Abstract
Several isoforms of superoxide dismutase (SOD) with a high isoelectric point (pI) have been identified by isoelectric focusing chromatography in protein extracts from Scots pine (Pinus sylvestris) needles. One of these isoforms, a CuZn-SOD with a pI of about 10 and thus denoted hipI-SOD, has been isolated and purified to apparent homogeneity. A cDNA encoding the hipI-SOD protein was cloned and sequenced. Northern hybridization of mRNA isolated from different organs and tissues showed that hipI-SOD has a markedly different pattern of expression compared with chloroplastic and cytosolic SOD. Furthermore, the transcript levels of hipI-SOD and cytosolic SOD were found to respond differently to mechanical wounding, treatment with oxidized glutathione, paraquat, and ozone. Immunogold electron microscopy localized the hipI-SOD in the plasma membrane of sieve cells and the Golgi apparatus of albuminous cells. Moreover, high protein density was also detected in extracellular spaces such as secondary cell wall thickenings of the xylem and sclerenchyma and in intercellular spaces of parenchyma cells.
Superoxide dismutases (SODs; E.C. 1.15.1.1) belong to a family of metalloenzymes which catalyze the disproportionation of superoxide anion (O2−) radicals to yield molecular oxygen and hydrogen peroxide (H2O2). The main function of SOD is to scavenge O2− radicals generated in various physiological processes, thus preventing the oxidation of biological molecules, either by the radicals themselves, or by their derivatives (Liochev and Fridovich, 1994; Fridovich, 1995). A number of environmental stresses can lead to enhanced production of O2− within plant tissues, and plants are believed to rely on the enzyme SOD to detoxify this reactive oxygen species.
Plants possess three types of SODs with different prosthetic metal groups: CuZnSOD (Cannon et al., 1987; Perl-Treves et al., 1988; Bowler et al., 1992), MnSOD (White and Scandalios, 1988; Bowler et al., 1989), and FeSOD (van Camp et al., 1991). Although only a single isoform of CuZnSOD has been detected in chloroplasts, several cytosolic isoforms have been described (Perl-Treves et al., 1988; Scioli and Zilinskas, 1988, Streller et al., 1994).
Immunoelectron microscopic studies have demonstrated that the cytosolic (cyt)-CuZnSOD is present in the cytosol adjacent to the vacuole in plant cells (Ogawa et al., 1996). Apoplastic isoforms of SOD have been identified in extracellular washing fluids of Scots pine (Pinus sylvestris) needles (Streller and Wingsle, 1994). Several isoforms of SOD have also been detected by isoelectric focusing (IEF) chromatography of protein extracts from Scots pine needles in the high-pI range (Schinkel et al., 1998). In the study described in this paper, we purified one of these isoforms and present here its cDNA sequence, expression pattern, redox regulation, and subcellular localization as determined by immunoelectron microscopy.
RESULTS
Purification of hipI-SOD
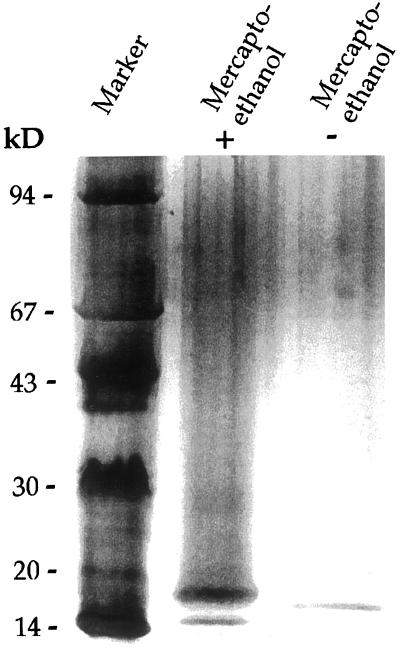
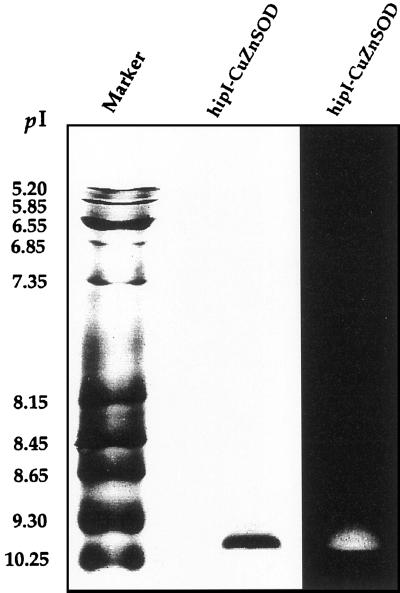
An SOD isoform with a specific activity of 245,000 units mg protein−1 has been purified from homogenates of Scots pine needles to apparent homogeneity (Table I). The enzymatic activity of the purified enzyme corresponds to 6,360 units mg protein−1 in the McCord and Fridovich xanthine oxidase/cytochrome c assay (Marklund, 1985). Overall, a 1,360-fold purification was obtained, with a recovery of 0.8% of the initial total SOD activity and a yield of 360 μg protein. Both the chromatographic and gel electrophoretic properties of the protein indicate that this SOD is a dimeric enzyme. The final preparation contained a single protein band at a position corresponding to a size of 16 kD after SDS-PAGE when denaturated without addition of mercaptoethanol (Fig. 1). However, when denaturated in the presence of mercaptoethanol, one band at 17.5 kD together with a possible minor degradation product of a lower Mr was found (Fig. 1). The shift toward a higher Mr indicates the intrachain disulfide bond between Cys-58 and Cys-147 (Fig. 2; Bordo et al., 1994). The SOD was eluted in size exclusion chromatography at a position corresponding to a molecular mass of approximately 24.8 kD. The pI of the active enzyme, as determined by IEF on a pH gradient gel of 6.5 to 10.5 was about 10.2, close to the pI of the cytochrome c marker (Fig. 3). The presence of either 2 mm KCN or 5 mm H2O2 in the direct KO2 assay completely abolished the SOD activity (data not shown). The amounts of enzymatic activity of this new SOD form in leaves in relation to total SOD activity was estimated to be around 2%. Raising the salt concentration in the extraction buffer (to 1 m NaCl) increased the relative level of extracted hipI-SOD from Scots pine needles by approximately 30% (data not shown).
Table I.
Purification of hpI-SOD from Scots pine
| Purification Step | Total Activity | Specific Activity |
|---|---|---|
| units ×10−3 | units mg protein−1 × 10−3 | |
| 90% (NH4)SO4 | 11,420 | 0.18 |
| DEAE-cellulose | 8,200 | 0.22 |
| Phenyl-sepharose | 4,500 | 7 |
| DEAE-cellulose | 310 | 26 |
| Sepharose S | 165 | 245 |
Figure 1.
SDS-PAGE of purified hipI-SOD from Scots pine needles. Gel showing mobility of 25-ng hipI-SOD with and without addition of mercaptoethanol. Molecular mass calibration proteins in kilodaltons are indicated to the left: phosphorylase b (94 kD), albumin (67 kD), ovalbumin (43 kD), carbonic anhydrase (30 kD), trypsin inhibitor (20.1 kD), and α-lactalbumin (14.4 kD).
Figure 2.
The deduced amino acid sequence hipI-SOD from Scots pine. The comparison of the deduced amino acid sequence of hipI-SOD with cytosolic CuZnSOD from Scots pine, tomato (Lycopersicon esculentum), and maize (Zea mays; Cannon and Scandalios, 1987; Perl-Treves et al., 1988; Karpinski et al., 1992) and chloroplastic CuZnSOD from pea, tomato, and Scots pine (Scioli and Zilinskas, 1988; Perl-Treves et al., 1988; Karpinski et al., 1992). Strictly homologous amino acid residues are shaded. Underlined amino acids indicate regions sequenced by the Edman degradation method. Asterisks show the residues binding Cu and Zn (Bordo et al., 1994). The GenBank accession no. for the hipI-SOD sequence is AJ307586.
Figure 3.
Determination of pI of hipI-SOD from Scots pine by IEF, pH 6.5 to 10.5. Purified hipI-SOD of Scots pine (50 ng) was visualized by silver staining (middle lane) and by activity staining (right lane). Calibration standards (pI) are indicated to the left: γ-lactoglobulin (5.2), bovine carbonic anhydrase (5.85), human carbonic anhydrase (6.55), horse myoglobin (6.85), horse myoglobin (7.35), lentil lectin (8.15), lentil lectin (8.45), lentil lectin (8.65), trypsinogen (9.3), and cytochrome c (10.25).
Characterization of cDNA and Amino Acid Sequence Alignment
The full length of hipI-SOD cDNA encodes 156 amino acids. The extent of hipI-SOD homology to cyt-SOD from maize, tomato, and Scots pine was 70%, 71%, and 70%, respectively, whereas its homology to chloroplastic (cp)-SODs from pea (Pisum sativum), tomato, and Scots pine was only 62%, 63%, and 64%, respectively. The positions of amino acids in hipI-SOD have been compared with corresponding residues in the three-dimensional structures of CuZnSOD (Getzoff et al., 1983, 1992; Kitagawa et al., 1991) to investigate structural relationships and to deduce their possible function. The protein represents a CuZn-type of SOD enzyme and histidines and the Asp, which coordinates the Cu2+ and Zn2+ are conserved in the Scots pine SOD amino acid sequence (Fig. 2).
Transcript Levels of hipI-SOD in Different Organs and Tissues of Scots Pine
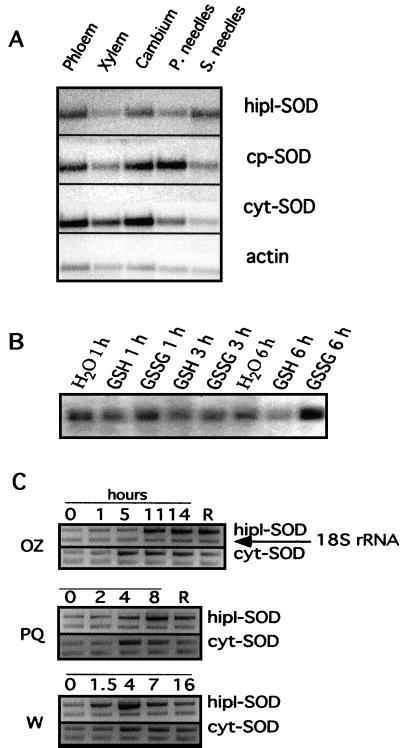
To characterize the expression of hipI-SOD in different organs and tissues of Scots pine, poly(A+) RNA isolated from the stem and needles (primary and secondary) was analyzed by northern hybridization using the 3′-non-translated region of hipI-SOD cDNA as a probe (Fig. 4A). A hipI-SOD transcript of about 1,000 bp has been found in samples isolated from stem and in both types of needles, with higher expression in secondary needles. Among stem tissues, the highest transcript level of hipI-SOD was found in phloem.
Figure 4.
Detection of SOD transcripts by RNA gel blot analysis and relative quantitative reverse transcriptase (RT)-PCR. A, Northern-blot hybridization of hipI-SOD, cyt-, and cp-SOD in different organs and stem tissues of Scots pine. Poly(A+) RNA (3.5 μg) was separated by gel electrophoresis, transferred to a filter, and hybridized to homologous, gene-specific hipI-, cyt-, and cp-SOD cDNA probes. P. needles, Primary needles; S. needles, secondary needles. B, Northern hybridization; poly(A+) RNA (10 μg per lane) isolated from needles of Scots pine shoots treated for 1, 3, and 6 h with water, 5 mm GSSG, or 5 mm GSH as described previously (Wingsle and Karpinski, 1996). The RNA was separated by gel electrophoresis, transferred to a filter, and hybridized to homologous hipI-SOD and cyt-SOD cDNA probes. C, Quantitative RT-PCR; separation of quantitative RT-PCR products in Scots pine cotyledons after 2, 4, and 8 h of treatment with 8 μm paraquat (PQ) and after 24 h of recovery (R). Seedlings were also fumigated with 0.5 μL L−1 ozone (OZ) and samples collected after 1.5, 11, and 14 h of treatment and after recovering for 24 h (R). Wounded seedlings (W) were collected after 1.5, 4, 7, and 16 h after crushing. To control the changes of mRNA levels during the experiment, non-treated needles were collected parallel with the experimental samples and used for RT-PCR (data not shown).
To show that the novel hipI-SOD differs in expression pattern from cyt- and cp-SODs, the same northern blot was used for hybridization with two other probes containing mostly 3′-non-translated regions of cyt- and cp-SOD cDNAs. The strongest signals using a cp-SOD probe were detected in primary needles and, to a lesser extent, in phloem and cambium layers. The highest level of cyt-SOD transcript was also found in both layers, but in contrast to hipI-SOD, quite high levels of this message were also found in the xylem.
Regulation of hipI-SOD in Response to Oxidative Stress Factors and Wounding
We have shown recently that the level of cyt-SOD transcript is reduced after treating Scots pine seedlings with reduced glutathione (GSH), but it does not change when the plants are treated with oxidized GSH (GSSG; Wingsle and Karpinski, 1996). To test the effects of GSH and GSSG on hipI-SOD transcript, we used mRNA obtained from this experiment. Similar to cyt-SOD, hipI-SOD transcript level was found to be reduced after treatment with GSH. However, in contrast to cyt-SOD, the level of hipI-SOD transcript was significantly increased after 6 h of GSSG treatment (Fig. 4B).
The response of hipI-SOD and cyt-SOD transcripts to wounding and treatment with paraquat and ozone was analyzed by quantitative RT-PCR relative to the 18S rRNA RT-PCR product. The results show that both transcripts were induced in response to mechanical wounding, ozone, and paraquat; however, the levels of increase and time to elicit the response differed between hipI-SOD and cyt-SOD (Fig. 4C)
Characterization of hipI-SOD Antibodies
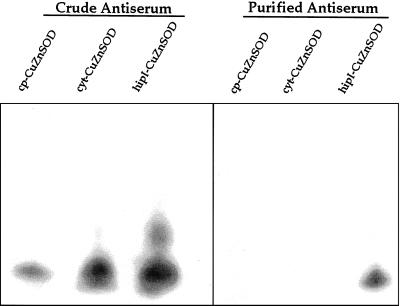
The IgG fraction of the antiserum against hipI-SOD after affinity purification on a column of immobilized CuZnSOD showed no cross reactivity with purified cp-SOD and cyt-SOD, but gave a strong signal with purified hipI-SOD on western blots (Fig. 5). In no case was the signal detected when pre-immune serum was used (data not shown).
Figure 5.
Western-blot analysis of hipI-SOD from Scots pine after SDS-PAGE. Western-blot analysis, using crude and purified hipI-SOD antiserum, of 25 ng of purified hipI-SOD, cyt-SOD, and cp-SOD.
Immunogold Localization of hipI-SOD
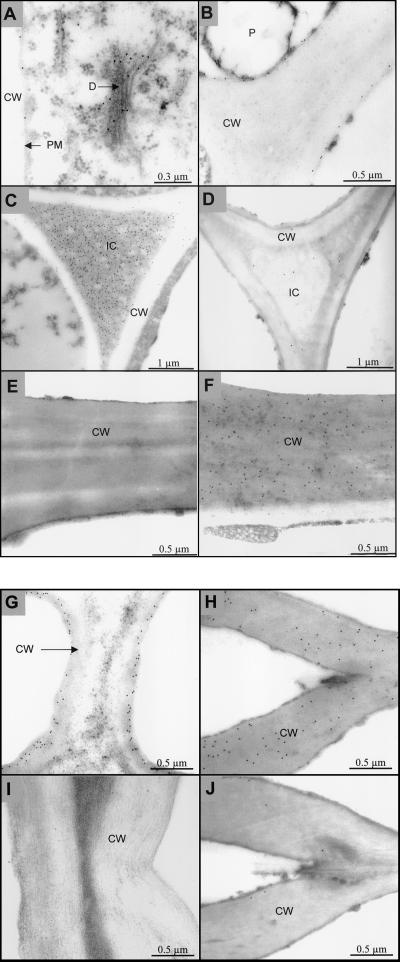
Immunogold electron microscopy was used to investigate the subcellular distribution of hipI-SOD in plants. Thin sections of cotyledons (8 d after germination), primary needles, and secondary needles of Scots pine were used for these studies. The protein was not detected in the cytoplasm, mitochondria or nuclei. A high density of label was found over dictyosomes in the albuminous cells of primary needles (Fig. 6A) and plasma membrane of sieve cells in cotyledons (Fig. 6B). Intense labeling with hipI-SOD antibodies was also observed in the extracellular matrix (apoplast), e.g. in cell walls (Fig. 6, F–H) and in intercellular spaces (IC; Fig. 6C). The cell walls of xylem tracheid (Fig. 6F), bordered pits of the xylem (Fig. 6H), and sclerenchyma cells (Fig. 6G) were densely stained with hipI-SOD antibodies. When pre-immune serum was used as a control, there was no significant immunogold labeling in the secondary cell wall of xylem (Fig. 6E). The cell wall of bordered pits in the xylem of secondary needles was densely stained, but the cell walls of bordered pits in transfusion tracheids of parenchyma were free of gold label (Fig. 6J). Intensive labeling was observed only in intercellular spaces and cell walls of young, developing cotyledons, whereas the mature and older needles showed no labeling. Dense staining could be detected between cells of developing parenchyma in young cotyledons (Fig. 6C), whereas spaces between mature cells of old needles were barely labeled (Fig. 6D). The secondary cell wall of developing sclerenchyma cells of young Scots pine cotyledons were significantly labeled (Fig. 6G), but only very weak labeling was observed in sclerenchyma cells of old Scots pine needles (Fig. 6I). Similar protein localization has been found in tobacco (Nicotiana tabacum) leaves using antibodies against Scots pine hipI-SOD (data not shown).
Figure 6.
Immunogold localization of hipI-SOD in cross-sections of Scots pine needles and cotyledons. A, Primary needle and dictyosome in albuminous cell; B, cotyledon and connection between two sieve cells; C, cotyledon, intercellular, and parenchyma cells; D, secondary needle, intercellular, and parenchyma cells; E, secondary needle, control with pre-immune sera, and xylem tracheid; F, secondary needle and xylem tracheid; G, cotyledon and developing sclerenchyma; H, secondary needle and bordered pit of xylem; I, secondary needle and mature sclerenchyma; J, secondary needle and pit of transfusion tracheid. CW, Cell wall; D, dictyosome; IC, intercellular space; PM, plasma membrane; P, plastid.
DISCUSSION
As has been shown previously, SODs with high pIs represent a separate group of SOD isoforms that migrate on IEF gel to ≥pI 7 and they are easily distinguished from cyt- and cp-SODs, which are detected at pI 5.5 (Schinkel et al., 1998). A cDNA corresponding to one of these isoforms has been isolated from Scots pine. The properties of the protein, together with its expression patterns and transcript regulation, strongly suggest that hipI-SOD represents a novel isoform of SOD, distinct from cyt- and cp-SODs. The difference between them is also indicated by the low amino acid sequence homology, estimated as only 70% to cyt-SOD and 64% to cp-SOD of Scots pine. Because the cyt-SODs are at least 90% homologous within any plant species, and at least 70% homologous when isoforms from different species are compared, the hipI-SOD shows quite low homology both to cyt-SODs from Scots pine and other species (Cannon and Scandalios, 1989; Kanematsu and Asada, 1991). Nevertheless, this homology is mostly due to the highly conserved C-terminal two-thirds of hipI-SOD. The N-terminal regions of four cyt-SOD proteins (of about 20 amino acids) have already been characterized in Scots pine (Wingsle et al., 1990; Streller et al., 1994). The homology among them ranges between 80% to 95%; however, they show only 45% to 50% homology to the corresponding portion of hipI-SOD.
As northern analysis shows, low transcript levels of hipI-SOD are detectable in primary needles, whereas higher transcript levels are found in secondary needles. Among stem tissues, high levels of expression of hipI-SOD are detected in phloem and cambium regions, but levels are very low in xylem. The higher levels of hipI-SOD transcript present in secondary needles as compared with primary ones could be due to the relatively high proportions of phloem cells in secondary needles. The opposite pattern is found if analyzing the transcript levels of cyt-SOD and cp-SOD, which are significantly higher in primary needles. The presence of CuZnSOD in vascular tissues has been shown previously by immunogold-electron microscopic analysis of spinach (Spinacia oleracea) leaves (Ogawa et al., 1997). Furthermore, the promoter of the gene encoding cytosolic CuZnSOD fused to β-glucuronidase reporter gene has been shown to be highly active in phloem cells of leaves and stem in tobacco (Hérouart et al., 1994).
The analysis of the regulation of hipI-SOD and cyt SOD transcripts shows that the transcription is modulated in response to a variety of factors. In contrast to the previously analyzed cyt-SOD (Wingsle and Karpinski, 1996), the transcript level of hipI-SOD in Scots pine needles was increased by GSSG treatment. This clearly indicates that there are differences in the redox regulation of hipI-SOD by GSH as compared with cyt-SOD. The results are also in contrast with the data of Hérouart et al. (1993), where the induction of the phloem-localized cyt-SOD has been observed by GSH but not by GSSG in tobacco. The discrepancy can be either due to the fact that different systems were used, e.g. protoplasts versus intact plant or the result of the differences in redox transduction pathways between angiosperms and gymnosperms.
Furthermore, quantitative RT-PCR analysis showed that the levels of hipI-SOD and cyt-SOD transcripts are induced in response to paraquat, ozone, and mechanical wounding; however, they differ in time required for eliciting the response and the level of induction. This may suggests a hierarchy of regulatory events acting at the transcription of SOD genes, unless the location of hipI-SOD in the plant is simply less accessible than cyt-SOD to redox reagents.
Because hipI-SOD represents a novel isoform of CuZnSOD, we have been interested in investigating the subcellular localization of this enzyme in plants using immunoelectron microscopy. HipI-SOD protein was detected in Golgi apparatus of albuminous cells and plasma membrane of Scots pine sieve cells (Fig. 6, A and B). It is well known that proteins for these cells are synthesized in neighboring cells (albuminous or companion cells) and are secreted into the sieve elements. The protein label found in the dictyosomes of albuminous cells suggests that hipI-SOD is likely translocated to the sieve elements via the Golgi secretory pathway. Moreover, the protein detected in cell walls of variety of cells must be also transported out of these cells. The classical pathway for secretion of extracellular proteins generally involves translocation of proteins with appropriate N-terminal signal sequences from the cytosol into the lumen of the ER (Galili et al., 1998), after which they are transported in vesicles through the Golgi apparatus toward the plasma membrane. However, there are a variety of proteins that lack traditional N-terminal secretory signal sequences, and yet are secreted out of the cells (Monroe et al., 1991; Zhou et al., 1992). It has been demonstrated that thioredoxin-H, one of the major proteins in rice (Oryza sativa) phloem sap, is translocated into sieve tubes, although no signal peptide can be identified in the protein sequence (Ishiwatari at al., 1995). Thus, in some cases the mechanism of intercellular protein transport between companion/albuminous cells and sieve elements is still unclear. HipI-SOD appears to be such a protein because its primary structure does not show any N-terminal ER targeting sequence.
Numerous studies demonstrate that the ROS, O2− and H2O2, are produced in vascular tissues, i.e. xylem and phloem (Hérouart et al., 1994; Ogawa et al., 1997). Wounded plant organs, in particular, can produce a burst of these oxygen intermediates (Salin and Bridges, 1981; Sekizawa et al., 1987). Recent results also show that H2O2 present in vascular tissues may function as a systemic signal in wounding responses and systemic acquired resistance in Arabidopsis (Orozco-Cardenas and Ryan, 1999; S. Karpinski, unpublished data). The fact that the transcript of hipI-SOD increased in response to mechanical wounding supports this hypothesis. Thus, it is possible that hipI-SOD in the phloem could act as a regulator of H2O2 pulses, being involved in the transmission of systemic signals in wounding or pathogen responses.
Dense immunolabelling also occurred in the extracellular matrix, e.g. in cell walls and the intercellular space in various cell types. The secondary cell wall of developing sclerenchyma cells of young Scots pine cotyledons were densely stained, whereas only very weak labeling was observed in sclerenchyma cells of mature Scots pine needles (Fig. 6, G and I). Intense labeling was detected between cells of developing parenchyma tissues, but spaces between mature cells were rarely labeled (Fig. 6, C and D). All these results hint at a developmental regulation of protein expression.
Substantial labeling was also observed in the cell walls of xylem tracheid and bordered pit of xylem (Fig. 6, F and H), which are also known to be highly lignified. However, the transfusion cell (not lignified) in parenchyma did not show any significant labeling (Fig. 6J; Esau, 1977). This indicates that SOD may participate in the lignification process by generation of H2O2. This conclusion is confirmed by the observation that the densely labeled cell wall of the bordered pit of xylem was also intensively stained with phloroglucinol-HCl, whereas unlabeled cell wall of bordered pit in transfusion tracheid was not. The contribution of SOD to lignification has been suggested previously on the basis of experiments showing colocalization of cytosolic CuZnSOD in vascular tissues of spinach hypocotyls and sites of H2O2 production (Ogawa et al., 1997).
The immunogold label found in extracellular compartments of a variety of cells suggests that hipI-SOD antibodies recognize other SOD proteins, closely related to hipI-SOD. The later suggestion can be supported by the fact that more partial sequences similar to hipI-SOD have been isolated both from the Scots pine cDNA library and cloned 5′- and 3′-RACE products. Moreover, the results of Southern-blot hybridization, showing at least two restriction fragments of DNA hybridizing to the N-terminal region of hipI-SOD, indicate the presence of other genes similar to hipI-SOD (data not shown).
In further experiments, we aim to characterize transgenic lines overexpressing hipI-SOD from Scots pine generated in Arabidopsis, and to generate lines with antisense inhibition hipI-SOD ortholog isolated from Arabidopsis. This approach will clarify the function and subcellular distribution of this novel type of CuZnSOD.
MATERIALS AND METHODS
Plant Material
Current and 1-year-old needles were collected in August from healthy 15-year-old field-grown Scots pine (Pinus sylvestris) trees. Scots pine seedlings were grown on fertilized peat in a greenhouse under an 18-h photoperiod comprising natural daylight supplemented with metal halogen lamps (HQI-TS, 400 W, Osram, Germany) giving a minimum quantum flux density of 150 μE m−2 s−1. The temperature was about 22°C during the day and approximately 17°C at night. Four-week-old seedlings were fumigated with 0.5 μL L−1 of ozone as described previously (Wingsle et al., 1992), treated with 8 μm paraquat as shown for GSSG/GSH treatment by Wingsle and Karpinski (1996), or wounded by crushing the surface for the analysis of the relative quantitative RT-PCR.
Purification and Analysis of hipI-(CuZn) SOD
Field-grown Scots pine needles (5 kg fresh weight) were frozen in liquid nitrogen and homogenized in a coffee mill to a fine powder. Purification steps including ammonium sulfate precipitation, the DEAE 52-cellulose, and phenyl-superose were performed as earlier described (Streller et al., 1994). Solid ammonium sulfate was added to the eluent from DEAE 52-cellulose to raise the concentration to 1.7 m.
The sample was batch adsorbed to 300 mL Phenyl-Sepharose Fast Flow (Pharmacia LKB, Uppsala), equilibrated with 50 mm Na-phosphate buffer (pH 7.0), containing 1.7 m ammonium sulfate and eluted with a gradient of 1.7 to 0 m ammonium sulfate in the phosphate buffer. After desalting, the active fraction from phenyl-sepharose separation was applied to a column (5 × 15 cm) of DEAE 52-cellulose, equilibrated with Tris-HCl buffer. The column was washed with 600 mL of the same buffer at a flow rate of 3 mL h−1 cm−2. The eluent was collected and concentrated using a macrosolute concentrator (Amicon, Danvers, MA) with a 10-kD cut-off filter, to a volume of 10 mL. The sample buffer was exchanged using PD-10 (Pharmacia LKB) columns, equilibrated with 50 mm 2-(N-morpholino)-ethanesulfonic acid-NaOH buffer (pH 6.5). The gel-filtered sample was then applied to a column (2.5 × 6 cm) of Sepharose S Fast Flow (Pharmacia LKB), equilibrated with the same buffer.
SOD activity was eluted by gravity flow with 20-column volumes of a 0 to 150 mm NaCl gradient in the buffer. SOD activity was determined by the direct KO2 assay (Marklund, 1985). Staining for SOD activity after IEF was performed as described by Beauchamp and Fridovich (1971). Protein content was estimated from the light absorbance of the sample at 280 nm, assuming that A = 1 corresponds to 1 mg of protein per mL. Purified hipI-SOD was hydrolyzed and analyzed for protein content by HPLC (Näsholm et al., 1987).
Preparation and Purification of Antibodies
Purified hipI-SOD was used to prepare antibodies by intramuscular injection into rabbit. The enzyme (200 μg) was emulsified with Freud's complete adjuvant before injection. Subsequent injections (50 μg) were given in the 3rd and 6th weeks, and after that, the last booster serum was collected. Horseradish SOD (5 mg, Sigma, St. Louis) was bound to 1.8 g CNBr-activated Sepharose 4B according to the manufacturer's recommendations (Pharmacia LKB). The Sepharose gel with coupled horseradish SOD (6.3 mL) was packed in a column (1.5 × 5 cm) and equilibrated with Tris-buffered saline (TBS; 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl) containing 0.25% (w/v) Triton X-100 (TBS-Triton). Approximately 200 μL anti-hipI-SOD antiserum, pH adjusted by addition of 20 μL 1 m Tris-HCl (pH 7.5), was applied to the affinity column and was incubated overnight with very gentle agitation on the column to promote immunological interaction. Purified anti-hipI-SOD antibody was collected by elution with 6 mL TBS-Triton. The affinity column was regenerated by a series of washes with 50 mm diethanolamine (pH 11.9) and 0.2 m Gly (pH 2.6), containing 0.5 m NaCl, and with 6 m urea.
Electrophoresis and Immunoblotting
SDS-PAGE, IEF and western blotting were performed at 15°C using a Phast system (Pharmacia LKB). SDS-PAGE was carried out on 10% to 15% (w/v) polyacrylamide gradient gels or on SDS-high-density gels (20% [w/v] polyacrylamide and 30% [w/v] ethylene glycol) for 60 or 120 AVh, respectively. IEF gels, pI 3 to 9 gels, and dry IEF gels were used for IEF. The dry gels were rehydrated in 2.5% (w/v) Pharmalyte (Pharmacia LKB), pI 6.5 to 9; 2.5% (w/v) Pharmalyte, pI 8 to 10.5; or 10% (w/v) Suc and 50 mm Arg for 30 min under nitrogen. Proteins were visualized by silver staining. Proteins resolved by IEF or SDS-PAGE were electroblotted onto polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA). The blotted membranes were blocked with 5% (w/v) milk powder in TBS containing 0.05% (w/v) nonidet P-40 (TBS-nonidet). After blocking, the membranes were incubated with Scots pine anti-hipI-SOD antiserum or purified Scots pine anti-hipI-SOD antibody in TBS containing 2% (w/v) milk powder and 0.25% (w/v) Triton X-100 (antibody buffer). The membranes were washed four times in TBS-nonidet for 10 min and then incubated in secondary antibody (anti-rabbit IgG-POD, Boehringer Mannheim, Ottweiler, Germany) in antibody buffer for 30 min. After washing in TBS-nonidet as before, the blots were immersed in detection solution and chemiluminescence was detected as described by the manufacturer (Boehringer Mannheim). Controls were run with rabbit serum to check for nonspecific adsorption of primary antibody.
Fixation, Substitution, Embedding, and Immunoelectron Microscopy
Leaf sections (1 mm2) were fixed, substituted, and embedded in Lowycryl HM20 resin (Plano GmbH, Marburg, Germany) as described previously (Teige et al., 1998). The cured blocks were cut into ultra-thin sections with a thickness of 70 to 90 nm on an ultramicrotome (Ultra cut F; Reichert, Wien, Austria) and mounted on copper grids, followed by immunogold labeling with 10 nm goldprotein-A, as previously described (Süss et al., 1993). Finally, specimens were stained with uranyl acetate prior to examination in a transmission electron microscope (CEM 902A, Zeiss, Jena, Germany) at 80 kV.
Amino Acid Sequence Analysis
Purified hipI-SOD was desalted by gel filtration using Sephadex G-25 columns. The Trp and Cys amino acid residues of the desalted hipI-SOD were modified by vapor phase S-pyridylethylation (Yamada et al., 1991). Approximately 200 pmol of modified hipI-SOD (3.4 μg protein) was treated with CNBr (Fontana and Gross, 1986) or digested with Protease V8 (Boehringer Mannheim) in ammonium carbonate buffer (pH 7.8; Homard and Drapeau, 1972). The resulting peptides were separated by SDS-high-density PAGE and were electrotransferred to polyvinylidene difluoride membranes. Electroblotted peptides were visualized by Coomassie G-250 dye binding (Bio-Rad). The NH2-terminal portions of the peptides were sequenced by the Edman degradation method (model 477 A protein/peptide sequencer coupled with the online model 120 A PTH Analyzer; Applied Biosystems, Foster City, CA).
Cloning and Sequencing and of Scots Pine hipI-SOD
On the basis of the amino acid sequence of Scots pine hipI-SOD, two degenerated, 29-mer oligonucleotides—5′-GAGGCTACAAAGGTTTTTGGI(A/C) G(ATGC) AT(CAT) AC-3′ (sense) coding EATKVFGRI and 5′-GGCCGCTTIA(A/G) AGGAAT(C/T) TTCCA(A/G) TC(C/T) TG-3′ (antisense) coding ELSIQDWKI—were synthesized and used for the amplification of an internal sequence of the hipI-SOD (231 bp) from a Scots pine lambda gt 10 cDNA library (Karpinski et al., 1992). The amplified cDNA library was grown in Escherichia coli strain C 600. Duplicate filters representing 500,000 plaques were prepared for screening with the 32P-dCTP-labeled 250-bp fragment (Ausubel et al., 1989). Filters were washed twice for 10 min in 1× SSC and 0.1% (w/v) SDS at room temperature and once for 10 min in 1× SSC and 0.1% (w/v) SDS at 65°C. After screening and rescreening, one positive clone (PST 101) was isolated and subcloned in the pGEM vector (Promega, Madison, WI).
DNA sequencing was carried out using an Applied Biosystems DNA sequencer model 373A (Applied Biosystems) with an ABI PRISM Dye-Terminator Cycle Sequencing Ready Reaction kit (Perkin Elmer, Foster City, CA) and specific primers, according to the manufacturer's instructions.
The full-length sequence has been obtained by 5′ and 3′ RACE using the SMART Race cDNA amplification kit (CLONTECH Laboratories, Inc., Palo Alto, CA).
The translation products of the SOD genes were aligned using the PILEUP program in the GCG package (Genetics Computer Group, Madison, WI).
Northern Hybridization
Primary and secondary needles were obtained from 3-week- to 1-year-old Scots pine seedlings. Vascular tissues were collected from the stem of a 6-year-old Scots pine tree by peeling the bark, and scraping the tissues from xylem, the cambial zone (with some of the phloem), and phloem. The fraction on the xylem side (denoted xylem) consisted of differentiating xylem with the primary wall xylem elements. The first, very gently scraped fraction taken from the bark side (denoted cambium) consisted of cambial zone cells, and differentiating and functional phloem. The second layer, scraped harder, contained nonfunctional phloem with phloem fibers.
Total RNA was prepared according to Chang et al. (1993). Poly(A+) RNA was obtained from the total RNA using Oligo (dT)25 Dynabeads (Dynal A.S., Oslo). Poly(A+) RNA (3.5 μg) was glyoxilized and electrophoretically separated on a 1% (w/v) agarose gel. The corresponding RNA gel blot was hybridized sequentially with fragments containing specific, 3′-terminal non-coding regions of cyt-SOD, cp-SOD, and hipI-SOD. Probes were labeled with [α-32P]-(dCTP) by the random primer method using the kit and protocol of the supplier (Pharmacia LKB). Hybridization was performed according to Church and Gilbert (1984), and filters were washed in stringent conditions at 65°C (0.1% [w/v] SSC and 0.1% [w/v] SDS). The equivalence of RNA loading among lanes on agarose gels was established by hybridization with a cDNA probe encoding the conserved region of the Scots pine actin. The filters were analyzed on a phosphor imaging system (GS-525 Molecular Imager, Storage Phospor Imaging Systems, Bio-Rad). The probe was removed between hybridizations by treatment of the nylon membrane (Hybond N, Amersham Pharmacia, Chalfont, UK) with boiling 0.1% (w/v) SDS solution.
Relative Quantitative RT-PCR
Relative quantitative RT-PCR was performed according to the Intermedica Instruction Manual. In general, 1 μg of total RNA (extracted as above) was converted to the first strand of cDNA with random primer mixture at 42°C in 1 h, using Superscript II RNAse H− reverse transcriptase (Life Technologies/Gibco-BRL, Cleveland). RNA samples were treated with DNAse I and RNAse-free (Boehringer Mannheim) and control PCR was run to check the absence of genomic DNA. 18SRNA was used as internal, constitutive control and co-amplified with hipI-SOD or cyt-SOD specific primers in the same reaction. The primers have been chosen to amplify only specific regions of cDNAs (hipI-SOD; forward, TGCCTTCAGGGAGCGAGAACG, reverse, CTGGAAGCCTTTATCCAGG and cyt-SOD; and forward, GAATGGTGCTGCTGATGTCAAGG, reverse, GCCAGCCTATAAATAACCAACTG). The number of cycles (usually 27–29) was established empirically to amplify the target cDNA and the control in the linear range.
ACKNOWLEDGMENTS
The authors are grateful to Jan-Erik Hällgren and Stanislaw Karpinski for critical reading of the manuscript.
Footnotes
This work was supported by the Swedish Councils for Forestry and Agricultural Research, Natural Sciences, and Strategic Research (grants to Swedish University of Agricultured Sciences).
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols of Molecular Biology. New York: Green Publishing Associates/Wiley Interscience; 1989. pp. 6.0.3–6.4.7. [Google Scholar]
- Beauchamp CO, Fridovich I. Superoxide dismutase: improved assay and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- Bordo D, Djinovic K, Bolognesi M. Conserved patterns in the Cu,Zn superoxide dismutase family. J Mol Biol. 1994;238:366–386. doi: 10.1006/jmbi.1994.1298. [DOI] [PubMed] [Google Scholar]
- Bowler C, Alliotte T, de Loose M, van Montague M, Inzé D. The induction of Mn-superoxide dismutase in response to stress in N. plumbaginifolia. EMBO J. 1989;8:31–38. doi: 10.1002/j.1460-2075.1989.tb03345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, van Montague M, Inzé D. Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:83–116. [Google Scholar]
- Cannon RE, Scandalios JG. Two cDNAs encode two nearly identical CuZn-superoxide dismutase proteins in maize. Mol Gen Genet. 1989;219:1–8. doi: 10.1007/BF00261150. [DOI] [PubMed] [Google Scholar]
- Cannon RE, White JA, Scandalios JG. Cloning of cDNA for maize superoxide dismutase 2 (SOD 2) Proc Natl Acad Sci USA. 1987;84:179–183. doi: 10.1073/pnas.84.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Rep. 1993;11:113–116. [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants. New York: John Wiley; 1977. [Google Scholar]
- Fontana A, Gross E. Fragmentation of polypeptides by chemical methods. In: Darbre A, editor. Practical Protein Chemistry. Chichester, UK: John Wiley; 1986. pp. 67–120. [Google Scholar]
- Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- Galili G, Sengupta-Gopalan C, Ceriotti A. The endoplasmic reticulum of plant cells and its role in protein maturation and biogenesis of oil bodies. Plant Mol Biol. 1998;38:1–29. [PubMed] [Google Scholar]
- Getzoff ED, Cabielli DE, Fisher CL, Parge HE, Viezzoli MS, Hallewell RA. Faster superoxide dismutase mutants designed by enhanced electrostatic guidance. Science. 1992;358:347–351. doi: 10.1038/358347a0. [DOI] [PubMed] [Google Scholar]
- Getzoff ED, Tainer JA, Weiner PK, Kollman PA, Richardson J, Richardson DC. Electrostatic recognition between superoxide and copper-zinc superoxide dismutase. Nature. 1983;306:287–290. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- Hérouart D, Van Montague M, Inzé D. Redox-activated expression of the cytosolic copper/zinc superoxide dismutase gene in Nicotiana. Proc Natl Acad Sci. 1993;90:3108–3112. doi: 10.1073/pnas.90.7.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérouart D, Van Montague M, Inzé D. Developmental and environmental regulation of Nicotiana plumbaginifolia cytosolic Cu/Zn-superoxide dismutase promoter in transgenic tobacco. Plant Physiol. 1994;104:873–880. doi: 10.1104/pp.104.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homard J, Drapeau GR. Staphylococal protease: a proteolytic enzyme specific for glutamyl bonds. Proc Natl Acad Sci USA. 1972;69:3506–3509. doi: 10.1073/pnas.69.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwatari Y, Honda C, Kawashima I, Nakamura S, Hirano H, Mori S, Fujiwara T, Hayashi H, Chino M. Thioredoxin-H is one of the major proteins in rice phloem. Planta. 1995;195:456–463. doi: 10.1007/BF00202605. [DOI] [PubMed] [Google Scholar]
- Kanematsu S, Asada K. Chloroplast and cytosol isozymes of CuZn-superoxide dismutase: their characteristic amino acid sequences. Free Radic Res Commun. 1991;12–13:383–390. doi: 10.3109/10715769109145808. [DOI] [PubMed] [Google Scholar]
- Karpinski S, Wingsle G, Olsson O, Hällgren J-E. Characterization of cDNAs encoding CuZn-superoxide dismutase in Scots pine. Plant Mol Biol. 1992;18:545–555. doi: 10.1007/BF00040670. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y, Tanaka N, Hata Y, Kusonoki M, Lee GP, Katsube Y, Asada K, Aibara S, Morita Y. Three-dimensional structure of CuZn-superoxide dismutase from spinach at 2 Å resolution. J Biochem. 1991;109:477–485. doi: 10.1093/oxfordjournals.jbchem.a123407. [DOI] [PubMed] [Google Scholar]
- Liochev S, Fridovich I. The role of superoxide anion radicals in the production of hydroxyl radicals: in vitro and in vivo. Free Radic Biol Med. 1994;16:29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Marklund SL. Direct assay with potassium superoxide. In: Greenwald R, editor. Handbook of Methods for Oxygen Radical Research. Boca Raton, FL: CRC Press; 1985. pp. 249–255. [Google Scholar]
- Monroe JD, Salminen MD, Preiss J. Nucleotide sequence of a cDNA clone encoding a β-amylase from Arabidopsis thaliana. Plant Physiol. 1991;97:1599–1601. doi: 10.1104/pp.97.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsholm T, Sandberg G, Ericsson A. Quantitative analysis of amino acids in conifer tissues by HPLC and fluorescence detection of their 9-fluorenylmethyl chloroformate-derivates. J Chromatogr. 1987;396:225–236. [Google Scholar]
- Ogawa K, Kanematsu S, Asada K. Intra- and extra-cellular localization of cytosolic CuZn-superoxide dismutase in spinach leaf and hypocotyl. Plant Cell Physiol. 1996;37:790–799. [Google Scholar]
- Ogawa K, Kanematsu S, Asada K. Generation of superoxide anion and localization of CuZn-superoxide dismutase in vascular tissue of spinach hypocotyls: their association with lignification. Plant Cell Physiol. 1997;38:1118–1126. doi: 10.1093/oxfordjournals.pcp.a029096. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M, Ryan CA. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA. 1999;96:6553–6557. doi: 10.1073/pnas.96.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl-Treves R, Nacmias B, Aviv D, Zeelon EP, Galun E. Isolation of two cDNA clones from tomato containing two different superoxide dismutase sequences. Plant Mol Biol. 1988;11:609–624. doi: 10.1007/BF00017461. [DOI] [PubMed] [Google Scholar]
- Salin ML, Bridges SM. Chemiluminescence in wounded root tissue: evidence for peroxidase involvement. Plant Physiol. 1981;67:43–46. doi: 10.1104/pp.67.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel H, Streller S, Wingsle G. Multiple forms of extracellular superoxide dismutase in needles, stem tissues and seedlings of Scots pine. J Exp Bot. 1998;49:931–936. [Google Scholar]
- Scioli JR, Zilinskas BA. Cloning and characterization of cDNA encoding chloroplastic copper/zinc-superoxide dismutase from pea. Proc Natl Acad Sci USA. 1988;85:7661–7665. doi: 10.1073/pnas.85.20.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekizawa Y, Haga M, Hirabayashi E, Takeuchi N, Takino Y. Dynamic behavior of superoxide generation in rice leaf tissue infected with blast fungus and its regulation by some substances. Agric Biol Chem. 1987;51:763–770. [Google Scholar]
- Streller S, Karpinski S, Hällgren J-E, Wingsle G. Four cytosolic CuZn-superoxide dismutases in germinating seeds of Pinus sylvestris (L.) Physiol Plant. 1994;92:443–450. [Google Scholar]
- Streller S, Wingsle G. Pinus sylvestris (L.) needles contain extracellular CuZn superoxide dismutase. Planta. 1994;192:195–201. [PubMed] [Google Scholar]
- Süss KH, Arkona C, Manteuffel R, Adler K. Calvin cycle multienzyme complexes are bound to chloroplast thylakoid membranes of higher plants in situ. Proc Natl Acad Sci USA. 1993;90:5514–5518. doi: 10.1073/pnas.90.12.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Melzer M, Süss KH. Purification, properties and in situ localization of the amphibolic enzymes D-ribulose-5-phosphate 3-epimerase and transketolase from spinach chloroplasts. Eur J Biochem. 1998;252:237–244. doi: 10.1046/j.1432-1327.1998.2520237.x. [DOI] [PubMed] [Google Scholar]
- van Camp W, Bowler C, Villaroel R, Tsang EWT, van Montague M, Inzé D. Characterization of iron superoxide dismutase cDNAs from plants obtained by genetic complementation in E. coli. Proc Natl Acad Sci USA. 1991;87:9903–9907. doi: 10.1073/pnas.87.24.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Moriya H, Tsugita A. Development of an acid hydrolysis method with high recovery of tryptophan and cysteine for microquantities of protein. Biochemistry. 1991;198:1–5. doi: 10.1016/0003-2697(91)90496-g. [DOI] [PubMed] [Google Scholar]
- White JA, Scandalios JG. Isolation and characterization of a cDNA for mitochondrial manganese superoxide dismutase of maize and its relation to other superoxide dismutases. Biochem Biophys Acta. 1988;951:61–70. doi: 10.1016/0167-4781(88)90025-5. [DOI] [PubMed] [Google Scholar]
- Wingsle G, Gardeström P, Hällgren J-E, Karpinski S. Isolation, purification, and subcellular localization of isozymes of superoxide dismutase from Scots pine (Pinus sylvestris L.) needles. Plant Physiol. 1990;95:21–28. doi: 10.1104/pp.95.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingsle G, Karpinski S. Differential redox regulation by glutathione of glutathione reductase and CuZn-superoxide dismutase gene expression in Pinus sylvestris (L.) needles. Planta. 1996;198:151–157. doi: 10.1007/BF00197598. [DOI] [PubMed] [Google Scholar]
- Wingsle G, Mattson A, Ekblad A, Hällgren J-E, Selstam E. Activities of glutathione reductase and superoxide dismutase in relation to changes of lipids and pigments due to ozone in seedlings of Pinus sylvestris (L) Plant Sci. 1992;82:167–178. [Google Scholar]
- Zhou J, Rumeau D, Showalter AM. Isolation and characterization of two wound-regulated tomato extensin genes. Plant Mol Biol. 1992;20:5–17. doi: 10.1007/BF00029144. [DOI] [PubMed] [Google Scholar]