Abstract
Two cDNA clones (NtmybAS1 and NtmybAS2) encoding MYB-related proteins with strong sequence similarity to petunia (Petunia hybrida) PhMYB3 were isolated from a tobacco (Nicotiana tabacum cv Samsun) pollen cDNA library. Northern blot and in situ hybridization revealed that NtmybAS transcripts are specifically expressed in both sporophytic and gametophytic tissues of the anther including tapetum, stomium, vascular tissue, and developing pollen. Random binding site selection assays revealed that NtMYBAS1 bound to DNA sequences closely resembling consensus MYB binding sites MBSI and MBSIIG, with a higher affinity for MBSI. Transient expression analyses of the N-terminal MYB domain demonstrated the presence of functional nuclear localization signals, and full-length NtMYBAS1 was able to activate two different phenylalanine ammonia-lyase promoters (PALA and gPAL1) in tobacco leaf protoplasts. Similar analysis of truncated NtmybAS1 cDNAs identified an essential, C-terminal trans-activation domain. Further in situ hybridization analyses demonstrated strict co-expression of NtmybAS and gPAL1 in the tapetum and stomium. Despite abundant expression of NtmybAS transcripts in mature pollen, gPAL1 transcripts were not detectable in pollen. Our data demonstrate that NtMYBAS1 is a functional anther-specific transcription factor, which is likely to be a positive regulator of gPAL1 expression and phenylpropanoid synthesis in sporophytic, but not in gametophytic, tissues of the anther.
Male gametogenesis provides a tractable system to investigate cell differentiation, cell-cell interactions, and the regulation of cell-specific gene expression (McCormick, 1993; Twell et al., 1998). The male gametophytes, or pollen grains, of flowering plants develop within the anther and depend on complex interactions between sporophytic and gametophytic cells. The vital role of sporophytic anther tissues during early microgametogenesis is directly illustrated by experiments in which tapetal cells were genetically ablated (Mariani et al., 1990), and from the large number of male-sterile mutants arising from aberrant tapetal cell development (van der Meer et al., 1992; Chaudhury, 1993; Preuss, 1995; Aarts et al., 1997). In a similar manner, gametophytic expression of certain pollen and anther-specific genes is essential for pollen development and fertility (Muschietti, et al., 1994; Xu et al., 1995). Although a significant number of genes specifically expressed in anthers and/or pollen have been identified, details of their regulation in sporophytic and gametophytic tissues remain largely unknown (Twell, 1994).
Given the central role of transcription factors in the regulation of development, the identification of anther-/pollen-specific genes encoding transcription factors is an important step toward understanding regulatory mechanisms that operate during male gametogenesis. Several genes encoding putative transcription factors specifically expressed in anthers and/or pollen have been isolated; however, their functional activity and/or putative target genes remain to be demonstrated. These include pollen-specific genes encoding zinc finger proteins of the LIM class from sunflower and tobacco (Nicotiana tabacum cv Samsun; Baltz et al., 1992; Sweetman et al., 2000); anther and pollen-specific MADS box transcription factors, DEFH125 (Zachgo et al., 1997) and ZmMADS2 (Heuer et al., 2000), from snapdragon (Antirrhinum majus) and maize (Zea mays), respectively; and a family of anther-specific zinc finger proteins from petunia (Petunia hybrida; Kobayashi et al., 1998).
The myb gene family represents the largest transcription factor gene family in plants, with about 100 different myb genes identified in Arabidopsis (Kranz et al., 1998). The MYB domain, which is composed of two or three imperfect repeats of about 50 amino acids with a predicted helix-turn-helix motif, binds DNA in a sequence-specific manner. Plant MYB proteins typically bind to one or more of two types of cis-elements known as MYB binding sites (MBSI, MBSII, or MBSIIG.), the consensus sequences of which are CNGTTR, GKTWGTTR, and GKTWGGTR, respectively (where n = A/T/C/G, R = A/G, K = G/T, and W = A/T; Romero et al., 1998).
Plant MYB proteins have been shown to regulate diverse developmental processes, as well as being involved in environmental signaling and secondary metabolism (Lipsick, 1996; Jin and Martin, 1999). The most well-established role of plant MYB proteins, however, is in the control of genes in the phenylpropanoid biosynthetic pathway (Quattrocchio et al., 1993; Franken et al., 1994; Martin and Paz-Ares, 1997). For example, PhMYB3 from petunia (Solano et al., 1995), AmMYB305 from snapdragon (Sablowski et al., 1994; Moyano et al., 1996), PcMYB26 from pea (Uimari and Strommer, 1997), and ZmMYB1 from maize (Franken et al., 1994) regulate levels of anthocyanins and flavonoids in flowers, whereas ZmMYBPL from maize (Cone et al., 1993) performs a similar function in vegetative tissues. None of these myb genes, however, are known to be expressed in anthers and/or pollen.
Phenylpropanoids are known to play an important role in pollen fertility (for review, see Taylor and Hepler, 1997). Mutants deficient in chalcone synthase (CHS) expression in anthers produce flavonol-deficient pollen that is defective in germination or pollen tube growth (Mo et al., 1992; van der Meer et al., 1992). This defect can be rescued by the application of specific flavonols, which are thought to play a structural or signaling role in pollen tube growth (Ylstra et al., 1994; Napoli et al., 1999).
Based on the hypothesis that myb family members may play important roles in male gametogenesis, we screened a mature pollen cDNA library of tobacco for myb-related sequences and isolated two cDNA clones (NtmybAS1 and NtmybAS2) encoding MYB proteins. These two genes were expressed specifically in sporophytic and gametophytic tissues of tobacco anthers. We show that NtmybAS1 encodes a functional regulatory protein, which binds both MBSI and MBSIIG, with a higher affinity for MBSI, and to MBS-containing fragments of tobacco PAL promoters. Transcriptional activation of gPAL1 by NtMYBAS1 in leaf protoplasts and co-expression of NtmybAS1 and gPAL1 transcripts in young anther tissues strongly suggests that NtMYBAS1 is a regulator of PAL expression and phenylpropanoid biosynthesis in sporophytic tissues of the anther.
RESULTS
Cloning of NtmybAS1/2 cDNAs
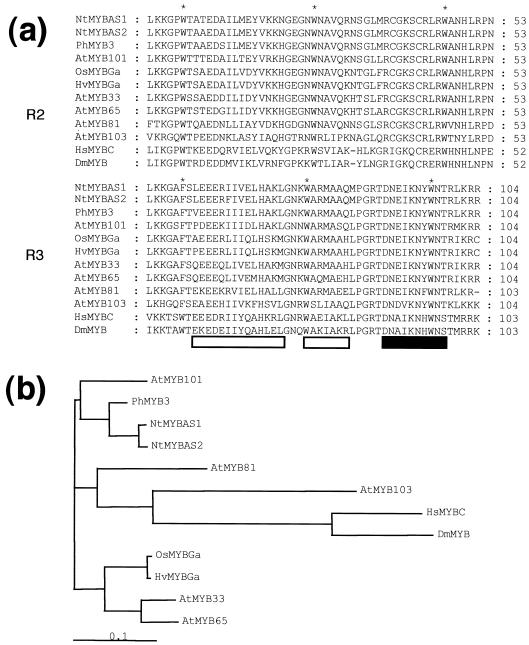
To identify putative pollen-expressed myb genes, we screened a tobacco mature pollen cDNA library with a mixture of two MYB domain consensus 42 mer oligonucleotides according to Jackson et al. (1991). Sequence comparison of the cDNA insert sequences of four positive clones revealed two unique cDNA sequences. NtmybAS1 contains a 1,409-bp open reading frame encoding a 470-amino acid protein with an estimated molecular mass of 52.2 kD, whereas NtmybAS2 contains a 1,421-bp open reading frame encoding a 474-amino acid protein with an estimated molecular mass of 52.7 kD. Comparison of NTMYBAS1 and NTMYBAS2 amino acid sequences revealed that they share 92% sequence identity overall. Within their two contiguous R2 and R3 MYB repeats, which together constitute the MYB domain, this figure rises to 98% (Fig. 1). Southern-blot analysis showed that both genes are single copy in tobacco, with the parental Nicotiana sylvestris and Nicotiana tomentosiformis genomes contributing one gene copy each (data not shown). Therefore, these genes are likely to be orthologues.
Figure 1.
Comparison of the amino acid sequence of NtMYBAS1/2 and PhMYB3. The R2 and R3 repeats of the MYB domains are boxed. Arrowheads correspond to conserved Trp residues in the MYB domain. The MYB protein subgroup 18 consensus motif (QRaGLPxYPxE/S; Kranz et al., 1998) is highlighted in bold. Identical amino acids are indicated below with asterisks.
The NtMYBAS1/2 proteins form a tightly related group with the petunia PhMYB3 protein (Avila et al., 1993). The 92% sequence identity in the R2 and R3 MYB repeats between NtMYBAS1/2 and PhMYB3 might suggest that they bind similar sequences. In addition to the N-terminal MYB domain, their C-terminal domains also shows regions of high identity (Fig. 1, marked by asterisks). Regions of high similarity exist between NtMYBAS1/2 and PhMYB3 in three potential trans-activation domains (amino acids 326–347, 357–378, and 413–432 in NtMYBAS1 and amino acids 330–351, 355–384, and 417–436 in NtMYBAS2) by virtue of their acidity and potential to be α-helical. Sequence similarity in this region of the proteins could indicate that NtMYBAS1/2 share a similar biological function to PhMYB3, which has been proposed to regulate CHS expression and thereby flavonoid biosynthesis in petunia petals (Avila et al., 1993; Solano et al., 1995).
According to Kranz et al. (1998), the MYB proteins in Arabidopsis were classified into 22 subgroups. NtMYBAS1/2 proteins belong to subgroup 18, which share the QRaGLPxYPxE/S motif outside the MYB domain (Fig. 1, bold characters). Aligning the R2 and R3 MYB repeats of NtMYBAS proteins with other MYB proteins, the three conserved Trp residues separated by 18 to 19 amino acids implicated in DNA binding (Saikumar et al., 1990; Ogata et al., 1994) are present in NtMYBAS1/2 sequences (Fig. 2a, marked as asterisks). As in other plant MYB domains, the first Trp of the R3 repeat in the NtMYBAS1/2 proteins is replaced by a Phe.
Figure 2.
Alignment (a) and dendrogram (b) of the deduced amino acid sequence of the MYB domain of NtMYBAS1/2 with those of related MYB proteins. Gene, genus nomenclatures, and accession nos. are as follows: NtMYBAS1, tobacco, AF198499; NtMYBAS2, tobacco, AF198498; PhMYB3, petunia, Z13998; AtMYB101, Arabidopsis, X90379; OsMYBGa, Oryza sativa, X98355; HvMYBGa, Hordeum vulgare, X87690; AtMYB33, Arabidopsis, AF062875; AtMYB65, Arabidopsis, AF062899; AtMYB81, Arabidopsis, AF062911; AtMYB103, Arabidopsis, AF048839; HsMYBC, Homo sapiens, M15024; and DmMYB, Drosophila melanogaster, X05939. Asterisks correspond to positions of conserved Trp residues. Boxes beneath the sequences indicate the three helices in each repeat including the recognition helix (black box). Sequence alignment and dendrogram were performed by the neighbor-joining algorithm of CLUSTALW, where the scale bar represents the degree of base pair substitution per million years (Thomson et al., 1994).
In Figure 2b, a dendrogram represents the degree of protein sequence similarity based on the alignments in Figure 2a. The NtMYBAS1/2 proteins are predicted to be relatively recently diverged from each other, and to cluster tightly with the PhMYB3 sequence relative to other plant sequences. HsMYBC and DmMYB from Homo sapiens and Drosophila melanogaster, respectively, form a separate deeply rooted branch in addition to the AtMYB101 and AtMYB81 proteins in this group (Kranz et al., 1998).
NtmybAS Expression Is Developmentally Regulated during Male Gametogenesis
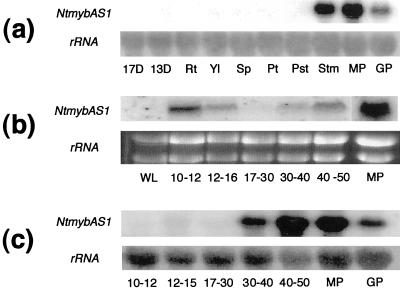
Given the likelihood of NtMYBAS1 and NtMYBAS2 being orthologous, we continued our investigation with NtMYBAS1 only. The organ-specific expression pattern of NtmybAS1 revealed by northern-blot analysis showed high expression of a 1.8-kb transcript in stamens and mature and germinating pollen (Fig. 3a). In contrast, young seedlings and other vegetative or floral organs did not show expression of NtmybAS1 transcripts.
Figure 3.
mRNA expression analysis of NtmybAS. Northern blots of tobacco total RNA probed with radiolabeled NtmybAS1 probe. a, Spatial expression. 13D and 17D, Seedlings 13 and 17 d after germination; Rt, root; Yl, young leaf; Sp, sepal; Pt, petal; Pst, pistil; Stm, stamen; MP, mature pollen; GP, germinating pollen. b, Temporal expression in anthers. WL, Wild- type leaf; MP, mature pollen. Flower bud lengths shown in millimeters. c, Temporal expression in isolated spores. MP, Mature pollen; GP, germinating pollen.
To determine the temporal expression pattern of NtmybAS transcripts, total RNA was extracted from anthers at five developmental stages (10–12-mm, 12–16-mm, 17–30-mm, 30–40-mm, and 40–50-mm bud lengths) and from mature pollen. NtmybAS1 transcripts were strongly detectable at early and late stages of anther development with maximum levels in mature pollen (Fig. 3b).
To investigate the gametophytic expression pattern of NtmybAS1, total RNA was extracted from purified populations of microspores and pollen from five different stages of development. NtmybAS1 transcripts were first detectable in late uninucleate microspores (12–15-mm buds). After pollen mitosis I (17–30-mm buds), the level of NtmybAS1 transcripts decreased to a basal level. Transcript abundance subsequently increased in mid- (30–40-mm buds) and late (40–50-mm buds) bicellular pollen, and decreased in germinating pollen. Therefore, NtmybAS1 shows a typical gametophytic pattern of expression similar to late pollen-specific genes in tobacco (Twell, 1994; Sweetman et al., 2000).
NtmybAS Transcripts Are Expressed in Sporophytic and Gametophytic Anther Tissues
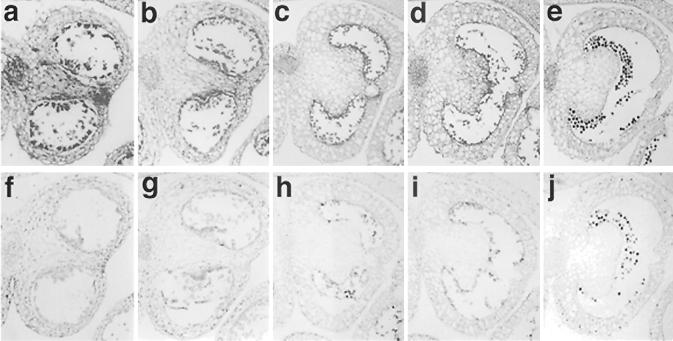
To define the cellular expression pattern of NtmybAS1, in situ hybridization was carried out on sections of flower buds from five developmental stages (8–9 mm, 10–12 mm, 13–15 mm, 16–20 mm, and 21–30 mm in length). The 243 bp (position 1,345–1,587) of NtmybAS1 cDNA was used as a probe.
The antisense NtmybAS1 probe produced strong signals in sporophytic tissues, including vascular tissue, stomium, and tapetum, at early stages of development (Fig. 4a). As the anthers matured there was a decrease in sporophytic and an increase in gametophytic (pollen) hybridization (Fig. 4, b–e). The reduced sporophytic hybridization coincided with the degeneration of the tapetum and stomium. No significant hybridization signals were observed using the NtmybAS1 sense probe except for mature pollen, which showed lower background staining (Fig. 4, f–j).
Figure 4.
Localization of NtmybAS mRNA during tobacco anther development. Paraffin-embedded sections were hybridized with digoxygenin-labeled NtmybAS1 probes and viewed under bright field. Scale bar = 400 μm. Magnification ×100 in 8- to 9-mm and 10- to 12-mm buds; and ×40 in 13- to 15-mm, 16- to 20-mm, and 21- to 30-mm buds. a through e, NtmybAS1 antisense probe with anthers from: a, 8 to 9 mm; b, 10 to 12 mm; c, 13 to 15 mm; d, 16 to 20 mm; and e, 21- to 30-mm buds. Staining represents hybridization signal. f through j, NtmybAS1 sense probe with anthers from: f, 8 to 9 mm; g, 10 to 12 mm; h, 13 to 15 mm; i, 16 to 20 mm; and j, 21- to 30-mm buds.
The hybridization signals at each stage were directly correlated with transcript levels observed in the northern analyses of anthers and isolated spores (Fig. 3). These data clearly demonstrate early sporophytic and late gametophytic expression of NtmybAS1 in developing anthers.
The N-Terminal MYB Domain Efficiently Targets Green Fluorescent Protein (GFP) to the Nucleus
The N-terminal MYB domain of NtMYBAS1 contains a basic motif (119-RLKRRQR-125) resembling a monopartite nuclear localization signal. To determine whether the MYB domain containing this motif plays a role in nuclear import of NtMYBAS1 in vivo, pollen was bombarded with a control plasmid, pLAT52-sGFP, and a plasmid, pLAT52-sGFPM1, harboring the N-terminal 126 amino acids of NtMYBAS1 fused to sGFP. When sGFP was expressed alone (pLAT52-sGFP), the majority of the fluorescence signal was localized in the pollen cytoplasm (Fig. 5a). In contrast, the recombinant protein (sGFPM1) was exclusively localized to the pollen nucleus, with some localized accumulation in the nucleolus (Fig. 5b). We conclude that the first 126 amino acids of NtMYBAS1 are sufficient to target and retain sGFP in the nucleus.
Figure 5.
Nuclear localization of NtMYBAS1-sGFP fusion proteins. Tobacco pollen grains bombarded with pLAT52-sGFP control plasmid (a) and pLAT52-sGFPM1-3 plasmids harboring NtMYBAS1-sGFP fusion constructs (b–d). a, The control, sGFP alone, remains predominantly in the cytoplasm. b, pLAT52-sGFPM1. N-terminal 126 amino acids of NtMybAS1 result in localization of sGFP exclusively to the pollen nucleus. c, pLAT52-sGFPM2. N-terminal 119 amino acids of NtMybAS1 result in a dramatic reduction in nuclear localization. d, pLAT52-sGFPM3. The MTRLKRRQRA motif alone is not sufficient for exclusive nuclear targeting of sGFP. R2R3, MYB repeats; TEV-L, tobacco etch virus 5′-untranslated region; C3′, cauliflower mosaic virus 35S transcript polyadenylation sequence; n, NcoI; B, BspHI; NB, NcoI/BsphI junction.
To investigate the role of specific sequences in nuclear localization within this N-terminal domain, the localization of two additional mybAS1-sGFP fusion proteins were examined. Specific removal of the RLKRRQR motif from the N-terminal Myb domain (pLAT52-sGFPM2) severely reduced nuclear localization of sGFP, demonstrating its involvement in nuclear targeting (Fig. 5c). However, enhanced accumulation of sGFPM2 in the nucleus relative to sGFP alone suggests that the N-terminal domain (amino acids 1–119) retains partial nuclear localization function. In a similar manner, the RLKRRQR motif domain alone (pLAT52-sGFPM3) was not sufficient to exclusively direct sGFP to the nucleus, although enhanced accumulation of sGFPM3 was observed in the nucleus and nucleolus (Fig. 5d). These results demonstrate that the RLKRRQR motif is necessary but not sufficient for nuclear localization, and suggest that additional upstream sequences within the N-terminal Myb domain are also required for efficient nuclear targeting.
NtMYBAS1 Binds to MBS Motifs
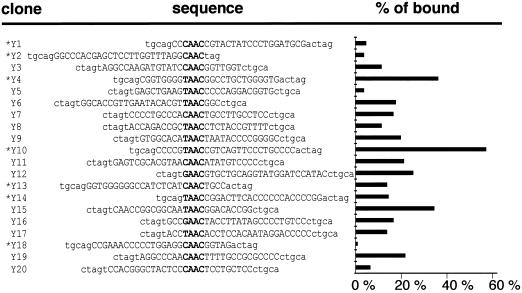
To determine NTMYBAS1 DNA-binding specificity, random binding site selection experiments were performed with a C terminally truncated derivative, NtMYBASΔC1. This truncated protein (amino acids 1–208) containing the intact MYB domain, was expressed as an N-terminal poly-His fusion protein in Escherichia coli. NtMYBASΔC1 was incubated with oligonucleotides containing a random 23-bp core and was separated from unbound oligonucleotides by electrophoretic mobility shift assay (EMSA). After six rounds of screening, bound oligonucleotides were cloned and sequenced.
All of the oligonucleotides selected by NtMYBASΔC1 contained at least one AAC core motif characteristic of MYB binding sites (Fig. 6). The deduced consensus, (T/C) AACNGC, is similar to MBSI, (T/C) AAC(G/C) G, which is bound by animal MYB and some plant MYB proteins (Romero et al., 1998). NtMYBASΔC1 bound to these selected oligonucleotides with differing affinities in separate EMSAs (Fig. 6), with Y10 and Y18 showing the highest and lowest percentages of bound oligonucleotide, respectively. Because all oligonucleotides contain the same AAC core motif, this suggests that sequence context is an additional factor affecting binding affinity.
Figure 6.
Oligonucleotide sequences selected by NtMYBASΔC1 and relative binding affinity. Sequences of 20 oligonucleotides isolated after six rounds of selection with NtMYBASΔC1. The core binding sequence is in bold; lowercase letters represent the conserved oligonucleotide ends. Asterisks indicate that the sequence has been inverted. The percentage of bound oligonucleotide was quantified using a phosphorimager and expressed as the percentage of bound/total radioactivity in each lane.
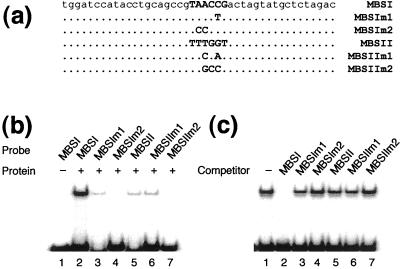
To confirm the binding site specificity of NtMYBASΔC1, we tested the ability of NtMYBAS1 to bind to oligonuclecotides containing either MBSI, or MBSIIG, and to mutated forms of these sequences. NtMYBASΔC1 bound strongly to MBSI oligonucleotides containing the sequence TAACCG (Fig. 7b). Extensive mutation of this sequence to TCCCCG in MBSIm2 abolished binding (Fig. 7b, lane 4), whereas changes outside the core sequence to TAACTG as in MBSIm1 resulted in a low level of binding (Fig. 7b, lane 3). This demonstrates that the core MYB binding site motif, AAC, is critical for NtMYBAS binding.
Figure 7.
EMSA with purified recombinant NtMYBASΔC1 protein. a, The sequences of double-stranded oligonucleotides used as probes. b, The NtMYBASΔC1 protein binds preferentially to MBSI (GTAACCG). The binding reaction fractionated in the first lane did not contain NtMYBASΔC1, whereas those in other lanes contained 200 ng NtMYBASΔC1. c, Competition for binding of NtMYBASΔC1 to the MBSI probe. The binding assay was performed by pre-incubating with unlabeled competitor before addition of 1 ng of 32P-labeled MBSI probe. The reaction fractionated in lane 1 contained no competitor, whereas those in lanes 2 through 7 contained 200 ng of competitor.
NtMYBASΔC1 also bound to the sequence GTTTGGT, an MBSIIG motif (Fig. 7b, lane 5). Extensive mutation of this sequence as in MBSIIm2 (TTGCCT) abolished binding (Fig. 7b, lane 7), whereas a 2-bp mutation to TTCGAT in MBSIIm1 had no effect (Fig. 7b, lane 6). The intensity of the shifted band in Figure 7b implies that NtMYBASΔC1 can bind both types of MBS sequences, but with a preference for MBSI. To further demonstrate preferential binding of NtMYBASΔC1 to MBSI, competition experiments were performed. Binding of NtMYBASΔC1 to MBSI was effectively reduced by the addition of unlabeled MBSI (Fig. 7c). In contrast, competition with MBSII, and with mutated derivatives of MBSI and MBSII, had little or no effect on binding.
NtMYBAS1 Binds Tobacco PAL Promoters in Vitro
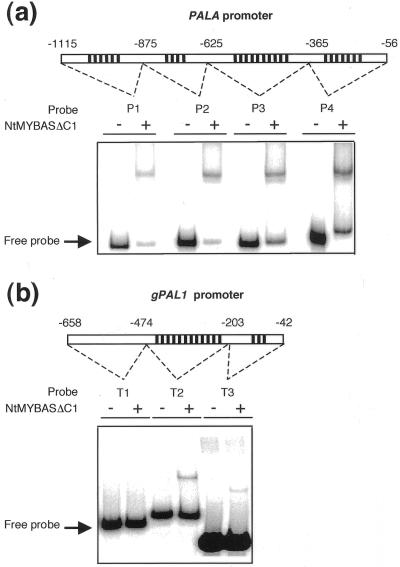
Because several MYB proteins are known to play important roles in transcriptional regulation of phenylpropanoid biosynthetic genes (Martin and Paz-Ares, 1997) including trans-activation of the Phe ammonia lyase (PAL) promoter (Sablowski et al., 1994), we postulated that NtMYBAS1 similarly may regulate PAL expression specifically in anthers. To test whether NtMYBAS1 was capable of binding to a PAL gene promoter, EMSA was performed with two tobacco PAL genes, PALA (Pellegrini et al., 1994) and gPAL1 (Tomoko et al., 1996), which contain MBSs in their promoters. Four fragments (P1, P2, P3, and P4) of the PALA promoter, each containing multiple MBSs, were bound by NtMYBASΔC1 (Fig. 8a). Two of the gPAL1 promoter fragments (T2 and T3) were also bound, whereas T1, which has no MBSs, failed to produce a shifted band (Fig. 8b). We conclude that NtMYBAS1 specifically binds tobacco PAL promoter regions containing MBSs.
Figure 8.
EMSA with plant promoter fragments. Binding of the NtMYBASΔC1 to PALA (a) and gPAL1 promoter fragments (b). The locations of the fragments in the promoters relative to the transcriptional start site are given as follows: PALA P1, −1,115 to −876; P2, −875 to −626; P3, −625 to −366; P4, −365 to −356; gPAL1 T1, −658 to −475; T2, −474 to −204; and T3, −203 to −42. ψ represents the presence of MBSs.
NtMYBAS1 Activates Transcription of PAL Promoters in Vivo
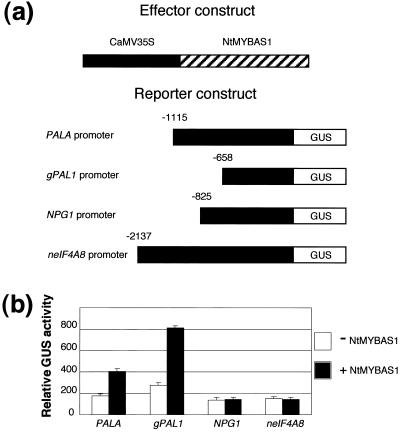
To investigate whether the NtMYBAS1 protein could transcriptionally activate PAL, we cotransfected tobacco leaf protoplasts with an effector plasmid (35S::NtMYBAS1) harboring NtmybAS1 under the control of CaMV35S promoter and four different reporter plasmids. Reporter plasmids contained the tobacco PALA (Pellegrini et al., 1994) or gPAL1 (Tomoko et al., 1996) promoters, and two tobacco pollen-specific promoters, NPG1 (Tebbutt et al., 1994) and neIF4A8 (Brander and Kuhlemeier, 1995) fused to the GUS reporter gene. NtMYBAS1 specifically trans-activated both the PALA and gPAL1 promoters, whereas NPG1 and neIF4A8 promoters were not activated above basal levels of the reporter plasmids alone (Fig. 9b).
Figure 9.
Trans-activation of tobacco promoters by NtMYBAS1. a, Effector and reporter constructs used in cotransfection experiments. The 35S::NtMYBAS1 effector plasmid consisted of the coding region of NtmybAS1 under control of the CaMV 35S promoter. The reporter plasmids consist of two tobacco PAL promoters, PALA (Pellegrini et al., 1994) and gPAL1 (Tomoko et al., 1996); tobacco polygalacturonase NPG1 promoter (Tebbutt et al., 1994) and tobacco neIF4A8 promoter (Brander and Kuhlemeier, 1995) fused to the GUS reporter gene. b, Response of reporter constructs to NtMYBAS1 in transient expression assays. Tobacco leaf protoplasts were cotransfected with each reporter plasmid together with 35S::NtMYBAS2. Error bars represent se (n > 7).
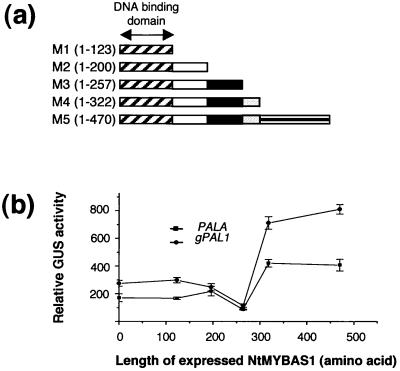
To identify the putative trans-activation domain within NtMYBAS1, we constructed five effector plasmids (M1–M5), harboring either the full-length or C-terminal truncations of NtMYBAS1 (Fig. 10a). These were tested for their ability to trans-activate a GUS reporter gene under the control of the two tobacco PAL promoters. The truncated protein M3 (amino acids 1–257) produced a significant decrease in the trans-activation of both PAL promoters, whereas truncation M4 (amino acids 1–322) did not affect trans-activation (Fig. 10b). Thus, sequences between 257 and 322 define an important transcriptional activation domain.
Figure 10.
Effect of C-terminal deletions on NtMYBAS1 trans-activation of PAL genes. a, The NtmybAS1 cDNAs containing deleted regions, as indicated by numbers of amino acid residues, were inserted between the CaMV 35S promoter and the NOS terminator. Effector plasmids were cotransfected into tobacco leaf protoplasts with reporter plasmids, containing gPAL1 and PALA, respectively. b, Transfection and assay of the reporter activity are as in Figure 9. The error bars represent the se (n > 10).
gPAL1 Expression Coincides with NtmybAS1 Expression in Anthers
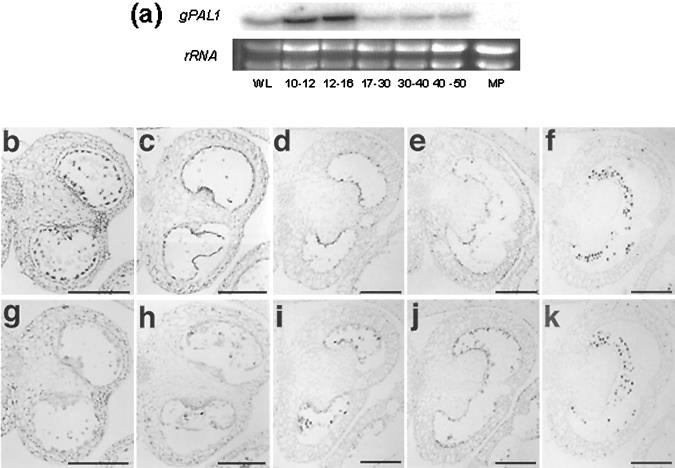
From the DNA binding assays and transient expression analyses carried out it is possible to propose NtMYBAS1 as a regulator of PAL expression in anthers. To investigate this hypothesis, we studied the expression pattern of PAL in anthers and pollen. A northern blot previously probed with NtmybAS1 (Fig. 3b) was reprobed with a gPAL1 probe. gPAL1 was strongly expressed during the same early stages of anther development at which NtmybAS1 transcripts were highly expressed (Fig. 11a). The transcript abundance in 10- to 16-mm buds was higher than in leaves and at other stages of anther development (Fig. 11a). In contrast to NtmybAS1, there was no detectable gPAL1 expression in pollen.
Figure 11.
Expression of gPAL1 mRNA during tobacco anther development. a, Temporal northern analysis of gPAL1 expression in tobacco anthers. WL, Wild-type leaf; MP, mature pollen. Flower bud lengths shown in millimeters. b through k, In situ hybridization with gPAL1 antisense (b–f) and sense (g–k) probes. Bud lengths: b and g, 8 to 9 mm; c and h, 10 to 12 mm; d and i, 13 to 15 mm; e and j, 16 to 20 mm; f and k, 21 to 30 mm. Black bar = 400 μm. b, c, g, and h, magnification ×100; d through f and i through k, magnification ×40.
In situ hybridization of anther sections with a gPAL1-specific probe (Fig. 11, b–k) revealed sporophytic expression in stomium and tapetum at early stages (8–9 mm) of anther development, in which NtmybAS1 is highly active (Fig. 4). At later stages, gPAL1 signal was absent from the stomium, but still detectable in the tapetum (Fig. 11c), before decreasing upon tapetal degeneration (Fig. 11, d–f). In contrast, gPAL1 hybridization was not detected in gametophytic tissues. These data demonstrate that NtmybAS1 and gPAL1 are co-expressed in sporophytic tissues of the anther, but only NtmybAS1 is expressed in maturing pollen.
DISCUSSION
We have isolated two cDNAs clones (NtmybAS1 and NtmybAS2) from tobacco encoding MYB-related proteins, expressed specifically in sporophytic and gametophytic anther tissues. The 98% amino acid sequence identity between NtMYBAS1 and NtMYBAS2 within their N-terminal DNA binding domains suggests that their biological roles may be similar. Furthermore, based on the finding that in tobacco both NtmybAS1/2 are single copy, and inherited from the genome of each parental line, we speculate that these genes are likely to be orthologous. When compared with all other available MYB protein sequences, NtMYBAS1/2 show the highest similarity to petunia PhMYB3. According to the classification of MYB proteins from Arabidopsis (Kranz et al., 1998), this places NtMYBAS1/2 in subgroup 18, although the closest MYB family members in Arabidopsis are relatively divergent. PhMYB3 is expressed in petal epidermal cells and is able to trans-activate the promoter from a CHS gene, suggesting an involvement in the regulation of flavonoid biosynthesis (Solano et al., 1995). The function of the other MYB proteins in this group remains unknown.
Northern-blot analysis revealed that NtmybAS transcripts are initially abundant in young anthers when the tapetum is intact and subsequently decrease in line with tapetal cell degeneration (Koltunow et al., 1990). However, NtmybAS transcripts also accumulate gametophytically in mature and germinating pollen similar to the “late class” of pollen-expressed genes (Mascarenhas, 1990; Twell, 1994). In situ hybridization revealed strong sporophytic expression in tapetum, stomium, and vascular tissues of young anthers, and confirmed abundant expression in maturing pollen. These data suggest that NtMYBAS1/2 are likely to regulate gene expression in both sporophytic anther tissues and in maturing pollen.
Several anther- and pollen-specific genes encoding putative transcription factors have been isolated(Baltz et al., 1992; Zachgo et al., 1997; Chung et al., 1998; Kobayashi et al., 1998; Schiefthaler et al., 1999; Yang et al., 1999; Heuer, et al., 2000), including a tapetum-specific myb-related gene from Arabidopsis (Li et al., 1999). NtmybAS1/2 appear to be unique in that they show both sporophytic and gametophytic expression. However, there are several examples of anther-specific genes, which show this overlap in expression. For example, Bcp1 is expressed in tapetum and microspores and is functionally important in both cell types (Xu et al., 1995). Several lipid biosynthetic genes are also expressed both in tapetal cells and pollen grains (Piffanelli et al., 1997). Therefore, such sporophytic/gametophytic overlap of gene expression in the anther may be regulated by transcription factors that are expressed in the same manner as NtMYBAS1/2.
The N-Terminal Myb Domain of NtmybAS1 Contains a Functional Nuclear Localization Signal (NLS)
Localization of plant Myb proteins in the nucleus has previously been shown by immunocytochemical analysis for petunia PhMyb3 (Avila et al., 1993) and by transient expression for Arabidopsis CCA1 (Wang et al., 1997). However, in neither case has the signal responsible for nuclear targeting been identified.
Transient expression analysis demonstrated that the N-terminal 126 amino acids of NTMYBAS1 localized sGFPM1 exclusively to the pollen nucleus. A putative NLS (119-RLKRRQR-125) in NtMybAS1 resembling monopartite NLSs (Raikhel, 1992; Merkle and Nagy, 1997) is present within this region. Deletion of the RLKRRQR motif from this region (sGFPM2) severely reduced, but did not abolish, the nuclear localization of sGFP, demonstrating that this motif is necessary for efficient nuclear import. Analysis of surface probability (MacVector, Oxford Molecular Group PLC, Oxford) revealed that the Arg at position 122 (within the 119-RLKRRQR-125 motif) is predicted to have the highest surface probability along the length of NTMYBAS1. This implies that the six amino acids surrounding this Arg have the highest probability of being exposed and recognized by the nuclear import machinery.
The partial nuclear accumulation of sGFPM2 suggests the presence of additional sequences that contribute to nuclear localization. Further analysis demonstrated that the RLKRRQR motif alone also was not sufficient for exclusive nuclear localization of sGFP, but showed enhanced accumulation in the nucleolus. These data suggest that the NTMYBAS1 NLS is not simply monopartite but depends on the cooperative function of the RLKRRQR motif with an upstream element in the DNA binding domain. The cooperative functions of bipartite and multiple NLSs have been reported for the potyviral NIa protein (Carrington et al., 1991) and B-Myb (Takemoto et al., 1994). Two separate NLSs, a monopartite (NLS1) in the central region and a bipartite (NLS2) in the C-terminal basic region were shown to be necessary for efficient nuclear targeting of B-Myb (Takemoto et al., 1994).
It is interesting that the basic RLKRRQR motif resembles the nucleolar targeting signals of the RNA-binding rat ribosomal protein L31 (RLSRKR; Quaye et al., 1996). Furthermore, Arg-rich motifs are known to bind RNA and their binding affinity is dramatically enhanced by the presence of a single Gln (Q) or Asn (N; Tan and Frankel, 1998). This raises the possibility that the partial localization of sGFPM3 (containing the RLKRRQR motif alone) to the nucleolus may be mediated by a similar mechanism via RNA binding.
NtMYBAS1 as a Regulator of gPAL1 Expression
DNA binding assays showed that NtMYBAS1 binds preferentially to a type I MYB binding site, but that it could also bind weakly to a type IIG site (Romero et al., 1998). Random binding site selection has been successful in deriving consensus target sequences for MYB proteins, which are very similar to their presumptive native targets (Solano et al., 1995; Gubler et al., 1999). Therefore, the results described here may reflect the in vivo binding properties of NtMYBAS1.
PAL gene promoters typically contain multiple potential MBS elements, which are important for expression of these genes (Hatton et al., 1996). NtMYBAS1 was shown to bind specifically to fragments from two tobacco PAL gene promoters that contain MBSI and MBSII sites. In addition, NtMYBAS1 was able to trans-activate the same PAL gene promoters in tobacco protoplasts, whereas it failed to activate two tobacco pollen-specific promoters. Transient expression of truncated NtMYBAS1 derivatives revealed an internal region (amino acids 257–322) necessary for trans-activation. In contrast, other MYB proteins have been shown to contain acidic C-terminal domains that are required for trans-activation (Facchinetti et al., 1997; Gubler et al., 1999). The internal trans-activation region in NtMYBAS1 is not acidic and is not predicted to form α-helix. It is interesting that this region contains putative protein kinase C (TTR, amino acids 264–266) and caseine kinase C phosphorylation sites (SIPE, amino acids 293–296; TSSD, amino acids 305–308; and SSDD, amino acids 306–309), which could be important posttranslational regulatory sites. These results collectively suggest NtMYBAS1 is a good candidate for a positive regulator of PAL transcription in tobacco anthers.
NtMYBAS1/2 are most closely related to PhMYB3 from petunia, which binds both MBSI and MBSII sites and weakly to MBSIIG (Solano et al., 1995, 1997). The evidence thus far suggests PhMYB3 to be involved in phenylpropanoid metabolism, possibly in anthocyanin biosynthesis due to its ability to trans-activate a CHS gene promoter in tobacco protoplasts. We have not tested the ability of NtMYBAS1/2 to trans-activate the CHS promoter, nor has PhMYB3 been tested against a PAL promoter, but this may be evidence for structurally similar proteins from different species not necessarily activating the same target genes. This situation is true for the highly conserved MYBs ZmMYBC1 and PhMYBAN2, which have been shown genetically to control different genes in maize and petunia, respectively (Quattrochio et al., 1999). This also may not be too surprising given that the introduction of a single amino acid change in the R3 repeat of PhMYB3 has the effect of changing the binding site preference from MBSI and MBSII, to MBSI alone in the mutated form (Solano et al., 1997). On the other hand, structurally dissimilar MYB proteins can bind to similar DNA sequences (Solano et al., 1997; Romero et al., 1998) and therefore could have potentially similar target genes and functions.
NtMYBAS1 is not the only MYB protein to be shown to trans-activate a PAL gene in vivo. AmMYB305 from snapdragon, an evolutionarily distant MYB protein to NtMYBAS1/2, will trans-activate a PAL gene promoter in tobacco protoplasts (Sablowski et al., 1994). AmMYB305 is expressed in carpels of young snapdragon flowers but unlike NtMYBAS1/2 preferentially binds to MBSII and MBSIIG sites in vitro and not to MBSI (Solano et al., 1997; Romero et al., 1998). AmMYB305 can also trans-activate two other genes involved in flavonoid metabolism (Moyano et al., 1996). The R2R3 myb gene family in plants is a large complex one, with an estimated 100 members in Arabidopsis (Kranz et al., 1998; Romero et al., 1998) and more than 80 in maize (Rabinowicz et al., 1999). The function of most of these proteins and therefore the reason for such diversity is at present unknown (Jin and Martin, 1999).
gPAL1 Expression in Tapetum as a Regulator of Phenylpropanoid Synthesis
Several lines of evidence demonstrate that specific flavonols derived from the phenylpropanoid pathway play an essential role in pollen fertility (for review, see Taylor and Hepler, 1997). PAL, which catalyzes the first committed step in the phenylpropanoid pathway, is an important regulatory point (Hahlbrock et al., 1976; Bate et al., 1994). PAL activity in the tapetum therefore is expected to be an important regulator of flavonol synthesis in the anther. In this regard, PAL enzyme activity was maximal in immature anthers and immunolocalization showed that PAL was predominantly localized to the tapetal cells. PAL activity in immature anthers has also been positively correlated with pollen fertility in male-fertile and cytoplasmic male-sterile strains of Brassica oleracea (Kishitani et al., 1993). Furthermore, tapetum-specific sense and antisense expression of a sweet potato PAL cDNA in tobacco resulted in partial male sterility and abnormal pollen grains devoid of flavonols (Matsuda et al., 1996). In sharp contrast, flavonoids do not play an essential role in pollen fertility in Arabidopsis (Burbulis et al., 1996), so the flavonoid requirement for male gametophyte development can differ between species.
Given the importance of PAL in providing phenylpropanoid precursors for flavonol synthesis in the tapetum, identification of transcriptional regulators of PAL is equally important. The demonstration of binding of NtMYBAS1 to PAL promoters in vitro and trans-activation of PAL-GUS in protoplasts strongly supports the hypothesis that NtMYBAS1 may be a regulator of PAL expression and therefore phenylpropanoid biosynthesis in tobacco anthers.
The colocalization of NtmybAS and gPAL1 in the tapetum at early stages of anther development provides further compelling evidence for the role of NtMYBAS1 as an activator of gPAL1 expression in the tapetum. The onset of repression of gPAL1 expression in anthers at mid-bicellular stage anthers coincided precisely with the suppression of NtmybAS expression and the decay of the tapetal cells. However, the strong induction of NtmybAS mRNA in pollen at later stages of anther development further suggests that NtMYBAS1/2 might perform other regulatory functions in mature pollen.
Based on the proposal that NtMYBAS1 acts as a positive regulator of gPAL1, we can postulate potential mechanisms that could account for the specific activation of gPAL1 in sporophytic anther tissues. First, transcription of gPAL1 in sporophytic tissues may be mediated by binding of NtMYBAS1 to MBSs in the gPAL1 promoter. In gametophytic cells, including mature pollen, suppression of gPAL1 may result from the presence of a repressor binding either to the gPAL1 promoter or to NtMYBAS1. Because some MYB proteins are known to cooperate with other transcription factors (Tice-Baldwin et al., 1989; Burk et al., 1993), NtMYBAS1-mediated induction of gPAL1 in sporophytic tissues could also be indirect via a second factor, and the absence of this factor in pollen could prevent gPAL1 transcription. Posttranslational modifications such as phosphorylation and acetylation, which are known to regulate MYB protein activity, could also modulate NtMYBAS1 binding to the gPAL1 promoter in a tissue-specific manner (Johnson et al., 1999; Tomita et al., 2000). Among plant MYB proteins CCA1 has been shown to interact with, and is phosphorylated by, a regulatory subunit (CKB3) of the Ser/Thr protein kinase CK2 (Sugano et al., 1998).
Confirmation of the primary role of NtMYBAS1 as a positive regulator of PAL expression in sporophytic tissue of the anther and a demonstration of its other regulatory functions in the anther and pollen requires further work. One approach to this will be to examine the effect of tissue-specific reduction of NtMYBAS expression in tapetum and pollen and to map the structurally important cis-elements in the tobacco gPAL1 promoter.
MATERIALS AND METHODS
Plant Materials
Tobacco (Nicotiana tabacum cv Samsun) plants were grown under standard greenhouse conditions (16-h day, 18°C–28°C).
cDNA Library Construction and Screening
RNA isolation and construction of a UniZAPII cDNA library from mature pollen poly(A+) RNA of tobacco were carried out as previously described (Sweetman et al., 2000). Approximately 4 × 105 pfu were screened using a mixture of two 42 mer oligonucleotides, O1 and O2 (Jackson et al., 1991). Oligonucleotide labeling and hybridization procedures performed as previously described (Jackson et al., 1991).
DNA Sequencing and Analysis
Sequencing was performed using the automated ABI Prism procedure (Applied Biosystems, San Jose, CA). Sequence data was analyzed using the Sequence Editor v 1.0.3, GeneJockey II (Biosoft, Cambridge, UK) and protein secondary structure predictions were compiled using the Chou Fasman and Robson Garnier algorithm (MacVector, Oxford Molecular Group PLC).
The nucleotide sequences of cDNA clones NtmybAS1 and NtmybAS2 are available in GenBank as accession nos. AF198499 and AF198498, respectively. Clones are freely available from the corresponding author (twe@le.ac.uk) for non-commercial research purposes subject to a material transfer agreement with the University of Leicester.
Northern and Southern Analysis
Genomic DNA and total RNA isolations, gel-blot studies, and hybridization were carried out as previously described (Sweetman et al., 2000).
RNA in Situ Hybridization
For in situ hybridization, probes were labeled with digoxigenin-11-UTP using a nucleic acid labeling kit (Boehringer, Indianapolis). The 243-bp (position 1,345–1,587) NtmybAS1 cDNA was amplified using two oligonucleotide primers, 5′-GGATGCCACCCCAATAAAGCACTCTGAA-3′ and 5′ GATTTTGAGCAATGTGATTGTATTTGGT-3′. The 229-bp (position 85–313) gPAL1 cDNA was amplified using two oligonucleotide primers, 5′-GCTGAATCCTTAAGAGGGAGTC-ATTTGG-3′ and 5′-CAGTTCCTTTATTCATACTGTCCAT-AAC-3′. The PCR-amplified cDNAs were cloned into pGEM-T (Promega, Madison, WI) and used as templates for synthesizing digoxigenin-labeled RNA probes. Sense and antisense NtmybAS1 probes were made using T7 and SP6 RNA polymerases after digestion with SacI and ApaI, respectively. Sense and antisense gPAL1 probes were made using SP6 and T7 RNA polymerases after digestion with ApaI and SacI, respectively.
Flower buds at five developmental stages (8–9 mm, 10–12 mm, 13–15 mm, 16–20 mm, and 21–30 mm in length) were fixed and paraffin embedded according to Dixon et al. (1995). Sectioning, hybridization, and detection of hybridization signals were performed as described by Sung et al. (1999).
Construction of sGFP and NLS Fusion Plasmids
The control plasmid pLAT52-sGFP, used to direct expression of GFP in pollen, consisted of the pollen-specific lat52 promoter (Twell et al., 1989b), the tobacco etch virus translational enhancer sequence (TEV-L), the sGFP coding sequence (Chiu et al., 1996), and the cauliflower mosaic virus 35S polyadenylation/terminator sequence (C3′) in pUC19 (precise construction details available on request). An NtMYBAS1-GFP fusion plasmid (pLAT52-sGFPM1) was constructed as follows. A 392-bp fragment of NtmybAS1 cDNA (position 176–567) encoding the N-terminal 126 amino acids of NtMYBAS1 was amplified using oligonucleotide primers (5′-GGAATCATGACACCAGATGGAGGAG-3′ and 5′- GGCATCATGACTCTTTGTCTTCTTTTTAGC-3′) with BspHI restriction sites at both ends. The amplified fragment was digested with BspHI, gel purified, and cloned into the NcoI site of pLAT52-sGFP fusing the first 126 amino acids of the NtMYBAS1 protein (position 181–558) to sGFP.
The fusion plasmid pLAT52-sGFPM2 was constructed to determine the effect of removing the putative NTMYBAS1 NLS (RLKRRQR motif) on nuclear targeting of sGFP. A 371-bp fragment of the NtmybAS1 cDNA (position 176–546) encoding the N-terminal 119 amino acids of NtMybAS1 lacking the RLKRRQR motif was amplified by PCR. The same forward primer used in the construction of pLAT52-sGFPM1 and a reverse primer (5′-CTTTTCATGATTGTG- TTCCAGTAATTC-3′) with a BspHI site at 3′ end were used. The PCR-amplified fragment was digested with BspHI, gel purified, and cloned into the compatible NcoI site of pLAT52-sGFP.
The fusion plasmid pLAT52-sGFPM3 was constructed to investigate the ability of the RLKRRQR motif alone to direct sGFP to the pollen nucleus. A 30-bp fragment of the NtmybAS1 cDNA encoding the MTRLKRRQRA motif (position 529–558) was synthesized using two oligonucleotide primers, 5′-CATGACAAGGCTAAAAAGAAGACAAAGAGC3′and 5′-CATGGCTCTTTGTCTTCTTTTTAGCCTTGT-3′. BspHI and NcoI sites at 5′ and 3′ ends of these oligonucleotides were introduced to avoid replacement of Thr (T) and Ala (A) residues, at positions 2 and 10, with other amino acids. Oligonucleotides were kinased, annealed, and the double-stranded fragment cloned into the compatible NcoI site of pLAT52-sGFP.
Particle Bombardment and Microscopy
Plasmids were bombarded into pollen as described by Twell et al. (1989a) with some modifications. The pollen suspension (30 mg mL−1) was prepared in pollen germination medium (Tupy et al., 1991) and 0.4 mL of the suspension was spread onto the surface of a 2-cm Hybond membrane square. Each bombardment was performed in duplicate and the experiment repeated five times. Localization of sGFP was visualized after 16 h using a Nikon Optiphot microscope with a 470- to 490-nm excitation filter and a 480-nm barrier filter. Images were captured directly using a CCD camera (JVC KYF55B) and Imagegrabber software (Neotec, Southampton, UK).
Production and Purification of Recombinant Protein
To introduce restriction enzyme sites at 5′ and 3′ ends of the coding region of NtmybAS1, two 27 mer oligonucleotides were synthesized. Forward primer containing a BamHI site (TGCGGATCCAATGGCACCAGATGGAGG) and reverse primer containing a SalI site (GCGGTCGACGCCTGATTTTGAGCAATG) were used for PCR amplification of the NtmybAS1 cDNA. The PCR product was ligated into the BamHI/SalI site of the pET 24b(+) vector (Novagen, Madison, WI). As material for EMSA, part of the sequence (amino acids 209–470) encoding the potential activator domain was removed. To remove this region, pET:NtmybAS1 was cut with HindIII. The HindIII site was religated such that the intact MYB domain and part of the activation domain remained. This recombinant protein, NtMYBASΔC1, was purified from IPTG-induced Escherichia coli cells over an Ni+-agarose column under native conditions according to the manufacturer's instructions (Qiagen, Valencia, CA). Purified protein was eluted from the column using elution buffer (1× Tris-buffered saline with 500 mm imidazole) and dialysis was done in a buffer containing 50% (w/v) glycerol, 1× Tris-buffered saline, and 1 mm phenylmethylsulfonyl fluoride at 4°C overnight.
PCR-Assisted Binding Site Selection
Binding site selection was performed essentially according to Blackwell et al. (1990). The oligonucleotide 5′-TCATGGATCCATACCTGCAGN23AGTAGTATGCTCTAG ACGCT-3′ was synthesized and used for PCR. Approximately 2 pmol of radiolabeled double-stranded DNA was incubated with 100 ng of purified recombinant protein in binding buffer [10 mm Tris-Cl (pH 7.6), 50 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, 5% (w/v) glycerol, 0.05 μg mL−1 phenylmethylsulfonyl fluoride, 12.5 μg mL−1 poly R478, and 2.5% (w/v) 3-(3 chloramidopropyl)-dimethylamino-1-pranesulphonate]. To prevent nonspecific binding, 1 μg of poly (dI-dC) was included in the reaction buffer. The purified recombinant proteins were first incubated in binding buffer for 15 min on ice and then with 10,000 cpm of labeled probe for 30 min at room temperature. Bound and free DNA was separated by electrophoresis on a 5% (w/v) polyacrylamide gel containing 1× Tris-borate-EDTA. After 3 h at 150 V, the wet gel was wrapped in Saran wrap and exposed to film for up to 3 h. The retarded band was excised and the DNA eluted by heating for 5 min in Tris-EDTA (10 mm Tris-Cl and 1 mm EDTA [pH 8.0]), extracted with phenol/chloroform, precipitated with ethanol, and resuspended in 20 μL distilled water. Five microliters of eluted DNA was used as a template for PCR (annealing temperature 55°C) with constant sequence primers end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After 20 cycles, the amplified DNA was purified using PAGE (8% [w/v] acrylamide and 1× Tris-borate-EDTA), visualized by auto-radiography, followed by elution from the gel piece as described above. The purified radiolabeled DNAs were used in the next round of binding and DNA protein complex purification. After six rounds of NtMYBASΔC1 binding and gel purification, the selected oligonucleotides bound by NtMYBASΔC1 were PCR amplified for 40 cycles and cloned into TOPO-PCR2.1 (Invitrogen, Carlsbad, CA).
EMSAs
Oligonucleotide probes were end labeled with [○-32P]ATP by T4 polynucleotide kinase. DNA binding reactions and electrophoresis were carried out as detailed above for PCR-assisted binding site selection (Grotewold et al., 1994). Binding reactions with NtMYBAS1-selected oligos were repeated four times with qualitatively similar results in each experiment. Relative binding affinities presented in Figure 6 represent mean values. For the optimization of competition assays, a range of excesses (50× to 500×) of unlabeled specific competitor were used. A 200× excess was selected because this concentration was sufficient for complete and effective competition for MBSI. Three independent experiments were performed with qualitatively similar results. A representative example is shown in Figure 7. Binding assays with PAL promoters were also repeated three times and gave qualitatively consistent results. A representative example is shown in Figure 8.
Construction of Effector Plasmids
Effector plasmids used in transfection were generated using PCR-based construction. C-terminal deleted cDNA fragments were amplified from full-length NtmybAS1 cDNA using the following synthetic oligonucleotides containing a BamHI or SacI site at their 5′ ends: 5′-TGCGGATCCAA-TGGCACCAGATGGAGG-3′ (amino acid 1), 5′-GCGGAG-CTCTCTTCTTTTTAGCCTTGT-3′ (amino acid 123), 5′-GCGGAGCTCGTTGTGAGATATGGAGAA-3′ (amino acid 200), 5′-GCGGAGCTCTATGTTGAAGTGGTGGCA-3′ (amino acid 257), 5′-GCGGAGCTCTCCTCGCGATAATCCA-GG-3′ (amino acid 322), and 5′-GCGGAGCTCGCCTGA-TTTTGAGCAATG-3′ (amino acid 470).
PCR fragments were digested with BamHI and SacI and purified fragments cloned into pBI221 (CLONTECH, Palo Alto, CA) by replacing the BamHI/SacI fragment of the GUS coding region.
Construction of Reporter Plasmids
To construct reporter plasmids, plant promoters were amplified from tobacco genomic DNA using the following synthetic oligonucleotides containing HindIII or XbaI sites at their 5′ and 3′ ends, respectively: gPAL1 promoter region, 5′-GCGAAGCTTGATCCGGACAAGAATGCA-3′ (for position −1,115) and 5′-GCGTCTAGATGTTAAAGGTTGTGAGGA-3′ (for position −56); PALA promoter region, 5′-GCGAAGCTTGTCGACCTGCAGGTCAAC-3′ (for position −659) and 5′-GCGTCTAGAGTGTAAAGGGGTTGGTTT-3′ (for position −42); neIF4A8 promoter region, 5′-GCGAAGCTTAAGCTTTCTAAATCCTGG-3′ (for position −2,138) and 5′-GCGTCTAGAGACTGTAATGTACGTACT-3′ (for position −2); and NPG1 promoter region, 5′-GCGAAGCTTGTCGACCTTTTAGTTGTG-3′ (for position −825) and 5′-GCGTCTAGATATGCCCCTCCAACCTCC-3′ (for position −6).
PCR products were digested with HindIII and XbaI and purified fragments cloned into pBI221 (CLONTECH) replacing the CaMV 35S promoter region.
Protoplast Isolation and Transfection
Isolation and transfection of tobacco leaf protoplasts was performed according to Goodall et al. (1990). Expression of reporter genes was monitored using the fluorimetric assay for GUS activity (Jefferson, 1987). Protein concentrations were determined using a Bradford Protein Assay Kit (Bio-Rad, Mississauga, ON, Canada).
ACKNOWLEDGMENTS
We thank Cathie Martin for providing samples of oligonucleotides O1 and O2 and Graham Benskin and June Saddington for maintaining plants.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (studentship to J.P.S.) and by Iran's Agricultural Research, Education, and Extension Organization (to S.A.). Work carried out in Long Ashton was supported by the British Council (fellowship to S.Y.).
LITERATURE CITED
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 1997;12:615–623. doi: 10.1046/j.1365-313x.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- Avila J, Nieto C, Canas L, Benito MJ, Paz-Ares J. Petunia hybrida genes related to the maize regulatory C1 gene and to animal myb proto-oncogenes. Plant J. 1993;3:553–562. doi: 10.1046/j.1365-313x.1993.03040553.x. [DOI] [PubMed] [Google Scholar]
- Baltz R, Domon C, Pillay DT, Steinmetz A. Characterization of a pollen-specific cDNA from sunflower encoding a zinc finger protein. Plant J. 1992;2:713–721. [PubMed] [Google Scholar]
- Bate NJ, Orr J, Ni W, Meromi A, Nadler-Hassar T, Doerner PW, Dixon RA, Lamb CJ, Elkind Y. Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc Natl Acad Sci USA. 1994;91:7608–7612. doi: 10.1073/pnas.91.16.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;23:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Brander KA, Kuhlemeier C. A pollen-specific DEAD-box protein related to translation initiation factor eIF-4a from tobacco. Plant Mol Biol. 1995;27:637–649. doi: 10.1007/BF00020219. [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell. 1996;8:1013–1025. doi: 10.1105/tpc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk O, Mink S, Ringwald M, Klempnauer KH. Synergistic activation of the chicken mim-1 gene by v-myb and C/EBP transcription factors. EMBO J. 1993;12:2027–2038. doi: 10.1002/j.1460-2075.1993.tb05852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Freed DD, Leinicke AJ. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell. 1991;3:953–962. doi: 10.1105/tpc.3.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM. Nuclear genes controlling male fertility. Plant Cell. 1993;5:1277–1283. doi: 10.1105/tpc.5.10.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;1:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Chung YY, Lee KJ, An G. Characterization of a tobacco MADS-box gene homologous to AGL2. Mol Cell. 1998;8:764–769. [PubMed] [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell. 1993;5:1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon CD, Cutt JR, Klessig DF. In situ hybridization for detection of RNA in plant tissues. In: Maliga P, Klessig DF, Cashmore AR, Gruissem W, Varner JE, editors. Methods in Plant Molecular Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 111–137. [Google Scholar]
- Facchinetti V, Loffarelli L, Schreek S, Oelgeschlager M, Luscher B, Introna M, Golay J. Regulatory domains of the A-Myb transcription factor and its interaction with the CBP/p300 adaptor molecules. Biochem J. 1997;15:729–736. doi: 10.1042/bj3240729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Schrell S, Peterson PA, Saedler H, Wienand U. Molecular analysis of protein domain function encoded by the myb-homologous maize genes C1, Zm1 and Zm38. Plant J. 1994;6:21–30. doi: 10.1046/j.1365-313x.1994.6010021.x. [DOI] [PubMed] [Google Scholar]
- Goodall GJ, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;17:1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Knoblock KH, Kreuzler F, Potts JRM, Wellman E. Coordinated induction and subsequent activity changes of two groups of metabolically interrelated enzymes. Eur J Biochem. 1976;61:119–206. doi: 10.1111/j.1432-1033.1976.tb10012.x. [DOI] [PubMed] [Google Scholar]
- Hatton D, Smith C, Bevan M. Tissue-specific expression of the PAL3 promoter requires the interaction of two conserved cis sequences. Plant Mol Biol. 1996;31:393–397. doi: 10.1007/BF00021800. [DOI] [PubMed] [Google Scholar]
- Heuer S, Lorz H, Dresselhaus T. The MADS box gene ZmMADS2 is specifically expressed in maize pollen and during maize pollen tube growth. Sex Plant Reprod. 2000;13:21–27. [Google Scholar]
- Jackson D, Culianez-Marcia F, Prescott AG, Roberts K, Martin C. Expression patterns of myb genes from Antirrhinum flowers. Plant Cell. 1991;3:115–125. doi: 10.1105/tpc.3.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Johnson TK, Schweppe RE, Septer J, Lewis RE. Phosphorylation of B-Myb regulates its trans-activation potential and DNA binding. J Biol Chem. 1999;274:36741–36749. doi: 10.1074/jbc.274.51.36741. [DOI] [PubMed] [Google Scholar]
- Kishitani S, Yomoda A, Konno N, Tanaka Y. Involvement of phenylalanine ammonia-lyase in the development of pollen in broccoli (Brassica oleracea L.) Sex Plant Reprod. 1993;6:244–248. [Google Scholar]
- Kobayashi A, Sakamoto A, Kubo K, Rybka Z, Kanno Y, Takatsuji H. Seven zinc-finger transcription factors are expressed sequentially during the development of anthers in petunia. Plant J. 1998;13:571–576. doi: 10.1046/j.1365-313x.1998.00043.x. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C. Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Li SF, Higginson T, Parish RW. A novel MYB-related gene from Arabidopsis thaliana expressed in developing anthers. Plant Cell Physiol. 1999;40:343–347. doi: 10.1093/oxfordjournals.pcp.a029548. [DOI] [PubMed] [Google Scholar]
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- Mariani C, De Beuckeleer M, Truettner J, Leemans J, Goldberg RB. Induction of male-sterility in plants by a chimeric ribonuclease gene. Nature. 1990;347:737–741. [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP. Gene activity during pollen development. Annu Rev Plant Physiol. 1990;41:317–338. [Google Scholar]
- Matsuda N, Tsuchiya T, Kishitani S, Tanaka Y, Toriyama K. Partial male sterility in transgenic tobacco carrying antisense and sense PAL cDNA under the control of a tapetum-specific promoter. Plant Cell Physiol. 1996;37:215–222. [Google Scholar]
- McCormick S. Male gametophyte development. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle T, Nagy F. Nuclear import of proteins: putative import factors and development of in vitro import systems in higher plants. Trends Plant Sci. 1997;2:458–464. [Google Scholar]
- Mo YY, Nagel C, Taylor LP. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA. 1992;89:7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano E, Martinez GJ, Martin C. Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell. 1996;8:1519–1532. doi: 10.1105/tpc.8.9.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S. LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 1994;6:321–338. doi: 10.1046/j.1365-313x.1994.06030321.x. [DOI] [PubMed] [Google Scholar]
- Napoli CA, Fahy D, Wang HY, Taylor LP. white anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol. 1999;120:615–622. doi: 10.1104/pp.120.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the MYB DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Rohfritsch O, Fritig B, Legrand M. Phenylalanine ammonia-lyase in tobacco. Plant Physiol. 1994;106:877–886. doi: 10.1104/pp.106.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ. Intra-and extra-cellular lipid composition and associated gene expression patterns during pollen development in Brassica napus. Plant J. 1997;11:549–562. doi: 10.1046/j.1365-313x.1997.11030549.x. [DOI] [PubMed] [Google Scholar]
- Preuss D. Being fruitful: genetics of reproduction in Arabidopsis. Trends Genet. 1995;11:147–153. doi: 10.1016/s0168-9525(00)89029-0. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, Van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin 2 gene of petunia and its role in the evolution of flower colour. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaye IKE, Toku S, Tanaka T. Sequence requirement for nucleolar localization of rat ribosomal protein L31. Eur J Cell Biol. 1996;69:151–155. [PubMed] [Google Scholar]
- Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold I. Maize R2R3 Myb genes: sequence analysis reveals amplification in the higher plants. Genetics. 1999;153:427–444. doi: 10.1093/genetics/153.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel N. Nuclear targeting in plants. Plant Physiol. 1992;100:1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C. A flower-specific MYB protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikumar P, Murali R, Reddy EP. Role of tryptophan repeats and flanking amino acids in MYB-DNA interactions. Proc Natl Acad Sci USA. 1990;87:8452–8456. doi: 10.1073/pnas.87.21.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Fuertes A, Sanchez-Pulido L, Valencia A, Paz-Ares J. A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J Biol Chem. 1997;272:2889–2895. doi: 10.1074/jbc.272.5.2889. [DOI] [PubMed] [Google Scholar]
- Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB-Ph3) from Petunia hybrida. EMBO J. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung SK, Yu GH, An G. Characterization of MdMADS2, a member of the SQUAMOSA subfamily of genes, in apple. Plant Physiol. 1999;120:969–978. doi: 10.1104/pp.120.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman J, Spurr C, Eliasson A, Gass N, Steinmetz A, Twell D. Isolation and characterization of two pollen-specific LIM domain protein cDNAs from Nicotiana tabacum. Sex Plant Reprod. 2000;12:339–345. [Google Scholar]
- Takemoto Y, Tashiro S, Handa H, Ishii S. Multiple nuclear localization signals of the B-myb gene product. FEBS Lett. 1994;350:55–60. doi: 10.1016/0014-5793(94)00733-0. [DOI] [PubMed] [Google Scholar]
- Tan R, Frankel AD. A novel glutamine-RNA interaction identified by screening libraries in mammalian cells. Proc Natl Acad Sci USA. 1998;95:4247–4252. doi: 10.1073/pnas.95.8.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Hepler PK. Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- Tebbutt SJ, Rogers HJ, Lonsdale DM. Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol Biol. 1994;25:283–297. doi: 10.1007/BF00023244. [DOI] [PubMed] [Google Scholar]
- Thomson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice-Baldwin K, Fink GR, Arndt KT. BAS1 has a MYB motif and activates HIS4 transcription only in combination with BAS2. Science. 1989;246:931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- Tomita A, Towatari M, Tsuzuki S, Hayakawa F, Kosugi H, Tamai K, Miyazaki T, Kinoshita T, Saito H. c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene. 2000;19:444–451. doi: 10.1038/sj.onc.1203329. [DOI] [PubMed] [Google Scholar]
- Tomoko F, Shain-dow K, John CW. Phenlyalanine ammonia-lyase gene, structure, expression, and evolution in Nicotiana. Plant Mol Biol. 1996;30:711–722. doi: 10.1007/BF00019006. [DOI] [PubMed] [Google Scholar]
- Tupy J, Ribhova L, Zarsky V. Production of fertile tobacco pollen from microspores in suspension culture and its storage for in situ pollination. Sex Plant Reprod. 1991;4:284–287. [Google Scholar]
- Twell D. The diversity and regulation of gene expression in the pathway of male gametophyte development. In: Scott RJ, Stead AD, editors. Molecular and Cellular Aspects of Plant Reproduction. Seminar Series 55, Society for Experimental Biology. Cambridge, UK: Cambridge University Press; 1994. pp. 83–135. [Google Scholar]
- Twell D, Klein TM, Fromm ME, McCormick S. Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiol. 1989a;91:1270–1274. doi: 10.1104/pp.91.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D, Park SK, Lalanne E. Asymmetric division and cell-fate determination in developing pollen. Trends Plant Sci. 1998;3:305–310. [Google Scholar]
- Twell D, Wing RA, Yamaguchi J, McCormick S. Isolation and expression of an anther-specific gene from tomato. Mol Gen Genet. 1989b;217:240–245. doi: 10.1007/BF02464887. [DOI] [PubMed] [Google Scholar]
- Uimari A, Strommer J. Myb26: a MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 1997;12:1273–1284. doi: 10.1046/j.1365-313x.1997.12061273.x. [DOI] [PubMed] [Google Scholar]
- van der Meer IM, Stam ME, van Tunen AJ, Mol JN, Stuitje AR. Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell. 1992;4:253–262. doi: 10.1105/tpc.4.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Knox RB, Taylor PE, Singh MB. Bcp1, a gene required for male fertility in Arabidopsis. Proc Natl Acad Sci USA. 1995;92:2106–2110. doi: 10.1073/pnas.92.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra B, Busscher J, Franken J, Hollman PCH, Mol JNM, van Tunen AJ. Flavonols and fertilization in Petunia hybrida: localization and mode of action during pollen-tube growth. Plant J. 1994;6:201–212. [Google Scholar]
- Zachgo S, Saedler H, Schwarz-Sommer Z. Pollen-specific expression of DEFH125, a MADS-box transcription factor in Antirrhinum with unusual features. Plant J. 1997;11:1043–1050. doi: 10.1046/j.1365-313x.1997.11051043.x. [DOI] [PubMed] [Google Scholar]