Abstract
Microarray analysis of gene expression during the yeast division cycle has led to the proposal that a significant number of genes in Saccharomyces cerevisiae are expressed in a cell-cycle-specific manner. Four different methods of synchronization were used for cell-cycle analysis. Randomized data exhibit periodic patterns of lesser strength than the experimental data. Thus the cyclicities in the expression measurements in the four experiments presented do not arise from chance fluctuations or noise in the data. However, when the degree of cyclicity for genes in different experiments are compared, a large degree of non-reproducibility is found. Re-examining the phase timing of peak expression, we find that three of the experiments (those using α-factor, CDC28 and CDC15 synchronization) show consistent patterns of phasing, but the elutriation synchrony results demonstrate a different pattern from the other arrest-release synchronization methods. Specific genes can show a wide range of cyclical behavior between different experiments; a gene with high cyclicity in one experiment can show essentially no cyclicity in another experiment. The elutriation experiment, possibly being the least perturbing of the four synchronization methods, may give the most accurate characterization of the state of gene expression during the normal, unperturbed cell cycle. Under this alternative explanation, the observed cyclicities in the other three experiments are a stress response to synchronization, and may not reproduce in unperturbed cells.
INTRODUCTION
When a result or experiment is recognized as fundamental to a field, and is cited by many as a key result that future work should look to, it is important that the experiments be beyond reproach and unconditionally acceptable. We wish to analyze such an experiment and express our concerns regarding the experiments, the analysis and the results. We do this so that the field of cell cycle studies can rest on results that are correct and not subject to major revision. We hope either that the concerns raised are answered in the future or that they lead to a different view of the eukaryotic cell cycle.
Microarray analysis has been used to identify a large number of genes in Saccharomyces cerevisiae that are proposed to be expressed in a cell-cycle-specific manner. In one set of experiments (1) cells were synchronized three different ways (α-factor arrest, temperature arrest of a temperature sensitive mutant, elutriation synchronization). The mRNA was extracted at a number of points following synchronization, and the expression levels of approximately 6000 genes were determined using a two-color microarray protocol. Genes expressed in a cell-cycle-specific manner were identified using a Fourier fitting algorithm (1). 800 genes were classified as being expressed in a cell-cycle-specific manner, with 300 classified as G1-phase genes, 71 as S-phase genes, 121 as G2-phase genes, 195 as M-phase genes and 113 as M/G1-phase genes.
An independent analysis of S.cerevisiae gene expression during the division cycle used two temperature-sensitive mutants to synchronize cells (2). Using Affymetrix microarrays, gene expression following synchronization was determined at a sequence of points in nearly the same set of transcripts that were measured in the two-color experiments. Cyclic expression patterns were identified by subjective or visual analysis of the microarray expression measurements. The CDC28 synchronization results from the Affymetrix experiments are included in the analysis of yeast genes (1) to give four experiments that can be compared.
At the time of writing, the data on gene expression during the S.cerevisiae cell cycle (1) has been referenced by more than 293 published papers. These microarray data have spawned a large body of analytical work on gene expression during the cell cycle of yeast (3–17). Because of the enormous interest in understanding which (or whether) specific genes are expressed at particular times during the cell cycle, it is important that any uncharacterized sources of experimental variation or reasonable alternative explanations for the patterns in the experimental data be brought to light. We have reconsidered the expression results, and find that while the α-factor, CDC15 and CDC28 experiments are in agreement, the elution results are non-conforming. This discrepancy was very likely the reason that the elution results were not used for the main argument that there are large numbers of genes in S.cerevisiae with cell-cycle-coordinated expression.
We suggest an alternative explanation and conclusion, namely that the results of the four experiments are what should be expected. Since the α-factor, CDC15 and CDC28 synchronizations may perturb cells, release from arrest would be expected to have a dramatic influence on gene expression. This perturbation may lead to cyclic variations in gene expression. The elution-based synchronization, being less perturbing, does not produce such cycles. This analysis has implications regarding whether the observed periodicities in expression may be interpreted as being representative of gene expression in cells growing under normal, unperturbed conditions.
Besides performing a statistical reanalysis of the microarray results, we shall also raise basic biological issues relating to the experimental analysis, and thereby argue that it is important, irrespective of the microarray results, to re-examine the proposal of cycle-specific gene expression in yeast.
MATERIALS AND METHODS
Data
The raw microarray measurements are available from two websites. The two-color data (1) were obtained from the website http://genome-www.stanford.edu/cellcycle. The Affymetrix CDC28 data (2) are available in raw form from the website http://171.65.26.52/yeast_cell_cycle/cellcycle.html. The Affymetrix CDC28 values were processed (1) to allow direct comparison with the results of the two-color analysis (1). In the analysis presented here, we use both the raw two-color data as well as the processed CDC28 data (1).
Normalization of data
The data were normalized (1) to enable direct comparison across experiments, across genes and across the two different types of microarray. In addition to the processing carried out as described in Spellman et al. (1) we applied a logarithmic transformation to each measurement, then centered the genes within each experiment.
Sampling interval and interdivision time
Since different synchronization methods required different growth conditions, the nominal interdivision times varied across the experiments. Additionally, RNA was collected at different sampling intervals in different experiments, and different numbers of samples were obtained in different experiments. Based on the information reported in the primary papers, Table 1 summarizes the values used for the sampling intervals, total experiment times, interdivision times and number of samples obtained. All of the analyses presented here have also been done using the values for these parameters chosen by Aach and Church (4) (Table 1) and the results obtained are qualitatively similar to the results shown below.
Table 1. Experimental sampling values for the published experimental data (1).
| Sampling interval (min) | Total experiment time (min) | Interdivision time (min) | Number of points sampled | |
|---|---|---|---|---|
| Alpha | 7 | 120 | 66 (67.5)b | 18 |
| CDC15 | 10 | 290 | 110 (119) | 24 |
| CDC28a | 10 | 160 | 85 | 17 |
| Elution | 30 | 390 | 390 (422.5) | 14 |
aCDC28 data is from Cho et al. (2).
bThe values in parentheses are the values used by Aach and Church (4) in their analysis of the data (1).
Numerical characterization of sinusoidal expression
For each gene, the measured time points were fit using least squares to two basis curves. The first basis curve has the form S(t) = sin(2πt/T) and the second basis curve has the form C(t) = cos(2πt/T), where T is the nominal interdivision time. Suppose Yi(t) denotes the measured expression for transcript i at time t. The vector Yi(t) was regressed against S(t) and C(t) leading to the decomposition Yi(t) = aiS(t) + biC(t) + Ri(t), where Zi(t) = aiS(t) + biC(t) represents the periodic component of expression with T min period, and Ri(t) represents the component of expression that is either aperiodic, or that has a period substantially different from T min. The proportion of variance explained by the Fourier basis (Fourier-PVE) is the ratio mi = var[Zi(t)]/var[Yi(t)], which lies between 0 and 1. Values closer to 1 indicate greater sinusoidal expression with a T min period, while values closer to zero indicate a lack of periodicity, or periodicity with a period that is substantially different from T min.
The fitted waveform Zi(t) is proportional to a shifted sine wave of the form sin(πU + 2πt/T), where –1 < U < 1 is the phase. Phases close to 0, 1 or –1 are sine like, in that at time zero they take on an intermediate value, whereas phases close to 1/2 or –1/2 are cosine like, in that at time zero they are close to their maximum or minimum value.
Randomization of data
It was important to determine whether the level of sinusoidal cell-cycle-specific expression patterns in the different experiments could be explained as arising from chance arrangements of random fluctuations in the measurements (i.e. variation arising from biological or technical sources that have an equal influence on all time points). Therefore an artificial data set was constructed that was compatible with the observed data in terms of the overall variation at each time point, but which lacked any special tendency to exhibit periodic or sinusoidal expression patterns. To construct this data set, a random permutation of the observed values for a given gene across the time points was generated in such a way that any permutation was as likely to appear as any other. In other words, an artificial experiment was constructed by sampling uniformly and without replacement from the measured values for each gene in an actual experiment.
RESULTS
Our statistical analysis is framed as a response to three questions. First, we ask whether the number of cyclic genes is sufficiently high that one may conclude that at least some of the apparent cyclicity in gene expression does not arise from coincidental arrangements of measurement error and non-cyclic biological variation. To this, we answer yes. Then we ask whether the same genes exhibit high cyclicity across the four synchronization methods. Here the answer is more complex, as there is only a weak positive correlation of cyclicities between different experiments. Our third question is whether, among the cyclic genes, the peak expression occurs at the same relative point within the cell cycle in all four experiments. The answer to this question depends on the methods, with the elution experiment being different from the other three experiments, which are in substantial agreement. The statistical analysis of the experimental data is then followed by an analysis of the biology of S.cerevisiae, the importance of the pattern of growth and division for cell-cycle analysis, and an analysis of specific gene expression patterns in different experiments.
Comparison of experimental data with random data
An initial question was whether the observed cyclical expression in S.cerevisiae identified using microarrays could be explained by purely statistical considerations. Given the existence of measurement error as well as biological or experimental variation in expression that is not due to the cell cycle, it is possible that even if none of the genes were truly expressed in synchrony with the cell cycle, a certain number of genes with apparently cyclic expression might be found due to chance fluctuations and non-cyclic biological variation. Put another way, it is generally agreed that a substantial number of genes present on the microarray do not possess cell-cycle-specific expression. In any given experiment, many of these genes will exhibit cyclic expression due to the chance arrangement of non-cyclic, random fluctuations. We were interested in determining whether cyclic variation in expression can be explained as arising from these random fluctuations.
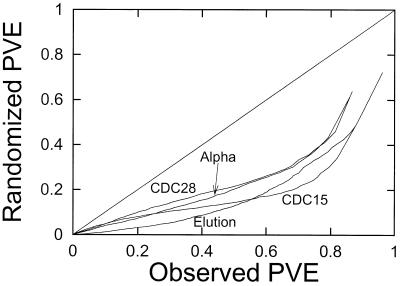
An analysis of the yeast data is presented in Figure 1. For each rank, r = 1, 2, …, the cyclicity (quantified as Fourier-PVE) of the rth most cyclic gene in the randomized data is plotted against the cyclicity of the rth most cyclic gene in the observed data. The data used in Figure 1 are from the 1000 genes with the highest standard deviation. This selection process eliminates from consideration a large number of genes with negligible variation. Points below the diagonal line indicate a level of cyclicity in the observed data that cannot be explained by measurement error or non-cyclic biological variation. For example, in the α-factor synchronization experiment in Figure 1, a number of genes have a measured cyclicity greater than 0.8, but one would have to go down to a cyclicity threshold of approximately 0.5 to acquire the same number of genes in the randomized data. Similar results are presented for the three other synchronization methods.
Figure 1.
Cyclicity of expression following different synchronization methods. For each experiment, the 1000 genes with the highest standard deviation in experimental values (using the raw data) were determined. For each of these 1000 genes the Fourier-PVE was calculated. These values were also determined for a randomized data set derived from the experimental values. The randomized data set (construction described in Materials and Methods) was produced by a randomization of the values for each gene. This produces two lists of numbers, both of which were sorted from least to greatest. The cyclicity values for each rank of gene in order is plotted for the experimental values against the randomized values. The cyclicity in each of the four experiments is therefore compared with the cyclicity in its randomized counterpart. Since the points fall below the diagonal line, this graph shows that there is more cyclicity present in the experimental values than can be explained as arising from chance fluctuations in the measurement process.
Figure 1 indicates that the periodicities observed in gene expression under all four synchronizations of S.cerevisiae cells cannot be accounted for by the chance arrangement of random fluctuations in the measurements. These patterns can be attributed to genuine periodicities in gene expression in the synchrony experiments. This result is consistent with the original analysis (1), where a similar randomization comparison led to the conclusion that the false positive rate lies between 3 and 10%.
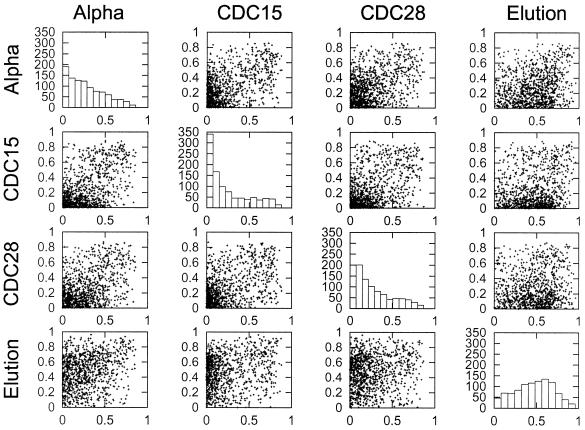
Reproducibility of the cyclicity values
To determine whether genes that have cyclic expression in one experiment also tend to have cyclic expression in the other experiments, we looked at the reproducibility of the Fourier-PVE across the four experiments. In Figure 2, the cyclicities (quantified as Fourier-PVE) between each pair of distinct synchronization methods is shown as a scatter plot. As in Figure 1, only the 1000 genes with greatest average standard deviation across the four experiments are included. Visual inspection of Figure 2 indicates that there is a very small positive association between elution results and any of the other three synchronization methods. The association between the α-factor, CDC15 and CDC28 experiments is slightly stronger, but also appears to be quite weak. Much of this apparent lack of reproducibility may arise from the fact that even very cyclic patterns may not conform to a sine wave, as is predicated by the Fourier analysis. We do realize that the choice of a sine wave fit is only one of many possible curves that could have been used, and future work may require additional analysis using different idealizations of cell-cycle-specific gene expression.
Figure 2.
Reproducibility of cyclicity between different experiments. For each distinct pair of synchronization methods used to analyze the cell cycle (1), the cyclicity levels for the 1000 genes with the highest standard deviations for one of the experiments are presented as a scatter plot. Thus, for the first line of four boxes, the 1000 genes in the α-factor experiment were identified, and their cyclicities in the α-factor experiment are compared with the cyclicities in the other three experiments. The four diagonal graphs are histograms showing the marginal level of cyclicity in each experiment for all 1000 genes for the selected experiment (listed at the left).
The distribution of cyclicity values for the different experiments (Fig. 2, histograms) also shows that the overall level of cyclicity in the 1000 selected genes is different for the elutriation experiment compared with the three arrest methods of synchronization.
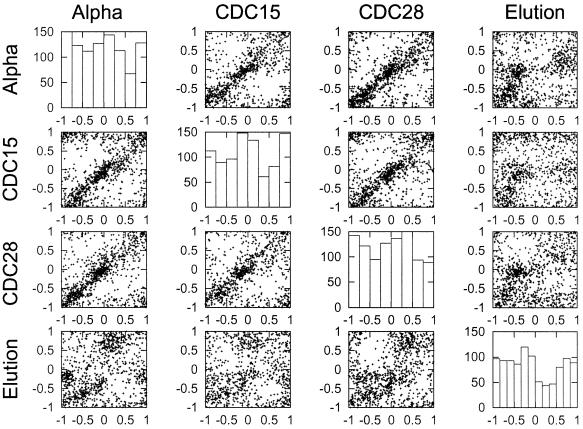
Reproducibility of the time of peak expression
For each of the four synchronization methods, we selected the top 1000 genes based on Fourier-PVE and determined whether the location of peak expression was consistent between the experiment used to do the selection and the other three experiments. We used the phase values (described in Materials and Methods) to identify the time of peak expression in each gene. In Figure 3, each row shows scatter plots of the timing of peak expression in the experiment used to do the gene selection against the timing of gene expression in the other three experiments.
Figure 3.
Correlation of peak timing between different experiments. For each set of genes in an experiment the 1000 genes with the highest cyclicities were identified. The phase locations of these genes were then compared in a scatter plot against the phase location in the other three experiments. The phase location was determined for each gene using the fitted Fourier pattern. The graphs on the diagonal are histograms summarizing the frequency of the 1000 genes with the highest cyclicities having peaks of expression at particular times during the cell cycle.
With the exception of the elution experiment (see below), there is a high level of reproducibility in the relative timing of peak expression. This suggests that within the α-factor, CDC15 and CDC28 experiments, although the Fourier-PVE shown in Figure 2 may intrinsically be somewhat variable, the consistency in peak location is far too high to be explained by chance. We draw two conclusions from this finding. First, we conclude that the Fourier analysis is a suitable method for estimating the timing of peak expression, even while the cyclicity values given by Fourier-PVE should be treated as lying on a coarse scale. More importantly, we conclude that the α-factor, CDC15 and CDC28 synchronization methods give a strong and reproducible signal, whereas the elution experiment gives a different pattern.
The phase pattern in the elution experiment is not completely unrelated to the pattern under arrest/release synchronization methods. The relationship is not indicated by a clear diagonal, as is the case with the other comparisons. It is possible that the data would be consistent with differences in the relative phases in the elutriation and the arrest/release methods of cell cycle analysis. A resolution of the question of phase of expression will probably require experiments using non-perturbing methods of cell-cycle analysis to determine whether there is a particular order of gene expression during the yeast cell cycle.
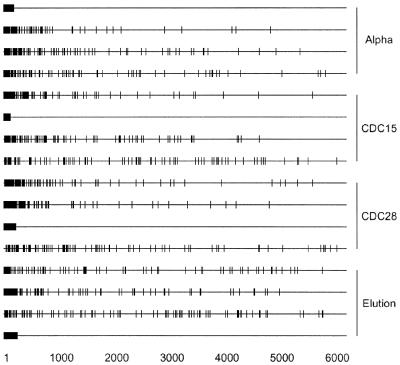
Reproducibility of particular gene cyclicities
Another way of looking at reproducibility between different synchronization experiments is presented in Figure 4, where the 100 most cyclically expressed genes under each of the four synchronization methods are displayed based on their cyclicity levels. The cyclicity levels are expressed as ranks within the roughly 6200 genes, with a low rank corresponding to a high cyclicity. Each block of four horizontal lines displays the cyclicity ranks in all four experiments for the 100 most cyclic genes in a given experiment. Numerous genes have no apparent reproducibility of gene cyclicity between different experiments. Genes that are periodic under one synchronization procedure are not necessarily periodic under a different synchronization procedure. In fact, some genes that are very periodic in one experiment are at the other extreme end of the cyclicity scale in another experiment, showing essentially no reproducibility. This result indicates that numerous periodicities observed in some experiments are not clearly due to an innate, cell-cycle-related expression pattern. Rather, this result (Fig. 4) suggests that the observed cyclicities may possibly be due to a biological—but not a cell-cycle driven or related—response to the experimental treatments. If cell cycle expression were innate, then the cyclicity results would be expected to be consistent in all synchronization experiments. Furthermore, this result can be used to conclude that at least some of the synchronization methods used to study cell-cycle-dependent gene expression do not truly synchronize cells.
Figure 4.
Reproducibility of cyclicity for specific genes for different synchronization experiments. In each panel of four lines are displayed the 100 most cyclic genes from a given synchronization method (1). Each vertical dash indicates the rank cyclicity for a single gene in a single experiment. The first line is the location of the 100 most cyclic genes for the α-factor experiment. Lines 2–4 are the particular genes from line 1 and their relative cyclicity for synchronization by CDC15 arrest, CDC28 arrest and elution. Thus rank cyclicities of genes selected for high cyclicity for one synchronization method are shown for all four synchronization methods. Similarly, the remaining 12 lines are comparative analyses of the data for high cyclicity genes from the CDC15, CDC28 and elution experiments compared with the same genes analyzed by different methods.
A fourth way of looking at the reproducibility of cyclicity across experiments is to determine how often a particular gene appears near the top of a list of genes sorted by cyclicity in a given synchronization experiment. A tabulation of these findings is presented in Table 2. For each of the four cell-cycle experiments (1), the top 50, 100, 200 and 300 genes in terms of cyclicity were determined. If no gene appeared in the top group of genes for more than a single experiment, then one would have 200, 400, 800 or 1200 genes listed. Because some genes are present in more than one list, fewer distinct genes are found in the aggregate list. Specifically, 175, 322, 595 and 883 genes are found for the four choices of list size. In Table 2 the distribution of these duplications, triplications and quadruplications are presented. Even when retaining the top 300 genes in each experiment, 73% of the genes are not present in more than one experiment; that is, they are present as single representatives. Only nine genes are present among the top 300 for all four experiments. If one examines only the top 100 genes for each of the four experiments, then there are no genes that are present in all four lists. This again indicates that the level of reproducibility of cyclic gene expression over different synchronization experiments is of a low order.
Table 2. Frequency of high cyclicity values for different synchronization methods.
| Genes selected from each synchronization experiment | 300 | 200 | 100 | 50 |
| Total analyzed after elimination of duplicates | 883 | 595 | 322 | 175 |
| Expected with no duplicates |
1200 |
800 |
400 |
200 |
| Number of times a given gene is found |
Frequency of finding a gene either 1, 2, 3 or 4 times among the top cyclical genes in an experiment |
|||
| 4 | 9 | 3 | 0 | 0 |
| 3 | 59 | 40 | 12 | 2 |
| 2 | 172 | 116 | 54 | 21 |
| 1 | 643 | 436 | 256 | 152 |
| Percentage singles | 73% | 73% | 80% | 87% |
DISCUSSION
In the analysis of yeast gene expression during the division cycle (1) various choices were made that may have had the effect of emphasizing data supportive of cyclic gene expression. These choices thus de-emphasized data that might not be supportive of cyclic gene expression.
In calculating the aggregate cyclicity value for each gene, the individual values for the α-factor, CDC28 and CDC15 synchronization experiments were included, whereas the results for the elution experiment were excluded. The reason that the elutriation data was not included was that ‘it was not possible to calculate a [value] that maximized the value of more than a handful of the known genes’ (1). Notably, the CDC28 data were more similar to the α-factor and CDC15 data than were the elution measurements, even though the CDC28 data were generated by a different group using a different type of microarray.
It is interesting in this context to note that in a paper devoted to analyzing the extant yeast cell cycle (5) a classification of genes according to function was different from the original (1) classification. The explanation given (5) was that ‘This may be due to the poor quality of the elutriation expression data, as synchronization by elutriation was not very effective in this experiment. For the α-factor-synchronized cell cycle expression there is much better agreement between the two classifications.’
One possible explanation for the different classification results, and the lack of conformity of the elutriation data with the α-factor, CDC15 and CDC28 results, is that the elutriation data may be closer to the expression pattern in unperturbed cells. From this point of view, one might conclude that there is actually little variation in gene expression during the division cycle.
Conversely, the α-factor-synchronized cell cycle data may give stronger periodicities that lead to a more robust classification scheme. If this were the case, one would expect reproducible results for the α-factor experiments but less reproducible results for the elutriation data. Again, this reproducibility and strength of periodicity should not be taken as an indication that a particular gene is expressed in a cell-cycle-specific manner during the normal, unperturbed cell cycle.
To put this more clearly, the results from the α-factor synchronization (as well as CDC28 and CDC15 synchronization) may lead to more defined and reproducible analyses than the data from the elutriation experiment. Such reproducibility may be useful for gene expression analysis. But utility is not a proper criterion for accepting data that may be affected by potential perturbations or artifacts. It is possible that the weaker cyclicities in the elutriation data are closer to the natural situation in growing cells. For this reason one should be wary of extending the batch synchronization methods (whether by α-factor or by temperature sensitive arrest synchronization) to determine the pattern of gene expression during the division cycle.
Beyond the issue that a selection bias may have led to the aggregation of a subset of experimental results that support a particular prior point of view, another more fundamental point must be made about the elution results. The elution synchronization approach is the only one of the four methods that has a clear theoretical basis. [The problems with the current view of arrest synchronization methods have been clearly described (18) and will not be further detailed here.] Small cells are younger cells. Large cells are cells later in the division cycle. Selection of small cells selects cells that are preferentially young cells. The theoretical basis for synchronization by the other three methods is less clear. The three inhibition methods assume that cells arrest at a point in the cell cycle. It is further assumed that when released from inhibition the arrested cells resume growth from that point to produce synchronized growth. These assumptions are debatable.
We favor the elutriation results over the results from the other three inhibition methods of synchronization. To be fair, we point out that the elutriation results, in contrast to the other results, cover only one cell cycle (Table 1). This aspect of the design leaves open the possibility that a more complete set of elution measurements may turn out to show a greater consistency with the inhibition methods.
The question remains whether the observed periodicities are truly representative of the normal cell cycle in unperturbed cells. It has been argued that the batch synchronization methods (e.g. α-factor arrest, temperature sensitive inhibition, nocodazole) used to analyze the cell cycle of S.cerevisiae do not actually synchronize cells but merely align cells for particular cell properties (18). But equally importantly, it is generally accepted that such inhibition methods may lead to introduction of artifacts or anomalous periodicities that were not present in the unperturbed cells (19).
Problems associated with aggregating several experiments
In another approach to analyzing cyclicity, an aggregate cyclicity value was determined by combining the values for three different experiments (1). Cyclicity levels in individual experiments were not reported, and the elutriation was dropped for reasons that may be considered arbitrary. In order to assess whether the Affymetrix data using CDC28 synchronization, which had been produced by a different laboratory, was strongly influencing the results, the aggregation analysis was repeated without including this experiment. Although it is argued that the results are not significantly impacted by the inclusion or exclusion of the Affymetrix data (1), it is worth noting that the Affymetrix measurements contribute only one-third of the aggregate score, so it is difficult to assess whether the influence is in fact large. We suggest that while an aggregate score may be a useful way to borrow strength across several experiments, relying solely on an aggregated cyclicity measure may serve to mask problems in the individual experiments. We propose that before an aggregate score is considered, an effort should be made to demonstrate that there is a reasonable strength of signal in the individual experiments, and a reasonable level of reproducibility between the individual experiments.
Clustering of genes according to function, cyclicity and control DNA sequences
A crucial point regarding the interpretation of the CDC28 experiments resides in a subsequent analysis of the original data (16). The CDC28 data were analyzed using a clustering algorithm. Clusters of genes were produced that exhibited similar expression patterns. The DNA sequences associated with the genes in each cluster were also analyzed. Two types of clusters were observed. First, three clusters that represented three different functional categories (replication and DNA synthesis, organization of the centrosome, and budding and cell polarity) gave cyclical patterns that were associated with expression at particular times during the cell cycle (G1, S and G2). More significantly, a search of the DNA sequences associated with these genes revealed a surprising level of consensus among the DNA sequences within the clusters. The consensus sequences associated with a given cluster were rarely found outside the cluster.
A second type of cluster gave groups of gene expression patterns with no reproducibility between the first and second cycles of gene expression. For example, the ribosome cluster gave no peak in the first cycle after synchronization and a single peak in G1 in the second cycle. The methionine and sulfur metabolism cluster gave a peak in S–G2 of the first cycle and no peak in the second cycle. Carbohydrate metabolism genes were low in the first cycle and high in the second cycle. This lack of reproducibility between two successive cycles is an indication that the cells are perturbed. Non-reproducibility of expression levels between two cycles for any gene group means that there is a perturbation of the cells, such that there is either an induction or a repression of expression in the first cycle that is not present in the second cycle.
What about the interesting correlation of expression patterns and the upstream DNA sequence motifs that were common to members of each cluster? One explanation for the reproducibility of expression among members of a cluster (even if that pattern is not related to the cell cycle) and upstream DNA sequences is the possibility that the CDC28 synchronization method affects genes in the cluster through the common sequence motifs. However, this finding does not imply anything more than that the stresses applied as part of the synchronization process produced perturbations that were mediated through the common upstream sequences. The correlation of cluster pattern and upstream gene sequences does not mean that the observed cyclicities are related to the normal, unperturbed cell cycle.
Problems with continuous variation in cell-cycle expression patterns
One of the more striking results on cyclic gene expression during the yeast division cycle is that the times of peak gene expression appeared to be uniformly distributed throughout the division cycle (1). When the genes were sorted vertically according to peak expression time, it appeared that at each point in the division cycle approximately the same number of genes exhibited their peak expression. The color coding used to demonstrate this effect might obscure precise timing of peak gene expression. Therefore the timing of peak expression was quantified using the Fourier phase analysis and the distribution of phases during the cell cycle was determined (Fig. 3, histograms). While there is some evidence of clustering in the timing of gene expression, the main result is that peak expression appears to be able to occur at any time during the division cycle.
The finding of a continuous range for the timing of peak gene expression during the yeast cell cycle actually raises more problems than it solves. While the idea of sequential activation of genes as cells pass through the cell cycle may be attractive, this proposal raises questions as to whether each individual gene in the sequence of activation possesses a particular signal that initiates its expression at that particular time. This model would entail each gene being associated with a timing signal that is specific to that gene. From a design point of view, this seems to be inordinately complex relative to a more parsimonious solution in which a few global regulators turn on sets of genes at a relatively small number of key transition points in the cell cycle. At present, for reasons that may be due to the limited temporal resolution of the microarray experiments, there is little evidence that such transition points are being detected.
The rate of false positives
The analysis presented here is consistent with the original proposed 3–10% false positive rate. From this point of view, the proposed 3–10% false positive rate (1) can be interpreted as referring to false positives in identifying genes with a cyclic response to the synchronizing perturbation, and not necessarily to false positives in identifying genes with innate cyclic expression driven by the cell cycle.
Implications for analysis of cell cycle regulation
The analysis presented here is independent of specific views or models of the cell cycle. Our motivation in preparing this analysis stems from a desire to ensure that the quantitative data used to support cell-cycle-specific gene expression is treated in a rigorous and objective manner.
Synchronization experiments where the cells are synchronized by a batch procedure (i.e. those where all cells are treated equally, such as with starvation, inhibition, or temperature arrest) are subject to two criticisms. First, there is the criticism that the cells are not synchronized at all. Cells may be aligned for a particular property (e.g. G1-phase DNA content), but this does not mean the cells are representative of any particular cell age during the division cycle, nor does it mean the cells are synchronized (18).
A second and more generally accepted critique is that such starvation/inhibition synchronization procedures may affect the experimental observations by introducing artificial periodicities that appear as cell-cycle-specific gene expression. Thus, there may be periodicities in the data, but these periodicities are not necessarily present in the original, unperturbed cells. These artificial periodicities could be considered artifacts that are unrelated to the real, underlying pattern of gene expression during the division cycle.
Classical identification of cell-cycle expression
In support of the microarray data it was pointed out that from a list of 104 genes that were previously identified as being expressed in a cell-cycle-specific manner, the microarray analysis identified 96 of them (1). That the microarray data confirmed 92% of the genes that were identified as cell-cycle specific using more classical methods is important. These 104 genes are identified in 77 papers. (The complete list can be found at ‘Word document’ at the website http://cellcycle-www.stanford.edu/data/rawdata/.) We have looked at almost all of the papers that were available and merely point out that most of the previous analysis was based on α-factor synchronization, temperature sensitive arrest synchronization, raffinose–galactose arrest–regrowth, feed–starve synchronization, nocodazole synchronization and hydroxyurea arrest, with some of the papers also using elutriation. The repetition of cyclicity in the microarray analysis may very likely be due to the use of the same methods that were used to synchronize or align cells in the previously published papers.
Some of the previously published work using standard methods to analyze the cell cycle is quite impressive. The issue raised here is only whether one can use certain synchronization methods in conjunction with microarray analysis to identify cell-cycle-specific expression. Even more importantly, it is not argued here that this reanalysis of the microarray data demonstrates that there are no cell-cycle-specific patterns of gene expression in yeast. We merely propose that the microarray data using four different synchronization methods should be looked at with caution.
On the choice of numerical threshold for cyclical expression
The threshold chosen as the cut-off value for cell-cycle-specific expression was actually chosen to include as many of the ‘known’ genes that had been previously proposed to be cyclically expressed using classical methods. This argument in support of the microarray method as one that can find cyclically expressed genes has a degree of circularity to it. The finding that 92% of the known genes are included in the microarray set is related to lowering the threshold to include these genes. There was no independent choice of threshold value that can be tested for its ability to include the ‘known’ set of cyclically expressed genes.
More problematically, genes that are expected to be non-cyclic in their expression during the division cycle have been found to be cyclic. For example, an extended discussion is presented for the tubulin genes and the methionine genes which are not expected to be cyclical in expression but which are included in the cyclical gene group. Some ad hoc explanations are presented to justify the found cyclicity. We merely point out that these genes could be used as arguments to raise the threshold to exclude genes that are known, suspected or believed to be non-cyclically expressed. Raising the threshold to exclude known non-cyclic genes could suggest that genes proposed to be cyclic are weakly supported by the microarray data. Of course, it is possible that the tubulin genes are expressed in a cyclic manner as indicated by the microarray data. In the absence of independent data indicating that this is the case, we can only point out that the threshold determination should consider the expectations of non-cyclicity as well as cyclicity of gene expression.
Databanks and data analysis
The availability of the raw data on the web allowed a re-examination of the proposal of cell-cycle-specific expression in yeast. If the data were not publicly available, it would not have been possible to test whether there were reproducibility or other problems with the data. This analysis and conclusions presented here therefore emphasize the need for the raw data to be available for further study and analysis. In the original yeast paper (1) the authors conclude by writing ‘we hope that our colleagues in the scientific community will find this paper to be valuable not as only a description of our results but also as a resource for data for some time to come.’ We agree with and support this hope, but also expect that any deficiencies in the data be recognized and taken into account in future analyses.
G1 phase and the cell cycle
Although the analysis presented here is independent of particular models of cell cycle control, it is of interest to point out that the impetus for this study is the proposal that there are no G1 phase-specific controls. The finding that there are a significant number of patterns that were attributed to the G1 phase (1) led to this re-examination of the microarray data. This view of the G1 phase has been reviewed and applied to a number of experimental results (18,20–28).
Caveats and limitations on the analysis
In fairness to the analysis of Spellman et al. (1), it is important to raise one argument against the analysis presented in Figure 3. In Figure 3, the time of peak expression during the division cycle for various genes is correlated between experiments. For many comparisons, there is an obvious diagonal slope to the points. The interpretation of this diagonal is that it implies that the relative timing of peak expression for genes is relatively reproducible between any two block/release experiments. When the elutriation experiment is correlated with the block/release experiments, this obvious diagonal is missing. Our interpretation was that the elutriation result may be the correct result, and stress-induced variations in the block/release experiment are artifacts of the experimental treatment. It should be pointed out, in opposition to this interpretation, that cells from the block/release experiments are grown in glucose, have a relatively short interdivision time, and so have short G1 phases. In contrast, the elutriated cells were grown in ethanol, have a longer interdivision time (∼300 min) and so have a longer G1 phase. It is possible that the peak phase-times are different for the cells with different growth rates. Thus, halfway through the cycle for the glucose-grown, block/release cells is late S or possibly G2 phase. But half way through the cycle for the elutriated cells may just be past the midpoint of the G1 phase. This phase problem could have prevented the inclusion of results from the elutriation experiment in the aggregate, numerical results. To put together the elutriation results and the block/release results, there is need for an independent analysis of where the cells are in the cell cycle, so that one can match up the two very differently timed cell cycles.
There is another problem with analyzing the elution results. The elution experiment is the one experiment among the four synchronization methods studied that is restricted to one cycle. Stress responses to synchronization would be expected to primarily affect the first cycle of a multi-cycle-synchronized culture (although it is not clear that such a restriction must be in place). The multi-cycle cyclicity analysis would tend to identify genes that are cyclic over more than one cycle. This would tend to eliminate genes cyclical over one cycle, which could therefore eliminate many stress-induced variations. This distinction cannot be made for the elutriation results, as there is only one cycle for analysis. This may lead to the inclusion, in the elutriation data, of more cyclical patterns produced by stress effects.
Biological problems regarding cell-cycle expression analysis
Beyond the statistical analysis of the published data on gene expression in cells proposed to be synchronized, we wish to point out some specifically biological problems.
We first must ask ‘what is the purpose of analyzing gene expression in synchronized cells?’ To this question we feel the answer is ‘to understand gene expression in the normal cell cycle’. Specifically, the analysis is not to understand expression in the particular experimental conditions used to analyze gene expression, but to describe what happens during the division cycle of unstarved, unperturbed, exponentially growing cells.
That this is problematic is indicated by the fact that in the three arrest procedures the cells continue to grow so that they are much larger than normal cells. How can one say that whatever happens in an inhibition/release experiment is a reflection of the normal cell cycle? If the cytoplasm/nuclear ratio is much larger after inhibition of the cells, how can one be assured that the results obtained reflect the pattern of synthesis in cells with a smaller cytoplasm/nuclear ratio?
More troubling is the fact that when multiple cycles of S.cerevisiae are studied, the cycles beyond the first are composed of two different cells. One cell is the ‘mother’ cell, which is larger than the smaller ‘daughter’ cells that budded from the mother cell. We find it difficult to understand the multiple cycle experiments when cells have this basic pattern of division. It is possible that larger cells could produce both mother and daughter cells that are large enough at division to be equivalent regarding the cell cycle. However, until this problem is worked out in detail, it is important to be cautious when analyzing the S.cerevisiae cell cycle.
Gene expression during the division cycle of human cells
A parallel study of microarray analysis of gene expression during the division cycle of human cells has shown that similar problems exist in this study. Microarray analysis of gene expression patterns for thousands of human genes has led to the proposal that a large number of genes are expressed in a cell-cycle-specific manner (29). The identification of cyclically expressed genes was based on Affymetrix microarray analysis of gene expression following double-thymidine block synchronization. A statistical re-analysis (30) of the original data leads to three principal findings. (i) Randomized data exhibit periodic patterns of similar or greater strength than the experimental data. This suggested that all apparent cyclicities in the expression measurements may arise from chance fluctuations. (ii) The presence of cyclicity and the timing of peak cyclicity in a given gene are not reproduced in two replicate experiments. This suggested that there is an uncontrolled source of experimental variation that is stronger than the innate variation of gene expression in cells over time. (iii) The amplitude of peak expression in the second cycle is not consistently smaller than the corresponding amplitude in the first cycle. This finding placed doubt on the assumption that the cells are actually synchronized. We have proposed that the microarray results do not support the proposal that there are numerous cell-cycle-specifically expressed genes in human cells (30).
Acknowledgments
ACKNOWLEDGEMENTS
We benefited greatly from comments by Drs Paul Spellman and Raymond Cho. This work was supported by a grant from the University of Michigan Cancer Committee. Correspondence related to detailed mathematical and statistical methods should be addressed to K.S. Correspondence related to general analysis of the mammalian division cycle should be addressed to S.C. Further readings on the alternative view of the cell cycle may be found at http://www.umich.edu/~cooper.
REFERENCES
- 1.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho R.J., Campbell,M.J., Winzeler,E.A., Steinmetz,L., Conway,A., Wodicka,L., Wolfsberg,T.G., Gabrielian,A.E., Landsman,D., Lockhart,D.J. et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- 3.Aach J., Rindone,W. and Church,G.M. (2000) Systematic management and analysis of yeast gene expression data. Genome Res., 10, 431–445. [DOI] [PubMed] [Google Scholar]
- 4.Aach J. and Church,G.M. (2001) Aligning gene expression time series with time warping algorithms. Bioinformatics, 17, 495–508. [DOI] [PubMed] [Google Scholar]
- 5.Alter O., Brown,P.O. and Botstein,D. (2000) Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl Acad. Sci. USA, 97, 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Issel-Tarver,L., Kasarskis,A., Scafe,C.R., Sherlock,G., Binkley,G., Jin,H. et al. (2000) Integrating functional genomic information into the Saccharomyces Genome Database. Nucleic Acids Res., 28, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ball C.A., Jin,H., Sherlock,G., Weng,S., Matese,J.C., Andrada,R., Binkley,G., Dolinski,K., Dwight,S.S., Harris,M.A. et al. (2001) Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res., 29, 80–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Futcher B. (2000) Microarrays and cell cycle transcription in yeast. Curr. Opin. Cell Biol., 12, 710–715. [DOI] [PubMed] [Google Scholar]
- 10.Getz G., Levine,E., Domany,E. and Zhang,M. (2000) Super-paramagnetic clustering of yeast gene expression profiles. Physica A, 279, 457–464. [Google Scholar]
- 11.Holter N.S., Mitra,M., Maritan,A., Cieplak,M., Banavar,J.R. and Fedoroff,N.V. (2000) Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc. Natl Acad. Sci. USA, 97, 8409–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holter N.S., Maritan,A., Cieplak,M., Fedoroff,N.V. and Banavar,J.R. (2001) Dynamic modeling of gene expression data. Proc. Natl Acad. Sci. USA, 98, 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 14.Hughes T.R., Marton,M.J., Jones,A.R., Roberts,C.J., Stoughton,R., Armour,C.D., Bennett,H.A., Coffey,E., Dai,H., He,Y.D. et al. (2000) Functional discovery via a compendium of expression profiles. Cell, 102, 109–126. [DOI] [PubMed] [Google Scholar]
- 15.Stevenson L.F., Kennedy,B.K. and Harlow,E. (2001) A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl Acad. Sci. USA, 98, 3946–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavazoie S., Hughes,J.D., Campbell,M.J., Cho,R.J. and Church,G.M. (1999) Systematic determination of genetic network architecture. Nature Genet., 22, 281–285. [DOI] [PubMed] [Google Scholar]
- 17.Wolfsberg T.G., Gabrielian,A.E., Campbell,M.J., Cho,R.J., Spouge,J.L. and Landsman,D. (1999) Candidate regulatory sequence elements for cell cycle-dependent transcription in Saccharomyces cerevisiae. Genome Res., 9, 775–792. [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper S. (1998) Mammalian cells are not synchronized in G1-phase by starvation or inhibition: considerations of the fundamental concept of G1-phase synchronization. Cell Prolif., 31, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper S. (1991) Bacterial Growth and Division. Academic Press, San Diego, CA.
- 20.Cooper S. (2000) The continuum model and G1-control of the mammalian division cycle. Prog. Cell Cycle Res., 4, 27–39. [DOI] [PubMed] [Google Scholar]
- 21.Cooper S., Yu,C. and Shayman,J.A. (1999) Phosphorylation-dephosphorylation of retinoblastoma protein is not necessary for passage through the mammalian division cycle. IUBMB Life, 1, 225–230. [DOI] [PubMed] [Google Scholar]
- 22.Cooper S. (1998) On the proposal of a G0 phase and the restriction point. FASEB J., 12, 367–373. [DOI] [PubMed] [Google Scholar]
- 23.Cooper S. (1998) On the interpretation of the shortening of the G1-phase by overexpression of cyclins in mammalian cells. Exp. Cell Res., 238, 110–115. [DOI] [PubMed] [Google Scholar]
- 24.Cooper S. (1982) The continuum model: statistical implications. J. Theor. Biol., 94, 783–800. [DOI] [PubMed] [Google Scholar]
- 25.Cooper S. (1988) The continuum model and c-myc synthesis during the division cycle. J. Theor. Biol., 135, 393–400. [DOI] [PubMed] [Google Scholar]
- 26.Cooper S. (1979) A unifying model for the G1 period in prokaryotes and eukaryotes. Nature, 280, 17–19. [DOI] [PubMed] [Google Scholar]
- 27.Cooper S. and Shayman,J.A. (2001) Revisiting retinoblastoma protein phosphorylation during the mammalian cell cycle. Cell. Mol. Life Sci., 58, 580–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper S. (2001) Revisiting the relationship of the mammalian G1 phase to cell differentiation. J. Theor. Biol., 208, 399–402. [DOI] [PubMed] [Google Scholar]
- 29.Cho R.J., Huang,M., Dong,H., Steinmetz,L., Sapinoso,L., Hampton,G., Elledge,S.J., Davis,R.W., Lockhart,D.J. and Campbell,M.J. (2001) Transcriptional regulation and function during the human cell cycle. Nature Genet., 27, 48–54. [DOI] [PubMed] [Google Scholar]
- 30.Shedden K. and Cooper,S. (2002) Analysis of cell-cycle-specific gene expression in human cells as determined by microarrays and double-thymidine block synchronization. Proc. Natl Acad. Sci. USA, 99, 4379–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]