Abstract
Activating transcription factor 3 (ATF3) is a member of the ATF/CREB family of transcription factors and its expression is increased by various pathophysiological conditions and in several cancer cells. In this study, we describe two alternatively spliced ATF3ΔZip mRNAs: ATF3ΔZip2a and ATF3ΔZip2b. Both variants encoded the same truncated protein of 135 amino acids, which lacked the leucine zipper domain and was incapable of binding to the ATF/CRE motif. The ATF3ΔZip2 protein was shown to be localized in the nuclei and counteracted the transcriptional repression by the full-length ATF3. Western blot analysis showed that ATF3ΔZip2 was expressed in cells exposed to A23187. Further study showed that, similar to the full-length ATF3, the expression of ATF3ΔZip2 was induced by a wide range of stress stimuli. However, its expression was not detectable in cancer cells that constitutively over-expressed ATF3. Taken together, our results suggest that ATF3ΔZip2, a protein derived from alternatively spliced mRNAs, is induced by various stress signals and may modulate the activity of the full-length ATF3 protein during stress response.
INTRODUCTION
Alternative splicing of pre-mRNA encoding transcription factors is one of the common mechanisms for generating the complexity and diversity of gene regulation (1–3). It produces a variety of functionally distinct isoforms from a single gene by use of different combinations of splice junctions. For example, alternative splicing within the DNA binding domain of Pax-6 (4) and Wilm’s tumor-associated protein 1 (5) alters their DNA binding specificity. Alternative splicing of the transactivation domains in Pax-8 (6), the POU homeodomain family protein Pit-1 (7), and the zinc finger transcription factor GATA-5 (8) results in isoforms possessing different activation potencies. Deletion of the activation domain in AML1a (9) and CREB (10) by splicing produces variants with dominant negative activity. Thus, additional genetic complexity and diversity might be accomplished by altering regulatory properties through expression of alternatively spliced isoforms of transcription factor.
Transcriptional repressor activating transcription factor 3 (ATF3) is one of the immediate early response genes (11–14). It is induced upon exposure of cells to a variety of physiological and pathological stimuli, including seizure, toxic chemicals such as carbon tetrachloride (15), anti-cancer drugs (16), proteasome inhibitor (17), genotoxic agents (18), homocysteine (19) and ischemia-coupled with reperfusion (20). ATF3 is also rapidly induced in regenerating liver (12), or in cells treated by growth-stimulating factors such as serum (21,22). Furthermore, it is over-expressed in murine melanoma cells with high metastatic potentials (23), or esophageal cancer cells (24). Thus, ATF3 functions in both cellular stress response and cell proliferation.
ATF3 is composed of 181 amino acids and the region from amino acid 40 to 84 has been reported to have the transcriptional repression activity (25). The basic/leucine zipper (bZip) domain from 88 to 147 amino acids is required for dimer formation and specific DNA binding (21,25). ATF3 forms homodimer with itself and the resulting dimer represses transcription from various promoters with ATF sites (21). In addition, this ATF3 homodimer has been demonstrated to repress tumor necrosis factor (TNF)-α-induced E-selectin gene expression (26), and arsenite-induced GADD153 gene expression (27,28). ATF3 also forms dimers with other bZip proteins. Heterodimers of ATF3 with c-Jun and JunB have been reported to activate transcription in transient transfection assays (25). Interestingly, adenovirus E1A induces the expression of ATF3 and leads to the formation of c-Jun/ATF3 heterodimer. Significantly, this activity of E1A is thought to play a role in its ability to transform cells (29). Therefore, ATF3 can activate or repress target genes by forming homo- or heteromeric complexes, and some of these activities have been implicated in significant biological functions.
Previously, an alternative spliced isoform ATF3ΔZip was isolated from HeLa cells stimulated by serum (21). It lacks the leucine zipper domain and does not bind to the ATF/CRE sequence, but stimulates transcription of reporter driven by promoters with or without ATF/CRE motif, presumably by sequestering co-inhibitory factors away from the promoters (21). Thus, ATF3ΔZip regulates the expression of target genes. However, the functional significance of the alternative splice variants, or the presence of other forms of truncated ATF3 remains largely unknown.
In this report, we describe a novel form of alternatively spliced ATF3ΔZip2 and characterized its expression in stress response and in cancer cells. ATF3ΔZip2 expression was highly induced by various stress stimuli, but not in cancer cells over-expressing ATF3. It may regulate gene expression of cells in response to stress stimuli.
MATERIALS AND METHODS
Nucleotide sequence accession numbers
Nucleotide sequences for cDNA of ATF3ΔZip2a and b have been deposited in DDBJ/EMBL/GenBank with accession numbers AB078026 and AB078027, respectively.
Reagents
A23187 was purchased from Wako Pure Chemical Co (Osaka, Japan). Thapsigargin, tunicamycin, dl-homocysteine and colchicine were obtained from Sigma. Proteosome inhibitor MG132, Z–Leu–Leu–Leu–CHO, was from BIOMOL Research Laboratories, and methyl methanesulfonate (MMS) was purchased from nacalai tesque (Kyoto, Japan). Recombinant TNF-α was purchased from Genzyme. Anti-glutathione S-transferase (GST) and anti-hemagglutinin (HA) monoclonal antibody were purchased from Santa Cruz Biotechnology and Boehringer Mannheim, respectively. Anti-ATF3 antibody was generated by immunizing rabbit with ATF3 produced in Escherichia coli (21). Texas Red-conjugated goat anti-mouse IgG (H+L) was from Molecular Probes. 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) was from Nacalai Tesque (Kyoto, Japan). Mammalian expression plasmid for human ATF3, pCIATF3, was as described (19). Reporter plasmid pLuc-4xATF was as described (30) and contained four repeats of ATF/CRE site. Plasmids for expressing the Flag-tagged full-length ATF3 and ATF3ΔZip2, pME-Flag-ATF3 and pME-Flag-ATF3ΔZip2, respectively, were prepared by inserting each fragment between EcoRI and NotI sites of pME (31).
Cell culture, stimulation and RNA preparation
Primary human umbilical vein endothelial cells (HUVECs) were obtained from Clonetics Corp., and grown in EBM-2 medium (Clonetics, Corp.) as described (19). Saos 2 cells were cultured in DMEM containing 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. After the cells (5 × 105 cells) were stimulated with various agents for the time indicated, total RNA was isolated by an acid guanidinium-thiocyanate-phenol-chloroform extraction method using Isogen (Nippon Gene, Japan). The amount of RNA was quantified by measuring optical density at 260 nm. Human esophageal cancer cell lines over-expressing ATF3 were cultured as described (24).
Reverse transcriptase–polymerase chain reaction (RT–PCR) analysis
Total RNA (1 µg) was assayed for ATF3 and ATF3ΔZip2 mRNA by RT–PCR using a kit from Takara, Japan. The RT reaction was performed using oligo(dT) primer at 42°C for 15 min using avian myeloblastosis virus RT. Following the reaction, PCR was performed at 95°C for 1 min, at 60°C for 1 min, and at 72°C for 2 min for 24–28 cycles. Primers used for the analysis are shown in Figure 1A. To amplify the full-length ATF3 and ATF3ΔZip2, a pair of primer 5′ in exon B, 5′-ATGATGCTTCAACACCCAGGC-3′ and primer 3′ in exon E, 5′-TTAGCTCTGCAATGTTCCTTC-3′ was employed. For analysis of ATF3ΔZip2, a pair of primer 5′ and primer Zip3 in exon D, 5′-GCTCATCAAGCTGCGAGAGG-3′, or a pair of primer Zip4 in exon D′, 5′-CTCCCAAGGCCCTTTTGGG-3′ and primer Zip3 were employed. mRNA for glyceraldehyde 3-phosphate dehydrogenase was also amplified using a primer pair of 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′and 5′-CATGTGGGCCATGAGGTCCACCAC-3′. Reaction products were separated on 2% agarose gel in Tris–acetate EDTA buffer, and stained with ethidium bromide.
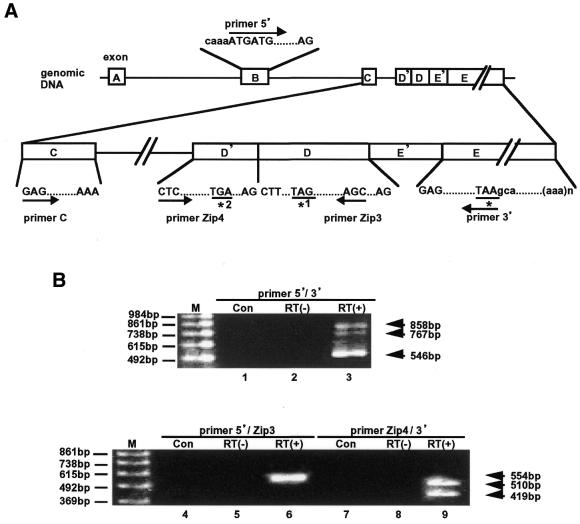
Figure 1.
RT–PCR analysis of ATF3ΔZip2 mRNA in A23187-treated HUVECs. (A) Primers used in this study and their locations in the human ATF3 gene are shown. Exons are indicated by boxes, and labeled as A, B, C, D and E as in Liang et al. (33). Exons D′ and E′ are novel ones described in the text. Initiation codon ATG in exon B is shown under primer 5′. Stop codons for the full-length ATF3 (*), ATF3ΔZip (*1) and ATF3ΔZip2 (*2) are shown. (B) HUVECs (3 × 105 cells) were treated with 10 µM A23187 for 4 h, and total RNA (1 µg) was assayed for RT–PCR using primer 5′ and primer 3′ (lanes 1–3), primer 5′ and primer Zip3 (lanes 4–6), or primer Zip4 and primer 3′ (lanes 7–9). Lanes 1, 4 and 7 are derived from untreated cells; all the other lanes are from A23187-treated cells with (lanes 3, 6 and 9) or without (lanes 2, 5 and 8) RT. Bands of 546, 858 and 767 bp represented products of full-length ATF3, ATF3ΔZip2a and ATF3ΔZip2b, respectively (lane 3). 554 bp was the product derived from ATF3ΔZip2a and b (lane 6); note that the primer set used in this experiment (primer 5′ and primer Zip3) does not distinguish ATF3ΔZip2a from ATF3ΔZip2b. 510 and 419 bp represented ATF3ΔZip2a and ATF3ΔZip2b, respectively (lane 9). M, the 123 bp DNA size marker.
Cloning and sequencing of ATF3ΔZip2 cDNA
Total RNA (1 µg) from the A23187-treated HUVECs was subjected to RT–PCR using primer pairs for ATF3ΔZip2 as in Figure 1. The reaction products were subcloned into TA vector (Invitrogen) and the sequences of at least four clones of each transformant were determined by the dideoxy sequencing method. Genomic DNA fragments encompassing the intron C or D of the human ATF3 gene were PCR-amplified and sequenced. Primer pairs used were primer C, 5′-GTAGCCCCTGAAGAAGATGAAA-3′ and primer Zip3 for intron C and primer Zip4 and primer 3′ for intron D, respectively, as depicted in Figure 1. The sequence of the intron C and D was also compared with that reported in the NCBI DNA data bank.
Western blot analysis
HUVECs (1 × 106 cells) were treated with 10 µM A23187 for 8 h and whole-cell extract was prepared as described (19). The amount of protein was quantified by the Lowry method using bovine serum albumin as standard (32). Whole-cell extracts (20 µg protein) of HUVECs were separated on an SDS–polyacrylamide gel, and subjected to western blot analysis as described (19). The band was visualized by chemiluminescence as in the protocol of the ECL kit from Amersham.
Expression of recombinant ATF3 and ATF3ΔZip2 fused to GST in E.coli
Expression plasmids encoding ATF3 and ATF3ΔZip2 fused to GST were prepared by subcloning the EcoRI–NotI fragment of ATF3 and the EcoRI fragment of ATF3ΔZip2 into pGEX-4T-1 and pGEX-2TK (Pharmacia), respectively. Recombinant protein was expressed in BL21 cells harboring each plasmid after induction by 1 mM IPTG, and purified on a glutathione–Sepharose column as per the supplier’s protocol.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as described previously (19). Briefly, GST–ATF3, GST–ATF3ΔZip2 and GST protein (2.5–5 µg) were incubated in 20 µl of binding buffer (10 mM HEPES–KOH, pH 7.9, 60 mM KCl, 0.5 mM EDTA, 5 mM MgCl2, 0.1 mM PMSF, 5 mM β-mercaptoethanol) containing 0.1 µg of poly(dI–dC) and 0.5 ng of radiolabeled DNA probe at room temperature for 30 min. The DNA probe was the ATF/CRE motif at –99 to –69 of the human ATF3 gene promoter (19,33), and radiolabeled with 25 µCi [γ-32P]ATP (6000 Ci/mmol) and 10 U polynucleotide kinase. The mutant oligonucleotide used for competition experiment was as described (19). The binding mixture was applied onto a 5% polyacrylamide slab gel in Tris–borate EDTA buffer. After electrophoresis, the gel was dried on a 3MM Whatmann paper and visualized by a Fuji Bas 2500 image analyzer.
Expression and immunocytochemical analysis of ATF3 and ATF3ΔZip2 in mammalian cells
cDNA fragment encoding ATF3ΔZip2 was PCR-amplified and the EcoRI–SalI and SalI–NotI fragments were subcloned into pEGFP-C1 (Clontech) and pHA vector containing RSV promoter, respectively. Resultant expression plasmids, pEGFP-ATF3ΔZip2 and pHA-ATF3ΔZip2, encoded the ATF3ΔZip2 protein that was N-terminally fused to GFP and HA peptide, respectively. Each plasmid (1 µg) for ATF3ΔZip2 and pCIATF3 (1 µg) for full-length ATF3 was transfected into Cos-7 cells using Superfect (Qiagen) as described (19). At 24 h post-transfection, ATF3ΔZip2 and ATF3 were detected by GFP fluorescence or immunostaining using mouse monoclonal anti-HA antibody and anti-ATF3 antibody, respectively. For counter-staining of nuclei, cells were also stained by DAPI.
Reporter assay
Cos-7 cells (3 × 105 cells) grown in RPMI medium to 70–80% confluency in a 35 mm dish were cotransfected with pLuc-4xATF and effector plasmids as indicated. At 24 h post-transfection, the supernatant of cell extracts was assayed for firefly and sea pansy luciferase activity using a dual luciferase reporter assay system (Promega) as described (19). pRL-CMV (Toyo Ink, Tokyo, Japan) containing the sea pansy luciferase gene was used as an internal control of transfection and expression.
Statistical analysis
Quantitative data were expressed as mean ± SD. Statistical analysis was performed with Student’s t-test. Differences were considered significant when probability values were <0.05.
RESULTS
Isolation of alternatively spliced ATF3ΔZip2
We previously reported that ATF3 expression is induced in HUVECs by homocysteine and various stimuli (19). During our analysis by RT–PCR using a pair of primers, primer 5′ and primer 3′ indicated in Figure 1A, we observed two bands of 858 and 767 bp in addition to the 546 bp full-length ATF3 mRNA in HUVECs after treatment with A23187 (Fig. 1B, lane 3). To test whether these are alternative spliced mRNAs containing the exon D described previously (21), we performed PCR using primer 5′ and primer Zip3 localized in exon D (Fig. 1A). As shown in Figure 1B, lane 6, a band of 554 bp was produced, suggesting the presence of an alternatively spliced mRNA containing exon D. However, because two alternatively spliced isoforms were expected from the two bands observed above (858 and 767 bp), we suspected that this pair of primers failed to distinguish the isoforms. Sequence analysis of this PCR product (554 bp) revealed a previously unknown exon, designated as exon D′ in this report (Figs 1A and 2). We then designed a primer called primer Zip4 from the new exon, and performed PCR using primer Zip4 and primer 3′. As shown in Figure 1B, lane 9, two bands of 510 and 419 bp were amplified. These bands were produced after reverse transcription using oligo-dT primer, suggesting that the bands were alternative splice variants. Therefore, we determined the sequence of these RT–PCR products and compared them with that of the human ATF3 gene. Results showed that the additional two bands in Figure 1B represented alternatively spliced isoforms of ATF3 mRNA. We named them ATF3ΔZip2a and b. As shown in Figure 2A, both isoforms utilized a novel 3′-acceptor site in the intron C that is at 76 bases upstream from exon D. Therefore, this 76 base sequence was designated as exon D′ in this study. ATF3ΔZip2a and b, however, differed in the splicing between exons D and E. Splicing of intron D occurred in ATF3ΔZip2b as in the full ATF3, but no splicing was observed in ATF3ΔZip2a, which was amplified as a 91 bp larger fragment than ATF3ΔZip2b in RT–PCR analysis (Figs 1B and 2A). This 91 bp sequence was the same as the intron D sequence in the human ATF3 gene (BAC RP11–248, accession no. AL606913 in the BAC draft sequence).
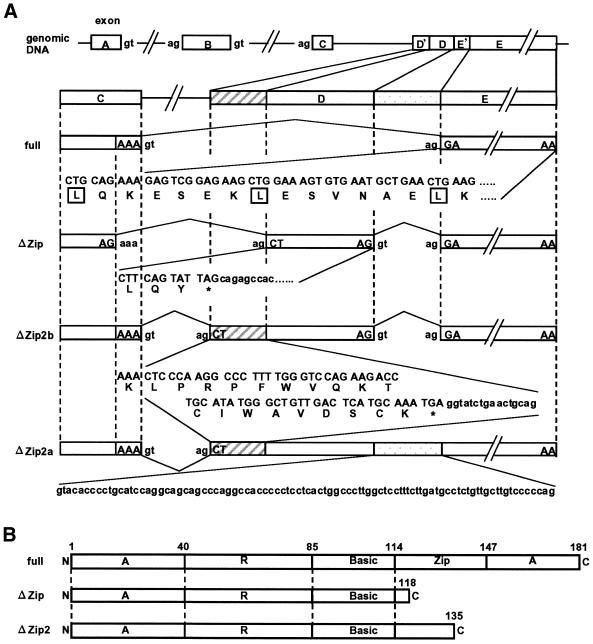
Figure 2.
Splicing events for generating ATF3ΔZip2a and b and comparison of their nucleotide and amino acid sequences. (A) Splicing events for generating ATF3ΔZip2a and b and their nucleotide and amino acid sequences are compared with those for full-length ATF3 and ATF3ΔZip as described in Liang et al. (33). Exon D′ is comprised of a 76 base sequence of intron C (shaded squares) that precedes exon D. Exon E′ is an original intron D of 91 bp (dotted square) and transcribed into ATF3ΔZip2a mRNA. Leucine residues in the zipper region were marked. (B) Schematic representations of the full-length ATF3, ATF3ΔZip and ATF3ΔZip2 are shown. Basic, Zip, A and R represent the basic, leucine zipper, activation, repression region, respectively (21).
Structure of ATF3ΔZip2
In Figure 2, the nucleotide and predicted amino acid sequence of ATF3ΔZip2a and b were aligned and compared with those of full-length ATF3 and ATF3ΔZip. Both variants of ATF3ΔZip2 encoded the same protein composed of 135 amino acids, among which 1–115 amino acids are common to those of full-length ATF3 and ATF3ΔZip (Fig. 2B). At the C-terminal, the ATF3ΔZip2 protein contained the basic region but deleted the leucine zipper domain, which was replaced by a novel sequence of 20 amino acids. Therefore, ATF3ΔZip2 is a C-terminally truncated protein.
Relative expression of ATF3ΔZip2 and full-length ATF3 mRNA in A23187-treated HUVECs
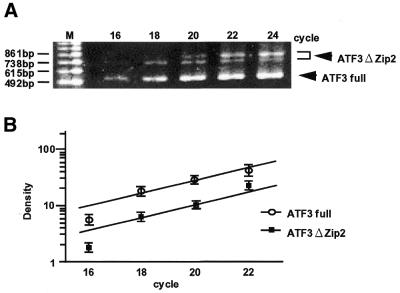
As the first step to understand the significance of ATF3ΔZip2 in vivo, we performed semi-quantitative analysis of the relative expression of ATF3ΔZip2. Figure 3 shows a representative RT–PCR analysis of the A23187-treated HUVECs using primer 5′ and primer 3′. The amount of PCR products was analyzed at every two cycles from the 16th to the 24th cycle, and the density of each band was compared. The combined amount of mRNAs for ATF3ΔZip2a and b was 34 ± 4% compared with that of full-length ATF3, based on the data at linear range of amplification from the 18th to the 22nd cycle. This indicates that the induction of ATF3ΔZip2 is approximately one- third of that for full-length ATF3. We further examined whether ATF3ΔZip, which is induced in serum-stimulated HeLa cells (21), was present in the A23187-treated cells. RT–PCR using a pair of primer 5′ and Zip3 for amplifying both ATF3ΔZip and ΔZip2, followed by sequence determination, could not detect ATF3ΔZip (data not shown). Thus, A23187 treatment selectively induced the expression of ATF3ΔZip2, but not of ATF3ΔZip. These data indicate that ATF3ΔZip2 is the major alternatively spliced isoform in the A23187-treated HUVECs.
Figure 3.
Semi-quantitative measurement of ATF3ΔZip2 and full ATF3 mRNA. Total RNA (1 µg) from the A23187-treated HUVECs was subjected to RT–PCR using primer 5′ and primer 3′ and the expression of full ATF3 and ATF3ΔZip2 was measured at various cycles of PCR. A typical experiment is shown in (A). In (B), the logarithm of density of each band was plotted against the number of PCR cycles, and the ratio of expression of ATF3ΔZip2 to full ATF3 was determined from data at the linear amplification range of PCR. Results were means ± SE bars of three independent experiments. The amount of ATF3ΔZip2 represents a total of ATF3ΔZip2a and b.
ATF3ΔZip2 protein is expressed in the A23187-treated HUVECs
We next examined whether ATF3ΔZip2 protein was expressed in the A23187-treated HUVECs. Western blot analysis showed that the full-length ATF3 and a protein of lower molecular weight were induced after treatment with A23187 (Fig. 4, lane 3), while these bands were not detected in control cells (lane 2). The lower band showed the same migration mobility as the Flag-tagged ATF3ΔZip2 (lanes 3 and 4). We note that the anti-ATF3 antibody used in this experiment was generated against the full-length ATF3 protein (21). It could recognize the N-terminal portion of ATF3 and thus both the full-length ATF3 and the ATF2ΔZip2 proteins. It also detected a non-specific band around 35 kDa in HUVEC cell extracts. The anti-ATF3 antibody C-19 (from Santa Cruz) recognizes the C-terminal region of ATF3 and could not detect the ATF3ΔZip2 protein (data not shown). These results indicate that ATF3ΔZip2 protein is expressed in the A23187-treated HUVECs.
Figure 4.
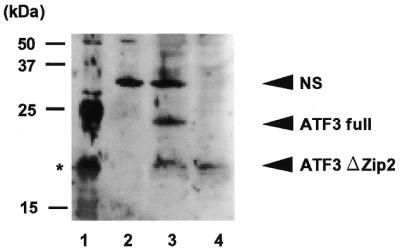
Western blot analysis of ATF3ΔZip2 protein in A23187-treated HUVECs. HUVECs (3 × 105 cells) were treated with 10 µM A23187 for 8 h. Whole-cell extracts (20 µg protein) were prepared and separated on a 15% SDS–PAGE and subjected to western blot analysis using anti-ATF3 antibody (21) as described in Materials and Methods. Lane 1, bacterially expressed full-length His-tagged ATF3. The asterisk denotes a proteolytic artifact. Lane 2, control untreated HUVECs; lane 3, A23187-treated HUVECs; lane 4, Flag-tagged ATF3ΔZip2 expressed in Cos-7 cells. NS, the non-specific 35 kDa protein in HUVECs.
ATF3ΔZip2 is not capable of binding to the ATF/CRE motif, but is localized in nuclei
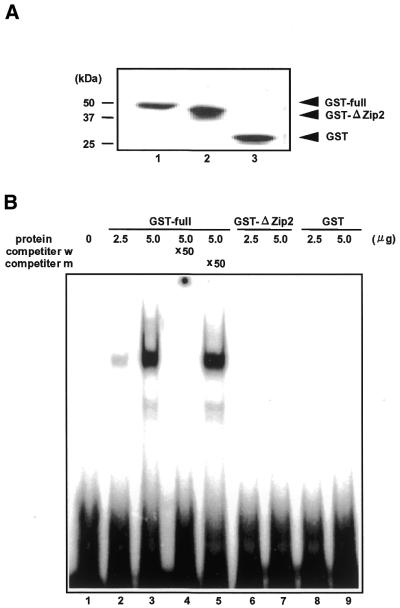
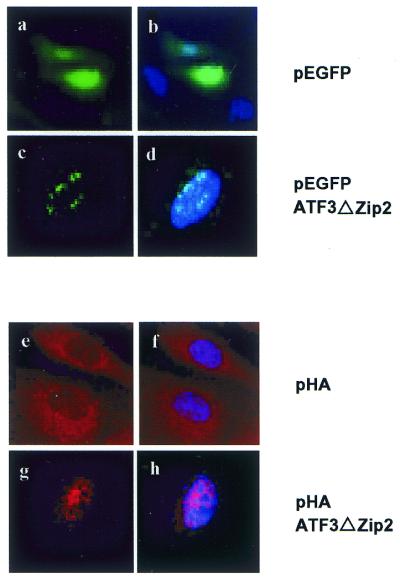
To obtain evidence for possible gene regulation by ATF3ΔZip2, we expressed it as GST fusion protein in E.coli, and tested its DNA binding activity. Figure 5 shows that the full-length ATF3–GST fusion protein formed the protein–DNA complex that was specifically abolished by the wild-type ATF/CRE motif, but not by the mutant sequence (lanes 2–5). In contrast, GST–ATF3ΔZip2 did not yield a DNA–protein complex, indicating that ATF3ΔZip2 protein is not capable of binding to the ATF/CRE sequence. Next, we sought to determine subcellular localization of ATF3ΔZip2 protein. For this purpose, we transiently expressed ATF3ΔZip2 in Cos-7 cells and examined its localization by immunostaining. Figure 6 shows that ATF3ΔZip2 was localized in nuclei when it was fused to GFP or HA epitope. Full-length ATF3 and ATF3ΔZip were also localized in nuclei in this assay (data not shown). These data clearly indicate that ATF3ΔZip2 protein is present in nuclei.
Figure 5.
EMSA of recombinant ATF3ΔZip2. Full-length ATF3 and ATF3ΔZip2 were expressed as GST-fusion proteins as described in the Materials and Methods. (A) Purified GST–ATF3 fusion protein was analyzed by western blot using monoclonal anti-GST antibody. Lane 1, GST-full ATF3; lane 2, GST–ATF3ΔZip2; lane 3, GST. (B) Recombinant ATF3 proteins were assayed for their DNA binding activity by EMSA as detailed in the Materials and Methods. Lane 1, probe only; lane 2, 2.5 µg GST–full-length ATF3; lanes 3–5, 5 µg GST–full-length ATF3 in the absence (lane 3) or presence of wild-type ATF/CRE sequence (lane 4) and mutated sequence (lane 5), respectively; lanes 6 and 7, 2.5 and 5 µg GST–ATF3ΔZip2; lanes 8 and 9, 2.5 and 5 µg GST.
Figure 6.
Nuclear localization of ATF3ΔZip2. ATF3ΔZip2 protein N-terminally fused to GFP (pEGFPATF3ΔZip2) or HA-tag (pHAATF3ΔZip2) was transiently expressed in Cos-7 cells as described in the Materials and Methods. At 24 h post-transfection, GFP was detected in cells transfected with empty vector [pEGFP (a and b] or pEGFPATF3ΔZip2 (c and d). HA-tagged ATF3 protein was detected by monoclonal anti-HA antibody in cells transfected with empty vector [pHA (e and f)] or pHAATF3ΔZip2 (g and h). Nuclei were also stained by DAPI in (b, d, f and h).
ATF3ΔZip2 counteracts the transcriptional repression by full-length ATF3
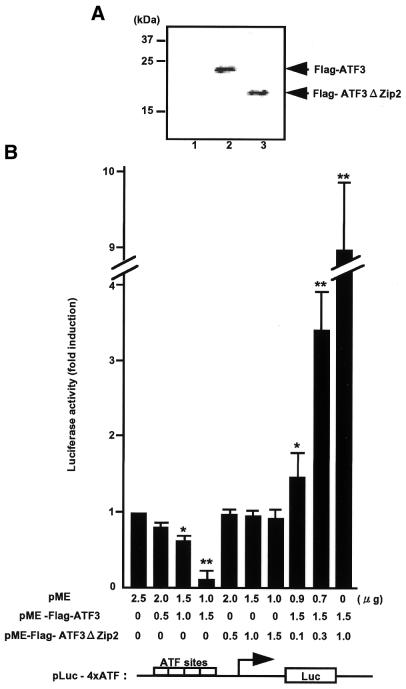
We next determined whether ATF3ΔZip2 is capable of regulating gene transcription since ATF3ΔZip is reported to stimulate transcription, presumably by sequestering co-inhibitory factors away from the promoters (21). Thus, we tested whether ATF3ΔZip2 could affect gene expression in a reporter assay. Figure 7 shows that full-length ATF3 repressed the expression of reporter gene containing four repeats of the ATF/CRE motif, while ATF3ΔZip2 alone had no effect. However, when ATF3ΔZip2 was co-expressed with full ATF3, it remarkably counteracted the repression by the full-length ATF3 and further stimulated the reporter activity. Since there was no significant difference between the raw and normalized luciferase activity, this result is consistent with that for ATF3ΔZip (21). The data clearly indicate that ATF3ΔZip2 is capable of regulating the ATF/CRE-dependent gene expression.
Figure 7.
Effects of ATF3ΔZip2 on transcriptional repression by ATF3. (A) Cos-7 cells were transfected with pME (lane 1), pME-Flag-ATF3 (lane 2) or pME-Flag-ATF3ΔZip2 (lane 3) and their whole-cell extract were analyzed by western blot using monoclonal anti-Flag antibody. (B) Cos-7 cells were co-transfected with pLuc-4xATF and increasing amounts of pME-Flag-ATF3 or pME-Flag-ATF3ΔZip2. At 24 h post-transfection, cell extracts were assayed for luciferase activity as detailed in the Materials and Methods. Results represent mean ± SE from three independent experiments. Statistically significant inhibition or activation are indicated (*P < 0.05; **P < 0.005).
ATF3ΔZip2 is induced in response to stress stimuli, but not expressed in ATF3-overexpressing cancer cells
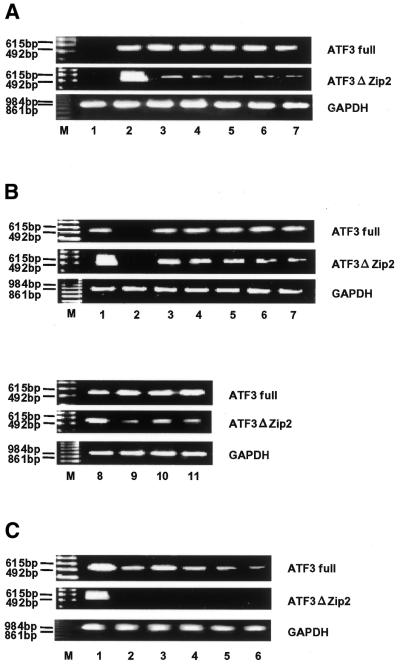
The expression of ATF3 is induced by a wide range of pathophysiological stimuli (13,15). Thus, we investigated whether the expression of ATF3ΔZip2 is also induced by stress stimuli other than A23187. Figure 8A shows that both full-length ATF3 and ATF3ΔZip2 were induced in HUVECs after treatment by several stimuli including homocysteine, thapsigargin, tunicamycin, hydrogen peroxide and TNF-α, although the level of ATF3ΔZip2 mRNA was variable and most significant in the A23187-treated cells. In Saos2 cells, these stimuli also induced ATF3ΔZip2 (Fig. 8B, lanes 3–7). We further examined the effect of MMS, MG132 and colchicine, because these agents can induce ATF3 (16–18). As shown in Figure 8B, lanes 9–11, these agents induced both ATF3 and ATF3ΔZip2. It is also reported that the expression of ATF3 is elevated in several cancer cells and that its expression correlates with high metastatic potency (23,24). Thus, several esophageal cancer cells that exhibit high expression of ATF3 were examined. Figure 8C shows that the ATF3ΔZip2 mRNA was not detectable in these cancer cells. Taken together, our results indicate that, although ATF3ΔZip2 is co-induced with the full-length ATF3 by many stress stimuli, it is not co-expressed with ATF3 in these esophageal cancer cells.
Figure 8.
Induced expression of ATF3ΔZip2 in response to various stimuli. Cells were stimulated with various agents and their total RNA (1 µg) was isolated and subjected to RT–PCR. The full-length ATF3 mRNA was assayed using primer 5′ plus primer 3′, and ATF3ΔZip2 mRNA using primer 5′ plus primer Zip3. The cycle numbers of PCR for full-length ATF3 and ATF3ΔZip2 were 24 and 28 cycles, respectively. (A) HUVECs were untreated (lane 1) or stimulated with 10 µM A23187 (lane 2), 3 mM homocysteine (lane 3), 2 µM thapsigargin (lane 4), 10 µg/ml tunicamycin (lane 5), 0.05 mM hydrogen peroxide (lane 6), 10 ng/ml TNF-α (lane 7) for 2 h. (B) Saos2 cells were untreated (lane 2), or treated with A23187 (lane 3), homocysteine (lane 4), thapsigargin (lane 5), tunicamycin (lane 6) and TNF-α (lane 7) as in (A), and with 0.1 µg/ml MMS for 4 h (lane 9), 10 µM MG132 for 14 h (lane 10), and 2.5 µM colchicine for 1.5 h (lane 11). Lanes 1 and 8, A23187-treated HUVECs as the positive control. (C) Esophageal cancer cell lines over-expressing ATF3 (29) were analyzed. Lane 1, A23187-treated HUVECs as the positive control; lanes 2–6, KYSE70, KYSE150, KYSE170, KYSE190 and KYSE200 cell lines, respectively.
DISCUSSION
Alternative splicing is a powerful and versatile regulatory mechanism that affects gene expression and results in functional diversification of proteins. The primary transcript encoding the ATF3 protein is alternatively spliced to produce ATF3ΔZip2, which is produced through the activation of alternative splicing between the 5′ donor site of intron C and the novel 3′ acceptor site in intron C (Fig. 2). We also showed that the truncated ATF3 protein that had the same mobility as ATF3ΔZip2 was expressed in response to stress stimuli (Fig. 4), providing evidence that ATF3ΔZip2 mRNA could be translated into a protein. This is of particular importance since mRNA harboring a premature stop codon might be subject to nonsense-mediated decay, resulting in the translational repression of mRNA (34,35). Nonsense-mediated decay of mRNA is most likely to occur when the premature termination codon is present upstream of the most 3′ exon–exon junction (34). We speculate that ATF3ΔZip2a mRNA is not a target of decay since its termination codon is present downstream of the most 3′ exon–exon junction (Fig. 2). In contrast, ATF3ΔZip2b mRNA could be a target of decay since the nonsense codon is present upstream of junction of exon D and E. However, more investigation is required to determine whether ATF3ΔZip2 mRNA is subject to the decay system.
The expression of ATF3ΔZip2 was induced in cells exposed to various stimuli. These include a wide range of agents, such as genotoxic agents, proteasome inhibitors, oxidative stress, agents that perturb calcium homeostasis, inhibitors of N-glycosylation, and proinflammatory cytokines. Some of these agents, A23187, thapsigargin, tunicamycin and homocysteine, are inducers of endoplasmic reticulum stress and cause the unfolded protein response. Since all the stress stimuli described in this study also induced the full-length ATF3, what is the biological function of co-inducing ATF3ΔZip2 during stress response? ATF3ΔZip2 lacks the leucine zipper domain and is incapable of binding to the ATF/CRE motif. Thus, it is defective in forming heteromeric complexes with other factors, such as c-Jun, JunB, ATF2 or GADD153. As shown in Figure 7, ATF3ΔZip2 rescued the reporter activity repressed by the full-length ATF3 in a transient transfection assay. However, when the amount of DNA encoding ATF3ΔZip2 was increased, it stimulated the reporter activity. This is consistent with the inhibitory co-factor model proposed previously (21). In that model, ATF3 represses promoters with the ATF/CRE sites by stabilizing the inhibitory co-factors at the promoters. When not bound to DNA, however, ATF3 could sequester the inhibitory co-factors away from the promoters, resulting in stimulation of transcription (21). Since ATF3ΔZip2 is similar to ATF3ΔZip in that it does not bind to the ATF/CRE sites, our results of stimulation of transcription by ATF3ΔZip2 may be consistent with the model. It is thus possible that one function of co-inducing ATF3ΔZip2 is to modulate the transcriptional effects of the full-length ATF3 during stress. Co-induction of alternatively spliced mRNAs that encode proteins with opposite functions has been described previously. One example is the co-induction of ΔFosB with full-length FosB (36). ΔFosB lacks the C-terminal region of FosB and inhibits the function of FosB and other Fos proteins (36). The counteracting effect of ATF3ΔZip2 on ATF3 brings up an important question. What is the physiological significance of ATF3 expression during stress response? Although the answer is not clear, one notion is that ATF3 contributes to the detrimental consequences of stress response (13,15). This notion is mainly derived from the ectopic expression approach. Transgenic mice expressing ATF3 in selective tissues have been demonstrated to have malfunction in the target tissues. As an example, transgenic mice expressing ATF3 in the pancreas have a defect in endocrine function and develop symptoms characteristic of insulin-dependent diabetes (37). Furthermore, transgenic mice expressing ATF3 in the heart have conduction abnormality and contractile dysfunction (38). Therefore, ATF3 appears to be a part of the cellular response that leads to detrimental outcomes. Due to the intrinsic limitations of the transgenic approach, experiments using different approaches are required to substantiate this notion. However, if the full-length ATF3 protein is indeed a detrimental component of stress response, then the co-induction of ATF3ΔZip2 may be a part of the efforts to alleviate the detrimental consequence.
In this context, we note that ATF3 is also induced by signals that induce cell proliferation (12–14) and is expressed in certain cancer cells (23,24). Recently, ATF3 was shown to partially transform chick embryo fibroblasts by promoting growth factor-independent proliferation (39). It is intriguing that ATF3 expression correlates with high potentials of metastasis in murine melanoma cells (23), and its down-regulation by anti-sense oligonucleotides effectively suppressed the growth of human colon cancer cells in mice (40). In our study, very little or no expression of ATF3ΔZip2 was observed in cancer cells that over-express the full-length ATF3. Therefore, the lack of a built-in mechanism to modulate the activities of the full-length ATF3 protein may be a part of the differences that distinguish cancer cells from cells eliciting stress response. However, at present, it is not known whether this discrepancy of expression of ATF3 and ATF3ΔZip2 has a functional role in cell proliferation.
In summary, our demonstration of a novel alternative spliced ATF3ΔZip2 provides a molecular basis for diverse functions of the immediate early response gene ATF3, and strengthens the pivotal role of alternative splicing during the stress response. Furthermore, this might provide a clue to investigating the molecular device for controlling the stress response and cell proliferation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr J. Inoue (Keio University, Japan) for kindly supplying pME-Flag plasmid. This work was supported in part by a Grant-in-Aid for Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and from NASDA and Japan Space Forum to S.K. T.H. acknowledges the support of the National Institute of Health (Eso8690).
DDBJ/EMBL/GenBank accession nos AB078026, AB078027
REFERENCES
- 1.Lopez J.A. (1995) Developmental role of transcription factor isoforms generated by alternative splicing. Dev. Biol., 172, 396–411. [DOI] [PubMed] [Google Scholar]
- 2.Cooper T.A. and Mattox,W. (1997) The regulation of splice-site selection and its role in human disease. Am. J. Hum. Genet., 61, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 4.Chen C.-Y. and Schwartz,R.J. (1996) Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol. Cell. Biol., 16, 6372–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickmore W.A., Oghene,K., Little,M.H., Seawright,A., VanHeyningen,V. and Hastie,N.D. (1992) Modulation of DNA binding specificity by alternative splicing of the Wilms tumor wt1 gene transcript. Science, 257, 235–238. [DOI] [PubMed] [Google Scholar]
- 6.Kozmik Z., Kurzbauer,P., Dorfler,P. and Busslinger,M. (1993) Alternative splicing of Pax-8 gene transcripts is developmentally regulated and generates isoforms with different transactivation properties. Mol. Cell. Biol., 13, 6024–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris A.E., Kloss,B., McChesney,R.E., Bancroft,C. and Chasin,L.A. (1992) An alternatively spliced Pit-1 isoform altered in its ability to transactivate. Nucleic Acids Res., 20, 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macneil C., Ayres,B., Laverriere,A.C. and Burch,J.E. (1997) Transcripts for functionally distinct isoforms of chicken GATA-5 are differentially expressed from alternative first exon. J. Biol. Chem., 272, 8396–8401. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T., Tanaka,K., Ogawa,S., Kurokawa,M., Mitani,K., Nishida,J., Shibata,Y., Yazaki,Y. and Hirai,H. (1995) An acute myeloid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternatively spliced forms. EMBO J., 14, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulkes N.S. and Sassone-Corsi,P. (1992) More is better: activators and repressors from the same gene. Cell, 68, 411–414. [DOI] [PubMed] [Google Scholar]
- 11.Hai T., Liu,F., Coukos,W.J. and Green,M.R. (1989) Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev., 3, 2083–2090. [DOI] [PubMed] [Google Scholar]
- 12.Hsu J,-C., Laz,T., Mohn,K.L. and Taub,R. (1991) Identification of LRF-1, a leucine-zipper protein that is rapidly and highly induced in regenerating liver. Proc. Natl Acad. Sci. USA, 88, 3511–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hai T., Wolfgang,C.D., Marsee,D.K., Allen,A.E. and Sivaprasad,U. (1999) ATF3 and stress responses. Gene Expr., 7, 321–335. [PMC free article] [PubMed] [Google Scholar]
- 14.Hartman M.G. and Hai,T. (2001) The molecular biology and nomenclature of the ATF/CREB family of transcription factors: ATF proteins and homeostasis. Gene, 273, 1–11. [DOI] [PubMed] [Google Scholar]
- 15.Chen B.P.C., Wolfgang,C.D. and Hai,T. (1996) Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell. Biol., 16, 1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shtil A.A., Mandlekar,S., Yu,R., Walter,R.J., Hagen,K., Tan,T.H., Roninson,I.B. and Kong,A.-N.T. (1999) Differential regulation of mitogen-activated protein kinases by microtubule-binding agents in human breast cancer cells. Oncogene, 18, 377–384. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann J., Erdmann,D., Lalande,I., Grossenbacher,R., Noorani,M. and Furst,P. (2000) Proteasome inhibitor induced gene expression profiles reveal overexpression of transcriptional regulators ATF3, GADD153 and MAD1. Oncogene, 19, 2913–2920. [DOI] [PubMed] [Google Scholar]
- 18.Amundson S.A., Bittner,M., Chen,Y., Trent,J., Meltzer,P. and Fornace,A.J. (1999) Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene, 18, 3666–3672. [DOI] [PubMed] [Google Scholar]
- 19.Cai Y., Zhang,C., Nawa,T., Aso,T., Tanaka,M., Oshiro,S., Ichijo,H. and Kitajima,S. (2000) Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH2 terminal kinase and promoter response element. Blood, 96, 2140–2148. [PubMed] [Google Scholar]
- 20.Yin T., Sandhu,G., Wplfgang,C.D., Burrier,A., Webb,R.L., Rigel,D.F., Hai,T. and Whelan,J. (1997) Tissue-specific pattern of stress kinase activation in ischemic/reperfused heart and kidney. J. Biol. Chem., 272, 19943–19950. [DOI] [PubMed] [Google Scholar]
- 21.Chen B.P.C., Liang,G., Whelan,J. and Hai,T. (1994) ATF3 and ATF3 delta Zip. Transcriptional repression versus activation by alternatively spliced isoform. J. Biol. Chem., 269, 15819–15826. [PubMed] [Google Scholar]
- 22.Iyer V.R., Eisen,M.B., Ross,D.T., Schuler,G., Moore,T., Lee,J.C.F., Trent,J.M., Staudt,L.M., Hudson,J., Boguski,M.S., Lashkari,D., Shalon,D., Botstein,D. and Brown,P.O. (1999) The transcriptional program in the response of human fibroblasts to serum. Science, 283, 83–87. [DOI] [PubMed] [Google Scholar]
- 23.Ishiguro T., Nakajima,M., Naito,M., Muto,T. and Tsuruo,T. (1996) Identification of genes differentially expressed in B16 murine melanoma sublines with different metastatic potentials. Cancer Res., 56, 875–879. [PubMed] [Google Scholar]
- 24.Pimkhaokham A., Shimada,Y., Fukuda,Y., Kurihara,N., Imoto,I., Yang,Z.-Q., Imamura,M., Nakamura,Y., Amagasa,T. and Inazawa,J. (2000) Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon. Jpn J. Cancer Res., 91, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu J.-C., Bravo,R. and Taub,R. (1992) Interactions among LRF-1, JunB, c-Jun and c-Fos define a regulatory program in the G1 phase of liver regeneration. Mol. Cell. Biol., 12, 4654–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nawa T., Nawa,T.M., Cai,Y., Zhang,C., Uchimura,I., Narumi,S., Numano,F. and Kitajima,S. (2000) Repression of TNF-α-induced E-selectin expression by PPAR activators: involvement of transcriptional repressor LRF-1/ATF3. Biochem. Biophys. Res. Commun., 275, 406–411. [DOI] [PubMed] [Google Scholar]
- 27.Fawcett T.W., Martindale,J.L., Guyton,K.Z., Hai,T. and Holbrook,N.J. (1999) Complexes containing ATF/CREB interact with the C/EBP-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J., 339, 135–141 [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfgang C.D., Chen,B.P.C., Martindale,J.L., Holbrook,N.J. and Hai,T. (1997) gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell. Biol., 17, 6700–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagmeyer B.M., Duyndam,M.C., Angel,P., de Groot,R.P., Verlaan,M., Elfferich,P., van der Eb,A.J. and Zantema,A. (1996) Altered AP-1/ATF complexes in adenovirus-E1-transformed cells due to E1A-dependent induction of ATF3. Oncogene, 12, 1025–1032. [PubMed] [Google Scholar]
- 30.Tamaru M. and Narumi,S. (1999) E-selectin gene expression is induced synergistically with the coexistence of activated classic protein kinase C and signals elicited by interleukin-1β but not tumor necrosis factor-α. J. Biol. Chem., 274, 3753–3763. [DOI] [PubMed] [Google Scholar]
- 31.Tsukamoto N., Kobayashi,N., Azuma,S., Yamamoto,T. and Inoue,J. (1999) Two differently regulated nuclear factor κB activation pathways triggered by the cytoplasmic trail of CD40. Proc. Natl Acad. Sci. USA, 96, 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowry O.H., Rosebrough,N.J., Farr,A.L. and Randall,R.J. (1951) Protein measurement with folin phenol reagent. J. Biol. Chem., 193, 265–275. [PubMed] [Google Scholar]
- 33.Liang G., Wolfgang,C.D., Chen,B.P.C., Chen,T.-H. and Hai,T. (1996) ATF3 gene. genome organization, promoter and regulation. J. Biol. Chem., 271, 1695–1701. [DOI] [PubMed] [Google Scholar]
- 34.Lykke-Anderson J., Shu,M.-D. and Steiz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- 35.Maquat L.E. and Carmichael,G.G. (2001) Quality control of mRNA function. Cell, 104, 173–176. [DOI] [PubMed] [Google Scholar]
- 36.Nakabeppu Y. and Nathans,D. (1991) A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell, 64, 751–759. [DOI] [PubMed] [Google Scholar]
- 37.Allen-Jennings A.E., Hartman,M.G., Kociba,G.J. and Hai,T. (2001) The roles of ATF3 in glucose homeostasis: a transgenic mouse model with liver dysfunction and defects in endocrine pancreas. J. Biol. Chem., 276, 29507–29514. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto Y., Chaves,A., Chen,J., Kelley,R., Jones,K., Weed,H.G., Gardner,K.L., Gangi,L., Yamaguchi,M., Klomkleaw,W., Nakayama,T., Hamlin,R.L., Carnes,C.A., Altschuld,R.A., Bauer,J.A. and Hai,T. (2001) Transgenic mice expressing ATF3 in the heart have conduction abnormalities and contractile dysfunction. Am. J. Pathol., 159, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez S., Vial,E., van Dam,H. and Castellazzi,M. (2001) Transcription factor ATF3 partially transforms chick embryo fibroblasts by promoting growth factor-independent proliferation. Oncogene, 20, 1135–1141. [DOI] [PubMed] [Google Scholar]
- 40.Ishiguro T., Nagawa,H., Naito,M. and Tsuruo,T. (2000) Inhibitory effect of ATF3 antisense oligonucleotide on ectopic growth of HT29 human colon cancer cells. Jpn J. Cancer Res., 91, 833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]