Abstract
Ribozyme activity in vivo depends on achieving high-level expression, intracellular stability, target colocalization, and cleavage site access. At present, target site selection is problematic because of unforeseeable secondary and tertiary RNA structures that prevent cleavage. To overcome this design obstacle, we wished to engineer a ribozyme that could access any chosen site. To create this ribozyme, the constitutive transport element (CTE), an RNA motif that has the ability to interact with intracellular RNA helicases, was attached to our ribozymes so that the helicase-bound, hybrid ribozymes would be produced in cells. This modification significantly enhanced ribozyme activity in vivo, permitting cleavage of sites previously found to be inaccessible. To confer cleavage enhancement, the CTE must retain helicase-binding activity. Binding experiments demonstrated the likely involvement of RNA helicase(s). We found that attachment of the RNA motif to our tRNA ribozymes leads to cleavage in vivo at the chosen target site regardless of the local RNA secondary or tertiary structure.
Hammerhead ribozymes (Rz) are small, naturally occurring catalytic RNAs that can be used as tools for basic research and show promise as therapeutic agents (1–3). Such RNAs can cleave oligoribonucleotides at specific sites (4) and have been used successfully to suppress gene expression in several different organisms (5–14). However, despite extensive efforts, the efficiency of Rz in vivo usually is not high enough to achieve the desired biological effect(s) (15). Successful gene inactivation by Rz in vivo depends strongly on the design of the expression vector. The design can determine both the level of expression and the half-life of the expressed Rz (12). In previous studies we found that RNA polymerase III-mediated expression of Rz as tRNA fusions resulted in highly expressed stable Rz (8–10, 12–14).
However, even these improved Rz were sometimes ineffective, probably because the Rz was unable to locate its target. One potential explanation for this ineffectiveness is that the rate-limiting step in vivo for the cleavage of phosphodiester bonds is the annealing/association of the Rz with its target site (16). In general, the regions that interact with DNA of DNA-cleaving restriction enzymes, such as EcoRI and EcoRV, are positively charged so that they can search for their target sites by sliding along the polynucleotide chain (Fig. 1A; see refs. 17 and 18 for details of linear diffusion and the sliding mechanism). As a result, wherever such a restriction enzyme binds to DNA, it can locate the cleavage site efficiently by sliding along the DNA. Kinetically unfavorable and repetitive association/dissociation events can be avoided during the search for the target site. Indeed, the cleavage efficiency of restriction enzymes increases with the length of the substrate DNA up to several thousand base pairs, a phenomenon that indicates that, once the restriction enzyme has bound to DNA, it can slide along the DNA strand for a few thousand base pairs before it dissociates from the DNA (17). In contrast, RNA-cleaving Rz are negatively charged, as are their RNA substrates. Therefore, Rz cannot slide along the RNA chain, and they have to search for their target sites in a series of kinetically unfavorable repetitive association/dissociation events (Fig. 1B). As a result, the longer the target RNA, the lower the efficiency of cleavage by the Rz. Furthermore, in a long RNA chain, significant numbers of target sites are not accessible to the Rz, because they are hidden within secondary or tertiary structures (15). This problem is often critical in attempts to exploit Rz activity, particularly for in vivo reactions.
Figure 1.
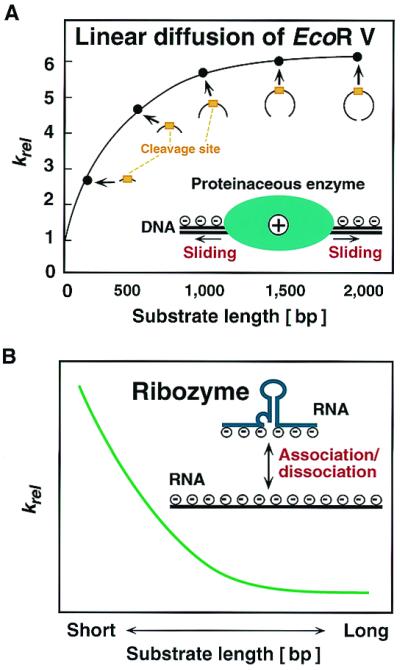
Schematic representation of differences between mechanisms of target recognition between a proteinaceous enzyme (A) and a Rz (B).
To overcome the problem of accessibility, computer-generated predictions of secondary structure (19) are typically used to identify targets that are most likely to have an open conformation. However, these predictions are often inaccurate because of unpredictable RNA–protein interactions that change the structure of RNA in cells. To circumvent this limitation, sometimes an unwieldy systematic approach involving huge numbers of candidate antisense molecules is used (20, 21). To avoid being dependent on either of these approaches, we sought to develop a Rz that would be able to access any chosen target site regardless of local secondary structure.
We reasoned that it would be useful to design a Rz that could recruit a protein that could, in turn, relieve any interfering secondary structure, thereby making any site accessible to the Rz. To create this Rz, we tried to link a Rz to an RNA helicase, a member of a class of proteins demonstrated to have nonspecific RNA binding, sliding, and unwinding activities (22–25). We introduced an RNA motif, the constitutive transport element (CTE) (Fig. 2A), that appears to interact with RNA helicases in vitro and in vivo (26–34). The CTE was discovered as a cytoplasmic transport signal for D-type retroviral RNA (35, 36). We hypothesized that an RNA helicase coupled to a Rz might efficiently guide the Rz to its target site by resolving any inhibitory mRNA structures, thereby leading to efficient substrate cleavage.
Figure 2.
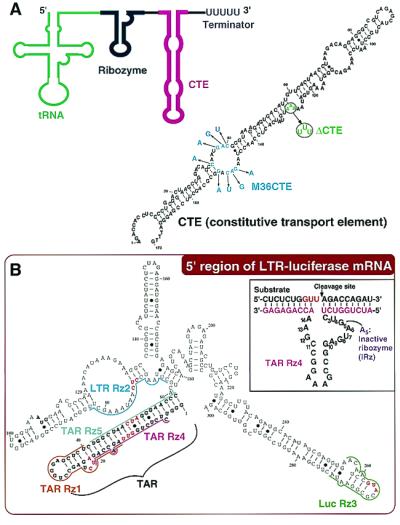
CTE-Rz design. (A) Schematic representation of a CTE-Rz (Left) and the secondary structure (predicted by MulFold; ref. 19) of the constitutive transport element (CTE) (Right). The sites of mutation that generate M36CTE (27, 28) and ΔCTE (26, 31) are indicated by blue and green letters, respectively. (B) The secondary structure (predicted by MulFold; ref. 19) of the 5′ region of long terminal repeat (LTR)-luciferase mRNA targeted by the indicated Rz. The sequence of the TAR Rz 4 is shown as a typical example. Complete loss of the catalytic activity of hammerhead Rz is achieved by a point mutation of G5 to A5 in the catalytic core.
Materials and Methods
Plasmids.
The construction of Rz expression vectors derived from the pUC-dt plasmid was described previously (12, 13). To generate CTE-Rz expression vectors, we inserted the CTE (SRV CTE-1; refs. 26, 27, 32, and 36) sequence derived from the simian type D retrovirus (Fig. 2A). pUC-dt was double-digested by Csp 45I and SalI and each Rz sequence (Fig. 2B), with KpnI and EcoRV sites and the terminator sequence TTTTT at the 3′ end, was cloned into this plasmid (Fig. 2A). The KpnI and EcoRV sites were used for subsequent insertion of the CTE sequence. The pRcCMV-mychDbp5 vector encodes the gene for human Dbp5 helicase (hDbp5) with the myc tag at the N terminus (32). The pcDNA3 RHA-HA vector encodes the gene for human RNA helicase A (RHA) with the hemagglutinin epitope (HA) tag at the N terminus (26, 31).
Measurement of Activities of Rz in LTR-Luc HeLa Cells.
Luciferase activity was monitored basically as described elsewhere (13, 37). LTR-Luc HeLa cells in 12-well plates were cotransfected with 4 μg of each Rz expression plasmid and 150 ng of Tat-expressing plasmid (pCD-SRα/tat; 37) with the use of 4 μl of Lipofectin reagent (GIBCO/BRL, Rockville, MD). In the case of the assay of dominant-negative activity, indicated amounts of plasmid were used for transfection. Cells were cultured for 36 h after the transfection.
Luciferase activity was measured with a PicaGene kit (Toyo-inki, Tokyo, Japan) as described elsewhere (13, 37). To normalize the efficiency of transfection by reference to β-galactosidase activity, cells were cotransfected with 50 ng of the pSVβ-galactosidase control vector (Promega), as described elsewhere (13).
Analysis of the Cleavage Activity of Rz in Vitro.
Each Rz, with or without the CTE sequence, and substrate RNA, the 5′-region of the LTR-Luc mRNA (300 nt) (Fig. 2B), were prepared in vitro with the use of T7 RNA polymerase. Assays of Rz activity in vitro were performed, in 10 mM MgCl2 and 50 mM Tris⋅HCl (pH 7.5) at 37°C, under enzyme-saturating (single-turnover) conditions (10 μM Rz, 2 nM substrate), as described elsewhere (10, 13).
Western Blotting Analysis.
NIH 3T3 cells that had been transfected with each Rz expression vector were harvested. Fifty micrograms of protein per lane was loaded onto an SDS/15% polyacrylamide gel. After electrophoresis, bands of protein were transferred to a polyvinylidene difluoride membrane (Amersham Pharmacia). The membrane was probed with rabbit polyclonal antibodies against CPP32 and rabbit polyclonal antibodies against actin, as described elsewhere (10). After incubation of the membrane with FITC-conjugated antibodies against IgG as the second antibody, bands were detected with a Fluoro-Image Analyzer (Molecular Dynamics). Blocking and detection were performed basically as described elsewhere (10).
Northern Blotting Analysis.
Cytoplasmic RNA and nuclear RNA were isolated from LTR-Luc HeLa cells that had been transfected with individual Rz expression vectors as described previously (10, 12, 13). Thirty micrograms of total RNA per lane was loaded onto a 3.0% NuSieve 3:1 agarose gel (FMC). After electrophoresis, bands of RNA were transferred to a Hybond-N nylon membrane (Amersham Pharmacia). The membrane was probed with synthetic oligonucleotides that were complementary to the sequences of the various Rz as described elsewhere (12). A synthetic probe complementary to the sequence of the CTE was used to determine the localization and the steady-state level of CTE RNA.
Detection of Interaction in Cells Between CTE-Rz and RNA Helicases.
Coimmunoprecipitation of CTE-Rz RNA and hDbp5 or RHA was done by transiently transfecting HeLa S3 cells with the CTE-Rz expressing vector and either pRcCMV-mychDbp5 (32) or pcDNA3 RHA-HA (26, 31), as described elsewhere (31). Antibodies specific for either the c-myc tag (CLONTECH) or the HA tag (Roche Molecular Biochemicals) were used for the immunoprecipitations. Cell extracts were incubated overnight at 4°C, with each antibody conjugated to protein A-agarose beads (Amersham Pharmacia). The extracted RNA was subjected to RT-PCR with the Rz-specific primer.
Precipitation of Proteins That Interact with in Vitro Synthesized CTE-Rz.
Precipitation of proteins that interact with in vitro synthesized CTE-Rz was carried out basically as described elsewhere (31). Biotin-labeled RNA was synthesized with an AmpliScribe T7 transcription kit (Epicentre Technologies, Madison, WI). The molar ratio of biotin-21-UTP (CLONTECH) to UTP in the reaction was 1:5. As a control, a biotinylated transcript without tRNA, Rz, or CTE sequences (MCS) was prepared with pBluescript (Stratagene) as a template. Two hundred microliters of cell extract from 2 × 107 HeLa S3 cells transfected with either pRcCMV-mychDbp5 or pcDNA3 RHA-HA was mixed with 70 μg of biotinylated RNA, incubated on ice for 10 min, and then adjusted to 1 ml with binding buffer (20 mM Tris⋅HCl, pH 7.5/60 mM KCl/2.5 mM EDTA/0.1% Triton X-100). To this sample was added 70 μl of streptavidin-conjugated agarose beads (GIBCO/BRL), which were washed twice with binding buffer, suspended in 100 μl of binding buffer, and kept on ice. After incubation overnight at 4°C, the beads were washed three times with washing buffer (20 mM Tris⋅HCl, pH 7.5/350 mM KCl/0.01% Nonidet P-40) and resuspended in 20 μl of binding buffer. Proteins were eluted by boiling and separated by SDS/PAGE (7% polyacrylamide). For immunodetection of each RNA helicase, proteins were transferred to a polyvinylidene difluoride membrane by the standard procedure and then were probed with the antibodies mentioned above. As controls, whole-cell lysates from HeLa cells transfected with pRcCMV-mychDbp5 or pcDNA3 RHA-HA were also subjected to Western blotting.
Proteins that interact with in vitro synthesized CTE-Rz were also precipitated from HeLa S3 cells in the same way and dissolved in 50 μl of binding buffer. The effect of the addition of the precipitated proteins on the cleavage of the RNA substrate in vitro by the CTE-Rz was investigated. Twenty microliters of the reaction mixtures containing 6 μl of the proteins, 5 μM Rz, 2 nM 5′-32P-labeled substrate, 20 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 50 mM KCl, 1 mM DTT, 5% glycerol, and 5% RNase inhibitor was incubated at 37°C for 45 min in the presence or absence of 2 mM ATP and 0.2 mM GTP. The substrate and the products of each reaction were separated by electrophoresis as described above.
Results
Design of Hybrid Rz (CTE-Rz) and Their Effects on the Expression of the LTR-Luciferase Chimeric Gene.
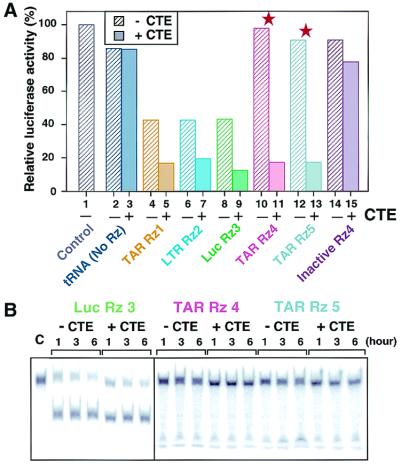
To achieve high levels of Rz expression, we previously developed an RNA polymerase III-mediated expression system that embeds a hammerhead Rz within the 3′ terminus of the human tRNAVal gene (8–10, 12–14). Thus, the Rz expressed is preceded by a partially modified human tRNA (Fig. 2A). When the attached tRNA maintains a cloverleaf-like structure, the transcript is stabilized and efficiently transported to the cytoplasm, where the target mRNA can be found (10, 12, 13). To further improve this tRNA-Rz system, the CTE sequence from the simian type D retrovirus was attached to the 3′ end (CTE-Rz) (Fig. 2A). We quantitatively evaluated the intracellular activities of Rz and CTE-Rz directed against the TAR region of the long terminal repeat (LTR) from HIV-1. This challenging target was chosen because of its extensive secondary structure (Fig. 2B). The target gene was stably expressed in HeLa cells and consisted of the LTR of HIV-1 and a luciferase gene (13, 37). Because this reporter is Tat-dependent, an expression plasmid for Tat was transiently transfected along with the Rz expression vectors.
To test the efficacy of the CTE-Rz design, we made five CTE-connected and unconnected Rz aimed at specific targets. Three Rz, namely, TAR Rz 1, LTR Rz 2, and Luc Rz 3, were designed to target relatively accessible sites located in predicted loop regions of the LTR-luciferase chimeric mRNA (Fig. 2B). As anticipated, these Rz significantly reduced expression of the reporter (Fig. 3A, lanes 4, 6, and 8).
Figure 3.
Activities of the CTE-Rz. (A) Suppression of LTR-driven luciferase activity by CTE-Rz. Red stars indicate results obtained with Rz targeted to relatively inaccessible sites of the TAR. LTR-Luc HeLa cells were transiently transfected with Tat alone (lane 1) or with Tat and the indicated tRNA-based Rz constructs (13, 37). Luciferase activity as an indicator of Rz activity is reported as a percentage of the Tat-only control. The presented values are the mean of at least three data points. Because the assays used transient transfections, there is statistical variability. The error bars are within 10% when data are taken on the same day and are within 10–25% when taken on different days. However, in all cases, the CTE-Rz always had significantly greater activity than the non-CTE-Rz. (B) The cleavage activity of the CTE-Rz in vitro. Each Rz at 10 μM was incubated with a small amount of the 5′-32P-labeled substrate under enzyme-saturated conditions, in 10 mM MgCl2/50 mM Tris⋅HCl (pH 7.5) at 37°C for the indicated hours.
TAR Rz 4 and TAR Rz 5 were designed to target sites predicted, and later confirmed (see below), to be inaccessible within the well-documented stable stem structure of the TAR region (Fig. 3A, lanes 10 and 12, as indicated by stars). These Rz without the CTE (non-CTE Rz) were unable to affect luciferase reporter activity levels (Fig. 3A, lanes 10 and 12). When the CTE was attached, these Rz became to inhibit remarkably the expression of the luciferase gene (Fig. 3A, lanes 11 and 13), resulting in an 80% reduction in reporter activity. Furthermore, these CTE-coupled Rz were more active than non-CTE Rz designed to target “open” sites (TAR Rz 1, LTR Rz 2, Luc Rz 3). Attachment of CTE to other Rz also enhanced their activity. Importantly, TAR CTE-Rz 4 and TAR CTE-Rz 5 achieved suppression levels similar to those seen for TAR CTE-Rz 1, LTR CTE-Rz 2, and Luc CTE-Rz 3 (Fig. 3A, lanes 5, 7, and 9). This suppression suggests that addition of the CTE moiety enables all Rz to efficiently attack its target site.
To demonstrate that the observed inhibitory effects were actually due to Rz-mediated cleavage, we tested an inactivated TAR Rz 4 that contains a single mutation (G5 to A5; see Fig. 2B) in the catalytic site (Fig. 3A, lanes 14 and 15). The lack of cleavage demonstrates that inhibition by TAR CTE-Rz4 is due to Rz-mediated cleavage, not antisense effects. Also, when fused to abundantly expressed tRNA, the CTE did not have a nonspecific effect on luciferase reporter activity (Fig. 3A, lanes 3 and 15).
To verify the computer structure prediction that the TAR region is difficult to access in vivo because of RNA folding and not because of RNA-bound protein(s), cleavage assays in vitro were performed (Fig. 3B). Very little cleavage of the TAR substrate was observed for TAR Rz 4 and TAR Rz 5, whether the CTE was present or not. In contrast, Luc Rz 3 directed toward a relatively accessible site (Fig. 3B) achieved substantial substrate cleavage. These data suggest that the inherent folding of the TAR region is responsible for its inaccessibility. Furthermore, non-CTE- and CTE-Rz directed against sites predicted to be accessible displayed similar high in vitro activity levels (Fig. 3B). Therefore, it seems that the CTE-mediated enhancement that occurs in cells is dependent on cellular factors, perhaps a helicase(s) that can resolve RNA secondary structure, as originally hypothesized.
General Applicability of Hybrid Rz.
To test the general applicability of the CTE-Rz, we targeted endogenous mouse procaspase-3 (CPP32) at five sites, including one predicted to be inaccessible (Fig. 4A). Mouse NIH 3T3 cells were transfected with the Rz expression plasmids, and procaspase-3 expression levels were determined by Western blotting (Fig. 4B) and quantitated (Fig. 4C). Actin expression levels were used as controls. As seen previously for the LTR-luc reporter, CTE-containing Rz were more effective than their non-CTE counterparts. In particular, CPP CTE-Rz 10 had a significant inhibitory effect (Fig. 4C, lane 13), whereas its parental Rz had virtually no effect (lane 12). None of the CTE-Rz interfered with actin expression. Similar results have been obtained for several other endogenous targets (data not shown). These results demonstrate the specificity, potent activity, and general utility of CTE-Rz.
Figure 4.
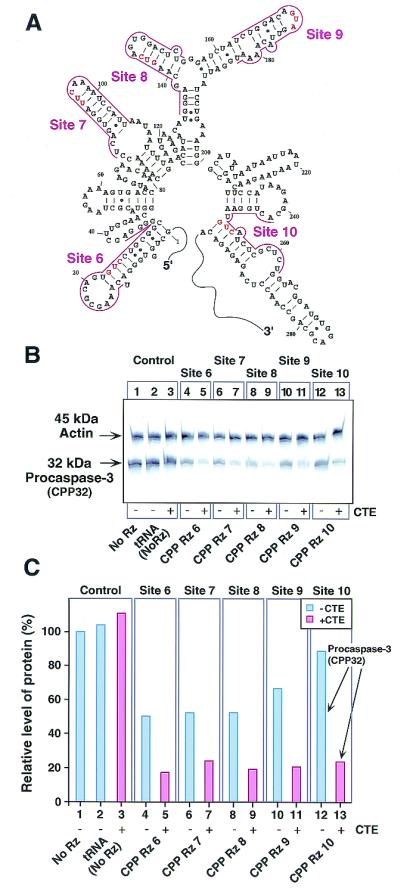
Inhibition of procaspase-3 (CPP32) gene expression by CTE-Rz. (A) The secondary structure predicted by MulFold (19) of the 5′ region of procaspase-3 mRNA targeted by Rz. (B) Detection of procaspase-3 and actin proteins by Western blotting (10). Mouse NIH 3T3 cells were transfected with the indicated Rz constructs. (C) The results in B presented as a histogram. FITC-labeled antibodies against rabbit IgG were used as the secondary antibody, and the band intensities for actin and procaspase-3 were quantitated. Procaspase-3 protein levels were normalized to actin protein levels. The normalized level of protein recorded when cells were untransfected with the Rz-expressing vector was taken as 100% (lane 1).
The Level of Expression and Localization of the Hybrid Rz.
Rz expression levels, stability, and localization are important determinants of Rz efficacy in vivo (5, 12, 13). As described previously, we expected the CTE to enhance Rz activity by resolving RNA structure. The CTE is known to be a signal for the transport of D-type retrovirus RNA to the cytoplasm (26–36); it seemed possible that the CTE might also function by enhancing cytoplasmic transport of Rz, thereby leading to higher activities. Or, because the CTE has a stem structure, it might stabilize the Rz-containing transcript. To determine the cellular expression levels and localization of a CTE-Rz, we performed Northern blot analyses of RNA from fractionated cells transfected with TAR Rz 4 and TAR CTE-Rz 4 (Fig. 5A). In our previous studies, in all cases examined, non-CTE-Rz with high activity under the control of the human tRNAVal promoter were found exclusively in the cytoplasm (8–10, 12, 13). As anticipated, TAR Rz 4 and TAR CTE-Rz 4 were expressed at similar levels and were localized to the cytoplasm, not to the nucleus (Fig. 5A). Therefore, CTE-mediated enhancement of Rz activity does not occur because of increased expression levels, as a consequence of either increased stability or transport efficiency.
Figure 5.
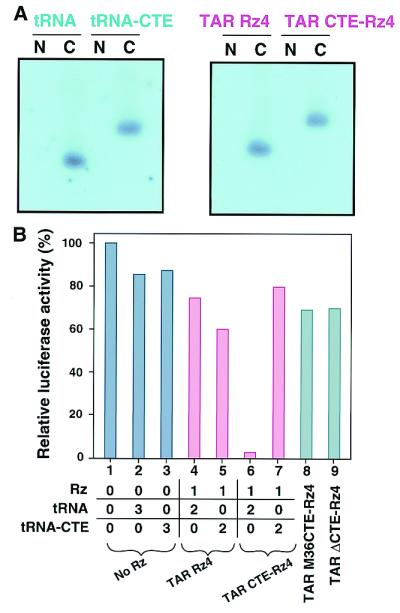
Effects of the CTE on Rz expression and mutant CTE on Rz activity. (A) Expression, stability, and intracellular localization of Rz transcripts in HeLa cells. RNA prepared from HeLa cells transfected with the indicated expression constructs was fractionated. N and C, nuclear and cytoplasmic fractions, respectively. (B) Dominant-negative effect of tRNA-CTE alone (compare lanes 6 and 7) and the effects of mutations in the CTE sequence on the activity of CTE-Rz (compare lanes 8 and 9). The amount (in μg) of plasmid that encoded each RNA used for transfection is indicated. The presented values are the mean of at least three data points. Because the assays used transient transfections, there is statistical variability. The error bars are within 10% when data are taken on the same day and are within 10–25% when taken on different days.
Importance of the CTE Sequence for Enhanced Efficiency.
Because the improved efficacy of the CTE-Rz appeared to be attributable to properties other than enhanced transcript stability and intracellular transport, it seemed likely that the ability of CTE to interact with a helicase(s) was responsible. To test for involvement of a helicase(s), we performed competition experiments. Coexpressing TAR CTE-Rz 4 with tRNA-CTE, which contains no Rz, reduced the efficacy of TAR CTE-Rz 4 cleavage of the LTR-luc substrate (Fig. 5B, lanes 6 and 7). That excess tRNA-CTE (Fig. 5B, lane 7), and not excess tRNA alone (no Rz and no CTE) (Fig. 5B, lane 6), interferes with TAR CTE-Rz 4 cleavage suggests that tRNA-CTE and TAR CTE-Rz 4 were competing for some limiting factor(s).
The CTE is known to interact with RHA and hDbp5 RNA helicase (hDbp5) (26–34). Two mutant forms of CTE (Fig. 2A), ΔCTE (26, 31, 33, 34) and M36CTE (27, 28), prevent the CTE from interacting with RHA and hDbp5, respectively. When the wild-type CTE of TAR CTE-Rz 4 was replaced by either of these mutants, Rz enhancement activity was lost (Fig. 5B, lanes 8 and 9). The results obtained with these CTE mutants underscore the importance of the CTE and strongly support the participation of RNA helicase(s) in the increased efficiency of CTE-Rz activity.
Interaction Between the Hybrid Rz and RNA Helicases.
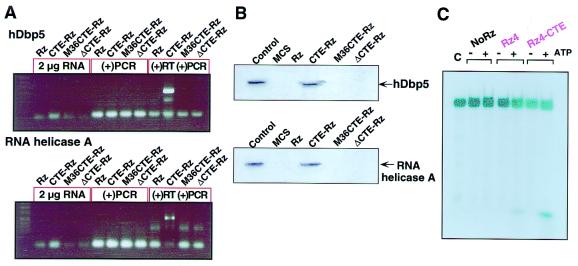
To test directly whether either of these RNA helicases does interact with our CTE-Rz, coimmunoprecipitations were carried out with hDbp5 and RHA. First, TAR CTE-Rz 4 was cotransfected with either c-myc-tagged hDbp5 (32) or HA-tagged RHA (26) into HeLa cells. Then, cell lysates were subjected to immunoprecipitation with the use of either c-myc or HA antibodies. The resulting precipitates were evaluated for the presence of TAR CTE-Rz 4 by RT-PCR. TAR CTE-Rz 4 was clearly found to be in the hDbp5 precipitate, thereby indicating that TAR CTE-Rz 4 and hDbp5 interact in vivo (Fig. 6A). TAR CTE-Rz 4 also associates with RHA, but this interaction appears to be weaker than that observed for hDbp5 (Fig. 6A). When TAR Rz 4, TAR M36CTE-Rz 4, and TAR ΔCTE-Rz 4 are used, no interaction is observed with hDbp5 and RHA, thereby confirming that the interaction between TAR CTE-Rz 4 and either of the helicases is dependent on the presence of a functional CTE.
Figure 6.
Interaction in cells between tRNAVal-driven CTE-Rz and RNA helicases. (A) Coimmunoprecipitation of CTE-Rz RNA with hDbp5 and RHA (31). Either c-myc-hDbp5 (32) or HA-RHA (26, 31) was transiently cotransfected into HeLa S3 cells with the indicated versions of the TAR Rz 4 expression vectors. Immunoprecipitates obtained with the use of the appropriate tag were subjected to RT-PCR [(+)RT (+)PCR]. The CTE-Rz was detected only when wild-type CTE was present. As controls, the RNA was analyzed before RT-PCR (2 μg RNA) and after being subjected to PCR without RT treatment [(−)RT (+)PCR]. (B) RNA helicase hDbp5 and RHA interact with CTE-Rz. The indicated versions of TAR Rz4 and a multiple cloning site of pBluescript (MCS) were synthesized in vitro with the use of biotinylated UTP and then mixed with HeLa S3 cell extract from cells transfected with either c-myc-hDbp5 or HA-RHA (31). Streptavidin beads were used to precipitate the biotinylated RNAs and associated proteins. Western blotting with antibodies recognizing the appropriate tag revealed that only TAR CTE-Rz 4 interacted with the helicases. Control, whole-cell lysate from transfected cells. (C) Stimulated cleavage by CTE-Rz upon the addition of proteins precipitated with the synthesized CTE-Rz. Proteins that interact with in vitro synthesized CTE-Rz were precipitated from HeLa cells and were added to the reaction mixtures. The reaction mixtures were incubated at 37°C for 45 min in the presence or absence of 2 mM ATP and 0.2 mM GTP. Only when ATP was added to the reaction mixture was TAR CTE-Rz able to stimulate cleavage of the RNA substrate. The lesser extent of the stimulated cleavage upon the addition of the proteins and ATP was also observed, even in the case of the TAR Rz 4. It seems to be a potential enhancement by the RNA-unwinding activity of RNA helicases itself. The enhancement of cleavage by the precipitated proteins strongly suggests the presence of cellular factors responsible for the enhanced suppression by CTE-Rz.
The reciprocal precipitation confirmed these results. In this case, in vitro transcribed biotinylated Rz were mixed with cell lysates derived from HeLa cells transfected with either c-myc-hDbp5 (32) or HA-RHA (26, 31, 33, 34). The four Rz constructs used were TAR Rz 4, TAR CTE-Rz 4, TAR M36CTE-Rz 4, and TAR ΔCTE-Rz 4. To control for nonspecific interactions, a biotinylated transcript without tRNA, Rz, or CTE sequences was used (MCS, Fig. 6B). Proteins were precipitated with the use of avidin-conjugated agarose beads that recognize biotin. The precipitates were probed for c-myc-hDbp5 or HA-RHA by Western blotting. Only TAR CTE-Rz 4 was found to be complexed with c-myc-hDbp5 and HA-RHA, verifying that the CTE enhances CTE-Rz activity by preferentially interacting with hDbp5 and/or RHA.
Moreover, to corroborate our hypothesis that a certain cellular factor, probably RNA helicase, is responsible for the CTE-mediated enhancement, we demonstrate in vitro the enhanced cleavage by the CTE-Rz. Proteins were precipitated from HeLa cells by probing with a synthesized TAR CTE-Rz 4. The precipitated proteins were added to either TAR Rz 4- or TAR CTE-Rz 4-mediated cleavage reaction mixtures, and the effect on the in vitro cleavage of RNA by CTE-Rz was measured (Fig. 6C). The activity of the RNA helicases is NTP (preferably ATP) dependent. Only when ATP was added to the reaction mixture was TAR CTE-Rz able to stimulate cleavage of the RNA substrate. It seems to be a potential enhancement by the RNA-unwinding activity of RNA helicases itself. The enhancement in cleavage by the precipitated proteins strongly suggests the presence of cellular factors responsible for the enhanced suppression by CTE-Rz. Furthermore, the ATP dependency of the enhancement is consistent with the involvement of RNA helicases.
Discussion
By adding the CTE sequence to our tRNA-Rz, we were able to efficiently cleave sites that had previously been refractory because of local RNA folding. The presence of the CTE also improved cleavage activity for Rz that were already functional. It is also noteworthy that TAR CTE-Rz 4 and TAR CTE-Rz 5 were able to cleave TAR even in the presence of Tat, a protein known to bind TAR. When CTE mutants defective for RNA helicase interaction were substituted for wild-type CTE, the resulting TAR ΔCTE-Rz 4 and TAR M36CTE-Rz 4 completely lost activity. Therefore, to be effective, the CTE must be present in cis and must retain the capacity to interact with an RNA helicase(s).
The increased activity mediated by the CTE does not appear to involve increased stability, expression, or transport of the Rz. Rather, the CTE seems to mediate its effects by interacting with RNA helicases. Two RNA helicases (hDbp5 and RHA) have been demonstrated previously to interact with CTE (26–34). We demonstrated that our CTE-Rz indeed interacted with these RNA helicases [at least with RNA helicase hDbp5, and to a lesser extent, with RHA (Fig. 6A)] in mammalian cells. RNA helicase hDbp5 has been shown to interact with an adapter protein known as Tip-associated protein (27, 28, 30). This interaction is crucial for hDbp5-CTE interaction. By using cells devoid of Tip-associated protein (QCl-3 cells; ref. 30), we have obtained preliminary evidence indicating that Tip-associated protein is essential for CTE-Rz activity (data not shown), thereby further implicating hDbp5 in CTE-Rz activity. Because RNA helicases, including hDbp5, have been shown to possess RNA-unwinding activity (22–25, 32), we hypothesize that the CTE is recruiting this helicase(s) to the target site, where it unwinds inhibitory structures. It is attractive to consider that the helicase may even be able to slide the tRNA-Rz along a transcript, consistent with the sliding mechanisms of action demonstrated for several RNA helicases (Fig. 7; refs. 22–25, 32). The key aspect of our CTE-Rz is that it overcomes a major obstacle of Rz use, thereby greatly increasing the general utility of Rz. In particular, attachment of the CTE has made it possible to suppress the expression of genes that were previously found to be recalcitrant to Rz cleavage.
Figure 7.
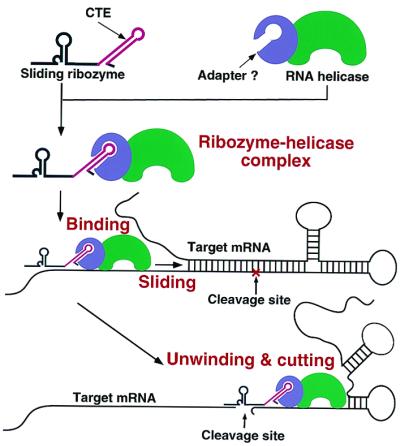
Schematic representation of how a CTE-Rz coupled to an RNA helicase might cleave a hidden target site upon the unwinding of the local secondary structure.
Despite recent advances in Rz technology that generally improved the utility of Rz, Rz design continues to be problematic (8–14). For a Rz to be successful it must have an easily accessible target sequence. Until now, such a target site was identified based on computer-aided structural predictions of the target RNA or by unwieldy trial-and-error experiments. To overcome this limitation we sought to construct a Rz that would be able to access any target site regardless of its local secondary or tertiary structure. Hoping to take advantage of the natural ability of RNA helicases to modulate RNA structure, we decided to connect a Rz to an RNA helicase. This connection was made by linking our Rz to a sequence, the CTE, that has been shown to interact with an RNA helicase(s).
For all mRNAs tested (five as of now), our CTE-Rz suppressed expression much more efficiently than the parental, non-CTE Rz. Most importantly, they were able to cleave the target mRNA at any site, regardless of the predicted secondary or tertiary structure. All of the CTE-Rz displayed robust activity in cell culture and in many cases were able to work when the parental, non-CTE Rz were inactive. Therefore, we believe the hybrid Rz has broad applicability because it is extremely easy to design and use, whereas previous Rz technology required specialized skills. Furthermore, its improved efficacy makes it even more suitable for a wide range of applications. Having improved the efficacy of Rz by eliminating target site selection constraints, we believe that the CTE-Rz is a powerful tool with both basic research and therapeutic utility.
Acknowledgments
We thank Prof. E. Izaurralde for the gift of pRcCMV-mychDbp5 and Prof. B. R. Cullen for the gift of QCl-3 cells. We thank Dr. B. Spain for helpful comments and discussions.
Abbreviations
- Rz
ribozyme(s)
- CTE
constitutive transport element
- HA
hemagglutinin epitope
- LTR
long terminal repeat
- RHA
human RNA helicase A
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Symons R H. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- 2.Eckstein F, Lilley D M J, editors. Catalytic RNA, Nucleic Acids and Molecular Biology. Berlin: Springer; 1996. [Google Scholar]
- 3.Zhou D-M, Taira K. Chem Rev. 1998;98:991–1026. doi: 10.1021/cr9604292. [DOI] [PubMed] [Google Scholar]
- 4.Shimayama T, Nishikawa S, Taira K. Biochemistry. 1995;34:3649–3654. doi: 10.1021/bi00011a020. [DOI] [PubMed] [Google Scholar]
- 5.Sullenger B A, Cech T R. Science. 1993;262:1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- 6.Yu M, Leavitt M C, Maruyama M, Yamada O, Young D, Ho A D, Wong-Staal F. Proc Natl Acad Sci USA. 1995;92:699–703. doi: 10.1073/pnas.92.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee N S, Good P, Chatterjee S, Grange T, Pictet R, Kohn D, et al. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 8.Kawasaki H, Eckner R, Yao T P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Nature (London) 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 9.Kuwabara T, Warashina M, Orita M, Koseki S, Ohkawa J, Taira K. Nat Biotechnol. 1998;16:961–965. doi: 10.1038/nbt1098-961. [DOI] [PubMed] [Google Scholar]
- 10.Kuwabara T, Warashina M, Tanabe T, Tani K, Asano S, Taira K. Mol Cell. 1998;2:617–627. doi: 10.1016/s1097-2765(00)80160-4. [DOI] [PubMed] [Google Scholar]
- 11.Plehn-Dujowich D, Altman S. Proc Natl Sci Acad USA. 1998;95:7327–7332. doi: 10.1073/pnas.95.13.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koseki S, Tanabe T, Tani K, Asano S, Shioda T, Nagai Y, Shimada T, Ohkawa J, Taira K. J Virol. 1999;73:1868–1877. doi: 10.1128/jvi.73.3.1868-1877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuwabara T, Warashina M, Nakayama A, Ohkawa J, Taira K. Proc Natl Acad Sci USA. 1999;96:1886–1891. doi: 10.1073/pnas.96.5.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanabe T, Kuwabara T, Warashina M, Tani K, Taira K, Asano S. Nature (London) 2000;406:473–474. doi: 10.1038/35020190. [DOI] [PubMed] [Google Scholar]
- 15.Birikh K R, Heaton P A, Eckstein F. Eur J Biochem. 1997;245:1–16. doi: 10.1111/j.1432-1033.1997.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- 16.Kato, Y., Kuwabara, T., Warashina, M., Toda, H. & Taira, K. (2001) J. Biol. Chem.276, in press. [DOI] [PubMed]
- 17.Jeltsch A, Wenz C, Stahl F, Pingoud A. EMBO J. 1996;15:5104–5111. [PMC free article] [PubMed] [Google Scholar]
- 18.Young M C, Weitzel S E, von Hippel P H. J Mol Biol. 1996;264:440–452. doi: 10.1006/jmbi.1996.0652. [DOI] [PubMed] [Google Scholar]
- 19.Jaeger J A, Turner D H, Zuker M. Methods Enzymol. 1989;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 20.Milner N, Mir K U, Southern E M. Nat Biotechnol. 1997;15:537–541. doi: 10.1038/nbt0697-537. [DOI] [PubMed] [Google Scholar]
- 21.Patzel V, Sczakiel G. Nat Biotechnol. 1998;16:64–68. doi: 10.1038/nbt0198-64. [DOI] [PubMed] [Google Scholar]
- 22.Lee C-G, Zamore P D, Green M R, Hurwitz J. J Biol Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 23.Wagner J D O, Jankowsky E, Company M, Pyle A M, Abelson J N. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Cruz J, Kressler D, Linder P. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 25.Jankowsky E, Gross C H, Shuman S, Pyle A M. Nature (London) 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 26.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 27.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 28.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodge C A, Colot H V, Stafford P, Cole C N. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang Y, Cullen B R. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Tang H, Mullen T M, Westberg C, Reddy T R, Rose D W, Wong-Staal F. Proc Natl Acad Sci USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues J P, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, Izaurralde E. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westberg C, Yang J P, Tang H, Reddy T R, Wong-Staal F. J Biol Chem. 2000;275:21396–21401. doi: 10.1074/jbc.M909887199. [DOI] [PubMed] [Google Scholar]
- 34.Tang H, Wong-Staal F. J Biol Chem. 2000;275:32694–32700. doi: 10.1074/jbc.M003933200. [DOI] [PubMed] [Google Scholar]
- 35.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjold M-L. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koseki S, Ohkawa J, Yamamoto R, Takebe Y, Taira K. J Controlled Release. 1998;53:159–173. doi: 10.1016/s0168-3659(97)00250-2. [DOI] [PubMed] [Google Scholar]