Abstract
Background and aims
The prevalence of metabolic dysfunction associated steatotic liver disease (MASLD), formerly known as nonalcoholic fatty liver disease (NAFLD), has become a significant public health concern with an increased atherosclerotic cardiovascular disease risk. This study investigates the impact of NAFLD-related single nucleotide polymorphisms (SNPs) on carotid atherosclerosis development in a Japanese population without diabetes, dyslipidemia, and hypertension.
Methods
The prospective observational study, part of the Kyushu and Okinawa Population Study (KOPS), included 945 participants (median age 55 [47, 63]) without carotid atherosclerosis, increased alcohol intake, diabetes, dyslipidemia, hypertension, or chronic hepatitis at baseline. NAFLD-related SNPs (GCKR, NCAN, and PNPLA3) were genotyped, and carotid intima-media thickness (cIMT) was measured using ultrasonography. Univariate and multivariate regression analyses were performed to assess the association of NAFLD-related SNPs on newly developed carotid atherosclerosis over five years.
Results
After five years, 125 (13.2 %) participants developed carotid atherosclerosis. The NCAN (rs2228603) T allele was associated with a lower incidence rate of carotid atherosclerosis (4.7 % in NCAN CT/TT genotype vs. 13.9 % in CC genotype; p = 0.04), and NCAN T allele carriers exhibited a favorable lipid profile. These associations were not altered by either recruiting area or obese. The GCKR T allele and PNPLA3 C allele were associated with low carotid atherosclerosis development rates but were not significant.
Conclusions
Our results suggested that some NAFLD-related SNPs may influence atherosclerosis through lipid metabolism among Japanese individuals without metabolic syndrome.
Keywords: NAFLD, SNP, NCAN, Atherosclerosis, Carotid intima-media thickness
Highlights
-
•
Association between atherosclerosis and nonalcoholic fatty liver disease (NAFLD) related SNPs is limited.
-
•
We evaluated the impact of NAFLD related SNPs on carotid atherosclerosis incident over five years in a general population.
-
•
The carotid atherosclerosis development rate was lower in NCAN (rs2228603) minor allele carriers.
-
•
NCAN minor allele carriers also exhibited a favorable lipid profile especially in remnant-like particle cholesterol.
Non-standard abbreviations and acronyms
- ASCVD
atherosclerotic cardiovascular disease
- BMI
body mass index
- cIMT
carotid intima-media thickness
- FPG
fasting plasma glucose
- GCKR
glucokinase regulator
- GWAS
genome-wide association study
- HbA1c
hemoglobin A1c
- HDL
high-density lipoprotein
- hs-CRP
high-sensitivity C-reactive protein
- KOPS
Kyushu and Okinawa Population Study
- LDL
low-density lipoprotein
- MASLD
metabolic dysfunction associated steatotic liver disease
- MASH
metabolic dysfunction associated steatohepatitis
- NAFLD
nonalcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- NCAN
neurocan
- PNPLA3
patatin-like phospholipase domain-containing protein 3
- RLP
remnant-like particle
- SNP
single nucleotide polymorphism
- TG
triglycerides
- TM6SF2
trans-membrane 6 superfamily member 2
- VLDL
very low-density lipoprotein
1. Introduction
The number of metabolic dysfunction associated steatotic liver disease (MASLD), formerly known as nonalcoholic fatty liver disease (NAFLD), patients have been increasing rapidly, and MASLD has become the most common chronic liver disease and a significant global health concern [1]. MASLD can progress to metabolic dysfunction associated steatohepatitis (MASH), cirrhosis, and hepatocellular carcinoma [2]. Moreover, MASLD is also known to be a risk factor for atherosclerotic cardiovascular disease (ASCVD), which includes cardiovascular disease and stroke [3,4]. ASCVD remains the leading cause of mortality worldwide, claiming over 15 million lives annually and accounting for 85 % of deaths related to cardiovascular dysfunction [5]. The pathogenes of MASLD are fat accumulation, chronic inflammation, oxidative stress, and fibrosis progression in the liver, similar to that of atherosclerotic changes in vascular walls. Thus, there would be common genetic prepositions for the pathogenesis between MASLD and ASCVD.
Several studies have reported on the relationship between NAFLD-related SNPs and atherosclerosis. PNPLA3, a representative genetic variant for NAFLD, has been shown to influence the severity of carotid atherosclerosis in patients with NAFLD [6]. GCKR has been reported to affect lipid metabolism in NAFLD patients [7]. Similarly, genetic polymorphisms in NCAN and TM6SF2 have also been reported to influence lipid metabolism in patients with NAFLD [8,9]. Moreover, many genetic risk loci have also been identified for NAFLD, some related to lipid metabolism, which would be a significant risk factor for ASCVD [[10], [11], [12], [13]]. However, few reports have examined the direct impact of NAFLD-related single nucleotide polymorphism (SNP) on atherosclerosis development independent of NAFLD or MASLD.
In this pilot study, we selected four NAFLD-related SNPs and assessed their impact on the development of carotid atherosclerosis, which is defined as the combination of either a newly mean carotid intima-media thickness (cIMT) exceeding 1.1 mm or the formation of new plaques, a focal thickening over 1.1 mm, among the community-dwelling Japanese population without increased alcohol intake, diabetes, dyslipidemia, hypertension, or chronic hepatitis.
2. Patients and methods
2.1. Study population and design
This prospective observational study was part of the Kyushu and Okinawa Population Study (KOPS), a prospective community-based study investigating the epidemiology of lifestyle-related diseases and cancer in Japan since 2004 [14]. KOPS consists of two populations: wave 1 and wave 2. Wave 1 includes 10,697 participants who visited free public examinations between 2004 and 2007, and wave 2 includes 6380 participants between 2009 and 2012. KOPS has a total of 17,077 participants, 3006 of whom visited both wave 1 and 2 [14]. All participants were aged ≥20 at the examination, and the population was almost entirely Japanese.
In the present study, the 945 analyzed participants were selected by the following criteria: 1) underwent carotid ultrasonography at wave 1 and 2 surveys, 2) without increased alcohol intake (average daily alcohol intake 30g or more for males and 20g and more for females), hepatic dysfunction, hypertension, diabetes, and dyslipidemia at wave 1, 3) without chronic hepatitis including viral hepatitis, and 4) without adrenocorticosteroid therapy. All participants provided blood samples after an overnight fast and underwent physical examinations, including measurements of blood pressure, height, and weight, as part of their participation in this study. All participants also provided their medical history, use of all medications, and lifestyle habits.
Each participant provided informed consent prior to enrolment. All studies were conducted in accordance with the principles of the Declaration of Helsinki, as revised in 2008 and approved by the Kyushu University Hospital Ethics Committee before data collection (approval number: 590-04).
2.2. SNP genotyping
Genomic DNA was isolated from peripheral blood leukocytes. The four NAFLD-related SNPs were genotyped with TaqMan PCR using an ABI TaqMan allelic discrimination kit (QuantStudio 5 Real‐Time PCR Systems; Applied Biosystems). We analyzed the following four NAFLD-related SNPs, which have been reported to have a minor allele frequency above a certain level in the Japanese population: rs1260326 (C/T) on chromosome 2 in glucokinase regulator (GCKR), rs2228603 (C/T) on chromosome 19 in neurocan (NCAN), rs58542926 (C/T) on chromosome 19 in trans-membrane 6 superfamily member 2 (TM6SF2), and rs738409 (G/C) on chromosome 22 in patatin-like phospholipase domain-containing protein 3 (PNPLA3). All four SNPs passed genotyping quality control. The genotype call rate was 97.5 %.
2.3. Laboratory measurements
All blood samples were collected at least 8 h after overnight fasting. Aliquots of whole blood and fresh serum and plasma samples from each participant were immediately separated. Plasma fasting plasma glucose (FPG), high-density lipoprotein (HDL) cholesterol, and triglyceride (TG) levels were measured using standard enzymatic methods; in addition, direct LDL cholesterol, small dense LDL cholesterol, LDL-TG, remnant-like particle (RLP) cholesterol, and lipoprotein(a) were measured on an Olympus AU400 automated chemistry analyzer using assay kits obtained from Denka Corporation (Niigata, Japan), as previously described [[14], [15], [16]]. Hemoglobin A1c (HbA1c) levels were measured in fresh whole blood samples using a coherent immune method (RAPIDIA Auto HbA1c, Fujirebio) and were expressed as US National Glycohemoglobin Standardization Program levels (%). Serum C-reactive protein (CRP) levels were measured using nephelometry (N Latex CRP II; Siemens Healthcare Diagnostics Inc., Tokyo, Japan). The coefficient of variation for all assays was <5 %.
2.4. Assessment of carotid intima-media thickness
The cIMT levels were assessed using high-resolution B-mode ultrasonography with a 10-MHz linear array probe (UF-400AX®, Fukuda Denshi Co., Ltd, Tokyo, Japan. Well-trained staff doctors of our department performed all ultrasound examinations according to the Japanese Carotid Artery Ultrasound Guidelines [17]. Carotid ultrasonography was performed focusing on an area 20 mm proximal to the bulb of far walls of the bilateral common carotid arteries, and obtained images were analyzed offline using automated digital edge-detection IMT measurement software (Intima-Scope; MEDIA CROSS Co., Ltd., Tokyo, Japan) as previously described [18,19]. Analysis of cIMT levels using Intima-Scope was performed by a well-trained technician who was blinded to the clinical information. Carotid atherosclerosis was defined as 1) carotid thickening as a mean cIMT ≥1.1 mm or 2) the presence of a plaque, a focal thickening >1.1 mm at the level of common carotid arteries or bulbs. These criteria in Japan have been proposed by the Joint Committee of the Japan Academy of Neurosonology and the Japan Society of Embolus Detection and Treatment on Guideline for Nuerosonology [17].
2.5. Statistical analysis
A total of 945 participants were analyzed in this study. Data of continuous variables are expressed as median values with the 25th and 75th percentile values. Categorical variables are reported as frequencies and percentages. Univariate analyses were conducted using the Mann-Whitney U and Kruskal-Wallis tests for continuous variables or the chi-square test for categorical variables. Multivariate regression analysis was performed to compare the body mass index, systolic blood pressure, and lipoproteins between the two groups. Analysis of variance was performed to compare those continuous variables across three groups. Univariate and multivariate logistic regression analyses were conducted to estimate the influence of NAFLD-related SNPs on the development of carotid atherosclerosis. In these analyses, we calculated the number of alleles present in each genotype. In multivariate analyses, we added the participants' residential areas (Ishigaki, Iki, Hoshino, and Kasuya) as a confounding factor. In addition, we conducted interaction analyses to assess the robustness of our findings. We assessed the interactions between the NAFLD-related genetic polymorphisms and the following three factors: 1) the participants’ residential areas, 2) obese, and 3) sex. Obese was defined as a BMI of 25 kg/m2 or higher. The p-values for trends in multivariate adjustment factors were calculated using the general linear regression model. All statistical analyses were performed using SAS (version 9.4; SAS Institute Inc., Cary, NC, USA). A p-value of <0.05 was considered statistically significant.
3. Results
3.1. Baseline characteristics
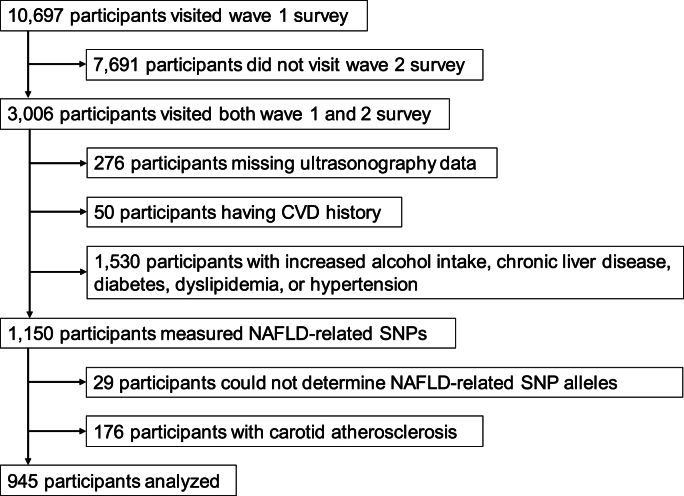
Fig. 1 shows the flowchart of subject selection for this study. This analysis included 945 individuals. Table 1 presents the characteristics of the 945 analyzed participants without carotid atherosclerosis at baseline. Among 945 participants, 125 (13.2 %) developed carotid atherosclerosis after five years. The 125 who developed carotid atherosclerosis had significantly older age, a higher proportion of males, and elevated systolic blood pressure. No significant differences were observed in BMI or smoking and alcohol consumption habits. Those 125 also had significantly lower HDL cholesterol and higher HbA1c levels. While not statistically significant, those 125 showed higher levels of triglycerides, LDL cholesterol, small dense LDL cholesterol, and lipoprotein(a).
Fig. 1.
The study flowchart. In this study, we selected participants without a history of cardiovascular disease, increased alcohol intake, chronic liver disease, diabetes, dyslipidemia, or hypertension from a large-scale epidemiological study in Japan.
Table 1.
Characteristics of participants classified by the atherosclerosis development in the carotid artery over 5 years.
| With atherosclerosis development (n = 125) | Without atherosclerosis development (n = 820) | P value | |
|---|---|---|---|
| Demographic | |||
| Age – years | 59 [51, 66] | 53 [44.5, 60.5] | <0.001 |
| Sex – no. (women/men) | 77 (61.6)/48 (38.4) | 640 (78.1)/180 (21.9) | <0.001 |
| Body mass index – kg/m2 | 23.0 [20.7, 25.0] | 22.3 [20.4, 24.6] | 0.13 |
| Obese – no. (yes/no) | 31 (24.8)/94 (75.2) | 170 (21.0)/640 (79.0) | 0.33 |
| Smoking habit – no. (current/past/never) | 19 (16.5)/13 (11.3)/83 (72.2) | 89 (11.4)/103 (13.2)/590 (75.5) | 0.22 |
| Alcohol drinking habit – no. (yes/no) | 42 (36.5)/73 (63.5) | 300 (38.4)/481 (61.6) | 0.68 |
| Systolic blood pressure – mmHg | 120 [112, 130] | 114 [106, 124] | <0.001 |
| NAFLD risk SNP alleles | |||
| GCKR – C/T | 130 (52.0)/120 (48.0) | 767 (47.5)/873 (52.5) | 0.14 |
| NCAN – C/T | 247 (98.8)/3 (1.2) | 1578 (96.3)/62 (3.8) | 0.04 |
| TM6SF2 – C/T | 228 (91.2)/22 (8.8) | 1520 (92.7)/120 (7.3) | 0.44 |
| PNPLA3 – C/G | 134 (53.6)/116 (46.4) | 921 (56.2)/719 (43.8) | 0.45 |
| Laboratory data | |||
| HDL cholesterol – mg/dl | 63 [54, 73] | 66 [57, 76] | 0.01 |
| Triglycerides – mg/dl | 84 [65, 107] | 79 [60.5, 106] | 0.36 |
| LDL cholesterol – mg/dl | 87 [72, 104] | 82 [68, 100] | 0.14 |
| Small dense LDL cholesterol – mg/dl | 19.0 [13.8, 25.7] | 17.1 [12.8, 23.2] | 0.08 |
| LDL-triglyceride – mg/dl | 6.4 [4.5, 9.4] | 6.4 [4.3, 9.4] | 0.64 |
| RLP cholesterol – mg/dl | 9.05 [4.5, 15.3] | 10.1 [6.5, 17.45] | 0.06 |
| Lipoprotein (a) – mg/dl | 8.0 [4.4, 14.5] | 6.9 [4.0, 12.8] | 0.14 |
| Fasting plasma glucose – mg/dl | 92 [86, 99] | 91 [86, 97] | 0.46 |
| Hemoglobin A1c – % | 5.4 [5.2, 5.7] | 5.3 [5.1, 5.5] | <0.001 |
| C reactive protein – mg/dl | 0.0315 [0.014, 0.0735] | 0.029 [0.012, 0.062] | 0.41 |
Data are shown as median [1st quartile, 3rd quartile] or number (%).
GCKR, glucokinase regulatory protein; NAFLD, nonalcoholic fatty liver disease; NCAN, nurocan; TM6SF2, transmembrane 6 superfamily member 2; PNPLA3, patatin-like phospholipase domain containing 3 protein; HDL, high-density lipoprotein; LDL, low-density lipoprotein; RLP, remnant-like particle.
The genetic frequencies of the polymorphisms tested in this study are reported in Table 1. Regarding NAFLD-related SNPs, the 125 participants who developed carotid atherosclerosis had a significantly higher prevalence of C allele carriers in NCAN. Of the four polymorphisms, GCKR, NCAN, and PNPLA3 fit with the Hardy-Weinberg equilibrium (χ2 = 0.08 and p = 0.78 for GCKR rs1260326; χ2 = 0.01 and p = 0.91 for NCAN rs2228603; χ2 = 0.04 and p = 0.85 for PNPLA3 rs738409, respectively). TM6SF2 rs58542926 did not fit the Hardy-Weinberg equilibrium (χ2 = 4.77 and p = 0.03). Thus, we excluded TM6SF2 from the further analyses.
3.2. Association between carotid atherosclerosis development over 5 years and NAFLD-related SNPs
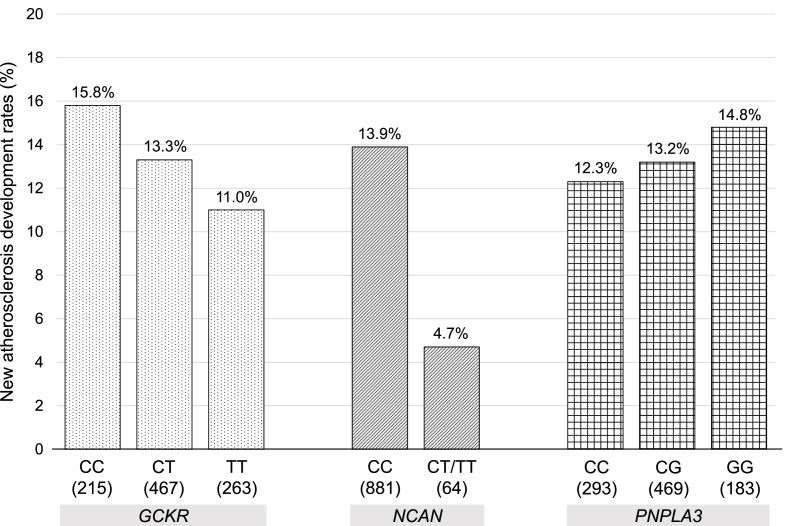
Fig. 2 illustrates the five-year development rates of carotid atherosclerosis according to the genotype of NAFLD-related SNPs. The overall five-year development rate of carotid atherosclerosis in the analyzed population was 13.2 %. Due to limited sample sizes in NCAN genotype TT, genotype CT and TT were combined for this analysis. Regarding NCAN, the development rate was significantly lower at 4.7 % for genotype CT or TT compared to 13.9 % for genotype CC. For GCKR, the TT genotype had the lowest development rate at 11.0 %, while the CC genotype had the highest rate at 15.8 %. For PNPLA3, the CC genotype had the lowest development rate at 12.3 %, while the CC genotype had the highest rate at 14.8 %.
Fig. 2.
Development rates of newly carotid atherosclerosis according to NAFLD-related SNP genotypes.
Newly carotid atherosclerosis development rates are shown according to NAFLD-related SNP genotypes. The development of new carotid atherosclerosis was defined as the mean carotid intima-media thickness (cIMT) level less than 1.1 mm without plaque, a focal thickening over 1.1 mm, at wave 1 (baseline) and the cIMT level ≥1.1 mm or plaques observed at wave 2 (five-year later).
cIMT, carotid intima-media thickness; GCKR, glucokinase regulator; NAFLD, nonalcoholic fatty liver disease; NCAN, neurocan; PNPLA3, Patatin-like phospholipase domain-containing protein 3; SNP, single nucleotide polymorphism.
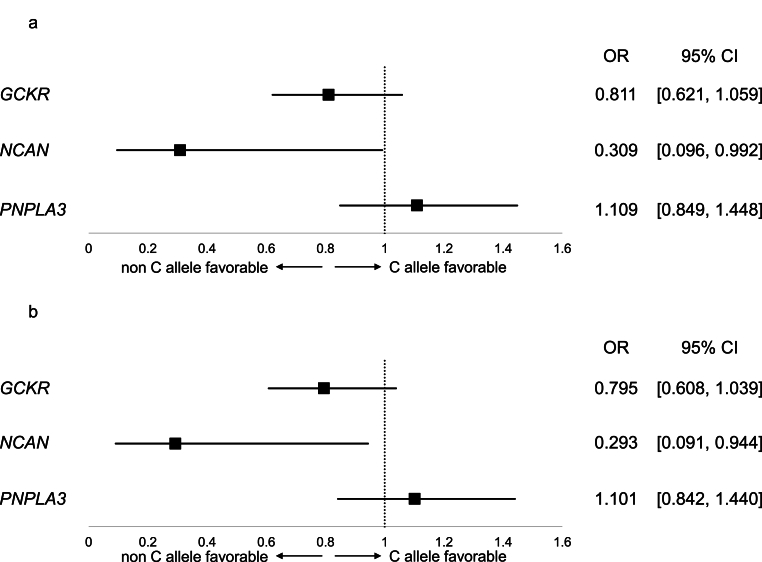
Fig. 3 depicts the impact of each NAFLD-related SNPs on the development of carotid atherosclerosis. Univariate regression analysis showed that the odds ratio of the T allele of NCAN was 0.309 against the C allele, suggesting that the T allele of NCAN is protective for the development of carotid atherosclerosis (Fig. 3a). Multivariate regression analysis, adjusting for the participants’ residential area, also indicated the protective nature of the T allele of NCAN for the development of carotid atherosclerosis, with a multivariate-adjusted odds ratio of 0.293 (Fig. 3b). However, the p-values for these two regression analyses were 0.04, which did not meet the significance level after the Bonferroni correction (p < 0.017). Variants of GCKR and PNPLA3 did not exhibit significant trends in the univariate or multivariate analysis.
Fig. 3.
Univariate and multivariate analyses of NAFLD-related genetic polymorphisms for newly carotid atherosclerosis development.
The results of univariate (Fig. 3a) and multivariate (Fig. 3b) logistic regression analyses for carotid atherosclerosis development and MASLD-related SNP alleles are shown. In multivariate analysis, participants' residential area was included.
CI, confidential interval; GCKR, glucokinase regulator; NAFLD, nonalcoholic fatty liver disease; NCAN, neurocan; OR, odds ratio; PNPLA3, Patatin-like phospholipase domain-containing protein 3; SNP, single nucleotide polymorphis.
3.3. Association between risk factors for atherosclerosis and NAFLD-related SNPs
We examined the association between polymorphisms of NAFLD-related SNPs and risk factors for atherosclerosis. Regarding NCAN SNPs, the genotype CT/TT group exhibited significantly lower RLP cholesterol than the genotype CC group. In addition, the genotype CT/TT group showed higher HDL cholesterol and lower triglycerides, small dense LDL cholesterol, LDL-TG, systolic blood pressure, and BMI than the genotype CC group (Table 2). For GCKR, the genotype TT group showed significantly higher LDL-TG and lower FPG than the CC and CT groups (Table 3). In PNPLA3, the genotype GG group had higher RLP cholesterol than the CG and CC groups, but not significant (Table 4). These associations did not alter when adjusted with the participants’ residential area.
Table 2.
Characteristics of participants classified by the NCAN genotype.
| NCAN CC (n = 881) | NCAN non-CC (n = 64) | P value | |
|---|---|---|---|
| Demographic | |||
| Carotid artery atherosclerosis development – no. (%) | 122 (13.9) | 3 (4.7) | 0.04 |
| Age – years | 54 [45, 61] | 51 [45, 58.5] | 0.13 |
| Sex – no. (women/men) | 669 (75.9)/212 (24.1) | 48 (75.0)/16 (25.0) | 0.87 |
| Body mass index – kg/m2 | 22.5 [20.5, 24.6] | 21.5 [20.1, 24.4] | 0.22 |
| Obese – no. (yes/no) | 191 (21.9)/682 (78.1) | 10 (16.1)/52 (83.9) | 0.29 |
| Smoking habit – no. (current/past/never) | 101 (12.1)/104 (12.5)/629 (75.4) | 7 (11.1)/12 (19.1)/44 (69.8) | 0.61 |
| Alcohol drinking habit – no. (yes/no) | 318 (38.2)/515 (61.8) | 24 (38.1)/39 (61.9) | 0.68 |
| Systolic blood pressure – mmHg | 116 [106, 126] | 112 [107, 122] | 0.36 |
| Laboratory data | |||
| HDL cholesterol – mg/dl | 65 [57, 76] | 67.5 [59, 76] | 0.47 |
| Triglycerides – mg/dl | 79 [61, 106] | 77.5 [59, 103] | 0.38 |
| LDL cholesterol – mg/dl | 82 [69, 101] | 83 [66, 98.5] | 0.69 |
| Small dense LDL cholesterol – mg/dl | 17.4 [13.1, 23.4] | 16.55 [11.9, 22.8] | 0.34 |
| LDL-triglyceride – mg/dl | 6.4 [4.3, 9.4] | 6.25 [4.9, 8.45] | 0.89 |
| RLP cholesterol – mg/dl | 10.2 [6.1, 17.5] | 7.55 [5.0, 10.9] | 0.02 |
| Lipoprotein (a) – mg/dl | 7.1 [4.1, 13.1] | 5.95 [4.1, 15.0] | 0.99 |
| Fasting plasma glucose – mg/dl | 91 [86, 97] | 90.5 [85, 97] | 0.75 |
| Hemoglobin A1c – % | 5.3 [5.1, 5.5] | 5.3 [5.1, 5.55] | 0.59 |
| C reactive protein – mg/dl | 0.029 [0.013, 0.064] | 0.023 [0.009, 0.053] | 0.20 |
Data are shown as median [1st quartile, 3rd quartile] or number (%).
Obese was defined as body mass index ≥25 kg/m2.
Since only 2 participants had the NCAN TT genotype, the CT and TT genotypes were combined and categorized as the non-CC genotype.
NCAN, nurocan; HDL, high-density lipoprotein; LDL, low-density lipoprotein; RLP, remnant-like particle.
Table 3.
Characteristics of participants classified by the GCKR genotype.
| GCKR CC (n = 215) | GCKR CT (n = 467) | GCKR TT (n = 263) | P value | |
|---|---|---|---|---|
| Demographic | ||||
| Carotid artery atherosclerosis development – no. (%) | 34 (15.8) | 62 (13.3) | 29 (11.0) | 0.12 |
| Age – years | 55 [47, 62] | 54 [45, 61] | 54 [45, 61] | 0.64 |
| Sex – no. (women/men) | 162 (75.4)/53 (24.6) | 350 (75.0)/117 (25.0) | 205 (78.0)/58 (22.0) | 0.48 |
| Body mass index – kg/m2 | 22.7 [20.5, 24.8] | 22.5 [20.4, 24.7] | 22.2 [20.6, 24.2] | 0.47 |
| Obese – no. (yes/no) | 47 (22.1)/166 (77.9) | 109 (23.4)/357 (76.6) | 45 (17.6)/211 (82.4) | 0.21 |
| Smoking habit – no. (current/past/never) | 27 (13.0)/22 (10.6)/159 (76.4) | 56 (12.6)/66 (14.9)/321 (72.5) | 25 (10.2)/28 (11.4)/193 (78.4) | 0.41 |
| Alcohol drinking habit – no. (yes/no) | 74 (35.6)/134 (64.4) | 164 (37.1)/278 (62.9) | 104 (42.3)/142 (57.7) | 0.10 |
| Systolic blood pressure – mmHg | 119 [107, 126] | 116 [107, 124] | 114 [106, 125] | 0.55 |
| Laboratory data | ||||
| HDL cholesterol – mg/dl | 66 [56, 77] | 66 [57, 76] | 64 [58, 75] | 0.97 |
| Triglycerides – mg/dl | 74 [56, 102] | 79.5 [63, 106] | 81 [61, 111] | 0.07 |
| LDL cholesterol – mg/dl | 85 [70, 100] | 81 [68, 101] | 83.5 [69, 101] | 0.81 |
| Small dense LDL cholesterol – mg/dl | 17.5 [13.1, 22.7] | 17.1 [13.1, 23.0] | 17.95 [12.8, 24.8] | 0.53 |
| LDL-triglyceride – mg/dl | 5.8 [3.9, 8.7] | 6.5 [4.2, 9.5] | 6.6 [4.7, 9.8] | 0.03 |
| RLP cholesterol – mg/dl | 10.5 [6.0, 17.5] | 9.8 [6.1, 17.0] | 10.75 [5.95, 16.65] | 0.79 |
| Lipoprotein (a) – mg/dl | 7.0 [3.75, 12.9] | 7.2 [3.9, 13.2] | 7.0 [4.6, 13.3] | 0.47 |
| Fasting plasma glucose – mg/dl | 93 [88, 99] | 91 [86, 97] | 90 [84, 97] | <0.01 |
| Hemoglobin A1c – % | 5.4 [5.2, 5.6] | 5.3 [5.1, 5.5] | 5.3 [5.1, 5.5] | 0.09 |
| C reactive protein – mg/dl | 0.029 [0.014, 0.064] | 0.024 [0.011, 0.0615] | 0.033 [0.014, 0.067] | 0.18 |
Data are shown as median [1st quartile, 3rd quartile] or number (%).
Obese was defined as body mass index ≥25 kg/m2.
GCKR, glucokinase regulatory protein; HDL, high-density lipoprotein; LDL, low-density lipoprotein; RLP, remnant-like particle.
Table 4.
Characteristics of participants classified by the PNPLA3 genotype.
| PNPLA3 CC (n = 293) | PNPLA3 CG (n = 469) | PNPLA3 GG (n = 183) | P value | |
|---|---|---|---|---|
| Demographic | ||||
| Carotid artery atherosclerosis development – no. (%) | 36 (12.3) | 62 (13.2) | 27 (14.8) | 0.45 |
| Age – years | 54 [45, 61] | 54 [46, 61] | 54 [43, 62] | 0.95 |
| Sex – no. (women/men) | 222 (75.8)/71 (24.2) | 363 (77.4)/106 (22.6) | 132 (72.1)/51 (27.9) | 0.48 |
| Body mass index – kg/m2 | 22.7 [20.6, 24.7] | 22.1 [20.3, 24.6] | 22.6 [20.6, 24.6] | 0.43 |
| Obese – no. (yes/no) | 62 (21.4)/228 (78.6) | 101 (21.8)/363 (78.2) | 38 (21.0)/143 (79.0) | 0.95 |
| Smoking habit – no. (current/past/never) | 37 (13.3)/38 (13.6)/204 (73.1) | 52 (11.6)/56 (12.4)/342 (76.0) | 19 (11.3)/22 (13.1)/127 (75.6) | 0.45 |
| Alcohol drinking habit – no. (yes/no) | 109 (39.1)/170 (60.9) | 158 (35.2)/291 (64.8) | 75 (44.6)/93 (55.4) | 0.42 |
| Systolic blood pressure – mmHg | 114 [106, 124] | 116 [107, 125] | 120 [107, 128] | 0.30 |
| Laboratory data | ||||
| HDL cholesterol – mg/dl | 66 [57, 76] | 64.5 [57, 76] | 66 [56, 75] | 0.97 |
| Triglycerides – mg/dl | 80 [63, 110] | 78.5 [59, 103] | 79 [61, 110] | 0.30 |
| LDL cholesterol – mg/dl | 82 [68, 103] | 83 [67.5, 100] | 82 [69, 100] | 0.87 |
| Small dense LDL cholesterol – mg/dl | 17.35 [13.0, 24.3] | 17.3 [12.7, 22.8] | 17.65 [13.8, 23.9] | 0.46 |
| LDL-triglyceride – mg/dl | 6.45 [4.6, 9.4] | 6.3 [4.2, 9.1] | 6.55 [4.1, 9.7] | 0.59 |
| RLP cholesterol – mg/dl | 9.8 [5.7, 17.4] | 9.6 [5.95, 16.3] | 11.9 [7.6, 18.6] | 0.054 |
| Lipoprotein (a) – mg/dl | 7.1 [4.3, 11.7] | 6.95 [3.9, 13.5] | 6.9 [4.4, 14.7] | 0.92 |
| Fasting plasma glucose – mg/dl | 91 [86, 98] | 91 [86, 97] | 92 [87, 98] | 0.06 |
| Hemoglobin A1c – % | 5.3 [5.1, 5.6] | 5.3 [5.1, 5.5] | 5.3 [5.2, 5.55] | 0.58 |
| C reactive protein – mg/dl | 0.033 [0.014, 0.062] | 0.029 [0.011, 0.064] | 0.0275 [0.014, 0.0665] | 0.42 |
Data are shown as median [1st quartile, 3rd quartile] or number (%).
Obese was defined as body mass index ≥25 kg/m2.
PNPLA3, patatin-like phospholipase domain containing 3 protein; HDL, high-density lipoprotein; LDL, low-density lipoprotein; RLP, remnant-like particle.
3.4. Interaction analyses
We conducted interaction analyses to assess whether the associations between NAFLD-related genes and newly developed carotid atherosclerosis were influenced by 1) participants' residential area, 2) obesity, and 3) sex. These three interaction analyses did not show any significance.
4. Discussion
In this study, we investigated the impact of NAFLD-related genetic polymorphisms on carotid atherosclerosis in 945 Japanese community-dwelling individuals without chronic liver diseases, excessive alcohol intake, diabetes, dyslipidemia, hypertension, or atherosclerosis in carotid arteries at baseline. After five years, 125 individuals (13.2 %) developed carotid atherosclerosis. Among the analyzed three NAFLD-related SNPs (GCKR, NCAN, and PNPLA3), the minor allele of NCAN demonstrated a protective effect against the development of carotid atherosclerosis.
NCAN is a gene variant reported in GWAS in neurologic diseases and is considered a risk factor for neuropsychiatric disorders [20,21]. Neurocan, encoded by the NCAN gene, is a chondroitin sulfate proteoglycan. Neurocan is expressed not only in neuronal tissue but also in the liver and may be involved in the transport processes of the liver. The NCAN genetic variant (rs2228603: C > T) leads to an amino acid exchange (proline to serine) at position 92 and alters protein structure and function. It has been reported that NCAN polymorphism influences the development of hepatocellular carcinoma in alcoholic liver disease [22] and the development of NAFLD [11,23]. Among the NAFLD patients undergoing bariatric surgery, individuals with the NCAN T allele showed a high prevalence of fibrosis and inflammation in their liver [23]. However, this study also reported that the NCAN T allele group had lower LDL cholesterol and triglycerides levels. This seemingly paradoxical phenomenon may occur due to either increased fatty acids uptake or decreased lipolysis; consequently, intrahepatic lipid accumulation is promoted, and lipid secretion from the liver decreases. Similar to this study of European descendants, studies with Asian populations reported that NCAN T allele carriers exhibited a favorable lipid profile [8,24]. In our study, targeting the non-metabolic syndrome Japanese population, NCAN T allele carriers showed a lower risk of progression of carotid atherosclerosis regardless of the participants’ residential area, obesity, and sex. In addition, NCAN T allele carriers exhibited a favorable lipid profile; RLP cholesterol levels were significantly lower. RLP cholesterol is a remnant particle derived from triglyceride-rich lipoproteins [25]. Elevated levels of RLP cholesterol have been associated with an increased risk of cardiovascular diseases, as these particles may contribute to atherosclerosis [26]. Similar to RLP cholesterol, apolipoproteins containing ApoB, such as LDL cholesterol and small dense LDL cholesterol, are substantial risk factors for cardiovascular disease and atherosclerosis. Although NAFLD-related SNPs have been reported to be associated with lipid metabolism, most studies have been conducted in patients with NAFLD [27,28]. The participants of this study did not have hypertension, diabetes, or dyslipidemia, allowing us to observe the effects of NAFLD-related SNPs on lipid metabolism directly. The NCAN minor allele carriers also exhibited lower levels of small dense LDL cholesterol and lipoprotein(a), suggesting that genetic polymorphisms of NCAN may influence lipid metabolism. Another hypothesis regarding the influence of NCAN genetic polymorphisms on lipid metabolism involves the interaction between the gut, liver, and brain, potentially modulating dietary fat intake. Further research is necessary to understand how NCAN genetic polymorphisms impact blood vessels and central control of lipid metabolism.
The glucokinase regulatory protein encoded by the GCKR gene acts as an inhibitor of glucokinase. Glucokinase is the principal hexokinase in the liver and participates in glucose metabolism and hepatic insulin sensitivity, playing a pivotal role in developing NAFLD. The GCKR genetic variant (rs1260326: C > T) is associated with lower FPG, elevated triglycerides, and increased risk of NAFLD, indicating its pleiotropic function [13,29]. In this study, the GCKR T allele carriers had elevated triglycerides and LDL-TG levels, in line with previous studies. However, there was no significant difference in the development rate of carotid atherosclerosis between GCKR alleles. Further large-scale studies are necessary to elucidate the impact of GCKR polymorphism on carotid atherosclerosis.
PNPLA3 is a representative genetic variant associated with NAFLD across multiple ethnicities [10,30] and was also reported to be associated with the development of hepatocellular carcinoma [31]. PNPLA3 is localized in the endoplasmic reticulum and lipid droplet membranes in human hepatocytes, and the variant is considered to induce lipid accumulation in the liver [32]. While MASLD and NAFLD are often associated with obesity, some studies with non-obese Asian populations demonstrated that carriers of the PNPLA3 GG genotype increased the risk of the development of fatty liver [33,34]. In addition, a study in Italian NAFLD patients illustrated that the PNPLA3 GG genotype is associated with a higher severity of carotid atherosclerosis, especially in younger patients [6]. Our study, which consisted of a Japanese population without diabetes, dyslipidemia, and hypertension, described that the PNPLA3 GG genotype carriers have higher RLP cholesterol levels and a higher prevalence of newly developed carotid atherosclerosis, in line with previous studies in NAFLD participants.
Several limitations need to be addressed in this study. First, we could not analyze behavioral changes, such as dietary habits, since we did not correct these data. Second, the number of subjects was relatively small because this study design was based on epidemiological data. Based on the data from previous studies among NAFLD patients, we conducted sample size analyses under the following assumption: an odds ratio of 1.2 for newly developed carotid atherosclerosis associated with NAFLD-related SNPs and a prevalence of carotid atherosclerosis of 15 % in the general population. When the risk allele frequency is 5 %, such as NCAN, the required sample size is approximately 5100 participants. If the risk allele frequency is 50 %, such as PNPLA3 and GCKR, the required sample size is 1200 participants. In addition, due to the small sample size, we could not examine the interactions or effect sizes of the three SNPs. Moreover, we included only Japanese participants. Larger-scale research, including a more diverse population across different ethnicities, will be necessary to validate the findings of this study. However, the strength of this study was that all participants were without any chronic hepatitis, excessive alcohol intake, diabetes, dyslipidemia, or hypertension.
5. Conclusions
In conclusion, our results suggest that NAFLD-related SNPs, especially rs2228603 NCAN, may influence newly developed carotid atherosclerosis among Japanese individuals without metabolic disorders. Our data also show that NAFLD-related SNPs may affect lipid metabolism.
Financial Support
This study was supported by Grants-in-Aid for Scientific Research for Priority Areas of Cancer (No. 17015018), Innovative Areas (No. 221S0001), and by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant (No. 20K17155, No. 16H06277 and 22H04923 [CoBiA]; No. 21H04824) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology. H.I. was supported by the Japan Heart Foundation/Bayer Yakuhin Research Grant Abroad Program, Tokyo, Japan and the research grant of the Japanese Society of Hospital General Medicine, Fukuoka, Japan. These funding sources have no role in this study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors gratefully acknowledge the participants and the staff doctors who contributed to this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.athplu.2024.12.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Riazi K., Azhari H., Charette J.H., Underwood F.E., King J.A., Afshar E.E., Swain M.G., Congly S.E., Kaplan G.G., Shaheen A.A. The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2022;7(9):851–861. doi: 10.1016/S2468-1253(22)00165-0. [DOI] [PubMed] [Google Scholar]
- 2.Rinella M.E., Lazarus J.V., Ratziu V., Francque S.M., Sanyal A.J., Kanwal F., Romero D., Abdelmalek M.F., Anstee Q.M., Arab J.P., Arrese M., Bataller R., Beuers U., Boursier J., Bugianesi E., Byrne C.D. Castro narro ge, chowdhury A, cortez-pinto H, cryer dr, cusi K, el-kassas M, Klein S, Eskridge W, fan J, Gawrieh S, Guy CD, Harrison SA, Kim SU, Koot BG, Korenjak M, Kowdley KV, Lacaille F, Loomba R, Mitchell-Thain R, Morgan TR, Powell EE, Roden M, Romero-Gómez M, Silva M, Singh SP, Sookoian SC, Spearman CW, Tiniakos D, Valenti L, Vos MB, Wong VW, Xanthakos S, Yilmaz Y, Younossi Z, Hobbs A, Villota-Rivas M, newsome PN; NAFLD Nomenclature consensus group. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J Hepatol. 2023;79(6):1542–1556. doi: 10.1016/j.jhep.2023.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., Bjornsson E.S., Charatcharoenwitthaya P., Mills P.R., Keach J.C., Lafferty H.D., Stahler A., Haflidadottir S., Bendtsen F. Liver fibrosis, but No Other Histologic Features, is associated with Long-term Outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149(2):389–397. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duell P.B., Welty F.K., Miller M., Chait A., Hammond G., Ahmad Z., Cohen D.E., Horton J.D., Pressman G.S., Toth P.P. American Heart association council on arteriosclerosis, thrombosis and vascular biology; council on hypertension; council on the kidney in cardiovascular disease; council on lifestyle and cardiometabolic health; and council on peripheral vascular disease. Nonalcoholic fatty liver disease and cardiovascular risk: a scientific statement from the American Heart association. Arterioscler Thromb Vasc Biol. 2022;42(6):e168–e185. doi: 10.1161/ATV.0000000000000153. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization Who Cardiovascular diseases (CVDs). Available at. 2021. https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds
- 6.Petta S., Valenti L., Marchesini G., Di Marco V., Licata A., Cammà C., Barcellona M.R., Cabibi D., Donati B., Fracanzani A., Grimaudo S., Parrinello G., Pipitone R.M., Torres D., Fargion S., Licata G., Craxì A. PNPLA3 GG genotype and carotid atherosclerosis in patients with non-alcoholic fatty liver disease. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y.J., Hong S.C., Yin R.X., Yang Q., Cao X.L., Chen W.X. Polymorphisms in the GCKR are associated with serum lipid traits, the risk of coronary artery disease and ischemic stroke. Int J Clin Exp Med. 2015;8(7):10678–10686. [PMC free article] [PubMed] [Google Scholar]
- 8.Deng G.X., Yin R.X., Guan Y.Z., Liu C.X., Zheng P.F., Wei B.L., Wu J.Z., Miao L. Association of the NCAN-TM6SF2-CILP2-PBX4-SUGP1-MAU2 SNPs and gene-gene and gene-environment interactions with serum lipid levels. Aging (Albany NY) 2020;12(12):11893–11913. doi: 10.18632/aging.103361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmen O.L., Zhang H., Fan Y., Hovelson D.H., Schmidt E.M., Zhou W., Guo Y., Zhang J., Langhammer A., Løchen M.L., Ganesh S.K., Vatten L., Skorpen F., Dalen H., Zhang J., Pennathur S., Chen J., Platou C., Mathiesen E.B., Wilsgaard T., Njølstad I., Boehnke M., Chen Y.E., Abecasis G.R., Hveem K., Willer C.J. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46(4):345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L.A., Boerwinkle E., Cohen J.C., Hobbs H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., Nalls M.A., Clark J.M., Mitchell B.D., Shuldiner A.R., Butler J.L., Tomas M., Hoffmann U., Hwang S.J., Massaro J.M., O'Donnell C.J., Sahani D.V., Salomaa V., Schadt E.E., Schwartz S.M., Siscovick Ds, Nash Crn, GIANT Consortium; MAGIC Investigators; Voight BF, Carr JJ, Feitosa MF, Harris TB, Fox CS, Smith AV, Kao WH, Hirschhorn JN, Borecki IB; GOLD Consortium. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits PLoS Genet. 2011;7(3) doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carim-Todd L., Escarceller M., Estivill X., Sumoy L. Cloning of the novel gene TM6SF1 reveals conservation of clusters of paralogous genes between human chromosomes 15q24-->q26 and 19p13.3-->p12. Cytogenet Cell Genet. 2000;90(3–4):255–260. doi: 10.1159/000056784. [DOI] [PubMed] [Google Scholar]
- 13.Krawczyk M., Liebe R., Lammert F. Toward genetic prediction of nonalcoholic fatty liver disease trajectories: PNPLA3 and beyond. Gastroenterology. 2020;158(7):1865–1880. doi: 10.1053/j.gastro.2020.01.053. [DOI] [PubMed] [Google Scholar]
- 14.Ikezaki H., Furusyo N., Nakashima R., Umemoto M., Yamamoto K., Matsumoto Y., Ohta A., Yamasaki S., Hiramine S., Takayama K., Ogawa E., Toyoda K., Murata M., Shimono N., Hayashi J. Kyushu and Okinawa Population Study (KOPS): a large prospective cohort study in Japan. BMJ Open. 2021;11(12) doi: 10.1136/bmjopen-2021-053763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikezaki H., Lim E., Cupples L.A., Liu C.T., Asztalos B.F., Schaefer E.J. Small dense low-density lipoprotein cholesterol is the most atherogenic lipoprotein parameter in the prospective framingham offspring study. J Am Heart Assoc. 2021;10(5) doi: 10.1161/JAHA.120.019140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer E.J., Ikezaki H., Diffenderfer M.R., Lim E., Liu C.T., Hoogeveen R.C., Guan W., Tsai M.Y., Ballantyne C.M. Atherosclerotic cardiovascular disease risk and small dense low-density lipoprotein cholesterol in men, women, African Americans and non-African Americans: the pooling project. Atherosclerosis. 2023;367:15–23. doi: 10.1016/j.atherosclerosis.2023.01.015. [DOI] [PubMed] [Google Scholar]
- 17.The Joint committee of "the Japan Academy of Neurosonology" and "the Japan society of Embolus detection and treatment" on guideline for Nuerosonology. Carotid ultrasound examination. Neurosonology. 2006;19(2):49–67. in Japanese. [Google Scholar]
- 18.Ikezaki H., Furusyo N., Yokota Y., Ai M., Asztalos B.F., Murata M., Hayashi J., Schaefer E.J. Small dense low-density lipoprotein cholesterol and carotid intimal medial thickness progression. J Atherosclerosis Thromb. 2020;27(10):1108–1122. doi: 10.5551/jat.54130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanase T., Nasu S., Mukuta Y., Shimizu Y., Nishihara T., Okabe T., Nomura M., Inoguchi T., Nawata H. Evaluation of a new carotid intima-media thickness measurement by B-mode ultrasonography using an innovative measurement software, intimascope. Am J Hypertens. 2006;19(12):1206–1212. doi: 10.1016/j.amjhyper.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Cichon S., Mühleisen T.W., Degenhardt F.A., Mattheisen M., Miró X., Strohmaier J., Steffens M., Meesters C., Herms S., Weingarten M., Priebe L., Haenisch B., Alexander M., Vollmer J., Breuer R., Schmäl C., Tessmann P., Moebus S., Wichmann H.E., Schreiber S., Müller-Myhsok B., Lucae S., Jamain S., Leboyer M., Bellivier F., Etain B., Henry C., Kahn J.P., Heath S. Bipolar disorder genome study (BiGS) consortium; hamshere M, O'Donovan MC, owen MJ, craddock N, schwarz M, vedder H, kammerer-ciernioch J, reif A, sasse J, bauer M, hautzinger M, Wright A, Mitchell PB, Schofield PR, Montgomery GW, Medland SE, Gordon SD, Martin NG, Gustafsson O, Andreassen O, Djurovic S, Sigurdsson E, Steinberg S, Stefansson H, Stefansson K, Kapur-Pojskic L, Oruc L, Rivas F, Mayoral F, Chuchalin A, Babadjanova G, Tiganov AS, Pantelejeva G, Abramova LI, Grigoroiu-Serbanescu M, Diaconu CC, Czerski PM, Hauser J, Zimmer A, Lathrop M, Schulze TG, Wienker TF, Schumacher J, Maier W, Propping P, Rietschel M, Nöthen MM. Genome-Wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88(3):372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dannlowski U., Kugel H., Grotegerd D., Redlich R., Suchy J., Opel N., Suslow T., Konrad C., Ohrmann P., Bauer J., Kircher T., Krug A., Jansen A., Baune B.T., Heindel W., Domschke K., Forstner A.J., Nöthen M.M., Treutlein J., Arolt V., Hohoff C., Rietschel M., Witt S.H. NCAN cross-disorder risk variant is associated with limbic gray matter deficits in healthy subjects and major depression. Neuropsychopharmacology. 2015;40(11):2510–2516. doi: 10.1038/npp.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nischalke H.D., Lutz P., Krämer B., Söhne J., Müller T., Rosendahl J., Fischer J., Berg T., Hittatiya K., Fischer H.P., Soyka M., Semmo N., Nattermann J., Sauerbruch T., Strassburg C.P., Stickel F., Spengler U. A common polymorphism in the NCAN gene is associated with hepatocellular carcinoma in alcoholic liver disease. J Hepatol. 2014;61(5):1073–1079. doi: 10.1016/j.jhep.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Gorden A., Yang R., Yerges-Armstrong L.M., Ryan K.A., Speliotes E., Borecki I.B., Harris T.B., Chu X., Wood G.C., Still C.D., Shuldiner A.R., Gerhard G.S., GOLD Consortium. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity Hum Hered. 2013;75(1):34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M.J., Yuan C., Lu L.L., An B.Q., Xuan S.Y., Xin Y.N. Role of NCAN rs2228603 polymorphism in the incidence of nonalcoholic fatty liver disease: a case-control study. Lipids Health Dis. 2016;15(1) doi: 10.1186/s12944-016-0367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirao Y., Nakajima K., Machida T., Murakami M., Ito Y. Development of a novel homogeneous assay for remnant lipoprotein particle cholesterol. J Appl Lab Med. 2018;3(1):26–36. doi: 10.1373/jalm.2017.024919. [DOI] [PubMed] [Google Scholar]
- 26.Saeed A., Feofanova E.V., Yu B., Sun W., Virani S.S., Nambi V., Coresh J., Guild C.S., Boerwinkle E., Ballantyne C.M., Remnant-Like Particle Cholesterol Hoogeveen RC., Lipoprotein Triglycerides Low-Density, Disease Incident Cardiovascular. J Am Coll Cardiol. 2018;72(2):156–169. doi: 10.1016/j.jacc.2018.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., Nalls M.A., Clark J.M., Mitchell B.D., Shuldiner A.R., Butler J.L., Tomas M., Hoffmann U., Hwang S.J., Massaro J.M., O'Donnell C.J., Sahani D.V., Salomaa V., Schadt E.E., Schwartz S.M., Siscovick Ds, Nash Crn GIANT Consortium; MAGIC Investigators; Voight BF, Carr JJ, Feitosa MF, Harris TB, Fox CS, Smith AV, Kao WH, Hirschhorn JN, Borecki IB; GOLD Consortium. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3) doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krawczyk M., Liebe R., Lammert F. Toward genetic prediction of nonalcoholic fatty liver disease trajectories: PNPLA3 and beyond. Gastroenterology. 2020;158(7):1865–1880. doi: 10.1053/j.gastro.2020.01.053. [DOI] [PubMed] [Google Scholar]
- 29.Raimondo A., Rees M.G., Gloyn A.L. Glucokinase regulatory protein: complexity at the crossroads of triglyceride and glucose metabolism. Curr Opin Lipidol. 2015;26(2):88–95. doi: 10.1097/MOL.0000000000000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagenknecht L.E., Palmer N.D., Bowden D.W., Rotter J.I., Norris J.M., Ziegler J., Chen Y.D., Haffner S., Scherzinger A., Langefeld C.D., Association of PNPLA3 with non-alcoholic fatty liver disease in a minority cohort: the Insulin Resistance Atherosclerosis Family Study Liver Int. 2011;31(3):412–416. doi: 10.1111/j.1478-3231.2010.02444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta A., Ogawa E., Murata M., Matsumoto Y., Yamasaki S., Ikezaki H., Furusyo N. Impact of the PNPLA3 genotype on the risk of hepatocellular carcinoma after hepatitis C virus eradication. J Med Virol. 2022;94(10):5007–5014. doi: 10.1002/jmv.27904. [DOI] [PubMed] [Google Scholar]
- 32.He S., McPhaul C., Li J.Z., Garuti R., Kinch L., Grishin N.V., Cohen J.C., Hobbs H.H. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285(9):6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oniki K., Saruwatari J., Izuka T., Kajiwara A., Morita K., Sakata M., Otake K., Ogata Y., Nakagawa K. Influence of the PNPLA3 rs738409 polymorphism on non-alcoholic fatty liver disease and renal function among normal weight subjects. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin H., Wong G.L., Whatling C., Chan A.W., Leung H.H., Tse C.H., Shu S.S., Chim A.M., Lai J.C., Yip T.C., Wong V.W. Association of genetic variations with NAFLD in lean individuals. Liver Int. 2022;42(1):149–160. doi: 10.1111/liv.15078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.