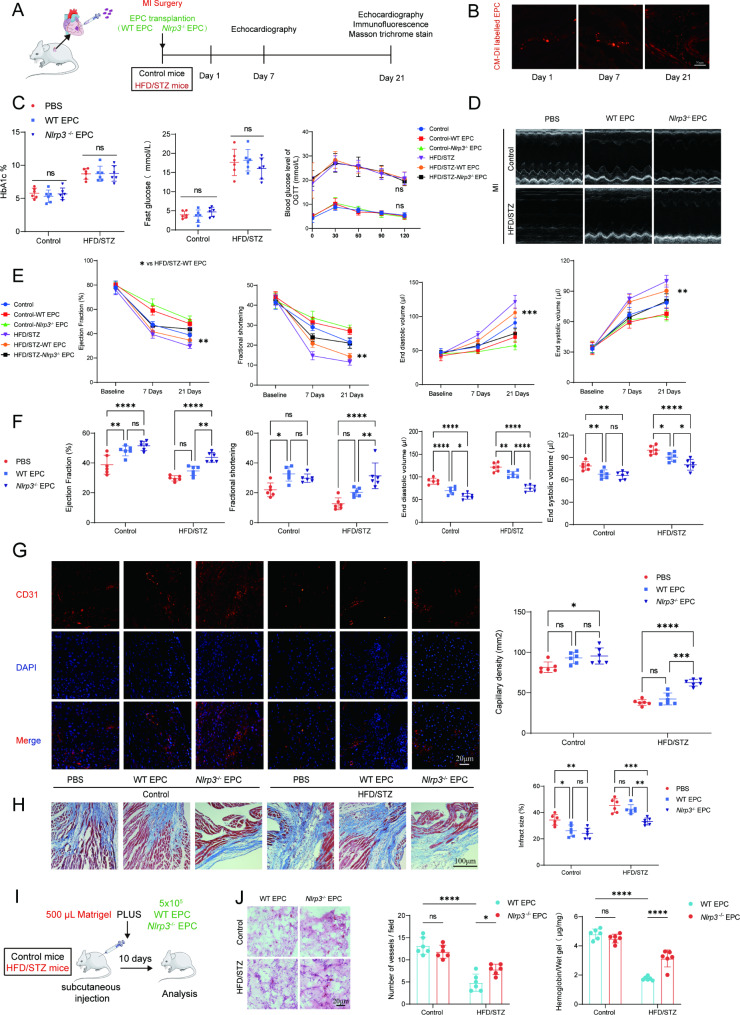
IntramyocardialinjectionNlrp3knockout EPC improved cardiac function and restrained left ventricular remodeling after permanent myocardial ischemia in HFD/STZ-induced diabetic mice
To assess EPC dysfunction under diabetic condition, WT EPC, Nlrp3−/−EPC, or PBS were administered intramyocardially to the border zone of myocardial infarction in AMI mouse model. The schedule of experimental procedures were presented in Fig. 3A and Supplementary Fig. 3A. To examine EPC incorporation at post-transplantation time points, heart tissue from mice injected with Dil-labeled EPC was subjected to frozen sections (Fig. 3B). In addition, intramyocardial injection of EPC did not alter the glucose and hemoglobin levels in the HFD/STZ-induced diabetic mice (Fig. 3C). Our data revealed that compared with PBS, WT-EPC or Nlrp3−/− EPC treatment significantly improved left ventricle cardiac function (Fig. 3D) as evident from increased percentage ejection fraction and fractional shortening on both day 7 and day 21 post-MI and a significant difference in end diastolic volume and end systolic volume in MI mice without diabetes (Fig. 3E and 3F). However, for HFD/STZ-induced diabetic mice with MI, WT EPC intramyocardial injection exerts a limited therapeutic benefit, Nlrp3−/− EPC intramyocardial injection significantly improved cardiac function. Furthermore, overexpression of Nlrp3 reversed the therapeutic effects of EPC on myocardial infarction (Supplementary Fig. 3C and 3D). Intramyocardial injection Nlrp3 overexpression EPC decreased 30 days survival in diabetic myocardial ischemia mice model (Supplementary 3B). Immunofluorescence and histological analysis on day 21 post-MI heart samples indicated that Nlrp3−/− EPC treatment significantly decreased infarction size and promoted angiogenesis compared to WT-EPC in HFD/STZ-induced diabetic mice with MI (Fig. 3G and 3H). Overexpression of Nlrp3 also decreased angiogenesis and increased infarction size in HFD/STZ-induced diabetic mice with MI (Supplementary Fig. 3E and 3F). Matrigel plug results showed significantly reduced hemoglobin content and blood vessel infiltration in HFD/STZ-induced diabetic mice 10 days after injection (Fig. 3I and 3J). Similarly, we found overexpression of Nlrp3 failed to increase angiogenesis in Matrigel plug from HFD/STZ-induced diabetic mice (Supplementary Fig. 3G and 3H). Taken together, these data suggested that diabetes can exacerbate myocardial infarction in vivo by impairing the angiogenic function of the EPC through the NLRP3 inflammasome.
Fig. 3.
Intramyocardial injectionNlrp3 knockout EPC improved cardiac function and restrained left ventricular remodeling after permanent myocardial ischemia in HFD/STZ-induced diabetic mice. (A) a diagram of the transplanted experimental protocol. (B) Representative images showing that the fluorescence signal observed in the heart of myocardial ischemia mice on days 1, 7 and 21 after injection of Dil-labeled EPC. (C) Fasting glucose, oral glucose tolerance test and HbA1c in HFD/STZ-induced diabetic myocardial infarction mice followed by intramyocardial injection of EPC. (D) Representative short-axis M-mode echocardiograms of the left ventricle at day 21 in the control and HFD/STZ-induced diabetic groups. (E) Left ventricular function after injection of Nlrp3 knockout EPC was assessed by measurements of ejection fraction, LV fractional shortening, end diastolic volume, and end systolic volume at day 7 and day 21 after MI (n = 6 mice/group). (F) Left ventricular function was assessed by measurements of ejection fraction, LV fractional shortening, end diastolic volume, and end systolic volume at day 21 after MI (n = 6 mice/group). (G) Representative endothelial CD31 staining at the infarction border zone sections. (H) Representative images and quantitative infarct size in Masson’s trichrome stained mice hearts on day 21 after MI (n = 6 mice/group). (I) a diagram of the matrigel plug experimental protocol. (J) Representative HE photographs of matrigel plugs removed from mice 10 days after injection. The relative amounts of hemoglobin extracted from the excised matrigel plugs and the number of blood vessels in matrigel plugs. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Similar results were obtained from three independent experiments
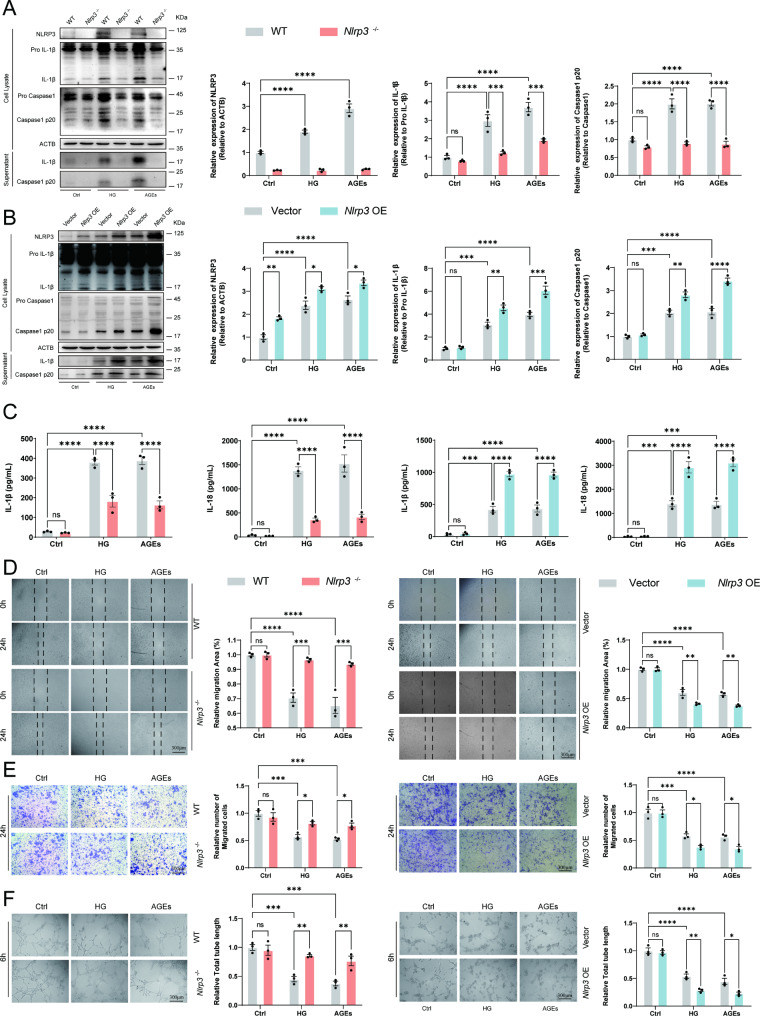
NLRP3 inflammasome activation impaired angiogenesis function of EPC in response to HG or AGEs
Next, we further explored how diabetes, as an independent factor, caused inflammatory responses in the EPC and affected the angiogenic function of the EPC. To confirm the role of HG or AGEs in activating inflammatory response in EPC in vitro, EPC from wildtype or Nlrp3−/− mice was stimulated with HG or AGEs for 72 h. The immunoblotting analysis showed that Nlrp3−/− EPC responded less efficiently than WT EPC to HG or AGEs. HG or AGEs induced content of IL-1β and Caspase1 p20 in supernatants which indicated the activation of NLRP3 inflammasome (Fig. 4A). Our results suggest that both HG or AGEs effectively activated NLRP3 inflammasome in EPC. Nlrp3 knockout effectively inhibited the NLRP3 inflammasome activation under stimulated conditions with HG or AGEs. In addition, overexpression of Nlrp3 dramatically activated the NLRP3 inflammasome in EPC primed by stimulation with HG or AGEs for 72 h (Fig. 4B). Further, we found the secretion of Il-1β and Il-18 was significantly decreased in Nlrp3−/− EPC primed by stimulation with HG or AGEs for 72 h (Fig. 4C). Nlrp3 overexpression significantly increased the level of Il-1β and Il-18 in the supernatant (Fig. 4C).
Fig. 4.
Regulated angiogenesis function of EPC by NLRP3 inflammasome in response to HG or AGEs. (A) Western blot analysis of NLRP3, Caspase1, and IL-1β cleavage in EPC from WT or Nlrp3−/− mice following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. The supernatant was collected directly after a 72-hour incubation period with high glucose and AGEs. (B) EPC from wild-type mice were transfected with Vector or Nlrp3. Western blot analysis of NLRP3, Caspase1, and IL-1β cleavage in EPC transfected with Vector or Nlrp3 following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. (C) ELISA analysis of IL-18, and IL-1β in supernatants of EPC from WT or Nlrp3−/− mice and EPC transfected with Vector or Nlrp3 following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. (D) Wound healing of EPC response to HG (30 mM) or AGEs (200 µg/ml) in vitro. (E) Migration assay with EPC treated with HG (30 mM) or AGEs (200 µg/ml) was performed. (F) The angiogenic function of EPC was evaluated by tube formation assay. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Representative results from three biologically independent experiments
To investigate whether the HG or AGEs induced a decrease in angiogenesis activity of EPC, we carried out the vitro angiogenesis assays using WT, Nlrp3−/− EPC and Nlrp3 overexpressed EPC following stimulation with HG or AGEs for 72 h. Our data show that the knockout of Nlrp3 in EPC abrogated HG or AGEs induced a decrease in tube formation, and chemotactic migration activity, suggesting that HG or AGEs diminish the angiogenesis capacity of EPC through NLRP3 inflammasome activation. However, no change was observed in the EPC from WT or Nlrp3−/− mice without the following stimulation with HG or AGEs (Fig. 4D and F). Respectively, overexpression of Nlrp3 significantly aggravated impairment of angiogenesis in EPC response to HG or AGEs while no significant angiogenesis was observed without the stimulation of HG or AGEs (Fig. 4D and F).
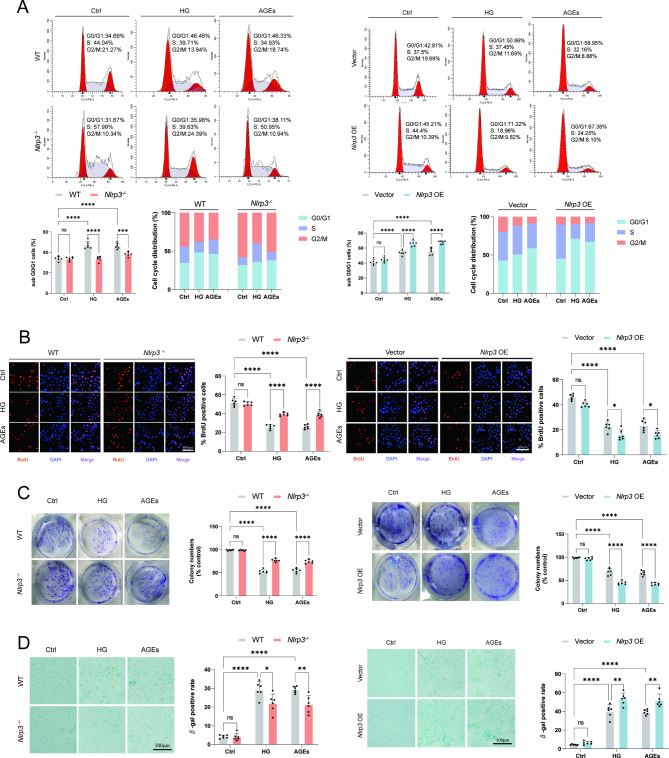
Regulated proliferation function of EPC by NLRP3 inflammasome in response to HG or AGEs
Next, we investigated whether NLRP3 inflammasome could modulate the proliferation capacity of EPC. Cell cycle analyses showed fewer EPC in the G0/G1 phase and more in the S phase in Nlrp3−/− cells, suggesting that Nlrp3 knockout promotes G1/S transition in EPC following stimulation with HG or AGEs (Fig. 5A). Additional experiments were performed to examine the incorporation of BrdU into EPC following stimulation with HG or AGEs, Nlrp3 knockout effectively increases the BrdU positive cell ratio (Fig. 5B). Consistently, overexpression of Nlrp3 significantly inhibited the incorporation of BrdU and G1/S transition, which resulted in cell cycle arrest in the G1 phase. Chronic low-grade inflammation adversely affects the functionality of stem cells, including their capacity for proliferation and differentiation. HG or AGEs treatment significantly decreased the number of clones formed in EPC. Knockout of Nlrp3 partially restored the proliferative capacity impaired by HG and AGEs (Fig. 5C). Senescence-associated β-galactosidase (SA-β-gal) is an enzyme that exhibits increased expression in senescent cells. Its activity is relatively low under normal physiological pH conditions but escalates in aging cells. Nlrp3 knockout significantly reduced the increase in the number of senescent EPC cells caused by high HG and AGEs (Fig. 5D). However, overexpression of NLRP3 not only significantly reduced the number of colonies under HG and AGEs treatment but also significantly increased the proportion of senescent EPC following stimulation with HG and AGEs (Fig. 5C and D). Collectively, these results indicated that HG or AGEs negatively impaired angiogenesis and proliferation function by activating the NLRP3 inflammasome in vitro.
Fig. 5.
Regulated proliferation function of EPC by NLRP3 inflammasome in response to HG or AGEs. (A) Flow cytometry analysis was conducted on EPC following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. The representative pattern of the cell cycle distribution of EPC in synchrony after stimulation. The representative pattern of the cell cycle distribution of EPC in synchrony and after stimulation. (B) Representative images of BrdU incorporation status after stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h in EPC. Images were generated by merging the DAPI and BrdU channels. (C) Clone formation assay results showed the effects of NLRP3 inflammasome on the proliferation of EPC. (D) Senescence-associated beta-galactosidase staining of EPC following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Similar results were obtained from three independent experiments
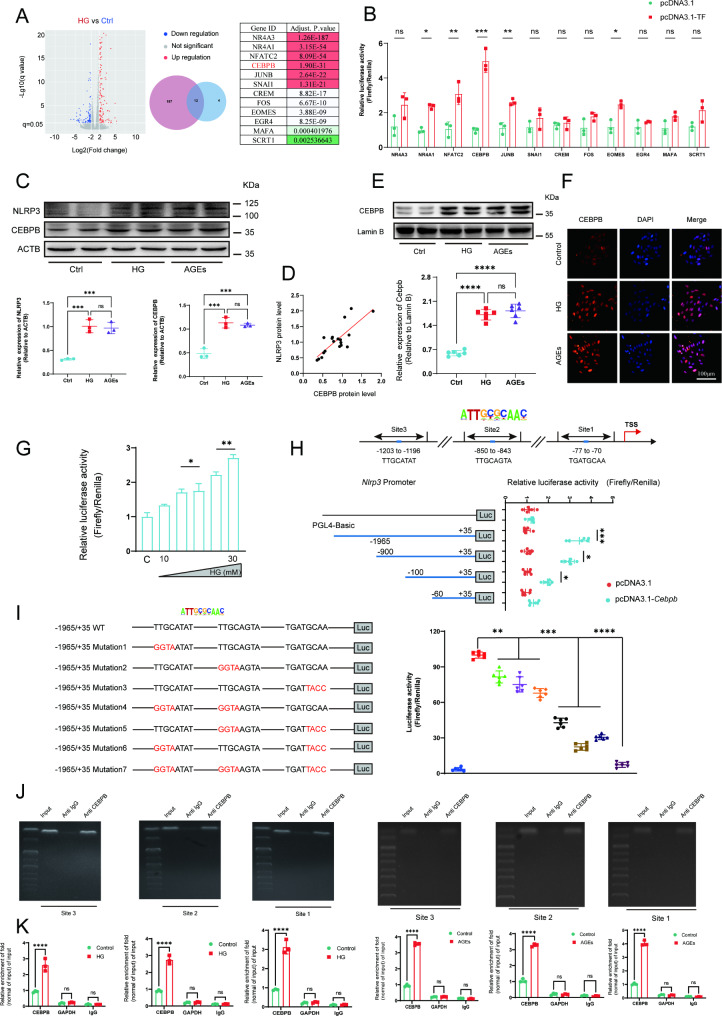
CEBPB initiated theNlrp3 transcription by directly binding to the promoter region of Nlrp3in EPC
The upstream and downstream regulation mechanism of the Nlrp3 gene in EPC remains unclear. We conducted RNA sequencing on normal EPC as well as EPC stimulated with high glucose. We obtained 12 transcription factors by taking intersections between upregulated differential genes and transcription factors that may regulate Nlrp3 gene transcription (Fig. 6A). Through conducting luciferase reporter gene assays in EPC, we identified CEBPB as the most significant transcription factor regulating the NLRP3 promoter. (Fig. 6B). The CCAAT/enhancer binding protein (C/EBP) transcription factor family is associated with inflammation [25]. These transcription factors have binding sites in the promoter and enhancer regions of many cytokines and other pro-inflammatory genes. To confirm the role of CCAAT/enhancer binding protein in activating inflammatory response in EPC, protein expression levels of NLRP3 and CEBPB were measured. Western blotting confirmed that there was about a 2 to 4-fold increase in NLRP3 and CEBPB expression in EPC primed by stimulation with HG or AGEs for 72 h (Fig. 6C). Indeed, a positive correlation was observed between the protein levels of NLRP3 and CEBPB in EPC treated with HG or AGEs (Fig. 6D).
Fig. 6.
CEBPB initiated the transcription ofNlrp3by directly binding to the promoter region ofNlrp3. (A) The volcano map of DEGs in EPC at the transcriptional level following stimulation with HG (30 mM) or not for 72 h (∣log2fold change∣ > 1 and FDR < 0.05). Venn diagram based on the overlapping DEGs between upregulated genes and mouse transcription factor. Differentially expressed transcription factors and adjust P.value in HG vs. Control group. (B After transfection with various transcription factors, luciferase reporter gene assays using the Nlrp3 promoter were performed in EPC. (C) Relative NLRP3 and CEBPB protein levels in EPC following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. (D) Correlation between NLRP3 levels and CEBPB levels in EPC. (E) Western blot of nuclear extracts obtained from EPC following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h was performed to investigate the expression of CEBPB. Each density of CEBPB was normalized with that of LAMIN B. (F) Representative immunofluorescence imaging of CEBPB nuclear translocation after stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. (G) Luciferase reporter assay in EPC using Nlrp3 promoter after stimulation with different concentrations of glucose (10, 15, 20, 25, 30mM) for 72 h. (H) Putative binding sites of CEBPB in the promoter region of Nlrp3. TSS, transcriptional start site. Luciferase reporter assay in EPC using different fragments of Nlrp3 promoter after transfection with Cebpb. (I) A series of luciferase reporter constructs harbouring site-directed mutations in the CEBPB binding sites of the Nlrp3 promoter region were assayed for promoter activity in EPC. Luciferase reporter assay in EPC using different mutations on site1, site2 and site3 respectively. (Site3: TTGCATAT to GGTAATAT, Site2: TTGCAGTA to GGTAAGTA, Site1: TGATGCAA to TGATTACC). (J) ChIP-PCR analysis of EPC following stimulation with HG (30 mM) or AGEs (200 µg/ml) for 72 h. PCR primers were designed to surround the predicted CEBPB-binding sites 1–3 in the Nlrp3 promoter. Nonspecific IgG was used as a control. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. These experiments were repeated independently three times with similar results
To determine whether Nlrp3 was transcriptionally regulated by CEBPB in EPC, we performed western blotting and immunofluorescence and found that HG or AGEs treatment could increase CEBPB nuclear translocation (Fig. 6E and F). Luciferase reporters containing the 5′ -promoter regions (–1965to + 35 bp) of Nlrp3 were expressed in Cebpb-overexpressing EPC and luciferase activities were notably altered (Fig. 6G). These results indicate that HG or AGEs promoted the transcription of Nlrp3 through CEBPB.
Three putative CEBPB-binding sites were identified on the Nlrp3 promoter. Thus, a set of pGL4-derived constructs were transiently transfected in EPC. All constructs showed activity in these cells, suggesting the presence of the elements necessary for the transcriptional activity of the Nlrp3 promoter region. Fragment full-length (–1965to + 35 bp), which harbors all putative CEBPB-binding sites, exhibited the highest luciferase activity (Fig. 6H). We next conducted site-directed mutagenesis to disrupt CEBPB binding sites in selected Nlrp3 promoter constructs. Promoter activity of mutated con-structs was determined in EPC and expressed relative to that of the wild type con-struct. Single mutation of the CEBPB site however resulted in 25 ± 7% reductions respectively. A double mutation reduced promoter activity by 69 ± 9%, whereas mutation of all sites reduced activity by 92%. Luciferase reporter genes indicate that CEBPB may bind in all three predicted regions (Fig. 6I). Furthermore, a ChIP assay was conducted to analyze the interaction between CEBPB and the putative binding sites on the chromatin of EPC. The antibody to CEBPB precipitated proteins bound in vivo to the amplified DNA sequence with three CEBPB putative binding sites (sites 1–3) (Fig. 6J). Collectively, these results strongly indicated that CEBPB may bind to sites 1–3 in the Nlrp3 promoter and upregulate the transcription of the Nlrp3 gene. Furthermore, ChIP-qPCR results demonstrated a significant enrichment of CEBPB at the Nlrp3 promoter region following HG treatment. Similarly, CEBPB enrichments within the Nlrp3 promoter region was also significantly increased by AGEs treatment in EPC (Fig. 6K). These findings suggest that CEBPB plays a crucial role in regulating Nlrp3 expression in response to HG or AGEs treatment.
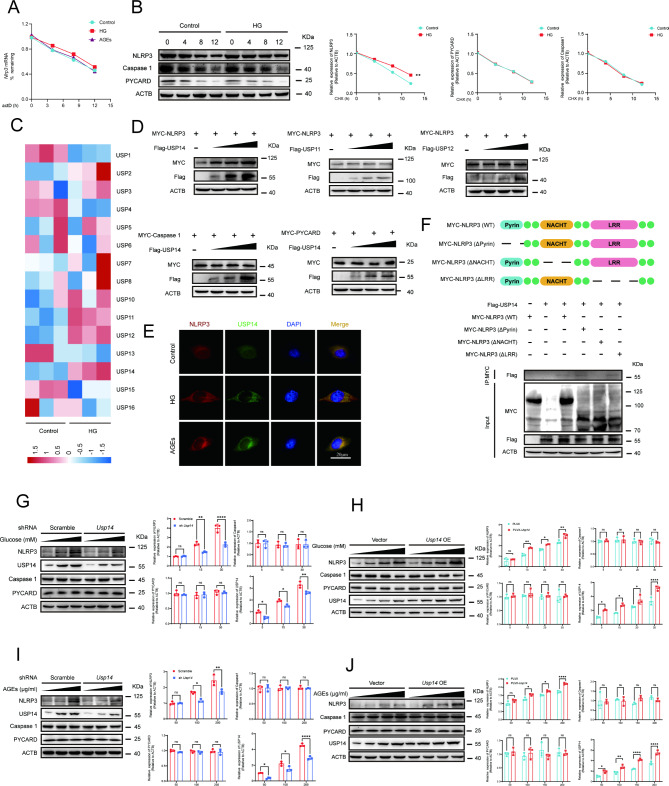
USP14 interacted with NLRP3 and inhibited NLRP3 protein degradation in EPC
Gene expression is regulated through multiple steps at both transcriptional and post-transcriptional levels. Considering the positive regulation of CEBPB on Nlrp3 at the transcriptional level, we examined whether HG or AGEs could modulate NLRP3 at the post-transcriptional level. Actinomycin D is widely used in mRNA stability assays to inhibit new mRNA synthesis and to assess mRNA decay by measuring mRNA abundance after transcriptional repression [26]. Our results suggested that there was no significant difference in the degradation of Nlrp3 mRNA between normal conditions and stimulation with HG or AGEs (Fig. 7A). To directly confirm whether HG or AGEs regulated NLRP3 protein post-translational modification, EPC were first stimulated with HG or AGEs for 72 h before the treatment of protein synthesis inhibitor cycloheximide (CHX) for different times. We found the NLRP3 protein degradation was attenuated in HG or AGEs treated EPC compared to that in control EPC. However, no differences in CASPASE1 and PYCARD protein degradation were observed between control EPC and EPC stimulated with HG (Fig. 7B). These data suggested that HG increased their levels not only through transcriptional mechanisms but also by reducing the degradation of NLRP3 protein. Ubiquitination is a key post-translational modification (PTM) that regulates NLRP3 inflammasome activation [27]. The heatmap showed the expression of USP11, USP12, and USP14 was relatively high expression in USP family genes identified in control EPC vs. HG-treated EPC (Fig. 7C). Our results showed that USP11 and USP12 did not significantly affect NLRP3 expression, while USP14 significantly affected NLRP3 protein expression (Fig. 7D). Next, we determined whether USP14 regulates NLRP3 protein expression. The expression of myc-NLRP3 was evaluated in the presence of graded expression levels of flag-USP14. And there was no impact on the expression of CASPASE1 and PYCARD protein levels (Fig. 7D). Additionally, we conducted further experiments to assess the endogenous levels of NLRP3 upon overexpression of USP14. The results indicated that the endogenous protein levels of NLRP3 were significantly elevated following the overexpression of Usp14 (Supplementary Fig. 4A). Furthermore, Usp14 overexpression enhanced HG or AGEs induced NLRP3 expression and had no effect on the expression of two pattern recognition receptors. These data indicated that USP14 selectively controlled NLRP3 expression at the protein level (Fig. 7H and J). USP14 is a major regulator of the proteasome by deubiquitinating proteasome-bound substrates that are ubiquitinated at multiple sites [28]. The confocal analysis demonstrated that co-localization between USP14 and NLRP3 was up-regulated upon HG or AGEs stimulation (Fig. 7E).
Fig. 7.
USP14 interacted with NLRP3 and inhibited NLRP3 protein degradation. (A) The expression of Nlrp3 mRNA at 0,4,8 and 12 h post-stimulation was determined by qRT-PCR and normalized to the 18 S rRNA. (B) Immunoblot analysis of extracts from EPC stimulated with HG (30 mM) for 72 h, followed by treatment with cycloheximide for various times (CHX, 100 ug/mL). NLRP3, PYCARD, and Caspase1 expression levels were quantitated by measuring band intensities and normalized to ACTB. (C) Heatmap showing the expression of USP family genes identified in control EPC vs. HG (30 mM) treated EPC. (D) MYC-Nlrp3 was transfected in EPC, as well as graded amounts of FLAG-Usp14, FLAG-Usp11 and FLAG-Usp12. The protein level of NLRP3 were detected by western blot. MYC-Caspase1, and MYC-Pycard were transfected in EPC, as well as graded amounts of FLAG-Usp14. The protein level of Caspase1, and PYCARD were detected by western blot. (E) EPC was stimulated with HG (30 mM) or AGEs (200 µg/ml) for 72 h, then fixed and incubated with a secondary antibody. Colocalization between USP14 and NLRP3 was examined by confocal microscopy. Confocal imaging results are representative of three independent experiments. (F) Schematic diagram of NLRP3 and its truncation mutants. MYC-tagged Nlrp3 or its mutants along with Flag-Usp14 were individually transfected into EPC. The cell lysates were immunoprecipitated with anti-MYC antibodies and then immunoblotted with anti-Flag antibodies. Similar results were obtained from three independent experiments. (G) Western blot analysis of NLRP3, USP14, Caspase1, and PYCARD in wildtype (WT) or Usp14 knockdown EPC following stimulation with a gradient concentration of glucose (5, 15,30mM) for 72 h. (H) Western blot analysis of NLRP3, USP14, Caspase1, and PYCARD in EPC transfected with Vector or Nlrp3 following stimulation with a gradient concentration of glucose (5, 10, 20,30mM) for 72 h. (I) Western blot analysis of NLRP3, USP14, Caspase1, and PYCARD in wildtype (WT) or Usp14 knockdown EPC following stimulation with a gradient concentration of AGEs (50, 100, 200 µg/ml) for 72 h. (J) Western blot analysis of NLRP3 USP14, Caspase1, and PYCARD in EPC transfected with Vector or Nlrp3 following stimulation with a gradient concentration of AGEs (50, 100, 150, 200 µg/ml) for 72 h. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. These experiments were repeated independently at least three times with similar results
To further find the domain of NLRP3 responsible for the interaction with USP14, a series of MYC-tagged NLRP3 truncation mutants were constructed. NLRP3 wild-type (WT), LRR domain deletion mutant (ΔLRR), NACHT domain deletion mutant (ΔNACHT), and PYD domain deletion mutant (ΔPYD) were co-transfection with Flag-USP14 into EPC. Co-IP results showed that NLRP3 mutant with NACHT domain deletion lost the ability to interact with USP14 (Fig. 7F). Taken together, these observations suggested that USP14 interacts with the NLRP3 protein through the USP motif on USP14 and the NACHT domain on NLRP3.
To investigate the potential role of USP14 in NLRP3 inflammasome activation, we examined the effects of USP14 knockdown in EPC stimulated with HG or AGEs. In line with the previous findings, the knockdown of Usp14 significantly reduced NLRP3 protein expression, while it did not affect the expression of CASPASE1 and PYCARD proteins in EPC stimulated with HG or AGEs (Fig. 7G and I).
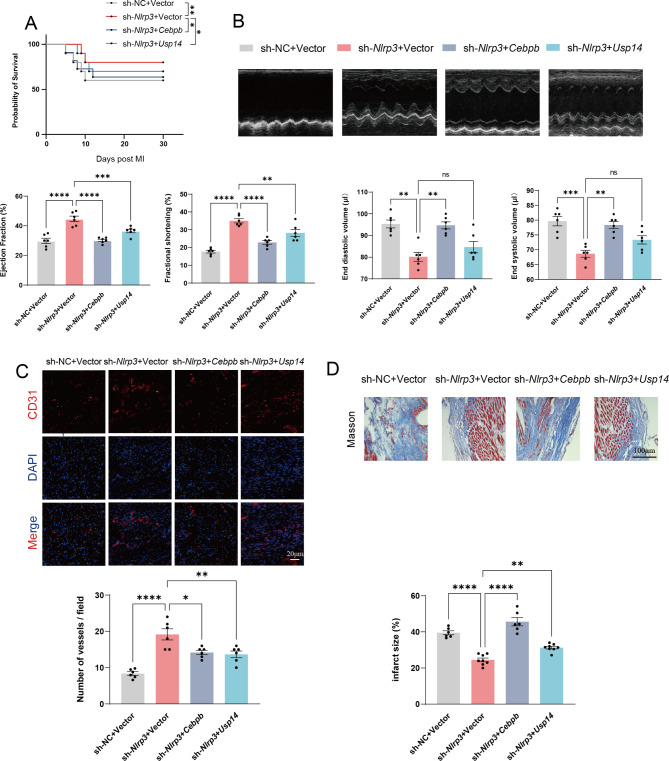
CEBPB and USP14 regulated cardiac function by modulating NLRP3 inflammasome in vivo
In vivo, our experiments showed that overexpression of Cebpb or Usp14 decreased the 30-day survival compared to Nlrp3 knockdown EPC (Fig. 8A). In addition, echocardiography showed that overexpression of Cebpb reversed the therapeutic effect of intramyocardial injection. Nlrp3 knockdown EPC, as evident from decreased percentage ejection fraction and fractional shortening and a significant increase in end diastolic volume and end systolic volume (Fig. 8B). Similarly, Usp14 overexpression weakened the therapeutic effect compared to Nlrp3 knockdown EPC. Immunofluorescence and histological analysis indicated that overexpression of Cebpb or Usp14 in Nlrp3 knockdown EPC significantly increased infarction size and suppressed angiogenesis (Fig. 8C and D). Taken together, the impact of CEBPB and USP14 on NLRP3 is likely minimal due to the knockdown of Nlrp3. The improvement in survival and cardiac function by CEBPB and USP14 suggests the involvement of pathways or mechanisms independent of NLRP3 activity that are responsible for the ameliorations mediated by CEBPB and USP14.
Fig. 8.
CEBPB and USP14 regulated cardiac Function by modulating NLRP3 inflammasome in vivo. (A) Short-term survival curve after myocardial infarction and the indicated treatments. (B) Representative short-axis M-mode echocardiograms of the left ventricle at baseline and day 21 in the HFD/STZ-induced diabetic mice with myocardial infarction groups. Left ventricular function was assessed by measurements of ejection fraction, LV fractional shortening, end diastolic volume, and end systolic volume. (C) Representative endothelial CD31 staining at the infarction border zone sections. (D) Representative images and quantitative infarct size in Masson’s trichrome stained mice hearts on day 21 after MI (n = 6 mice/group). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. These experiments were repeated independently at least three times with similar results
NLRP3 inflammasome activation inhibited angiogenesis via PI3K/Akt/mTOR pathway in EPC
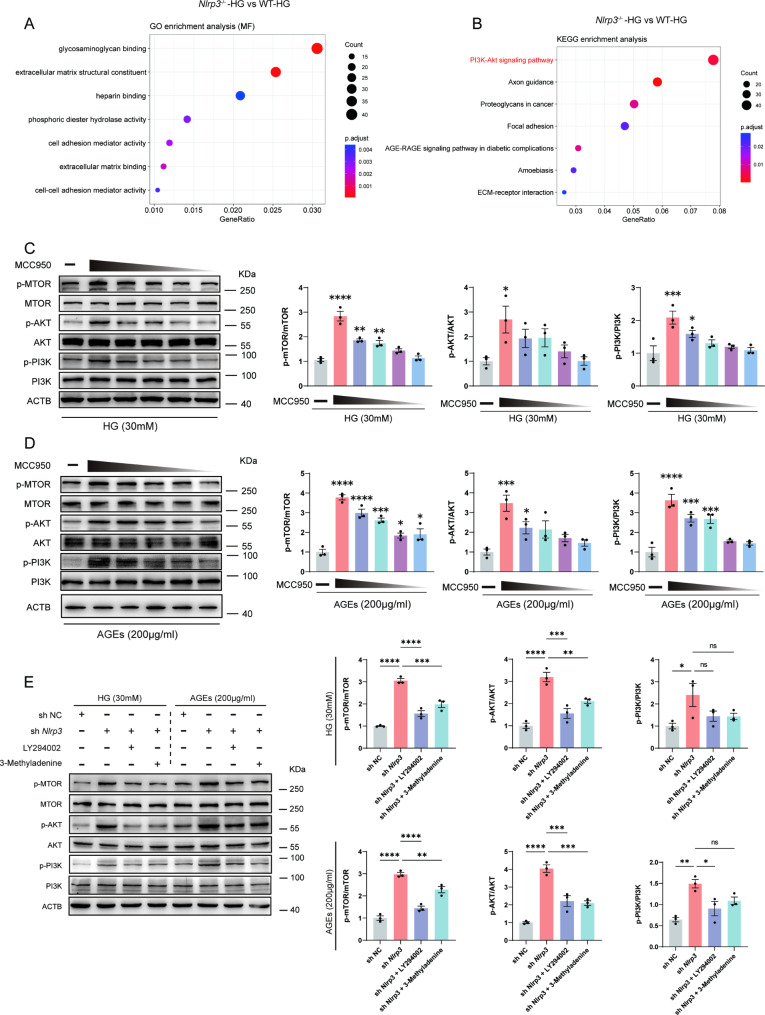
To further clarify how NLRP3 inflammasome activation induces dysfunction in the angiogenic capabilities of EPC, we treated both wild-type and Nlrp3 knockout EPC with high glucose and performed RNA sequencing. The differentially expressed genes were then analyzed using Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis to identify the downstream biological processes and metabolic pathways affected by Nlrp3 knockout in EPC under HG stimulation. Cell adhesion activity and extracellular matrix structural constituent were significantly altered in EPC with high glucose following Nlrp3 knockout (Fig. 9A). The extracellular matrix (ECM) not only provides a scaffold for cellular attachment and migration but also modulates the availability and activity of growth factors involved in angiogenesis. Alterations in the ECM can influence angiogenic signaling pathways, affecting the process of new blood vessel formation. In addition, our results suggested that PI3K/ Akt/mTOR signaling showed strikingly significant upregulation of the entire pathway in Nlrp3 knockout EPC stimulated with HG (Fig. 9B). MCC950 is a potent and well-studied inflammasome lead compound that inhibits NLRP3-dependent ASC oligomerization. PI3K/ Akt/mTOR signaling pathway was significantly inhibited in EPC stimulated with HG, while MCC950 was shown to inhibit its effects in a concentration-dependent manner (Fig. 9C and D). LY294002 and 3-Methyladenine, selective PI3K inhibitors, were applied to our mechanism rescue experiment. In vitro, Nlrp3 knockdown significantly restored the inhibition of the PI3k signaling pathway in response to HG or AGEs. However, the inhibitor of the PI3K pathway inhibited the effectiveness of Nlrp3 knockdown in HG or AGEs conditions (Fig. 9E).
Fig. 9.
NLRP3 inflammasome activation inhibited angiogenesis via PI3K/Akt/mTOR pathway in EPC. (A) GO enrichment analysis of the known molecular function in wild-type and Nlrp3 knockout EPC stimulated with high glucose (30mM) for72h. (B) KEGG enrichment analysis of the known oncogenic or metabolic pathways in wild-type and Nlrp3 knockout EPC stimulated with high glucose (30mM) for72h. (C) Western blot analysis of p-MTOR, MTOR, p-AKT, AKT, p-PI3K, PI3K protein levels in EPC pretreated with MCC950 (10µM, 8µM, 4µM, 2µM, 1µM) for 6 h in response to HG (30 mM). (D) Western blot analysis of p-MTOR, MTOR, p-AKT, AKT, p-PI3K, PI3K protein levels in EPC pretreated with MCC950 (10µM, 8µM, 4µM, 2µM, 1µM) for 6 h in response to AGEs (200 µg/ml). (E) Western blot analysis of p-MTOR, MTOR, p-AKT, AKT, p-PI3K, PI3K protein levels in EPC pretreated with shNlrp3, LY294002 (20µM) for 12 h, and 3-methyladenine (5 Mm) for 12 h. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. Control. These experiments were repeated independently three times with similar results
Discussion
Type 2 diabetes mellitus is one of the major chronic cardiovascular diseases. T2DM and its complications have become a critical worldwide public health problem [29]. Myocardial infarction is the leading cause of death in patients with T2DM. Patients with type 2 diabetes are more likely to suffer a myocardial infarction and have a worse outcome than those without diabetes [30]. Moreover, T2DM is an independent risk factor for recurrent myocardial infarction, all-cause mortality, and a composite endpoint in patients with prior myocardial infarction [31]. Current treatments for post-myocardial infarction heart failure are limited and treatment is needed to improve the clinical condition by replacing the damaged heart cells [32]. In recent years, stem cells have becoming the most important tool in the application of “regenerative medicine” and clinical work in cardiac regeneration has focused on bone marrow-derived cells, mesenchymal stem cells, and putative cardiac progenitor cells [33]. Despite the great promise of regenerative approaches, clinical work involving the transplantation of stem and progenitor cells has largely failed to materialize [32].
Endothelial progenitor cells can be generated from the bone marrow or peripheral blood in response to ischemic signals and thus be mobilized to the site of injury. EPC has been shown to play an active role in models of skin injury, hindlimb ischemia, and cardiac infarction and is shown to improve vessel formation as well as to accompany stable integration into the forming vasculature [34, 35]. Bone marrow and peripheral blood-derived EPC are also practiced for ischemic heart disease in clinical. Although studies on injection therapy using EPC have shown the overall feasibility and safety of cell therapy, the improvement in cardiac function remains modest [36]. When exposed to chronic inflammatory diseases, EPC experience stress-induced cellular senescence as well as impaired EPC function and angiogenic capacity. Interestingly, Tousoulis D found that inflammation appears to play a dual role in EPC mobilization. Low-level inflammation induces EPC mobilization, whereas high and prolonged inflammatory stimulation has the opposite effect [37]. However, a prolonged inflammatory state appears to worsen the recovery of cardiac function after myocardial infarction and may alter autologous progenitor cell angiogenesis, thereby impairing the therapeutic effect of EPC transplantation.
The NLRP3 inflammasome is activated by diverse stimuli, affecting regulated cells’ biological functions. Our previous studies have shown that activation of NLRP3 inflammasome leads to endothelial and smooth muscle cell dysfunction in diabetic mice, thus leading to impaired vascular function [38, 39]. Recent studies suggested that AGEs targeting the NLRP3 inflammasome signaling pathway decreased the level of NO, and the expression of eNOS, and reduced EPC numbers [40]. The selective NLRP3-inflammasome inhibitor MCC950 improves cardiac function and inhibits left ventricular remodeling after permanent myocardial ischemia [41]. After MI, T2DM mice showed increased infarct size and myocardial necrosis, as well as decreased overall survival time. However, the contribution of NLRP3 in EPC repair under myocardial infarction is still not elaborated. In this study, we further explored the effect of NLRP3 deficiency on EPC function and the mechanistic basis of inflammation-induced stem cell dysfunction both in vitro and in vivo.
The current results indicate that intramyocardial injection EPC can improve cardiac function in a clinically relevant model of myocardial infarction mice. Importantly, the repair capacity of WT EPC under myocardial infarction with diabetes is significantly deficient. However, intramyocardial injection Nlrp3 knockout EPC did partially reverse the impairment of angiogenic function by diabetes. HG or AGES could activate NLRP3 inflammasome, leading to inhibition of EPC proliferative capacity, EPC angiogenic function, and promotion of its aging phenotype in vitro. These effects could also explain the clear inhibition of the impaired angiogenesis function after the suppression of NLRP3 expression in EPC. However, how diabetes causes NLRP3 inflammasome activation is not clearly articulated.
CEBPB, a member of the CCAAT/enhancer binding protein (C/EBP) family of transcription factors, has been found to play an important role in multiple inflammatory diseases observed in neurodegenerative diseases and brain injury [42]. The promoters of many pro-inflammatory genes contain putative C/EBPβ shared sequences, and C/EBPβ levels in macrophages are also upregulated in response to pro-inflammatory stimulation [43]. There is a correlation between CEBPB and HFD-induced inflammation in macrophages and adipose tissue in mice [44]. However, the function of CEBPB in NLRP3 inflammasome pathways as an immunity regulator has not been studied. In this paper, we investigate the function of CEBPB in NLRP3 inflammasome signaling for the first time.
The mouse Nlrp3 gene promoter also has predicted CEBPB-binding sites, and our studies examining EPC stimulated with HG or AGEs expressing high CEBPB have also been detected. Based on these findings, the authors speculate that Nlrp3 transcription is directly upregulated through the binding of CEBPB at the Nlrp3 gene promoter in EPC. In this study, ChIP-PCR, a luciferase reporter gene system was used to confirm that CEBPB directly transcriptionally regulates Nlrp3.
USPs, the largest family of DUBs, are essential in the regulation of ubiquitin signaling [45, 46] and are associated with human diseases such as cancer, central nervous system disorders, autoimmune diseases and infections [47]. As a key modulator of the proteasome and one of the three proteasome-related deubiquitinating enzymes, USP14 is critical in the regulation of protein homeostasis, inflammation, neurodegeneration, and tumorigenesis [47, 48]. In our study, we investigated the function of USP14 in NLRP3 inflammasome signaling by maintaining protein homeostasis. We showed that USP14 interacts with NLRP3 through the USP motif and the NACHT domain on NLRP3. In addition, the USP14 did not affect the expression of two pattern recognition receptors. Based on the above results we can assume that USP14 maintains homeostasis by specifically targeting NLRP3. Recent studies have considered USP as a major pro-inflammatory signal, including the nuclear factor-κB (NF-κB) and transformed growth factor-BETA (TGF-β) pathways. The NF-κB signaling pathway is critical for the induction of pro-inflammatory gene expression, regulation of the inflammasome and activation of inflammation, and innate immune cell differentiation in inflammatory disease. Similar to our study, USP14 forms a complex with TRIM14 and promotes inflammatory responses by inhibiting KDM4D autophagic degradation. Our study provides new insights into USP14 in inflammatory diseases through regulating the NLRP3 inflammasome.
To further clarify how NLRP3 inflammasome activation affects EPC angiogenesis, transcriptome sequencing was performed to identify differentially expressed genes and conduct pathway enrichment analysis. Recent studies showed that the PI3K/Akt/mTOR pathway plays a critical role in many cellular functions, including proliferation, adhesion, migration, invasion, and survival [49]. In this study, we found that the PI3K/Akt/mTOR pathway is mediated by NLRP3 inflammasome response in diabetic EPC. NLRP3 specific inhibitor (MCC950) almost completely rescued phosphorylation of PI3K and expression of its downstream genes under HG and AGEs treatment conditions. We used two selective PI3K inhibitors and found that two inhibitors attenuated the benefits of NLRP3 knockout, and we hypothesize that NLRP3 inflammasome activation may inhibit the PI3K/Akt/mTOR pathway by acting on the PI3K component of this signaling cascade. Importantly, activation of the PI3K/Akt pathway was accompanied by almost complete attenuation of the negative effects of NLRP3 inflammasome activation on EPC survival and angiogenic function under diabetic conditions. Vascular growth factor (VEGF) whose effects on endothelial cells are mediated in part by the PI3K pathway, is an essential mediator of angiogenesis.
Conclusion
In summary, our results reveal that diabetic condition regulates NLRP3 inflammasome activation through pre-transcriptional as well as post-translational modifications, thereby inhibiting the PI3K signaling pathway, leading to the impaired angiogenic function of EPC and exacerbating myocardial infarction. Therefore, it may be concluded that the inhibition of NLRP3 inflammasome activation is a potential therapeutic strategy for the treatment of myocardial infarction in T2DM patients, and raises the possibility of cell therapy for ischemic heart repair (Table 1).
Table 1.
The sequence of primer for qRT-PCR
| Gene | Sequence |
|---|---|
| Nlrp3 | Forword: 5’-ATTACCCGCCCGAGAAAGG-3’ |
| Reverse: 5’-TCGCAGCAAAGATCCACACAG-3’ | |
| Nlrp1 | Forword: 5’-GCTGAATGACCTGGGTGATGGT-3’ |
| Reverse: 5’-CTTGGTCACTGAGAGATGCCTG-3’ | |
| Nlrp6 | Forword: 5’-CTGGCGTCATTGTGGAACCTCT-3’ |
| Reverse: 5’-TCTCACTCAGCTCCACAGAGGT-3’ | |
| Nlrc4 | Forword: 5’-CTCACCACGGATGACGAACAGT-3’ |
| Reverse: 5’-TGTCATCCAGTATGAGTCTCTCG-3’ | |
| Aim2 | Forword: 5’-GAAAACTGCTCTGCTGCCT-3’ |
| Reverse: 5’-TGCCACCATCTGTTTCTTCTGA-3’ | |
| Caspase1 | Forword: 5’-GGCACATTTCCAGGACTGACTG-3’ |
| Reverse: 5’-GCAAGACGTGTACGAGTGGTTG-3’ | |
| Il-1b | Forword: 5’-TGGACCTTCCAGGATGAGGACA-3’ |
| Reverse: 5’-GTTCATCTCGGAGCCTGTAGTG-3’ | |
| 18 S rDNA | Forword: 5’-AGTCCCTGCCCTTTGTACACA-3’ |
| Reverse: 5’-CGATCCGAGGGCCTCACTA-3’ | |
| Actb | Forword: 5’-CATTGCTGACAGGATGCAGAAGG-3’ |
| Reverse: 5’-TGCTGGAAGGTGGACAGTGAGG-3’ |
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge the Public platform of State Key Laboratory of Natural Medicines for the use of analytical instrumentation facilities(FUJIFILM VisualSonics Vevo 3100 LT), and also thank L. Guo for her assistance in taking images. All authors have read the journal’s authorship agreement and the manuscript has been reviewed by and approved by all named authors.
Abbreviations
- AGEs
advanced glycation end products
- AMI
acute myocardial infarction
- CHX
cycloheximide
- CVD
cardiovascular disease
- EPC
endothelial progenitor cells
- GO
gene ontology
- HG
high glucose
- KEGG
kyoto encyclopedia of genes and genomes
- SA-β-gal
senescence-associated β-galactosidase
- T2DM
type 2 diabetes mellitus
- VEGF
vascular growth factor
Author contributions
Jia-Peng Li and Shu Qiu performed the study concept and design; Guang-Jie Tai and Wei Wei performed the development of methodology and writing, review, and revision of the paper; Meng-Meng Fu, Pan-Qi Fang, and Xiao-Xue Li provided acquisition, analysis, and interpretation of data, and statistical analysis; Ming Xu provided technical and material support. All authors read and approved the final paper.
Funding
This work was supported by Grant 82373870 from the National Natural Science Foundation of China, and Xinjiang Nature Fund key project (2022D01D50).
Data availability
The experimental data sets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Consent for publication
All authors consent for the publication of this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jia-Peng Li and Shu Qiu contributed equally to this work.
References
- 1.Kivimaki M, Steptoe A. Effects of stress on the development and progression of cardiovascular disease. Nat Rev Cardiol. 2018;15(4):215–29. [DOI] [PubMed] [Google Scholar]
- 2.Nemet I, Saha PP, Gupta N, et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell. 2020;180(5):862–77. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Waes JA, Nathoe HM, De Graaff JC, et al. Myocardial injury after noncardiac surgery and its association with short-term mortality. Circulation. 2013;127(23):2264–71. [DOI] [PubMed] [Google Scholar]
- 4.Smilowitz NR, Gupta N, Guo Y, et al. Perioperative acute myocardial infarction associated with non-cardiac surgery. Eur Heart J. 2017;38(31):2409–17. [DOI] [PubMed] [Google Scholar]
- 5.Smilowitz NR, Gupta N, Ramakrishna H, et al. Perioperative major adverse cardiovascular and cerebrovascular events associated with noncardiac surgery. JAMA Cardiol. 2017;2(2):181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiser TG, Haynes AB, Molina G, et al. Estimate of the global volume of surgery in 2012: an assessment supporting improved health outcomes. Lancet. 2015;385(Suppl 2):S11. [DOI] [PubMed] [Google Scholar]
- 7.Joseph JJ, Deedwania P, Acharya T, et al. Comprehensive Management of Cardiovascular Risk factors for adults with type 2 diabetes: a Scientific Statement from the American Heart Association. Circulation. 2022;145(9):e722–59. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Hu X, Lee SH, et al. Diabetes exacerbates myocardial Ischemia/Reperfusion Injury by down-regulation of microRNA and up-regulation of O-GlcNAcylation. JACC Basic Transl Sci. 2018;3(3):350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quevedo HC, Hatzistergos KE, Oskouei BN, et al. Allogeneic mesenchymal stem cells restore cardiac function in chronic ischemic cardiomyopathy via trilineage differentiating capacity. Proc Natl Acad Sci U S A. 2009;106(33):14022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon YS, Wecker A, Heyd L, et al. Clonally expanded novel multipotent stem cells from human bone marrow regenerate myocardium after myocardial infarction. J Clin Invest. 2005;115(2):326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehman J, Li J, Orschell CM, et al. Peripheral blood endothelial progenitor cells are derived from monocyte/macrophages and secrete angiogenic growth factors. Circulation. 2003;107(8):1164–9. [DOI] [PubMed] [Google Scholar]
- 12.Tian T, Chen B, Xiao Y, et al. Intramyocardial autologous bone marrow cell transplantation for ischemic heart disease: a systematic review and meta-analysis of randomized controlled trials. Atherosclerosis. 2014;233(2):485–92. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Lin L, Zong W. Bone marrow mononuclear cells transfer for patients after ST-Elevated myocardial infarction: a Meta-analysis of Randomized Control Trials. Yonsei Med J. 2018;59(5):611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rathinam VA, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell. 2016;165(4):792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16(7):407–20. [DOI] [PubMed] [Google Scholar]
- 17.Prochnicki T, Latz E. Inflammasomes on the crossroads of innate immune recognition and metabolic control. Cell Metab. 2017;26(1):71–93. [DOI] [PubMed] [Google Scholar]
- 18.Mangan MSJ, Olhava EJ, Roush WR, et al. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17(9):688. [DOI] [PubMed] [Google Scholar]
- 19.Vandanmagsar B, Youm YH, Ravussin A, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17(2):179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youm YH, Grant RW, Mccabe LR, et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 2013;18(4):519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol. 2014;14(7):463–77. [DOI] [PubMed] [Google Scholar]
- 22.Luo M, Guan X, Luczak ED, et al. Diabetes increases mortality after myocardial infarction by oxidizing CaMKII. J Clin Invest. 2013;123(3):1262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki T, Itoh T, Sunaga D, et al. Effects of diabetes on myocardial infarct size and cardioprotection by preconditioning and postconditioning. Cardiovasc Diabetol. 2012;11:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garikipati VN, Krishnamurthy P, Verma SK, et al. Negative regulation of miR-375 by Interleukin-10 enhances bone marrow-derived progenitor cell-mediated myocardial repair and function after myocardial Infarction. Stem Cells. 2015;33(12):3519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi A, Adachi S, Morita M, et al. Post-transcriptional stabilization of Ucp1 mRNA protects mice from diet-induced obesity. Cell Rep. 2015;13(12):2756–67. [DOI] [PubMed] [Google Scholar]
- 27.Shim DW, Lee KH. Posttranslational regulation of the NLR family pyrin domain-containing 3 inflammasome. Front Immunol. 2018;9:1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee BH, Lu Y, Prado MA, et al. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature. 2016;532(7599):398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Standl E, Khunti K, Hansen TB, et al. The global epidemics of diabetes in the 21st century: current situation and perspectives. Eur J Prev Cardiol. 2019;26(2suppl):7–14. [DOI] [PubMed] [Google Scholar]
- 30.Lago RM, Nesto RW. Type 2 diabetes and coronary heart disease: focus on myocardial infarction. Curr Diab Rep. 2009;9(1):73–8. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Li M, Gao C, et al. Impact of type 2 diabetes mellitus on recurrent myocardial infarction in China. Diab Vasc Dis Res. 2016;13(6):395–404. [DOI] [PubMed] [Google Scholar]
- 32.Cahill TJ, Choudhury RP, Riley PR. Heart regeneration and repair after myocardial infarction: translational opportunities for novel therapeutics. Nat Rev Drug Discov. 2017;16(10):699–717. [DOI] [PubMed] [Google Scholar]
- 33.Kloner RA. Current state of clinical translation of cardioprotective agents for acute myocardial infarction. Circ Res. 2013;113(4):451–63. [DOI] [PubMed] [Google Scholar]
- 34.Yoon CH, Hur J, Park KW, et al. Synergistic neovascularization by mixed transplantation of early endothelial progenitor cells and late outgrowth endothelial cells: the role of angiogenic cytokines and matrix metalloproteinases. Circulation. 2005;112(11):1618–27. [DOI] [PubMed] [Google Scholar]
- 35.Yoder MC, Mead LE, Prater D, et al. Redefining endothelial progenitor cells via clonal analysis and hematopoietic stem/progenitor cell principals. Blood. 2007;109(5):1801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdel-Latif A, Bolli R, Tleyjeh IM, et al. Adult bone marrow-derived cells for cardiac repair: a systematic review and meta-analysis. Arch Intern Med. 2007;167(10):989–97. [DOI] [PubMed] [Google Scholar]
- 37.Tousoulis D, Andreou I, Antoniades C, et al. Role of inflammation and oxidative stress in endothelial progenitor cell function and mobilization: therapeutic implications for cardiovascular diseases. Atherosclerosis. 2008;201(2):236–47. [DOI] [PubMed] [Google Scholar]
- 38.Wei W, Li XX, Xu M. Inhibition of vascular neointima hyperplasia by FGF21 associated with FGFR1/Syk/NLRP3 inflammasome pathway in diabetic mice. Atherosclerosis. 2019;289:132–42. [DOI] [PubMed] [Google Scholar]
- 39.Li JP, Wei W, Li XX, et al. Regulation of NLRP3 inflammasome by CD38 through cADPR-mediated ca(2+) release in vascular smooth muscle cells in diabetic mice. Life Sci. 2020;255:117758. [DOI] [PubMed] [Google Scholar]
- 40.Bai B, Yang Y, Wang Q, et al. NLRP3 inflammasome in endothelial dysfunction. Cell Death Dis. 2020;11(9):776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Hout GP, Bosch L, Ellenbroek GH, et al. The selective NLRP3-inflammasome inhibitor MCC950 reduces infarct size and preserves cardiac function in a pig model of myocardial infarction. Eur Heart J. 2017;38(11):828–36. [DOI] [PubMed] [Google Scholar]
- 42.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365(Pt 3):561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia Y, Wang ZH, Zhang J, et al. C/EBPbeta is a key transcription factor for APOE and preferentially mediates ApoE4 expression in Alzheimer’s disease. Mol Psychiatry. 2021;26(10):6002–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahman SM, Janssen RC, Choudhury M, et al. CCAAT/enhancer-binding protein beta (C/EBPbeta) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J Biol Chem. 2012;287(41):34349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–92. [DOI] [PubMed] [Google Scholar]
- 46.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–63. [DOI] [PubMed] [Google Scholar]
- 47.Harrigan JA, Jacq X, Martin NM, et al. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17(1):57–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Poot S, a H, Tian G, Finley D. Meddling with fate: the proteasomal deubiquitinating Enzymes. J Mol Biol. 2017;429(22):3525–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bader AG, Kang S, Zhao L, et al. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5(12):921–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental data sets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.