Abstract
Background
Bone marrow mesenchymal stem cells (BMSCs) are a crucial component of the tumor microenvironment (TME), with hypoxic conditions promoting their migration to tumors. Exosomes play a vital role in cell-to-cell communication within the TME. Hypoxic TME have a great impact on the release, uptake and biofunctions of exosomes. This study aims to elucidate the communication between BMSC-derived exosomal miRNA and triple-negative breast cancer (TNBC) in a hypoxic environment.
Methods
Exosomes were isolated via ultracentrifugation and identified using scanning electron microscopy (SEM), nanoparticle tracking analysis (NTA) and western blot. A range of bioinformatics approaches were used to screen exosomal miRNAs and the target mRNAs of miRNAs and predict the possible signaling pathways. Expression levels of genes and proteins were assessed by quantitative real-time PCR and western blot. Cell proliferation, apoptosis, migration and invasion were analyzed using CCK-8 assay, EDU assay, transwell migration, wound healing assay and invasion assay, respectively. Dual luciferase reporter gene assay was conducted to confirm the binding between miRNAs and the target mRNAs. The impact of hypoxic BMSC-derived exosomal miRNA on TNBC progression in vivo was evaluated using tumor xenograft nude mouse models. Furthermore, the impact of patients’ serum exosomal miRNA on TNBC was implemented.
Results
Exosomes derived from hypoxic BMSCs promotes the proliferation, migration, invasion and epithelial-mesenchymal transition of TNBC and suppresses the apoptosis of TNBC. The expression of miR-210-3p in BMSC-derived exosomes is markedly elevated in hypoxic conditions. Exosome-mediated transfer of miR-210-3p from hypoxic BMSCs to TNBC targets NFIX and activates Wnt/β-Catenin signaling in TNBC. Deletion of miR-210-3p in hypoxic BMSC-derived exosomes attenuates TNBC in vivo. Additionally, human exosomal miR-210-3p from the serum of TNBC patients promotes TNBC progression. Moreover, we notably observed a marked downregulation of NFIX expression levels in cancerous tissues compared to paracancerous tissues.
Conclusions
Hypoxic BMSC-derived exosomal miR-210-3p promotes TNBC progression via NFIX-Wnt/β-catenin signaling axis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05947-5.
Keywords: Triple-negative breast cancer, Exosomes, Hypoxia, miR-210-3p, NFIX-Wnt/β-catenin axis
Background
Breast cancer is the most prevalent malignant tumor in women, contributing significantly to the global cancer burden [1–4]. It is highly heterogeneous, with variations in clinical presentation and prognoses both among different molecular subtypes and within patients of the same subtype [5]. Among these subtypes, triple-negative breast cancer (TNBC) is particularly challenging, characterized by the absence of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (Her-2) expression [6]. Compared to hormone receptor-positive and Her-2 positive breast cancer, TNBC is associated with the highest rates of recurrence and mortality, primarily due to its aggressive biological behavior and the absence of effective targeted therapies [7–9].
The hypoxic microenvironment plays a pivotal role in enhancing tumor aggressiveness and heterogeneity, increasing the risk of metastasis, and promoting resistance to various therapeutic approaches in TNBC [10]. Hypoxia triggers a range of alterations in tumor cells, including the suppression of apoptosis pathways by decreasing the bax/bcl-2 ratio, reducing cytochrome c release, and inhibiting caspase-3 activity [11]. Moreover, hypoxia facilitates epithelial-mesenchymal transition (EMT) by downregulating the expression of E-cadherin and upregulating the expression of transcription factors such as Slug, Snail, and Twist [12]. Importantly, hypoxic conditions not only enhance resistance to chemotherapy but also contribute to therapeutic failure by activating drug efflux pumps, such as P-glycoprotein, or by limiting drug efficacy due to insufficient oxygenation [13]. A deeper understanding of the intricate interactions between tumor cells and other cells within the tumor microenvironment (TME) under hypoxic conditions is essential for addressing the complex biological characteristics of TNBC and its therapeutic resistance [14].
Exosomes, small lipid-bilayer encapsulated vesicles, facilitate intercellular communication by transporting bioactive molecules such as proteins, DNA, mRNA/miRNA/lncRNA and lipids between cancer cells, neighboring stromal cells, mesenchymal stem cells (MSCs), and immune cells [15–17]. Under hypoxic conditions, tumor cells produce an increased number of exosomes to meet their nutritional demands and facilitate intercellular communication within the TME [18, 19]. Additionally, the hypoxic environment also increases the heterogeneity of exosomes, further complicating tumor biology and contributing to the complexity of tumor progression and resistance mechanisms [20, 21]. Bone marrow-derived mesenchymal stem cells (BMSCs), a type of pluripotent stem cells, are recruited into TME and contribute to tumor progression and metastasis [22, 23]. Hypoxia has been shown to promote the migration of these stem cells to tumors, with exosomes being a key mediator in the paracrine mechanisms involved in tumor development and resistance to treatment [24–28].
MicroRNAs are small non-protein coding RNA molecules, typically consisting of about 22 nucleotides, which regulated the expression of multiple genes by binding to the 3’ untranslated regions of target mRNAs, leading to mRNA silencing [15, 29]. Previous studies have shown that miRNAs affect proliferation, migration and invasion of breast cancer [30]. Exosomal miRNAs, secreted in large quantities by various cell types and easily accessible, have emerged as promising diagnostic tools and potential therapeutic targets for breast cancer [31–33]. This study aims to investigate the relationship between BMSC-derived exosomal miRNAs and TNBC in the hypoxic microenvironment, and to explore the underlying mechanisms involved. Further, we validated the findings in serum exosomes from patients with TNBC.
Materials and methods
Ethics statement
The study protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiao tong University Shaanxi Province (Xi’an, China). The study purpose and experimental procedures were conveyed to all participants and informed consent was obtained. All experiments were approved by the Health Science Center of Xi’an Jiao tong University Approval for Research Involving Animals.
Patients and specimens
The serum was gained from five patients with pathologically-confirmed TNBC who underwent surgery at the Comprehensive Breast Care Center, formerly known as Department of Oncology (Second Affiliated Hospital of Xi’an Jiaotong University) and five healthy women who visited the medical examination center at the Second Affiliated Hospital of Xi’an Jiaotong University for a check-up. Concurrently, we procured cancerous and paracancerous tissues from five patients diagnosed with TNBC. And, specimen collection was informed by all participants with written consent.
Tumor xenografts in nude mice
Female BALB/c nude mice (weight 18–22 g) were acquired from Experimental Animal Center of Xi’an Jiaotong University (Xi’an, China). BALB / c nude mice were randomly divided into five groups. PBS was used as the control group. MDA-MB-231 cells, treated with normal exosomes group, hypoxic exosomes group, hypoxic exosomes scramble group, miR-210-3p knockdown in hypoxic exosomes group, respectively, were resuspended in 0.25 mL of PBS at a concentration of 2.0 × 106 cells (1 × 106) subcutaneously injected into each nude mouse. Tumor volumes were measured every 6 days. Measurements of length, width and thickness were obtained using calipers. On day 30 post-inoculation, mice were euthanised by inhalation of CO2. Apoptosis was detected by TUNEL assay (AccuRef Scientific, Xi’an, China). Ki67 was detected by histochemical stain and western blot was used to detect the expression of molecular markers related to apoptosis, migration and invasion.
Cell culture
BMSCs purchased from American Type Culture Collection (ATCC) were cultured in a conditioned medium (HUXMA-03011-440) at 37 °C, 90% humidity, and 5% CO2. Hypoxia induction was then performed. Cells were cultured in a gas incubator with 20% oxygen (normal oxygen). Hypoxic cells were maintained in a gas incubator with 1% oxygen (1% oxygen + 5% carbon dioxide + 94% nitrogen).
Breast cancer cell lines MDA-MB-231 and Hs578T purchased from ATCC. MDA-MB-231 were cultured in Leibovitz medium containing 10% fetal bovine serum (FBS). Hs578T were cultured in DEME medium containing 0.01 mg/ml insulin, 10% FBS and 1% penicillin-streptomycin. All cells cultured at 37 °C, 90% humidity, and 5% CO2.
Human embryonic kidney 293T cells (HEK293T) purchased from ATCC were cultured under the same conditions as Hs578T.
Extraction of exosomes and identification
Ultracentrifugation was used to extract exosomes [28, 34]. FBS was centrifuged at 100,000 × g at 4 °C for 18 h to remove extracellular vesicles (EVs) and filtrated through 0.22 μm filter. BMSCs were cultured in the medium with FBS (no EVs). The culture supernatant was collected by ultracentrifugation after 72 h: 300 ×g (10 min, 4 °C), 2000 ×g (15 min, 4 °C) and 12,000 ×g (30 min, 4 °C). Then, the precipitated exosomes were filtrated through 0.22 μm filter and purified by centrifugation according to the above conditions. BCA protein quantitative kit used to detect exosome concentration.
The blood samples (10 mL) were collected in ethylenediaminetetraacetic acid tubes and centrifuged to obtain serum. The process for extracting exosomes were as described previously.
The morphology of exosomes was recorded by scanning electron microscopy (SEM) and the grain size distribution was described by nanoparticle tracking analysis (NTA). exosomes were identified using CD63, TSG 101, HSP 70 and Calnexin by western blot.
Exosome-derived miR-210-3p isolation and RNA analyses
Total RNA was extracted from normal exosomes and hypoxia exosomes using TRIzol reagent. NanoDrop™ 2000 used to measure the concentration and quality of total RNA. quantitative real-time PCR (RT-qPCR) examined the expression of miR-210-3p.
Cell transfection
Scramble, miR-210-3p KD, mimic-NC and miR-210-3p mimic were obtained from RiboBio, China, and their sequences are shown in Table S1. MDA-MB-231 and Hs578T cells were seeded into 6-well plates and grown to 70% confluence. Transient transfection with RNA was performed using Lipofectamine 3000 (Invitrogen, UK) in accordance with the manufacturer’s instructions.
Quantitative real-time PCR
TRIzol (AccuRef Scientific, Xi’an, China) reagent and was used to isolate the total RNA. The quality of RNA was evaluated by measuring RNA concentration and purity using Nanodrop 2000 (DHS Life Science & Technology Co., Ltd, China). All RNA samples were stored at -80 °C refrigerator. Accuref 1st Strand cDNA Synthesis Kit (AccuRef Scientific, Xi’an, China) was used to reverse transcription. Table S2 contains a list of all primer sequence used in this study.
CCK8 assay
MDA-MB-231 and Hs578T cells were planted into 96-well plates one day before treatment. Exosomes (20 µg/mL) were directly added into cells when the cells grew at about 70% of confluent. The specific experimental groupings were PBS, normal exosomes, hypoxia exosomes, hypoxia exosomes scramble and hypoxia exosomes miR-210-3p knockdown (PBS vs. Normal Exo vs. Hypoxia Exo; Hypoxia Exo vs. Hypoxia Exo/ scramble vs. Hypoxia Exo/ miR-210-3p KD).
5-Ethynyl-20-deoxyuridine (EDU) assay
MDA-MB-231 and Hs578T (after different conditions of intervention) were incubated with 10 mM EDU medium diluent (AccuRef Scientific, Xi’an, China) in 96-well plates for 2 h. Following being fixed and permeabilized, 0.5 mL ApolloVR reaction cocktail was employed to react with the EDU for 30 min. Cell nuclei were stained with DAPI. Finally, EDU positive cells were imaged by the fluorescence microscopy and analyzed by Image J software.
Western blot analysis
RIPA lysis buffer (AccuRef Scientific, Xi’an, China) was used for protein extraction. Protein concentration was detected by BCA Protein assay kit (AccuRef Scientific, Xi’an, China). The cell protein samples were separated on 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes via electrophoresis. The primary antibody solution was blocked in 5% skim milk powder and incubated overnight at 4 °C, followed by incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. Anti-β-actin antibody (ab8227, Abcam) was used as an internal control. The chemiluminescent signals were captured by HRP-ECL.
Cell migration and invasion assays
Transwell migration was performed using a transwell chamber. 1 × 105 cells were suspended with 100 µL serum-free medium and loaded into the upper chamber. Meanwhile, 100 µL medium containing 10% FBS was spread over the lower chamber. After 24 h, we removed chambers and flicked the upper layer of the filter membrane with a cotton swab to remove excess cells, and inverted the chambers to air dry. 500 µL of 0.1% crystal violet was added to the culture plate. The chambers were put into the culture wells to ensure the filter membrane is completely submerged in the staining solution. Remove the chamber carefully with tweezers after standing for 30 min at 37 °C. Migrated cell images were imaged under microscope. And, cell migration was quantitated by counting in 4 random fields on the lower membrane surface.
Transwell invasion assay was performed in the presence of Matrigel. And, the remaining process was the same as Transwell migration assay.
Wound healing assay
Cells were inoculated into 6-well plates and resuspended by 0.25% trypsin digestion after growing to 90%. The resuspended cells were seeded to the 6-well plates at a density of 5 × 105/well. And, 2 ml of culture medium was added to each well. Vertical plate was scribed with 200 µL pipette tip after 24 h. The migrated ability into the gap of cells was captured by microphotography at 0 h and 24 h. The wound healing rate was quantified using Image J.
Data collection and differential expression analysis of miRNAs
The RNA-sequence and miRNA datasets of breast invasion carcinoma (BRCA) were downloaded from the UCSC Xena website (https://xena.ucsc.edu/, derived from the TCGA). We identified differentially expressional miRNAs by R packages “DESeq2”. Volcano plot was utilized to visualize the differentially expressed miRNAs. The exosomal miRNAs related overall survival of BRCA were obtained from ExoBCD database (https://exobcd.liumwei.org/) in June 2023 [35]. Venn diagrams were constructed using the jvenn online tools (https://jvenn.toulouse.inrae.fr/app/example.html) to identify and select the miRNA in the intersecting regions as the target miRNAs [36]. R package “Heatmap” was used to draw the heatmap of the target miRNAs. “ggplot2” and “forcats” were used to draw boxplot.
Prediction of target mRNAs of miRNAs
The target mRNAs of the candidate exosomal miRNAs were predicted by using TargetScan (https://www.targetscan.org/vert_80/) [37] and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) [38] databases in June 2023. After merging the target mRNAs predicted by the two tools, target mRNAs potentially involved in regulation of BRCA progression by exosomal miRNAs were acquired by intersection of the differentially expressed mRNAs and target mRNAs.
Identification of target genes of miRNA
TargetScan and miRWalk bioinformatics analysis online tools were used to analyze and predict the downstream genes of miR-210-3p (our study defined three mRNAs as candidate mRNAs). HEK-293T cells were transfected with NC mimic and miR-210-3p mimic. And, the expression levels of target genes were detected by RT-qPCR. NFIX was identified as the potential targets of miR-210-3p. NFIX 3’-UTR sequence containing the binding site of miR-210-3p (NFIX-WT) and mutated miR-210-3p binding site (NFIX-MUT) were constructed. Lipofectamin™ 3000 was used to transfer the reporter vectors containing NFIX-WT or NFIX-MUT with mimic NC or miR-210-3p mimic were co-transfected into MDA-MB-231 cells. The luciferase activities of renin and firefly were measured using a dual-luciferase reporter assay system (Promega) after 48 h.
Signaling pathways involved in NFIX-induced breast cancer progression
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets were obtained from the Gene Set Enrichment Analysis website (GSEA, https://www.gsea-msigdb.org/gsea/downloads.jsp) in June 2023 [39]. GO and KEGG functional annotations and enriched pathways associated with NFIX were analyzed using the R packages “Deseq2” and “clusterProfiler”.
Statistical analysis
The experimental results were presented as mean ± standard deviation (± SD). Comparison between two groups was analyzed by Student’s t-test. Statistical differences between multiple groups were compared using one-way analysis of variance (one-way ANOVA). Dunn Multiple Comparison Test was used to determine significant difference between groups. SPSS 21.0 was used for statistical analysis, with a test level of α = 0.05 and P < 0.05 indicating statistical significance. Statistical significance is represented as follows: *p < 0.05, **p < 0.01, ***p < 0.001. The graphs generated by GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA) and combined by Adobe Photoshop 2020. All experimental data were replicated three times.
Results
Hypoxic BMSC-derived exosomes promote TNBC progression
To investigate the impact of hypoxic BMSC-derived exosomes on TNBC, we isolated exosomes from the cell culture medium of normoxic BMSCs and hypoxic BMSCs, and treated MDA-MB-231 and Hs578T cells with them, respectively. The exosomes exhibited a characteristic saucer-like bilayer membrane structure and showed a narrow size distribution with a peak value at 70 nm (Figure S1A). The exosomes were positive for CD63, CD81, and HSP70, typical biomarkers for exosomes, and negative for calnexin (Figure S1B).
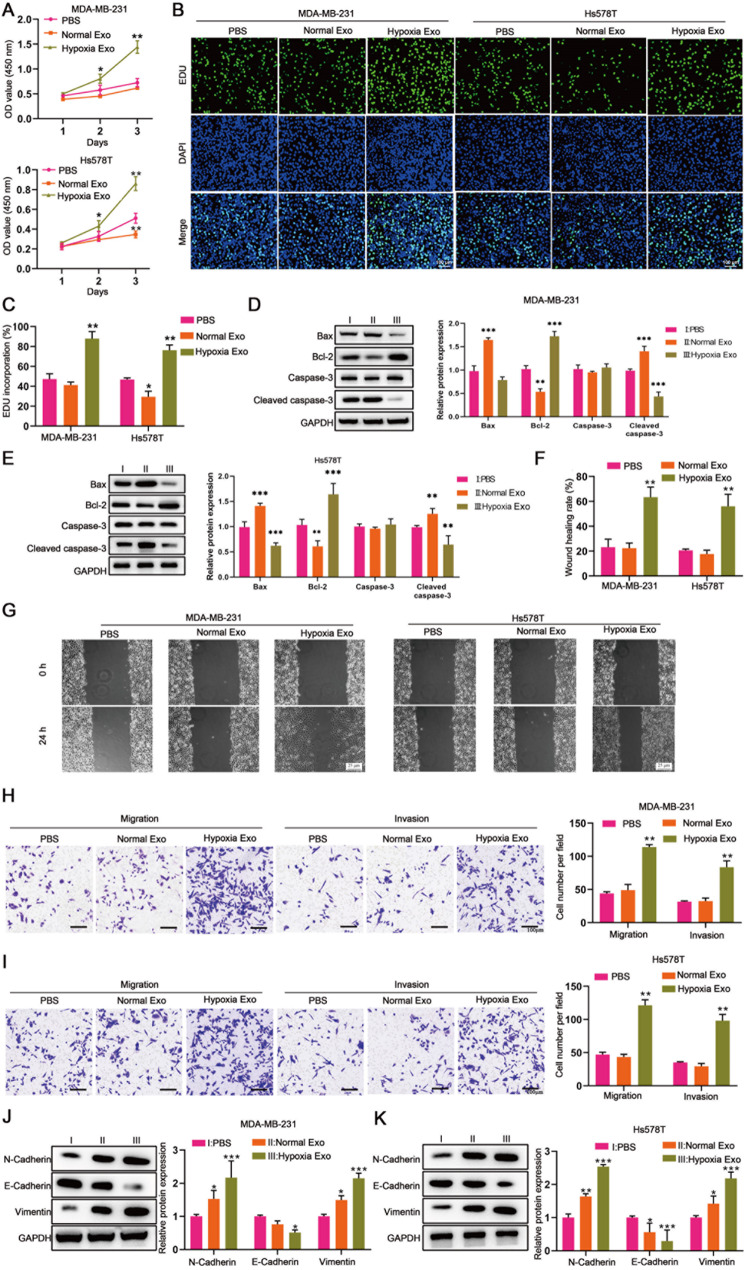
As the results shown, MDA-MB-231 and Hs578T treated with hypoxic BMSC-derived exosomes showed enhanced cell viability and proliferation compared to those treated with normoxic BMSC-derived exosomes (Fig. 1A, B and C). In addition, hypoxic BMSC-derived exosomes significantly repressed apoptosis of MDA-MB-231 and Hs578T, as indicated by decreased Bax and cleaved caspase-3 and increased Bcl-2 expression compared to normoxic BMSC-derived exosomes (Fig. 1D and E). Wound healing assay and transwell assay revealed that hypoxic BMSC-derived exosomes promoted migration and invasion of MDA-MB-231 and Hs578T cells (Fig. 1F, G, H and I). We further measured epithelial and mesenchymal markers of MDA-MB-231 and Hs578T. It was shown that the epithelial marker E-cadherin was lower and the mesenchymal markers, including N-cadherin and vimentin, were higher in MDA-MB-231 and Hs578T treated with hypoxic BMSC-derived exosomes compared to those treated with normoxic BMSC-derived exosomes (Fig. 1J and K), indicating that hypoxic BMSC-derived exosomes promote the EMT of TNBC.
Fig. 1.
Hypoxic BMSC-derived exosomes promote the TNBC progression. MDA-MB-231 and Hs578T cells were co-cultured with PBS, normoxic BMSC-derived exosomes and hypoxic BMSC-derived exosomes for 24, 48 and 72 h, respectively [PBS (I)vs. Normal Exo (II)vs. Hypoxia Exo (III)]. (A) CCK8 assay detected the cell viability. (B) EDU assay detected the cell proliferation. (D and E) The expression of apoptosis-related proteins (Bax、Bcl-2、Caspase-3、Cleaved caspase-3) was measured by western blot. (F and G) Wound healing assay revealed the cell migration. (H and I) Transwell assay revealed the migration and invasion of MDA-MB-231 and Hs578T cells. (J and K) The expression of epithelial and mesenchymal markers (N-Cadherin、E-Cadherin、Vimentin) was measured by western blot
miR-210-3p is upregulated in hypoxic BMSC-derived exosomes
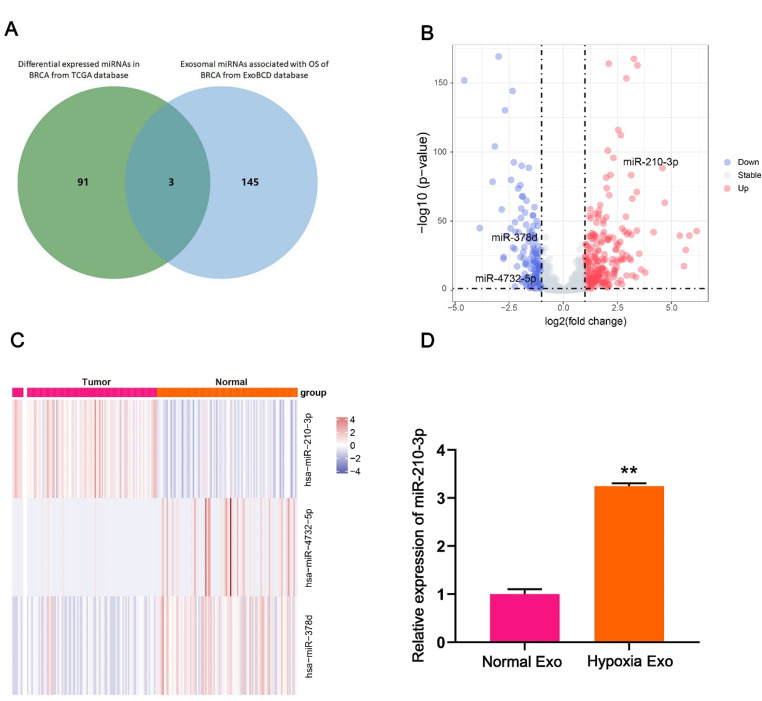
To identify miRNAs implicated in hypoxic BMSCs in breast cancer, we screened 148 exosomal miRNAs associated with breast cancer prognosis from the ExoBCD database. Among these, miR-210-3p, miR-4732-5p and miR-378d were differentially expressed in breast cancer tissues compared to normal tissues (Fig. 2A). Notably, miR-210-3p was the most obviously upregulated in breast cancer, while miR-4732-5p and miR-378d were downregulated (Fig. 2B). What’s more, miR-210-3p exhibited more significant differences in 102 paired samples of breast cancer compared to miR-4732-5p and miR-378d (Fig. 2C, Figure S2). Further analysis confirmed that miR-210-3p expression was increased in hypoxic BMSC-derived exosomes compared to normoxic BMSC-derived exosomes (Fig. 2D).
Fig. 2.
miR-210-3p is upregulated in hypoxic BMSC-derived exosomes. (A) Venn diagram showed the overlap of shared the miRNAs in breast invasion carcinoma (BRCA) and exosomes. (B) miRNAs (miR-210-3p, miR-4732-5p and miR-378d) differentially expressed in breast cancer tissue and normal tissue (derived from the TCGA). (C) Heatmap of the miR-210-3p, miR-4732-5p and miR-378d in 102 paired samples of breast cancer (derived from the TCGA). (D) Expression miR-210-3p in normoxic BMSC-derived exosomes and hypoxic BMSC-derived exosomes was detected by RT-qPCR
Exosomal miR-210-3p from hypoxic BMSCs promotes TNBC progression
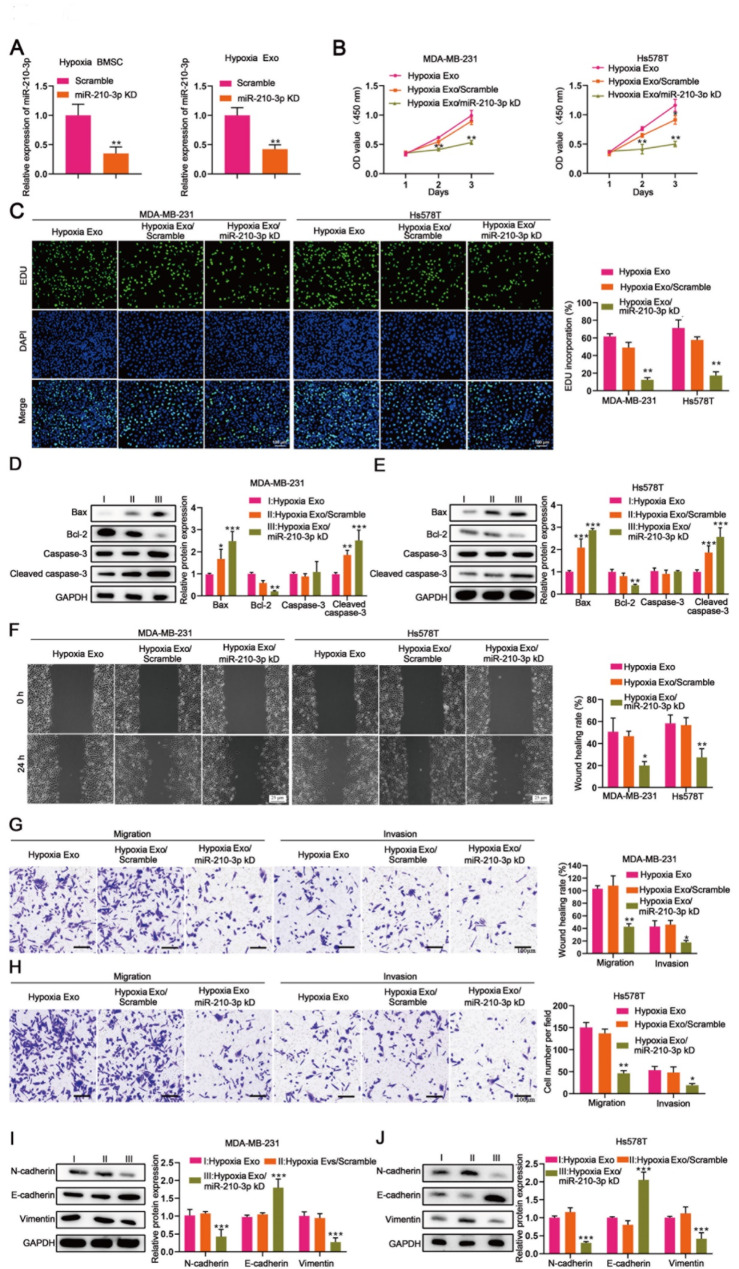
To elucidate the role of exosomal miR-210-3p from hypoxic BMSCs in TNBC progression, we knocked down miR-210-3p in hypoxia BMSCs (Fig. 3A), and co-cultured the exosomes with MDA-MB-231 and Hs578T cells. The knockdown of exosomal miR-210-3p reduced cell viability and proliferation in both MDA-MB-231 and Hs578T cells (Fig. 3B and C), while increasing apoptosis, as evidenced by upregulation of Bax and cleaved caspase-3 and downregulation of Bcl-2 (Fig. 3D and E). Additionally, miR-210-3p-deficient exosomes reduced migration and invasion of MDA-MB-231 and Hs578T compared to scramble control exosomes and hypoxia exosomes (Fig. 3F, G and H). Elevated E-cadherin and decreased N-cadherin and vimentin in these cells treated with miR-210-3p-knockdown exosomes further confirmed the role of exosomal miR-210-3p in promoting EMT in TNBC (Fig. 3I and J).
Fig. 3.
Exosomal miR-210-3p from hypoxic BMSCs promotes TNBC progression. MDA-MB-231 and Hs578T cells were co-cultured with hypoxia exosomes, hypoxia exosomes scramble and hypoxia exosomes miR-210-3p knockdown for 24, 48 and 72 h, respectively [Hypoxia Exo (I) vs. Hypoxia Exo/scramble (II)vs. Hypoxia Exo/miR-210-3p KD (III)]. (A) miR-210-3p expression in hypoxic BMSCs and hypoxic BMSC-derived exosomes transduced with scramble and miR-210-3p KD measured by RT-qPCR. (B) CCK8 assay detected the cell viability. (C) EDU assay detected the cell proliferation. (D and E) The expression of apoptosis-related proteins (Bax、Bcl-2、Caspase-3、Cleaved caspase-3) was measured by western blot. (F) Wound healing assay revealed the cell migration. (G and H) Transwell assay revealed the migration and invasion of MDA-MB-231 and Hs578T cells. (I and J) The expression of epithelial and mesenchymal markers (N-Cadherin、E-Cadherin、Vimentin) was measured by western blot
Deletion of miR-210-3p in hypoxic BMSC-derived exosomes attenuates TNBC in vivo
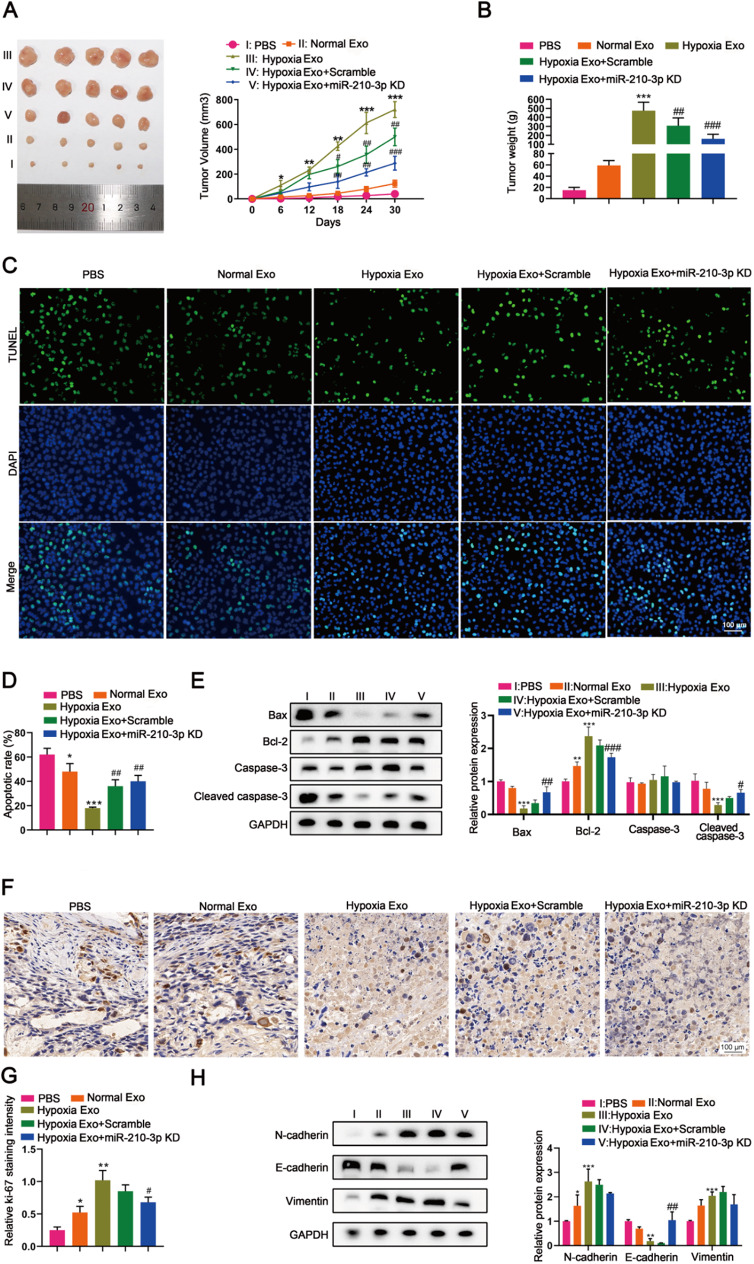
The role of exosomal miR-210-3p derived from hypoxic BMSCs in tumor progression in vivo was further evaluated using tumor xenograft nude mouse models. Mice injected with hypoxic BMSC-derived exosomes exhibited increased tumor volume and tumor weight compared to those receiving normoxic BMSC-derived exosomes (Fig. 4A and B). Notably, mice treated with miR-210-3p-deficient exosomes showed reduced tumor volume and weight compared to those with scramble control exosomes (Fig. 4A and B). TUNEL staining revealed that hypoxic BMSC-derived exosomes inhibited tumor apoptosis (Fig. 4C and D), which was further confirmed by western blot showing decreased Bax and cleaved caspase-3 and increased Bcl-2 expression (Fig. 4E). Hypoxic BMSC-derived exosomes also promoted tumor proliferation evidenced by Ki67 immunostaining (Fig. 4F and G). Importantly, miR-210-3p deletion significantly induced apoptosis and suppressed proliferation of tumors in mice (Fig. 4C, D, E, F and G). Furthermore, the deletion of miR-210-3p in exosomes mitigated tumor EMT promotion induced by hypoxic BMSC-derived exosomes, as indicated by changes in E-cadherin, N-cadherin and vimentin expression (Fig. 4H).
Fig. 4.
Deletion of miR-210-3p in hypoxic BMSC-derived exosomes attenuates TNBC in vivo. Female BALB/c nude mice were injected with PBS, MDA-MB-231 cells treated with normal BMSC-derived exosomes, hypoxic BMSC-derived exosomes, hypoxic BMSC-derived exomes scramble, hypoxic BMSC-derived exomes miR-210-3p KD, respectively [PBS (I) vs. normal Exo (II) vs. Hypoxia Exo (III) vs. Hypoxia Exo/scramble (IV)vs. Hypoxia Exo/miR-210-3p KD (V)]. (A) Tumor volume of BALB/c nude mice. (B) Tumor weight of BALB/c nude mice. (C and D) TUNEL assay was used to detect apoptosis. (E) Apoptosis-related proteins (Bax、Bcl-2、Caspase-3、Cleaved caspase-3) was measured by western blot. (F and G) Evaluation of cell proliferation in tumor tissue using Ki-67 staining (histochemical stain). (H) The expression of epithelial and mesenchymal markers (N-Cadherin、E-Cadherin、Vimentin) was measured by western blot
Exosomal miR-210-3p from hypoxic BMSCs targets NFIX/Wnt signaling in TNBC
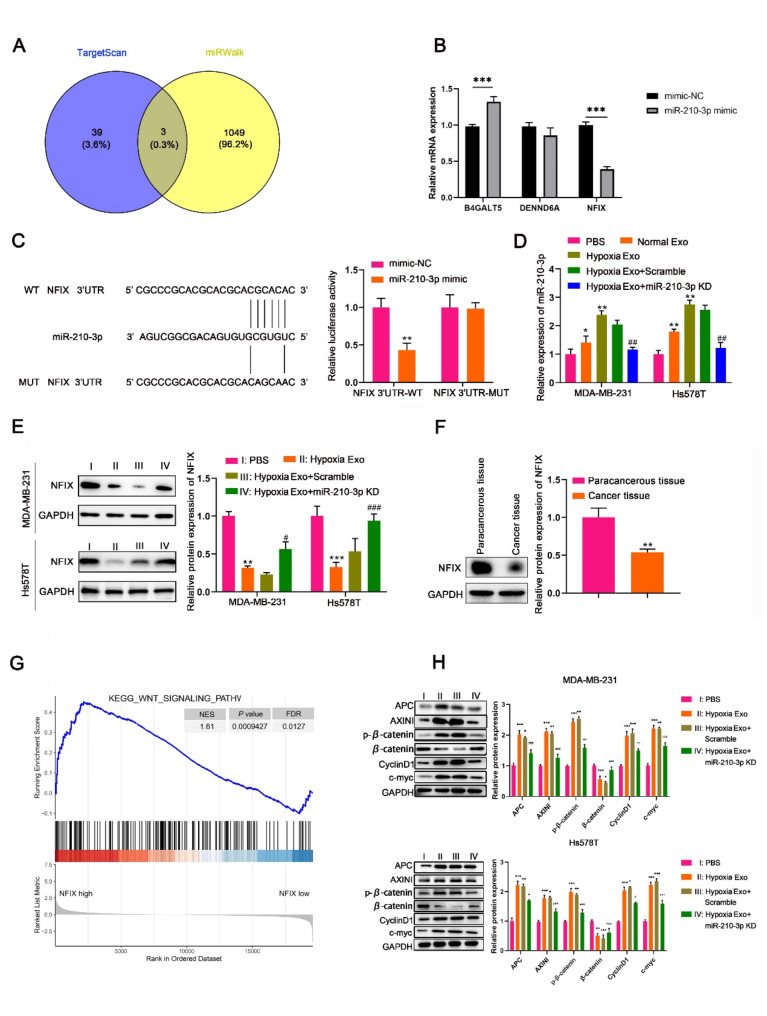
We then utilized TargetScan and miRWalk databases to predict the downstream target genes of miR-210-3p. Three candidate genes (B4GALT5, DENND6A and NFIX) were identified (Fig. 5A). Specifically, NFIX expression was reduced in HEK-293T cells transfected with miR-210-3p mimic (Fig. 5B). Dual luciferase reporter gene assay confirmed that miR-210-3p mimic specifically inhibited luciferase activity in cells with wild-type NFIX 3’-UTR, establishing NFIX as a direct target of miR-210-3p (Fig. 5C). Further western blot analysis demonstrated that NFIX expression was decreased significantly in MDA-MB-231 and Hs578T cells treated with hypoxic BMSC-derived exosomes, while increased when stimulated with exosomes with miR-210-3p knockdown (Fig. 5D and E), indicating the regulation of exosomal miR-210-3p on NFIX expression in TNBC. Furthermore, we investigated NFIX expression in clinical samples from TNBC patients and observed a notable reduction in NFIX expression levels in the cancerous tissues compared to paracancerous tissues (Fig. 5F).
Fig. 5.
Exosomal miR-210-3p from hypoxic BMSCs targets regulates NFIX/Wnt signaling in TNBC. MDA-MB-231 and Hs578T cells were co-cultured with PBS, hypoxia exosomes, hypoxia exosomes scramble and hypoxia exosomes miR-210-3p knockdown, respectively [PBS (I) vs. Hypoxia Exo (II) vs. Hypoxia Exo/scramble (III) vs. Hypoxia Exo/miR-210-3p KD (IV)]. (A) Venn diagram showed the overlap of shared the mRNAs of miR-210-3p in database from TargetScan and miRWalk. (B) Expression of three target mRNAs (B4GALT5, DENND6A and NFIX) in HEK-293T cells transfected with miR-210-3p mimic measured by RT-qPCR. (C) Plasmids containing NFIX-WT or NFIX-MUT with mimic NC or miR-210-3p mimic were co-transfected into MDA-MB-231 cells, and luciferase activity of the reporter vector was assayed in MDA-MB-231 cells. (D) miR-210-3p expression in normal BMSC-derived exosomes, hypoxic BMSC-derived exosomes transduced with scramble and miR-210-3p KD measured by RT-qPCR. (E) Expression levels of NFIX protein in MDA-MB-231 and Hs578T cells was detected by western blot. (F) Expression levels of NFIX protein in the cancerous tissues compared to paracancerous tissues from TNBC patients. (G) GO and KEGG functional annotations and enriched pathways associated with NFIX (Wnt signaling). (H) Western blot analysis of Wnt/β-catenin signaling protein markers (APC、AXINI、p-β-catenin、β-catenin、CyclinD1、c-myc) in MDA-MB-231 and Hs578T cells
To further elucidate the mechanism underlying miR-210-3p/NFIX in TNBC progression, we analyzed the GO functional annotations and KEGG pathways associated with NFIX and investigated 25 potential NFIX-associated signaling pathways in breast cancer. The data revealed a close association between NFIX and Wnt signaling pathway (Fig. 5G). We then assessed whether exosomal miR-210-3p from hypoxic BMSCs modulates Wnt signaling in TNBC. Hypoxic BMSC-derived exosomes increased the expression of APC, AXINI, p-β-catenin, CyclinD1 and c-myc in MDA-MB-231 and Hs578T cells (Fig. 5H). Notably, these protein levels were reduced in cells treated with miR-210-3p-deficient exosomes compared to those treated with scramble control exosomes, highlighting the critical role of exosomal miR-210-3p in activating the Wnt/β-Catenin pathway of TNBC (Fig. 5G and H).
Human exosomal mir-210-3p drives progression of TNBC
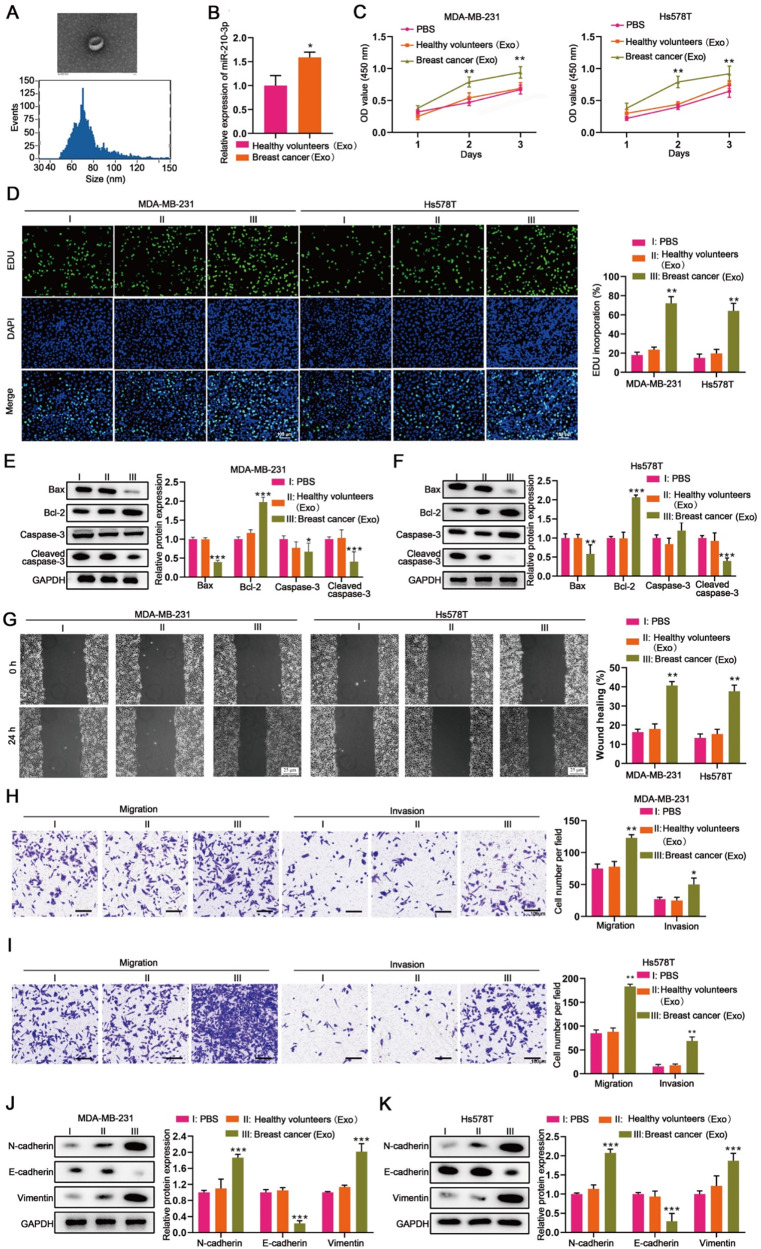
We then isolated exosomes from the serum of TNBC patients and healthy individuals, which were identified by SEM (Fig. 6A). The expression of miR-210-3p was upregulated in exosomes from TNBC patients compared to healthy individuals (Fig. 6B). To further investigate the role of circulating miR-210-3p in TNBC progression, we co-cultured MDA-MB-231 and Hs578T cells with these exosomes. Compared to the cells treated with exosomes from healthy individuals, the cells treated with exosomes from TNBC patients exhibited enhanced cell viability and proliferation (Fig. 6C and D), and reduced apoptosis, indicated by downregulated Bax and cleaved caspase-3 and upregulated Bcl-2 expression (Fig. 6E and F). Moreover, exosomes from TNBC patients promoted migration and invasion of MDA-MB-231 and Hs578T cells (Fig. 6G and I). Decreased E-cadherin and increased N-cadherin and vimentin in these cells treated with exosomes from TNBC patients further confirmed the role of circulating miR-210-3p in promoting EMT in TNBC (Fig. 6J and K).
Fig. 6.
Serum exosomal miR-210-3p from patients promotes TNBC progression. MDA-MB-231 and Hs578T cells were co-cultured with PBS, healthy individual-derived exosomes and patient-derived exosomes for 24, 48 and 72 h, respectively [PBS (I) vs. healthy individuals Exo (II) vs. breast cancer Exo (III)]. (A) Identifying exosomes secreted from TNBC patients’ serum (by TEM and NTA analysis). (B) miR-210-3p expression in serum exosomes from TNBC patients and healthy individuals measured by RT-qPCR. (C) CCK8 assay detected the cell viability. (D) EDU assay detected the cell proliferation. (E and F) The expression of apoptosis-related proteins (Bax、Bcl-2、Caspase-3、Cleaved caspase-3) was measured by western blot. (G) Wound healing assay revealed the cell migration. (H and I) Transwell assay revealed the migration and invasion of MDA-MB-231 and Hs578T cells. (J and K) The expression of epithelial and mesenchymal markers (N-Cadherin、E-Cadherin、Vimentin) was measured by western blot
Discussion
TNBC is the most challenging subtype of breast cancer, characterized by its heterogeneity and the lack of effective therapies beyond chemotherapy, leading to poor patient outcomes [8]. Hypoxia alters the tumor microenvironment, subsequently regulating the critical metabolic, developmental, and survival pathways in TNBC [13, 40]. Increasing evidence highlights the significant role of EVs, particularly exosomes, as key mediators of intercellular communication in the hypoxic TME [20]. The miRNA content within exosomes has been shown to influence various aspects of breast cancer biology, including tumor growth, angiogenesis, invasion, migration, metastasis, metabolism and drug resistance [29, 41–44]. Unlike miRNAs circulating in the blood, exosomal miRNAs are shielded by the exosomal membrane, protecting them from external interference [45, 46]. Therefore, miRNAs within exosomes demonstrate enhanced stability and accuracy, making them more suitable as novel biomarkers for early tumor diagnosis, as well as for longitudinal monitoring of recurrence, metastasis, and prognosis [47, 48].
A multicenter prospective cohort study identified three miRNAs, namely miR-421, miR-128-1 and miR-128-2, in peripheral blood vesicles as independent factors with poor prognosis in breast cancer patients [49]. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d have been shown to promote breast cancer stemness and lead to chemoresistance through activation of the zeste homolog 2 (EZH2)/ signal transducer and activator of transcription 3(STAT3) signaling pathway. Additionally, combining chemotherapeutic agents combined with an EZH2 inhibitor effectively reversed exosome-induced resistance in a nude mouse tumor xenograft model [50]. In our study, we screened 148 hypoxia-regulated miRNAs within exosomes associated with breast cancer prognosis from the ExoBCD database (https://exobcd.liumwei.org/) [35]. Among these, miR-210-3p was found to be highly expressed in breast cancer tissues.
miR-210-3p is the most prominent hypoxia-regulated miRNA, with elevated expression levels observed in various malignancies, including breast cancer [51–55]. Previous studies have demonstrated that miR-210-3p inhibits electron transport chain complexes and promotes cell migration and angiogenesis, thereby contributing to tumor progression [56]. Du et al. identified miR-210-3p as a key regulator of metabolic reprogramming in TNBC glycolysis. It targets GPD1L to stabilize HIF-1α and mediates the inhibition of p53 activity through CYGB, ultimately regulating the downstream glycolytic genes of HIF-1α and p53 to promote aerobic glycolysis [51]. Additionally, exosomal miR-210 from hypoxic breast cancer alters the expression of genes involved in vascular remodeling within the TME, including Ephrin A3 and PTP1B, thereby promoting angiogenesis [57]. In our study, we observed elevated levels of exosomal miR-210-3p in the serum of TNBC patients and further demonstrated its role in driving the progression of TNBC.
In addition, we found that the expression of miR-210-3p in BMSC-derived exosomes was significantly upregulated under hypoxic conditions. Inhibition of miR-210-3p expression in hypoxic BMSC-derived exosomes impaired the proliferation, migration, and invasive ability of TNBC cells and promoted the apoptosis of TNBC. BMSCs, multipotent stromal cells within the TME, have been shown to play an essential role in cancer progression and metastasis, partly due to the paracrine factors they secrete [23]. Exosomes released by BMSCs are considered an important mechanism through which these cells exert their effects on tumor cells. The role of BMSC-derived exosomal miRNAs in cancer progression has been shown to be bidirectional [58]. For example, miR-208a in BMSC-derived exosomes promotes osteosarcoma cell proliferation, migration and invasion by downregulating PDCD4 and activating the ERK1/2 signaling pathway [59], while miR-16-5p suppresses breast cancer progression by inhibiting NF-κB signaling and decreasing EPHA1 expression [28]. Hypoxia influences exosome release, cargo composition, and biofunction. miR-301a-3p, which is upregulated in gastric cancer-derived exosomes under hypoxia, was demonstrated to enhance HIF-1α stability by targeting PHD3, promoting invasiveness and metastasis of gastric cancer [60]. Hypoxic BMSC-derived exosomal miRNAs, including miR-193a-3p, miR-210-3p and miR-5100, promoted the metastasis of lung cancer cells via STAT3-induced EMT [61].
To elucidate the mechanism by which BMSC-derived exosomal miR-210-3p influences TNBC, we employed bioinformatics analysis and dual-luciferase reporter assays, which confirmed that miR-210-3p targets NFIX and downregulates its expression in TNBC cells. NFIX, a member of the nuclear factor I (NFI) family of transcription factors, plays an important role in the development of central nervous and muscle development system [62]. It has been shown to be dysregulated in various cancers, frequently promoting pro-tumorigenic functions, such as leading to proliferation, differentiation, and migration [63]. While NFIX downregulation has been shown to reduce proliferation and cell viability in lung cancer [64], its role can differ depending on the cancer type, as it has been found to promote proliferation in both endometrial cancer [65] and glioblastoma [66].
Accumulating evidence underscores the critical role of Wnt signaling in various aspects of breast cancer, including proliferation, metastasis, immune microenvironment regulation, stem cell maintenance, treatment resistance, and phenotype modulation [67]. Given its central role in tumorigenesis, several inhibitors targeting components of the Wnt signaling pathway have been developed as potential anti-tumor therapies [68, 69]. In our study, we identified a close association between NFIX and Wnt signaling pathway. Furthermore, we demonstrated that hypoxic BMSC-derived miR-210-3p activates the NFIX-Wnt/β-catenin signaling axis, thereby further promoting progression of TNBC (Figure S3).
Although this study reveals the potential role of hypoxic BMSC-derived exosomes and their miR-210-3p in the progression of TNBC, several limitations remain. Firstly, while we focused on key tumor progression processes such as cell proliferation, apoptosis, migration, and invasion, the critical process of angiogenesis, essential for tumor growth, was not thoroughly investigated. Therefore, future studies should expand to evaluate the specific impact of hypoxic BMSC-derived exosomes on TNBC angiogenesis. Secondly, miR-210-3p promotes TNBC progression by activating the NFIX-Wnt/β-catenin signaling axis, yet the precise interaction and activation mechanisms between miR-210-3p and this pathway are not fully elucidated, requiring further investigation, particularly at the molecular level. Moreover, the repurposing of existing drugs presents new possibilities for cancer treatment [70] and it is highly worthwhile to investigate whether these drugs can modulate miR-210-3p-mediated signaling pathways. Future research should focus on assessing the impact of existing anti-cancer drugs on miR-210-3p-related signaling pathways and explore their potential applications in TNBC therapy.
Conclusion
Our research demonstrated that hypoxic BMSC-derived exosomal miR-210-3p promotes progression of TNBC by activating the Wnt/β-catenin pathway via down-regulating NFIX. In addition, the miR-210-3p derived from patients’ serum exosomes also plays a significant role in facilitating the progression of TNBC. These observations reveal exosomal miR-210-3p and NFIX as promising non-invasive biomarkers and therapeutic targets for TNBC progression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- TNBC
Triple-negative breast cancer
- TME
Tumor microenvironment
- MSCs
Mesenchymal stem cells
- BMSCs
Bone marrow-derived mesenchymal stem cells
- EVs
Vesicles
- SEM
Scanning electron microscopy
- NTA
Nanoparticle tracking analysis
- RT-qPCR
Quantitative real-time PCR
- EDU
5-Ethynyl-20-deoxyuridine
- BRCA
Breast invasion carcinoma
- EMT
Eepithelial mesenchymal transition
- EZH2
Zeste homolog 2
- STAT3
Signal transducer and activator of transcription 3
Authors contributions
Yi Zheng, Guochao Mao and Shai Lin is responsible for data analysis of the database. Meng Wang, Qian Hao and Zhen Zhai completes cell and animal experiments. Huafeng Kang and Xiaobin Ma provided patients. Baobao Liang collected the samples. Zhijun Dai, Huafeng Kang and Xiaobin Ma guided experiments. Meng Wang, Yi Zheng and Shai Lin analyzed and interpreted the data. Meng Wang finished manuscript editing. Xiaobin Ma reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by International Science and Technology Cooperation Program Project of Shaanxi Province, China (Grant/Award Number: 2023-GHZD-24) and The Freedom to Explore Program of the Second Affiliated Hospital of Xi’an Jiaotong University (Grant/Award Number: 2020YJ(ZYTS)625).
Data availability
The RNA-sequence and miRNA datasets of breast invasion carcinoma (BRCA) were downloaded from the UCSC Xena website (https://xena.ucsc.edu/, derived from the TCGA). The exosomal miRNAs related overall survival of BRCA were obtained from ExoBCD database (https://exobcd.liumwei.org/). The target mRNAs of the candidate exosomal miRNAs were predicted by using TargetScan (https://www.targetscan.org/vert_80/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/). All data generated or analyzed during this study are contained in this paper or supplementary materials. Processed data are available from the corresponding author upon reasonable request.
Declarations
Ethics committee approval and patient consent
The study protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiao tong University Shaanxi Province (Xi’an, China) (approval number: 2024YS128). The study purpose and experimental procedures were conveyed to all participants and informed consent was obtained. All experiments were approved by the Health Science Center of Xi’an Jiao tong University Approval for Research Involving Animals (approval number: XJTUAE2024-184).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huafeng Kang, Email: kanghuafeng1973@126.com.
Xiaobin Ma, Email: binbinmxb@xjtu.edu.cn.
References
- 1.Newman L. Oncologic anthropology: global variations in breast cancer risk, biology, and outcome. J Surg Oncol. 2023;128(6):959–66. 10.1002/jso.27459. [DOI] [PubMed] [Google Scholar]
- 2.Katsura C, Ogunmwonyi I, Kankam HK, Saha S. Breast cancer: presentation, investigation and management. British journal of hospital medicine (London, England: 2005). 2022;83(2):1–7. 10.12968/hmed.2021.0459 [DOI] [PubMed]
- 3.Zheng RS, Chen R, Han BF, Wang SM, Li L, Sun KX et al. [Cancer incidence and mortality in China, 2022]. Zhonghua Zhong Liu Za Zhi [Chinese journal of oncology]. 2024;46(3):221–31. 10.3760/cma.j.cn112152-20240119-00035 [DOI] [PubMed]
- 4.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. Cancer J Clin. 2023;73(1):17–48. 10.3322/caac.21763. [DOI] [PubMed]
- 5.Mahmoud MM, Sanad EF, Elshimy RAA, Hamdy NM. Competitive endogenous role of the LINC00511/miR-185-3p Axis and miR-301a-3p from Liquid Biopsy as molecular markers for breast Cancer diagnosis. Front Oncol. 2021;11:749753. 10.3389/fonc.2021.749753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang YF, Huang KC, Chen HY, Hamdy NM, Huang TC, Chang HY, et al. Hinokitiol inhibits breast Cancer cells in Vitro Stemness-Progression and Self-Renewal with apoptosis and autophagy modulation via the CD44/Nanog/SOX2/Oct4 pathway. Int J Mol Sci. 2024;25(7). 10.3390/ijms25073904. [DOI] [PMC free article] [PubMed]
- 7.Leon-Ferre RA, Goetz MP. Advances in systemic therapies for triple negative breast cancer. BMJ (Clinical Res ed). 2023;381:e071674. 10.1136/bmj-2022-071674. [DOI] [PubMed] [Google Scholar]
- 8.Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat Reviews Clin Oncol. 2022;19(2):91–113. 10.1038/s41571-021-00565-2. [DOI] [PubMed] [Google Scholar]
- 9.Kudelova E, Smolar M, Holubekova V, Hornakova A, Dvorska D, Lucansky V, et al. Genetic heterogeneity, Tumor Microenvironment and Immunotherapy in Triple-negative breast Cancer. Int J Mol Sci. 2022;23(23). 10.3390/ijms232314937. [DOI] [PMC free article] [PubMed]
- 10.Srivastava N, Usmani SS, Subbarayan R, Saini R, Pandey PK. Hypoxia: syndicating triple negative breast cancer against various therapeutic regimens. Front Oncol. 2023;13:1199105. 10.3389/fonc.2023.1199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S, Shukla N, Singh SS, Kushwaha S, Shrivastava R. Mechanism of interaction between autophagy and apoptosis in cancer. Apoptosis: Int J Program cell Death. 2021;26(9–10):512–33. 10.1007/s10495-021-01687-9. [DOI] [PubMed] [Google Scholar]
- 12.Lin YT, Wu KJ. Epigenetic regulation of epithelial-mesenchymal transition: focusing on hypoxia and TGF-β signaling. J Biomed Sci. 2020;27(1):39. 10.1186/s12929-020-00632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhise K, Gavande NS, Iyer AK. Leveraging hypoxia in triple-negative breast cancer as a promising treatment strategy. Drug Discovery Today. 2023;28(11):103761. 10.1016/j.drudis.2023.103761. [DOI] [PubMed] [Google Scholar]
- 14.Elanany MM, Mostafa D, Hamdy NM. Remodeled tumor immune microenvironment (TIME) parade via natural killer cells reprogramming in breast cancer. Life Sci. 2023;330:121997. 10.1016/j.lfs.2023.121997. [DOI] [PubMed] [Google Scholar]
- 15.Lakshmi S, Hughes TA, Priya S. Exosomes and exosomal RNAs in breast cancer: A status update. European journal of cancer (Oxford, England: 1990). 2021;144:252 – 68. 10.1016/j.ejca.2020.11.033 [DOI] [PubMed]
- 16.Guo W, Qiao T, Dong B, Li T, Liu Q, Xu X. The Effect of Hypoxia-Induced exosomes on Anti-tumor Immunity and its implication for Immunotherapy. Front Immunol. 2022;13:915985. 10.3389/fimmu.2022.915985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai B, Clark AM, Wells A. Mesenchymal stem cell-secreted exosomes and Soluble signals regulate breast Cancer metastatic dormancy: current Progress and Future Outlook. Int J Mol Sci. 2024;25(13). 10.3390/ijms25137133. [DOI] [PMC free article] [PubMed]
- 18.Shao X, Hua S, Feng T, Ocansey DKW, Yin L. Hypoxia-regulated tumor-derived exosomes and Tumor Progression: a focus on Immune Evasion. Int J Mol Sci. 2022;23(19). 10.3390/ijms231911789. [DOI] [PMC free article] [PubMed]
- 19.Möller A, Lobb RJ. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer. 2020;20(12):697–709. 10.1038/s41568-020-00299-w. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, Deep G. Hypoxia in tumor microenvironment regulates exosome biogenesis: molecular mechanisms and translational opportunities. Cancer Lett. 2020;479:23–30. 10.1016/j.canlet.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 21.He G, Peng X, Wei S, Yang S, Li X, Huang M, et al. Exosomes in the hypoxic TME: from release, uptake and biofunctions to clinical applications. Mol Cancer. 2022;21(1):19. 10.1186/s12943-021-01440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buschhaus JM, Rajendran S, Chen S, Wharram BL, Bevoor AS, Cutter AC, et al. Bone marrow mesenchymal stem cells induce metabolic plasticity in Estrogen receptor-positive breast Cancer. Mol cancer Research: MCR. 2023;21(5):458–71. 10.1158/1541-7786.Mcr-22-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi C, Shi H, Fan M, Chen W, Yao H, Jiang C, et al. Microvesicles from bone marrow-derived mesenchymal stem cells promote Helicobacter pylori-associated gastric cancer progression by transferring thrombospondin-2. Cell Communication Signaling: CCS. 2023;21(1):274. 10.1186/s12964-023-01127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong L, You S, Zhang Q, Osuka S, Devi NS, Kaluz S, et al. Arylsulfonamide 64B inhibits Hypoxia/HIF-Induced expression of c-Met and CXCR4 and reduces primary Tumor Growth and Metastasis of Uveal Melanoma. Clin cancer Research: Official J Am Association Cancer Res. 2019;25(7):2206–18. 10.1158/1078-0432.Ccr-18-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Li L, Rong Y, Qian D, Chen J, Zhou Z, et al. Hypoxic mesenchymal stem cell-derived exosomes promote bone fracture healing by the transfer of miR-126. Acta Biomater. 2020;103:196–212. 10.1016/j.actbio.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 26.Lin Z, Wu Y, Xu Y, Li G, Li Z, Liu T. Mesenchymal stem cell-derived exosomes in cancer therapy resistance: recent advances and therapeutic potential. Mol Cancer. 2022;21(1):179. 10.1186/s12943-022-01650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding B, Lou W, Fan W, Pan J. Exosomal miR-374c-5p derived from mesenchymal stem cells suppresses epithelial-mesenchymal transition of hepatocellular carcinoma via the LIMK1-Wnt/β-catenin axis. Environ Toxicol. 2023;38(5):1038–52. 10.1002/tox.23746. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Lai X, Yue Q, Cao F, Zhang Y, Sun Y, et al. Bone marrow mesenchymal stem cells-derived exosomal microRNA-16-5p restrains epithelial-mesenchymal transition in breast cancer cells via EPHA1/NF-κB signaling axis. Genomics. 2022;114(3):110341. 10.1016/j.ygeno.2022.110341. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Jin LJ, Zhang XY. Exosomal miRNA-205 promotes breast cancer chemoresistance and tumorigenesis through E2F1. Aging. 2021;13(14):18498–514. 10.18632/aging.203298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anilkumar KV, Rema LP, John MC, Vanesa John T, George A. miRNAs in the prognosis of triple-negative breast cancer: a review. Life Sci. 2023;333:122183. 10.1016/j.lfs.2023.122183. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee S, Dhar R, Jonnalagadda S, Gorai S, Nag S, Kar R et al. Exosomal miRNAs and breast cancer: a complex theranostics interlink with clinical significance. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2023;28(6):502–18. 10.1080/1354750x.2023.2229537 [DOI] [PubMed]
- 32.Hong F, Li N, Feng Z, Zheng Y, Zhu C, Zhang F. Exosomal microRNAs as novel diagnostic biomarkers in breast cancer: a systematic evaluation and meta-analysis. Asian J Surg. 2023;46(11):4727–36. 10.1016/j.asjsur.2023.05.115. [DOI] [PubMed] [Google Scholar]
- 33.Singh T, Kaushik M, Mishra LC, Behl C, Singh V, Tuli HS. Exosomal miRNAs as novel avenues for breast cancer treatment. Front Genet. 2023;14:1134779. 10.3389/fgene.2023.1134779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang YT, Huang YY, Zheng L, Qin SH, Xu XP, An TX, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med. 2017;40(3):834–44. 10.3892/ijmm.2017.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Chai Z, Pan G, Hao Y, Li B, Ye T, et al. ExoBCD: a comprehensive database for exosomal biomarker discovery in breast cancer. Brief Bioinform. 2021;22(3). 10.1093/bib/bbaa088. [DOI] [PubMed]
- 36.Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. Jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15(1):293. 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4. 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed]
- 38.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by walking the genes of three genomes. J Biomed Inform. 2011;44(5):839–47. 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deepak KGK, Vempati R, Nagaraju GP, Dasari VR, Rao SN. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res. 2020;153:104683. 10.1016/j.phrs.2020.104683. [DOI] [PubMed] [Google Scholar]
- 41.Samuels M, Jones W, Towler B, Turner C, Robinson S, Giamas G. The role of non-coding RNAs in extracellular vesicles in breast cancer and their diagnostic implications. Oncogene. 2023;42(41):3017–34. 10.1038/s41388-023-02827-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang M, Zhang W, Zhang R, Liu P, Ye Y, Yu W, et al. Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene. 2020;39(24):4681–94. 10.1038/s41388-020-1322-4. [DOI] [PubMed] [Google Scholar]
- 43.Zhao S, Pan T, Deng J, Cao L, Vicencio JM, Liu J, et al. Exosomal transfer of miR-181b-5p confers senescence-mediated doxorubicin resistance via modulating BCLAF1 in breast cancer. Br J Cancer. 2023;128(4):665–77. 10.1038/s41416-022-02077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park EJ, Jung HJ, Choi HJ, Jang HJ, Park HJ, Nejsum LN, et al. Exosomes co-expressing AQP5-targeting miRNAs and IL-4 receptor-binding peptide inhibit the migration of human breast cancer cells. FASEB Journal: Official Publication Federation Am Soc Experimental Biology. 2020;34(2):3379–98. 10.1096/fj.201902434R. [DOI] [PubMed] [Google Scholar]
- 45.Qiao P, Du H, Guo X, Yu M, Zhang C, Shi Y. Serum exosomal miR-200c is a potential diagnostic biomarker for breast cancer. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2024:1–8. 10.1080/1354750x.2024.2406520 [DOI] [PubMed]
- 46.Henning RJ. Cardiovascular exosomes and MicroRNAs in Cardiovascular Physiology and Pathophysiology. J Cardiovasc Transl Res. 2021;14(2):195–212. 10.1007/s12265-020-10040-5. [DOI] [PubMed] [Google Scholar]
- 47.Shu L, Li X, Liu Z, Li K, Shi A, Tang Y, et al. Bile exosomal miR-182/183-5p increases cholangiocarcinoma stemness and progression by targeting HPGD and increasing PGE2 generation. Hepatology (Baltimore MD). 2024;79(2):307–22. 10.1097/hep.0000000000000437. [DOI] [PubMed] [Google Scholar]
- 48.Davidson CL, Vengoji R, Jain M, Batra SK, Shonka N. Biological, diagnostic and therapeutic implications of exosomes in glioma. Cancer Lett. 2024;582:216592. 10.1016/j.canlet.2023.216592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao S, Hu T, Liu J, Su J, Sun J, Ming Y, et al. Genomic instability-derived plasma extracellular vesicle-microRNA signature as a minimally invasive predictor of risk and unfavorable prognosis in breast cancer. J Nanobiotechnol. 2021;19(1):22. 10.1186/s12951-020-00767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Q, Zhao S, Shi Z, Cao L, Liu J, Pan T, et al. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J Experimental Clin cancer Research: CR. 2021;40(1):120. 10.1186/s13046-021-01901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Y, Wei N, Ma R, Jiang S, Song D. A mir-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis. 2020;11(9):731. 10.1038/s41419-020-02952-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powell BH, Turchinovich A, Wang Y, Gololobova O, Buschmann D, Zeiger MA, et al. miR-210 expression is strongly Hypoxia-Induced in anaplastic thyroid Cancer cell lines and is Associated with Extracellular vesicles and Argonaute-2. Int J Mol Sci. 2023;24(5). 10.3390/ijms24054507. [DOI] [PMC free article] [PubMed]
- 53.Arora L, Patra D, Roy S, Nanda S, Singh N, Verma AK, et al. Hypoxia-induced mir-210-3p expression in lung adenocarcinoma potentiates tumor development by regulating CCL2 mediated monocyte infiltration. Mol Oncol. 2024;18(5):1278–300. 10.1002/1878-0261.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Huang X, He Z, Xiong Y, Fang Q. miRNA-210-3p regulates trophoblast proliferation and invasiveness through fibroblast growth factor 1 in selective intrauterine growth restriction. J Cell Mol Med. 2019;23(6):4422–33. 10.1111/jcmm.14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gamal-Eldeen AM, Raafat BM, Alrehaili AA, El-Daly SM, Hawsawi N, Banjer HJ, et al. Anti-hypoxic effect of Polysaccharide Extract of Brown Seaweed Sargassum dentifolium in Tongue squamous cell carcinoma. Front Nutr. 2022;9:854780. 10.3389/fnut.2022.854780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shao MX, Qu AZ, Wang YQ, Zhong YY. Expression level of miRNA-210-3p in cervical cancer and its prognostic potential. Eur Rev Med Pharmacol Sci. 2020;24(12):6583–8. 10.26355/eurrev_202006_21643. [DOI] [PubMed] [Google Scholar]
- 57.Jung KO, Youn H, Lee CH, Kang KW, Chung JK. Visualization of exosome-mediated miR-210 transfer from hypoxic tumor cells. Oncotarget. 2017;8(6):9899–910. 10.18632/oncotarget.14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao R, Chen X, Song H, Bie Q, Zhang B. Dual role of MSC-Derived exosomes in Tumor Development. Stem Cells Int. 2020;2020:8844730. 10.1155/2020/8844730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin F, Tang H, Zhang Y, Zhang Z, Huang P, Zhu J. Bone marrow-derived mesenchymal stem cell-derived exosomal microRNA-208a promotes osteosarcoma cell proliferation, migration, and invasion. J Cell Physiol. 2020;235(5):4734–45. 10.1002/jcp.29351. [DOI] [PubMed] [Google Scholar]
- 60.Xia X, Wang S, Ni B, Xing S, Cao H, Zhang Z, et al. Hypoxic gastric cancer-derived exosomes promote progression and metastasis via MiR-301a-3p/PHD3/HIF-1α positive feedback loop. Oncogene. 2020;39(39):6231–44. 10.1038/s41388-020-01425-6. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. 2019;18(1):40. 10.1186/s12943-019-0959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davila RA, Spiller C, Harkins D, Harvey T, Jordan PW, Gronostajski RM, et al. Deletion of NFIX results in defective progression through meiosis within the mouse testis†. Biol Reprod. 2022;106(6):1191–205. 10.1093/biolre/ioac049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ribeiro V, Martins SG, Lopes AS, Thorsteinsdóttir S, Zilhão R, Carlos AR. NFIXing Cancer: the role of NFIX in oxidative stress response and cell fate. Int J Mol Sci. 2023;24(5). 10.3390/ijms24054293. [DOI] [PMC free article] [PubMed]
- 64.Rahman NIA, Abdul Murad NA, Mollah MM, Jamal R, Harun R. NFIX as a Master Regulator for Lung Cancer Progression. Front Pharmacol. 2017;8:540. 10.3389/fphar.2017.00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Yang Z, Yao R, Li Y, Liu Z, Chen X, et al. miR-210 promotes progression of endometrial carcinoma by regulating the expression of NFIX. Int J Clin Exp Pathol. 2018;11(11):5213–22. [PMC free article] [PubMed] [Google Scholar]
- 66.Ghadami E, Gorji A, Pour-Rashidi A, Noorbakhsh F, Kabuli M, Razipour M, et al. CircZNF609 and circNFIX as possible regulators of glioblastoma pathogenesis via miR-145-5p/EGFR axis. Sci Rep. 2024;14(1):13551. 10.1038/s41598-024-63827-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu X, Zhang M, Xu F, Jiang S. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Mol Cancer. 2020;19(1):165. 10.1186/s12943-020-01276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13(1):165. 10.1186/s13045-020-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatterjee A, Paul S, Bisht B, Bhattacharya S, Sivasubramaniam S, Paul MK. Advances in targeting the WNT/β-catenin signaling pathway in cancer. Drug Discovery Today. 2022;27(1):82–101. 10.1016/j.drudis.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 70.Hamdy NM, Suwailem SM, El-Mesallamy HO. Influence of vitamin E supplementation on endothelial complications in type 2 diabetes mellitus patients who underwent coronary artery bypass graft. J Diabetes Complicat. 2009;23(3):167–73. 10.1016/j.jdiacomp.2007.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequence and miRNA datasets of breast invasion carcinoma (BRCA) were downloaded from the UCSC Xena website (https://xena.ucsc.edu/, derived from the TCGA). The exosomal miRNAs related overall survival of BRCA were obtained from ExoBCD database (https://exobcd.liumwei.org/). The target mRNAs of the candidate exosomal miRNAs were predicted by using TargetScan (https://www.targetscan.org/vert_80/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/). All data generated or analyzed during this study are contained in this paper or supplementary materials. Processed data are available from the corresponding author upon reasonable request.