Abstract
In addition to its role in energy storage, adipose tissue also accumulates cholesterol. Concentrations of cholesterol and triglycerides are strongly correlated in the adipocyte, but little is known about mechanisms regulating cholesterol metabolism in fat cells. Here we report that antidiabetic thiazolidinediones (TZDs) and other ligands for the nuclear receptor PPARγ dramatically upregulate oxidized LDL receptor 1 (OLR1) in adipocytes by facilitating the exchange of coactivators for corepressors on the OLR1 gene in cultured mouse adipocytes. TZDs markedly stimulate the uptake of oxidized LDL (oxLDL) into adipocytes, and this requires OLR1. Increased OLR1 expression, resulting either from TZD treatment or adenoviral gene delivery, significantly augments adipocyte cholesterol content and enhances fatty acid uptake. OLR1 expression in white adipose tissue is increased in obesity and is further induced by PPARγ ligand treatment in vivo. Serum oxLDL levels are decreased in both lean and obese diabetic animals treated with TZDs. These data identify OLR1 as a novel PPARγ target gene in adipocytes. While the physiological role of adipose tissue in cholesterol and oxLDL metabolism remains to be established, the induction of OLR1 is a potential means by which PPARγ ligands regulate lipid metabolism and insulin sensitivity in adipocytes.
Introduction
The adipocyte is the major site of fatty acid storage in the body and plays a critical role in maintaining normal glucose and lipid homeostasis. In a healthy person, excess fat is stored as triglycerides in the adipose tissue, and fatty acids are released into the bloodstream only in response to an increased energy requirement, for example, during fasting. If the capacity of the adipocyte to store lipids is exceeded, it can no longer regulate the release of FFAs into the circulation, which ultimately leads to the abnormal accumulation of lipid in nonadipose depots. A buildup of triglycerides in the liver, pancreatic islets, and the muscle is thought to lead to metabolic dysregulation of these tissues (1); in particular, increased plasma FFA levels and elevated intramyocellular lipids are highly correlated with insulin resistance (2, 3).
Obesity can be viewed as a state of long-term lipid disequilibrium that is marked by massive adipocyte hypertrophy and is a major risk factor for developing insulin resistance and type 2 diabetes. When compared with small fat cells from lean controls, enlarged adipocytes isolated from obese animals or humans demonstrate a decreased ability to store triglycerides (4), increased insulin resistance (5), and increased secretion of leptin and TNF-α (6). Interestingly, adipose tissue from ob/ob mice also exhibits an increase in cholesterol biosynthesis (7), and hypertrophied adipocytes from 2 obese rodent models showed elevated mRNA levels of SREBP-2, 3-hydroxy-3-methylglutaryl-CoA (HMG CoA) reductase, and the LDL receptor (8, 9), which suggests that these cells are relatively cholesterol deficient. Adipocytes normally contain a significant amount of free cholesterol and express a number of cholesterol receptors at the plasma membrane, such as scavenger receptor–B1 (SR-B1) and CD36 (10). There is evidence to suggest that, as triglyceride storage increases, cholesterol is redistributed from the plasma membrane to the surface of the lipid droplet (11, 12) and adipocyte cholesterol levels increase in proportion to the triglyceride content (13, 14). Numerous studies have demonstrated that the perturbation of cholesterol levels in adipocytes alters metabolic activity. For example, fatty acid uptake is markedly reduced in cholesterol-depleted 3T3-L1 adipocytes (15). Furthermore, sterol depletion of 3T3-L1 adipocytes leads to an inhibition of insulin-stimulated glucose oxidation, upregulation of TNF-α and IL-6 mRNA, and decreased expression of glucose transporter 4 (GLUT4) (9). Taken together, these data suggest an important role for cholesterol in the ability of the adipocyte to efficiently respond to insulin and to properly metabolize fatty acids and glucose.
The nuclear receptor peroxisome PPARγ is the key transcriptional regulator of adipogenesis and directly activates many genes involved in adipocyte lipid storage (16). PPARγ is expressed at its highest levels in white adipose tissue and is required for adipocyte differentiation (17–20). It binds DNA as an obligate heterodimer with retinoid X receptor α (RXRα) to a peroxisome proliferator–activated response element (PPRE), consisting of 2 direct repeats of the consensus nuclear receptor half-site separated by 1 base pair (17). This motif, known as a direct repeat 1 (DR-1) element, is found in the promoters of many genes involved in lipid storage, such as the fatty acid binding protein aP2 and the cholesterol and fatty acid transporter FATP/CD36 (4). Upon ligand binding to PPARγ, gene expression is activated by the release of the corepressors nuclear receptor corepressor (N-CoR) (21) and silencing mediator for retinoid and thyroid receptors (SMRT) (22) and the recruitment of coactivators such as the p160/steroid receptor coactivator (p160/SRC) family, the mediator complex including PPARγ binding protein (PBP), and histone acetyltransferases such as CREB binding protein (CBP) and p300 (23).
The antidiabetic drugs thiazolidinediones (TZDs) were found to be high-affinity ligands for PPARγ (24). Because the in vitro binding affinity of different TZDs to PPARγ corresponds to their antidiabetic activity in vivo (25, 26), and as PPARγ is expressed predominantly in white fat, it is thought that TZDs exert many of their therapeutic effects by activating PPARγ in the adipose tissue; however, the mechanism by which this occurs is unclear. While originally discovered because of their potent insulin-sensitizing and glucose-lowering activity, TZDs were also found to stimulate adipogenesis by upregulating many of the PPARγ target genes involved in fatty acid metabolism and storage (27). In fact, numerous studies in rodent models and in humans have shown that TZD treatment causes weight gain (28–30). Thus, a paradox emerged: how do TZDs improve insulin sensitivity and glucose metabolism while also causing increased adiposity, a state generally associated with insulin resistance?
PPARγ appears to regulate 2 distinct yet overlapping gene programs in the adipocyte: the adipogenic and the antidiabetic. Previous work has demonstrated that the effects of TZD treatment are not equivalent to those of PPARγ activation in adipocytes (31). For example, resistin is upregulated during adipogenesis but downregulated by rosiglitazone (32), while glycerol kinase is present only at low levels in mature adipocytes but is strongly induced by rosiglitazone (33). Both of these genes have been shown to play a role in whole-body glucose and lipid metabolism (34–36), which suggests that the identification and characterization of genes that are differentially regulated during adipogenesis versus TZD treatment could help to explain how TZDs work. Here we report that oxidized low-density lipoprotein receptor 1 (OLR1) is present at very low levels in the mature fat cell but dramatically upregulated only upon treatment of mouse adipocytes with PPARγ ligand. OLR1 is the major scavenger receptor for oxidized LDL (oxLDL) in endothelial cells (37), but its expression in adipocytes has never been characterized. We show that OLR1 is a direct PPARγ target gene and that its transcriptional regulation is controlled by 4 distinct PPREs that are differentially bound by corepressors and coactivators in the presence of rosiglitazone. Intriguingly, we also demonstrate that PPARγ ligands increase oxLDL uptake and total cholesterol levels in adipocytes and that OLR1 plays a significant role in mediating these effects. In vivo studies show that OLR1 is also induced by TZD treatment in the white adipose tissue of both lean and obese animal models and that serum oxLDL levels are reduced by PPARγ ligands. Increasing evidence suggests that cholesterol balance in the fat cell is an important regulator of metabolic activity and triglyceride storage capability. Although the physiological role of adipose tissue in cholesterol and oxLDL metabolism remains to be established, these data suggest a novel role for PPARγ, antidiabetic TZD ligands, and oxLDL in the control of adipocyte cholesterol content through the induction of OLR1.
Results
OLR1 is a novel gene regulated by PPAR γligands in 3T3-L1 adipocytes.
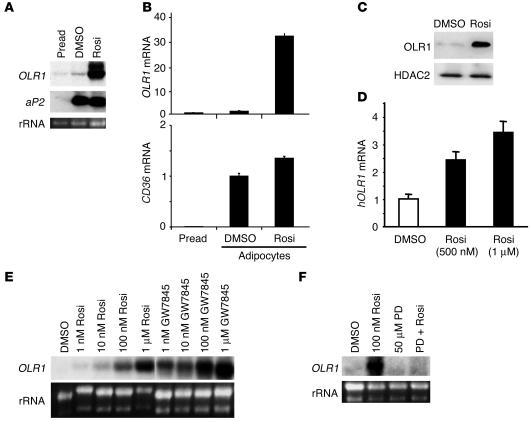
TZDs have been shown to induce the differentiation of preadipocytes by activating PPARγ and upregulating many of its target genes (18, 38, 39). However, the effects of TZDs in the fully differentiated adipocyte, which continues to express high levels of PPARγ, is much less well understood. Using microarray analysis to identify novel genes that are differentially regulated by PPARγ ligands in the mature mouse 3T3-L1 adipocyte, we identified OLR1 as a gene that was markedly upregulated by rosiglitazone treatment. Northern blot analysis demonstrated that while fully differentiated adipocytes expressed very little OLR1 mRNA, treatment with rosiglitazone dramatically upregulated its expression (Figure 1A). This pattern of expression is in stark contrast to the expression of aP2, which was upregulated during adipogenesis but did not increase significantly with rosiglitazone treatment (Figure 1A). OLR1 mRNA expression increased by more than 30-fold in adipocytes treated with rosiglitazone compared with untreated adipocytes (Figure 1B). CD36, the most well-characterized receptor for oxLDL, is also known to be upregulated by TZDs (40), but the magnitude of this increase (1.4-fold) in adipocytes was considerably less than that seen for OLR1 (Figure 1B). The increase in OLR1 mRNA expression led to a corresponding increase in OLR1 protein expression in rosiglitazone-treated cells (Figure 1C). We also observed increased OLR1 mRNA expression in primary human adipocytes exposed to rosiglitazone (Figure 1D), which suggests that the induction of OLR1 by TZDs occurs across species.
Figure 1.
OLR1 is induced by PPARγ ligands in 3T3 L1 adipocytes. (A) Comparison of OLR1 and aP2 mRNA levels in 3T3-L1 preadipocytes (Pread), adipocytes treated with vehicle (DMSO), and rosiglitazone-treated adipocytes (Rosi). (B) Quantitation of OLR1 compared with CD36 mRNA induction by rosiglitazone, using real-time PCR. Results are normalized to mRNA levels in vehicle-treated adipocytes. Data expressed as mean ± SEM (n = 4). (C) Rosiglitazone induces OLR1 protein expression in 3T3-L1 adipocytes according to Western blot analysis. Histone deacetylase 2 (HDAC2) protein levels show equal loading. (D) Rosiglitazone (500 nM or 1 μM for 4 days) induces OLR1 mRNA expression in human adipocytes. Real-time PCR results expressed as mean ± SEM (n = 4). hOLR1, human OLR1. (E) Dose response for OLR1 induction by rosiglitazone and the non-TZD PPARγ ligand GW7845. (F) Induction of OLR1 by rosiglitazone (100 nM) is blocked by the PPARγ antagonist PD068235 (PD; 50 μM). rRNA, ribosomal RNA.
A potent non-TZD PPARγ ligand, GW7845, also upregulated OLR1 gene expression (Figure 1E). The EC50s for rosiglitazone and GW7845 were in agreement with their in vitro binding affinities for PPARγ (41), which suggests that the upregulation of OLR1 by these ligands was a direct result of PPARγ activation. Consistent with this, a specific competitive PPARγ antagonist, PD068235, abrogated the induction of OLR1 by rosiglitazone (42) (Figure 1F). Together, these data suggest that TZDs induce OLR1 expression through the activation of PPARγ.
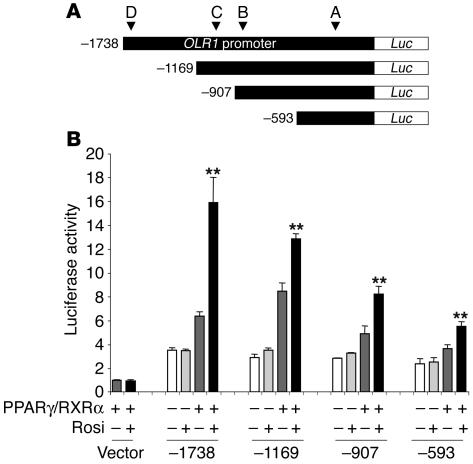
OLR1 is a direct transcriptional target gene of PPARγ. To investigate the transcriptional mechanism by which TZDs induce OLR1, we fused 5′ fragments of the OLR1 promoter upstream of a luciferase reporter and cotransfected them with PPARγ and RXRα into 293T cells in the presence or absence of rosiglitazone (Figure 2A). Rosiglitazone markedly activated the reporter gene containing 1,738 bp of the 5′ flanking sequence (Figure 2B), which is consistent with the existence of a PPARγ response element in the OLR1 promoter. Upon serial truncations (from −1,738 bp upstream of the start site to −1,169, −907, and −593 bp), the OLR1 promoter retained a transcriptional response to PPARγ/RXRα/rosiglitazone, although the magnitude of activation diminished with increasing truncation length, which raises the possibility that there could be multiple PPREs distributed throughout the OLR1 promoter that each contributed to the activation by rosiglitazone.
Figure 2.
The OLR1 promoter-enhancer is TZD-responsive. (A) Schematic of reporter constructs containing serial truncations of the OLR1 promoter-enhancer fused to luciferase (Luc). (B) Luciferase activity of OLR1 reporter constructs cotransfected with PPARγ/RXRα with or without 1 μM rosiglitazone. Results were normalized to β-galactosidase activity. Data represent the fold induction of luciferase activity over the basal activity of the empty pGL2 vector and are expressed as mean ± SEM (n = 3 per condition). **P < 0.01 compared with transfected cells treated with vehicle. RLU, relative luciferase units.
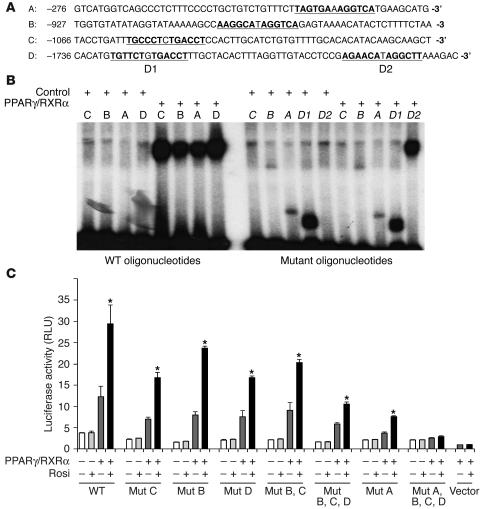
Inspection of the sequence of the OLR1 promoter revealed 4 potential PPARγ binding sites (designated A, B, C, and D), each consisting of an imperfect direct repeat of the consensus nuclear receptor half-site separated by 1 base pair (Figure 3A). DNA mobility shift assays demonstrated that each of the 4 sites was capable of binding the PPARγ/RXRα heterodimer (Figure 3B). None of the sites were bound by either PPARγ or RXRα alone (data not shown). Mutations within the half-sites completely abolished binding to the PPARγ/RXRα heterodimer (Figure 3B), while a full mobility shift was maintained in an oligonucleotide probe with mutations made in an irrelevant site (D2) (Figure 3B, last lane), which indicates that the binding of PPARγ/RXRα to DNA is site and sequence specific. In addition, PPARγ binding was effectively competed away by increasing concentrations of an oligonucleotide containing a wild-type, but not a mutated, PPRE (data not shown).
Figure 3.
The proximal PPARγ binding site in the OLR1 promoter is the predominant PPRE. (A) Identification of putative PPARγ binding sites (underlined) in the OLR1 promoter. Letters A–D correspond to arrowheads in Figure 2A. (B) DNA mobility shift assay showing the binding of PPARγ/RXRα to wild-type and mutant oligonucleotide probes corresponding to the putative PPARγ binding sites underlined in A. Control lane reactions did not contain PPARγ/RXRα. Mutant oligonucleotides contain point mutations in the putative PPARγ binding site; for specific mutations, see Methods. (C) Luciferase activity of OLR1 reporter constructs containing mutations in the PPARγ binding sites. Site-specific mutations were made in the longest OLR1 reporter construct (–1,738; Figure 2A). Data represent the fold induction of luciferase activity over the basal activity of the empty pGL2 vector and are expressed as mean ± SEM (n = 3 per condition). *P < 0.01 compared with transfected cells treated with vehicle.
To determine which of the 4 PPARγ binding sites was responsible for transcriptional activation by rosiglitazone, each site was mutated either individually or in combination in the luciferase reporter containing the –1,738-bp fragment of the OLR1 promoter. Luciferase assays demonstrated that single mutations in the 3 distal sites (B, C, and D) only modestly affected the ability of rosiglitazone to increase reporter activity (Figure 3C), although the mutations in combination reduced activation by rosiglitazone by approximately 50%. However, mutations in the most proximal PPARγ binding site (site A) reduced transcriptional activation by rosiglitazone to a much greater extent (Figure 3C). Mutations in all 4 binding sites completely abolished the transcriptional response to rosiglitazone. These data strongly suggest that the proximal PPARγ binding site is the predominant PPRE in the OLR1 promoter, with the 3 other DR-1 PPARγ binding sites making small contributions.
Rosiglitazone regulates the recruitment of corepressors and coactivators to the endogenous OLR1 promoter.
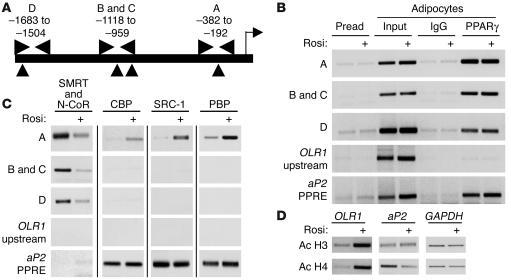
We next used chromatin immunoprecipitation (ChIP) to study the transcriptional regulation of the endogenous OLR1 gene in 3T3-L1 adipocytes. Primer sets were designed to span the PPARγ binding sites, and chromatin was sheared to an average fragment size of 300–400 bp so that each PPRE could be assessed separately using ChIP assays (Figure 4A; a single PCR primer set was used to investigate sites B and C, which are closely spaced). ChIP analysis demonstrated that adipocyte PPARγ bound in the regions of each of the PPREs in the OLR1 promoter in the presence and absence of rosiglitazone, while a region approximately 500 bp upstream of the most distal PPRE showed no binding of PPARγ (Figure 4B). As expected, the region of the aP2 promoter containing its PPRE was also bound by PPARγ (Figure 4B).
Figure 4.
Rosiglitazone regulates the recruitment of corepressors and coactivators to the endogenous OLR1 promoter. (A) Schematic of the OLR1 promoter showing the location of the PCR primers (horizontal arrowheads) used for ChIP analysis in relation to the PPARγ binding sites (vertical arrowheads) identified in Figure 3. Horizontal arrowheads indicate the region of PCR; vertical arrowheads indicate the locations of the PPARγ binding sites shown in Figure 3. (B) ChIP analysis of PPARγ association with the OLR1 promoter-enhancer in 3T3-L1 preadipocytes and adipocytes with or without rosiglitazone treatment (1 μM for 48 hours). The OLR1 upstream PCR primer set is located approximately 500 bp upstream of the PPRE D site and is used as a negative control. (C) ChIP analysis of the association of the corepressors N-CoR and SMRT and coactivators CBP, SRC-1, and PBP with the OLR1 and aP2 promoters in adipocytes with or without rosiglitazone treatment. (D) ChIP analysis of histone H3 and H4 acetylation (Ac H3 and Ac H4) at the OLR1, aP2, and GAPDH promoters in adipocytes with or without rosiglitazone treatment. For the OLR1 promoter, the PCR primer set for the PPRE A site was used.
The dramatic increase in OLR1 expression caused by TZDs is particularly striking because it is normally expressed at such low levels in fully differentiated adipocytes. In contrast, aP2, another PPARγ target gene, is upregulated during adipogenesis and remains expressed at a high level, with only a small increase upon TZD treatment in the mature fat cell. We hypothesized that the OLR1 gene is normally repressed by PPARγ due to corepressor recruitment. Indeed, corepressors SMRT and N-CoR were not detected on the active aP2 promoter in mature adipocytes in the presence or absence of TZD, whereas in untreated adipocytes, SMRT and N-CoR were recruited to the 3 distinct regions in the OLR1 promoter containing PPREs (Figure 4C). Rosiglitazone treatment released corepressor binding in all 3 regions. In contrast, the coactivators CBP, SRC-1, and PBP were robustly recruited only to the proximal PPRE, consistent with its functional predominance (Figure 4C). Binding of these coactivators to the OLR1 gene was greatly stimulated by rosiglitazone, whereas the coactivators bound in the region of the aP2 PPRE independently of ligand treatment, consistent with the regulation of the genes. Similarly, treatment with rosiglitazone substantially increased acetylation of histones H3 and H4 at the OLR1 promoter, whereas the levels of acetylated histones remained constant at the promoters of the aP2 and GAPDH genes (Figure 4D).
Rosiglitazone increases oxLDL uptake in adipocytes by upregulating OLR1.
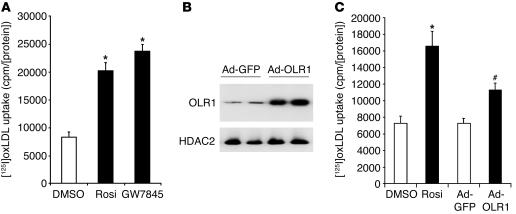
Since OLR1 functions as a receptor for oxLDL in endothelial cells, we next determined the effects of PPARγ ligand treatment on the uptake of oxLDL in 3T3-L1 adipocytes. Both rosiglitazone and the non-TZD PPARγ ligand GW7845 markedly stimulated the adipocyte uptake of [125I]oxLDL (Figure 5A). To test the specificity of this effect for oxidatively modified LDL, excess amounts of both oxLDL and native LDL were used to compete for binding of [125I]oxLDL. We found that [125I]oxLDL binding was reduced approximately 40% by native LDL competition (data not shown), which suggests that the rosiglitazone effect was relatively specific for oxLDL. oxLDL receptors include OLR1, CD36, SR-B1, and macrosialin (43). CD36 is a PPARγ target gene, although, as shown in Figure 1B, rosiglitazone induced OLR1 approximately 30-fold but only induced CD36 by approximately 1.4-fold. Moreover, rosiglitazone did not appreciably induce the expression of SR-B1 and macrosialin (data not shown).
Figure 5.
PPARγ ligands and ectopic expression of OLR1 increase oxLDL uptake in adipocytes. (A) Rosiglitazone (1 μM) and GW7845 (100 nM) increase oxLDL uptake in 3T3-L1 adipocytes. Results are expressed as mean ± SEM (n = 9). *P < 0.001 for each treatment compared with DMSO. (B) Adenoviral expression of OLR1 (Ad-OLR1) and GFP (Ad-GFP; negative control) in day 10 adipocytes. Western blot of HDAC2 is shown as a loading control. (C) Overexpression of OLR1 increased oxLDL uptake into adipocytes. For all uptake experiments, results were corrected for nonspecific binding measured by cold competition with unlabeled oxLDL and normalized for protein concentration. Data are expressed as mean ± SEM (n = 6). *P < 0.001, rosiglitazone-treated cells compared with DMSO; #P < 0.001, adenoviral cells expressing OLR1 compared with adenoviral cells expressing GFP.
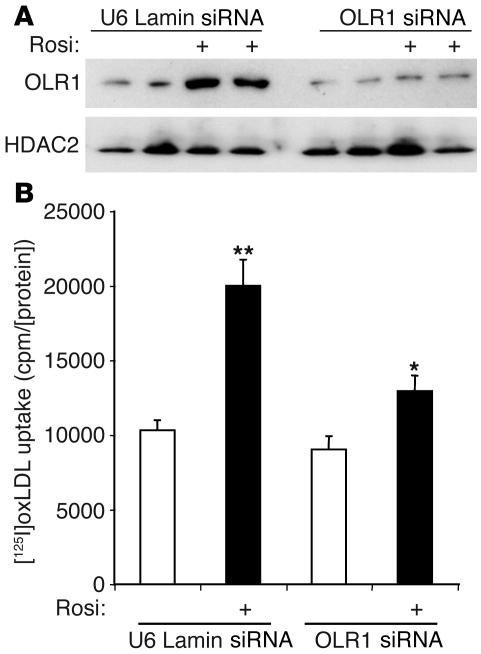
The potential role of OLR1 as the mediator of rosiglitazone-stimulated adipocyte oxLDL uptake was tested by adenoviral expression of OLR1 at approximately the same level induced by rosiglitazone treatment (Figure 5B). The induction in OLR1 expression caused an increase in oxLDL uptake that was approximately 50% of that resulting from rosiglitazone stimulation (Figure 5C). This suggests that induction of OLR1 could account for a significant proportion of TZD-stimulated oxLDL uptake. To further investigate the role of OLR1, we reduced levels of the endogenous protein by lentiviral expression of a short interfering RNA (siRNA) directed against OLR1. Both basal and rosiglitazone-induced OLR1 protein levels were decreased (Figure 6A). Rosiglitazone-stimulated oxLDL uptake was reduced by 50–60% in adipocytes in which OLR1 levels were diminished (Figure 6B). By contrast, basal oxLDL uptake, presumably mediated by CD36 (40), was only minimally affected (Figure 6B). These data strongly suggest that rosiglitazone stimulates oxLDL uptake in large part through the induction of OLR1.
Figure 6.
Induction of OLR1 is required for stimulation of adipocyte oxLDL uptake by rosiglitazone. (A) Reduction of OLR1 protein levels by lentivirally delivered siRNA for OLR1 (OLR1 siRNA) or lamin A/C (U6 Lamin siRNA) in adipocytes with or without 1 μM rosiglitazone. (B) Knockdown of OLR1 levels reduced rosiglitazone enhancement of oxLDL uptake into adipocytes by approximately 60%. Adipocytes were infected with lentivirus siRNA for OLR1 or lamin A/C (negative control) for 4 days, then treated with DMSO or 1 μM rosiglitazone for 48 hours. Uptake assays were corrected for nonspecific binding measured by cold competition with unlabeled oxLDL and normalized for protein concentration. Data are expressed as mean ± SEM (n = 6). *P < 0.05, rosiglitazone-treated vs. DMSO in OLR1 siRNA cells; **P < 0.01, rosiglitazone-treated vs. DMSO in U6 Lamin siRNA cells. Data were analyzed using ANOVA and post-hoc statistical tests.
Rosiglitazone upregulation of OLR1 increases adipocyte cholesterol content.
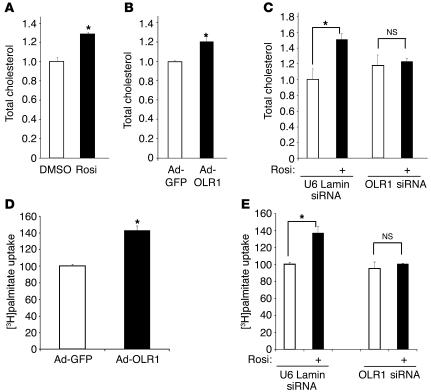
OxLDL is a rich source of cholesterol. Remarkably, rosiglitazone treatment significantly increased the total cholesterol content of adipocytes (Figure 7A). Adipocytes treated with rosiglitazone showed no significant increase in mRNA levels of SREBP-2 or HMG CoA reductase (data not shown), which are 2 key regulators in de novo cholesterol synthesis. We therefore explored the possibility that this increase in cholesterol levels resulted from an influx of exogenous cholesterol sources through the upregulation of OLR1. Indeed, adenoviral overexpression of OLR1 was sufficient to raise total cholesterol levels in adipocytes (Figure 7B). Moreover, lentiviral knockdown of endogenous OLR1 abolished most of the rosiglitazone-induced increase in total adipocyte cholesterol (Figure 7C). These data suggest that OLR1 induction is necessary for a significant portion of the rosiglitazone-stimulated increase in adipocyte cholesterol.
Figure 7.
Rosiglitazone upregulation of OLR1 increases adipocyte cholesterol content and enhances fatty acid uptake. (A) Rosiglitazone treatment (1 μM for 48 hours) increases adipocyte cholesterol content. (B) Adenoviral overexpression of OLR1 increases adipocyte cholesterol content. (C) Specific knockdown of OLR1 abrogates the rosiglitazone-stimulated increase in adipocyte cholesterol content. For all cholesterol assays, lipids were extracted in isopropanol, and total cholesterol levels were measured and normalized to protein concentration. Data were all normalized to cholesterol levels in vehicle-treated cells, and differences are expressed as fold changes for all experiments. (D) Adenoviral overexpression of OLR1 increases the initial uptake of [3H]palmitate into adipocytes. (E) Knockdown of OLR1 abrogates the rosiglitazone-induced increase in [3H]palmitate uptake into adipocytes. Data were all normalized to protein concentration. Results are expressed as the percentage of palmitate uptake compared with vehicle-treated or adenoviral cells expressing GFP control cells. All results are shown as mean ± SEM (n = 4). *P < 0.01 compared with vehicle using Student’s t test or ANOVA with post-hoc statistical tests, as appropriate.
OLR1 induction is required for rosiglitazone enhancement of fatty acid uptake.
We and others have previously demonstrated that rosiglitazone increases fatty acid uptake and retention in the form of triglycerides (30, 33, 44). These pathways involve the coordinated induction of several genes involved in fatty acid uptake and metabolism in adipocytes, which leads to a net flux of FFAs from peripheral tissues and the circulation into adipocytes. This change in lipid partitioning has been implicated in the insulin-sensitizing effects of TZDs, through a combination of reduced lipotoxicity and circulating fatty acid levels (45). Increasing evidence also suggests that adipocyte cholesterol levels are directly proportional to its triglyceride content and are involved in the regulation of FFA uptake and storage (10). Since OLR1 increases cholesterol content, we hypothesized that it might subsequently regulate fatty acid metabolism in adipocytes. Indeed, ectopic expression of OLR1 in adipocytes stimulated the initial uptake of palmitic acid (Figure 7D). Moreover, enhancement of palmitate uptake in adipocytes due to rosiglitazone treatment was largely abrogated by knockdown of OLR1 (Figure 7E). These data suggest that TZD-mediated stimulation of cholesterol and fatty acid accumulation in adipocytes are coupled via the induction of OLR1.
OLR1 is induced in vivo by genetic and pharmacological activation of PPAR γand by obesity.
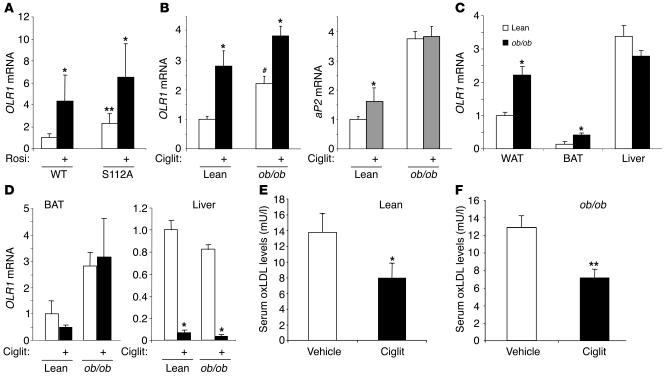
We next investigated the role of PPARγ activation in the regulation of adipocyte OLR1 gene expression in vivo. Treatment of wild-type mice with rosiglitazone led to a marked induction of OLR1 mRNA in white adipose tissue (Figure 8A). We also studied the expression of OLR1 in the adipose tissue of mice homozygous for the S112A mutation, which modestly activates endogenous PPARγ by preventing its phosphorylation (46). Adipose tissue OLR1 mRNA expression was increased in PPARγ-S112A compared with wild-type mice and was further induced by rosiglitazone (Figure 8A). These data suggest that genetic or pharmacological activation of PPARγ induces OLR1 in adipose tissue in vivo. We also assessed the effect of obesity on OLR1 gene expression in white adipose tissue. Notably, OLR1 expression was increased approximately 2-fold in the adipose tissue of ob/ob mice compared with lean littermates (Figure 8B). Ciglitazone treatment induced adipose OLR1 mRNA to similar levels in lean and ob/ob mice (Figure 8B). In comparison, expression of the classic PPARγ target gene aP2 was induced only modestly by ciglitazone in lean mice and was not increased by TZD treatment in ob/ob mice (Figure 8B), consistent with the different mechanisms by which OLR1 and aP2 are transcriptionally regulated.
Figure 8.
OLR1 is induced in vivo by genetic or pharmacological activation of PPARγ and by obesity. (A) Rosiglitazone treatment (4 mg/kg/d for 4 days) of lean C57BL/6 mice induces OLR1 mRNA in white adipose tissue by real-time PCR. OLR1 is increased in white adipose tissue (WAT) of PPARγ-S112A mice and is further induced by rosiglitazone treatment. Wild type, n = 6; S112A, n = 10. *P < 0.05 vs. vehicle; **P < 0.01 vs. wild type. (B) OLR1 mRNA is increased in white adipose tissue of ob/ob mice relative to lean littermates and is further induced by ciglitazone treatment (Ciglit; 100 mg/kg/d for 4 days). Results for aP2 mRNA are shown for comparison. n = 4 per condition. *P < 0.05 vs. vehicle; #P < 0.05 vs. lean. (C) Comparison of OLR1 mRNA levels in ob/ob mice and lean littermates in white adipose tissue, brown adipose tissue (BAT), and liver. n = 4 per condition. * P < 0.05 vs. lean. (D) The induction of OLR1 by TZDs in vivo is specific to white adipose tissue. Brown adipose tissue expression of OLR1 mRNA in lean or obese mice treated with ciglitazone does not significantly change, while OLR1 expression in liver is reduced with TZD treatment. n = 4 per condition. *P < 0.01 vs. vehicle-treated. (E) Serum oxLDL levels are reduced in lean mice treated with ciglitazone. n = 4 per treatment. *P < 0.05 vs. vehicle. (F) Serum oxLDL levels are reduced in ob/ob mice treated with ciglitazone. n = 4 per treatment. *P < 0.01 vs. vehicle-treated.
We next investigated OLR1 expression levels in various tissues of lean and ob/ob mice. In comparison with OLR1 mRNA levels in white adipose tissue of lean mice, OLR1 was expressed at much lower levels in brown adipose tissue but was induced in obese mice (Figure 8C). By contrast, liver OLR1 gene expression was similar in ob/ob and lean mice (Figure 8C). Furthermore, following treatment of lean and ob/ob mice with ciglitazone, OLR1 mRNA levels did not change significantly in brown adipose tissue and actually decreased in liver (Figure 8D). These data suggest that the induction of OLR1 by TZDs in vivo is specific for white adipose tissue.
Serum oxLDL levels are decreased by TZD treatment in lean and ob/ob mice.
Having shown that the induction of OLR1 in 3T3-L1 adipocytes enhances oxLDL uptake, we hypothesized that TZD treatment would lower serum oxLDL levels in vivo. Indeed, ciglitazone treatment of both lean C57BL/6 mice (Figure 8E) and ob/ob mice (Figure 8F) significantly decreased circulating oxLDL levels. These data strongly correlate with the effect of OLR1 in cultured 3T3-L1 adipocytes and suggest that adipose OLR1 may play a role in the uptake of oxLDL in response to TZD treatment in vivo.
Discussion
We have discovered that the scavenger receptor OLR1, also known as lectin-like oxidized low-density lipoprotein receptor-1, is dramatically upregulated in adipocytes upon treatment with PPARγ ligands; shown that this is a direct effect of PPARγ; and identified the molecular basis for PPARγ repression and activation of this gene. To our knowledge, this is the first report of OLR1 expression in adipose tissue. Metabolic studies reveal that OLR1 expression is sufficient to stimulate oxLDL uptake, cholesterol accumulation, and fatty acid uptake into adipocytes. Moreover, we have shown that OLR1 induction is largely required for TZD regulation of these aspects of adipocyte lipid metabolism.
OLR1, like CD36, is a scavenger receptor that binds, endocytoses, and proteolytically degrades oxLDL (37). It was originally identified as the major receptor for oxLDL in endothelial cells in large arteries (37) but is also expressed in macrophages and smooth muscle cells (47). OLR1 is upregulated in vascular pathologies, such as hypertension, diabetes, and atherosclerosis; in fact, the induction of OLR1 and subsequent binding and internalization of oxLDL have been hypothesized to cause endothelial cell dysfunction and apoptosis, as well as foam cell formation (47). Of note, OLR1 has been shown to recognize various modified forms of LDL, including mildly and fully oxLDL (47).
The oxidative modification of LDLs is generally believed to play a pathogenic role in the development of atherogenic lesions (48–50). Although the true in vivo nature and clinical relevance of oxLDL has remained obscure, mounting evidence in the literature demonstrates that not only do various oxidatively modified forms of LDL exist, but they can actually be measured in the circulation using antibodies directed against oxidation-specific epitopes. For example, an antibody generated against an oxidized phosphatidylcholine moiety was used to show that oxidized forms of LDL were present even in the serum of healthy subjects and that levels were increased by 2- to 4-fold in patients with vascular diseases (43, 49). A separate study used an antibody directed against malondialdehyde-LDL, another oxidized form of LDL, to demonstrate approximately 2- to 5-fold increases in oxLDL levels in the plasma of patients with coronary artery disease and myocardial infarcts compared with control patients (51). OxLDL most likely exists in vivo as a heterogeneous mixture of particles, ranging from minimally modified LDL (52) to myeloperoxidase-treated LDL (53); importantly, these forms of oxidatively modified LDL were shown to induce strong cellular responses in macrophages and endothelial cells to a level similar to oxLDL prepared by copper oxidation in vitro (54–56). OxLDL levels in the circulation are low relative to the total amount of LDL (48), due to efficient clearance by the hepatic reticuloendothelial system (57), blood antioxidants, and immune responses generated by autoantibodies against oxLDL (58). However, it is now widely accepted that oxidized forms of LDL do exist in vivo and at detectable levels in the circulation, even in healthy patients. While the role of oxLDL in adipocytes has not been well studied, adipocytes are known to express scavenger receptors, such as SR-B1 and CD36 (59, 60), and are capable of recognizing and degrading oxLDL in part through CD36 (61). Our discovery that OLR1 is upregulated in adipocytes by TZDs provides further evidence that oxLDL may play an important, though as-yet-undefined, role in fat cell metabolism. Adipocytes are poor at de novo cholesterol biosynthesis (13) and obtain a majority of their sterols from lipoproteins in the circulation (10). Since oxLDL is a rich source of cholesterol, scavenger receptors could be essential for adipocyte uptake of the cholesterol required for normal signal transduction, lipid raft function, and vesicular trafficking. Sufficient cholesterol levels have also been shown to be essential for efficient insulin signaling, glucose transport, and FFA uptake in adipocytes (9, 15).
Alternatively, since the adipocyte is designed to store large amounts of lipids, it seems logical that the adipose tissue could also serve as a major site of oxLDL detoxification, thus removing it from the circulation and potentially inhibiting the formation of atherosclerotic lesions. Increasing clinical evidence has shown that rosiglitazone treatment significantly improves factors associated with coronary artery disease, including endothelial cell activity, inflammatory processes, and dyslipidemia (62–64). PPARγ ligands have also been demonstrated to reduce atherosclerosis in mouse models (65, 66); 1 mechanism for this may involve the induction of a cholesterol efflux pathway, which would prevent foam cell formation (67, 68). In this study, we have demonstrated that TZD treatment specifically induces OLR1 in adipose tissue and reduces serum oxLDL levels in lean and obese animals. The trapping of circulating oxLDL by adipocytes may thus be a novel pathway by which TZDs mediate their antiatherosclerotic effects, and this hypothesis should be further explored in vivo using mouse models of atherosclerosis. Interestingly, while we have shown that TZDs induce OLR1 in adipocytes to promote oxLDL uptake, PPARγ ligands were shown to have a beneficial effect on atherosclerosis by inhibiting OLR1 expression in endothelial cells (69), which demonstrates the cell-specific and cardioprotective effects of TZDs on oxLDL uptake.
The regulation of OLR1 expression in adipocytes by TZDs provides further insight into the mechanisms by which these drugs regulate lipid metabolism and insulin sensitivity. Found at very low levels in mature adipocytes but markedly induced by TZD treatment, OLR1’s pattern of expression is distinct from that of the classical PPARγ target gene aP2, which is upregulated during adipogenesis with only a mild further increase after TZD treatment. In the same adipocyte milieu, PPARγ is bound to PPREs in the promoters of both genes, but there is a differential recruitment of cofactors: the aP2 gene is bound by coactivators, while the OLR1 gene is occupied by corepressors N-CoR and SMRT. Upon addition of a potent pharmacological PPARγ ligand, corepressors are released from the OLR1 promoter, which leads to the recruitment of coactivators to the most proximal PPRE, histone hyperacetylation, and active transcription of the gene. Other genes repressed by PPARγ in a manner that is reversed by TZD treatment of adipocytes are glycerol kinase (33) and GLUT4 (70). We have recently reported a similar molecular switch underlying PPARγ regulation of glycerol kinase expression in adipocytes (71). The present data demonstrate that gene-specific corepressor recruitment is a generalized phenomenon and suggest that OLR1 may be more highly regulated than glycerol kinase due to the presence of multiple PPREs within 2 kb of the promoter to which corepressors are recruited in the absence of synthetic PPARγ ligand. The specific component of the adipocyte milieu that regulates the differential requirement for synthetic ligand to activate PPARγ on the OLR1 and aP2 genes remains to be determined.
Although results from tissue-specific knockout models suggest direct roles of PPARγ in liver and muscle (72–74), PPARγ is expressed predominantly in adipocytes, and TZDs are less effective in the absence of adipose tissue (29). Thus, it is likely that TZDs improve whole-body insulin sensitivity through their actions on PPARγ in adipocytes (75). The present data support the notion that PPARγ regulates 2 distinct gene programs in adipocyte, one required for adipogenesis, the other involving nonadipogenic gene regulation that is responsible for the effects of TZDs on lipid metabolism and insulin sensitivity. This appears to involve the regulation of adipocyte-secreted proteins — including leptin, TNF-α, resistin, and adiponectin (76) — and adipocyte lipid metabolism (77). TZDs promote fatty acid uptake and triglyceride storage in adipose tissue to reduce the level of plasma FFAs and to promote the flux of fatty acids away from the liver and muscle; high levels of plasma FFAs and of fatty acid in liver and muscle correlate with insulin resistance (78, 79).
Adipose tissue is an ideal “sink” for lipids, but there must exist a balance among the different lipid components in the cell that permits the storage of large amounts of triglycerides yet allows the adipocyte to respond to lipolytic and hormonal signals. There are increasing data to suggest that the ability of adipocytes to efficiently store triglycerides is correlated with the cholesterol balance in the cells and that cholesterol levels affect FFA uptake (15) and the triglyceride content of the lipid droplet (10). Our results show that the upregulation of OLR1 increases the total cholesterol content and FFA uptake in adipocytes. One explanation for this result is that the increased levels of adipocyte cholesterol caused by OLR1 induction create a cellular environment that promotes FFA uptake and enhances lipid storage. This is consistent with previous work showing that cholesterol loading of adipocytes enhances long-chain fatty acid uptake (15) and supports the concept that adipocyte cholesterol uptake and lipid storage may be coupled processes (10). However, another possibility is that OLR1 could be a direct transporter for FFAs as well as for cholesterol. The identification of OLR1 as a gene induced by TZDs suggests a novel role for PPARγ in the control of cholesterol metabolism in the adipocyte, although further experiments are needed to elucidate the mechanism by which this occurs. It should be noted that TZDs might induce HMG CoA enzyme activity, although HMG CoA reductase mRNA was not changed by treatment with rosiglitazone, and ectopic expression of OLR1 was sufficient to increase adipocyte cholesterol content.
TZDs have been reported to cause weight gain in patients due to increased adiposity (80), despite the fact that excess fat is often correlated to insulin resistance. This clinical observation is consistent with experiments showing that TZDs increase the triglyceride content of adipose tissue (30) and stimulate adipocyte differentiation (81). Thus, weight gain due to increased storage of FFAs and triglycerides in adipocytes may protect the liver and muscle from lipotoxicity and thereby promote insulin sensitivity. Increased lipid storage in mature adipocytes due to TZD treatment, promoted by elevated cholesterol levels mediated largely through OLR1, would be expected to increase their size, and larger adipocytes tend to be insulin resistant (30). However, the overall effects of TZDs tend to mitigate this effect. TZDs cause the differentiation of new, small adipocytes and promote apoptosis of older, larger adipocytes (30). In addition, TZDs induce PGC-1 and mitochondrial biogenesis in adipocytes, increasing lipid oxidation (82).
Furthermore, hypertrophic, insulin-resistant adipocytes from obese rodents are relatively cholesterol deficient (8), and cholesterol-depleted adipocytes exhibit decreased insulin-stimulated glucose oxidation, reduced GLUT4 mRNA levels, and upregulated TNF-α and IL-6 mRNA (9). Our observation that adipose OLR1 expression is modestly increased in obesity may thus represent a physiological response; the relative cholesterol deficiency seen in hypertrophic adipocytes causes the upregulation of genes involved in cholesterol biosynthesis (8) and may also upregulate cholesterol receptors such as OLR1. In addition, obesity is associated with increased circulating levels of FFAs (78), which act as weak PPARγ ligands and could submaximally increase transcription of OLR1. Treatment with TZDs taps into this pathway pharmacologically and induces OLR1 to an even greater magnitude. The corresponding replenishment of cholesterol in differentiated adipocytes caused by TZDs may also contribute to enhanced FFA uptake and improved insulin sensitivity.
In sum, we have identified OLR1 as a novel PPARγ-regulated gene in adipocytes, thus uncovering a potential new role for oxLDL in adipocyte metabolism. Rosiglitazone treatment increases oxLDL uptake and total cholesterol levels in the adipocyte, and OLR1 plays an important role in mediating these effects. TZDs also reduce serum oxLDL levels in lean and obese mouse models, potentially by inducing OLR1 in white adipose tissue. Furthermore, adipocytes in which the level of total cholesterol has been increased, whether by rosiglitazone treatment or by increased expression of OLR1, also exhibit an increased uptake of palmitic acid. Consistent with this, rosiglitazone has been shown to increase FFA uptake in 3T3-L1 adipocytes (33) and adipose tissue (83). We propose that cholesterol provides an essential link between TZD treatment and increased FFA uptake in adipocytes. While the physiological role of adipose tissue in cholesterol and oxLDL metabolism remains to be defined, our data support a critical role for OLR1 in the regulation of adipocyte lipid metabolism and, potentially, insulin sensitivity, by PPARγ and its antidiabetic ligands.
Methods
Materials.
All cell culture reagents were from Life Technologies Inc. Human insulin, dexamethasone, isobutyl methylxanthine, and fatty acid–free BSA were from Sigma-Aldrich. BRL 49653 (rosiglitazone) was from BIOMOL. GW7845 was from GlaxoSmithKline, and the PPARγ antagonist PD068235 was from Pfizer.
Microarray analysis.
3T3-L1 adipocytes were maintained and differentiated as previously described (18). On day 8, cells were treated with either vehicle, 1 μM rosiglitazone, or 100 nM GW7845. Total RNA was harvested with TRIzol (Invitrogen Corp.) 48 hours later, and biotin-labeled cRNA was prepared according to the Affymetrix technical manual. Hybridization to the murine genome U74Av2 array was performed at the University of Pennsylvania Microarray facility, and expression data was analyzed using GeneSpring software (Silicon Genetics; Agilent Technologies).
RNA analysis.
3T3-L1 adipocytes were maintained and differentiated as described (84). On day 8 after differentiation, cells were treated with appropriate reagents (see figure legends) for 48 hours. Total RNA was extracted from 3T3-L1 cells, human adipocytes (Zen-Bio), and mouse epididymal fat pads using TRIzol. For Northern blotting, the OLR1 probe was prepared using PCR with 3T3-L1 cDNA with the following primers: forward, 5′-CCAAGCGAACCTTACTCAGC-3′; reverse, 5′-CCTGCTCTTTGGATTTCTCG-3′. The aP2 probe was prepared with PstI digestion of an expression plasmid. Total RNA (10 μg) was analyzed on a 1% formaldehyde agarose gel and transferred to Hybond-N membranes (Amersham Biosciences). Hybridization of the 32P-labeled probes was done using QuikHyb solution (Stratagene). For quantitative PCR, RNA was subjected to DNase digestion followed by RT-PCR (Invitrogen Corp.). mRNA transcripts were quantitated by the fluorogenic probe method using a Prism 7700 sequence detector system (Applied Biosystems) and normalized to 36B4. TaqMan Gene Expression Assays (Applied Biosystems) were used for mouse CD36 and human OLR1. Primer and probe sequences used were as follows: murine OLR1 (mOLR1) forward, 5′- CTGCACTCCTTCTTCCCCTTT-3′; mOLR1 reverse, 5′-GCCTGCACTTGAGGAGGATTT-3′; mOLR1, probe, 5′-CCTGCCTGACCTGGCCATGCTT-3′; 36B4 forward, 5′-TCATCCAGCAGGTGTTTGACA-3′; 36B4 reverse, 5′-GGCACCGAGGCAACAGTT-3′; 36B4 probe, 5′-AGAGCAGGCCCTGCACTCTCG-3′.
In vivo experiments.
C57BL/6J and ob/ob mice (Jackson Laboratory), aged 11 weeks, were housed (n = 4 per cage) under 12-hour light/12-hour dark cycles at 23°C with ad libitum access to food and water. Animals were mock dosed for 4 days with 0.25% methylcellulose before receiving either vehicle, 4 mg/kg/d rosiglitazone, or 100 mg/kg/d ciglitazone via oral gavage for 4 days. Epididymal adipose tissue was dissected, and total RNA was extracted for Northern blot analysis. Animal care and procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Luciferase reporter assays.
The promoter fragments were generated using PCR of mouse genomic DNA (BD Biosciences — Clontech) and cloned into the pCR Blunt II TOPO vector using the Zero Blunt TOPO PCR cloning kit (Invitrogen Corp.). The following primer sequences were used: –1,738 forward, 5′-ACATGTGTTCTGTGACCTTTGC-3′; –1,169 forward, 5′-CAGAGATAGGAGGGAATCTGGA-3′; –907 forward, 5′-AGCCAAGGCATAGGTCAGAGTA-3′; –593 forward, 5′-CTATTTCTACTGGGCTGCTGCT-3′; +34 reverse, 5′-ATCAAAAGTCATTTCCAAATTCATGCTA-3′.
After sequencing for mutations and orientation, the promoter fragments were excised with either SacI and XbaI (correct orientation) or EcoRV and SacI (reversed) and ligated into the pGL2 Basic vector (Promega) digested with either SacI and NheI (for correctly oriented inserts) or SmaI and SacI (for reversed inserts). Point mutations were generated in the longest reporter plasmid using the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol, and all constructs were confirmed by sequencing. Transfections were performed in HEK 293T cells (ATCC) with FuGENE 6 (Roche Diagnostics Corp.) using 0.2 μg each of pCMX-PPARγ2 and pCMX-RXRα and 0.4 μg luciferase reporter per well of a 12-well plate. β-Galactosidase plasmid (0.1 μg per well) was used for transfection normalization. All assays were analyzed using the Luciferase Reporter Assay system (Promega). Experiments were performed in triplicate multiple times.
EMSAs.
Double-stranded DNA oligomers were designed with 5′ EcoRI and 3′ BamHI overhangs and annealed in STE buffer (20 mM TRIS HCI ph 7.4, 0.15 M NaCl, 1mM EDTA, 1% aprotinin). The annealed oligomers were ligated into pCMX, and the probe was excised with XhoI and NheI digestion, then end-labeled with [α-32P]dATP using the large fragment of E. coli polymerase I (Klenow fragment). The complete probe sequences are shown in Figure 3; the wild-type and mutated PPREs were as follows (PPREs are underlined): A, 5′-GTTTCTTAGTGAAAGGTCATGAAGC-3′; mutated A, 5′-GTTTCTTCCTGAAACCTAATGAAGC-3′; B, 5′-AAAGCCAAGGCATAGGTCAGAGTAA-3′; mutated B, 5′-AAAGCCACCGCATACCTAAGAGTAA-3′; C, 5′-CTGATTTGCCCTCTGACCTCCACTT-3′; mutated C, 5′-CTGATTTGCAATCTCAAATCCACTT-3′; D, 5′- CACATGTGTTCTGTGACCTTTGCTA-3′; mutated D, 5′-CACATGTGTAATGTCAAATTTGCTA-3′.
In vitro translation of pCMX-PPARγ2 and pCMX-RXRα was performed using the TnT-coupled reticulocyte lysate system (Promega) as recommended by the manufacturer. Recombinant proteins (2 μl of each) were incubated with 0.2 μg of radiolabeled probe in a reaction buffer consisting of 40 mM KCl, 10 mM Tris-HCl (pH 7.5), 0.2 mM EDTA, 1 mM DTT, 6% (vol/vol) glycerol, 0.1% (vol/vol) NP-40, and 2 μg poly (dI-dC) (Roche Diagnostics Corp.) in a total volume of 20 μl. These reactions were allowed to proceed for 20 minutes at room temperature, then loaded onto a 6% polyacrylamide nondenaturing gel and resolved in ×0.5 tris-borate-EDTA buffer (TBE). For the cold competition experiments, an unlabeled oligonucleotide containing the wild-type PPRE DR-1 sequence was added to the reaction buffer (2 μl of 200 nM, 2 μM, and 20 μM stock solutions) and preincubated for 20 minutes at room temperature before the radiolabeled probe was added.
ChiP assays.
The ChIP assay was modified from a published protocol (85) as follows. 3T3-L1 adipocytes were treated on day 8 with 1 μM rosiglitazone for 48 hours. Chromatin was sonicated to an average size of 300–400 bp, and 400 μg/ml of total DNA was used per immunoprecipitation. Ten micrograms of the following antibodies were used: normal rabbit IgG, PPARγ, CBP, SRC-1, and PBP (Santa Cruz Biotechnology Inc.), acetylated histone H3 and H4 (Upstate Biotechnology), and SMRT and N-CoR (ABR — Affinity Bioreagents). All assays were repeated at least 3 times, with similar results. Primers used for ChIP PCR were as follows: A forward, 5′-TGAACAAACAAGTCGAACCATC-3′; A reverse, 5′-TGTGGGTGGGGAGAATTATATC-3′; B forward, 5′-AGCCAAGGCATAGGTCAGAGTA-3′; B reverse, 5′-ATGTTCCAATGGCCAGGTATAG-3′; C forward, 5′-CAGAGATAGGAGGGAATCTGGA-3′; C reverse, 5′-TATGTGTGCAAAACACAGATGC-3′; D forward, 5′-ACATGTGTTCTGTGACCTTTGC-3′; D reverse, 5′-CTGGGACTTTTCTCTTGTGCTT-3′; aP2 forward, 5′-ATGTCACAGGCATCTTATCCACC-3′; aP2 reverse, 5′-AACCCTGCCAAAGAGACAGAGG-3′.
Adenoviral overexpression of OLR1.
The open reading frame of OLR1 was cloned and FLAG-tagged using PCR of 3T3-L1 adipocyte cDNA with the following primers: forward, 5′-CCACCATGGATTACAAGGATGACGACGATAAGACTTTTGATGACAAGATG-3′; reverse, 5′-CTTCTCCAGAATCTTTAGATTCACTAAATTTGCAAATGA-3′. The OLR1 ORF was cloned into pCR2.1-TOPO using the TOPO TA Cloning Kit (Invitrogen Corp.) and sequence verified. The insert was excised using KpnI and EcoRV digestion and ligated into the KpnI and EcoRV sites of pCMX. FLAG-OLR1 recombinant protein expression was confirmed by transfection into HEK 293T cells and Western blotting of the protein lysates with an anti-FLAG antibody (Sigma-Aldrich) and an anti-OLR1 antibody (Y-21; Santa Cruz Biotechnology Inc.). Adenovirus expressing OLR1 was generated using the BD Adeno-X Expression System (BD Biosciences — Clontech) according to the manufacturer’s protocol. Briefly, the FLAG-OLR1 cDNA insert from pCR2.1-TOPO was excised with BamHI and XbaI digestion and cloned into the same sites in the pDNR-CMV (BD Biosciences — Clontech) vector. Recombinants were sequence verified and transfected into HEK 293T cells to ensure that the protein was being expressed under the new CMV promoter. The insert was then recombined into the Adeno-X vector using Cre recombinase. The adenoviral expression vector containing FLAG-OLR1 was linearized with PacI and transfected into HEK 293 cells (ATCC) using FuGENE. After 14 days, cells were harvested and subjected to freeze-thaw lysis; the cell lysate was then used to infect fresh HEK 293 cells. The cell lysates were harvested after 4 days and used to infect 3T3-L1 adipocytes. Adenovirus expressing EGFP was generated in parallel as a control, and the 2 adenoviruses were titered using the BD Adeno-X Rapid Titer Kit (BD Biosciences — Clontech). Adenovirus infection of 3T3-L1 adipocytes was performed as described previously (86). Equal titers of adeno-GFP and adeno-OLR1 were used, which resulted in a greater than 95% infection efficiency. Cells were infected on day 5 after differentiation, and experiments were performed on days 8–10.
Western blotting.
3T3-L1 cells were treated as described, and whole-cell extracts were made in RIPA buffer. Equal amounts of protein were subjected to SDS-PAGE and transferred to PVDF membranes. OLR1 primary antibody (Y-21) and anti-goat secondary antibody (Santa Cruz Biotechnology Inc.) were used to probe for OLR1 protein, and ECL Plus (Amersham Biosciences) was used for detection.
Lentiviral RNA interference of OLR1.
The BLOCK-iT Lentiviral siRNA Expression System (Invitrogen Corp.) was used to construct siRNA vectors for OLR1 according to the manufacturer’s protocol. The siRNA sequence for OLR1, 5′-AAGTCATGTGGCAAGAAGCCT-3′, was designed as a hairpin sequence and cloned into the U6 promoter Entry Vector (Invitrogen Corp.), then recombined into the pLenti6/BLOCK-iT siRNA vector. This vector was sequence verified and used for lentivirus production at the Vector Core of the University of Pennsylvania. A lentivirus siRNA vector against human lamin A/C provided in the kit was used as a negative control. Equal titers of lentivirus were mixed with DMEM containing 10% FBS and 6 μg/ml hexadimethrine bromide (Sigma-Aldrich), and 250 μl of the virus media was added to each well of a 24-well plate of 3T3-L1 adipocytes on day 4 after differentiation. The plate was spin-infected at 300 g for 1 hour at 30°C immediately and once again 24 hours later. Fresh media was added on day 6, and the cells were treated with DMSO or 1 μM rosiglitazone on day 9 for 48 hours. Reduction of OLR1 levels in the cells was confirmed by Western blotting.
[125I]oxLDL uptake assays.
Uptake assays were performed essentially as described previously (61). [125I]oxLDL and unlabeled human LDL and oxLDL were purchased from BTI Inc. A standard protocol was used by the manufacturer, in which oxLDL was prepared by exposing LDL to 20 μM CuSO4 for 24 hours at 37°C. Thiobarbituric acid–reactive substances were determined using malondialdehyde as a standard to evaluate the degree of oxidation (oxLDL is 200- to 300-fold more reactive than starting LDL), and oxLDL was evaluated for receptor binding to macrophages. Day 10 3T3-L1 adipocytes treated for 48 hours with vehicle or rosiglitazone were serum starved for 1 hour in DMEM with 2% fatty acid–free BSA (Sigma-Aldrich). [125I]oxLDL (0.25–1 μCi) was added to each well of a 12-well plate, and the cells were incubated at 37°C for 4–5 hours. Nonspecific binding was measured in the presence of a 20-fold excess of unlabeled oxLDL, and results were subtracted from the raw values. The cells were washed 3 times in PBS plus 0.5% BSA and 3 times in PBS, then lysed with 0.1% SDS in PBS. Radioactivity was assessed in a gamma counter. All experiments were repeated at least 3 times.
Total cholesterol assays.
Day 8 3T3-L1 adipocytes in 24-well plates were incubated in DMEM containing 10% FBS and oxLDL (1–10 μg/ml) for 48–72 hours. Cells were washed with PBS, then lipids were extracted in 250 μl isopropanol for 6–12 hours. Total cholesterol levels were assayed with a colorimetric kit (Wako Chemicals USA Inc.). All experiments were performed in quadruplicate at least 3 times.
Fatty acid uptake assays.
Assays were performed essentially as described previously (44). Briefly, day 8 3T3-L1 adipocytes were incubated in DMEM with 10% FBS and oxLDL (1–10 μg/ml) for 48 hours, then preincubated for 4 hours in serum-free DMEM with 0.2% BSA (fatty acid–free). Media was changed to Krebs-Ringer phosphate (KRP) buffer with 0.2% BSA for 1 hour. A 100 μM 1:1 palmitate/BSA mixture in KRP with 5 mM glucose, containing trace amounts of [3H]palmitic acid (PerkinElmer), was added to each well, and uptake was performed for 5 minutes at 37°C. The media was then aspirated and the wells were washed 3 times with cold PBS containing 0.1% BSA and 200 μM phloretin. Cells were lysed with 0.1% SDS/PBS, and radioactivity was quantitated in a scintillation counter. Experiments were performed in triplicate multiple times.
Measurement of serum oxLDL levels.
Assays were performed using the Oxidized LDL ELISA immunoassay kit as described by the manufacturer (Mercodia).
Statistical analysis.
All results are expressed as mean ± SEM. Statistical significance was determined using either the 2-tailed Student’s t test or ANOVA, as appropriate, and P < 0.05 was deemed significant.
Acknowledgments
We thank D. Baldwin of the Penn Microarray facility, and the Functional Genomics Core and Viral Vector Core of the Penn Diabetes Center (DK19525). We also thank T. Willson and GlaxoSmithKline for providing GW7845. This work was funded by NIDDK R01 DK49780, and an unrestricted Bristol Myers Squibb Freedom to Discover Award in Metabolic Research (M.A. Lazar). P.C. Chui was funded by the Medical Scientist Training Program of the University of Pennsylvania.
Footnotes
Nonstandard abbreviations used: CBP, CREB binding protein; ChIP, chromatin immunoprecipitation; DR-1, direct repeat 1; GLUT4, glucose transporter 4; HMG CoA, 3-hydroxy-3-methylglutaryl-CoA; N-CoR, nuclear receptor corepressor; OLR1, oxidized LDL receptor 1; oxLDL, oxidized LDL; PBP, PPARγ binding protein; PPRE, peroxisome proliferator–activated response element; RXRα, retinoid X receptor α; SR-B1, scavenger receptor–B1; siRNA, short interfering RNA; SMRT, silencing mediator for retinoid and thyroid receptors; SRC, steroid receptor coactivator; TZD, thiazolidinedione.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Camp HS, Ren D, Leff T. Adipogenesis and fat-cell function in obesity and diabetes. Trends Mol. Med. 2002;8:442–447. doi: 10.1016/s1471-4914(02)02396-1. [DOI] [PubMed] [Google Scholar]
- 2.Boden G, Lebed B, Schatz M, Homko C, Lemieux S. Effects of acute changes of plasma free fatty acids on intramyocellular fat content and insulin resistance in healthy subjects. Diabetes. 2001;50:1612–1617. doi: 10.2337/diabetes.50.7.1612. [DOI] [PubMed] [Google Scholar]
- 3.Kuhlmann J, et al. Intramyocellular lipid and insulin resistance: a longitudinal in vivo 1H-spectroscopic study in Zucker diabetic fatty rats. Diabetes. 2003;52:138–144. doi: 10.2337/diabetes.52.1.138. [DOI] [PubMed] [Google Scholar]
- 4.Larsen TM, Toubro S, Astrup A. PPARgamma agonists in the treatment of type II diabetes: is increased fatness commensurate with long-term efficacy? [review] Int. J. Obes. Relat. Metab. Disord. 2003;27:147–161. doi: 10.1038/sj.ijo.802223. [DOI] [PubMed] [Google Scholar]
- 5.Friedman JM. Obesity in the new millennium. Nature. 2000;404:632–634. doi: 10.1038/35007504. [DOI] [PubMed] [Google Scholar]
- 6.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 7.Hahn P. Cholesterol metabolism in obese mice. Can. J. Biochem. 1980;58:1258–1260. doi: 10.1139/o80-168. [DOI] [PubMed] [Google Scholar]
- 8.Boizard M, et al. Obesity-related overexpression of fatty-acid synthase gene in adipose tissue involves sterol regulatory element-binding protein transcription factors. J. Biol. Chem. 1998;273:29164–29171. doi: 10.1074/jbc.273.44.29164. [DOI] [PubMed] [Google Scholar]
- 9.Le Lay S, et al. Cholesterol, a cell size-dependent signal that regulates glucose metabolism and gene expression in adipocytes. J. Biol. Chem. 2001;276:16904–16910. doi: 10.1074/jbc.M010955200. [DOI] [PubMed] [Google Scholar]
- 10.Dugail I, et al. New insights into how adipocytes sense their triglyceride stores. Is cholesterol a signal? Horm. Metab. Res. 2003;35:204–210. doi: 10.1055/s-2003-39475. [DOI] [PubMed] [Google Scholar]
- 11.Guerre-Millo M, Guesnet P, Guichard C, Durand G, Lavau M. Alteration in membrane lipid order and composition in metabolically hyperactive fatty rat adipocytes. Lipids. 1994;29:205–209. doi: 10.1007/BF02536730. [DOI] [PubMed] [Google Scholar]
- 12.Storch J, Shulman SL, Kleinfeld AM. Plasma membrane lipid order and composition during adipocyte differentiation of 3T3F442A cells. Studies in intact cells with 1-[4-(trimethylamino)phenyl]-6-phenylhexatriene. J. Biol. Chem. 1989;264:10527–10533. [PubMed] [Google Scholar]
- 13.Kovanen PT, Nikkila EA, Miettinen TA. Regulation of cholesterol synthesis and storage in fat cells. J. Lipid Res. 1975;16:211–223. [PubMed] [Google Scholar]
- 14.Schreibman PH, Dell RB. Human adipocyte cholesterol. Concentration, localization, synthesis, and turnover. J. Clin. Invest. 1975;55:986–993. doi: 10.1172/JCI108028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pohl J, et al. Long-chain fatty acid uptake into adipocytes depends on lipid raft function. Biochemistry. 2004;43:4179–4187. doi: 10.1021/bi035743m. [DOI] [PubMed] [Google Scholar]
- 16.Auwerx J. PPARγ, the ultimate thrifty gene. Diabetologia. 1999;42:1033–1049. doi: 10.1007/s001250051268. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 18.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor γ (PPARγ): adipose predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 19.Barak Y, et al. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 20.Rosen ED, et al. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 21.Horlein AJ, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 22.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 23.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 24.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for the nuclear peroxisome proliferator-activated receptor γ (PPARγ) J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 25.Willson TM, et al. The structure-activity relationship between peroxisome proliferator-activated receptor γ and the antihyperglycemic activity of thiazolidinediones. J. Med. Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 26.Berger J, et al. Thiazolidinediones produce a conformational change in peroxisome proliferator-activated receptor-γ: binding and activation correlate with antidiabetic actions in db/db mice. Endocrinology. 1996;137:4189–4195. doi: 10.1210/endo.137.10.8828476. [DOI] [PubMed] [Google Scholar]
- 27.Way JM, et al. Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor gamma activation has coordinate effects on gene expression in multiple insulin-sensitive tissues. Endocrinology. 2001;142:1269–1277. doi: 10.1210/endo.142.3.8037. [DOI] [PubMed] [Google Scholar]
- 28.de Souza CJ, et al. Effects of pioglitazone on adipose tissue remodeling within the setting of obesity and insulin resistance. Diabetes. 2001;50:1863–1871. doi: 10.2337/diabetes.50.8.1863. [DOI] [PubMed] [Google Scholar]
- 29.Chao L, et al. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. J. Clin. Invest. 2000;106:1221–1228. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamauchi T, et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J. Biol. Chem. 2001;276:41245–41254. doi: 10.1074/jbc.M103241200. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Lazar MA. Differential gene regulation by PPARγ agonist and constitutively active PPARγ. Mol. Endocrinol. 2002;16:1040–1048. doi: 10.1210/mend.16.5.0825. [DOI] [PubMed] [Google Scholar]
- 32.Steppan CM, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 33.Guan HP, et al. A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nat. Med. 2002;8:1122–1128. doi: 10.1038/nm780. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee RR, et al. Regulation of fasted blood glucose by resistin. Science. 2004;303:1195–1198. doi: 10.1126/science.1092341. [DOI] [PubMed] [Google Scholar]
- 35.Tordjman J, et al. Thiazolidinediones block fatty acid release by inducing glyceroneogenesis in fat cells. J. Biol. Chem. 2003;278:18785–18790. doi: 10.1074/jbc.M206999200. [DOI] [PubMed] [Google Scholar]
- 36.Patsouris D, et al. PPARalpha governs glycerol metabolism. J. Clin. Invest. 2004;114:94–103. doi:10.1172/JCI200420468. doi: 10.1172/JCI20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawamura T, et al. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386:73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- 38.Kletzien RF, Clarke SD, Ulrich RG. Enhancement of adipocyte differentiation by an insulin-sensitizing agent. Mol. Pharmacol. 1992;41:393–398. [PubMed] [Google Scholar]
- 39.Takamura T, Nohara E, Nagai Y, Kobayashi K. Stage-specific effects of a thiazolidinedione on proliferation, differentiation and PPARgamma mRNA expression in 3T3-L1 adipocytes. Eur. J. Pharmacol. 2001;422:23–29. doi: 10.1016/s0014-2999(01)01053-6. [DOI] [PubMed] [Google Scholar]
- 40.Moore KJ, et al. The role of PPAR-gamma in macrophage differentiation and cholesterol uptake. Nat. Med. 2001;7:41–47. doi: 10.1038/83328. [DOI] [PubMed] [Google Scholar]
- 41.Nugent C, et al. Potentiation of glucose uptake in 3T3-L1 adipocytes by PPAR gamma agonists is maintained in cells expressing a PPAR gamma dominant-negative mutant: evidence for selectivity in the downstream responses to PPAR gamma activation. Mol. Endocrinol. 2001;15:1729–1738. doi: 10.1210/mend.15.10.0715. [DOI] [PubMed] [Google Scholar]
- 42.Camp HS, Chaudhry A, Leff T. A novel potent antagonist of peroxisome proliferator-activated receptor gamma blocks adipocyte differentiation but does not revert the phenotype of terminally differentiated adipocytes. Endocrinology. 2001;142:3207–3213. doi: 10.1210/endo.142.7.8254. [DOI] [PubMed] [Google Scholar]
- 43.Itabe H. Oxidized low-density lipoproteins: what is understood and what remains to be clarified [review] Biol. Pharm. Bull. 2003;26:1–9. doi: 10.1248/bpb.26.1. [DOI] [PubMed] [Google Scholar]
- 44.Frohnert BI, Hui TY, Bernlohr DA. Identification of a functional peroxisome proliferator-responsive element in the murine fatty acid transport protein gene. J. Biol. Chem. 1999;274:3970–3977. doi: 10.1074/jbc.274.7.3970. [DOI] [PubMed] [Google Scholar]
- 45.Wyne KL. Free fatty acids and type 2 diabetes mellitus. Am. J. Med. 2003;115(Suppl. 8A):29S–36S. doi: 10.1016/j.amjmed.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 46.Rangwala SM, et al. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev. Cell. 2003;5:657–663. doi: 10.1016/s1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- 47.Chen M, Masaki T, Sawamura T. LOX-1, the receptor for oxidized low-density lipoprotein identified from endothelial cells: implications in endothelial dysfunction and atherosclerosis. Pharmacol. Ther. 2002;95:89–100. doi: 10.1016/s0163-7258(02)00236-x. [DOI] [PubMed] [Google Scholar]
- 48.Tsimikas S, Witztum JL. Measuring circulating oxidized low-density lipoprotein to evaluate coronary risk. Circulation. 2001;103:1930–1932. doi: 10.1161/01.cir.103.15.1930. [DOI] [PubMed] [Google Scholar]
- 49.Ehara S, et al. Elevated levels of oxidized low density lipoprotein show a positive relationship with the severity of acute coronary syndromes. Circulation. 2001;103:1955–1960. doi: 10.1161/01.cir.103.15.1955. [DOI] [PubMed] [Google Scholar]
- 50.Steinberg D, Parthasarathy S, Carew TE, Khoo JC, Witztum JL. Beyond cholesterol. Modifications of low-density lipoprotein that increase its atherogenicity. N. Engl. J. Med. 1989;320:915–924. doi: 10.1056/NEJM198904063201407. [DOI] [PubMed] [Google Scholar]
- 51.Holvoet P, Vanhaecke J, Janssens S, Van de Werf F, Collen D. Oxidized LDL and malondialdehyde-modified LDL in patients with acute coronary syndromes and stable coronary artery disease. Circulation. 1998;98:1487–1494. doi: 10.1161/01.cir.98.15.1487. [DOI] [PubMed] [Google Scholar]
- 52.Berliner JA, et al. Minimally modified low density lipoprotein stimulates monocyte endothelial interactions. J. Clin. Invest. 1990;85:1260–1266. doi: 10.1172/JCI114562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinecke JW. Oxidants and antioxidants in the pathogenesis of atherosclerosis: implications for the oxidized low density lipoprotein hypothesis. Atherosclerosis. 1998;141:1–15. doi: 10.1016/s0021-9150(98)00173-7. [DOI] [PubMed] [Google Scholar]
- 54.Navab M, et al. Monocyte transmigration induced by modification of low density lipoprotein in cocultures of human aortic wall cells is due to induction of monocyte chemotactic protein 1 synthesis and is abolished by high density lipoprotein. J. Clin. Invest. 1991;88:2039–2046. doi: 10.1172/JCI115532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vora DK, et al. Induction of P-selectin by oxidized lipoproteins. Separate effects on synthesis and surface expression. Circ. Res. 1997;80:810–818. doi: 10.1161/01.res.80.6.810. [DOI] [PubMed] [Google Scholar]
- 56.Podrez EA, Schmitt D, Hoff HF, Hazen SL. Myeloperoxidase-generated reactive nitrogen species convert LDL into an atherogenic form in vitro. J. Clin. Invest. 1999;103:1547–1560. doi: 10.1172/JCI5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling W, et al. Oxidized or acetylated low density lipoproteins are rapidly cleared by the liver in mice with disruption of the scavenger receptor class A type I/II gene. J. Clin. Invest. 1997;100:244–252. doi: 10.1172/JCI119528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shoji T, et al. Inverse relationship between circulating oxidized low density lipoprotein (oxLDL) and anti-oxLDL antibody levels in healthy subjects. Atherosclerosis. 2000;148:171–177. doi: 10.1016/s0021-9150(99)00218-x. [DOI] [PubMed] [Google Scholar]
- 59.Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 1993;268:17665–17668. [PubMed] [Google Scholar]
- 60.Acton SL, Scherer PE, Lodish HF, Krieger M. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J. Biol. Chem. 1994;269:21003–21009. [PubMed] [Google Scholar]
- 61.Kuniyasu A, Hayashi S, Nakayama H. Adipocytes recognize and degrade oxidized low density lipoprotein through CD36. Biochem. Biophys. Res. Commun. 2002;295:319–323. doi: 10.1016/s0006-291x(02)00666-6. [DOI] [PubMed] [Google Scholar]
- 62.Roberts AW, Thomas A, Rees A, Evans M. Peroxisome proliferator-activated receptor-gamma agonists in atherosclerosis: current evidence and future directions. Curr. Opin. Lipidol. 2003;14:567–573. doi: 10.1097/00041433-200312000-00004. [DOI] [PubMed] [Google Scholar]
- 63.Gilling L, Suwattee P, DeSouza C, Asnani S, Fonseca V. Effects of the thiazolidinediones on cardiovascular risk factors. Am. J. Cardiovasc. Drugs. 2002;2:149–156. doi: 10.2165/00129784-200202030-00002. [DOI] [PubMed] [Google Scholar]
- 64.Sidhu JS, Cowan D, Kaski JC. The effects of rosiglitazone, a peroxisome proliferator-activated receptor-gamma agonist, on markers of endothelial cell activation, C-reactive protein, and fibrinogen levels in non-diabetic coronary artery disease patients. J. Am. Coll. Cardiol. 2003;42:1757–1763. doi: 10.1016/j.jacc.2003.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Li AC, et al. Peroxisome proliferator-activated receptor gamma ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J. Clin. Invest. 2000;106:523–531. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z, et al. Troglitazone inhibits atherosclerosis in apolipoprotein E-knockout mice: pleiotropic effects on CD36 expression and HDL. Arterioscler. Thromb. Vasc. Biol. 2001;21:372–377. doi: 10.1161/01.atv.21.3.372. [DOI] [PubMed] [Google Scholar]
- 67.Chawla A, et al. A PPARγ-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 68.Li AC, et al. Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma. J. Clin. Invest. 2004;114:1564–1576. doi:10.1172/JCI200418730. doi: 10.1172/JCI18730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiba Y, Ogita T, Ando K, Fujita T. PPARgamma ligands inhibit TNF-alpha-induced LOX-1 expression in cultured endothelial cells. Biochem. Biophys. Res. Commun. 2001;286:541–546. doi: 10.1006/bbrc.2001.5361. [DOI] [PubMed] [Google Scholar]
- 70.Armoni M, et al. Peroxisome proliferator-activated receptor-gamma represses GLUT4 promoter activity in primary adipocytes, and rosiglitazone alleviates this effect. J. Biol. Chem. 2003;278:30614–30623. doi: 10.1074/jbc.M304654200. [DOI] [PubMed] [Google Scholar]
- 71.Guan HP, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 2005;19:453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gavrilova O, et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 73.Hevener AL, et al. Muscle-specific Pparg deletion causes insulin resistance. Nat. Med. 2003;9:1491–1497. doi: 10.1038/nm956. [DOI] [PubMed] [Google Scholar]
- 74.Norris AW, et al. Muscle-specific PPARgamma-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J. Clin. Invest. 2003;112:608–618. doi:10.1172/JCI200317305. doi: 10.1172/JCI17305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cock TA, Houten SM, Auwerx J. Peroxisome proliferator-activated receptor-gamma: too much of a good thing causes harm [review] EMBO Rep. 2004;5:142–147. doi: 10.1038/sj.embor.7400082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee CH, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors [review] Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 77.Rangwala SM, Lazar MA. Peroxisome proliferator-activated receptor gamma in diabetes and metabolism. Trends Pharmacol. Sci. 2004;25:331–336. doi: 10.1016/j.tips.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 78.Boden G. Role of fatty acids in the pathogenesis of insulin resistance and NIDDM. Diabetes. 1997;46:1–10. [PubMed] [Google Scholar]
- 79.Danforth E., Jr Failure of adipocyte differentiation causes type II diabetes mellitus? [letter] Nat. Genet. 2000;26:13. doi: 10.1038/79111. [DOI] [PubMed] [Google Scholar]
- 80.Fonseca V. Effect of thiazolidinediones on body weight in patients with diabetes mellitus. Am. J. Med. 2003;115(Suppl. 8A):42S–48S. doi: 10.1016/j.amjmed.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 81.Okuno A, et al. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J. Clin. Invest. 1998;101:1354–1361. doi: 10.1172/JCI1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson-Fritch L, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Invest. 2004;114:1281–1289. doi:10.1172/JCI200421752. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye JM, et al. Direct demonstration of lipid sequestration as a mechanism by which rosiglitazone prevents fatty-acid-induced insulin resistance in the rat: comparison with metformin. Diabetologia. 2004;47:1306–1313. doi: 10.1007/s00125-004-1436-1. [DOI] [PubMed] [Google Scholar]
- 84.Chawla A, Lazar MA. Peroxisome proliferator and retinoid signalling pathways coregulate preadipocyte phenotype and survival. Proc. Natl. Acad. Sci. U. S. A. 1994;91:1786–1790. doi: 10.1073/pnas.91.5.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hartman HB, Hu X, Tyler KX, Dalal CK, Lazar MA. Mechanisms regulating adipocyte expression of resistin. J. Biol. Chem. 2002;277:19754–19761. doi: 10.1074/jbc.M201451200. [DOI] [PubMed] [Google Scholar]
- 86.Orlicky DJ, Schaack J. Adenovirus transduction of 3T3-L1 cells. J. Lipid Res. 2001;42:460–466. [PubMed] [Google Scholar]