Abstract
Colon cancer remains a significant health burden globally, necessitating deeper investigation. Identification and targeting of prognostic markers can significantly improve the current therapeutic approaches for colon cancer. The differential nuclear transport (import and export) of cellular proteins, plays an important role in tumor progression. Exportins, critical mediators of nuclear export, have emerged as potential players in cancer pathogenesis. However, their precise roles and prognostic significance in colon adenocarcinoma remain elusive. This study was designed to comprehensively analyse the expression and prognostic significance of all seven exportins in Colon Adenocarcinoma (COAD) using the online public database. We used public databases UALCAN, C-Bio portal, Human Protein Atlas (HPA), and DAVID, to investigate exportins in COAD patients. Kaplan–Meier plotter, Gene ontology (GO), TIMER, STRING, and KEGG were used to analyse data and draw conclusions. Our observations showed a significant correlation of exportins expression with clinical parameters, used to predict a patient's prognosis in general, such as advancing tumor stage, overall/relapse-free survival, and immune cell infiltrations. Mutation analysis showed the presence of amplifications, missense mutations in XPO2 and XPO4, and deep deletions in XPO7 genes contributing to disease progression and patients survival. This study highlights the potential use of exportins as novel prognostic biomarkers and therapeutic targets for colon adenocarcinoma progression and management.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-025-01748-4.
Keywords: Colon adenocarcinoma (COAD), Exportins, Expression, Mutation, Prognosis
Introduction
The colon and rectum are parts of the large intestine, and their cancers are known as colon cancer and rectal cancer, respectively. These cancers are often collectively referred to as colorectal cancer. Colorectal cancer ranks among the most prevalent cancers worldwide, standing as the third most diagnosed globally and second leading cause of cancer-related deaths globally, contributing to 10% of all cancer cases, as per the latest WHO report. The predominant type of colorectal cancer is colorectal adenocarcinoma which usually emerges from the glandular epithelial cells of the large intestine, accounting for 95% of all cases [1, 2]. Similarly, the predominant type of colon cancer is Colon adenocarcinoma.
Cellular protein functions in defined subcellular compartment or cellular regions within the cell within the cell. This process of protein recruitment is made feasible with the help of transporter proteins. A well-identified transporters that transport the proteins and RNA across the nuclear membrane are exportins and importins. Importins facilitate the entry of cargo proteins into the nucleus while exportins mediate the exit of cargo proteins and RNA, such as mRNA, tRNA, and rRNA, from the nucleus. Additionally, these proteins also play crucial roles in other cellular processes including mitosis and transcription. Therefore, it is important to analyse the differential expression levels of these proteins in diseases including cancer, regardless of their function as transporter or non-transporter. In the current study, we have focused on the role of exportins and its association with colon adenocarcinoma. Exportins are known to play significant role in cellular processes such as cell cycle regulation, cellular differentiation, and metastasis necessitating its investigation specially expression status in cancer progression and management [3].
Till today, total seven exportins have been identified (XPO1, XPO2, XPO4, XPO5, XPO6, XPO7, XPOT) which export various cellular proteins and RNA molecules from nucleus to cytoplasm. XPO4 and XPO7 act as bidirectional transporters, facilitating both the import and export of proteins [4, 5]. Major exportins studied till date are XPO1 (CRM1), XPO2 (CSE1L), XPO5, and XPOT however, very limited information is available on XPO4, XPO6, and XPO7 in cancer progression.
XPO1 is a well-characterized exportin that has been known to carry cargoes related to centrosome proteins, vesicle coat complexes, translation, and the metabolism of cytoplasmic mRNA [6–8]. It also has been implicated in the progression of several cancer types, and being targeted for cancer therapeutics [8–11]. XPO2 is known to export of Imp α and it expression level play a very important role in gene silencing and reactivation [8]. XPO2 is found to be significantly overexpressed in a variety of cancers including oral cancer, gastric cancer, lung cancer, colorectal cancer, laryngeal cancer, breast cancer, osteosarcoma, chronic myeloid leukaemia, ovarian cancer, hepatocellular carcinoma [12–20]. XPO4 mediates the export of proteins such as eIF5A and Smad3, while also importing transcription factors like Sox2 and SRY [21]. XPO4 is upregulated in prostate cancer and downregulated in hepatocellular carcinoma [22, 23]. XPO5 adopts a ring-shaped closed conformation, primarily consisting of HEAT repeat segments, and plays a key role in transporting precursor miRNA and eukaryotic elongation factor 1A (eEF1A) out of the nucleus [24, 25]. XPO5 dysregulation is observed in colorectal cancer, breast cancer, non-small cell lung carcinoma, prostate cancer, and thyroid cancer [12, 26–29]. XPOT exports mature tRNA molecules from the nucleus. XPOT is upregulated in hepatocellular carcinoma, neuroblastoma, and triple-negative breast cancer [30–32]. XPO6 is mainly responsible for the export of protein belongs to actin family [33]. It has been exclusively identified as an oncogene in prostate and breast cancer [34, 35]. XPO7 is also bidirectional transporter, majorly working as exporter and plays a crucial role in erythropoiesis, causing nuclear condensation and histone loss, and disrupting terminal erythroid differentiation [5]. XPO7 was also found to be dysregulated in ovarian cancer, prostate cancer, liver cancer, and breast cancer [32, 36–38]. Inhibitors of nucleocytoplasmic transport are continuously being investigated in cancer [39, 40], including nuclear export inhibitors (NEIs) and selective inhibitors of nuclear export (SINEs), a promising class of cancer therapeutics. These agents effectively block the transport of proteins from the nucleus to the cytoplasm, disrupting key cellular processes involved in cancer progression [41, 42]. The most noticeable is LMB (Leptomycin B) which inhibits the interaction of XPO1 with its cargo protein [43, 44]. LMB induces apoptosis in cancer cell lines, but cell cycle arrest in normal lung fibroblast [41]. Seervi et al. demonstrated that inhibiting XPO1 significantly reduced the recovery of apoptotic cells in anastatic cancer cells [45]. Anastatic cancer cells are cells that resist apoptosis even after the strong apoptotic signal. Hence targeting exportin has significance in cancer therapeutics. However, its use has been poorly explored in colon cancer progression and management.
To our knowledge, no studies have addressed exportins expression as a prognostic marker in colorectal cancer or colon adenocarcinoma, except for one reported by Wang et al. in 2022. Wang et al. demonstrated that high expression of XPO2 (CSE1L) was a risk factor for colorectal cancer patients with poor prognosis [46]. The present study has comprehensively addressed the expression pattern and prognostic significance of all major exportins (XPO1, XPO2, XPO4, XPO5, XPO6, XPO7 and XPOT) in colon adenocarcinoma. Our analysis includes expression profiling, genetic alterations, and, immune infiltration to evaluate the prognostic significance of exportins in colon adenocarcinoma. Our observations suggest that exportins are differentially expressed, have frequent genetic alterations, and have several functions as oncogenes in colon adenocarcinoma. Their correlations with the patient's clinical parameters pose them to be potential prognostic factors in colon adenocarcinoma.
Materials and methods
Data collection
We used the COAD dataset for expression at the mRNA level and protein level from The Cancer Genome Atlas (TCGA) and Human Protein Atlas (HPA) databases respectively. A total of 45 normal and 274 tumor patient samples from the TCGA database were utilised in ULCAN analysis. A total of 1624 patients' data for overall survival and 1148 patients' data for relapse-free survival was used in KAPLAN–MEIER PLOTTER analysis from multiple databases including GEO, EGA, and TGCA (Supplementary Table 1). TCGA Firehose Legacy module of the cBioPortal database was used for genetic alteration analysis. It contains 640 samples (392 colon adenocarcinoma, 169 rectal adenocarcinoma, 66 mucinous colon adenocarcinoma, and 13 mucinous rectal adenocarcinoma samples). Our analysis was restricted to the 392 colon adenocarcinoma samples. The COAD dataset of the TIMER database was used to analyse the correlation between exportin expression and tumor immune cell infiltration in colon adenocarcinoma cancer.
UALCAN analysis
This interactive web resource enables the analysis of cancer omics data. It primarily uses the data from The Cancer Genome Atlas (TCGA) database. TCGA provides clinical patient data and allows to analyse of expression, survival, methylation, correlation, and pan-cancer view of 33 different cancer datasets [47]. Gene expression levels were quantified as transcripts per million (TPM), providing a relative measure that facilitates comparison across samples. Statistical significance was determined at a threshold of P < 0.05.
KAPLAN–MEIER PLOTTER analysis
It is an online tool used for patient survival analysis, primarily focusing on gene expression data to assess the impact of specific genes on the survival of patients. It integrates data from multiple databases, including GEO, EGA, and TGCA [48]. We utilized this to examine the prognostic relevance of Overall Survival (OS) and Relapse-Free Survival (RFS) by analysing the exportins mRNA expression database. Evaluation of Hazard ratio with a confidence interval of 95% and log-rank P-value was used to present overall survival/ and relapse-free survival between patients with low and high exportin expression. Differences were considered statistically significant with P < 0.05.
HUMAN PROTEIN ATLAS (HPA) analysis
It is an online database to map all human proteins in cells, tissue, and organs using omics technology, antibody-based imaging, mass spectrometry-based imaging, and transcriptomics [49]. Using the pathology section of HPA we compared the protein expression of exportins between normal and colon adenocarcinoma patients.
cBioPortal for cancer genomics
It is a web-based resource that provides visualization, analysis, and exploration tools for multidimensional cancer genomics data sets. It allows us to interactively explore genetic alterations, and other molecular characteristics across various cancer types and patient cohorts [50]. Here in this study we analysed the mutations of 7 different exportins in the COAD dataset.
TIMER (tumor immune estimation resource) analysis
TIMER is a comprehensive online resource for systemically analysing immune cell infiltrates in several cancer types. It is a powerful tool for the investigation of tumor-immune interactions, utilizing data from The Cancer Genome Atlas (TCGA) [51]. Using TIMER we analysed the correlation of exportins expression in tumors with the level of different immune cell infiltration. Statistical significance was determined at a threshold of P < 0.05.
STRING analysis
The STRING database is a comprehensive online resource that provides information about protein–protein interactions. It aims to collect, store, and integrate available sources including experimental data, computational predictions, and publicly available databases [52]. The ten most highly co-expressed genes for each exportin were collected from the co-expression module in the cBioPortal database. Then these gewere were used for STRING analysis to draw the network of protein–protein interaction. Spearman's correlation was used with P < 0.001.
DAVID database for gene ontology and KEGG pathway analysis
It is an online database for functional annotations and enrichment analysis [53]. We performed gene ontology (GO) and KEGG pathway analysis of exportins and their 70 co-expressed genes in DAVID. Statistical significance was determined at a threshold of P < 0.05.
Results
Exportins expression in colon adenocarcinoma patients
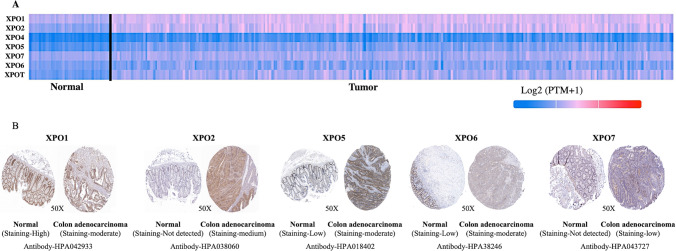
To explore the exportins expression in colon adenocarcinoma patients, we conducted UALCAN analysis on seven exportins (XPO1, XPO2, XPO4, XPO5, XPO6, XPO7, XPOT). We observed overexpression of all the exportins analysed in colon adenocarcinoma patients compared to normal individuals (Fig. 1A and Supplementary Fig. 1A). We also analysed the differential expression of all exportins using the TNMplot database and observed similar results (Supplementary Fig. 1B and Fig. 2 A & B). Protein expression of various exportins was analysed using the Human Protein Atlas (HPA) database. We compared the protein expression of exportins in COAD tissue samples with normal human colon tissue samples. Moderate staining for XPO2, XPO5, and XPO6 was observed in COAD samples, while normal samples exhibited low or undetectable staining. In COAD samples, low staining of XPO7 was observed, whereas normal tissues showed no staining. Conversely, XPO1 exhibited high staining in normal samples compared to COAD samples (Fig. 1B). Protein expression data for XPO4 and XPOT were not available in the HPA. Overall, this expression analysis reveals an upregulation of exportin proteins in COAD, except for XPO1 at the protein level, as illustrated in Fig. 1.
Fig. 1.
A Differential mRNA Expression of exportins in Colon Adenocarcinoma using TCGA database: A Heatmap from UALCAN database shows the mRNA expression of exportins. B Immunohistochemistry analysis of exportins in Colon Adenocarcinoma using HPA (Human Protein Atlas) database: Representative picture showing expression of XPO1, XPO2, XPO5, XPO6, XPO7 in Colon Adenocarcinoma and control tissue
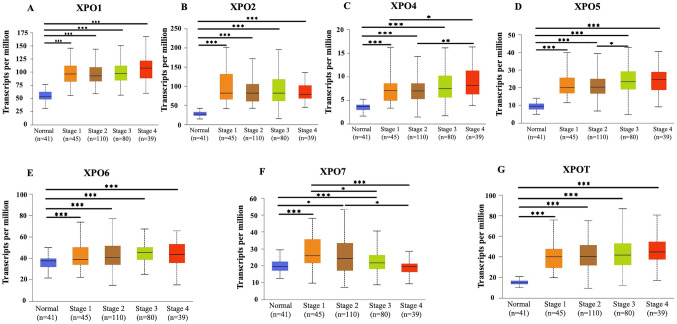
Exportins expression and its association with tumor stage and patient survival
We conducted UALCAN analysis to demonstrate the correlation of clinical parameters such as tumor stages and patient survival with the expression of exportins in COAD patients to understand the role of exportins in the prognosis of COAD patients. As shown in Fig. 2, we observed that stage 4 tumors show the highest median expression levels of exportins XPO1, XPO4, XPO5, and XPOT, suggesting that their expression increases with cancer progression. For XPO4, expression levels are significantly higher in stage 4 compared to stage 2, while XPO5 shows significantly higher expression in stage 3 than in stage 2 (Fig. 2). Notably, XPO7 exhibits a consistent and significant decrease in expression with advancing tumor stage, suggesting a negative correlation between tumor stage and its expression levels in COAD patients. The expression of XPO2, XPO6, and XPOT did not exhibit a conclusive trend with increasing tumor stage.
Fig. 2.
Box plot showing a correlation between exportins (XPO1, XPO2, XPO4, XPO5, XPO6, XPO7, and XPOT) mRNA expression and stage of tumor in Colon Adenocarcinoma using TGCA database in ULCAN analysis. Significant differences in expression between tumor and normal tissues, and between tumor stages are indicated by asterisks (*P < .05, **P < .01, ***P < .001)
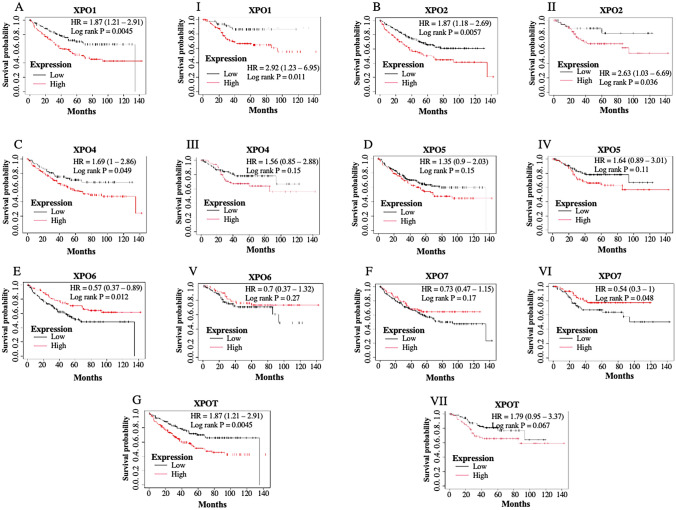
Further, we divided cancer patients into two groups (high expression level and low expression level of particular exportin based on their median expression level) and performed a survival analysis using the Kaplan–Meier Plotter. We analysed the relationship between mRNA expression of exportins and both overall survival and relapse-free survival of COAD patients. Among the exportins analysed, high expression levels of XPO1, XPO2, XPO4, and XPOT were significantly and negatively associated with the overall survival of the patients. High expression of XPO5 was also negatively associated with overall survival but differences were not significant (Fig. 3, upper panel; Table 1). Contrary to this the high expression of XPO6 and XPO7 was positively associated with the overall survival of the patients, although the differences were not significant for XPO7. On the other hand, among the exportin analysed expression levels of XPO1, XPO2, XPO4, XPO5, and XPOT were negatively associated with the relapse-free survival of the patients but the differences were significant only for the XPO1 and XPO2. Contrary to this the high expression of XPO6 and XPO7 was positively associated with the relapse-free survival of the patients, although the differences were marginally significant for XPO7 and not significant for XPO6 (Fig. 3, lower panel; Table 1).
Fig. 3.
Overall Survival (OS) and Relapse Free Survival (RFS) analysis using Kaplan–Meier Plotter Database in Colon Adenocarcinoma patients: Figure shows the overall survival (A–G) and relapse-free survival (I-VII) of patients with high and low expression levels. Compared to the median expression value, of various exportins (XPO1, XPO2, XPO4, XPO5, XPO6, XPO7, and XPOT). A P-value of < .05 has been considered as statistically significant
Table 1.
Hazard Ratios (HR) with 95% Confidence Intervals (CI) for overall survival and relapse-free survival of Colon Adenocarcinoma patients with high and low expression levels of exportins: The HR values indicate the relative risk of adverse outcomes associated with the expression levels (high or low compared to their median expression level of respective exportin) of each exportin
| Exportins | Overall survival | Relapse free survival | ||||
|---|---|---|---|---|---|---|
| Cases | HR (95% confidence interval) | P value | CASES | HR (95%confidence interval) | P value | |
| XPO1 | 232 | 1.78 (1.18–2.69) | 0.0052 | 164 | 2.92 (1.23–6.95) | 0.011 |
| CSE1L | 232 | 1.69 (1–2.86) | 0.049 | 164 | 2.63 (1.03–6.69) | 0.036 |
| XPO4 | 232 | 1.49 (0.97–2.31) | 0.07 | 164 | 1.56 (0.85–2.88) | 0.15 |
| XPO5 | 232 | 1.35 (0.9–2.03) | 0.15 | 164 | 1.64 (0.89–3.01) | 0.11 |
| XPO6 | 232 | 0.57 (0.37–0.89) | 0.012 | 164 | 0.7 (0.37–1.32) | 0.27 |
| XPO7 | 232 | 0.73 (0.47–1.15) | 0.17 | 164 | 0.54 (0.3–1) | 0.048 |
| XPOT | 232 | 1.87 (1.21–2.91) | 0.0045 | 164 | 1.79 (0.95–3.37) | 0.067 |
An HR greater than 1 suggests a higher risk, while a HR less than 1 suggests a lower risk. The 95% CI provides a range within which the true HR is expected to fall, with 95% confidence
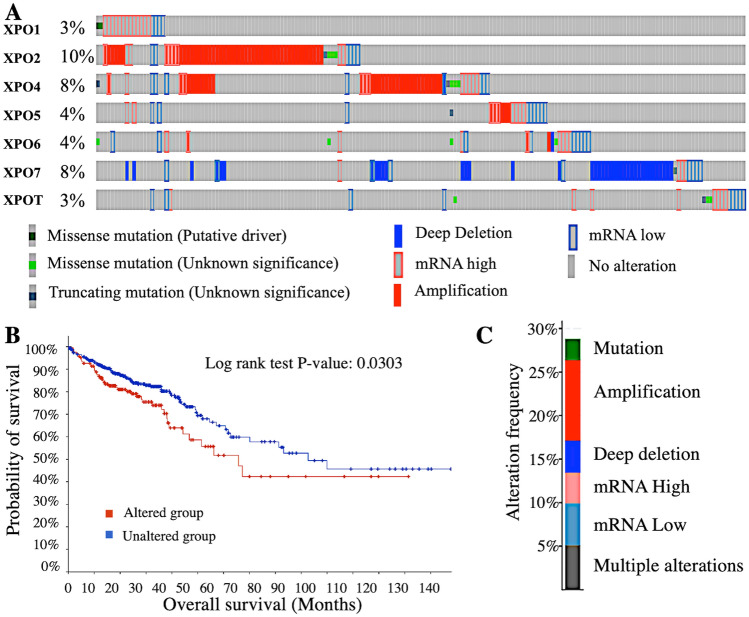
Genetic alterations in exportins gene
In the cBioPortal database, COAD patient samples with the TCGA Firehose Legacy module were selected, and the genetic alterations and mRNA microarray data of all exportins were analysed. As shown in Fig. 4A, the total genetic alterations rates, out of 634 cases, in each exportin were observed as follows: XPO1: 3% (11 cases), XPO2: 10% (39 cases), XPO4: 8% (31 cases), XPO5: 3% (11 cases), XPO6: 4% (16 cases), XPO7: 8% (31 cases), XPOT: 3% (11 cases). These genetic alterations are missense mutations, truncating mutations, amplifications, and deep deletions, as illustrated in Fig. 4A. Figure 4B shows the overall survival of patients with these genetic alterations compared to control. The overall survival of patients with genetic alterations is significantly lower than that of the control group (P = 0.03). Figure 4C depicts the contribution of exportins (XPO1, XPO2, XPO4, XPO5, XPO6, XPO7, and XPOT) to various genetic alterations and mRNA expression levels in COAD patients. These alterations levels are as follows: Mutations: 2.37% (9 cases), Amplifications: 9.15% (35 cases), Deep deletions: 3.63% (14 cases), mRNA upregulation: 3.47% (13 cases), mRNA downregulation: 4.73% (18 cases), Multiple alterations: 5.05% (19 cases). The major genetic alteration is amplification, particularly in XPO2, XPO4, and XPO5 (Fig. 4A). Deep deletions, which are associated with low mRNA expression, are most commonly seen in XPO7.
Fig. 4.
Genetic alteration analysis using cBioportal database in Colon Adenocarcinoma patients. A showing cBioportal onco-print with percentage of Genetic alteration in Colon Adenocarcinoma patient. B Survival analysis indicates that genetic alterations in exportins are associated with poor overall survival in patients. C Illustrating the types of mutations, mRNA expression levels, and their proportions contained in exportins (XPO1, XPO2, XPO4, XPO5, XPO6, XPO7, and XPOT)
Correlation of exportins expression with the level of immune cell infiltration in tumor:
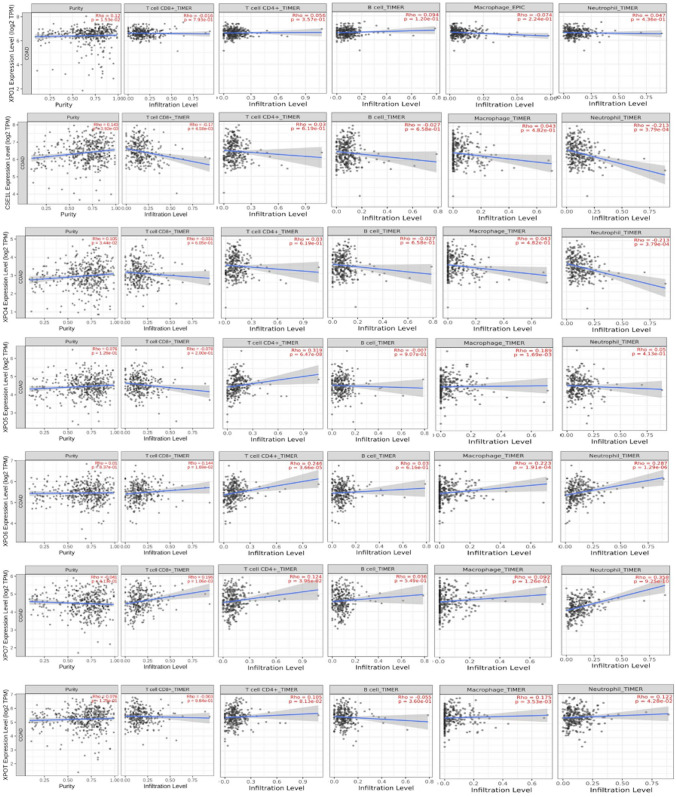
As we know, the tumor microenvironment plays a major role in cancer progression by providing a supportive environment for tumor growth. Consequently, immune cells in the tumor microenvironment are often suppressed or become tumor-associated. Bu et al. identified four tumor microenvironment-based rectal cancer subtypes and suggested a degenerate gene signature to predict the prognosis and immunotherapy response [54, 55]. It has been reported well that the level of infiltrated immune cells in the tumor microenvironment is indicative in predicting the prognosis of patients, thus we investigated it in this study [56, 57]. To investigate this, we examined the association of exportins expression with the level of immune cell infiltration in COAD patients' tumor microenvironment using the TIMER database. In the TIMER database, a p-value less than 0.05 indicates a statistically significant correlation between the gene and immune cell infiltration. As shown in Fig. 5: (1) XPO1: The only statistically significant correlation observed is between its mRNA expression and tumor purity, with a significant and positive correlation (Rho = 0.12, P = 0.0153). "tumor purity" refers to the proportion of cancer cells in a tumor sample relative to the total number of cells. There are no significant correlations between XPO1 expression and the infiltration levels of the different immune cells analysed such as CD8 + T cells (Rho = −0.016, P = 0.793), CD4 + T cells (Rho = 0.056, P = 0.357), B cells (Rho = −0.094, P = 0.120), macrophages (Rho = −0.074, P = 0.224), and neutrophils (Rho = −0.047, P = 0.436). Note that, although not significant., XPO1 is negatively correlated with the infiltration of CD8 + T cells, macrophages, and neutrophils. (2) XPO2: There is a statistically significant positive correlation between XPO2 expression and tumor purity (Rho = 0.143, P = 0.00392). A statistically significant negative correlation is observed with CD8 + T cell infiltration (Rho = −0.17, P = 0.00458), and neutrophil infiltration (Rho = −0.218, P = 0.000379). Correlation between XPO2 expression and infiltration of CD4 + T cell infiltration (Rho = 0.03, P = 0.619), B-cell (Rho = −0.027, P = 0.658), and macrophage (Rho = 0.043, P = 0.482) was not found to be statistically significant. (3) XPO4: There is a statistically significant positive correlation between XPO4 expression and tumor purity (Rho = 0.105, P = 0.0344), while neutrophil infiltration (Rho = 0.043, P = 0.475) showed significant negative correlation. Correlation between XPO4 expression and infiltration of CD8 + T (Rho = −0.031, P = 0.605), CD4 + T cell (Rho = 0.03, P = 0.619), B-cell (Rho = −0.027, P = 0.658) and macrophage (Rho = 0.043, P = 0.482) was not found to be statistically significant. (4) XPO5: A statistically significant positive correlation is observed between XPO5 expression and CD4 + T cell infiltration (Rho = 0.319, P = 6.47e-08), and macrophage infiltration (Rho = 0.189, P = 0.0069). However, no significant correlation is found with tumor purity (Rho = 0.076, P = 0.126), CD8 + T cell infiltration (Rho = −0.078, P = 0.200), B cell infiltration (Rho = −0.007, P = 0.907), and neutrophil infiltration (Rho = 0.05, P = 0.413). (5) XPO6: There is a statistically significant positive correlation between XPO6 expression with infiltration of CD8 + T cell (Rho = 0.144, P = 0.0169), CD4 + T cell (Rho = 0.240, P = 0.0000366), macrophage (Rho = 0.223, P = 0.000191), and neutrophil (Rho = 0.287, P = 1.29e-06). However, no significant correlation is found between tumor purity (Rho = 0.01, P = 0.837) and B cell infiltration (Rho = 0.029, P = 0.626). (6) XPO7: There is a statistically significant positive correlation between XPO7 expression with infiltration level of CD8 + T-cell (Rho = 0.196; P = 0.00106), CD4 + T-cell (Rho = 0.124, P = 0.0396), Neutrophil (Rho = 0.344, P = 2.50e-09). However, there is no significant correlation with tumor purity, (Rho = −0.041, P = 0.0413), infiltration level of B cell (Rho = 0.036, P = 0.549), and Macrophage (Rho = 0.092, P = 0.126). (7) XPOT: There is a statistically significant positive correlation between XPOT expression with infiltration level of CD4 + T-cell (Rho = 0.105, P = 0.0813), macrophage (Rho = 0.175; P = 0.00353) and Neutrophil (Rho = 0.122, P = 0.0428). However, there is no significant correlation between tumor purity, and infiltration level of CD8 + T-cell (Rho = −0.003; P = 0.0964), and B-cells (Rho = −0.055; P = 0.360) (Fig. 5).
Fig. 5.
Correlation of exportins expression with the level of infiltrated immune cell using TIMER database: The Y-axis of all the graphs shows the expression (Log2 TPM) of respective exportin and the X-axis shows the level of immune cells (CD8+, T-cell; CD4 + T-cell; B cell; Macrophage and Neutrophil) infiltration in tumor tissue of Colon Adenocarcinoma patients. The first graph in each panel shows the correlation of exportins expression with the purity of the tumor. "tumor purity" is the proportion of cancer cells out of the total number of cells
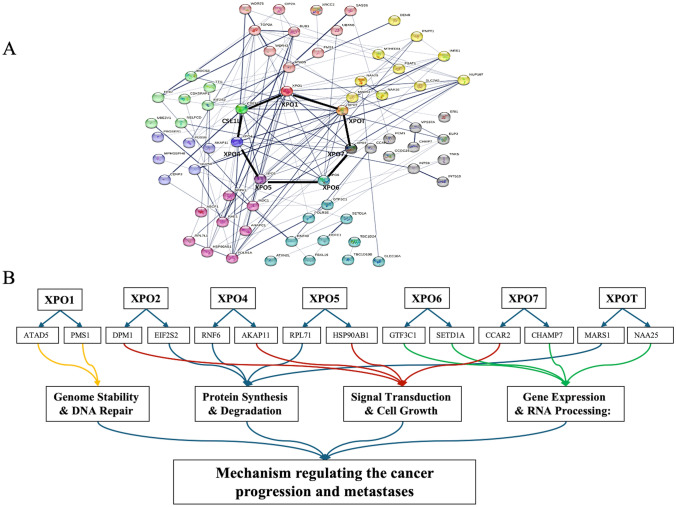
Co-expressed gene analysis and protein–protein interactions
We used the cBioPortal database to identify the top ten genes that are highly co-expressed with each exportin, using the co-expression module that showed the highest Spearman’s correlation (Supplementary Table 2). Out of these highly co-expressed genes, the top two co-expressed genes with the highest Spearman’s correlation coefficient for each exportin are as follows: XPO1: ATAD5 and PMS1; XPO2: DPM1 and EIF2S2; XPO4: RNF6 and AKAP11; XPO5: RPL71 and HSP90AB1; XPO6: GTF3C1 and SETD1A; XPO7: CCAR2 and CHAMP7; XPOT: MARS1 and NAA25. Further, using the STRING database and selecting the 10 highly co-expressed genes with the largest Spearman’s rank correlation values, we constructed a protein–protein interaction network for these genes. This analysis makes a map of interactions between exportins and their co-expressed genes (Fig. 6A). Further, using the DAVID database, we tried to categorize at least the top two co-expressed genes of each exportin (a total of 14 genes) based on their involvement in various biological processes. We observed that these genes are primarily involved in processes such as Genome Stability & DNA Repair, Protein Synthesis & Degradation, Signal Transduction & Cell Growth, and Gene Expression & RNA Processing (Fig. 7B; Supplementary Table 3). We also prepared a heatmap to show the co-expression of each exportin with the others (Supplementary Fig. 3).
Fig. 6.
Protein–protein interaction network using STRING analysis: A Protein–protein interaction network resulting from the total 70 co-expressed genes (10 of each exportin) analysed in STRING. B Categorising the top two highly co-expressed genes (with the highest Spearman correlation coefficient) to each exportin (total 14) in different biological processes involved in cancer progression
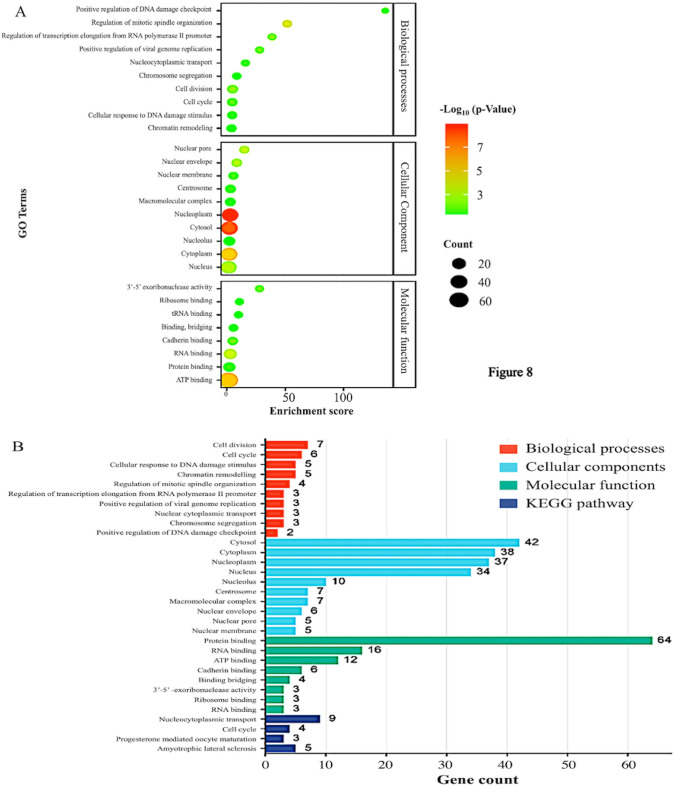
Fig. 7.
Gene Ontology functional analysis and KEGG pathway analysis: A total of 70 co-expressed genes (10 of each exportin) were analysed using the DAVID database. A GO enrichment analysis identifying and illustrating the significance of enriched GO terms associated with 70 co-expressed proteins. Each bubble represents a GO term. The size of the bubble corresponds to the number of genes (out of 70) associated with that particular GO term. B a visual representation of the relationships between GO terms. Gene Ontology graph of biological processes, cellular component, and molecular function, and KEGG pathway analysis
Gene ontology and KEGG pathway analysis using the DAVID database
We also performed ontology functional enrichment analysis using the DAVID database, focusing on the top ten biological processes, cellular components, and molecular functions associated with exportins and their co-expressed genes. Processes were considered significant with a p-value of < 0.05. As shown in Fig. 7A, the significant biological processes mediated by the exportins include: GO:0051301 (Cell cycle), GO:007049 (Cell division), GO:0006974 (Cellular response to DNA damage stimulus), GO:0006338 (Chromatin remodeling), GO:0060236 (Regulation of mitotic spindle organization), GO:0034243 (Regulation of transcription elongation from RNA polymerase II promoter), GO:0045070 (Positive regulation of viral genome replication), GO:0006913 (Nucleocytoplasmic transport), GO:0007059 (Chromosome segregation), and GO:0046601 (Positive regulation of DNA damage checkpoint). Significant cellular components identified include GO:0005829 (Cytosol), GO:0005737 (Cytoplasm), GO:0005654 (Nucleoplasm), GO:0005634 (Nucleus), GO:0005730 (Nucleolus), GO:0005813 (Centrosome), GO:0032991 (Macromolecular complex), GO:0005635 (Nuclear envelope), GO:0005643 (Nuclear pore), and GO:0031965 (Nuclear membrane). Significant molecular functions involving exportins include GO:0005515 (Protein binding), GO:0003723 (RNA binding), GO:0005524 (ATP binding), GO:0045296 (Cadherin binding), GO:0060090 (Binding, bridging), GO:0000175 (3'−5'-exoribonuclease activity), GO:0043022 (Ribosome binding), and GO:0000049 (tRNA binding). Figure 7B represents all gene ontology functions and KEGG pathway analyses. The pathways include hsa03013 (Nucleocytoplasmic transport), hsa04110 (Cell cycle), hsa04914 (Progesterone-mediated oocyte maturation), and hsa05014 (Amyotrophic lateral sclerosis). From these analyses, it is evident that the largest number of genes is associated with the term "Protein binding" under molecular functions (64 genes), indicating a significant role in binding activities. In the cellular component category, "Cytosol" involves the highest number of genes (42 genes), suggesting a high level of cellular activity in the cytosol. Important biological processes that involve exportins in colon adenocarcinoma are Cell division, Cell cycle, DNA damage checkpoints, RNA transport, 3’−5’ exoribonuclease activity, and nucleo-cytoplasmic. It is worth noting that the exportins analysed are also prominently involved in processes that are key targets in present-day cancer therapeutics such as DNA damage checkpoints. (Supplementary Table 4).
Discussion
Exportins are the cellular proteins that primarily facilitate the transport of molecules including oncogenic and tumor suppressor proteins from the nucleus to the cytoplasm and vice versa. Multiple studies have reported the association between exportins expression and tumor progression [58, 59]. It has also been reported that depending on the cell and tissue type, exportins can act either as tumor suppressors or tumor promoters [3]. Further investigation is needed to understand the role of exportins in cancer progression and management, especially in Colon Adenocarcinoma. The prognostic values of exportins have been highlighted previously in hepatocellular carcinoma and lung cancer [58, 59]. However, a comprehensive analysis of exportin expression and its prognostic significance in the context of Colon Adenocarcinoma has not been done previously. In this section of the manuscript, we will discuss (1) exportins and their association with tumor stage and patient survival, highlighting their prognostic significance, (2) the prognostic significance of exportins due to their association with level of immune cell infiltration, and (3) protein–protein interaction and exportins mediated pathway analysis.
XPO1 transports various proteins involved in tumor progression, and its expression is significantly higher in cancers. Aladhraei et al. have demonstrated the XPO1 is overexpressed in 52.5% of colorectal cancer tissue compared to the normal adjacent epithelium [60]. Further they suggested the prognostic significance of XPO1 in colorectal cancer as its overexpression was significantly associated with advanced tumor stages. The overexpression of XPO1 mRNA in our analysis is consistent with previous reports. XPO1 overexpression induces the export of p53 out of the nucleus, which facilitates tumor progression and lymph node metastases [61, 62]. However, our HPA data showed contradictory results for XPO1 expression at the protein level. There is no correlation between mRNA expression and protein levels for XPO1, which may be due to translation regulation or high protein degradation as previously described by Greenbaum et al. [63]. However, further analysis and validation with a larger sample size are needed to draw definitive conclusions. The decreased survival rate in patients with high XPO1 mRNA expression, along with its increased levels in advanced tumor stages, emphasize that Colon Adenocarcinoma patients with high expression of XPO1 would have poor survival. Similar to our observations, Chain et al. in hepatocellular carcinoma and Luo et al. in diffuse large B-cell lymphoma have shown that elevated XPO1 levels in advanced stages are consistent with poor patient prognosis [59, 64]. Our observations suggest that XPO1 can be used as a potential prognostic biomarker to identify the therapeutic effectiveness of Colon Adenocarcinoma therapeutics.
Consistent with our observation, Ma et al. and Ostasiewicz et al. have demonstrated the overexpression of XPO2 in colorectal cancer using RT-PCR [65, 66]. Further, Alnabulsi et al. and Wang et al. have shown that XPO2 is overexpressed in colorectal cancer at both the mRNA and protein levels [46, 67]. XPO2 has been shown to promote tumor growth in colorectal cancer, with its knockdown resulting in reduced cancer progression [20, 68]. Another study reports that XPO2 is not only overexpressed in colorectal cancer but also promotes metastasis and invasiveness [69]. Decreased patient survival with high levels of XPO2 highlights its prognostic significance in Colon Adenocarcinoma.
XPO4 is a bidirectional protein transporter involved in both importing and exporting cargo proteins between the nucleus and cytoplasm. Limited information is available regarding its prognostic role in cancer; however we observed a significant correlation between XPO4 gene expression and tumor stage. Notably, XPO4 expression was markedly elevated in advanced stages of colon adenocarcinoma, suggesting its prognostic significance. Furthermore, higher XPO4 expression was associated with poorer patient survival, reinforcing its potential as a prognostic marker in colon adenocarcinoma.
XPO5 is a nuclear export protein responsible for exporting small RNA molecules, particularly miRNA, and double-stranded RNA binding proteins [70]. XPO5 is overexpressed, both at mRNA and protein levels, in colorectal cancer compared with normal tissues suggesting it as an oncogene [71]. Some reports also suggest it acts as a tumor suppressor, thereby making it an intriguing candidate for cancer therapy [72]. Our observations suggest that XPO5 functions as an oncogene, with increased expression in advanced stage (stage III) compared to early stage, indicating its prognostic significance in colon adenocarcinoma, while its correlation with poor patient survival further highlights its prognostic value. In concordant to our observation, Shigeyasu et al. also demonstrated that increased XPO5 expression is associated with advanced stages of colorectal cancer and correlates with poor prognosis [71]. XPO6 has not been previously studied in colorectal cancer including colon adenocarcinoma, but its prognostic potential has been reported in prostate cancer recurrence [73]. The author showed that relatively high expression of XPO6 is significantly associated with poor patient prognosis. Contrary to this, our analysis reveals its overexpression with better overall survival of colon adenocarcinoma patients. Hence this needs further validation in more samples to better understand it and consider it as a prognostic biomarker.
Our study demonstrated significant mRNA overexpression of XPO7 in Colon Adenocarcinoma, suggesting its oncogenic potential. Previous reports have highlighted that XPO7 exhibits dual characteristics: it acts as a tumor suppressor in hepatocellular carcinoma and as an oncogene in prostate cancer [36, 38]. No studies have previously demonstrated the role and expression of XPO7 in Colon Adenocarcinoma. However, its prognostic significance has been reported in serous epithelial ovarian cancer [37]. Our study demonstrates that XPO7 is overexpressed, but its expression declines with advancing tumor stages. A significant positive correlation with relapse-free survival suggests that XPO7 may function as a potential tumor suppressor gene. Data from the Human Protein Atlas (HPA) showing weak expression in Colon Adenocarcinoma samples further support this. This decrease in protein expression, is concordant with the mRNA expression in advanced stage of tumor, however information about the stage of sample analysed is not available on HPA database. This pattern suggests three possible roles for XPO7 in Colon Adenocarcinoma: (1) Initiation of Cancer: XPO7 is likely involved primarily in the initial stage to trigger cancer initiation. (2) Tumor Suppressor Function: The decrease in XPO7 expression with tumor progression might be due to its tumor suppressor function, as previously reported [38]; (3) Oncogene-induced senescence: XPO7 probably promotes oncogene-induced cell senescence in cancer cells, leading to tumor suppression and better patient survival. This role has been previously reported by Andrew J. Innes et al. [38]. Senescence induction could limit tumor growth and improve clinical outcomes for patients. Andrew J. Innes et al. showed that XPO7 is a novel regulator of senescence and validated its function in replicative, and oncogene-induced senescence. As reported previously, oncogene-induced senescence primarily acts as a barrier to cancer development by inducing growth arrest in cancer cells, under certain conditions [74]. We speculate similarly in the case of XPO6, as very limited information is available on it. Thus, colon adenocarcinoma patients with high exportin expression are associated with worse overall survival and tumor relapse-free survival, implicating exportins in the aggressiveness and poor prognosis of the disease. XPOT has been identified as a prognostic predictor and potential therapeutic target in neuroblastoma, with its knockdown inhibiting cell proliferation and migration [31]. Concordant to this, our observation of increased XPOT expression in advanced tumor stages suggests its oncogenic role in colon adenocarcinoma and highlights its potential prognostic significance. Our above-mentioned observations validate exportins as potential biomarkers for prognosis and targets for therapeutic intervention in Colon Adenocarcinoma. A high number of genomic alterations in exportins suggests the presence of abnormal exportin protein, and its altered regulation at the genome level, majorly contributing to the cancer progression and patient survival.
The level of immune cell infiltration in the tumor microenvironment is crucial for the better prognosis of the patients [75, 76] CD8 + T-cells and neutrophils are key immune cells that play a major role in tumor cell death. The blood neutrophil-to-lymphocyte ratio (NLR) serves as a prognostic factor in solid tumors, being negatively associated with patient prognosis [77]. The presence of higher levels of infiltrating CD8 + T-cells in tumors is generally predictive of a better prognosis, while higher neutrophil levels are indicative of a worse prognosis. Our findings show a significant negative correlation between the expression of exportins (XPO2 and XPO4) and the levels of CD8 + T-cell and neutrophil infiltrations. It suggests that exportins could serve as prognostic markers as it is directly associated with the NLR in tumors. Similar to our previous discussion, the positive association of XPO 6 & 7 expressions with the level of CD8+ T-cell infiltrations suggests it is a tumor suppressor in Colon Adenocarcinoma. The association between XPO1, XPO5, and XPOT expression and level of CD8+ T-cells infiltration was not significant, so cannot be concluded in this study.
Most of the top co-expressed genes associated with the exportins, as shown in our STRING network analysis, are either directly or indirectly involved in major mechanisms regulating cancer progression and metastases. The co-expressed genes are correlated to other exportins too suggesting the complex functional interactions to regulate cancer progression and metastases. Considering the gene function information in STRING, the top 2 co-expressed genes, according to the Spearman coefficient, can be grouped primarily into four cellular processes as shown in Fig. 6B. These processes are predominantly involved in cancer progression. Gene ontology, functional enrichment analysis, and KEGG pathway analysis provide information that exportins analysed are related to the cell cycle, cell proliferation, signal transduction, and repair pathways, in addition to nucleo-cytoplasmic transports.
From our studies, we conclude that exportin expression can be used as a prognostic biomarker for colon adenocarcinoma progression and management. However, there are several open questions to elaborate on the role of exportins in cancer progression and other diseases such as; How genetic alterations affect the binding of cargo protein with exportins?, and the mechanism of XPO7 downregulation at different stages of cancer which alleviates oncogene-induced senescence. Whether it’s a transcription factor too for the genes involved in carcinogenesis. After all our comprehensive analysis opens an avenue to explore several aspects of exportins in cancer research and therapeutics.
Potential limitations of the study
Following are the limitations of the present study (1) Although the study uses bioinformatics tools to draw conclusions, it lacks experimental validation. This means, although the findings are significant but remain theoretical until proven experimentally. (2) Since the study was conducted using COAD patients from specific datasets, the findings may not be applicable to other subtypes of colon cancer. (3) The study primarily establishes correlations between exportin expression and clinical outcomes, but it doesn't conclusively demonstrate causation. (4) The databases (UALCAN, C-BioPortal, HPA, DAVID, etc.) used in this study often focuses on specific patient populations, and their results may not be generalizable to all.
Supplementary Information
Acknowledgements
We acknowledge DST INSPIRE, New Delhi, India for providing the fellowship and contingency grant to Punita Kalia.
Author contributions
PK: Data mining, analysis, and manuscript writing; RRN: Data analysis and revision of the manuscript for important intellectual content; SSY: Study conception and design, data analysis and interpretation, and manuscript writing.
Data availability
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rohini Ravindran Nair, Email: rohini.nair@gbu.edu.in.
Suresh Singh Yadav, Email: suresh4bhu@gmail.com.
References
- 1.Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003. 10.1097/01.MP.0000062859.46942.93. [DOI] [PubMed] [Google Scholar]
- 2.Mattiuzzi C, Sanchis-Gomar F, Lippi G. Concise update on colorectal cancer epidemiology. Ann Transl Med. 2019. 10.21037/atm.2019.07.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Çağatay T, Chook YM. Karyopherins in cancer. Curr Opin Cell Biol. 2018. 10.1016/j.ceb.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gontan C, Guttler T, Engelen E, Demmers J, Fornerod M, Grosveld FG, et al. Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J Cell Biol. 2009. 10.1083/jcb.200810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aksu M, Pleiner T, Karaca S, Kappert C, Dehne HJ, Seibel K, et al. Xpo7 is a broad-spectrum exportin and a nuclear import receptor. J Cell Biol. 2018. 10.1083/jcb.201712013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thakar K, Karaca S, Port SA, Urlaub H, Kehlenbach RH. Identification of CRM1-dependent nuclear export cargos using quantitative mass spectrometry. Mol Cell Proteomics. 2013. 10.1074/mcp.M112.024877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kırlı K, Karaca S, Dehne HJ, Samwer M, Pan KT, Lenz C, et al. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. Elife. 2015. 10.7554/eLife.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azmi AS, Uddin MH, Mohammad RM. The nuclear export protein XPO1 — from biology to targeted therapy. Nat Rev Clin Oncol. 2021. 10.1038/s41571-020-00442-4. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarti N, Boles A, Burzinski R, Sindaco P, Isabelle C, McConnell K, et al. XPO1 blockade with KPT-330 promotes apoptosis in cutaneous T-cell lymphoma by activating the p53–p21 and p27 pathways. Sci Rep. 2024;14:9305. 10.1038/s41598-024-59994-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azmi AS, Muqbil I, Wu J, Aboukameel A, Senapedis W, Baloglu E, et al. Targeting the nuclear export protein XPO1/CRM1 reverses epithelial to mesenchymal transition. Sci Rep. 2015. 10.1038/srep16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subhash VV, Yeo MS, Wang L, Tan SH, Wong FY, Thuya WL, et al. Anti-tumor efficacy of Selinexor (KPT-330) in gastric cancer is dependent on nuclear accumulation of p53 tumor suppressor. Sci Rep. 2018. 10.1038/s41598-018-30686-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int. 2019. 10.1186/s12935-019-0858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XY, Wang YH, Wang J, Quan JK, Li XD, Guan KP. The role of CSE1L silencing in the regulation of proliferation and apoptosis via the AMPK/mTOR signaling pathway in chronic myeloid leukemia. Hematology. 2023. 10.1080/16078454.2022.2161201. [DOI] [PubMed] [Google Scholar]
- 14.Yuksel UM, Dilek G, Dogan L, Gulcelik MA, Berberoglu U. The relationship between CSE1L expression and axillary lymph node metastasis in breast cancer. Tumori. 2015. 10.5301/tj.5000239. [DOI] [PubMed] [Google Scholar]
- 15.Tunccan T, Kılıc C, Duran AB, Ozlugedik S, Ant A, Alkan G. Role of CSE1L expression in determining recurrence and survival of laryngeal tumors. Eur Arch Otorhinolaryngol. 2022. 10.1007/s00405-021-07206-5. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Zhou Z, Li Y, Xu J, Shen Y, Luo S, et al. CSE1L silencing impairs tumor progression via MET/STAT3/PD-L1 signaling in lung cancer. Am J Cancer Res. 2021;11:4380. [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Yuan S, Liu J, Wang Y, Zhang Y, Chen X, et al. CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. J Cell Physiol. 2020. 10.1002/jcp.29107. [DOI] [PubMed] [Google Scholar]
- 18.Wang YS, Peng C, Guo Y, Li Y. CSE1L promotes proliferation and migration in oral cancer through positively regulating MITF. Eur Rev Med Pharmacol Sci. 2020. 10.26355/eurrev_202005_21327. [DOI] [PubMed] [Google Scholar]
- 19.Aboukameel A, Muqbil I, Baloglu E, Senapedis W, Landesman Y, Argueta C, et al. Down-regulation of AR splice variants through XPO1 suppression contributes to the inhibition of prostate cancer progression. Oncotarget. 2018. 10.18632/oncotarget.26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pimiento JM, Neill KG, Henderson-Jackson E, Eschrich SA, Chen DT, Husain K, et al. Knockdown of CSE1L gene in colorectal cancer reduces tumorigenesis in vitro. Am J Pathol. 2016. 10.1016/j.ajpath.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aksu M, Trakhanov S, Görlich D. Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat Commun. 2016. 10.1038/ncomms11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Wei S, Ning S, Jie Y, Ru Y, Gu Y. Evaluation of TGFβ, XPO4, elF5A2 and ANGPTL4 as biomarkers in HCC. Exp Ther Med. 2013. 10.3892/etm.2012.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haldrup J, Weiss S, Schmidt L, Sørensen KD. Investigation of enzalutamide, docetaxel, and cabazitaxel resistance in the castration resistant prostate cancer cell line C4 using genome-wide CRISPR/Cas9 screening. Sci Rep. 2023. 10.1038/s41598-023-35950-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamazawa R, Jiko C, Choi S, Park IY, Nakagawa A, Yamashita E, et al. Structural basis for selective binding of export cargoes by exportin-5. Structure. 2018. 10.1016/j.str.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, et al. A high-resolution structure of the pre-microrna nuclear export machinery. Science (1979). 2009. 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 26.Wen J, Gao Q, Wang N, Zhang W, Cao K, Zhang Q, et al. Association of microRNA-related gene XPO5 rs11077 polymorphism with susceptibility to thyroid cancer. Medicine. 2017. 10.1097/MD.0000000000006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Pu W, Sun HL, Zhou JK, Fan X, Zheng Y, et al. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. 2018. 10.1038/s41418-018-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, et al. Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006. 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor MicroRNAs in the nucleus of cancer cells. Cancer Cell. 2010. 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Lin J, Hou Y, Huang S, Wang Z, Sun C, Wang Z, et al. Exportin-T promotes tumor proliferation and invasion in hepatocellular carcinoma. Mol Carcinog. 2019. 10.1002/mc.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan LJ, Chen JL, Wu ZX, Wu YM. Exportin-T: a novel prognostic predictor and potential therapeutic target for neuroblastoma. Technol Cancer Res Treat. 2021. 10.1177/15330338211039132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehmood R, Jibiki K, Shibazaki N, Yasuhara N. Molecular profiling of nucleocytoplasmic transport factor genes in breast cancer. Heliyon. 2021. 10.1016/j.heliyon.2021.e06039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onuma A, Fujioka YA, Fujii W, Sugiura K, Naito K. Expression and function of exportin 6 in full-grown and growing porcine oocytes. J Reprod Dev. 2019. 10.1262/jrd.2019-040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu C, Kim SJ, Mooradian A, Wang F, Li Z, Holohan S, et al. Cancer-associated exportin-6 upregulation inhibits the transcriptionally repressive and anticancer effects of nuclear profilin-1. Cell Rep. 2021. 10.1016/j.celrep.2021.108749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Teng X, Lin Y, Jiang C, Chen X, Zhang Y. Targeting XPO6 inhibits prostate cancer progression and enhances the suppressive efficacy of docetaxel. Discov Oncol. 2023. 10.1007/s12672-023-00700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y, Zhan M, Xu B. Exportin XPO7 acts as an oncogenic factor in prostate cancer via upregulation of TCF3. J Cancer Res Clin Oncol. 2023. 10.1007/s00432-023-04705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cáceres-Gorriti KY, Carmona E, Barrès V, Rahimi K, Létourneau IJ, Tonin PN, et al. RAN nucleo-cytoplasmic transport and mitotic spindle assembly partners XPO7 and TPX2 are new prognostic biomarkers in serous epithelial ovarian cancer. PLoS ONE. 2014. 10.1371/journal.pone.0091000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Innes AJ, Sun B, Wagner V, Brookes S, McHugh D, Pombo J, et al. XPO7 p21CIP1is -dependent a tumor suppressor senescence regulating. Genes Dev. 2021. 10.1101/GAD.343269.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Guo L, Chen L, Gong B, Jia D, Sun Q. Nuclear transport proteins: structure, function, and disease relevance. Signal Transduct Target Ther. 2023. 10.1038/s41392-023-01649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kodiha M, Tran D, Morogan A, Qian C, Stochaj U. Dissecting the signaling events that impact classical nuclear import and target nuclear transport factors. PLoS ONE. 2009. 10.1371/journal.pone.0008420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mutka SC, Yang WQ, Dong SD, Ward SL, Craig DA, Timmermans PBMWM, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009. 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parikh K, Cang S, Sekhri A, Liu D. Selective inhibitors of nuclear export (SINE)-a novel class of anti-cancer agents. J Hematol Oncol. 2014. 10.1186/s13045-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998. 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 44.Schwarzerová K, Bellinvia E, Martinek J, Sikorová L, Dostál V, Libusová L, et al. Tubulin is actively exported from the nucleus through the Exportin1/CRM1 pathway. Sci Rep. 2019. 10.1038/s41598-019-42056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seervi M, Sumi S, Chandrasekharan A, Sharma AK, SanthoshKumar TR. Molecular profiling of anastatic cancer cells: potential role of the nuclear export pathway. Cell Oncol. 2019. 10.1007/s13402-019-00451-1. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Liufu J, Yang Q, Dai S, Wang J, Xie B. Identification and validation of a novel signature as a diagnostic and prognostic biomarker in colorectal cancer. Biol Direct. 2022. 10.1186/s13062-022-00342-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022. 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Győrffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharmacol. 2024. 10.1111/bph.16257. [DOI] [PubMed] [Google Scholar]
- 49.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science (1979). 2017. 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 50.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020. 10.1093/NAR/GKAA407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023. 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022. 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bu F, Zhao Y, Zhao Y, Yang X, Sun L, Chen Y, et al. Distinct tumor microenvironment landscapes of rectal cancer for prognosis and prediction of immunotherapy response. Cell Oncol. 2022. 10.1007/s13402-022-00725-1. [DOI] [PubMed] [Google Scholar]
- 55.Lei J, Luo J, Liu Q, Wang X. Identifying cancer subtypes based on embryonic and hematopoietic stem cell signatures in pan-cancer. Cell Oncol. 2024. 10.1007/s13402-023-00886-7. [DOI] [PubMed] [Google Scholar]
- 56.Pagès F, Galon J, Dieu-Nosjean MC, Tartour E, Sautès-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010. 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 57.Majid U, Bergsland CH, Sveen A, Bruun J, Eilertsen IA, Bækkevold ES, et al. The prognostic effect of tumor-associated macrophages in stage I-III colorectal cancer depends on T cell infiltration. Cell Oncol. 2024. 10.1007/s13402-024-00926-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan M, Huang P, Li L, Lei P, Fang L, Zhao L, et al. Comprehensive bioinformatics analysis on exportins in lung adenocarcinoma and lung squamous cell carcinoma. J Thorac Dis. 2023. 10.21037/jtd-23-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L, Huang Y, Zhou L, Lian Y, Wang J, Chen D, et al. Prognostic roles of the transcriptional expression of exportins in hepatocellular carcinoma. 2019. Biosci Rep. 10.1042/BSR20190827. [DOI] [PMC free article] [PubMed]
- 60.Aladhraei M, Suwannalert P, Al-Thobhani AK, Poungvarin N. Association of XPO1 overexpression with NF-κB and Ki67 in colorectal cancer. Asian Pac J Cancer Prev. 2019. 10.31557/APJCP.2019.20.12.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aladhraei M, Al-Salami E, Poungvarin N, Suwannalert P. The roles of p53 and XPO1 on colorectal cancer progression in Yemeni patients. J Gastrointest Oncol. 2019. 10.21037/jgo.2019.01.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Bill MA, Young GS, La Perle K, Landesman Y, Shacham S, et al. Novel small molecule XPO1/CRM1 inhibitors induce nuclear accumulation of TP53, phosphorylated MAPK and apoptosis in human melanoma cells. PLoS ONE. 2014. 10.1371/journal.pone.0102983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003. 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo B, Huang L, Gu Y, Li C, Lu H, Chen G, et al. Expression of exportin-1 in diffuse large B-cell lymphoma: immunohistochemistry and TCGA analyses. Int J Clin Exp Pathol. 2018;11:5547. [PMC free article] [PubMed] [Google Scholar]
- 65.Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y, et al. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging. 2018. 10.18632/aging.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ostasiewicz B, Ostasiewicz P, Duś-Szachniewicz K, Ostasiewicz K, Ziółkowski P. Quantitative analysis of gene expression in fixed colorectal carcinoma samples as a method for biomarker validation. Mol Med Rep. 2016. 10.3892/mmr.2016.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alnabulsi A, Agouni A, Mitra S, Garcia-Murillas I, Carpenter B, Bird S, et al. Cellular apoptosis susceptibility (chromosome segregation 1-like, CSE1L ) gene is a key regulator of apoptosis, migration and invasion in colorectal cancer. J Pathol. 2012. 10.1002/path.4031. [DOI] [PubMed] [Google Scholar]
- 68.Zhu JH, Hong DF, Song YM, Sun LF, Wang ZF, Wang JW. Suppression of cellular apoptosis susceptibility (CSE1L) inhibits proliferation and induces apoptosis in colorectal cancer cells. Asian Pac J Cancer Prev. 2013. 10.7314/APJCP.2013.14.2.1017. [DOI] [PubMed] [Google Scholar]
- 69.Tai CJ, Su TC, Jiang MC, Chen HC, Shen SC, Lee WR, et al. Correlations between cytoplasmic CSE1L in neoplastic colorectal glands and depth of tumor penetration and cancer stage. J Transl Med. 2013. 10.1186/1479-5876-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003. 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shigeyasu K, Okugawa Y, Toden S, Boland CR, Goel A. Exportin-5 functions as an oncogene and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017. 10.1158/1078-0432.CCR-16-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khan M, Khan Z, Uddin Y, Mustafa S, Shaukat I, Pan J, et al. Evaluating the oncogenic and tumor suppressor role of XPO5 in different tissue tumor types. Asian Pac J Cancer Prev. 2018. 10.22034/APJCP.2018.19.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hao J, Chiang YT, Gout PW, Wang Y. Elevated XPO6 expression as a potential prognostic biomarker for prostate cancer recurrence. Front Biosci Sch. 2016. 10.2741/S445. [DOI] [PubMed] [Google Scholar]
- 74.Ling LX, Ding J, Hua ML. Oncogene-induced senescence: a double edged sword in cancer. Acta Pharmacologica Sinica. 2018. 10.1038/aps.2017.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu B, Liu XY, Wang GP, Chen YX. The immune cell infiltration-associated molecular subtypes and gene signature predict prognosis for osteosarcoma patients. Sci Rep. 2024. 10.1038/s41598-024-55890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, Wang X, Zhang Z, Ma F, Wang F. A novel tumor-associated neutrophil gene signature for predicting prognosis, tumor immune microenvironment, and therapeutic response in breast cancer. Sci Rep. 2024. 10.1038/s41598-024-55513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu X, Gao X, Li X, Qi X, Ma M, Qin S, et al. Prognostic significance of neutrophil-to-lymphocyte ratio in prostate cancer: evidence from 16,266 patients. Sci Rep. 2016. 10.1038/srep22089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Chen L, Huang Y, Zhou L, Lian Y, Wang J, Chen D, et al. Prognostic roles of the transcriptional expression of exportins in hepatocellular carcinoma. 2019. Biosci Rep. 10.1042/BSR20190827. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.