Abstract
Chronic infection with cagA-positive Helicobacter pylori is associated with the development of atrophic gastritis, peptic ulcers, and gastric adenocarcinoma. The cagA gene product CagA is injected into gastric epithelial cells, where it undergoes tyrosine phosphorylation by Src family kinases. Translocated CagA disturbs cellular functions by physically interacting with and deregulating intracellular signaling transducers through both tyrosine phosphorylation-dependent and -independent mechanisms. To gain further insights into the pathophysiological activities of CagA in gastric epithelial cells, we executed a genome-wide screening of CagA-responsive genes by using DNA microarray and identified nuclear factor of activated T cells (NFAT) transcription factors whose binding sites were overrepresented in the promoter regions of CagA-activated genes. Results of reporter assays confirmed that CagA was capable of activating NFAT in a manner independent of CagA phosphorylation. Expression of CagA in gastric epithelial cells provoked translocation of NFATc3, a member of the NFAT family, from the cytoplasm to the nucleus and activated an NFAT-regulated gene, p21WAF1/Cip1. CagA-mediated NFAT activation was abolished by inhibiting calcineurin or phospholipase Cγ activity. Furthermore, treatment of cells with H. pylori VacA (vacuolating toxin), which inhibits NFAT activity in T lymphocytes, counteracted the ability of CagA to activate NFAT in gastric epithelial cells. These findings indicate that the two major H. pylori virulence factors inversely control NFAT activity. Considering the pleiotropic roles of NFAT in cell growth and differentiation, deregulation of NFAT, either positively or negatively, depending on the relative exposure of cells to CagA and VacA, may contribute to the various disease outcomes caused by H. pylori infection.
Keywords: nuclear factor of activated T cells, p21WAF1/Cip1, calcineurin, phospholipase Cγ
Infection with Helicobacter pylori is known to be associated with the development of chronic atrophic gastritis, peptic ulcers, and gastric adenocarcinoma (1–3). However, the molecular mechanisms that underlie the development of H. pylori-associated gastroduodenal diseases remain to be elucidated. Also, factors that determine the clinical outcome of an H. pylori-infected individual are not known. H. pylori can be subclassified into cagA-positive and cagA-negative strains based on the presence or absence of the cagA gene, which encodes a 120- to 145-kDa CagA protein (4). cagA is located at one end of the cag pathogenicity island (cagPAI), a 40-kb DNA fragment that also encodes molecules constituting the bacterial type IV injection system (5, 6).
Upon direct contact of H. pylori cagA-positive strains with gastric epithelial cells, CagA is injected from the bacteria into the host cells by means of the type IV injection system (7, 8). Translocated CagA localizes to the inner surface of the plasma membrane and undergoes tyrosine phosphorylation by the Src family of protein tyrosine kinases at its unique EPIYA motifs (8). Tyrosine-phosphorylated CagA specifically binds to the SH2 domain-containing protein tyrosine phosphatase 2 (SHP-2) and deregulates the phosphatase activity, thereby inducing an elongated cell shape termed the “hummingbird phenotype” (9, 10). SHP-2 has been shown to play a crucial role in both cell growth and movement, and gain-of function mutation of SHP-2 has been shown to be associated with a variety of human malignancies (11, 12). These findings indicate that SHP-2 is a bona fide human oncoprotein and suggest that deregulation of SHP-2 by CagA plays an important role in gastric carcinogenesis. CagA also interacts and activates the C-terminal Src kinase (Csk) in a phosphorylation-dependent manner, and this interaction has been suspected to attenuate CagA-SHP-2 signaling through inhibition of Src family kinases, which mediate CagA phosphorylation (13).
In addition to SHP-2 and Csk, CagA has been reported to bind Grb2, c-Met receptor, or phospholipase Cγ (PLCγ) in a manner independent of CagA tyrosine phosphorylation (14, 15). Interaction of CagA with these molecules also stimulates cell motility. Furthermore, CagA colocalizes with the tight junction protein ZO-1 and destroys cell–cell junctions in a manner independent of tyrosine phosphorylation (16). Collectively, these observations indicate that CagA functions as a bacterial adaptor/scaffolding protein that recruits multiple cellular proteins and deregulates a variety of cellular functions.
To systemically investigate cellular responses to H. pylori CagA, we analyzed changes in gene expression caused by ectopic expression of the cagA gene in gastric epithelial cells by using a DNA microarray. Analyses of gene sets that are specifically induced by CagA revealed the existence of the NFAT (nuclear factor of activated T cells) family of transcription factors, whose binding sites were significantly overrepresented in the promoters of CagA-activated genes when compared with the control gene set. CagA activated NFAT in gastric epithelial cells by inducing translocation of NFAT from the cytoplasm to the nucleus. Intriguingly, another H. pylori virulence factor, VacA (vacuolating toxin), counteracted the CagA activity for translocation of NFAT. We discuss the significance of NFAT deregulation by the H. pylori virulence factors, CagA and VacA, in both H. pylori pathogenicity and disease outcome of H. pylori infection.
Materials and Methods
Plasmid. Mammalian expression vectors for hemagglutinin-tagged WT CagA (ABCCC type WT-CagA) and its phosphorylation-resistant (PR) derivative (PR-CagA) are described in ref. 9. ΔABCCC CagA was made from WT-CagA by deleting the EPIYA-containing region (amino acids 869-1086). pNFAT-luc reporter plasmid was purchased from Stratagene. pGL3-p21-luc was made by inserting the SP-1 containing the p21WAF1/Cip1 promoter sequence into the pGL3 basic promoter (Promega) (17).
Cell Culture and Transfection. AGS human gastric epithelial cells were grown in RPMI medium 1640 supplemented with 10% FBS and were transfected by using Lipofectamine 2000 (Invitrogen) as described in ref. 10. At 18 h after transfection, cells were harvested or treated with 10 μg/ml cyclosporin A (CsA), 8 μM U73122, 100 nM phorbol 12-myristate 13-acetate (PMA), 0.5 μM calcium ionophore A23187, or 2.5 μg/ml VacA for an additional 6 h. VacA was purified from H. pylori ATCC49503 strain culture supernatant by using an anti-VacA antibody column as described in ref. 18.
Microarray Analysis. Total RNA was extracted from AGS cells by using TriZol Reagent (Invitrogen) and processed according to the protocol recommended by Affymetrix (Santa Clara, CA). Labeled cRNA samples were hybridized to the Human Genome Focus Array GeneChips (Affymetrix) (for details, see the legend of Table 1, which is published as supporting information of the PNAS web site).
Antibodies. Anti-hemagglutinin-epitope polyclonal antibody (Y-11), anti-SHP-2 polyclonal antibody (C-18), anti-p21WAF1/Cip1 polyclonal antibody (C-19), and anti-NFATc3 monoclonal antibody (F-1) were purchased from Santa Cruz Biotechnology. Anti-phosphotyrosine monoclonal antibody (4G10) was purchased from Upstate Biotechnology (Lake Placid, NY). Western blotting was performed as described in ref. 10.
Immunostaining. Cells were fixed and treated with anti-NFATc3 antibody at 18 or 24 h after transfection. Primary antibodies were localized by Alexa Fluor 488-conjugated anti-mouse IgG antibody (Invitrogen). The nuclei were stained with DAPI. Images were acquired by using a confocal microscope system (Fluoview, Olympus, Tokyo).
Luciferase Reporter Assay. Cells were transfected with various combinations of reporter plasmid (pNFAT-luc or pGL3-p21-luc), internal control pRL-TK (Promega), and expression plasmid. Cells were harvested at 18 h after transfection, and luciferase activities of the lysates were measured by using the Dual-Luciferase Reporter assay system (Promega) following the manufacturer's protocol.
EMSA. EMSA was performed as described in ref. 19.
Results
Changes in Gene Expression Caused by H. pylori CagA. To systemically investigate the effect of H. pylori CagA on gene expression of host cells, we transiently transfected an expression vector for WT-CagA (H. pylori strain NCTC11637-derived Western CagA) (10) in AGS human gastric epithelial cells. CagA protein was detectable 12 h after transfection by Western blotting (data not shown). Total RNAs were isolated from these CagA-transfected AGS cells at 12, 15, 18, and 24 h after transfection and were subjected to a genome-wide DNA microarray analysis by using GeneChip Human Genome Focus Array 8500 (Affymetrics). The results revealed that 339 of the 8,500 genes examined were activated and that 145 genes were repressed at least at one of the above-stated time points (see Table 1). Among those cagA-responsive genes, 71 genes whose mRNA levels were elevated at 12 h were subjected to further analysis, because changes at this time point were considered to represent direct effects of CagA on gene transcription (see Table 2, which is published as supporting information of the PNAS web site). Putative promoter sequences of the 71 identified genes, spanning between -1,000 and -1 from the transcription initiation sites, were screened for the presence of binding cis elements for a panel of transcription factors by the motif program (http://motif.genome.jp). The promoter analysis revealed that binding sites for the NFAT family of transcription factors were significantly overrepresented in the promoter regions of CagA-activated genes when compared with those in randomly selected control genes (see Table 3, which is published as supporting information of the PNAS web site).
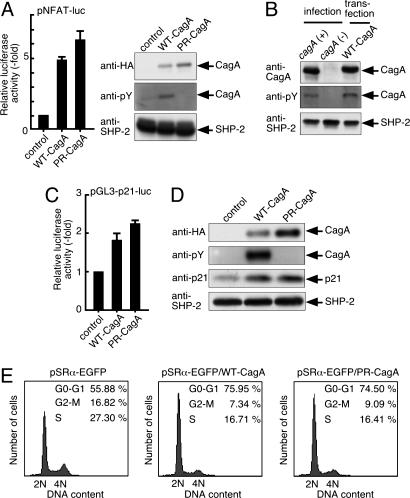
Activation of NFAT Transcription Factors by CagA. Given the above results, we examined whether CagA is capable of activating NFAT by using an NFAT-dependent luciferase reporter plasmid (pNFAT-luc). AGS cells were cotransfected with pNFAT-luc and WT-CagA expression vector. Luciferase assays of the cell lysates revealed that CagA specifically activated pNFAT-luc (Fig. 1A). The same result was also obtained when East Asian CagA (ABD type) was expressed or when another gastric epithelial cell line, MKN28, was used (data not shown). Because CagA elicits its biological activities both by tyrosine phosphorylation-dependent and -independent mechanisms, we next examined the effect of the PR-CagA, in which the tyrosine residues constituting the five EPIYA sites were substituted with alanines, and found that PR-CagA was also capable of activating pNFAT-luc (Fig. 1A). Thus, CagA activates NFAT-dependent gene transcription in a manner independent of its tyrosine phosphorylation in gastric epithelial cells. A stronger NFAT activation by PR-CagA than by WT-CagA may simply be due to differences in protein levels expressed. Alternatively, given that WT-CagA specifically binds SHP-2 and Csk in a phosphorylation-dependent manner, there is a possibility that WT-CagA molecules that have already formed complexes with SHP-2 or Csk cannot activate NFAT anymore. As shown in Fig. 1B, the level of transfected CagA in AGS cells was comparable with that of H. pylori-injected CagA. Thus, the cagA transfection system we used mimics naturally occurring CagA transduction by H. pylori infection.
Fig. 1.
CagA activates NFAT-dependent genes. (A) AGS cells were cotransfected with pNFAT-luc and WT-CagA or PR-CagA expression vector. Cell lysates were subjected to luciferase assay (Left) or immunoblotting (Right). The promoter activation was shown as a ratio of the luciferase activities in cells transfected with control empty vector (mean ± SD, n = 3 experiments). (B) AGS cells were transfected with WT-CagA expression vector for 18 h or infected with cagA-positive H. pylori strain NCTC11637 or its cagA-negative isogenic strain for 5 h at a multiplicity of infection of 50. Total cell lysates were subjected to immunoblotting. (C) AGS cells were cotransfected with pGL3-p21-luc and WT-CagA or PR-CagA expression vector. The promoter activation was shown as a ratio of the luciferase activities in cells transfected with control vector (mean ± SD, n = 3 experiments). (D) AGS cells were transfected with WT-CagA, PR-CagA, or control empty vector. Total cell lysates were subjected to immunoblotting. (E) AGS cells were transfected with EGFP, EGFP-tagged WT-CagA, or EGFP-tagged PR-CagA expression vector. After 36 h, cells were harvested, and GFP-positive cells were subjected to the cell-cycle analysis.
One of the few reported NFAT-target genes in nonimmune cells is p21WAF1/Cip1, which encodes a p21WAF1/Cip1 cyclin-dependent kinase inhibitor (hereafter abbreviated as p21) (17). Consistent with this, DNA microarray data showed that CagA expression resulted in an increase in p21 mRNA in AGS cells (see Table 2). To determine whether CagA transactivates p21 through NFAT, we synthesized the NFAT-responsive minimal promoter sequence of the p21 gene (from -56 to 28 of the p21 transcription initiation site) (17) and connected it upstream of the luciferase reporter gene (pGL3-p21-luc). Cotransfection experiments using pGL3-p21-luc showed that both WT-CagA and PR-CagA were capable of transactivating the p21 promoter (Fig. 1C). Given this, we next examined whether CagA increases the level of endogenous p21. Upon transient transfection of WT-CagA- or PR-CagA-expressing vector, p21 level was elevated in AGS cells (Fig. 1D). From these observations, we concluded that CagA activates NFAT and induces NFAT-dependent genes such as p21 in gastric epithelial cells. Consistent with the role of p21 in negative regulation of cell growth, AGS cells that express CagA, either WT or PR, exhibited G1 cell-cycle retardation (Fig. 1E).
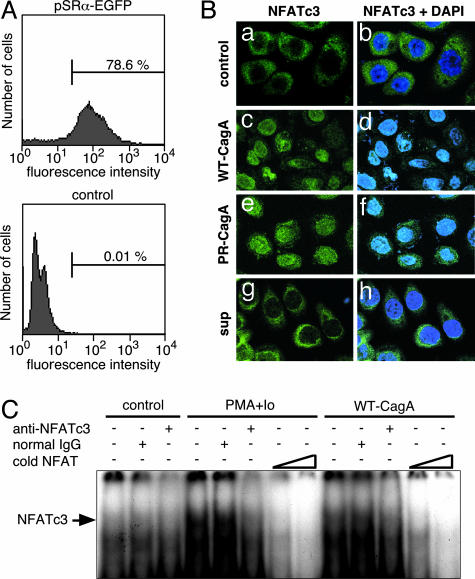
Nuclear Translocation of Cytoplasmic NFAT by CagA. In the absence of appropriate signals, NFAT is phosphorylated by glycogen synthase kinase-3β and is localized in the cytoplasm. Dephosphorylation of NFAT by calcineurin, a Ca2+/calmodulin-dependent serine/threonine phosphatase, elicits translocation of NFAT from the cytoplasm to the nucleus, where it transactivates NFAT-dependent genes (20). From the DNA microarray data, we noticed that NFATc3, a member of the NFAT family, is a major NFAT species expressed in AGS gastric epithelial cells. Anti-NFATc3 immunostaining revealed that NFATc3 was strictly localized to the cytoplasm in asynchronously proliferating AGS cells, and stimulation of cells with PMA and calcium ionophore A23187 elicited nuclear translocation of the cytoplasmic NFATc3 (data not shown). We then examined the effect of CagA on intracellular distribution of NFATc3. In our transient transfection experiment, the transfection efficiency was ≈80% in AGS cells (Fig. 2A). Transfection of WT-CagA expression vector resulted in nuclear staining of NFATc3 in a significant fraction of AGS cells (Fig. 2B c and d). Nuclear staining of NFATc3 was also observed in AGS cells transfected with PR-CagA expression vector (Fig. 2B e and f) but not in AGS cells transfected with control empty vector (Fig. 2B a and b). To exclude the possibility that CagA provoked NFAT translocation through paracrine mechanisms, we treated AGS cells with culture supernatants prepared from of CagA-transfected AGS cells. As shown in Fig. 2B g and h, the AGS culture supernatant did not induce nuclear translocation of NFATc3. Thus, CagA directly elicits nuclear translocation of cytoplasmic NFAT in gastric epithelial cells independent of CagA tyrosine phosphorylation. Nuclear translocation of NFAT by CagA was also investigated with the EMSA (Fig. 2C). A specific gel-shift band, which could be super-shifted by anti-NFATc3, was increased in nuclear extracts of AGS cells transfected with WT-CagA expression vector or treated with PMA plus A23187. The results of the EMSA confirmed that NFATc3 is a major NFAT species in AGS cells and that NFATc3 is translocated from the cytoplasm to the nucleus upon expression of CagA.
Fig. 2.
Nuclear translocation of NFATc3 by CagA. (A) AGS cells transfected with EGFP or control empty vector were harvested at 36 h after transfection and were subjected to flow cytometric analysis to calculate transfection efficiency. (B) AGS cells were transfected with WT-CagA (c and d), PR-CagA (e and f), or control empty vector (a and b) or were treated with the culture supernatant prepared from CagA-transfected AGS cells (g and h). After 18 h, cells were stained with anti-NFATc3 antibody and DAPI. (C) EMSA analysis of NFATc3 using the distal NFAT site of the human IL-2 promoter as a probe. The arrow indicates the position of the NFATc3-oligonucleotide complex.
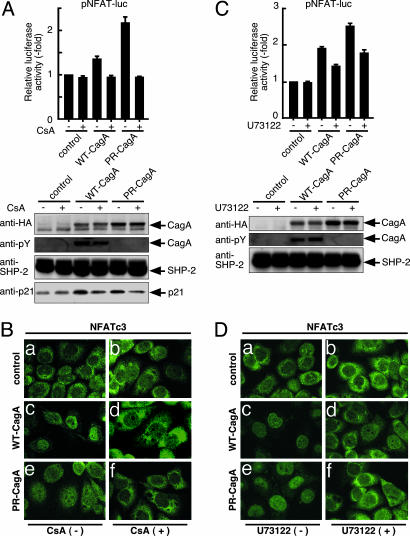
Requirement of Calcineurin or PLCγ Activity in CagA-Dependent Translocation and Activation of NFAT. Nuclear translocation of NFAT is physiologically induced by dephosphorylation of NFAT by calcineurin (20), which raised the possibility that CagA caused NFAT translocation by activating calcineurin. To address this, we examined the effect of CsA, a specific inhibitor of calcineurin, on CagA-mediated NFAT activation by transfecting pNFAT-luc together with WT-CagA or PR-CagA expression vector in AGS cells in the presence or absence of CsA. As shown in Fig. 3A, CsA abolished CagA-dependent NFAT activation as well as p21 induction. Consistently, nuclear translocation of NFAT by CagA was also inhibited by CsA (Fig. 3B). Because calcineurin is activated by cell-surface receptors coupled to store-operated Ca2+ entry by means of PLCγ (21), we next investigated whether PLCγ is involved in the NFAT activation by CagA. To do so, AGS cells cotransfected with pNFAT-luc and CagA expression vector were treated with a specific inhibitor of PLCγ, U73122, or were not treated. As shown in Fig. 3C, U73122 inhibited the CagA-dependent NFAT activation. Nuclear translocation of NFAT by CagA was also abolished by U73122 (Fig. 3D). From these observations, we concluded that CagA stimulates PLCγ, which in turn activates calcineurin. Activated calcineurin subsequently dephosphorylates cytoplasmic NFAT, resulting in its nuclear translocation.
Fig. 3.
Involvement of calcineurin and PLCγ in CagA-dependent NFAT activation. (A) AGS cells cotransfected with pNFAT-luc and WT-CagA or PR-CagA expression vector were cultured in the presence or absence of CsA, a specific inhibitor of calcineurin. (Upper) The promoter activation was shown as a ratio of the luciferase activities in cells transfected with control vector in the absence of CsA (mean ± SD, n = 3 experiments). (Lower) Total cell lysates were subjected to immunoblotting. (B) AGS cells transfected with WT-CagA, PR-CagA, or control empty vector were cultured in the presence (b, d, and f) or absence of CsA (a, c, and e). Cells were stained with anti-NFATc3 antibody. (C) AGS cells cotransfected with pNFAT-luc and WT-CagA or PR-CagA expression vector were cultured in the presence or absence of PLCγ inhibitor, U73122. (Upper) The promoter activation was shown as a ratio of the luciferase activities in cells transfected with control vector in the absence of U73122 (mean ± SD, n = 3 experiments). (Lower) Total cell lysates were subjected to immunoblotting. (D) AGS cells transfected with WT-CagA, PR-CagA, or control empty vector were cultured in the presence (b, d, and f) or absence (a, c, and e) of U73122. Cells were stained with anti-NFATc3 antibody.
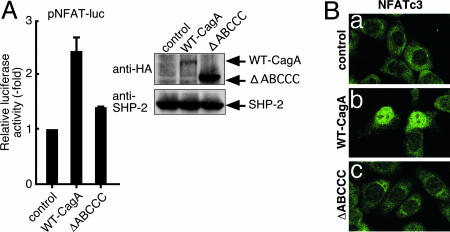
Delineation of the CagA Region Responsible for NFAT Activation. CagA is characterized by the presence of variations in the EPIYA-containing region, in which the EPIYA sites undergo tyrosine phosphorylation (8). SHP-2 and Csk bind specifically to CagA through the tyrosine-phosphorylated EPIYA sites (9, 13). To investigate whether the EPIYA-containing region is also involved in the NFAT activation by CagA, we cotransfected pNFAT-luc and WT-CagA or ΔABCCC CagA mutant that lacks the EPIYA-containing region (amino acids 869-1086) in AGS cells. The level of ΔABCCC CagA expressed was significantly higher than that of WT-CagA (Fig. 4A Right). However, the CagA mutant neither activated the pNFAT-luc reporter (Fig. 4A Left) nor induced nuclear translocation of cytoplasmic NFAT in AGS cells (Fig. 4B). Thus, the EPIYA-containing region is required for CagA to stimulate NFAT, although the CagA activity is independent of EPIYA tyrosine phosphorylation.
Fig. 4.
Requirement of the EPIYA-containing region in the activation of NFAT by CagA. (A) AGS cells were cotransfected with pNFAT-luc and WT-CagA or ΔABCCC CagA expression vector. Cell lysates were subjected to luciferase assay (Left) or immunoblotting (Right). The promoter activation was shown as a ratio of the luciferase activities in cells transfected with control empty vector (mean ± SD, n = 3 experiments). (B) AGS cells transfected with WT-CagA, ΔABCCC CagA or control empty vector were stained with anti-NFATc3 antibody.
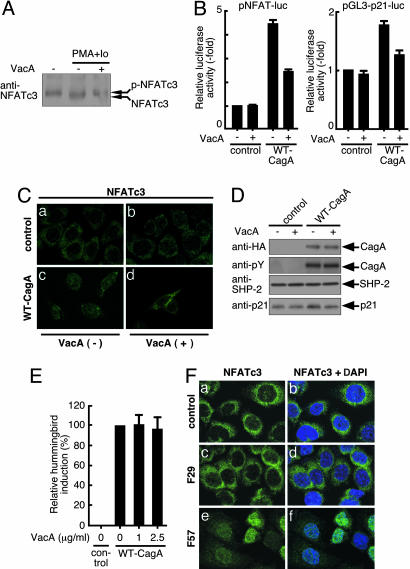
Effect of H. pylori VacA on CagA-Mediated NFAT Activation. Recent studies have shown that another H. pylori virulence factor, VacA, inhibits NFAT activity in T lymphocytes by preventing its nuclear translocation (22, 23). Because NFATc3 dephosphorylated by calcineurin migrates slightly faster than the phosphorylated form, we investigated the effect of VacA on the phosphorylation status of NFATc3 in AGS cells based on its migration positions. As expected, VacA abolished the appearance of a fast-migrating form of NFATc3 in AGS cells treated with PMA plus A23187 (Fig. 5A). This observation indicated that VacA prevents dephosphorylation of NFATc3 by calcineurin in gastric epithelial cells. Accordingly, we were interested in the functional interaction between CagA and VacA in the regulation of NFAT in gastric epithelial cells. To this end, AGS cells cotransfected with pNFAT-luc and WT-CagA expression vector were treated with VacA or were not treated. The VacA concentration used in the experiment (2.5 μg/ml) did not have any effect on the morphology of AGS cells during the course of the experiment. CagA-mediated activation of pNFAT-luc and pGL3-p21-luc was significantly reduced in AGS cells treated with VacA (Fig. 5B). Furthermore, VacA treatment abolished nuclear translocation of NFAT and subsequent p21 induction (Fig. 5 C and D) by CagA. These results indicate that VacA counteracts the CagA activity to activate NFAT. However, VacA did not prevent induction of the hummingbird phenotype by CagA (Fig. 5E). This result is consistent with the fact that the hummingbird phenotype depends on tyrosine phosphorylation of CagA (9).
Fig. 5.
Effect of VacA on CagA-dependent NFAT activation. (A) AGS cells were treated with PMA (33 nM) plus calcium ionophore A23187 (Io) (0.5 μM) for 30 min in the presence or absence of 2.5 μg/ml VacA. Cell lysates were immunoblotted with anti-NFATc3 antibody. p-NFATc3 indicates the phosphorylated form of NFATc3. (B) AGS cells cotransfected with WT-CagA expression vector and pNFAT-luc (Left) or pGL3-p21-luc (Right) were cultured in the presence or absence of VacA. The promoter activation was shown as a ratio of the luciferase activities in cells transfected with control vector (mean ± SD, n = 3 experiments). (C) AGS cells transfected with WT-CagA or control empty vector were cultured in the presence or absence of VacA. Cells were stained with anti-NFATc3. (D) AGS cells transfected with WT-CagA expression vector or control empty vector were cultured in the presence or absence of VacA. Cell lysates were subjected to immunoblotting. (E) AGS cells were transfected with WT-CagA expression vector in the presence or absence of VacA. After 18 h, cell morphology was examined by microscopy. Relative hummingbird induction was shown as the percentage ratio of the number of hummingbird cells induced by WT-CagA in the absence of VacA (mean ± SD, n = 3 experiments). (F) AGS cells were mock-infected or infected with H. pylori strain F29 or F57 at a multiplicity of infection of 50. After 5 h, cells were stained with anti-NFATc3 antibody and DAPI.
Activation of NFAT in Gastric Epithelial Cells Infected with cagA-Positive H. pylori. Finally, we investigated whether infection of gastric epithelial cells with cagA-positive H. pylori leads to activation of NFAT (Fig. 5F). Because VacA counteracts the CagA activity to stimulate NFAT, we wished to eliminate the effect of VacA during H. pylori infection. Consequently, we used H. pylori strains F29 and F57, both of which carry an inactivation mutation in the vacA gene (24). Furthermore, whereas F57 possesses an intact cagA gene, F29 contains a mutated cagA gene that encodes a CagA protein C-terminally truncated in the middle of the EPIYA-containing region (data not shown). As shown in Fig. 4, the EPIYA-containing region is crucial for the NFAT activation by CagA. In uninfected AGS cells and AGS cells infected with F29 strain, NFATc3 was present in the cytoplasm but was completely excluded from the nucleus (Fig. 5F a–d). In contrast, infection of cells with F57 strain resulted in the nuclear translocation of cytoplasmic NFATc3 (Fig. 5F e and f). Thus, NFAT is activated in gastric epithelial cells by infection with H. pylori carrying an intact cagA gene.
Discussion
Several groups have already reported results of genome-wide analyses of CagA-responsive host cell genes (25–28). However, those works were done by comparing data obtained from in vitro infection of gastric epithelial cells with cagA-positive or cagA-negative H. pylori strains. Considering the complicated H. pylori-host cell interactions that simultaneously stimulate a variety of H. pylori-specific and nonspecific cellular responses during infection, it seems extremely difficult to elucidate genes that respond specifically to CagA by using such an in vitro infection experiment. To avoid this potentially serious problem, we used gene transfection to express the CagA protein as a sole H. pylori component in gastric epithelial cells. By using DNA microarray data obtained from CagA-transfected cells, we found that CagA activates NFAT transcription factors in gastric epithelial cells by inducing nuclear translocation of cytoplasmic NFAT through the PLCγ-Ca2+-calcineurin pathway. Activation of NFAT required the EPIYA-containing region of CagA but was independent of CagA phosphorylation. We also found that activation of NFAT by CagA is antagonized by the H. pylori VacA toxin.
The NFAT family of transcriptional factors is composed of five related members, NFATc1–4 and NFAT5. The transcriptional activities of NFATs, except for NFAT5, are controlled by calcineurin, a Ca2+/calmodulin-dependent serine/threonine phosphatase (20, 21). In the absence of appropriate signals, NFAT proteins are phosphorylated by glycogen synthase kinase-3β and are localized exclusively in the cytoplasm. Upon activation by PLCγ-dependent Ca2+ mobilization, calcineurin dephosphorylates NFAT, allowing cytoplasmic NFAT to translocate into the nucleus. In the nucleus, NFAT binds to a specific DNA sequence and transactivates NFAT-responsive genes (20). We found that CagA stimulates nuclear translocation of cytoplasmic NFAT by means of activation of the PLCγ-Ca2+-calcineurin pathway. Interestingly, Churin et al. reported that CagA physically interacts with PLCγ (15). Thus, CagA might directly activate PLCγ upon complex formation.
Although originally identified as a key regulator of cytokine expression in T lymphocytes, recent studies have shown that NFAT plays a role in numerous cell types other than lymphoid cells. It is most likely that individual NFAT family members have both redundant and unique functions in cell differentiation and development (21, 29). Despite the diverse biological roles of NFAT, however, only a few genes such as COX2 have been identified as NFAT-target genes in nonlymphoid cells (30). Recently, Santini et al. demonstrated that NFAT transcriptionally activates p21 during keratinocyte differentiation (17). The NFAT-dependent p21 induction and subsequent cell-cycle withdrawal were essential pre-requisites for turning on the keratinocyte differentiation program. Accordingly, we speculate that CagA-induced p21 is not only involved in cell-cycle inhibition but also plays a role in the induction of intestinal metaplasia, a transdifferentiation from which gastric carcinoma arises (7).
VacA is a secreted H. pylori toxin that is capable of inducing the formation of vacuoles in cells in culture (31). It has recently been reported that VacA inhibits the activation of NFAT in T cells by preventing its nuclear translocation, thereby suppressing T cell immune responses (22, 23). VacA binds to the target cell membrane and is internalized to the cytoplasm, where it is associated with internal membrane-bound compartments (32, 33). In the membrane, VacA forms anion-specific channels, which can be blocked by chloride channel inhibitors (34). Opening of the abnormal anion channel may elicit deregulated membrane depolarization, which inhibits Ca2+ influx from the store-operated Ca2+ channels and thereby prevents activation of Ca2+-dependent phosphatase calcineurin. Hence, VacA is considered to block the signaling cascade that activates calcineurin, most probably through the inhibition of Ca2+ mobilization. Consistently, VacA blocked increases in intracellular Ca2+ by A23187 (22). Also, a constitutively active form of calcineurin abolished the activity of VacA to inhibit NFAT (23). These observations argue against the idea that VacA directly binds to and inhibits NFAT or calcineurin. We showed in this work that treatment of gastric epithelial cells with a low concentration of VacA, which does not have any toxic effects, including vacuolation, efficiently inhibits CagA-mediated nuclear translocation of NFAT. Our finding therefore reveals that CagA and VacA counteract one another by reciprocally regulating the PLCγ-Ca2+-calcineurin pathway in target cells. The opposing effects of the two H. pylori virulence factors on NFAT activity may have a significant implication in our understanding of gastric mucosal damage caused by cagA-positive H. pylori. H. pylori-injected CagA deregulates SHP-2 and other cellular target molecules that promote cell proliferation. Simultaneously, CagA activates NFAT and thereby induces NFAT-dependent genes such as p21. Elevated p21 then arrests gastric epithelial cells in G1 phase. Such G1-arrested cells subsequently undergo senescence, apoptosis, or intestinal transdifferentiation known as intestinal metaplasia. In contrast, when gastric epithelial cells simultaneously encounter VacA and CagA, VacA counteracts nuclear translocation of NFAT by CagA and thus abolishes induction of p21 in CagA-expressing cells. Accordingly, in the presence of adequate levels of VacA, CagA may stimulate deregulated cell growth, which eventually leads to transformation in gastric epithelial cells.
The vacA gene contains two variable regions, the s region (s1a, s1b, and s2 alleles) and the m region (m1 and m2 alleles). Production of VacA toxin is related to the allelic structure of vacA. The s1/m and s1/m2 genotype strains are high and moderate producers of VacA, respectively, whereas s2/m2 genotype strains do not produce VacA (35). Most H. pylori isolates from patients with peptic ulcer or gastric carcinoma coexpress CagA and VacA. Furthermore, there is a close genetic association between the presence of cagA and the s1 type vacA in H. pylori strains from patients with duodenal ulcer and gastric carcinoma (36). Thus, a combination of cagA with a particular vacA genotype may have an advantage in deregulation of the NFAT pathway by H. pylori, which results in more severe disease outcomes.
In conclusion, our present study revealed NFAT as a common cellular target of the two major H. pylori virulence factors, CagA and VacA. Deregulation of NFAT, either positively or negatively, may contribute to cellular dysfunctions that underlie diverged clinical manifestations caused by H. pylori infection.
Supplementary Material
Acknowledgments
We thank Y. Yamazaki and A. Yamakawa (both of University of Fukui, Fukui, Japan) for the H. pylori strains and H. Meguro (University of Tokyo, Tokyo) for help. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author contributions: K.Y., H.H., T.A., T.H., and M.H. designed research; K.Y., H.H., S.I., Y.F., S.K., H.K., A.W., H.A., and M.H. performed research; K.Y., H.H., S.I., T.A., A.W., T.H., H.A., and M.H. contributed new reagents/analytic tools; K.Y., H.H., S.I., Y.F., S.K., H.K., H.A., and M.H. analyzed data; and K.Y. and M.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NFAT, nuclear factor of activated T cells; VacA, vacuolating toxin; CsA, cyclosporin A; PR, phosphorylation-resistant; SHP-2, SH2 domain-containing protein tyrosine phosphatase 2; Csk, C-terminal Src kinase; PLCγ, phospholipase Cγ; PMA, phorbol 12-myristate 13-acetate.
References
- 1.Blaser, M. J., Perez-Perez, G. I., Kleanthous, H., Cover, T. L., Peek, R. M., Chyou, P. H., Stemmermann, G. N. & Nomura, A. (1995) Cancer Res. 55, 2111-2115. [PubMed] [Google Scholar]
- 2.Parsonnet, J., Friedman, G. D., Orentreich, N. & Vogelman, H. (1997) Gut 40, 297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uemura, N., Okamoto, S., Yamamoto, S., Matsumura, N., Yamaguchi, S., Yamakido, M., Taniyama, K., Sasaki, N. & Schlemper, R. J. (2001) N. Engl. J. Med. 345, 784-789. [DOI] [PubMed] [Google Scholar]
- 4.Covacci, A., Censini, S., Bugnoli, M., Petracca, R., Burroni, D., Macchia, G., Massone, A., Papini, E., Xiang, Z., Figura, N., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akopyants, N. S., Clifton, S. W., Kersulyte, D., Crabtree, J. E., Youree, B. E., Reece, C. A., Bukanov, N. O., Drazek, E. S., Roe, B. A. & Berg, D. E. (1998) Mol. Microbiol. 28, 37-53. [DOI] [PubMed] [Google Scholar]
- 7.Peek, R. M., Jr., & Blaser, M. J. (2002) Nat. Rev. Cancer. 2, 28-37. [DOI] [PubMed] [Google Scholar]
- 8.Hatakeyama, M. (2004) Nat. Rev. Cancer 4, 688-694. [DOI] [PubMed] [Google Scholar]
- 9.Higashi, H., Tsutsumi, R., Muto, S., Sugiyama, T., Azuma, T., Asaka, M. & Hatakeyama, M. (2002) Science 295, 683-686. [DOI] [PubMed] [Google Scholar]
- 10.Higashi, H., Tsutsumi, R., Fujita, A., Yamazaki, S., Asaka, M., Azuma, T. & Hatakeyama, M. (2002) Proc. Natl. Acad. Sci. USA 99, 14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tartaglia, M., Mehler, E. L., Goldberg, R., Zampino, G., Brunner, H. G., Kremer, H., van der Burgt, I., Crosby, A. H., Ion, A., Jeffery, S., et al. (2001) Nat. Genet. 29, 465-468. [DOI] [PubMed] [Google Scholar]
- 12.Bentires-Alj, M., Paez, J. G., David, F. S., Keilhack, H., Halmos, B., Naoki, K., Maris, J. M., Richardson, A., Bardelli, A., Sugarbaker, D. J., et al. (2004) Cancer Res. 64, 8816-8820. [DOI] [PubMed] [Google Scholar]
- 13.Tsutsumi, R., Higashi, H., Higuchi, M., Okada, M. & Hatakeyama, M. (2002) J. Biol. Chem. 278, 3664-3670. [DOI] [PubMed] [Google Scholar]
- 14.Mimuro, H., Suzuki, T., Tanaka, J., Asahi, M., Haas, R. & Sasakawa, C. (2002) Mol. Cell 10, 745-755. [DOI] [PubMed] [Google Scholar]
- 15.Churin, Y., Al-Ghoul, L., Kepp, O., Meyer, T. F., Birchmeier, W. & Naumann, M. (2003) J. Cell Biol. 161, 249-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amieva, M. R., Vogelmann, R., Covacci, A., Tompkins, L. S., Nelson, W. J. & Falkow, S. (2003) Science 300, 1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santini, M. P., Talora, C., Seki, T., Bolgan, L. & Dotto, G. P. (2001) Proc. Natl. Acad. Sci. USA 98, 9575-9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yahiro, K., Wada, A., Nakayama, M., Kimura, T., Ogushi, K., Niidome, T., Aoyagi, H., Yoshino, K., Yonezawa, K., Moss, J., et al. (2003) J. Biol. Chem. 278, 19183-19189. [DOI] [PubMed] [Google Scholar]
- 19.Zabel, M. D., Wheeler, W., Weis, J. J. & Weis, J. H. (2002) J. Immunol. 168, 3341-3350. [DOI] [PubMed] [Google Scholar]
- 20.Rao, A., Luo, C. & Hogan, P. G. (1997) Annu. Rev. Immunol. 15, 707-747. [DOI] [PubMed] [Google Scholar]
- 21.Crabtree, G. R. & Olson, E. N. (2002) Cell 109, S67-S79. [DOI] [PubMed] [Google Scholar]
- 22.Boncristiano, M., Paccani, S. R., Barone, S., Ulivieri, C., Patrussi, L., Ilver, D., Amedei, A., D'Elios, M. M., Telford, J. L. & Baldari, C. T. (2003) J. Exp. Med. 198, 1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebert, B., Fischer, W., Weiss, E., Hoffmann, R. & Haas, R. (2003) Science 301, 1099-1102. [DOI] [PubMed] [Google Scholar]
- 24.Ito, Y., Azuma, T., Ito, S., Suto, H., Miyaji, H., Yamazaki, Y., Kohli, Y. & Kuriyama, M. (1998) J. Infect. Dis. 178, 1391-1398. [DOI] [PubMed] [Google Scholar]
- 25.Bach, S., Makristathis, A., Rotter, M. & Hirschl, A. M. (2002) Infect. Immun. 70, 988-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox, J. M., Clayton, C. L., Tomita, T., Wallace, D. M., Robinson, P. A. & Crabtree, J. E. (2001) Infect. Immun. 69, 6970-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, J. W., Kim, H. & Kim, K. H. (2003) Int. J. Biochem. Cell Biol. 35, 1284-1296. [DOI] [PubMed] [Google Scholar]
- 28.Guillemin, K., Salama, N. R., Tompkins, L. S. & Falkow, S. (2002) Proc. Natl. Acad. Sci. USA 99, 15136-15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graef, I. A., Chen, F. & Crabtree, G. R. (2001) Curr. Opin. Genet. Dev. 11, 505-512. [DOI] [PubMed] [Google Scholar]
- 30.Duque, J., Fresno, M. & Iniguez, M. A. (2005) J. Biol. Chem. 280, 8686-8693. [DOI] [PubMed] [Google Scholar]
- 31.Cover, T. L. & Blanke, S. R. (2005) Nat. Rev. Microbiol. 3, 320-332. [DOI] [PubMed] [Google Scholar]
- 32.Garner, J. A. & Cover, T. L. (1996) Infect. Immun. 64, 4197-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massari, P., Manetti, R., Burroni, D., Nuti, S., Norais, N., Rappuoli, R. & Telford, J. L. (1998) Infect. Immun. 66, 3981-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szabo, I., Brutsche, S., Tombora, F., Moschioni, M., Satin, B., Telford, J. L., Rappuoli, R., Montecucco, C., Papini, E. & Zoratti, M. (1999) EMBO J. 18, 5517-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atherton, J. C., Cao, P., Peek, R. M., Jr., Tummuru, M. K., Blaser, M. J. & Cover, T. L. (1995) J. Biol. Chem. 270, 17771-17777. [DOI] [PubMed] [Google Scholar]
- 36.Van Doorn, L. J., Figueiredo, C., Megraud, F., Pena, S., Midolo, P., Queiroz, D. M., Carneiro, F., Vanderborght, B., Pegado, M. D., Sanna, R., et al. (1999) Gastroenterology 116, 823-830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.