Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease affecting motor neurons. Although genes causing familial cases have been identified, those of sporadic ALS, which occupies the majority of patients, are still elusive. In this study, we adopted machine learning to build binary classifiers based on the New York Genome Center (NYGC) ALS Consortium’s RNA-seq data of the postmortem spinal cord of ALS and non-neurological disease control. The accuracy of the classifiers was greater than 83% and 77% for the training set and the unseen test set, respectively. The classifiers contained 114 genes. Among them, 41 genes have been reported in previous ALS studies, and others are novel in this field. These genes are involved in mitochondrial respiration, lipid metabolism, endosomal trafficking, and iron metabolism, which may promote the progression of ALS pathology.
Keywords: ALS, Spinal cord, Machine learning, RNA-seq
Subject terms: Amyotrophic lateral sclerosis, Machine learning
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative condition that progresses over time, marked by the deterioration of both upper and lower motor neurons that regulate voluntary movements through the corticospinal tract1. While the majority of patients experience the onset of the disease in middle age, there exists significant clinical diversity in symptom initiation and the rate of disease advancement leading up to mortality2. Approximately 5–10% of ALS cases are hereditary, with familial instances, and the remaining cases are considered sporadic3. Although mutations in TDP-43, C9orf72, SOD1, TARDBP, FUS, NEK1, TBK1, and KIF5A have been identified in familial ALS population3, importantly, a significant number of non-familial ALS, i.e., the majority of ALS cases, i.e., the sporadic ALS, lack clear causative genetics.
The New York Genome Center (NYGC) ALS Consortium is a cooperation of 42 global institutes aiming to collect genetic information from several thousand samples to tackle ALS-causing genetics, which are of moderate impact and relatively rare in the population4. Since 2018, ALS research has been advanced based on the data collected by the ALS Consortium, as well as Project MinE5, including KIF5A6, DNAJC77, miR-2188, STMN29, UNC13A10, IL18RAP11, VPS3512, and ATXN313. However, the genetics underlying several clinical characteristics of ALS, including dysregulated energy metabolism14, lipid metabolism15, iron metabolism16, and intracellular transport, are still elusive.
One of the main difficulties in understanding and developing treatments for neurodegenerative diseases arises from the genetic heterogeneity present in these conditions17. This diversity in genetic factors can challenge the assumptions underlying traditional statistical methods like the T-test or ANOVA, which rely on normally distributed, independent samples and equal variance among groups18. When these assumptions are not met, the validity of such statistical approaches may be compromised. In contrast, machine learning classifiers are capable of identifying predictive patterns without needing to adhere to these assumptions19. For this reason, we have suggested that machine learning could be an effective supplementary method for studying diseases characterized by genetic heterogeneity. In fact, we have successfully employed machine learning techniques in our previous research on neurodegenerative conditions such as Alzheimer’s disease20 and Huntington’s disease21.
To uncover the ALS genetics, we applied machine learning on the ALS Consortium’s RNA-seq dataset of the postmortem spinal cord of ALS and non-neurological disease control in this study, and we report novel genes that may partly explain several clinical characteristics of ALS, such as dysregulated energy metabolism, lipid metabolism, iron metabolism, and intracellular transport.
Results
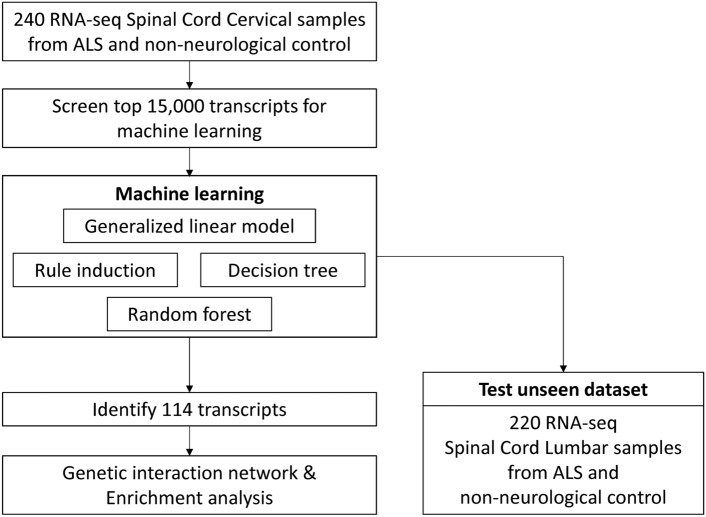
To better understand the pathological mechanisms in the ALS spinal cord that had previously been identified, we utilized RNA sequencing data from 240 cervical spinal cord samples, which included both ALS patients and non-neurological disease controls. This dataset served as the input for machine learning models designed to create binary classifiers capable of distinguishing between ALS and control samples. The entire workflow for building these classifiers is illustrated in Fig. 1. In our analysis, we employed four different machine learning algorithms: "Generalized Linear Model" (GLM), "Rule Induction," "Decision Tree," and "Random Forest." Each of these algorithms was carefully programmed, and their respective processes are depicted in Fig. 2A. To evaluate the robustness of the models, we conducted cross-validation, with the results for this process displayed in Fig. 2B.
Fig. 1.
Workflow of this study. Each block represents one step in the workflow, and arrows represent the direction of information flow. The first step is retrieving the ALS RNA-seq dataset. The second step is data cleansing. The third step is building ML classifiers using the training dataset. Two divergent steps follow: the ML model validation using the unseen dataset (the right branch), and the identifying of transcripts (the left branch). The final step is enrichment analysis and interaction network.
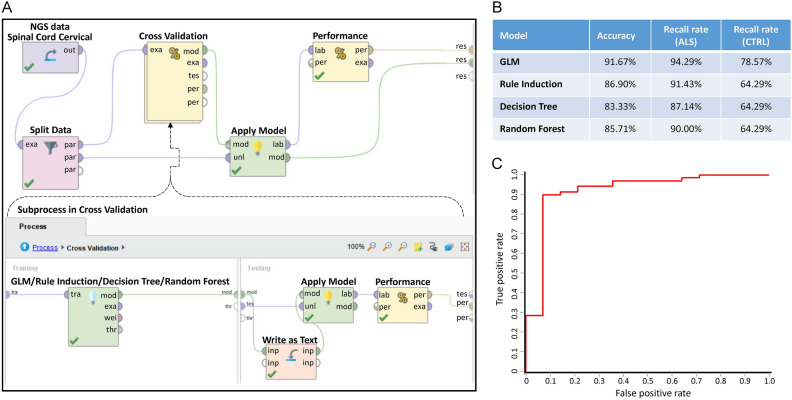
Fig. 2.
The building and performance of the binary classifiers of ALS. (A) The program setup for building binary classifiers of ALS. The subprocess inside the Cross-validation process is shown in the lower part. (B) The performance of the four established models. (C) ROC curve of the established binary classifier of ALS. This section focuses on the construction and evaluation of the binary classifiers used to distinguish between ALS and control samples. The process of building these classifiers involved several steps to ensure accuracy and robustness. In Figure A, we illustrate the overall program setup that was used for the creation of the binary classifiers targeting ALS. This diagram outlines the key components involved in developing the models, starting with data preprocessing, followed by feature selection and the subsequent model training. In (B), we present the performance metrics of the four different models that were established during the study. These models employed distinct machine learning algorithms, each optimized for binary classification tasks. The performance evaluation focused on key metrics such as accuracy and recall, all of which provided a comprehensive assessment of how well the classifiers were able to distinguish between ALS and control samples. (C) features the receiver operating characteristic (ROC) curve of the established binary classifier for ALS. The ROC curve is a crucial graphical representation of the classifier’s performance across different threshold settings. It plots the true positive rate (sensitivity) against the false positive rate (1-specificity) to evaluate the trade-offs between these two measures. A higher area under the ROC curve (AUC) indicates a better-performing model.
All four algorithms performed remarkably well, achieving an overall accuracy—defined as the total true positive rate—of more than 80%. This high accuracy suggests that the binary classifiers were able to effectively differentiate between the ALS and control samples. However, we did observe a bias in the recall rates of the models. Specifically, the recall rate for ALS predictions was higher compared to the recall rate for control predictions. This discrepancy in recall rates could likely be attributed to an imbalance in the sample sizes: the dataset contained 199 ALS samples compared to only 41 control samples, resulting in an approximately 5:1 ratio between ALS and control groups. Such an imbalance in the data could skew the classifiers to favor ALS predictions over control predictions.
Despite this bias, the performance of the classifiers remained robust, particularly for the GLM algorithm. As shown in Fig. 2C, the receiver operating characteristic (ROC) curve for GLM indicates that the model maintained strong performance, even when operating at lower confidence thresholds. This suggests that GLM is able to achieve good predictive accuracy while maintaining flexibility in its decision-making process, making it a reliable tool for identifying ALS-related patterns within the RNA-seq data.
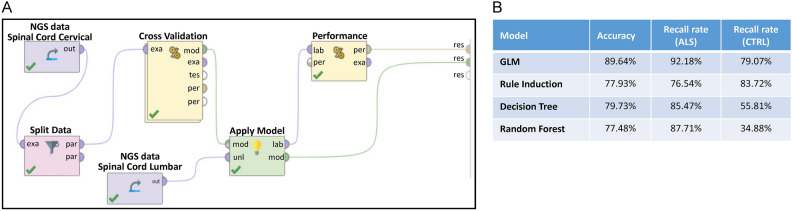
To further assess the effectiveness of our trained classifiers, we next applied them to an unseen dataset, comprising RNA sequencing data from 222 lumbar spinal cord samples. This dataset included both ALS patients and non-neurological disease controls, and it allowed us to independently validate the performance of the classifiers we had previously developed. The programming workflow for this validation process is illustrated in Fig. 3A, while the performance metrics of the classifiers are summarized in Fig. 3B.
Fig. 3.
The validation of the binary classifiers of ALS using an unseen dataset. (A) The program setup for validating the binary classifier of ALS. (B) The performance of the four established models. This figure details the validation process of the binary classifiers designed to distinguish ALS samples from controls, using an independent dataset that was not included in the model-building phase. This “unseen dataset” is critical for evaluating how well the classifiers generalize to new data and ensuring that their performance holds up outside the training environment. The unseen dataset contains samples that the models have not encountered before, which allows us to assess the true predictive power of the classifiers in real-world scenarios. This validation process is a crucial step in confirming that the binary classifiers are not overfitted to the training data and can reliably generalize to new, unseen datasets.
Across all four machine learning algorithms, the classifiers maintained a strong level of accuracy, exceeding 77% in all cases. This high accuracy suggests that the models were able to generalize well to new data and reliably distinguish between ALS and control samples. Furthermore, the recall rate for ALS predictions was consistently high, with all classifiers achieving a recall rate above 76%. This indicates that the classifiers were effective in correctly identifying ALS cases from the new dataset, which is crucial for ensuring that the models can be used to detect ALS with confidence.
However, the performance of the classifiers was more variable when it came to predicting control samples. The recall rate for controls ranged widely, from as low as 34% to as high as 83%, depending on the algorithm. This variation in recall rates for the control group may be due to several factors. The consistently high recall rate for ALS predictions suggests that the pathological genetic features associated with ALS are relatively similar across different parts of the spinal cord. In other words, the genetic patterns found in the cervical spinal cord, which were used to train the classifiers, appear to also be present in the lumbar spinal cord, making it easier for the models to detect ALS regardless of spinal cord region.
On the other hand, the lower recall rates for control samples in some classifiers could indicate that there are slight differences in the genetic background of the non-neurological disease controls between the cervical and lumbar regions of the spinal cord. These subtle genetic variations might make it more challenging for the classifiers to accurately identify control samples in the lumbar dataset, leading to a wider range of recall performance. This finding highlights the possibility that different parts of the spinal cord may exhibit distinct genetic characteristics, especially in non-neurological conditions, and it suggests that further refinement of the classifiers may be necessary to improve their performance in detecting control samples across diverse spinal cord regions.
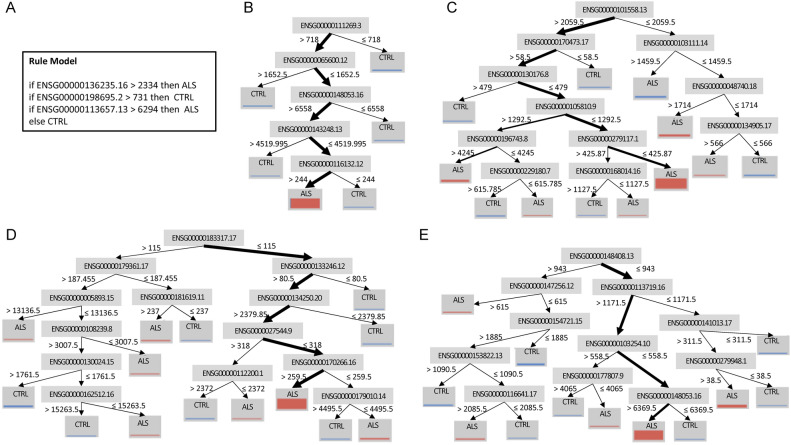
The classifiers that were developed and trained during our study are displayed in various figures and supplementary tables. Specifically, Fig. 4A illustrates the Rule Induction classifier, Fig. 4B shows the Decision Tree classifier, and Fig. 4C–E depict the Random Forest classifiers. Additionally, the Generalized Linear Model (GLM) is provided in Supplementary Table 1. Together, these four classifiers collectively identified 114 genes, which are listed in Supplementary Table 2. These genes represent key distinguishing features between ALS spinal cord samples and those from non-neurological disease controls.
Fig. 4.
The binary classifiers of ALS. (A) Rule induction. Rule induction systematically identifies patterns in the data and constructs a set of "if–then" rules that can be used to make predictions. For ALS classification, the rule induction classifier determines a series of logical conditions that differentiate ALS samples from control samples based on gene expression patterns. (B) Decision tree. The decision tree classifier uses the gene expression data to form a tree-like structure where each node represents a decision about a specific gene. The branches of the tree lead to different classifications—either ALS or control—based on the outcomes of these decisions. (C–E) Decision trees from Random forest. Each individual decision tree in a Random Forest is constructed from a random subset of the data, and the final classification is determined by aggregating the predictions from all the trees. In (B–E), the judgment criteria are noted near the splitting arrows, and the thickness of the arrows roughly represents the fraction of samples that fall in this criterion.
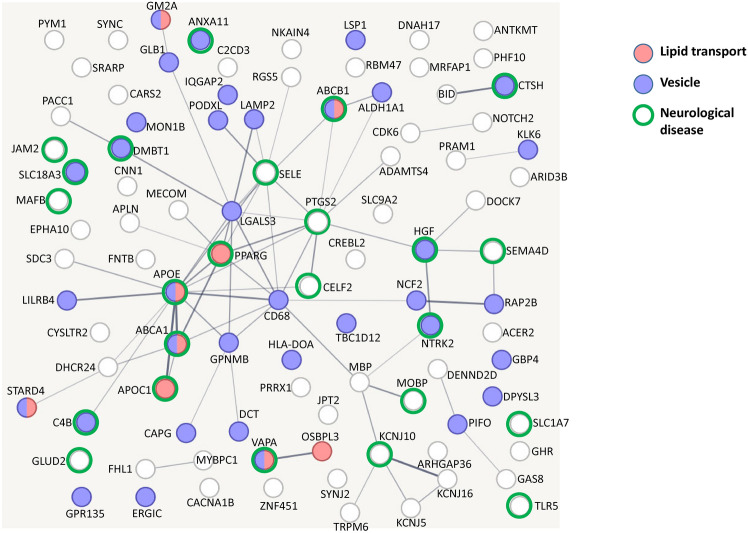
What is particularly noteworthy about these 114 genes is their emphasis on the composition of vesicles and lipid transport as the primary factors differentiating ALS from controls, as visualized in Fig. 5. This finding is important because it suggests that disruptions in vesicle formation and lipid movement may play a central role in the pathological mechanisms of ALS, offering a potential area for further investigation into the disease’s molecular underpinnings.
Fig. 5.
Interaction network of the genes used in the binary classifiers of ALS. The colors of the nodes denote their enriched biological processes. The thickness of the edges denotes the confidence of the connection between nodes.
Out of the 114 genes identified by the classifiers, 41 had been previously reported in other ALS research, as shown in Table 1. This overlap reinforces the relevance of these genes to ALS and suggests that our machine-learning approach was successful in pinpointing key genetic markers that have already been associated with the disease. However, the study also revealed 73 genes that are novel to ALS research, meaning they had not been reported in prior studies. This is a significant finding, as it expands our understanding of the genetic factors involved in ALS. Among these 73 newly identified genes, eight have been linked to other neurological diseases, as outlined in Table 2. This connection suggests that some genetic pathways may be shared across multiple neurological disorders, opening up potential avenues for cross-disease research.
Table 1.
Identified genes that have been reported in previous ALS studies. The first column contains the Ensembl transcript ID. The second column contains the gene symbol of the transcript. The third column shows the involvement of the transcript in the trained ML classifier. The fourth and fifth columns show the type of previous study and reference that identified the involvement of the gene in ALS.
| Accession | Symbol | Used in Classifiers | ALS study | References |
|---|---|---|---|---|
| ENSG00000165092.13 | ALDH1A1 | GLM | ALS association | 22 |
| ENSG00000130208.9 | APOC1 | GLM | ALS association | 23 |
| ENSG00000224389.9 | C4B | GLM | ALS association | 24 |
| ENSG00000042493.16 | CAPG | GLM | ALS association | 25 |
| ENSG00000103811.16 | CTSH | GLM | ALS association | 26 |
| ENSG00000187775.16 | DNAH17 | GLM | ALS association | 27 |
| ENSG00000268388.5 | FENDRR | GLM | ALS association | 28 |
| ENSG00000170266.16 | GLB1 | Random forest | ALS association | 29 |
| ENSG00000204252.14 | HLA-DOA | GLM | ALS association | 30 |
| ENSG00000145703.16 | IQGAP2 | GLM | ALS association | 13 |
| ENSG00000154721.15 | JAM2 | Random forest | ALS association | 31 |
| ENSG00000131981.16 | LGALS3 | GLM | ALS association | 32 |
| ENSG00000186818.12 | LILRB4 | GLM | ALS association | 33 |
| ENSG00000197971.15 | MBP | GLM | ALS association | 34 |
| ENSG00000116701.14 | NCF2 | GLM | ALS association | 35 |
| ENSG00000134250.20 | NOTCH2 | Random forest | ALS association | 36 |
| ENSG00000073756.12 | PTGS2 | GLM | ALS association | 37 |
| ENSG00000187714.7 | SLC18A3 | GLM | ALS association | 38 |
| ENSG00000162383.12 | SLC1A7 | GLM | ALS association | 39 |
| ENSG00000286159.1 | Antisense To PREX1 | GLM | ALS GWAS | 40 |
| ENSG00000122359.18 | ANXA11 | GLM | ALS GWAS | 41 |
| ENSG00000130203.10 | APOE | GLM | ALS GWAS | 42 |
| ENSG00000116133.13 | DHCR24 | GLM | ALS GWAS | 43 |
| ENSG00000113657.13 | DPYSL3 | Rule induction | ALS GWAS | 44 |
| ENSG00000113719.16 | ERGIC1 | Random forest | ALS GWAS | 45 |
| ENSG00000196743.8 | GM2A | Random forest | ALS GWAS | 46 |
| ENSG00000168314.17 | MOBP | GLM | ALS GWAS | 47 |
| ENSG00000132170.21 | PPARG | GLM | ALS GWAS | 48 |
| ENSG00000165029.16 | ABCA1 | GLM | ALS mechanism | 49 |
| ENSG00000085563.14 | ABCB1 | GLM | ALS mechanism | 50 |
| ENSG00000158859.10 | ADAMTS4 | GLM | ALS mechanism | 51 |
| ENSG00000147256.12 | ARHGAP36 | Random forest | ALS mechanism | 52 |
| ENSG00000129226.14 | CD68 | GLM | ALS mechanism | 53 |
| ENSG00000105810.9 | CDK6 | Random forest | ALS mechanism | 54 |
| ENSG00000187908.18 | DMBT1 | GLM | ALS mechanism | 55 |
| ENSG00000136235.16 | GPNMB | GLM, Rule induction | ALS mechanism | 56 |
| ENSG00000019991.17 | HGF | GLM | ALS mechanism | 57 |
| ENSG00000177807.9 | KCNJ10 | Random forest | ALS mechanism | 58 |
| ENSG00000148053.16 | NTRK2 | Decision tree, Random forest | ALS mechanism | 59 |
| ENSG00000187764.11 | SEMA4D | GLM | ALS mechanism | 60 |
| ENSG00000101558.13 | VAPA | Random forest | ALS mechanism | 61 |
Table 2.
Identified genes that are related to other neurological diseases. The first column contains the Ensembl transcript ID. The second column contains the gene symbol of the transcript. The third column shows the involvement of the transcript in the trained ML classifier. The fourth and fifth columns show the type of diseases and references that identified the involvement of the gene.
| Accession | Symbol | Used in classifiers | Diseases | References |
|---|---|---|---|---|
| ENSG00000048740.18 | CELF2 | Random forest | Encephalopathy | 62 |
| ENSG00000182890.4 | GLUD2 | GLM | Parkinson’s disease | 63 |
| ENSG00000167755.15 | KLK6 | GLM | Hydrocephalus | 64 |
| ENSG00000204103.4 | MAFB | GLM | Alzheimer’s disease | 65 |
| ENSG00000198763.3 | MT-ND2 | GLM | Leigh syndrome | 66 |
| ENSG00000198695.2 | MT-ND6 | GLM, rule induction | Leigh syndrome | 67 |
| ENSG00000007908.16 | SELE | GLM | Brain ischemia | 68 |
| ENSG00000187554.13 | TLR5 | GLM | Brain ischemia | 69 |
Additionally, 21 of the novel genes are involved in vesicle formation, ion channel function, or lipid transport, as detailed in Table 3. These genes are particularly intriguing because they point to new areas of investigation in ALS research. Their involvement in key cellular processes that are essential for neuron function and signaling suggests that further exploration of these pathways could yield valuable insights into how ALS develops at the molecular level. Given that these 21 genes have not been previously associated with ALS, their discovery offers a fresh perspective on the disease’s biology and could help to identify new therapeutic targets aimed at addressing these specific mechanisms.
Table 3.
Identified genes that are involved in vesicle, ion channel, or lipid transportation. The first column contains the Ensembl transcript ID. The second column contains the gene symbol of the transcript. The third column shows the involvement of the transcript in the trained ML classifier. The fourth column shows the biological pathway that the gene is involved in.
| Accession | Symbol | Used in classifiers | Involves in |
|---|---|---|---|
| ENSG00000177076.6 | ACER2 | GLM | Lipid metabolism |
| ENSG00000111269.3 | CREBL2 | Decision tree | Lipid genesis |
| ENSG00000070882.13 | OSBPL3 | GLM | Lipid transport |
| ENSG00000164211.13 | STARD4 | GLM | Lipid transport/vesicle |
| ENSG00000080166.16 | DCT | GLM | Vesicle |
| ENSG00000162654.9 | GBP4 | GLM | Vesicle |
| ENSG00000181619.11 | GPR135 | Random forest | Vesicle |
| ENSG00000005893.15 | LAMP2 | Random forest | Vesicle |
| ENSG00000130592.15 | LSP1 | GLM | Vesicle |
| ENSG00000103111.14 | MON1B | Random forest | Vesicle |
| ENSG00000173947.14 | PIFO | GLM | Vesicle |
| ENSG00000128567.17 | PODXL | GLM | Vesicle |
| ENSG00000181467.4 | RAP2B | GLM | Vesicle |
| ENSG00000078269.15 | SYNJ2 | GLM | Vesicle endocytosis |
| ENSG00000108239.8 | TBC1D12 | Random forest | Vesicle |
| ENSG00000148408.13 | CACNA1B | Random forest | Calcium channel |
| ENSG00000153822.13 | KCNJ16 | Random forest | Potassium channel |
| ENSG00000120457.12 | KCNJ5 | GLM | Potassium channel |
| ENSG00000101198.15 | NKAIN4 | GLM | Sodium–potassium pump |
| ENSG00000065600.12 | PACC1 | Decision tree | Chloride channel |
| ENSG00000115616.2 | SLC9A2 | GLM | Sodium antiporter |
To evaluate whether age, sex, and genetic variations correspond to specific RNA expression patterns in ALS, we re-run the analysis using age, sex, and the GGGGCC repeat size of C9orf72 or CAG repeat size of ATXN2 as the Machine-learning “labels” according to the clinical information of ALS samples.
For age as the ML label, the two strategies shown in Supplementary Fig. 1 were used. In the first strategy (the left flow in Supplementary Fig. 1), the label age was treated as continuous numbers, and the ML model of linear regression was trained using RNA expression. The resulting root-mean-square deviation (RMSD) of the regression model was 9.459 ± 3.604 years, which is not significantly different from the standard deviation of ALS samples of 10.05 years. This means the linear regression model could not effectively predict the age of ALS samples. In the second strategy (the right flow in Supplementary Fig. 1), the label age was treated as discrete data, i.e., label = 1 if age < 55, else label = 0, and the trained classification models were GLM, decision tree, random forest, and rule induction. The resulting AUC of the ROC curve for GLM, decision tree, random forest, and rule induction were 0.684, 0.500, 0.577, and 0.573, respectively, not much better than a 50% chance of flipping a coin. Meanwhile, the recall rate of age-under-55 ALS samples was 12.5%, 31.25%, 18.75%, and 12.5%, respectively. This means that trained classification models could not effectively predict the age of ALS samples. In sum, neither strategy could predict the age of ALS samples, i.e., we cannot claim the correlation between the age and the RNA expression pattern of ALS samples.
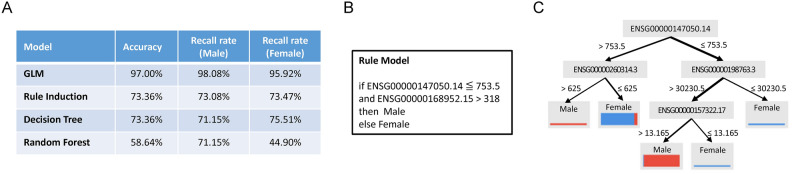
For sex as the ML label, i.e., the label for male = 1 and female = 0, we conducted two rounds of ML as shown in Supplementary Fig. 2. In the first round (the left flow in Supplementary Fig. 2), all RNA of ALS samples in the dataset were kept for the training of prediction models; as a result, we got 100% recall rate for both sexes. This result met our anticipation since Y chromosome genes were included. Therefore, in the second round (the right flow in Supplementary Fig. 2), Y chromosome genes were excluded from the training dataset, and the recall rate for both sexes was > 70% for three models (see Fig. 6A for detail, 6B for the rule induction, 6C for the decision tree, and Supplementary Table 6 for the GLM models). Interestingly, there is a common gene in the three models: ENSG00000147050.14 (KDM6A, lysine demethylase 6A).
Fig. 6.
The binary classifiers of the sex of ALS. (A) The performance of the four established models. (B) Rule induction. (C) Decision tree.
For genetic variation as the ML label, we set the label = 1 for those ALS samples carrying more than 30 GGGGCC repeats in C9ORF72 or intermediate (i.e., 30–33,) CAG repeats in ATXN2, and label = 0 for those ALS samples who did not meet the previous criteria. Using the workflow shown in Supplementary Fig. 3, the resulting AUC of the ROC curve for GLM, rule induction, decision tree, and random forest, were 0.508, 0.558, 0.508, and 0.454, respectively. Meanwhile, the recall rate of ALS samples carrying genetic mutation was 5.00%, 15.00%, 25.00%, and 30.00%, respectively. This means that trained classification models could not effectively predict the genetic mutation of ALS samples, i.e., we cannot claim the correlation between the genetic mutation and the RNA expression pattern of ALS samples.
Discussion
In this study, we generated several ALS-Control classifiers using an RNA-seq dataset from the cervical spinal cord and used the counterpart from the lumbar spinal cord as the unseen dataset for validation. The accuracy of classifiers was higher than 83% for the cross-validation during model building and 77% for the unseen dataset, which not only justifies the performance of the classifiers but also indicates the similarity of ALS transcriptomic signatures between different parts of the spinal cord. The relevance to previous ALS studies and the biological meaning of the novel findings are discussed below.
In the generated ALS- Control classifiers, 41 genes have been reported in previous ALS studies (Table 1), which cover different study types, including association studies, genome-wide association studies (GWAS), and mechanism research. We will not discuss these genes, but we have provided a reference list in Table 1 if more detail is needed. Notably, the rediscovery of the previously identified ALS genes strengthens the reliability of our study. In addition to those rediscovered ones, we identified 73 genes novel to ALS research. Among the novel genes, those eight genes in Table 2 have been mentioned in other neurological diseases, and those 21 genes in Table 3 involve critical biological functions of the spinal cord. Some of them may passively reflect the biological environment of ALS, but some may actively make progress in ALS pathology. We shall focus our discussion on the later part, which will be divided into four groups according to their biological functions, including mitochondrial respiration, lipid metabolism, endosomal trafficking, and ion channel.
The mitochondrial NADH dehydrogenase 2 (mt-ND2) and NADH dehydrogenase 6 (mt-ND6) are subunits of the NADH dehydrogenase, which is the largest electron transport chain complex in the mitochondrial inner membrane and responsible for mitochondrial ATP synthesis70. As shown in the Rule Induction classifier (Fig. 4A), a sample is classified as a non-ALS control if the expression of ENSG00000198695.2, i.e., mt-ND6, is greater than a certain level. This criterion indicates that the quantity of the NADH dehydrogenase subunit is fewer for the ALS spinal cord. Importantly, mt-ND6 is essential for the assembly of the membrane arm of the NADH dehydrogenase and is indispensable for mitochondrial respiratory function70. Interestingly, disrupted TCA cycle71 and increased glycolysis72 have been reported in ALS models. Thus, insufficient levels of mt-ND6 may directly limit the efficiency of mitochondrial respiration and indirectly force the utilization of glycolysis to fulfill the energy demand.
Altered lipid metabolism has been identified in animal models29,73 and cohorts74 of ALS. Interestingly, several rediscovered ALS genes in previous studies are relevant to lipid transport (Fig. 5), including APOE, APOC1, ABCA1, ABCB1, GM2A, PPARG, and VAPA. The novel ones are oxysterol-binding protein-related protein 3 (OSBPL3), StAR-related lipid transfer protein 4 (STARD4), and sphingolipid long chain base-responsive protein LSP1 (LSP1). OSBPL3 is located in the membrane contact site between plasma and endoplasmic reticulum (ER) membranes and forms a complex with VAPA75. OSBPL3 regulates plasma membrane phosphatidylinositol 4-phosphate (PI4P) levels and Ca2+ entry by exchanging phosphatidylcholine76. STARD4 regulates intracellular cholesteryl ester formation, sterol transport to the ER, and SREBP-2-mediated sterol sensing by SCAP/SREBP-277. LSP1 localizes at eisosomes and participates in lipid endocytosis78. The dysregulation of OSBPL3, STARD4, and LSP1 may exacerbate the altered lipid metabolism in ALS. In sum, the lipid transport pathway plays an important biological role in ALS, particularly in relation to how it affects neurons and their ability to function properly. Lipids are essential for maintaining the structure and function of cell membranes, particularly in neurons. They also serve as a source of energy. Lipid metabolism disruptions have been linked to ALS, with many patients experiencing altered lipid profiles, including elevated cholesterol and triglycerides15. These genes may affect how lipids are transported within neurons, leading to cellular stress or degeneration. Dysregulation of these genes can impair the normal trafficking and metabolism of lipids, disrupting cellular energy balance and membrane integrity. Defects in lipid transport can lead to neuronal dysfunction and contribute to motor neuron death, a hallmark of ALS.
The synaptic vesicle plays a crucial role in the pathology of ALS, primarily through its involvement in neurotransmitter release and neuronal communication79. Synaptic vesicles are small sacs that store neurotransmitters, which are chemicals used for communication between neurons. During synaptic transmission, vesicles fuse with the presynaptic membrane, releasing neurotransmitters into the synaptic cleft. This allows for the activation of postsynaptic neurons, facilitating communication between neurons, including motor neurons. Importantly, motor neurons, with their long axons, depend heavily on the efficient transport of synaptic vesicles from the cell body to the synapse. Disruptions in axonal transport mechanisms, often associated with ALS80, affect the delivery of synaptic vesicles, leading to synaptic dysfunction and degeneration of motor neuron connections. Endosomal trafficking is critical in maintaining the proper function of neurons, in the context of targeted transportation and protein recycling in the extremely asymmetric and complex intracellular space of a neuron81. Interestingly, endosomal trafficking is disrupted by either C9ORF7282 or SOD183 mutant in ALS. In this study, five identified genes involved in the endosome, including G-protein coupled receptor 135 (GPR135), Lysosome-associated membrane glycoprotein 2 (LAMP2), Vacuolar fusion protein MON1 homolog B (MON1B), Ras-related protein Rap-2b (RAP2B), and TBC1 domain family member 12 (TBC1D12). The dysregulation of these genes may contribute to the disruption of endosomal trafficking and promote neurodegeneration in ALS.
Abnormal accumulation of iron in CNS has been detected in neurodegenerative diseases, including ALS84. Iron level in the spinal cord is increased more than 1.5 fold in ALS85. Iron excess may induce oxidative stress86, ferroptosis87, and microglia activation88. In this study, we identified a proton-activated chloride channel (PACC1). PACC1 is a transmembrane protein that mediates the influx of chloride ions in response to extra-membrane acidic pH value89. Besides cellular membrane, PACC1 can translocate to endosomes and regulate transferrin receptor-mediated endocytosis90. As shown in Fig. 4B, lower PACC1, i.e., ENSG00000065600.12, predicts ALS, and according to the previous study89, PACC1 knockout results in increased transferrin uptake. Thus, the PACC1 down-regulation may promote neurodegeneration by mediating abnormal accumulation of iron in the spinal cord.
Men are generally more likely to develop ALS than women, particularly in younger age groups. However, this sex difference decreases with age. In older populations, the ratio between men and women diagnosed with ALS tends to even out91,92. Moreover, sporadic ALS (the most common form, making up 90–95% of cases) tends to occur more frequently in men, while familial ALS (accounting for 5–10% of cases) shows less of a sex disparity91,93. Although hormonal differences, especially related to estrogen, are considered a possible explanation for the sex disparity of ALS94, further investigations are needed to clarify this issue. In this study, we identified KDM6A as the common gene in sex classifiers of ALS, and a higher expression level of KDM6A predicts female ALS samples. KDM6A belongs to the family of histone demethylases, which modulate epigenetics during neurodevelopment and neurodegenerative diseases95. Interestingly, a previous study using microarray to probe blood RNA expression also identified higher expression levels of KDM6A in female than male ALS96. The exact role of KDM6A in ALS requires further investigation.
In conclusion, binary classifiers build by machine learning on spinal cord RNA-seq data successfully differentiate ALS and control samples. Besides, this study identified novel genes in mitochondrial respiration, lipid metabolism, endosomal trafficking, and iron metabolism, which may promote the progression of ALS pathology.
Methods
Source of NGS datasets
RNA-seq data of ALS and non-neurological control were retrieved from the Gene Expression Omnibus (GEO) database97 of the National Center for Biotechnology Information (NCBI) of the USA with the accession number GSE1539609, accessed on Sep 12th 2023, which contained RNA-seq data from the cervical and lumbar spinal cord, and the lumbar spinal cord samples were from the same cases/controls as the cervical spinal cord samples. For the development and cross-validation of the binary classifiers designed to differentiate between ALS and non-neurological conditions, we utilized data from 199 ALS samples and 41 non-neurological control samples derived from the cervical region of the spinal cord. This dataset provided the foundation for constructing the models and performing the necessary cross-validation to ensure the accuracy and reliability of the classifiers. Detailed information about this dataset, as well as the specifics of the model-building process, can be found in Supplementary Table 3. In addition to the training and validation performed on the cervical spinal cord dataset, we employed another independent set of data from the lumbar spinal cord to further test the generalizability of the classifiers. This unseen dataset consisted of RNA sequencing data from 179 ALS samples and 43 non-neurological control samples. By using this new dataset, we aimed to validate the performance of the classifiers on data that had not been used during the model training phase, ensuring that the classifiers could reliably predict ALS even when applied to samples from a different region of the spinal cord. This process of external validation helps to assess how well the classifiers can generalize to new data and different contexts, and the results of this validation, along with the details of the lumbar spinal cord dataset, are provided in Supplementary Table 4. By using distinct datasets from two different regions of the spinal cord—cervical for model building and cross-validation, and lumbar for independent testing—we were able to thoroughly evaluate the robustness and reliability of the classifiers. The inclusion of both regions ensures that the models are not overly specific to a single area of the spinal cord, increasing the likelihood that they will be applicable across different anatomical regions affected by ALS. This two-phase validation approach enhances the credibility of our findings, as it demonstrates that the models are capable of accurately predicting ALS across diverse sample sets, which is a critical step in advancing the potential for these classifiers to be used in broader clinical applications. The clinical information of ALS samples is listed in Supplementary Table 5.
Data cleansing
In the process of building the binary classifiers, we used the field labeled "Sample id alt" to represent the unique identifier for each sample, which was referred to as "ID." This “ID” allowed us to track and differentiate between individual samples throughout the analysis. Meanwhile, the field labeled “Group” served as the "Label," which functioned as the target variable for the binary classification task. The “Label” distinguished between the two groups of interest—ALS and non-neurological control—and was the outcome the classifiers were trained to predict. For the actual features, or input variables, used to train the classifiers, we relied on the “ENSEMBL ID” of transcripts, which was designated as the "Regular Attribute." This means that each transcript, identified by its unique ENSEMBL ID, was used as a predictive feature in the machine learning models. The transcripts represent the genetic expression data from the samples, and these were the variables that the models analyzed to learn patterns associated with ALS or control groups. To focus the learning tasks on the most relevant data, we applied a filtering step to reduce the number of transcripts included in the analysis. Specifically, we retained only the top 15,000 transcripts with the highest average read counts across the dataset. By keeping only these top transcripts, we ensured that the models were trained on the most informative and reliable genetic data, as transcripts with higher read counts are generally more robust and less prone to noise or variability. This step was crucial for improving the efficiency and accuracy of the learning tasks, as it allowed the classifiers to focus on the most significant genetic signals that could differentiate between ALS and control samples.
Machine learning
RapidMiner Studio version 9.5, running on a desktop PC with 16 GB RAM, was used to build and validate the binary classifiers of ALS. RNA-seq data of spinal cord cervical were split into 65% and 35% for model building and testing, respectively. Four algorithms were used, including the “Generalized Linear Model” (GLM), “Rule Induction”, “Decision Tree”, and “Random Forest”. The parameters are described as follows. Parameters of GLM: binomial family, IRLSM solver, use regularization, conduct lambda search, 47 lambdas, lambda min ratio of 0, use early stopping, 3 stopping rounds and stopping tolerance of 0.02.
Parameters of Rule Induction: criterion of information gain, sample ratio of 0.9, pureness of 0.9, and minimal prune benefit of 0.25.
Parameters of Decision Tree: criterion of gain ratio, with a maximal depth of 10, apply pruning with confidence of 0.1, apply prepruning with minimal gain of 0.01, minimal leaf size of 2, minimal split size of 4, and number of prepruning alternatives of 3.
Parameters of Random Forest: number of trees of 3, criterion of Gini index, maximal depth of 10, guess subset ratio, and voting strategy of confidence vote.
Interaction network
String-db version 12.098, accessed on Nov 20th, 2023, was used to generate the interaction network. The enrichment analysis was conducted using DAVID Bioinformatics Resources99, accessed on Nov 22nd, 2023. By utilizing String-db, we were able to visualize the potential relationships and interactions between different proteins, offering deeper insights into how these proteins might work together or influence one another in the context of ALS pathology. The use of such a resource significantly enhanced our ability to interpret the biological relevance of the identified genes, particularly in understanding how they may be functionally connected. In addition to building the interaction network, we performed an enrichment analysis to identify the biological pathways, functions, and processes that are overrepresented in the gene set. For this, we used the DAVID Bioinformatics Resources tool. Through this tool, we were able to explore which biological processes and molecular functions are significantly enriched in the genes identified in our study. By leveraging DAVID, we could link the identified genes to specific biological pathways, providing further context for their roles in ALS and other neurological conditions. This analysis enabled us to uncover patterns and commonalities among the genes, offering potential clues as to how genetic dysregulation might contribute to the progression of ALS.
Supplementary Information
Author contributions
W.Y.L. and H.P.L. initiated and supervised this study. J.C. and B.T.W. contributed to the acquisition, analysis, and interpretation of data. All authors discussed and drafted the manuscript.
Funding
This work was supported by grant from National Science and Technology Council of Taiwan (MOST 111-2314-B-039-017-MY3) and grants from China Medical University & Hospital (CMU111-MF-65, CMU112-MF-62, CMU113-MF-44, DMR-112-125, DMR-114-103).
Data availability
All data in this study are included in the supplementary data. The RNA-seq dataset can be accessed via the GEO database of the NCBI of the USA with the accession number GSE153960, with the website below. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153960.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jack Cheng and Bor-Tsang Wu contributed equally to this work.
Contributor Information
Hsin-Ping Liu, Email: hpliu@mail.cmu.edu.tw.
Wei-Yong Lin, Email: linwy@mail.cmu.edu.tw.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81315-z.
References
- 1.Kiernan, M. C. et al. Amyotrophic lateral sclerosis. Lancet377, 942–955 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Hardiman, O. et al. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers.3, 1–19 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Mathis, S., Goizet, C., Soulages, A., Vallat, J.-M. & Le Masson, G. Genetics of amyotrophic lateral sclerosis: A review. J. Neurol. Sci.399, 217–226 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Al-Chalabi, A., Van Den Berg, L. H. & Veldink, J. Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat. Rev. Neurol.13, 96–104 (2017). [DOI] [PubMed] [Google Scholar]
- 5.van der Spek, R. A. et al. The project MinE databrowser: bringing large-scale whole-genome sequencing in ALS to researchers and the public. Amyotroph. Lateral Scler. Frontotemp. Degener.20, 432–440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas, A. et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron97, 1268-1283.e1266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farhan, S. M. et al. Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein. Nat. Neurosci.22, 1966–1974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichenstein, I. et al. Human genetics and neuropathology suggest a link between miR-218 and amyotrophic lateral sclerosis pathophysiology. Sci. Transl. Med.11, eaav5264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prudencio, M. et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J. Clin. Investig.130 (2020). [DOI] [PMC free article] [PubMed]
- 10.Brown, A.-L. et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature603, 131–137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eitan, C. et al. Whole-genome sequencing reveals that variants in the Interleukin 18 Receptor Accessory Protein 3′ UTR protect against ALS. Nat. Neurosci.25, 433–445 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Torres, E. J. et al. Retromer dysfunction in amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci.119, e2118755119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphrey, J. et al. Integrative transcriptomic analysis of the amyotrophic lateral sclerosis spinal cord implicates glial activation and suggests new risk genes. Nat. Neurosci.26, 150–162 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Ahmed, R. M., Dupuis, L. & Kiernan, M. C. (BMJ Publishing Group Ltd, 2018).
- 15.Chaves-Filho, A. B. et al. Alterations in lipid metabolism of spinal cord linked to amyotrophic lateral sclerosis. Sci. Rep.9, 11642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petillon, C. et al. The relevancy of data regarding the metabolism of iron to our understanding of deregulated mechanisms in ALS; hypotheses and pitfalls. Front. Neurosci.12, 1031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young, A. L. et al. Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nat. Commun.9, 4273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou, Y., Zhu, Y. & Wong, W. K. Statistical tests for homogeneity of variance for clinical trials and recommendations. Contemp. Clin. Trials Commun. 101119 (2023). [DOI] [PMC free article] [PubMed]
- 19.Ij, H. Statistics versus machine learning. Nat. Methods15, 233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng, J., Liu, H.-P., Lin, W.-Y. & Tsai, F.-J. Machine learning compensates fold-change method and highlights oxidative phosphorylation in the brain transcriptome of Alzheimer’s disease. Sci. Rep.-UK11, 13704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng, J., Liu, H.-P., Lin, W.-Y. & Tsai, F.-J. Identification of contributing genes of Huntington’s disease by machine learning. BMC Med. Genom.13, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, H. et al. Aldehyde dehydrogenases 1A2 expression and distribution are potentially associated with neuron death in spinal cord of Tg (SOD1* G93A) 1Gur mice. Int. J. Biol. Sci.13, 574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, R. et al. A computational biology approach to identify potential protein biomarkers and drug targets for sporadic amyotrophic lateral sclerosis. Cell. Signal.112, 110915 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Goldknopf, I. L. et al. Complement C3c and related protein biomarkers in amyotrophic lateral sclerosis and Parkinson’s disease. Biochem. Biophys. Res. Commun.342, 1034–1039 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Dreger, M., Steinbach, R., Otto, M., Turner, M. R. & Grosskreutz, J. Cerebrospinal fluid biomarkers of disease activity and progression in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry93, 422–435 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, S. et al. Identification of molecular correlations between DHRS4 and progressive neurodegeneration in amyotrophic lateral sclerosis by gene co-expression network analysis. Front. Immunol.13, 874978 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziff, O. J. et al. Meta-analysis of the amyotrophic lateral sclerosis spectrum uncovers genome instability. medRxiv, 2022.2008. 2011.22278516 (2022).
- 28.Rey, F. et al. LncRNAs associated with neuronal development and oncogenesis are deregulated in SOD1-G93A murine model of amyotrophic lateral sclerosis. Biomedicines9, 809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández-Beltrán, L. C. et al. A transcriptomic meta-analysis shows lipid metabolism dysregulation as an early pathological mechanism in the spinal cord of SOD1 mice. Int. J. Mol. Sci.22, 9553 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eshima, J. et al. Molecular subtypes of ALS are associated with differences in patient prognosis. Nat. Commun.14, 95 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mamoor, S. Differential expression of JAM2 in amyotrophic lateral sclerosis. (2022).
- 32.Iridoy, M. O. et al. Neuroanatomical quantitative proteomics reveals common pathogenic biological routes between amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Int. J. Mol. Sci.20, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter, M. et al. Microglial transcriptome analysis in the rNLS8 mouse model of TDP-43 proteinopathy reveals discrete expression profiles associated with neurodegenerative progression and recovery. Acta Neuropathol. Commun.9, 1–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, J.-Y. et al. Blood–brain barrier dysfunction and myelin basic protein in survival of amyotrophic lateral sclerosis with or without frontotemporal dementia. Neurol. Sci. 1–10 (2022). [DOI] [PubMed]
- 35.Chen, H. et al. Differential expression and alternative splicing of genes in lumbar spinal cord of an amyotrophic lateral sclerosis mouse model. Brain Res.1340, 52–69 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Wan, M. et al. Intermediate-length GGC repeat expansion in NOTCH2NLC was identified in chinese patients with amyotrophic lateral sclerosis. Brain Sci.13, 85 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrés-Benito, P., Moreno, J., Aso, E., Povedano, M. & Ferrer, I. Amyotrophic lateral sclerosis, gene deregulation in the anterior horn of the spinal cord and frontal cortex area 8: implications in frontotemporal lobar degeneration. Aging (Albany NY)9, 823 (2017). [DOI] [PMC free article] [PubMed]
- 38.Sun, S. et al. Translational profiling identifies a cascade of damage initiated in motor neurons and spreading to glia in mutant SOD1-mediated ALS. Proc. Natl. Acad. Sci.112, E6993–E7002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, A. R. et al. Stratified gene expression analysis identifies major amyotrophic lateral sclerosis genes. Neurobiol. Aging36, 2006.e2001–2006.e2009 (2015). [DOI] [PubMed]
- 40.Lenzken, S. C. et al. In 2009 International Conference on Complex, Intelligent and Software Intensive Systems. 795–799 (IEEE). [DOI] [PMC free article] [PubMed]
- 41.Teyssou, E. et al. Genetic screening of ANXA11 revealed novel mutations linked to amyotrophic lateral sclerosis. Neurobiol. Aging.99, 102.e111–102.e120 (2021). [DOI] [PubMed]
- 42.Zetterberg, H., Jacobsson, J., Rosengren, L., Blennow, K. & Andersen, P. M. Association of APOE with age at onset of sporadic amyotrophic lateral sclerosis. J. Neurol. Sci.273, 67–69 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Hop, P. J. et al. Genome-wide study of DNA methylation in amyotrophic lateral sclerosis identifies differentially methylated loci and implicates metabolic, inflammatory and cholesterol pathways. medRxiv, 2021.2003. 2012.21253115 (2021).
- 44.Blasco, H. et al. A rare motor neuron deleterious missense mutation in the DPYSL3 (CRMP4) gene is associated with ALS. Hum. Mutat.34, 953–960 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Nakamura, R. et al. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun. Biol.3, 526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wainberg, M., Andrews, S. J. & Tripathy, S. J. Shared genetic risk loci between Alzheimer’s disease and related dementias, Parkinson’s disease, and amyotrophic lateral sclerosis. Alzheimer’s Res. Ther.15, 1–14 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liampas, I. et al. MOBP rs616147 polymorphism and risk of amyotrophic lateral sclerosis in a greek population: A case-control study. Medicina57, 1337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dash, B. P., Naumann, M., Sterneckert, J. & Hermann, A. Genome wide analysis points towards subtype-specific diseases in different genetic forms of amyotrophic lateral sclerosis. Int. J. Mol. Sci.21, 6938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dodge, J. C. et al. Neutral lipid cacostasis contributes to disease pathogenesis in amyotrophic lateral sclerosis. J. Neurosci.40, 9137–9147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qosa, H. et al. Astrocytes drive upregulation of the multidrug resistance transporter ABCB1 (P-Glycoprotein) in endothelial cells of the blood–brain barrier in mutant superoxide dismutase 1-linked amyotrophic lateral sclerosis. Glia64, 1298–1313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemarchant, S. et al. ADAMTS-4 promotes neurodegeneration in a mouse model of amyotrophic lateral sclerosis. Mol. Neurodegener.11, 1–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nam, H. et al. Critical roles of ARHGAP36 as a signal transduction mediator of Shh pathway in lateral motor columnar specification. Elife8, e46683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson, M. E. et al. Microglial CD68 and L-ferritin upregulation in response to phosphorylated-TDP-43 pathology in the amyotrophic lateral sclerosis brain. Acta Neuropathol. Commun.11, 1–22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, X., Li, D., Zhang, W., Guo, M. & Zhan, Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J.31, 4415–4427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkening, K., Keller, B. A., Leystra-Lantz, C. & Strong, M. J. RNA and protein interactors with TDP-43 in human spinal-cord lysates in amyotrophic lateral sclerosis (vol 17, pg 1712, 2018). J. Proteome Res.17, 2248–2248. 10.1021/acs.jproteome.8b00261 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, H. et al. The potential of GPNMB as novel neuroprotective factor in amyotrophic lateral sclerosis. Sci. Rep.-UK.2, 10.1038/srep00573 (2012). [DOI] [PMC free article] [PubMed]
- 57.Vallarola, A. et al. A novel HGF/SF receptor (MET) agonist transiently delays the disease progression in an amyotrophic lateral sclerosis mouse model by promoting neuronal survival and dampening the immune dysregulation. Int. J. Mol. Sci.21, 10.3390/ijms21228542 (2020). [DOI] [PMC free article] [PubMed]
- 58.Kaiser, M. et al. Progressive loss of a glial potassium channel (KCNJ10) in the spinal cord of the SOD1 (G93A) transgenic mouse model of amyotrophic lateral sclerosis. J. Neurochem.99, 900–912. 10.1111/j.1471-4159.2006.04131.x (2006). [DOI] [PubMed] [Google Scholar]
- 59.Pradhan, J., Noakes, P. G. & Bellingham, M. C. The role of altered BDNF/TrkB signaling in amyotrophic lateral sclerosis. Front. Cell. Neurosci.13, 368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leoni, E. et al. Combined tissue-fluid proteomics to unravel phenotypic variability in amyotrophic lateral sclerosis (vol 9, 4478, 2019). Sci. Rep.-UK10, 10.1038/s41598-020-74974-1 (2020). [DOI] [PMC free article] [PubMed]
- 61.Nakamichi, S., Yamanaka, K., Suzuki, M., Watanabe, T. & Kagiwada, S. Human VAPA and the yeast VAP Scs2p with an altered proline distribution can phenocopy amyotrophic lateral sclerosis-associated VAPB(P56S). Biochem. Biophys. Res. Commun.404, 605–609. 10.1016/j.bbrc.2010.12.011 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Itai, T. et al. De novo variants in CELF2 that disrupt the nuclear localization signal cause developmental and epileptic encephalopathy. Hum. Mutat.42, 66–76 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Zhang, W. et al. Functional validation of a human GLUD2 variant in a murine model of Parkinson’s disease. Cell Death Di.11, 897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, L. et al. Proteomics and functional study reveal kallikrein-6 enhances communicating hydrocephalus. Clin. Proteom.18, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson, A. G. et al. Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer’s disease-specific cis-regulatory elements. Cell Genom.3 (2023). [DOI] [PMC free article] [PubMed]
- 66.Mkaouar-Rebai, E. et al. Two new mutations in the MT-TW gene leading to the disruption of the secondary structure of the tRNATrp in patients with Leigh syndrome. Mol. Genet. Metab.97, 179–184 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Kishita, Y. et al. A high mutation load of m. 14597A> G in MT-ND6 causes Leigh syndrome. Sci. Rep.-UK11, 11123 (2021). [DOI] [PMC free article] [PubMed]
- 68.Ma, X. J. et al. E-selectin deficiency attenuates brain ischemia in mice. CNS Neurosci. Ther.18, 903–908 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu, L. et al. Impact of TLR5 rs5744174 on stroke risk, gene expression and on inflammatory cytokines, and lipid levels in stroke patients. Neurol. Sci.37, 1537–1544 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Bai, Y. & Attardi, G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J.17, 4848–4858 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veyrat-Durebex, C. et al. Disruption of TCA cycle and glutamate metabolism identified by metabolomics in an in vitro model of amyotrophic lateral sclerosis. Mol. Neurobiol.53, 6910–6924 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Valbuena, G. N. et al. Metabolomic analysis reveals increased aerobic glycolysis and amino acid deficit in a cellular model of amyotrophic lateral sclerosis. Mol. Neurobiol.53, 2222–2240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henriques, A. et al. Sphingolipid metabolism is dysregulated at transcriptomic and metabolic levels in the spinal cord of an animal model of amyotrophic lateral sclerosis. Front. Mol. Neurosci.10, 433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goutman, S. A. et al. Metabolomics identifies shared lipid pathways in independent amyotrophic lateral sclerosis cohorts. Brain145, 4425–4439 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weber-Boyvat, M. et al. OSBP-related protein 3 (ORP3) coupling with VAMP-associated protein A regulates R-Ras activity. Exp. Cell Res.331, 278–291 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Gulyás, G., Sohn, M., Kim, Y. J., Várnai, P. & Balla, T. ORP3 phosphorylation regulates phosphatidylinositol 4-phosphate and Ca2+ dynamics at plasma membrane–ER contact sites. J. Cell Sci.133, jcs237388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mesmin, B. et al. STARD4 abundance regulates sterol transport and sensing. Mol. Biol. Cell22, 4004–4015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walther, T. C. et al. Eisosomes mark static sites of endocytosis. Nature439, 998–1003 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Bauer, C. S. et al. An interaction between synapsin and C9orf72 regulates excitatory synapses and is impaired in ALS/FTD. Acta Neuropathol.144, 437–464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williamson, T. L. & Cleveland, D. W. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat. Neurosci.2, 50–56 (1999). [DOI] [PubMed] [Google Scholar]
- 81.Lasiecka, Z. M. & Winckler, B. Mechanisms of polarized membrane trafficking in neurons—focusing in on endosomes. Mol. Cell. Neurosci.48, 278–287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farg, M. A. et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet.23, 3579–3595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burk, K. & Pasterkamp, R. J. Disrupted neuronal trafficking in amyotrophic lateral sclerosis. Acta Neuropathol.137, 859–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rouault, T. A. Iron metabolism in the CNS: implications for neurodegenerative diseases. Nat. Rev. Neurosci.14, 551–564 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Kasarskis, E. J., Tandon, L., Lovell, M. A. & Ehmann, W. D. Aluminum, calcium, and iron in the spinal cord of patients with sporadic amyotrophic lateral sclerosis using laser microprobe mass spectroscopy: a preliminary study. J. Neurol. Sci.130, 203–208 (1995). [DOI] [PubMed] [Google Scholar]
- 86.Puntarulo, S. Iron, oxidative stress and human health. Mol. Aspects Med.26, 299–312 (2005). [DOI] [PubMed] [Google Scholar]
- 87.Chen, X., Yu, C., Kang, R. & Tang, D. Iron metabolism in ferroptosis. Front. Cell Dev. Biol.8, 590226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rathnasamy, G., Ling, E.-A. & Kaur, C. Consequences of iron accumulation in microglia and its implications in neuropathological conditions. CNS Neurol. Disord.-Drug Targets (Formerly Current Drug Targets-CNS & Neurological Disorders)12, 785–798 (2013). [DOI] [PubMed]
- 89.Yang, J. et al. PAC, an evolutionarily conserved membrane protein, is a proton-activated chloride channel. Science364, 395–399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osei-Owusu, J. et al. Proton-activated chloride channel PAC regulates endosomal acidification and transferrin receptor-mediated endocytosis. Cell Rep.34 (2021). [DOI] [PMC free article] [PubMed]
- 91.Curtis, A. F. et al. Sex differences in the prevalence of genetic mutations in FTD and ALS: A meta-analysis. Neurology89, 1633–1642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fontana, A. et al. Time-trend evolution and determinants of sex ratio in Amyotrophic Lateral Sclerosis: a dose–response meta-analysis. J. Neurol.268, 2973–2984 (2021). [DOI] [PubMed] [Google Scholar]
- 93.Manjaly, Z. R. et al. The sex ratio in amyotrophic lateral sclerosis: A population based study. Amyotroph. Lateral Scler.11, 439–442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Jong, S. et al. Endogenous female reproductive hormones and the risk of amyotrophic lateral sclerosis. J. Neurol.260, 507–512 (2013). [DOI] [PubMed] [Google Scholar]
- 95.Wang, H., Guo, B. & Guo, X. Histone demethylases in neurodevelopment and neurodegenerative diseases. Int. J. Neurosci. 1–11 (2023). [DOI] [PubMed]
- 96.Santiago, J. A., Quinn, J. P. & Potashkin, J. A. Network analysis identifies sex-specific gene expression changes in blood of amyotrophic lateral sclerosis patients. Int. J. Mol. Sci.22, 7150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barrett, T. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res.41, D991–D995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Szklarczyk, D. et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res.51, D638–D646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study are included in the supplementary data. The RNA-seq dataset can be accessed via the GEO database of the NCBI of the USA with the accession number GSE153960, with the website below. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153960.