Abstract
Disulfidptosis, which was recently identified, has shown promise as a potential cancer treatment. Nonetheless, the precise role of long non-coding RNAs (lncRNAs) in this phenomenon is currently unclear. To elucidate their significance in bladder cancer (BLCA), a signature of disulfidptosis-related lncRNAs (DRlncRNAs) was developed and their potential prognostic significance was explored. BLCA sample data were sourced from The Cancer Genome Atlas. A predictive signature comprising DRlncRNAs was formulated and subsequently validated. The combination of this signature with clinical characteristics facilitated the development of a nomogram with practical clinical utility. Additionally, enrichment analysis was conducted, the tumor microenvironment (TME) was assessed, the tumor mutational burden (TMB) was analyzed, and drug sensitivity was explored. Reverse transcription-quantitative PCR (RT-qPCR) was utilized to quantify lncRNA expression. The results revealed an eight-gene signature based on DRlncRNAs was established, and the predictive accuracy of the nomogram that incorporated the risk score [area under the curve (AUC)=0.733] outperformed the nomogram without it (AUC=0.703). High-risk groups were associated with pathways such as WNT signaling, focal adhesion and cell cycle pathways. The TME study revealed that high-risk patients had increased immune infiltration, whereas the TMB and tumor immune dysfunction and exclusion scores in low-risk patients indicated a potentially robust immune response. Drug sensitivity analysis identified appropriate antitumor drugs for each group. RT-qPCR experiments validated significant differences in DRlncRNAs expression between normal and BLCA cell lines. In conclusion, the prognostic risk signature, which includes the eight identified DRlncRNAs, demonstrates promise for predicting prognosis of patients with BLCA and guiding the selection of suitable immunotherapy and chemotherapy strategies.
Keywords: disulfidptosis, lncRNAs, BLCA, prognostic signature
Introduction
Bladder cancer (BLCA) accounts for over 550,000 new cases annually, ranking as the most common urinary malignancy (1). The primary treatment for BLCA involves radical cystectomy, combined with adjuvant cisplatin-based chemotherapy (2). Despite aggressive interventions including radiotherapy, surgery and chemotherapy, a considerable number of individuals experience recurrence or metastasis, resulting in poor 5-year survival outcomes (3). Therefore, there is a critical need for novel predictive biomarkers to improve prognostic accuracy and guide the management of patients with BLCA.
Disulfidptosis, a newly identified form of regulated cell death, was first described by the laboratory of BY Gan in 2023(4). Distinct from apoptosis, autophagy, ferroptosis and cuproptosis, disulfidptosis occurs in glucose-starved tumor cells. In this process, overexpression of SLC7A11 leads to significant depletion of nicotinamide adenine dinucleotide phosphate (NADPH), which subsequently triggers the abnormal accumulation of disulfide bonds (4). This accumulation disrupts the normal interactions between cytoskeletal proteins, causing conformational changes that ultimately result in rapid tumor cell death (5). Consequently, modulating cancer cell susceptibility to disulfidptosis may represent a promising therapeutic strategy.
lncRNAs are RNA transcripts >200 nucleotides that do not encode proteins (6). These molecules are involved in a wide range of regulatory functions, including the modulation of genome activity, protein modification and post-transcriptional regulation (7). lncRNAs play crucial roles in various cellular processes, such as gene expression control, chromatin remodeling, and cellular stress responses (8). In recent years, lncRNA-based signatures have gained significant attention for their prognostic potential in various cancers, including colorectal cancer (CRC) (9), nasopharyngeal carcinoma (10), and hepatocellular carcinoma (11). These signatures have shown promise in predicting disease outcomes and guiding treatment decisions. However, the development of prognostic signatures based on DRlncRNAs for BLCA remains limited. Given the emerging role of disulfidptosis in tumor cell death, exploring the relationship between DRlncRNAs and BLCA prognosis could offer new insights into therapeutic strategies.
As a result, a prognostic risk signature was developed and validated based on DRlncRNAs to forecast survival outcomes in patients with BLCA and the clinical applicability of this signature was explored.
Materials and methods
Gathering and processing data
The mRNA and lncRNA sequencing data of 394 BLCA samples and 19 controls were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). TCGA database was queried up to 24 January, 2024, to retrieve transcriptomic and clinical data (12). The dataset encompassed 413 patients with BLCA, including 394 tumor samples and 19 normal samples. For diverse data analyses, transcriptomic data in HTSeq-Counts and HTSeq-TPM formats were specifically chosen. Samples with incomplete clinical records or overall survival (OS) of <30 days were excluded.
Identification of DRlncRNAs essential for signature
Recent literature has identified several disulfidptosis-related genes (DRGs), including SLC7A11, SLC3A2, SLC2A1, NCKAP1, WASF2 and RAC1(4). A protein-protein interaction network for the six DRGs was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://cn.string-db.org/). Utilizing Pearson correlation analysis of these DRGs, a screening for DRlncRNAs (|R| >0.3, P<0.001) was conducted. Then ‘DESeq2’ package was performed for differential analysis (P<0.05, |log2-fold change| >1) (13).
Using a 7:3 ratio, the 364 BLCA samples were divided into training and testing cohorts. Initially, DRlncRNAs were selected through univariate Cox analysis of the training group (P<0.01). The selection was then refined using the least absolute shrinkage and selection operator (LASSO) algorithm to prevent overfitting of this signature. In the end, 8 DRlncRNAs were selected.
BLCA prognostic risk signature
The predictive risk score for each patient with BLCA was calculated using the following formula: Predicted risk score=Σ[(coefficient of lncRNAn) x (expression of lncRNAn)]. Subsequently, patients were stratified into two cohorts based on the median risk score derived from the prognostic signature: High-risk and low-risk groups. To investigate OS differences between these groups in each study cohort, the ‘survival’ and ‘survminer’ software tools were employed to construct Kaplan-Meier (K-M) curves. The ‘timeROC’ package was employed to calculate the area under the receiver operating characteristic (ROC) curves (AUC) to assess predictive accuracy. Additionally, principal component analysis (PCA) was conducted to assess the separation of risk groups (14). Variables that independently impact BLCA survival were identified through univariate and multivariate regression analyses. Finally, K-M survival curves were generated for subgroups with distinct clinical characteristics, providing a comprehensive assessment of the prognostic signature's clinical applicability.
BLCA prediction nomogram
Utilizing the ‘RMS’ package, a prognostic nomogram was constructed for predicting the OS of individual patients with BLCA. This comprehensive model integrates both the risk score and relevant clinical data (15). To validate its precision, ROC curves were employed and Decision Curve Analysis (DCA) was conducted. Furthermore, the nomogram's performance was visually evaluated by plotting calibration curves.
Enrichment analysis
Differentially expressed DRGs and their enrichment in biological signaling pathways were identified through Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Subsequently, the Gene Set Enrichment Analysis (GSEA) software (version 3.0; Broad Institute website (https://www.gsea-msigdb.org/)) was leveraged to scrutinize GOBP, KEGG and WIKIPATHWAYS gene sets (16). GSEA was conducted on risk-stratified gene expression profiles, utilizing 1,000 resamples, an upper limit of 5,000 genes, and a lower limit of 5. The significance thresholds were set at q-values <0.25 and P-values <0.05.
Tumor mutation burden (TMB) analysis
The ‘TCGAbiolinks’ package was utilized to aggregate somatic mutation data profiles in the format of mutation annotation. Subsequently, a comparative analysis of mutation profiles and TMB scores was conducted between the high- and low-risk groups, using the ‘maftools’ program (17).
Tumor microenvironment (TME) analysis
The ‘ESTIMATE’ software was utilized to analyze microenvironmental differences between the two subgroups. Additionally, seven algorithms (XCELL, CIBERSORT-ABS, TIMER, EPIC, QUANTISEQ, MCPCOUNTER and CIBERSORT) were used to investigate the correlation between risk scores and infiltrating immune cells (18). To evaluate infiltration scores in the BLCA microenvironment, the ‘GSVA’ tool was applied for single-sample GSEA (ssGSEA).
Disulfidptosis-related signature in immunotherapy and chemotherapy
The tumor immune dysfunction and exclusion (TIDE) platform was utilized to predict the response to immunotherapy in BLCA (19). Furthermore, the ‘oncoPredict’ package was harnessed to assess the IC50 of widely used chemotherapeutic agents and compare their sensitivity across different risk cohorts (20).
Reverse transcription-quantitative PCR (RT-qPCR) verification of lncRNAs in signature
The BLCA cell lines (T24 and 5637) (IMMOCELL; (http://www.immocell.com/), where T24 is generally considered to have a higher malignancy level compared with 5637, and the SV-HUC-1 cell line, derived from human normal bladder epithelial cells, were cultivated in a controlled incubator at 37˚C with 5% CO2, using RPMI-1640 medium (Thermo Fisher Scientific, Inc.). Total RNA was extracted from each sample using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific Inc.). cDNA was reverse-transcribed from the isolated RNA using the PrimeScript RT Reagent Kit (Takara Bio, Inc.), following the manufacturer's protocol. And the SYBR Green premixed qPCR kit (Hunan Accurate Bio-Medical Co., Ltd.) was used in a Roche LightCycler 480 II [Roche Diagnostics (Shanghai) Co., Ltd.]. Subsequently, RT-qPCR was performed. The thermocycling conditions were as follows: initial denaturation at 95˚C for 5 min, followed by 40 amplification cycles consisting of denaturation at 95˚C for 15 sec, annealing at 60˚C for 20 sec, and extension at 72˚C for 30 sec. A final extension step was performed at 72˚C for 5 min. The forward primer sequence for the reference gene GAPDH is 5'-GGAAGCTTGTCATCAATGGAAATC-3', and the reverse primer sequence is 5'-TGATGACCCTTTTGGCTCCC-3'. The relative expression of lncRNAs was quantified as a 2-ΔΔCq value after assessing gene expression levels via RT-qPCR (21). Experiments were conducted in triplicate, with primer details provided in Table SI. Furthermore, the protein expression profiles of the identified DRGs were examined and compared between normal and BLCA tissues using the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/).
Statistical analysis
Expression levels of DRlncRNAs in cell lines were detected using RT-qPCR. Statistical significance was determined by unpaired Student's t-test and one-way analysis of variance (ANOVA) to compare the expression levels between different cell lines. Should the ANOVA indicate a significant difference among the groups, the comparison between two groups was conducted using LSD method as the post hoc test. Data are presented as the mean ± standard deviation (SD) from at least three independent experiments. All statistical analyses were conducted using SPSS version 26.0 (IBM Corp.) and RStudio version 4.2.1 [PBC (http://www.rstudio.com/)]. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DRlncRNAs in patients with BLCA
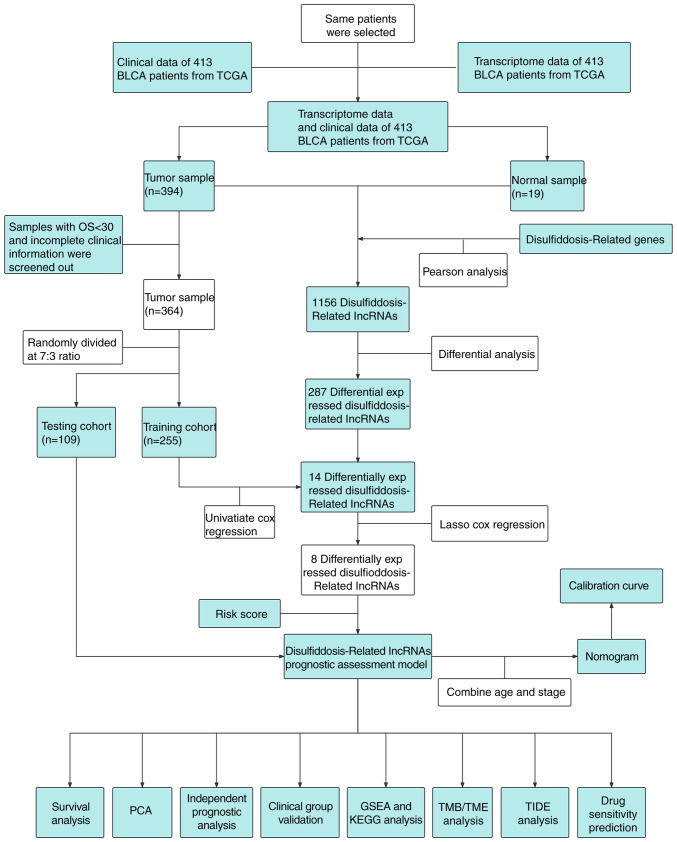
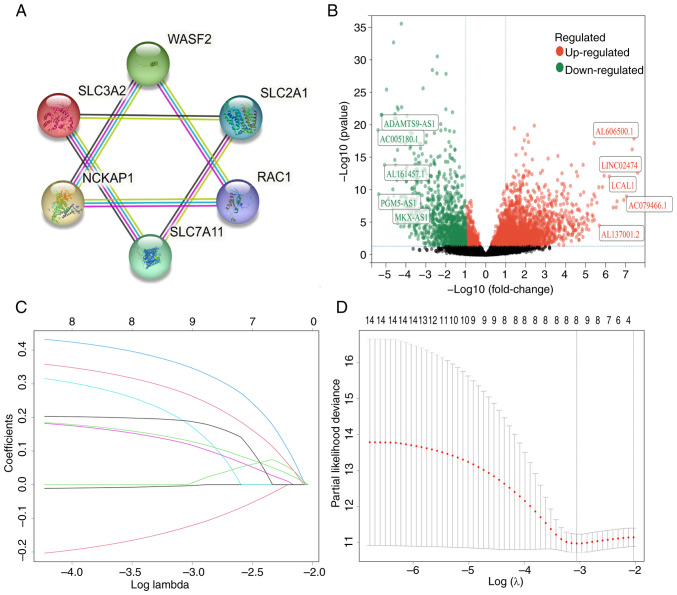
The schematic diagram representing the study is depicted in Fig. 1. In the initial phase, following the identification of six DRGs, the STRING database (https://cn.string-db.org/) was utilized to construct a protein-protein interaction network (Fig. 2A). Subsequently, through Pearson correlation analysis, 1,156 lncRNAs correlated with these DRGs were identified. Further differential analysis revealed that 287 lncRNAs exhibited differential expression between cancerous and normal tissues (Fig. 2B).
Figure 1.
Schematic diagram of the present study. BLCA, bladder cancer; lncRNA, long non-coding RNA; TCGA, The Cancer Genome Atlas; PCA, principal component analysis; TIDE, tumor immune dysfunction and exclusion; GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; TMB, tumor mutation burden; TME, tumor microenvironment.
Figure 2.
Identification of DRlncRNAs in patients with bladder cancer. (A) The protein-protein interaction network indicating the interactions among 6 disulfidptosis-related genes. (B) Volcano plot showing 287 differentially expressed DRlncRNAs. (C and D) Least absolute shrinkage and selection operator regression analysis. DRlncRNAs, disulfidptosis-related long non-coding RNAs.
The 364 patients were divided into two groups: A training group (n=255) and a test group (n=109). Detailed clinical information for each group is provided in Table I. Notably, 14 lncRNAs were identified in the training set through univariate analysis (Table SII). Subsequently, the LASSO algorithm refined this selection, resulting in the identification of 8 DRlncRNAs for the development of the signature (Fig. 2C and D; details in Table SIII). Additionally, the study also explored correlations among these final 8 DRlncRNAs (Fig. S1A) and their upregulation and downregulation patterns (Fig. S1B). Furthermore, the associations between these 8 lncRNAs and their associated genes were investigated (Fig. S1C).
Table I.
Clinical information of the patients in the test and training groups.
| Train cohort (n=255) | Test cohort (n=109) | Entire cohort (n=364) | ||||
|---|---|---|---|---|---|---|
| Characteristics | n | % | n | % | n | % |
| Age | ||||||
| <65 | 89 | 34.9 | 42 | 38.5 | 131 | 36.0 |
| >65 | 166 | 65.1 | 67 | 61.5 | 233 | 64.0 |
| Status | ||||||
| Alive | 142 | 55.7 | 57 | 52.3 | 199 | 54.7 |
| Dead | 113 | 44.3 | 52 | 47.7 | 165 | 45.3 |
| Sex | ||||||
| Female | 66 | 25.9 | 29 | 26.6 | 95 | 26.1 |
| Male | 189 | 74.1 | 80 | 73.4 | 269 | 73.9 |
| Stage | ||||||
| Stage I | 4 | 1.6 | 0 | 0 | 4 | 1.1 |
| Stage II | 70 | 27.5 | 25 | 22.9 | 95 | 26.1 |
| Stage III | 92 | 36.1 | 46 | 42.2 | 138 | 37.9 |
| Stage IV | 89 | 34.9 | 38 | 34.9 | 127 | 34.9 |
| T stage | ||||||
| T1 | 5 | 2.0 | 0 | 0 | 5 | 1.4 |
| T2 | 82 | 32.2 | 28 | 25.7 | 110 | 30.2 |
| T3 | 129 | 50.6 | 63 | 57.8 | 192 | 52.7 |
| T4 | 39 | 15.3 | 18 | 16.5 | 57 | 15.7 |
| M stage | ||||||
| M0 | 121 | 47.5 | 57 | 52.3 | 178 | 48.9 |
| M1 | 6 | 2.4 | 2 | 1.8 | 8 | 2.2 |
| Unknown | 128 | 50.2 | 50 | 45.9 | 178 | 48.9 |
| N stage | ||||||
| N0 | 148 | 58.0 | 65 | 59.6 | 213 | 58.5 |
| N1 | 31 | 12.2 | 12 | 11.0 | 43 | 11.8 |
| N2 | 52 | 20.4 | 23 | 21.1 | 75 | 20.6 |
| N3 | 3 | 1.2 | 2 | 1.8 | 5 | 1.4 |
| Unknown | 21 | 8.2 | 7 | 6.4 | 28 | 7.7 |
T stage, tumor stage; N stage, node stage; M stage, metastasis stage.
Developing and validating the risk score signature
Following the outlined steps, a prognostic signature was formulated for patients with BLCA, and the risk scores were established as specified: Predicted risk score = (-0.003727 x AL390719.2 expression) + (-0.126979 x ASMTL-AS1 expression) + (0.1324342 x AL031058.1 expression) + (0.3540896 x LINC02438 expression) + (0.1858756 x LINC01788 expression) + (0.1228996 x AC022613.2 expression) + (0.1901772 x RBMS3-AS3 expression) + (0.2711116 x AL122035.1 expression). Specific clinical data for both risk groups are provided in Table II.
Table II.
Clinical information for 364 patients in different risk categories.
| High-risk group (n=181) | Low-risk group (n=183) | |||
|---|---|---|---|---|
| Characteristics | n | % | n | % |
| Age | ||||
| <65 | 63 | 34.8 | 68 | 37.2 |
| >65 | 118 | 65.2 | 115 | 62.8 |
| Status | ||||
| Alive | 76 | 42.0 | 123 | 67.2 |
| Dead | 105 | 58.0 | 60 | 32.8 |
| Sex | ||||
| Female | 55 | 30.4 | 40 | 21.9 |
| Male | 126 | 69.6 | 143 | 78.1 |
| Stage | ||||
| Stage I | 1 | 0.6 | 3 | 1.6 |
| Stage II | 32 | 17.7 | 63 | 34.4 |
| Stage III | 79 | 43.6 | 59 | 32.2 |
| Stage IV | 69 | 38.1 | 58 | 31.7 |
| T stage | ||||
| T1 | 1 | 0.6 | 4 | 2.2 |
| T2 | 38 | 21.0 | 72 | 39.3 |
| T3 | 108 | 59.7 | 84 | 45.9 |
| T4 | 34 | 18.8 | 23 | 12.6 |
| M stage | ||||
| M0 | 81 | 44.8 | 97 | 53.0 |
| M1 | 4 | 2.2 | 4 | 2.2 |
| Unknown | 96 | 53.0 | 82 | 44.8 |
| N stage | ||||
| N0 | 108 | 59.7 | 105 | 57.4 |
| N1 | 28 | 15.5 | 15 | 8.2 |
| N2 | 36 | 19.9 | 39 | 21.3 |
| N3 | 3 | 1.7 | 2 | 1.1 |
| Unknown | 6 | 3.3 | 22 | 12.0 |
T stage, tumor stage; N stage, node stage; M stage, metastasis stage.
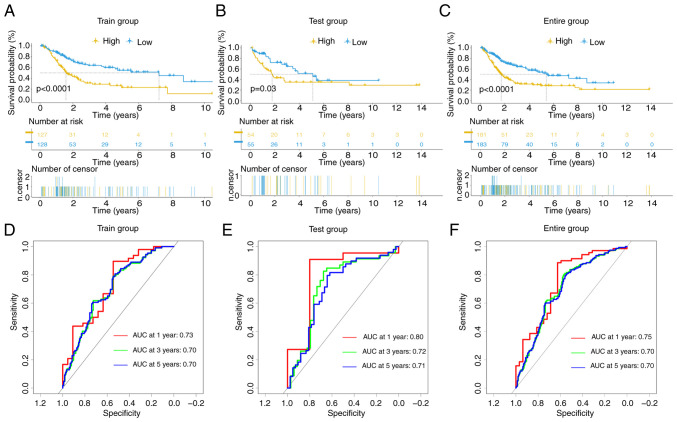
The prognostic outcomes of the high- and low-risk groups in the training (P<0.0001), test (P=0.03) and entire (P<0.0001) cohorts exhibited significant disparities, as evidenced by K-M curves (Fig. 3A-C). ROC curves revealed anticipated AUC values at different time intervals (1-, 3- and 5-years) of 0.73, 0.70 and 0.70, respectively, in the training cohort (Fig. 3D); 0.80, 0.72 and 0.71, respectively, in the test cohort (Fig. 3E); and 0.75, 0.70 and 0.70, respectively, in the entire cohort (Fig. 3F).
Figure 3.
Evaluation of the prognostic effectiveness of prognostic signature in the training group, test group and entire group. (A-C) The comparison of the Kaplan-Meier overall survival curves between low- and high-risk patients. (D-F) Receiver operating characteristic curves over one, three and five years. AUC, area under the curve.
These data affirm the outstanding predictive performance of the signature for patients with BLCA. Heatmaps depicting the expression of 8 DRlncRNAs, risk curves, risk survival status plots (Fig. S2A-C) and scatter dot plots (Fig. S2D-F) in each cohort vividly illustrate the unfavorable survival outcomes among high-risk patients.
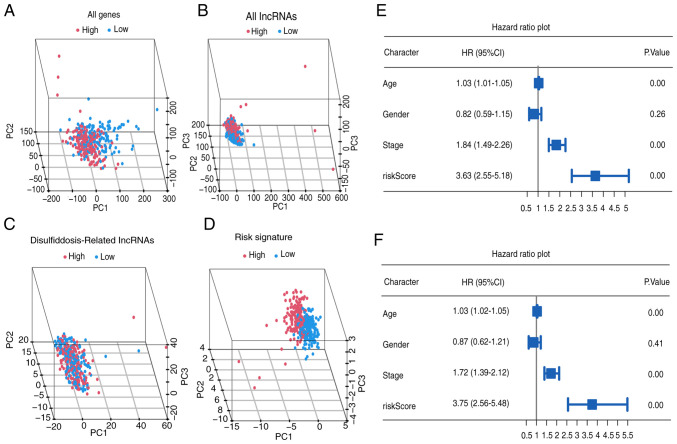
Furthermore, PCA underscores the superior discriminatory accuracy of the risk signature when distinguishing between the two groups of patients with BLCA, surpassing the discriminatory power of individual genes, lncRNAs and DRlncRNAs (Fig. 4A-D). Notably, univariate analysis identified age, tumor stage and risk score (all P<0.001) as significant prognostic factors (Fig. 4E). In the multivariate analysis, age, tumor stage and risk score (all P<0.001) independently demonstrated predictive significance (Fig. 4F).
Figure 4.
Independent prognostic analysis and PCA. (A-D) PCA analysis based on all genes, all lncRNAs, disulfidptosis-related lncRNAs and risk signature. (E and F) Univariate and multivariate analyses. PCA, principal component analysis; lncRNA, long non-coding RNA; HR, hazard ratio; CI, confidence interval.
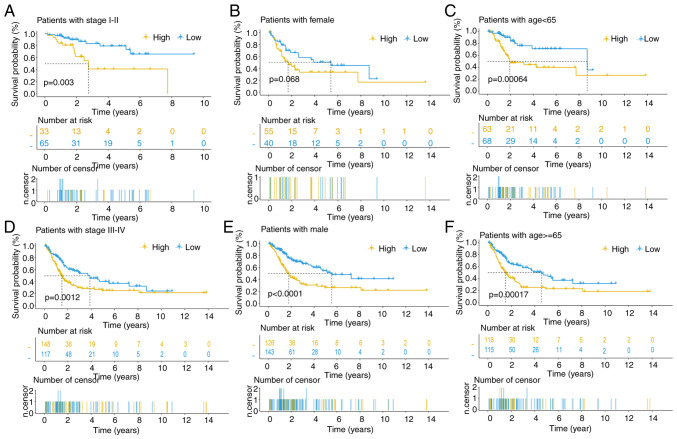
The heatmap visually portrays distinct expression patterns of the 8 lncRNAs alongside clinical features (tumor stage, age and sex) in both high- and low-risk patients (Fig. S3A). Scatter plots revealed that increasing risk scores associated with female sex (P=0.026) and mortality (P<0.001), while age and tumor stage do not exhibit significant associations with the risk score (Fig. S3B-E). Survival curves across diverse clinical subgroups underscore the robust predictive capacity of this signature, particularly among male patients across all tumor stages and age groups (Fig. 5A-F).
Figure 5.
Further validation of signature impacts. (A-F) Kaplan-Meier analysis of overall survival in various clinical characteristics groups.
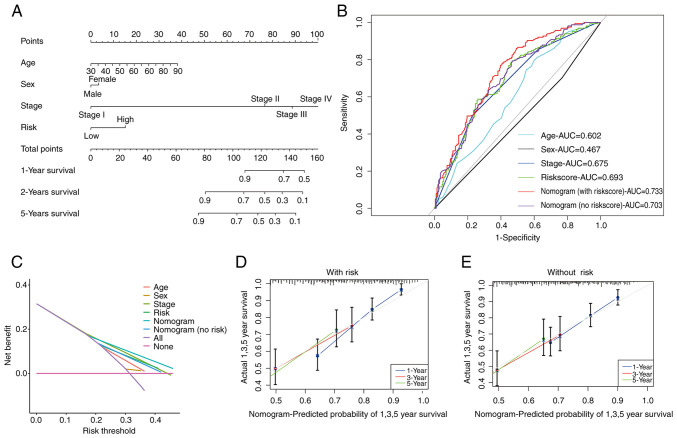
Nomogram in patients with BLCA
Following the aforementioned analyses, a prognostic nomogram that seamlessly integrates the risk score alongside other pertinent clinical features was constructed to predict the OS of patients with BLCA at 1, 3, and 5 years (Fig. 6A). Subsequent validation using data of patients with BLCA substantiated the effectiveness of this predictive tool (Fig. S4).
Figure 6.
Development of a nomogram and the predictive performance of the signature. (A) Forecast analysis of nomogram for 1-, 3- and 5-years. (B) Receiver operating characteristic curves that include diverse clinical data. (C) Decision curve analysis. (D and E) Calibration curves for 1-, 3- and 5-year nomogram (D) with risk score and (E) without risk score. AUC, area under the curve.
The AUC further attests to the accuracy of the risk score-based nomogram, with AUC values as follows: Risk score (0.693), age (0.602), sex (0.467) and stage (0.675). Notably, the nomogram incorporating the risk score (0.733) outperformed the nomogram without the risk score (0.703) and DCA confirmed the accuracy of the risk score-based nomogram (Fig. 6B and C). Additionally, calibration curves underscored the superior predictive capacity of the risk score-based nomogram (Fig. 6D and E).
Functional enrichment analysis
The salient pathways identified through KEGG pathway enrichment analysis are illustrated in Fig. S5A and B. This analysis, conducted across three libraries and examined using GSEA, yielded consistent outcomes. Remarkably, the pathways enriched in high-risk patients were linked to the cell cycle, focal adhesion and WNT signaling (Fig. S6A-C), while those in low-risk patients were predominantly related to substance metabolism (Fig. S6D). Comprehensive details of all pathways are available in Table SIV.
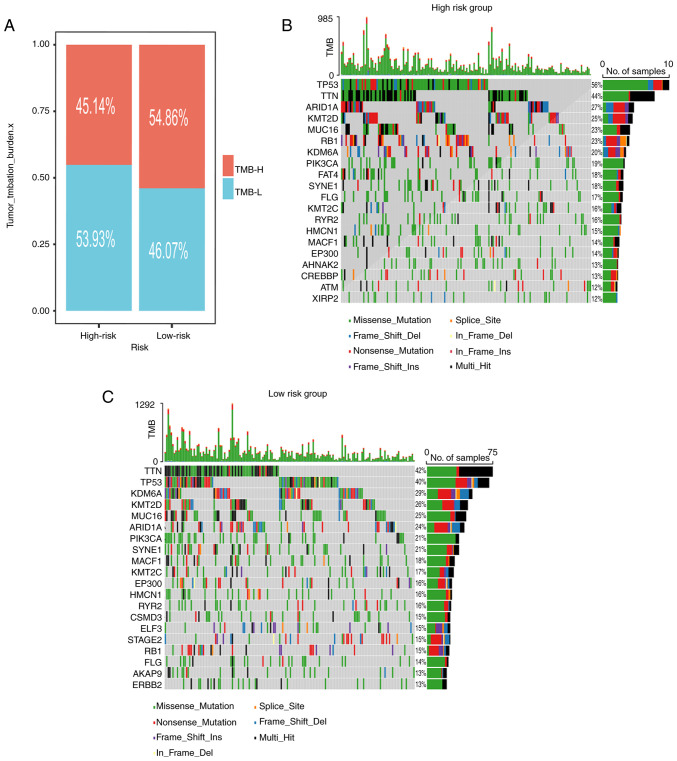
Somatic mutation landscape
As depicted in Fig. 7A, the low-risk subgroup exhibited an elevated TMB. The waterfall plots visually represented the 20 most frequently mutated genes in patients with BLCA (Fig. 7B and C). Notably, the top four mutated genes among high-risk patients were TP53 (56%), TTN (44%), ARID1A (27%) and KMT2D (25%). Meanwhile, among low-risk patients, the prominent mutated genes were TTN (42%), TP53 (40%), KDM6A (29%) and KMT2D (26%).
Figure 7.
TMB analysis. (A) Percentage bar graph. (B and C) Waterfall diagram of (B) high-risk groups and (C) low-risk groups. TMB, tumor mutation burden; H-, high; L-, low.
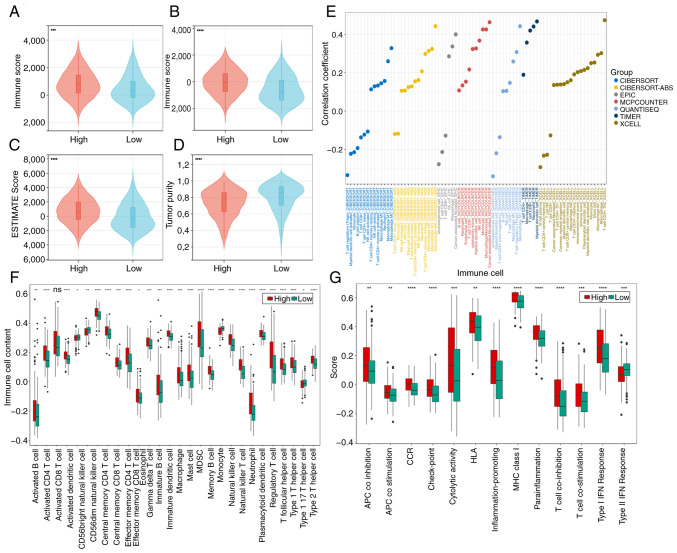
Immune infiltration landscape
In the cohort of high-risk patients, elevated stromal, immune and ESTIMATE scores were observed, alongside diminished tumor purity scores (Fig. 8A-D). These findings suggest a potential link between the unfavorable prognosis in high-risk patients and an immunosuppressive microenvironment that facilitates tumor immune evasion. The accompanying bubble chart delineates the associations of risk scores with immune cell populations (Fig. 8E).
Figure 8.
Immune infiltration landscape analysis. (A-D) Violin plots of differences in the microenvironment between the two groups. (E) The correlation between immune cells and risk scores under seven algorithms. (F) Single-sample gene set enrichment analysis evaluation of 28 immune cell infiltrations and (G) 13 immune function scores. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. ns, not significant (P>0.05).
Increased immune cell infiltration was revealed in high-risk patients, as discerned through ssGSEA analysis (Fig. 8F). Notably, immunosuppressive cell subsets [including myeloid-derived suppressor cells (MDSCs), Th2 cells (type 2 T helper cells), and regulatory T cells (Tregs; T follicular helper cells)] were significantly upregulated in the high-risk group. CD56dim natural killer cells and monocytes were more highly infiltrated in the low-risk group. For a comprehensive understanding of the intricate interplay, the detailed immune cell associations are presented in Fig. S7A-D. Overall, the high-risk cohort consistently exhibited augmented immune activity, as evidenced by the ssGSEA analysis of immune-related functions (Fig. 8G). Except for Type II IFN response, which was expressed more strongly in the low-risk group, the other 12 immune-related functions (APC co inhibition, APC co-stimulation, CCR, check-point, cytolytic activity, HLA, inflammation-promoting, MHC class I, para-inflammation, T cell co-inhibition, T cell co-stimulation and type I IFN response) were expressed more strongly in the high-risk group.
The exploration of treatment strategies for BLCA
In the exploration of therapeutic strategies for BLCA, it has been discerned that low-risk patients exhibit significantly lower TIDE scores, coupled with elevated Microsatellite instability scores, while T cell dysfunction scores remain relatively stable (Fig. S8A-D). These empirical findings posit that low-risk patients with BLCA may manifest reduced vulnerability to tumor immune subversion, thereby potentially augmenting the efficacy of immunotherapy.
Additionally, pharmaceutical agents to which high-risk patients evince heightened sensitivity are listed in Table SV, while drugs that elicit augmented responsiveness in low-risk patients are presented in Table SVI. The pharmacological agents whose sensitivity remains largely unaltered between the two patient cohorts are meticulously catalogued in Table SVII. A summary of antineoplastic drug target pathways is provided in Table SVIII. Noteworthy, compounds targeting the PI3K/mTOR signaling pathway appear to exert a more pronounced impact on high-risk populations, whereas those directed at apoptosis regulation exhibit heightened efficacy against low-risk populations.
Combined public data and in vitro validation of the prognostic signature
HPA, the valuable resource for understanding protein expression patterns, was meticulously explored to discern the protein expression disparities between BLCA and normal samples (Fig. S9). Analyzing DRGs protein expression may reveal their role in BLCA, and abnormal expression could serve as prognostic biomarkers or therapeutic targets. It was found that the proteins encoded by NCKAP1, RAC1, SLC2A1 and SLC3A2 have higher expression levels in tumors, while the protein encoded by WASF2 has higher expression levels in normal tissues. Survival analysis from the HPA indicated that high expression of proteins encoded by NCKAP1, RAC1, SLC2A1 and SLC3A2 is associated with a poor 5-year survival rate in BLCA, whereas WASF2 expression was not statistically significant. Detailed results are provided in Table SIX.
The expression levels of DRlncRNAs in the SV-HUC-1 cell line, which represents a normal bladder epithelial cell line, and the BLCA cell lines (T24 and 5637) were assessed using RT-qPCR (Fig. S10). The results revealed that ASMTL-AS1 expression is higher in the 5637 cell line compared with the T24 cell line, while the expression levels of the other seven genes do not show any statistically significant differences (Fig. S10). Additionally, as demonstrated in Fig. S11, AL390719.2, ASMTL-AS1, AL031058.1 and LINC02438 were upregulated in BLCA cell lines (T24 and 5637), whereas AC022613.2, RBMS3-AS3 and AL122035.1 were downregulated. No significant difference in expression was observed for LINC01788. In addition, to explore the potential roles of DRGs in the two cell lines, bar charts of the expression levels of DRGs in the two cell lines were obtained from HPA. The results indicated that RAC1, SLC7A11 and WASF2 were expressed higher in 5637 cells, while SLC2A1, NCKAP1 and SLC3A2 were expressed higher in T24 cells (Fig. S12).
Discussion
BLCA, the tenth most prevalent cancer worldwide, has presented an escalating incidence across several nations (22). Despite concerted endeavors in BLCA treatment, the 5-year survival rate remains disconcertingly low, hovering around a mere 14 months (2). Advances in molecular biology and deepening understanding of tumorigenesis have paved the way for personalized medicine in BLCA (23). As a result, the pursuit of novel biomarkers to improve patient prognosis prediction and advance BLCA therapy has become critical.
The process of disulfidptosis, characterized by the accumulation of disulfides and F-actin contraction, precipitates tumor cell demise (24). In light of this, DRlncRNAs have emerged as robust prognostic signatures for BLCA. Further investigations into the underlying molecular mechanisms and clinical applications are underway. The present data unequivocally demonstrated that the risk score functions independently as a potent prognostic indicator in patients with BLCA, exhibiting commendable predictive efficacy. Notably, higher risk scores correlate with poorer survival rates. The nomogram, too, showcases remarkable predictive performance. The nomogram also exhibits significant predictive performance, with the nomogram incorporating the risk score (AUC=0.733) showing greater accuracy compared with the nomogram without the risk score (AUC=0.703). Enrichment analysis revealed significant activity in the cell cycle, focal adhesion and the WNT signaling pathway among high-risk patients, while those enriched in low-risk patients were predominantly associated with substance metabolism. In addition, low-risk patients exhibit elevated TMB values and lower TIDE scores, hinting at improved outcomes with immunotherapy. There is also a differential sensitivity to immunotherapy and chemotherapy between the two risk groups.
Disulfidptosis, intricately linked to alterations in intracellular redox status, profoundly impacts cytoskeletal conformation, leading to tumor cell death (9). Prognostic signatures predicated upon disulfidptosis have consistently demonstrated robust predictive capabilities across diverse malignancies. Qi et al (25) validated the prognostic utility of a disulfidptosis-based risk signature in lung adenocarcinoma patients, yielding commendable results. Similarly, Wang et al (26) devised a disulfidptosis-related prognostic signature for predicting hepatocellular carcinoma survival, achieving favorable outcomes.
In the current investigation, transcriptomic data from patients with BLCA within the TCGA cohort were meticulously curated. Employing Pearson correlation tests and differential analyses, distinctively expressed DRlncRNAs were discerned. Subsequently, a training cohort was meticulously selected for subsequent univariate and LASSO analyses, culminating in the formulation of a robust prognostic signature that incorporates these DRlncRNAs. The outcome results of the high- and low-risk groups showed significant differences and vividly illustrate the poor outcome in high-risk patients. PCA emphasizes that prognostic signature has superior discriminative accuracy in distinguishing between two high-risk patient groups. Noteworthy, constituents of this signature include ASMTL-AS1, which orchestrates the release of miR-660 and miR-93-3p, thereby augmenting FOXO1 gene expression and effectively suppressing glycolysis and tumorigenesis (27). In the context of prostate cancer cells, overexpression of RBMS3-AS3 impedes cell proliferation and tumorigenesis (28). Furthermore, heightened expression of AL390719.2 in primary CRC is closely associated with an unfavorable prognosis, particularly in patients with CRC harboring KRAS mutations (29). However, the molecular functions of other lncRNAs in various cancers still need to be elucidated. In summary, our prognostic signature, comprising eight DRlncRNAs, emerges as a dependable tool for prognostic prediction in patients with BLCA. Finally, RT-qPCR experiments confirmed the significant differential expression of the seven DRlncRNAs in normal vs. cancer tissues.
In our efforts to elucidate relevant mechanisms, GSEA and TMB analyses were performed. Notably, the cell cycle, focal adhesion and WNT signaling pathways demonstrated significant enrichment in high-risk patients. Dysregulation of cell cycle checkpoints often leads to uncontrolled proliferation in cancer cells (30). Focal adhesion, a pivotal process integrating the extracellular matrix and cellular components, plays a significant role in tumor metastasis and invasion (31). For instance, upregulation of CircNIPBL can activate the WNT/β-catenin pathway, thereby inducing migration and invasion in BLCA (32). The observed inferior prognosis among high-risk patients may be attributed to the activation of these pathways. Those enriched in low-risk patients were predominantly associated with substance metabolism. Furthermore, an elevated TMB value was discerned in the low-risk group.
A comprehensive literature search revealed that high TMB in patients with BLCA who do not undergo immunotherapy correlates with prolonged OS and a favorable prognosis (33). Moreover, patients with high TMB BLCA generally exhibit more favorable outcomes when treated with immunotherapy, specifically anti-programmed cell death protein 1 therapy (34). It was posited that immunotherapy holds promise for achieving superior outcomes in the low-risk population. Among the high-risk group, the four most commonly mutated genes were TP53 (56%), TTN (44%), ARID1A (27%) and KMT2D (25%). TP53 mutations are common in BLCA, and TP53 function is impaired in 76% of cases, driving the progress of BLCA, affecting the prognosis and guiding the treatment (35). TTN is a common mutated gene in BLCA and can be used as a biomarker for predicting immune responses (36). ARID1A mutation is a truncal driven mutation that forms the basis for the development of a subgroup of urothelial carcinoma. When combined with other driving mutations, it leads to dysregulation of numerous key cellular processes (37). KMT2D mutations can lead to dysregulation of gene expression, thereby promoting tumor development, and its truncation mutations may serve as potential biomarkers for stratification of patients with BLCA (38).
Typically, lncRNAs are detected using RT-qPCR, RNA sequencing (RNA-seq), and in situ hybridization (ISH) (39). RT-qPCR is widely used in clinical settings due to its high sensitivity and specificity, while RNA-seq provides comprehensive lncRNA profiling and is increasingly integrated into clinical diagnostics, although its primary use remains in research. ISH enables the localization of lncRNAs within tissues, offering spatial context to their expression. In the present study, RT-qPCR was employed to precisely detect and quantify DRlncRNAs, with raw expression data normalized through established bioinformatics pipelines to reduce technical variability and ensure cross-sample comparability. The lncRNA signature developed in the present study facilitates effective risk stratification, classifying patients into high- and low-risk groups based on calculated risk scores, thereby guiding personalized therapeutic decisions. Additionally, combining the lncRNA signature with traditional clinical parameters, such as tumor stage and grade, significantly enhances the predictive accuracy of the model, providing a robust framework for prognosticating clinical outcomes in bladder cancer patients.
The current investigation has extended into the pivotal role of our distinctive signature in shaping both immunotherapeutic and chemotherapeutic strategies. Existing research has emphasized that MDSCs, triggered by transcription factors (NF-κB, STAT1, STAT3 and STAT6), impede T cell proliferation and induce an elevation in Tregs (40). Tregs infiltrating the tumor hinder the cytotoxic effect of CD8+ T cells on tumors by expressing CTLA-4 and competitively binding IL-2, thus fostering immune evasion within the TME (41). Additionally, Th2 cells, through the secretion of cytokines (such as IL-4 and IL-10), attenuate the immune response and promote tumor growth (42). The increased presence of these cells in high-risk populations may partially explain the unfavorable prognosis, providing valuable insights for immunotherapy strategies. CD56dim natural killer cells and monocytes were more highly infiltrated in the low-risk group. Furthermore, a decline in TIDE scores among low-risk patients indicates heightened immunotherapy efficacy (20). Consequently, it is advocated considering immunotherapy for patients at low risk. Notably, analyzing drug sensitivity offers essential guidance for clinical drug selection in patients with BLCA. By calculating and grouping risk scores, our risk scoring model can guide personalized drug therapy for patients with BLCA. High-risk patients exhibit increased responsiveness to medications targeting the PI3K/mTOR pathway, which is highly activated in BLCA and contributes to disease progression (43). For instance, AZD8186, as a PI3K β/δ inhibitor, which exhibits significant sensitivity in high-risk group. This heightened sensitivity in high-risk patients may be elucidated by the observed association. Moreover, patients with low risk demonstrate increased responsiveness to medications targeting the apoptosis regulation pathway. Targeting the apoptotic pathway of tumor cells represents a potent anticancer strategy, less likely to result in tumor recurrence. Several drugs directly targeting the intrinsic apoptotic pathway have received approval (44), reinforcing the inherent capability of our signature to guide clinical patients with BLCA in antineoplastic drug selection.
The present study exhibits several notable strengths. To bolster the scientific credibility of the current findings, the results were meticulously compared with those of similar studies. Compared with other similar models in the same industry, the present study has high accuracy in predicting patient prognosis. Unlike the DRlncRNA signature proposed by Sun et al (45), which underwent validation solely in the test group, our signature underwent rigorous validation across the entire cohort, rendering the results more universally applicable. Furthermore, compared with Lu et al (46), the present investigation was extended by incorporating TMB analysis to enhance the scientific robustness of our DRlncRNAs' signature, which already demonstrated commendable predictive value in patients with BLCA. Finally, the use of cell lines to validate DRlncRNAs reduced external interference and demonstrated significant expression differences. However, it is imperative to acknowledge the limitations inherent in the present study. Notably, the absence of further experiments exploring the functional roles of the DRlncRNAs within our signature constitutes a significant limitation. Additionally, the data utilized in the present study originates from the TCGA database rather than proprietary sources, potentially introducing inherent deviations. Consequently, future investigations, encompassing further in vitro or in vivo validated and clinical trials, remain essential to validate and refine the current findings. Future in vitro validation studies should include western blotting to quantify the levels of DRlncRNAs expression and corresponding protein markers in bladder cancer cell lines, and gene knockout/overexpression by utilizing RNA interference or CRISPR/Cas9 technology to downregulate or upregulate the expression of selected DRlncRNAs. Subsequently, to assess cell behavior, including proliferation, migration and apoptosis. To confirm in vitro findings, in vivo verification should be performed using animal models, such as xenotransplantation model (implanting bladder cancer cell lines with altered DRlncRNAs expression into immunodeficient mice to evaluate tumor growth and metastasis).
In conclusion, the present study has successfully established a prognostic risk scoring signature based on DRlncRNAs. The nomogram, incorporating risk scores and clinical characteristics, demonstrates significant predictive capability. Mechanistic analysis revealed that high-risk populations are enriched in the cell cycle, focal adhesion and WNT signaling pathways, while the low-risk subgroup shows elevated TMB levels and reduced TIDE scores, suggesting a more favorable response to immunotherapy. Drug sensitivity predictions indicate that targeting the PI3K/mTOR pathway offers greater efficacy for high-risk patients. Nevertheless, the limitations of the present study are acknowledged and the need for further validation through extensive basic and clinical research is emphasized.
Supplementary Material
Acknowledgements
The authors would like to thank professor Yiping Wei (Department of Thoracic Surgery, The Second Affiliated Hospital of Nanchang University) for his statistical advice.
Funding Statement
Funding: The present study was supported by the National Natural Science Foundation of China (grant no. 81560345).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
WZ had full access to all the data in the manuscript and takes responsibility for the integrity of the data and the accuracy of the data analysis. YL, HT, SJ, HW, LG, ZH, FL and WZ conceived and designed the study, and acquired, analysed, or interpreted data. YL, HT, SJ and HW performed experiments. YL, HT, SJ and WZ performed statistical analysis. YL, HT and WZ drafted the manuscript. YL, HT, WZ and FL supervised the study and critically revised the manuscript for important intellectual content. All authors read and approved the final version of the manuscript. YL and HT confirm the authenticity of all the raw data used in the present study and guarantee that the data has not been forged or tampered with. It is ensured that all data are actually obtained in the experiment and accurately reflect the experimental results.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49. doi: 10.3322/caac.21820. [DOI] [PubMed] [Google Scholar]
- 2.Witjes JA, Bruins HM, Cathomas R, Compérat EM, Cowan NC, Gakis G, Hernández V, Linares Espinós E, Lorch A, Neuzillet Y, et al. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: Summary of the 2020 guidelines. Eur Urol. 2021;79:82–104. doi: 10.1016/j.eururo.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 3.Tran L, Xiao JF, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104–121. doi: 10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Nie L, Zhang Y, Yan Y, Wang C, Colic M, Olszewski K, Horbath A, Chen X, Lei G, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. 2023;25:404–414. doi: 10.1038/s41556-023-01091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng Y, Chen X, Deng G. Disulfidptosis: A new form of regulated cell death for cancer treatment. Mol Biomed. 2023;4(18) doi: 10.1186/s43556-023-00132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 7.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond) 2021;41:109–120. doi: 10.1002/cac2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H, Wang L, Lu T, Zhang Y, Sun Z, Han X. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat Commun. 2022;13(816) doi: 10.1038/s41467-022-28421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang YL, Zhang Y, Tan XR, Qiao H, Liu SR, Tang LL, Mao YP, Chen L, Li WF, Zhou GQ, et al. A lncRNA signature associated with tumor immune heterogeneity predicts distant metastasis in locoregionally advanced nasopharyngeal carcinoma. Nat Commun. 2022;13(2996) doi: 10.1038/s41467-022-30709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19(77) doi: 10.1186/s12943-020-01188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutter C, Zenklusen JC. The cancer genome atlas: Creating lasting value beyond its data. Cell. 2018;173:283–285. doi: 10.1016/j.cell.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 13.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(550) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duforet-Frebourg N, Luu K, Laval G, Bazin E, Blum MGB. Detecting genomic signatures of natural selection with principal component analysis: Application to the 1000 genomes data. Mol Biol Evol. 2016;33:1082–1093. doi: 10.1093/molbev/msv334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Xie J, Wu W, Chen H, Li S, He H, Yu Y, Hu M, Li J, Zheng R, et al. A simple nomogram for predicting failure of non-invasive respiratory strategies in adults with COVID-19: A retrospective multicentre study. Lancet Digit Health. 2021;3:e166–e174. doi: 10.1016/S2589-7500(20)30316-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48 (W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeser D, Gruener RF, Huang RS. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22(bbab260) doi: 10.1093/bib/bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 23.Dobruch J, Oszczudłowski M. Bladder cancer: Current challenges and future directions. Medicina (Kaunas) 2021;57(749) doi: 10.3390/medicina57080749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng P, Zhou C, Ding Y, Duan S. Disulfidptosis: A new target for metabolic cancer therapy. J Exp Clin Cancer Res. 2023;42(103) doi: 10.1186/s13046-023-02675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi C, Ma J, Sun J, Wu X, Ding J. The role of molecular subtypes and immune infiltration characteristics based on disulfidptosis-associated genes in lung adenocarcinoma. Aging (Albany NY) 2023;15:5075–5095. doi: 10.18632/aging.204782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Guo K, Zhang D, Wang H, Yin J, Cui H, Wu W. Disulfidptosis classification of hepatocellular carcinoma reveals correlation with clinical prognosis and immune profile. Int Immunopharmacol. 2023;120(110368) doi: 10.1016/j.intimp.2023.110368. [DOI] [PubMed] [Google Scholar]
- 27.Feng Z, Chen R, Huang N, Luo C. Long non-coding RNA ASMTL-AS1 inhibits tumor growth and glycolysis by regulating the miR-93-3p/miR-660/FOXO1 axis in papillary thyroid carcinoma. Life Sci. 2020;244(117298) doi: 10.1016/j.lfs.2020.117298. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Z, Zhang Y, Chen X, Wu P, Chen D. Long noncoding RNA RBMS3-AS3 acts as a microRNA-4534 sponge to inhibit the progression of prostate cancer by upregulating VASH1. Gene Ther. 2020;27:143–156. doi: 10.1038/s41434-019-0108-1. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Huang QY, Ge CJ. Identification of prognostic long intergenic non-coding RNAs as competing endogenous RNAs with KRAS mutations in colorectal cancer. Oncol Lett. 2021;22(717) doi: 10.3892/ol.2021.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kar S. Unraveling cell-cycle dynamics in cancer. Cell Syst. 2016;2:8–10. doi: 10.1016/j.cels.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Lin X, Zhuang S, Chen X, Du J, Zhong L, Ding J, Wang L, Yi J, Hu G, Tang G, et al. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Mol Ther. 2022;30:688–702. doi: 10.1016/j.ymthe.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Kong Y, An M, Luo Y, Zheng H, Lin Y, Chen J, Yang J, Liu L, Luo B, et al. ZEB1-mediated biogenesis of circNIPBL sustains the metastasis of bladder cancer via Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2023;42(191) doi: 10.1186/s13046-023-02757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valero C, Lee M, Hoen D, Wang J, Nadeem Z, Patel N, Postow MA, Shoushtari AN, Plitas G, Balachandran VP, et al. The association between tumor mutational burden and prognosis is dependent on treatment context. Nat Genet. 2021;53:11–15. doi: 10.1038/s41588-020-00752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGrail DJ, Pilié PG, Rashid NU, Voorwerk L, Slagter M, Kok M, Jonasch E, Khasraw M, Heimberger AB, Lim B, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32:661–672. doi: 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyu Q, Lin A, Cao M, Xu A, Luo P, Zhang J. Alterations in TP53 are a potential biomarker of bladder cancer patients who benefit from immune checkpoint inhibition. Cancer Control. 2020;27(1073274820976665) doi: 10.1177/1073274820976665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, Zhou Y, Liu W, Dai J. A novel cuproptosis-related lncRNAs signature predicts prognostic and immune of bladder urothelial carcinoma. Front Genet. 2023;14(1148430) doi: 10.3389/fgene.2023.1148430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conde M, Frew IJ. Therapeutic significance of ARID1A mutation in bladder cancer. Neoplasia. 2022;31(100814) doi: 10.1016/j.neo.2022.100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Z, Aoi Y, Philips CN, Meghani KA, Gold SR, Yu Y, John LS, Qian J, Zeidner JM, Meeks JJ, Shilatifard A. Somatic mutations of MLL4/COMPASS induce cytoplasmic localization providing molecular insight into cancer prognosis and treatment. Proc Natl Acad Sci USA. 2023;120(e2310063120) doi: 10.1073/pnas.2310063120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang T, Tao T, Wu K, Liu L, Xu W, Zhou D, Fang H, Ding Q, Huang G, Wu S. Cancer-associated fibroblast-induced remodeling of tumor microenvironment in recurrent bladder cancer. Adv Sci (Weinh) 2023;10(e2303230) doi: 10.1002/advs.202303230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noyes D, Bag A, Oseni S, Semidey-Hurtado J, Cen L, Sarnaik AA, Sondak VK, Adeegbe D. Tumor-associated Tregs obstruct antitumor immunity by promoting T cell dysfunction and restricting clonal diversity in tumor-infiltrating CD8+ T cells. J Immunother Cancer. 2022;10(e004605) doi: 10.1136/jitc-2022-004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang MW, Tao LY, Jiang YS, Yang JY, Huo YM, Liu DJ, Li J, Fu XL, He R, Lin C, et al. Perineural invasion reprograms the immune microenvironment through cholinergic signaling in pancreatic ductal adenocarcinoma. Cancer Res. 2020;80:1991–2003. doi: 10.1158/0008-5472.CAN-19-2689. [DOI] [PubMed] [Google Scholar]
- 43.Meeks JJ, Lerner SP. Molecular landscape of non-muscle invasive bladder cancer. Cancer Cell. 2017;32:550–551. doi: 10.1016/j.ccell.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Z, Wang J, Fan Z, Yang Y, Meng X, Ma Z, Niu J, Guo R, Tran LJ, Zhang J, et al. Investigating the prognostic role of lncRNAs associated with disulfidptosis-related genes in clear cell renal cell carcinoma. J Gene Med. 2024;26(e3608) doi: 10.1002/jgm.3608. [DOI] [PubMed] [Google Scholar]
- 46.Lu H, Wu J, Liang L, Wang X, Cai H. Identifying a novel defined pyroptosis-associated long noncoding RNA signature contributes to predicting prognosis and tumor microenvironment of bladder cancer. Front Immunol. 2022;13(803355) doi: 10.3389/fimmu.2022.803355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.