Abstract
Exploitation of ribozymes in a practical setting requires high catalytic activity and strong specificity. The hammerhead ribozyme R32 has considerable potential in this regard since it has very high catalytic activity. In this study, we have examined how R32 recognizes and cleaves a specific substrate, focusing on the mechanism behind the specificity. Comparing rates of cleavage of a substrate in a mixture that included the correct substrate and various substrates with point mutations, we found that R32 cleaved the correct substrate specifically and at a high rate. To clarify the source of this strong specificity, we quantified the weak interactions between R32 and various truncated substrates, using truncated substrates as competitive inhibitors since they were not readily cleaved during kinetic measurements of cleavage of the correct substrate, S11. We found that the strong specificity of the cleavage reaction was due to a closed form of R32 with a hairpin structure. The self-complementary structure within R32 enabled the ribozyme to discriminate between the correct substrate and a mismatched substrate. Since this hairpin motif did not increase the Km (it did not inhibit the binding interaction) or decrease the kcat (it did not decrease the cleavage rate), this kind of hairpin structure might be useful for the design of new ribozymes with strong specificity and high activity.
INTRODUCTION
Hammerhead ribozymes are small catalytic RNAs that can cleave phosphodiester linkages at specific sites in RNA substrates. It is easy to design a ribozyme that can attack a specific target RNA and, therefore, it is anticipated that such ribozymes will be of considerable practical use (1–15). It is important, for practical applications, that hammerhead ribozymes should discriminate between matched and mismatched substrates and there have been several attempts to improve the specificity of hammerhead ribozymes (12,16–18).
Various important improvements have been achieved that have included truncation of substrate-binding arms (16,17), generation of self-complementary structures at substrate-binding sites (18) and creation of allosterically controllable ribozymes, known as maxizymes (12,13,19,20). In the first case, truncation of the substrate-binding arms enhanced the rate of dissociation of mismatched substrates from the ribozyme and, thus, mismatched substrates dissociated from the ribozyme–substrate complex before they could be cleaved. In the second case, self-complementary structures within the binding arms prevented mismatched substrates from binding to ribozymes. However, these modifications were introduced at the cost of unwelcome changes in kcat and Km, namely the rate of cleavage and binding affinity for the correct substrate(s) (16–21).
A 32mer hammerhead ribozyme, R32 (see Fig. 1A), was designed in our laboratory and its catalytic activity has been characterized in detail (4,5,22–30). R32 has high cleavage activity and the cleavage approaches completion at saturating concentrations of the ribozyme and trace levels of labeled substrate, i.e. under single-turnover conditions (4,5). Thus, populations of R32 appear to consist of fully active molecules (21–28,30). The cleavage activity of R32 is fairly high (21), with kcat > 4 min–1 at 25 mM MgCl2, 37°C and pH 8.0. Since R32 cleaves a matched substrate specifically, an analysis of its mechanism of discrimination should provide us with information that will be useful for the design of ribozymes that have high cleavage activity and sequence specificity.
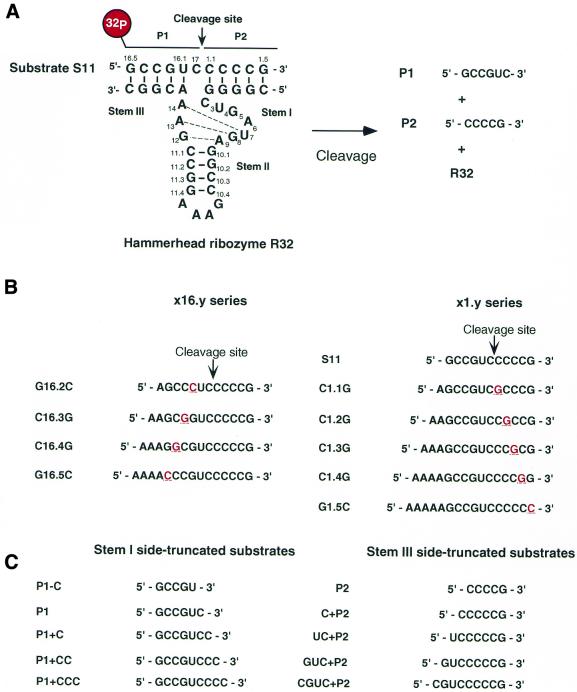
Figure 1.
Secondary structure of R32 and sequences of oligonucleotides used in this study. (A) Hammerhead ribozyme R32 and its substrate S11. An arrow indicates the cleavage site. (B) Substrate S11 and the point mutants used for analysis of the specificity of R32. Each underlined red letter indicates a single nucleotide mutation introduced into the substrate. (C) The truncated substrates used for investigations of binding affinity. Each substrate corresponds to S11 truncated at its 5′- or 3′-end. P1 and P2 refer to truncation on the 3′- and 5′-side of S11, respectively.
In this paper, we demonstrate the strong specificity of R32. We also present a detailed kinetic analysis of the weak interactions between R32 and various truncated substrates. The results of our analysis suggest a new method for designing hammerhead ribozymes with high activity and strong specificity.
MATERIALS AND METHODS
Synthesis and purification of RNAs
The hammerhead ribozyme R32 and its substrate, S11, were chemically synthesized with a DNA/RNA synthesizer (model 394; PE Applied Biosystems, Foster City, CA). The newly synthesized products were deprotected and purified by standard methods, as described elsewhere (22–25,30,31). The sequences of RNA oligomers with single point mutations, designated the x16.y and x1.y series and used for studies of the specificity of R32, are listed in Figure 1B. They were purchased from Genset Oligos (Paris, France). The sequences of the stem side-truncated substrates used for determinations of Ki are listed in Figure 1C. These substrates were synthesized as described above.
Measurements of the kinetics of reactions catalyzed by the hammerhead ribozyme
We performed cleavage experiments to examine the ability of R32 to distinguish between correct and mismatched substrates. R32 and a mixture of unlabeled and 5′-32P-labeled substrates (S11, x16.y and x1.y) were mixed in the reaction buffer without MgCl2 before reactions were initialized. Each reaction mixture contained an equal amount of each correct and mutated substrate. All reactions were started by addition of a MgCl2 solution and reactions were performed at 37°C in a buffer that contained 25 mM MgCl2 and 50 mM Tris–HCl, pH 8.0. Aliquots were removed from reaction mixtures at appropriate times and mixed with stop solution (100 mM EDTA, 7 M urea, 30% glycerol, 0.1% xylene cyanol and 0.1% bromophenol blue). Products and uncleaved substrates were separated by electrophoresis in a 20% polyacrylamide/7 M urea denaturing gel and were detected with an Image Analyzer (Storm 830; Molecular Dynamics, Sunnyvale, CA).
To determine the binding affinity of each truncated substrate, we performed cleavage experiments using each-truncated substrate as an inhibitor and calculated the inhibition constant (Ki) in each case. First, we performed multiple- turnover experiments. The concentration of S11 was varied from 10 to 5000 nM depending on the truncated substrate being tested. All reactions were performed with at least a 20-fold excess of substrate over ribozyme. The concentrations of truncated substrates were varied from 10 to 5000 nM. The ribozyme, substrate, truncated substrate and a trace amount of 5′-32P-labeled substrate were mixed together in the same reaction buffer. All reactions were started by addition of a MgCl2 solution and reactions were performed at 37°C in a buffer that contained 25 mM MgCl2 and 50 mM Tris–HCl, pH 8.0. Aliquots were removed from reaction mixtures at appropriate times, mixed with stop solution and analyzed as described above. Individual rate constants were obtained from initial rates of reactions. We calculated the apparent Km value (Km′) from Eadie–Hofstee and Lineweaver–Burk plots. We confirmed that inhibition by truncated substrates was competitive (see Results). Lineweaver–Burk plots at each concentration of a specific inhibitor had the same intercept on the y-axis (32–34). In competitive inhibition, the relationship between Km′ and the concentration of inhibitor is as follows:
Km′ = Km + (Km × [I])/Ki 1
where Km, Km′, Ki and [I] indicate the binding constant, the apparent binding constant, the inhibition constant and the concentration of truncated substrate, respectively. Using 1, we calculated Ki from Km′ and [I] profiles.
RESULTS AND DISCUSSION
The R32 ribozyme cleaved the correct substrate significantly more rapidly than the mismatched substrates
To investigate whether the hammerhead ribozyme R32 can effectively recognize and cleave its correct substrate specifically in a mixture of correct and mutated substrates, we performed cleavage experiments with a mixture that included the correct substrate and individual mutant substrates (Fig. 1A and B). In order that all reactions were performed under identical conditions, each mutant was attached to a sequence of A residues of different length so that all mutants could be included together and could be subjected to gel separation for subsequent quantification (Fig. 1B). Thus, the reaction mixture contained four or five labeled RNAs, including the stem I side- (x1.y series) or stem III side-mismatched (x16.y series) substrates, and the concentration of each substrate was kept the same in each reaction. The substrates shown in Figure 1B have systematic single nucleotide mutations at each binding site for the hammerhead ribozyme R32; for example, G16.2C denotes G16.2 with a G→C mutation.
If R32 has the ability to cleave the correct substrate specifically, the correct substrate should be cleaved much more rapidly than mutant variants in a mixture of correct and variant substrates. As shown in Figure 2, we found that under both single- (Fig. 2B) and multiple-turnover (Fig. 2A) conditions, the correct substrate S11 was specifically cleaved. During the 1 h for which results are shown, little cleavage of substrates with point mutations was detected and only very limited cleavage of mismatched substrates was detected after further incubation. In the case of G16.5C, 5′-r(AAA ACC CGU CCC CCG)-3′, and G1.5C, 5′-r(AAA AAG CCG UCC CCC C)-3′, in which a single mismatched nucleotide was placed at either the 5′- or the 3′-end of S11 with the remaining nine nucleotides making a continuous perfect match, significant cleavage was detected. Because of such cleavage, G16.5C and G1.5C were not used as competitors of S11 in subsequent analysis of the weak binding interactions between truncated substrates and R32.
Figure 2.
Autoradiographs of polyacrylamide gels showing the cleavage of each variant of S11 as a function of time. (A) Multiple:turnover experiment: 1 nM R32, 50 mM Tris–HCl, 25 mM MgCl2 and 40 nM total substrate at 37°C and pH 8.0. (B) Single:turnover experiment: 30 nM R32, 50 mM Tris–HCl, 25 mM MgCl2 and 20 nM total substrate at 37°C and pH 8.0. We could not distinguish between bands C1.4G and G1.5C on the gel since they were very close to each other. We exposed these mismatched substrates to alkaline hydrolysis and enzymatic hydrolysis and confirmed the assignments (data not shown).
Measurements of weak binding interactions between R32 and truncated substrates
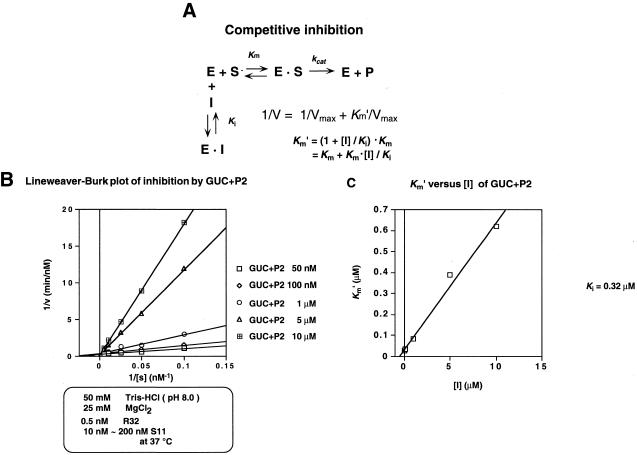
In order to investigate the origin of the high specificity of R32 in terms of its binding affinity for its correct substrate, we performed inhibition experiments with the systematically truncated substrates shown in Figure 1C. Each substrate corresponded to S11 truncated at its 5′- or 3′-end. In our nomenclature, P1+CCC and GUC+P2, for example, denote P1 with an additional 3′-CCC and P2 with an additional 5′-GUC, respectively. We anticipated that the rates of cleavage of these truncated substrates by the ribozyme would be too low to follow under our conditions and, therefore, that accurate measurement of kcat and Km for these truncated substrates would be difficult (35). Therefore, we evaluated the substrate-binding affinity as the inhibition constant for each truncated substrate, assuming that it acted as an inhibitor of the standard R32-catalyzed cleavage of S11. Under the conditions of our measurements we expected that the affinity of each truncated substrate for R32 could be estimated as the competitive inhibition constant Ki, as described by the cleavage reaction outlined in Figure 3A.
Figure 3.
Typical results of an inhibition experiment. (A) Reaction scheme for competitive inhibition. Km, Km′, Ki and [I] indicate the binding constant, apparent binding constant, inhibition constant and concentration of truncated substrate, respectively. (B) Lineweaver–Burk plots of inhibition by GUC+P2. (C) Graph of Km′ versus [I] plots.
We first confirmed that inhibition of the cleavage reaction was due to competitive inhibition, as follows. If each inhibitor were to act as a competitive inhibitor, the Lineweaver–Burk plot for each concentration of the inhibitor should have the same intercept on the y-axis as the others. As shown in Figure 3B, Lineweaver–Burk plots for GUC+P2, which was a stem III side-truncated substrate, demonstrated clearly that inhibition was indeed competitive.
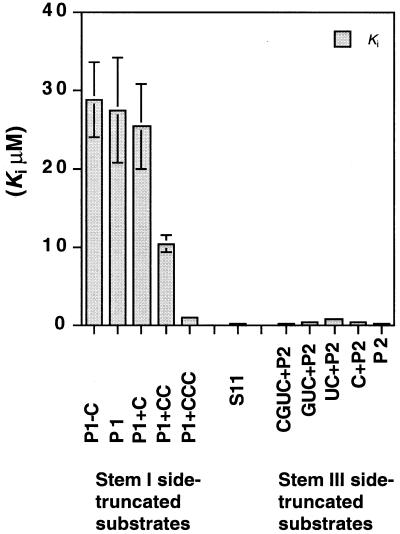
We confirmed that all the other truncated substrates acted as competitive inhibitors of S11 in the cleavage reaction (Fig. 4). In general, in competitive inhibition the correlation between the inhibition constant Ki and apparent binding constant Km′ can be described by 1 (Fig. 3A). Therefore, we plotted Km′ versus [I] (Figs 3C and 4) and calculated the inhibition constant Ki for each truncated substrate. The results are summarized in Figure 5, which clearly shows the difference in binding affinity between the stem I side- and stem III side-truncated forms of S11. The stem III side-truncated substrates all had smaller inhibition constants (indicative of stronger binding to R32) than the stem I side-truncated substrates. Moreover, in the case of stem I side-truncated substrates, as might have been anticipated, the shorter the truncated substrate, the larger its inhibition constant, clearly demonstrating weaker interactions of R32 with the shorter forms of S11. If we assume that R32 forms the simple structure shown in Figure 1A, the length of the stem should be related to the binding affinity. The stem I side-truncated substrates seemed to conform to this relationship. However, the large difference in terms of Ki values between the stem I side- and stem III side-truncated substrates cannot be explained by the assumption of the simple structure. The results shown in Figure 5 cannot be reconciled with such a simple open structure for R32.
Figure 4.
Results of inhibition experiments. Lineweaver–Burk plots and graphs of Km′ versus [I] plots are shown. All truncated substrates were confirmed to be competitive inhibitors of S11 in the R32-catalyzed cleavage reaction from Lineweaver–Burk plots.
Figure 5.
A summary of the results of inhibition experiments. The horizontal axis corresponds to truncated substrates, as indicated, and the vertical axis shows Ki for each truncated substrate except for S11, which is the correct substrate for R32. The value for S11 reflects Km.
A hairpin structure as the origin of the strong specificity of R32
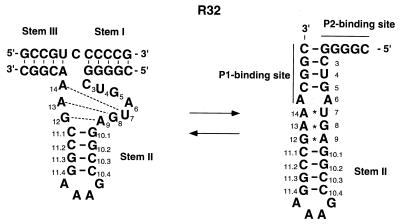
All our kinetic data indicate that, in the reaction mixture, the majority of R32 molecules are in a catalytically active form (4,5,22–29). However, we know, from NMR measurements, that R32 can form a closed hairpin structure (Fig. 6, right), in particular in the absence of divalent metal ions (25). In this hairpin structure, bases within the stem III region (P1 binding site) form a continuous pseudo-A-form helix by binding of G2.1–G5 around the uridine turn sequence to stem II, with resultant exposure of the binding region of stem I (P2 binding site) (25). The phenomenon shown in Figure 5 can be explained by such a hairpin structure, since the P2 binding site can act as a nucleation site for the interaction of R32 and its substrate/inhibitor.
Figure 6.
Open (left) and closed (right) structures of the hammerhead ribozyme R32. The closed form of R32 is a long hairpin structure, in which an additional intramolecular 4 bp duplex is formed.
The stem III side-truncated substrates retained regions that were complementary to the P2 binding site (nucleation site) and, thus, all of them had significant affinity (smaller Ki) for the ribozyme. In contrast, in the case of the stem I side-truncated substrates, the shorter the truncated substrate, the more difficult it was for the altered substrates to compete with the standard substrate S11 because the truncated substrates had shorter regions that could bind to the P2 binding site. Furthermore, the P1 binding site, which is embedded in the hairpin structure, must be opened up before the stem I side-truncated substrates can bind to R32. Thus, more energy is required for their binding to R32, as reflected in their larger Ki values.
Hammerhead ribozymes have the intrinsic ability to discriminate the correct substrate from an incorrect variant. Especially, when a substrate has a single mismatch next to the cleavage site, the cleavage efficiency of hammerhead ribozymes is significantly reduced (36). However, it is possible for an incorrect substrate with some mismatches to be cleaved when binding between the ribozyme and the incorrect substrate is strong enough or when the substrate dissociation rate is low enough. Therefore, binding affinity is one of the major parameters that determine specificity.
R32 appears to be an ideal ribozyme. Its cleavage rate is one of the highest among various known hammerhead ribozymes (4,5,21). In solution, the closed hairpin structure is in equilibrium with the open form and the addition of substrate shifts the equilibrium to the open form, with subsequent formation of the R32–substrate complex. Because of this rapid equilibrium, all molecules of R32 remain in an active conformation.
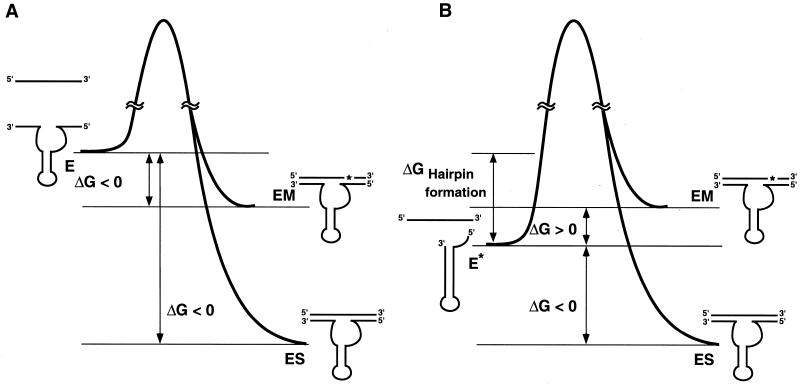
Figure 7 shows a hypothetical energy diagram for the binding of R32 to its substrate. Formation of the intramolecular hairpin structure stabilizes the ground state of the ribozyme itself. As a result, mismatched substrates that have formed complexes with R32 are more likely to dissociate from such complexes than substrates in complexes with the corresponding putative open complex. Earlier investigators examined the reaction either by shortening the binding arms (16) or by creating self-complementarity at each binding arm (18). In such cases, the relative energy level of the improved ribozyme and its substrate was reduced relative to the corresponding energy level of the authentic ribozyme and substrate, with resultant higher specificity. A similar approach is also applicable to antisense molecules (37).
Figure 7.
Hypothetical energy diagrams for the hammerhead ribozyme R32 in the open (A) and closed (B) forms. E, E*, ES and EM indicate open form R32, closed form R32, R32–substrate complex and R32–mismatched substrate complex, respectively.
In the putative case of the open form shown in Figure 7A, the free energy of free R32 and its substrate is higher than that of the ribozyme–substrate complex and also than that of the ribozyme–mismatched substrate complex. Therefore, it is possible that the ribozyme might form complexes with mismatched substrates and cleave them. In contrast, in the case of the closed form of the ribozyme (Fig. 7B), free R32 and its substrate become more stable as a result of formation of a hairpin structure and the free energy is reduced. Under optimal conditions, we can expect the closed form of the ribozyme to be more stable than the ribozyme–mismatched substrate complex and less stable than the ribozyme–substrate complex.
In contrast to other approaches, the advantage of the hairpin motif of R32 is the fact that, because of weak interaction within the stem region created by the P1 binding site, a relatively low Km is retained and the rapid equilibrium between the closed and open forms of R32 in solution does not create a strong inactive form of the ribozyme. Moreover, kcat is still large (21). This motif might be of limited utility because, for appropriate cleavage, the target RNA should include the 5′-YCCGUXZ-3′ sequence, where Y can be any base as long as it is complementary to Z and X can be any base except G. A rapid search for such a sequence within 1128 nt of the coding region of the mRNA for actin enabled us to identify two potential cleavage sites. Therefore, it may not be unrealistic to attempt to use such a motif for the highly specific and rapid cleavage of a target molecule.
REFERENCES
- 1.Uhlenbeck O.C. (1987) A small catalytic oligonucleotide. Nature, 328, 596–600. [DOI] [PubMed] [Google Scholar]
- 2.Haseloff J. and Gerlach,W.L. (1988) Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature, 334, 585–591. [DOI] [PubMed] [Google Scholar]
- 3.Symons R.H. (1992) Small catalytic RNAs. Annu. Rev. Biochem., 61, 641–671. [DOI] [PubMed] [Google Scholar]
- 4.Zhou D.-M. and Taira,K. (1998) The hydrolysis of RNA: from theoretical calculations to the hammerhead ribozyme-mediated cleavage of RNA. Chem. Rev., 98, 991–1026. [DOI] [PubMed] [Google Scholar]
- 5.Takagi Y., Warashina,M., Stec,W.J., Yoshinari,K. and Taira,K. (2001) Recent advances in the elucidation of the mechanisms of action of ribozymes. Nucleic Acids Res., 29, 1815–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson R.P. and Izant,J.G. (eds) (1992) Gene Regulation: Biology of Antisense RNA and DNA. Raven Press, New York, NY.
- 7.Murray J.A.H. (ed.) (1992) Antisense RNA and DNA. Wiley-Liss, New York, NY.
- 8.Rossi J.J. (1995) Controlled, targeted, intracellular expression of ribozymes: progress and problems. Trends Biotechnol., 13, 301–306. [DOI] [PubMed] [Google Scholar]
- 9.Eckstein F. and Lilley,D.M.J. (eds) (1996) Catalytic RNA: Nucleic Acids and Molecular Biology, Vol. 10. Springer-Verlag, Berlin, Germany.
- 10.Turner P.C. (ed.) (1997) Ribozyme Protocols: Methods in Molecular Biology, Vol. 74. Humana Press, Totowa, NJ.
- 11.Krupp G. and Gaur,R.K. (eds) (2000) Ribozyme: Biochemistry and Biotechnology. Eaton Publishing, Natick, MA.
- 12.Kuwabara T., Warashina,M., Tanabe,T., Tani,K., Asano,T. and Taira,K. (1998) A novel allosterically trans-activated ribozyme, the maxizyme, with exceptional specificity in vitro and in vivo. Mol. Cell, 2, 617–627. [DOI] [PubMed] [Google Scholar]
- 13.Tanabe T., Kuwabara,T., Warashina,M., Tani,K., Taira,K. and Asano,S. (2000) Oncogene inactivation in a mouse model. Nature, 406, 473–474. [DOI] [PubMed] [Google Scholar]
- 14.Kato Y., Kuwabara,T., Warashina,M., Toda,H. and Taira,K. (2001) Relationships between the activities in vitro and in vivo of various kinds of ribozyme and their intracellular localization in mammalian cells. J. Biol. Chem., 276, 15378–15385. [DOI] [PubMed] [Google Scholar]
- 15.Warashina M., Kuwabara,T., Kato,Y., Sano,M. and Taira,K. (2001) RNA-protein hybrid ribozymes that efficiently cleave any mRNA independently of the structure of the target RNA. Proc. Natl Acad. Sci. USA, 98, 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertel K.J., Herschlag,D. and Uhlenbeck,O.C. (1996) Specificity of hammerhead ribozyme cleavage. EMBO J., 15, 3751–3757. [PMC free article] [PubMed] [Google Scholar]
- 17.Werner M. and Uhlenbeck,O.C. (1995) The effect of base mismatches in the substrate recognition helices of hammerhead ribozymes on binding and catalysis. Nucleic Acids Res., 23, 2092–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohmichi T. and Kool,E.T. (2000) The virtues of self-binding: high sequence specificity for RNA cleavage by self-processed hammerhead ribozymes. Nucleic Acids Res., 28, 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwabara T., Warashina,M. and Taira,K. (2000) Allosterically controllable maxizymes cleave mRNA with high efficiency and specificity. Trends Biotechnol., 18, 462–468. [DOI] [PubMed] [Google Scholar]
- 20.Kuwabara T., Warashina,M. and Taira,K. (2000) Allosterically controllable ribozymes with biosensor functions. Curr. Opin. Chem. Biol., 4, 669–677. [DOI] [PubMed] [Google Scholar]
- 21.Stage-Zimmermann T.K. and Uhlenbeck,O.C. (1998) Hammerhead ribozyme kinetics. RNA, 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawata S., Shimayama,T., Komiyama,M., Kumar,P.K.R., Nishikawa,S. and Taira,K. (1993) Enhancement of the cleavage rates of DNA-armed hammerhead ribozymes by various divalent metal ions. Nucleic Acids Res., 21, 5656–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimayama T., Nishikawa,S. and Taira,K. (1995) Generality of the NUX rule: kinetic analysis of the results of systematic mutations in the trinucleotide at the cleavage site of hammerhead ribozymes. Biochemistry, 34, 3649–3654. [DOI] [PubMed] [Google Scholar]
- 24.Sawata S., Komiyama,M. and Taira,K. (1995) Kinetic evidence based on solvent isotope effects for the nonexistence of a proton-transfer process in reactions catalyzed by a hammerhead ribozyme: implication to the double-metal-ion mechanism of catalysis. J. Am. Chem. Soc., 117, 2357–2358. [Google Scholar]
- 25.Orita M., Vinayak,R., Andrus,A., Warashina,M., Chiba,A., Kaniwa,H., Nishikawa,F., Nishikawa,S. and Taira,K. (1996) Magnesium-mediated conversion of an inactive form of a hammerhead ribozyme to an active complex with its substrate. An investigation by NMR spectroscopy. J. Biol. Chem., 271, 9447–9454. [DOI] [PubMed] [Google Scholar]
- 26.Zhou D.-M., Zhang,L.-H. and Taira,K. (1997) Explanation by the double-metal-ion mechanism of catalysis for the differential metal ion effects on the cleavage rates of 5′-oxy and 5′-thio substrates by a hammerhead ribozyme. Proc. Natl Acad. Sci. USA, 94, 14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y., Morita,E.H., Hayashi,H., Kasai,Y., Tanaka,T. and Taira,K. (2000) Well-conserved tandem G·A pairs and the flanking C·G pair in hammerhead ribozymes are sufficient for capture of structurally and catalytically important metal ions. J. Am. Chem. Soc., 122, 11303–11310. [Google Scholar]
- 28.Yoshinari K. and Taira,K. (2000) A further investigation and reappraisal of the thio effect in the cleavage reaction catalyzed by a hammerhead ribozyme. Nucleic Acids Res., 28, 1730–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka Y., Kojima,C. Morita,E.H., Kasai,Y., Yamasaki,K. Ono,A. Kainosho,M. and Taira,K. (2002) Identification of the metal ion binding site on an RNA motif from hammerhead ribozymes using 15N NMR spectroscopy. J. Am. Chem. Soc., 124, 4595–4601. [DOI] [PubMed] [Google Scholar]
- 30.Takagi Y. and Taira,K. (2002) Detection of a proton-transfer process by kinetic solvent isotope effects in NH4+-mediated reactions catalyzed by a hammerhead ribozyme. J. Am. Chem. Soc., 124, 3850–3852. [DOI] [PubMed] [Google Scholar]
- 31.Nakamatsu Y. , Warashina,M., Kuwabara,T., Tanaka,Y., Yoshinari,K. and Taira,K. (2000) Significant activity of a modified ribozyme with N7-deazaguanine at G10.1: the double-metal-ion mechanism of catalysis in reactions catalysed by hammerhead ribozymes. Genes Cells, 5, 603–612. [DOI] [PubMed] [Google Scholar]
- 32.Fersht A. (ed.) (1977) Enzyme Structure and Mechanism. W.H. Freeman, San Francisco, San Diego, CA.
- 33.Purich D.L. (ed.) (1996) Contemporary Enzyme Kinetics and Mechanism. Academic Press, CA.
- 34.Cornish-Bowden A. (ed.) (1995) Fundamentals of Enzyme Kinetics. Portland Press, London, UK.
- 35.Hertel K.J., Peracchi,A., Uhlenbeck,O.C. and Herschlag,D. (1997) Use of intrinsic binding energy for catalysis by an RNA enzyme. Proc. Natl Acad. Sci. USA, 94, 8497–8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh K.K., Schluff,P., Lehnert,L. and Krupp,G. (1996) Design of hammerhead ribozymes to distinguish single base changes in substrate RNA. Antisense Nucleic Acid Drug Dev., 6, 165–168. [DOI] [PubMed] [Google Scholar]
- 37.Stocks M.R. and Rabbitts,T.H. (2000) Masked antisense: a molecular configuration for discriminating similar RNA targets. EMBO Rep., 1, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]