Abstract
Background
The biomedical community is developing new methods of data analysis to more efficiently process the massive data sets produced by microarray experiments. Systematic and global mathematical approaches that can be readily applied to a large number of experimental designs become fundamental to correctly handle the otherwise overwhelming data sets.
Results
The gene selection model presented herein is based on the observation that: (1) variance of gene expression is a function of absolute expression; (2) one can model this relationship in order to set an appropriate lower fold change limit of significance; and (3) this relationship defines a function that can be used to select differentially expressed genes. The model first evaluates fold change (FC) across the entire range of absolute expression levels for any number of experimental conditions. Genes are systematically binned, and those genes within the top X% of highest FCs for each bin are evaluated both with and without the use of replicates. A function is fitted through the top X% of each bin, thereby defining a limit fold change. All genes selected by the 5% FC model lie above measurement variability using a within standard deviation (SDwithin) confidence level of 99.9%. Real time-PCR (RT-PCR) analysis demonstrated 85.7% concordance with microarray data selected by the limit function.
Conclusion
The FC model can confidently select differentially expressed genes as corroborated by variance data and RT-PCR. The simplicity of the overall process permits selecting model limits that best describe experimental data by extracting information on gene expression patterns across the range of expression levels. Genes selected by this process can be consistently compared between experiments and enables the user to globally extract information with a high degree of confidence.
Background
The complete sequencing of several genomes, including that of the human, has signaled the beginning of a new era in which scientists are becoming increasingly interested in functional genomics; that is, uncovering both the functional roles of different genes, and how these genes interact with, and/or influence, each other. Increasingly, this question is being addressed through the simultaneous analysis of hundreds to thousands of unique genetic elements with microarrays. Already, analytical strategies have subdivided into distinct 'omic' domains, such as genomics, proteomics, and metabolomics. This enables researchers to examine not only genetic elements, but also the corresponding proteins and metabolites derived from these genes. All 'omic' technologies share the need for fresh, innovative looks at data analysis. To date, transcriptomics is the most widely studied molecular approach, enabling researchers to examine subtle differences in thousands of mRNA levels between experimental samples, medical biopsies, etc. Although mRNA is not the end product of a gene, the transcription of a gene is both critical and highly regulated, thereby providing an ideal point of investigation [1,2]. Development of microarrays has permitted global measurement of gene expression at the transcript level and provided a glimpse into the coordinated control and interactions between genes.
Presently, two technologies dominate the field of high-density microarrays: cDNA arrays and oligonucleotide arrays. The cDNA array has a long history of development [3] stemming from immunodiagnostic work in the 1980s; however, it has been most widely developed in recent years by Stanford University (California) researchers depositing cDNA tags onto glass slides, or chips, with precise robotic printers [4]. Labeled cDNA fragments are then hybridized to the tags on the chip, scanned, and differences in mRNA between samples identified and visualized using a variation of the red/green matrix originally introduced by Eisen and colleagues [5]. The light-generated oligonucleotide array, developed by Affymetrix, Inc. (Santa Clara, CA), involves synthesizing short 25-mer oligonucleotide probes directly onto a glass slide using photolithographic masks [6,7]. Sample processing includes the production of labeled cRNA, hybridization to a microarray, and quantification of the obtained signal after laser scanning. Regardless of the array used, the output can be readily transferred to commercially available data analysis programs for the selection and clustering of significantly modified genes.
Differentially expressed genes will be defined herein as gene data determined to be statistical outliers from some standard state, and which can not be ascribed to chance or natural variabilty. Various creative techniques have been proposed and implemented for the selection of differentially expressed genes; however, none have yet gained widespread acceptance for microarray analysis. Despite this, there remains a great impulse to develop new data analysis techniques, partly driven by the obvious need to move beyond setting simple fold change cut-offs which are out of context with the rest of the experimental and biological data at hand [8-11]. This has been the case for many studies, where the selection of differential gene expression is performed through a simple fold change cut-off, typically between 1.8 and 3.0. There is an inherent problem with this selection criterion, as genes of low absolute expression have a greater inherent error in their measured levels. These genes will then tend to numerically meet any given fold change cut-off even if the gene is not truly differentially expressed. The inverse also holds true, where highly expressed genes, having less error in their measured levels, may not meet an arbitrary fold-change cut-off of 2.0 even when they are truly differentially expressed [12]. Therefore, selecting differentially regulated genes based only on a single fold change across the entire range of experimental data preferentially selects lowly expressed genes [8]. This commonly used approach does not accommodate for background noise, variability, non-specific binding, or low copy numbers- characteristics typical of microarray data which may not be homogeneously distributed. Other approaches entail the use of standard statistical measures such as a student's t-test or ANOVA for every individual gene. However, due to the cost of repeating microarray experiments, the number of replicates usually remains low, leading to inaccurate estimates of variance [8]. Furthermore, due to the low number of replicates, the power of these "gene-by-gene" statistical tests to differentiate between regulated and non-regulated genes also remains very low.
The present article describes a model that considers both expression levels and fold changes for the identification of significant differentially expressed genes. This simple model allows the experimenter to estimate the relationship between these two parameters in the absence of large numbers of experimental replicates, where the inherent error of measures cannot be accurately estimated. Subsequently, gene transcripts determined to be outliers from the trend can be considered differentially expressed genes. An added strength to the model lies in its ease of application to any data set. This model should be considered a progressive and cyclical process, where the data analyst can quickly and globally identify a list of potentially differentially regulated genes with confidence, based on the inherent qualities of the data set under evaluation.
The model presented herein was developed with a data set from a nutritional experiment in a mouse model using Affymetrix Mu11K chips, where the effects of four diets were compared in a number of organs (pool of five mice for each sample in each organ): (1) control diet A in duplicate from the same pool; (2) diet B; (3) diet C; and (4) diet D. Details of the dietary treatments will be reported elsewhere. The present article will take only the data from the liver as an example for the development of a gene selection model. The model was validated by real-time polymerase chain reaction (RT-PCR) and indicates good concordance between the two experimental techniques.
Results and Discussion
Selection of differentially regulated genes and data analysis
The method developed herein includes: (A) determination of the upper X% of highest fold changes within narrow bins of absolute expression levels in order to generate a limit fold change (LFC) function; and (B) subsequent ranking of genes by a combined fold change/absolute expression calculation. The following discussions describe the development of the model within the context of our nutritional study; however, a generic protocol can be found in the Materials and Methods section.
(A) Selection of the upper X% of highest fold changes within binned absolute expression levels
The principal parameter for gene expression data stemming from a typical Affymetrix experiment is the average difference intensity (ADI), which is a representation of the absolute expression of a gene. As indicated in the literature, it is common practice to establish a minimal expression threshold below which data are considered to be noise. In the case of Affymetrix data, it is often necessary to discard minimal and negative ADIs, as these data are both biologically and mathematically difficult to interpret.
A number of previous reports have used an ADI threshold (At) value of 20 in the standard Affymetrix range [13-16], i.e. probe sets with ADI's of less than 20 would either be rejected or set to 20 as meaningful differences in gene expression can purportedly be evaluated above this level. Although empirically supported, an At of 20 is essentially an arbitrary selection and not all groups select the same threshold value. The exact setting of this lower At is not inherent to the LFC modeling process, and the reader is encouraged to set the At value based on additional criteria, such as that previously published by Gerhold et al. [17] and Dieckgraefe et al. [18]. However, an At of 20 will be used in the present work, for which the selection of differentially expressed genes in the context of ADI dependent variance is the central focus. Therefore, all ADI's less than 20 were set to 20 and any probe set with a value of 20 across all dietary treatments were discarded. After eliminating the probe sets which met these criteria there remained 9391 genes out of the original 13179 genes represented on the Mu11K GeneChip.
An additional parameter, highest fold change (HFC), was then applied to these remaining genes. HFC is defined as: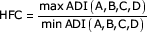
where A, B, C, and D represent the individual microarray results for each gene. The HFC is inherently a ratio metric of the maximum change in measured gene expression between any combination of experimental treatments. The present experiment has four dietary conditions with microarray data; however, it should be noted that the HFC equation could be expanded to any number of conditions or experimental treatments.
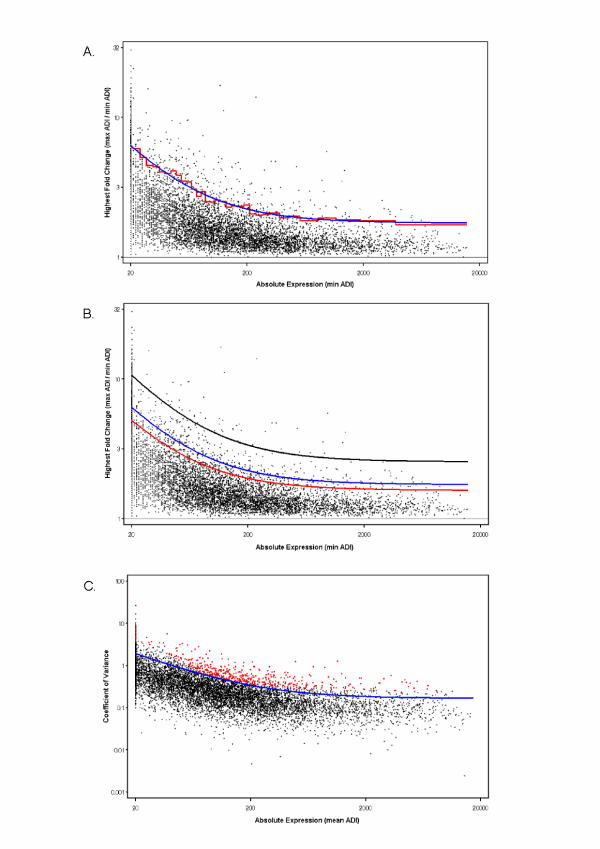
The determination of HFC is highly influenced by absolute expression, and trends can be readily observed in our data set where HFC is negatively correlated with absolute expression (Figure 1a). For example, with absolute expression values greater than 5000 it is of low probability to observe an HFC greater than 2. However, with absolute expression values near 50, an HFC of greater than 2 is readily seen. Although not shown in Figure 1a, this trend could be observed for any pair or triplet of experimental comparison in the current data set, i.e. AB, AC, AD, BC, BD, ABC, BCD. It has also been observed across multiple experiments examined in our laboratory (data not shown). This consistancy can be explained by the fact that there are very few genes out of the entire transcriptome which are differentially expressed due to treatment. Therefore, most measured gene transcripts display a typical coefficient of variation independent of treatment. The few genes which are differentially expressed do not unduly affect the overall trend. Therefore, the trend lends itself to characterization and may be used as a metric for determining differential gene expression across multiple experiments.
Figure 1.
The relationship between absolute value, limit fold change (LFC), and variance across the absolute expression range. A) The x-axis threshold indicates those genes that have a minimum ADI of 20. Genes in bins of 200 are examined for the top 5% highest fold changes (red horizontal lines indicate the 95th percentile for each bin). The line of best fit, drawn through each bin in blue, identifies the overall LFC cut-off and is described by the simple equation 5% LFC = 1.74 + 91.55/min ADI. B) Identifying the top 1% (black line) or 10% (red line) highest fold changes in each bin shifts the LFC curve, when compared to the 5% LFC model (blue line), and alters the severity for the selection of differentially expressed genes (1% LFC = 2.43 + 166.12/min ADI; 10% LFC = 1.59 + 69.47/min ADI). C) The upper 99.9% confidence limit (CL) of a robust estimation of the coefficient of variance (CV) for replicates (within-treatment variability) has been modeled as a function of absolute minimum expression of all treatments, as indicated by the blue line. Overlaying the 99.9% CL on the data selected by the 5% LFC model (red dots) ensures high confidence in the selected genes.
This empirically implies that natural variation, expressed here as HFC, tends to be much greater at low expression levels. This concept is supported in the literature [12] and questions the appropriateness of using a linear fold change cut-off in a system characterized by heterogenous variance.
As stated previously, the selection of differentially expressed genes is essentially a search for outliers, i.e. gene data lying outside some standard distribution of differences relative to a control state, and which cannot be ascribed to chance or natural variabilty. To determine those genes which are outliers, it is necessary to measure either the variability of the system or to make valid assumptions regarding the distribution of variability. In the present model we assume that: (1) as mentioned above, variability in the measurement of gene expression is related to the ADI; and (2) if a broad sampling of the transcriptome is measured, only a small number of genes will actually be outliers even in the harshest of experimental conditions. Assumption (1) is a fairly general analytical concept, i.e. the closer data is to the measurement threshold, the higher the variability is in that measurement [12,19]. Assumption (2) appears to be empiricaly valid when surveying the literature for high-density microarray experiments which evaluate severe biological events, from caloric restriction [20,21] to apoptosis [22,23]. In these experiments [20-23], regardless of the gene selection method used, less than 5% of the total number of genes probed were differentially regulated. Therefore, to develop the present model of gene selection, the validity of selecting outliers was evaluated for a range of highly variable genes using the top 5% as a benchmark. Model trends were then examined from 1% to 10%.
The model was developed by first binning gene expression data into tight classes across the entire range of absolute expression values, where genes with an equal absolute expression value were randomly ordered, and then selecting the upper 5% of HFC values for further consideration. Binning was carried out to divide the entire range of absolute expression values into bins containing an equal number, m, of genes, where m = 200. Therefore, bin widths (ADI) were not necessarily equal, yet the number of genes contained in each bin was equivalent. For the first round of analysis, the upper 5%, or 95th percentile, of HFC genes in each bin were selected for further consideration (Figure 1a). It was possible to search separately for the 5% of genes with the greatest HFCs in each class; however, in order to simplify the overall selection, we plotted the relationship between absolute expression, defined as min ADI (A,B,C,D), and HFC, in order to set the LFC function. Herein, the min ADI (A,B,C,D) will simply be referred to as min ADI. This relationship was then modeled using a simple equation of the form LFC = a + (b/min ADI), which is fitted to the 95th percentile of each bin (Figure 1a) to produce the LFC curve that best models the expression data. This modeled LFC curve (5% LFC model = 1.74 + 91.55/min ADI) fit the data well (R2 = 0.98) and further analysis indicated the residuals were randomly distributed (data not shown). The equation for the line of best fit contains two parameters that have various repercussions on gene selection, both of which can be defined in commercially available software using common "solve" functions (e.g. Microsoft Excel). First, a sets the asymptote, which corresponds to the minimum HFC value that can be observed at any given ADI. Second, b raises/lowers the limit function at a given ADI, and is therefore highly influenced by this latter value. For example, the smaller the ADI the greater the LFC, and vice versa. Figure 1b shows that as the selection criteria becomes more strict (top 5% → 1% of genes), the curve shifts (1% LFC model = 2.43 + 166.12/min ADI) and becomes more restrictive in the selection of differential genes, i.e. at any given absolute expression level a higher fold change must be observed for a gene to be considered differentially expressed. The opposite is true when the selection criteria becomes less strict (top 5% → 10% of genes), where the curve shifts (10% LFC model = 1.59 + 69.47/min ADI) and results in a more permissive selection of differential genes.
Using the aforementioned equations the selection of genes for further consideration becomes simple and 'global' (i.e. across the entire range of expression levels); where a gene is selected with the HFC approach if max ADI/min ADI > a+(b/min ADI). After applying the 10% LFC gene filter, 869 genes remained in the list out of the 9391 candidate genes selected from the original 13179 genes on the GeneChip. When interested in the top 5% and 1% of significant genes, the total number of genes that meet the LFC requirements is 471 and 82, respectively.
Lastly it should be noted that the LFC, i.e. the modeled trend of HFC vs. min ADI, is based on binned data of hundreds of genes across multiple conditions leading to a highly powerful characterization of a given threshold. In other words, there is a large amount of data available in order to accurately characterize the trend. The same argument holds for the generation of a modeled confidence interval based on low numbers of replicates, as will be described below. This is in contrast to the relatively low statistical power of conventional "gene-by-gene" tests such as the t-test or ANOVA, often used for the selection of differentially expressed genes [8].
(B) Assignment of gene rank
Following gene selection, a rank of 'importance' or 'interest level' was assigned to each selected gene. It should be noted that the LFC is not dependent on the rank calculation; rather rank simply lends relative 'importance' to selected genes by incorporating both the magnitude of fold change and absolute expression values. The rank number (RN) for each gene was determined by first calculating a rank value (RV), which can be defined as: RV = HFC * (max ADI – min ADI). After calculation of RV, gene lists were sorted and then assigned a simple RN of 1,2,3,4..., where a gene with a RN of 1 corresponds to the gene with the highest RV. The RV is an arbitrary value that simply lends importance to selected genes with both high fold changes and high differences in absolute expression. Both RV and RN aid in the discussion of differential gene effects by adding the concept of relative weight or importance amongst selected genes. This concept aids in the choice of genes for validation or follow-up studies, as detailed below.
(C) Model validation
Validation of the LFC model via characterization of measurement variability
Hess and colleagues have recently examined the concept that variability and absolute expression are related; however, they examined only the variability of replicate spots on a single slide [24]. Herein, we extended this concept to examine the variability between genes on different microarrays. Measurement variance was examined following the development of the LFC model, and was therefore used simply as a confirmation of this model. To further understand the nature of measurement variability within the current study, duplicate Mu11K Affymetrix microarrays for the controls were examined (see Materials and Methods section). A pooled RNA sample from mice (n = 5) fed the control diet was hybridized to two different chips, and the data was analyzed to characterize measurement variability. It was apparent from the trend that as absolute expression levels (ADI) increase, the coefficient of variation (CV= SD/MAE) decreases. The trendline was calculated as detailed in the Materials and Methods section. This trendline was overlayed on the entire data set, in addition to the 5% LFC selected data (shown in red), in Figure 1c. By overlaying the trendline of the within variability data on those genes determined to be significantly regulated by the LFC model, the CV upper confidence limit for these selected genes had a p value ≤ 0.001. Thus, the 5% LFC-selected data lies outside the 99.9% confidence interval surrounding measurement variability, reinforcing the validity of the results.
Real-time polymerase chain reaction (RT-PCR)
The results obtained from a microarray experiment are influenced by each step in the experimental procedure, from array manufacturing to sample preparation and application to image analysis [25]. The preparation of the cRNA sample is highly correlated to the efficiency of the reverse transcription step, where reagents and enzymes alike can influence the reaction outcome. These factors affect the representation of transcripts in the cRNA sample, necessitating the need for validations by complementary techniques. Analyses by northern blot and RNAse protection assays are commonly reported; however, the emerging 'gold-standard' validation technique is RT-PCR [26]. Microarrays tend to have a low dynamic range, which can lead to small yet significant under-representations of fold changes in gene expression [27]. As RT-PCR has a greater dynamic range, it is often used to validate the observed trends rather than duplicate the fold changes obtained by chip experiments [26,28,29].
Having chosen genes that lie across the range of RN, and therefore the range of model selection criteria, RT-PCR was performed in triplicate for each experimental condition (Diet A,B,C,D) using the same pooled stocks of liver RNA (5 mice per experiment). Genes were compared to the endogenous controls β-actin and GAPDH, which did not significantly change across the dietary treatments. As determined by our LFC selection model, the GeneChip microarrays indicated no significant differences amongst the 4 diets for either GAPDH or β-actin. Subsequent confirmation that both GAPDH and β-actin did not change was provided by RT-PCR, where a simple student's t-test with a predefined nominal α level of 0.05 indicated no significant differences between the experimental diets (B,C,D) and the control diet A. RT-PCR provided a means to confirm the effects of the 3 dietary treatments on 9 genes (Table 1) and the concordance between these 27 microarray and RT-PCR results was examined. Perfect concordance was not to be expected due to the inherent differences in sensitivity and dynamic range between the two techniques. However, a good overall concordance of 77.7% for differential gene expression was observed, i.e. the fold change for a given gene seen by microarray was directionally consistent with that seen by RT-PCR, regardless whether the results were significant by either the 5% LFC model (for microarray data) or a student's T-test (for RT-PCR data). When examining only those genes considered significantly changed by RT-PCR (α = 0.05, starred values in Table 1), concordance increases to 85.7%. Therefore, the value of 85.7% indicates the overall concordance between significantly changed genes seen by RT-PCR and those microarray pairwise comparisons (treatment vs. control) that meet the LFC model criteria (§ values in Table 1).
Table 1.
Concordance data between an Affymetrix 11MuK microarray and RT-PCR.
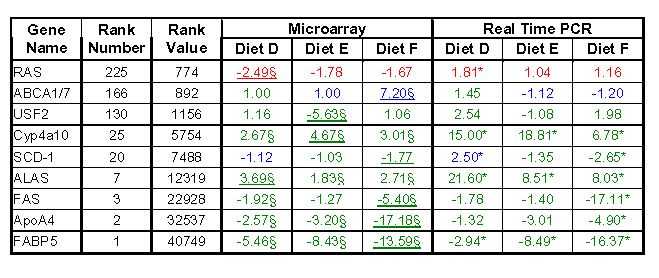 |
The fold changes observed with microarray and RT-PCR analysis are indicated, where a positive value indicates an increase in gene expression and a negative value a decrease in gene expression. Through the coloring scheme, validation (confirmation by RT-PCR of the direction of fold change seen with microarrays) of low RV genes is not achieved; however, as RV increases, concordance increases (red = genes with no concordance across the 3 diets; blue = genes with either one or two measurements in agreement; green = genes with 100% concordance). Overall concordance with the 5% LFC model was 77.7%, which includes measurements found to be both significant and non-significant by microarray analysis. Underlined numbers indicate the HFC that resulted in this gene being selected as significantly different by the 5% LFC model (77.7% concordance with RT-PCR results). Starred-numbers indicate significant fold changes, determined by a student's t-test using α = 0.05, seen by RT-PCR. § indicates those pairwise comparisons (treatment vs. control) that meet the 5% LFC model criteria. 85.7% concordance is seen when comparing significant fold changes by RT-PCR with significant fold changes using the 5% LFC model.
What is noticeable through the color scheme (Table 1) is genes with high RN (low RV) have relatively less concordance between the two techniques; where red indicates no concordance and blue indicates only one or two (out of three) of the results agreed. However, the majority of genes are colored in green, indicating perfect directional concordance. When specifically examining fatty acid synthase (FAS), a highly expressed gene, microarray fold changes of less than 2 can be corroborated between the two experimental techniques, reinforcing the strength of this fold change model. Furthermore, it is clear from the RT-PCR data that at very low expression levels, high fold changes are still problematic to verify and remain questionable. The present model takes this into account by raising the criteria appropriately at the low expression range, i.e. a higher fold change at low expression levels is required for a gene to be considered differentially expressed.
As the selection criteria with microarray data was that the HFC must be greater than the LFC model limits, the expectation was that the LFC function could be validated by RT-PCR (underlined values in Table 1 indicate HFC for each gene). This is predominantly the case across the full dynamic range of data selected by the model (77.7% / 85.7 % concordance), except for very lowly expressed genes such as the RAS oncogene. For genes with a slightly lower RN (higher RV), such as ABC1 member 7, some concordance is seen, indicating confidence is gaining as RV increases. For genes with an RN lower than 130 (RV > 1156; e.g. USF-2) concordance quickly approaches 100%, indicating high confidence when discussing gene trends or individual gene results. These results reinforce the concept that RN is correlated with confidence and validity when discussing the gene set produced by the LFC model.
Interestingly, one might expect that genes with an RN lower then 130 would be concentrated only at higher expression levels; however, when the spread of genes with an RN between 1-130 were examined, these genes were found to lie across the entire range of absolute expressions (data not shown). This indicates that a 5% LFC model is confidently selecting differentially regulated genes across the full range of absolute expression. Therefore, the 5% LFC model appears to be an appropriate selection criteria for the present experimental data set; however, the fold change percentage could easily be varied to meet other acceptable levels of risk, as is done with conventional hypothesis testing (e.g. α-, p-, and χ2-values). The X% selection criteria should then be re-evaluated for other experimental data sets in relation to the variance and validation data at hand.
Conclusions
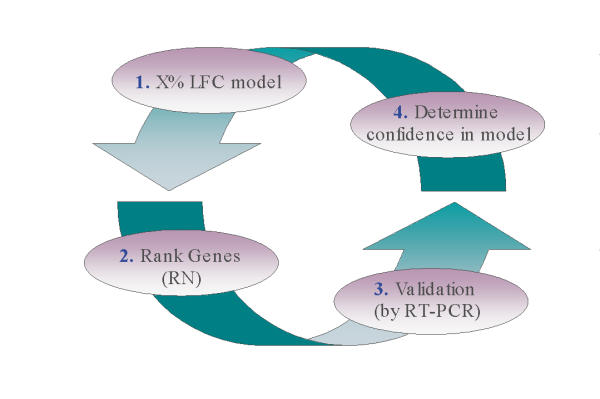
The analysis of microarray data is a developing field of study aimed at enabling the biomedical community to cope with the waves of large microarray data sets. Already, an evolution can be observed with respect to the methods for selecting significantly changed genes. Researchers are moving away from simple fold change cut-offs and incorporating the use of robust statistical concepts. The conclusion that highly expressed genes will rarely have a 2-fold change in mRNA levels and that lowly expressed genes will commonly have a greater than 2-fold change led to the development of a model that would accommodate for this real biological characteristic of gene expression measurements. The fold change model presented in this paper considers both the absolute expression level and fold change of every gene across the entire range of observed absolute expressions. In addition, the concept of increased variation in lowly expressed genes is incorporated into the selection model through the higher fold change requirements for differential gene selection at low expression levels. Following gene selection using an initial criterion of X%, gene rank was introduced as a basis for choosing genes to validate the model. Therefore, a limited but judicious choice of model parameters to select genes across a broad range of gene rank can then be used to reset the X% in order to correspond with the data at hand (Figure 2). The variance data characterizing measurement variability supports the selection model, indicating that selected genes lie outside measurement variability at very high confidence limits (> 99.9% CL). Further validation of this model in the current data set by RT-PCR confirmed these relationships, reinforcing that genes with fold changes even less than 1.8 can be measured, assuming adequate absolute expression levels. This demonstrates that biological changes in sample concentration of mRNA, even at low fold change levels, can be confidently determined.
Figure 2.
Schematic representation of the cyclical nature of the limit fold change (LFC) model. Selecting an initial X% LFC model (1) provides a starting point for the identification of those genes differentially regulated. Genes can then be ranked (2) by a calculation combining fold change and absolute expression in order to assign a degree of importance. Validation of the chosen LFC model by a complementary technique such as RT-PCR (3) and/or the characterization of variance (4) enables the analyst to reexamine the initial LFC model and determine the confidence level for the results. Depending on the data set, one could redefine the LFC model and repeat the cycle.
In summary, the X% LFC model enables one to define experiment specific selection stringency while maintaining simplicity and ensuring global coverage for the detection of differential gene selection.
Materials and Methods
Mice and feeding conditions
Mice were male Rj:NMRI mice from Elevage Janvier, Le Genest-Saint-Isle France, weighing 10-11 g at delivery and 33-51 grams on day 42, were housed 10 per cage. Mice received ad libitum quantities of bottled distilled water and purified powdered diets (7.5 g/mouse) in ceramic cups (10/group) for 42 d. Experimental diets will be described in detail in a biological follow up publication.
Dissection of mice
After administration of the aforementioned diets to 10 mice per group, 5 mice were randomly selected for inclusion in the gene expression analysis experiment. Organs were dissected according to standard protocols, then cut into 100-150 mg subsections, flash frozen in liquid nitrogen, and stored at -80°C until gene expression analysis.
Nucleic acid preparation
Tissue from each organ was extracted from 5 individual mice and extracted separately using Qiagen RNeasy mini-kits (Basel, Switzerland) according to manufacturer's instructions with one exception: During extractions, all RNeasy columns were impregnated with DNase I (Roche, Basel, Switzerland) to remove possible genomic DNA contamination. After extraction, equal amounts of material were pooled to obtain 10 μg total RNA per dietary group. RNA samples were quantified with the RiboGreen RNA Quantification Kit according to manufacturer's instructions (Molecular Probes, Eugene Oregon), and then analyzed via agarose gel electrophoresis for intact 18 and 28s rRNA. All samples included in the study were judged to contain high-quality RNA in sufficient amounts for hybridization.
Gene expression analysis using the murine 11k GeneChip
cRNA preparation
Total RNA (15 μg) was used as starting material for all samples. In all cases, a "test chip" provided by the manufacturer was run prior to using the Murine 11k GeneChip. In each case this confirmed that sufficient high quality RNA was present to detect gene expression in the various tissue samples. The first and second strand cDNA synthesis was performed using the SuperScript Choice System (Life Technologies) according to manufacturer's instructions, but using oligo-dT primer containing a T7 RNA polymerase binding site. Labeled cRNA was prepared using the MEGAscript, In Vitro Transcription kit (Ambion). Biotin labeled CTP and UTP (Enzo) was used together with unlabeled NTP's in the reaction. Following the in vitro transcription reaction, unincorporated nucleotides were removed using RNeasy columns (Qiagen).
Array hybridization and scanning
cRNA (10 μg) was fragmented at 94°C for 35 min in buffer containing 40 mM/L Tris-acetate pH 8.1, 100 mM/L KOAc, 30 mM/L MgOAc. Prior to hybridization, fragmented cRNA in a 6×SSPE-T hybridization buffer (1 M/L NaCl, 10 mM Tris pH 7.6, 0.005% Triton), was heated to 95°C for 5 min, cooled to 40°C, and then loaded onto the Affymetrix probe array cartridge. The probe array was incubated for 16 h at 40° C at 60 rpm. The probe array was washed 10× in 6×SSPE-T at 25°C followed by 4 washes in 0.5×SSPE-T at 50°C. The biotinylated cRNA was stained with a streptavidin-phycoerythrin conjugate, 10 g/ml (Molecular Probes) in 6×SSPE-T for 30 min at 25°C followed by 10 washes in 6×SSPE-T at 25°C. The probe arrays were scanned at 560 nm using a confocal laser-scanning microscope (made for Affymetrix by Hewlett-Packard). Readings from the quantitative scanning were analyzed with Affymetrix Gene Expression Analysis Software.
A step-by-step method to apply the LFC model to an experimental data set
The LFC-model follows a three-step approach. This approach is discussed below as a general protocol and illustrated with the current data set.
1. Data handling and 2-dimensional visualization
Overall, the values of all genes are compared across any number (p) of experimental conditions. The absolute expression value of the k-th gene for the j-th treatment is coded Zkj. When considering any given gene, the following data-handling rules are applied:
• All values below an ADI threshold (At) are set to At.
• If the values for gene k are At for all p treatments, the gene is defined as not expressed and isn't considered further.
• The absolute expression value for gene k is defined as min(Zk1, ..., Zkp).
• The highest fold change (HFC) of gene k is defined as the following: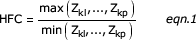
When visualizing all genes on a bivariate plot according to absolute expression and fold change, one obtains a data distribution similar to that of Figure 1a.
2. Modeling a discrete limit fold change model
The goal is to select the upper X% of genes with highest fold change across the entire range of expression levels. Therefore, the following rules are applied:
• Genes are ordered according to their absolute expression value min(Zk1, ..., Zkp), where equally expressed genes are randomly ordered.
• The overall expression range is divided into bins of different width, but containing an equal number m of genes.
• In each bin, the (1-X)-percentile fold change corresponding to a fold change that is exceeded by X% of genes in the bin is determined. For X% between 1% and 10%, m = 200 appears to be suitable.
When visualizing the (1-X)-percentile fold change in each bin, one obtains a data distribution similar to that seen in Figure 1a.
3. Modeling a continuous limit fold change (LFC) model
A continuous model is derived from the discrete one by relating the mean expressions of each bin with the corresponding (1-X)-limit fold change, using a least squares fit of the equation:
(1-X)-LFC = a + b/Z (minimum expression)
This equation appears to fit the data very well and, the interpretation of the parameters (a and b) is straightforward:
• Parameter a represents the asymptote of the curve. For very large expressions, the (1-X)-limit fold change tends to be equal to parameter a.
• Parameter b is proportional to the difference between the (1-X)-limit fold change of small and high expressions.
When visualizing this continuous limit fold change model, one obtains a curve similar to that observed in Figure 1a. In addition, increasing the (1-X)-percentile fold change shifts the curve up the y-axis and results in an increased stringency for gene selection, i.e. fewer genes meet the LFC requirement (Figure 1b).
Validation by Coefficient of Variance
For experiments that are performed without replicates, the LFC-model selects genes with the highest between-treatment variability (previously defined as fold change) at any expression level. If replicates are available, the inherent error of measures, the within-treatment variability, can be estimated. Therefore, it becomes possible to select the genes with the highest ratio of between-treatment-variability / within-treatment-variability.
In the data set that was used for illustrating the development of the LFC-model, duplicate measures were available for one of the four treatments. The within-treatment-variability appears to be highly dependent of the expression level of the gene, confirming the findings of Hess et al.[24].
In order to estimate the CV without taking into account extreme values of the duplicate we used a robust estimator, represented by the following equation: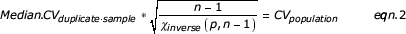
where the χinverse function returns the inverse of the one-tailed probability of the χ-squared distribution.
Applying the CV derived from replicate sample data (eqn. 2) to the quadruplicate diet data enabled the calculation of the CV upper confidence level (by bins of absolute expression level) using the following equation: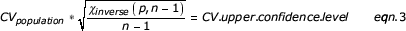
where the χinverse function returns the inverse of the one-tailed probability of the χ-squared distribution.
Eqn. 3 allows one to identify genes with a variance above measurement variability. This greater variability arose due to combined pool (biological) and treatment variabilities.
This confidence level could be raised or lowered according to the level of confidence desired by altering the p value. Therefore, modeling the variance data provides a complimentary method for examining the variation of genes across the complete range of absolute expression values.
The upper 99.9% confidence limit (CL) of a robust estimation of the coefficient of variance (CV) for replicates (within-treatment variability) has been modeled as a function of absolute minimum expression of all treatments using the following model:
Upper 99.9% CL = c + d/mean expression
The selected genes are now those for which the CV of treatment expressions (between-treatment variability) is larger than this limit (Figure 1c). By overlapping those genes selected by the LFC model (red dots) on the graph indicating the 99.9% CL (blue line), one observes that the LFC model is considerably more restrictive when selecting lowly expressed genes (Figure 1c).
Validation by real-time PCR (RT-PCR)
A subset of differentially expressed genes were selected to confirm the LFC model, where genes were selected across the range of absolute expressions and with varying fold changes. Although not discussed in the present manuscript, a good description of the technique and an example of an excellent experimental design can be found in previous publications [26,30], respectively. In brief, all genes were amplified in the Applied Biosystems 5700 instrument using SYBR® green (Molecular Probes), a dye that binds double-stranded DNA. Data represented means of triplicates for each experimental treatment using pooled RNA samples (n = 5). Amplification was performed using an ABI 5700 machine (Applied Biosystems, Foster City, CA, USA) with a hot start at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min for denaturation, annealing and elongation. Genes were normalized to either β-actin or GAPDH, and then experimental diets (B,C,D) were compared to the control diet (A). All fold changes were subjected to a student's t-test (α = 0.05) to ensure fold changes observed by RT-PCR were statistically significant. Comparisons between microarray data and RT-PCR were then performed.
Abbreviations
CV: coefficient of variation
FC: fold change
HFC: highest fold change
LFC: limit fold change function
MAE: mean absolute expression
RN: rank number
RT-PCR: real time polymerase chain reaction
RV: rank value
SD: standard deviation
Definitions
Average Difference Intensity (ADI): average measure of intensity of hybridization for a series of match and mismatch probe pairs tiled across a particular gene transcript. ADI is an indicator of the absolute expression of a gene.
Concordance: state of agreement between two complementary measurement techniques which is directionally consistent, e.g. two techniques determine that values are statistically significant and that they are both either positive or negative.
Author contributions
DM integrated the mathematical and biological interpretation of the experiment that resulted in the writing of this manuscript. AR and RM developed the mathematical formula describing the limit fold change model. AB and MR designed and carried out the DNA microarray studies in mice. MR initiated the development of robust mathematical techniques to evaluate microarray data at the Nestlé Research Center.
All authors read and approved the final manuscript.
Contributor Information
David M Mutch, Email: david.mutch@rdls.nestle.com.
Alvin Berger, Email: alvin.berger@rdls.nestle.com.
Robert Mansourian, Email: robert.mansourian@rdls.nestle.com.
Andreas Rytz, Email: andreas.rytz@rdls.nestle.com.
Matthew-Alan Roberts, Email: matthew-alan.roberts@rdls.nestle.com.
Acknowledgements
The authors would like to thank Professor Juan Medrano from the University of Davis, California for his critical review and discussion of this manuscript.
References
- Brazma A, Vilo J. Gene expression data analysis. FEBS Lett. 2000;480:17–24. doi: 10.1016/S0014-5793(00)01772-5. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Genes & Signals. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. 2002.
- Ekins RP. Ligand assays: from electrophoresis to miniaturized microarrays. Clin Chem. 1998;44:2015–2030. [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone AD, Beecher JE, Bury PA, Chen C, Doede T, Fidanza JA, McGall GH. Photolithographic synthesis of high-density oligonucleotide probe arrays. Nucleosides Nucleotides Nucleic Acids. 2001;20:525–531. doi: 10.1081/NCN-100002328. [DOI] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci U S A. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Woolf PJ, Wang Y. A fuzzy logic approach to analyzing gene expression data. Physiol Genomics. 2000;3:9–15. doi: 10.1152/physiolgenomics.2000.3.1.9. [DOI] [PubMed] [Google Scholar]
- Thomas JG, Olson JM, Tapscott SJ, Zhao LP. An efficient and robust statistical modeling approach to discover differentially expressed genes using genomic expression profiles. Genome Res. 2001;11:1227–1236. doi: 10.1101/gr.165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie JM. Computational methods for the identification of differential and coordinated gene expression. Hum Mol Genet. 1999;8:1821–1832. doi: 10.1093/hmg/8.10.1821. [DOI] [PubMed] [Google Scholar]
- Kersten S, Mandard S, Escher P, Gonzalez FJ, Tafuri S, Desvergne B, Wahli W. The peroxisome proliferator-activated receptor alpha regulates amino acid metabolism. Faseb J. 2001;15:1971–1978. doi: 10.1096/fj.01-0147com. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold D, Lu M, Xu J, Austin C, Caskey CT, Rushmore T. Monitoring expression of genes involved in drug metabolism and toxicology using DNA microarrays. Physiol Genomics. 2001;5:161–170. doi: 10.1152/physiolgenomics.2001.5.4.161. [DOI] [PubMed] [Google Scholar]
- Dieckgraefe BK, Stenson WF, Korzenik JR, Swanson PE, Harrington CA. Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol Genomics. 2000;4:1–11. doi: 10.1152/physiolgenomics.2000.4.1.1. [DOI] [PubMed] [Google Scholar]
- Baggerly KA, Coombes KR, Hess KR, Stivers DN, Abruzzo LV, Zhang W. Identifying differentially expressed genes in cDNA microarray experiments. J Comput Biol. 2001;8:639–659. doi: 10.1089/106652701753307539. [DOI] [PubMed] [Google Scholar]
- Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- Kayo T, Allison DB, Weindruch R, Prolla TA. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc Natl Acad Sci U S A. 2001;98:5093–5098. doi: 10.1073/pnas.081061898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer DW, Hirschberg DL, Xiao J, Lu Q, Roederer M, Lock CB, Herzenberg LA, Steinman L. Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc Natl Acad Sci U S A. 2000;97:2680–2685. doi: 10.1073/pnas.97.6.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo AK, Kruhoffer M, Leeman R, Orntoft T, Eizirik DL. Identification of novel cytokine-induced genes in pancreatic beta-cells by high-density oligonucleotide arrays. Diabetes. 2001;50:909–920. doi: 10.2337/diabetes.50.5.909. [DOI] [PubMed] [Google Scholar]
- Hess KR, Zhang W, Baggerly KA, Stivers DN, Coombes KR. Microarrays: handling the deluge of data and extracting reliable information. Trends Biotechnol. 2001;19:463–468. doi: 10.1016/S0167-7799(01)01792-9. [DOI] [PubMed] [Google Scholar]
- Rajeevan MS, Vernon SD, Taysavang N, Unger ER. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J Mol Diagn. 2001;3:26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider JV, Wechser MA, Lossos IS. Human disease characterization: real-time quantitative PCR analysis of gene expression. Drug Discov Today. 2001;6:1062–1067. doi: 10.1016/S1359-6446(01)01988-2. [DOI] [PubMed] [Google Scholar]
- Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV, Roninson IB. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc Natl Acad Sci U S A. 2000;97:4291–4296. doi: 10.1073/pnas.97.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayanil CS, George D, Freilich L, Miljan EJ, Mania-Farnell B, McLone DG, Bremer EG. Microarray analysis detects novel Pax3 downstream target genes. J Biol Chem. 2001;276:49299–49309. doi: 10.1074/jbc.M107933200. [DOI] [PubMed] [Google Scholar]
- Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin releasing hormone receptor-coupled gene gene network organization. J Biol Chem. 2001;276:47195–47201. doi: 10.1074/jbc.M108716200. [DOI] [PubMed] [Google Scholar]
- Kielar D, Dietmaier W, Langmann T, Aslanidis C, Probst M, Naruszewicz M, Schmitz G. Rapid quantification of human ABCA1 mRNA in various cell types and tissues by real-time reverse transcription-PCR. Clin Chem. 2001;47:2089–2097. [PubMed] [Google Scholar]