Abstract
Background
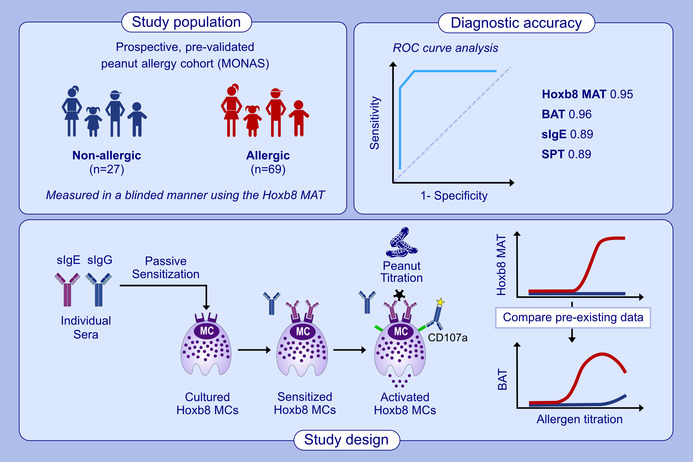
Peanut allergy is among the most severe and common food allergies. The diagnosis has a significant impact on the quality of life for patients and their families. An effective management approach depends on accurate, safe, and easily implementable diagnostic methods. We previously developed a cell‐based assay using Hoxb8 mast cells (Hoxb8 MCs) aimed at improving clinical allergy diagnosis. In this study, we assessed its diagnostic performance by measuring blinded sera from a prospectively enrolled and pre‐validated peanut allergy cohort.
Methods
Hoxb8 MCs were passively sensitized with sera from peanut‐allergic and peanut tolerant children and adolescents (n = 112). Degranulation of Hoxb8 MCs was quantified upon stimulation with dose‐titrated peanut extract by means of flow cytometry, using CD107a as activation marker. The results from the Hoxb8 mast cell activation test (Hoxb8 MAT) were compared to established diagnostic assays such as the skin prick test (SPT), specific IgE (sIgE) levels, and the basophil activation test (BAT). Additionally, serum samples from BAT nonresponders were assessed with the Hoxb8 MAT.
Results
Hoxb8 MAT displayed a robust dose‐dependent activation to peanut extract, with a cutoff value of ≤5.2% CD107a positive cells. The diagnostic accuracy was highest at allergen concentrations ≥100 ng/mL, with an area under the receiver operating characteristic curve (AUROC) of 0.97, 93% sensitivity, and 96% specificity, outperforming traditional SPT and sIgE tests. When compared to BAT, Hoxb8 MAT exhibited comparable diagnostic efficacy. Moreover, sera from BAT nonresponders were accurately classified into allergics and nonallergics by the Hoxb8 MAT.
Conclusions
The Hoxb8 MAT demonstrated a very good diagnostic precision in patients prospectively assessed for peanut allergy comparable to the fresh whole blood‐based BAT. Additionally, it demonstrated its value for accurate classification of BAT nonresponders into allergic and nonallergic individuals. Further investigations into its utility in the routine clinical setting are warranted.
Keywords: basophil activation test, Hoxb8 mast cell activation test, oral food challenge, peanut allergy diagnosis, specific IgE
The Hoxb8 MAT demonstrates high diagnostic accuracy, as determined by measuring blinded sera from a prospectively enrolled, prevalidated peanut allergy cohort. Similar to the BAT, the Hoxb8 MAT distinguishes between allergic and nonallergic patients who underwent clinically indicated OFCs to peanut. Additionally, it has the ability to accurately classify sera from BAT nonresponders.Abbreviations: BAT, basophil activation test; Hoxb8, homebox b 8; MAT, mast cell activation test; MCs, mast cells; OFCs, oral food challenges; sIgE/sIgG, specific IgE/IgG; SPT, skin prick test.
1. INTRODUCTION
Food allergies have become a significant global health concern. For some countries a prevalence as high as 10% has been reported, with mostly young children being affected. 1 Peanut allergy, in particular, is one of the most common conditions 2 , 3 and often manifests as severe, potentially life‐threatening reactions. 4 The burden of food allergies not only impacts affected individuals but has also broader implications for their families, healthcare system, and the food industry. 5 , 6 To effectively manage this debilitating disease, easily implementable diagnostic tools that are accurate, safe, and reliable are urgently required. 7 , 8
While the thorough assessment of a patient's clinical history builds the fundamental basis of a food allergy diagnosis, the oral food challenge (OFC) test in which the patient ingests the culprit allergen under professional supervision is still considered the diagnostic gold standard to achieve optimal diagnostic accuracy. However, access to OFCs is limited, as they require significant resources, and the procedure may be associated with major discomfort, anxiety, and the risk to develop a systemic allergic reaction. Therefore, the guidelines of food allergy testing 9 suggest a stepwise diagnostic workup in which a less elaborate in vivo diagnostic tool, the skin prick test (SPT), in conjunction with the serological quantification of allergen‐specific IgE (sIgE) should initially be performed. 9 , 10 , 11 SPT and sIgE have both been reported to feature high sensitivity but rather low specificity. 7 , 12 While these tests can be helpful to confirm a suspicion of IgE‐mediated food reactions, they are often not sufficient to diagnose a food allergy on their own and examination solely based on these tests would lead to overdiagnosis. 12 , 13 , 14 Especially sIgE measurement as a nonfunctional assay quantifies the level of sIgE antibodies in the patient's blood without providing further information on the potential of these antibodies to trigger an allergic reaction. Previous work has indicated that affinity and epitope specificity of sIgE antibodies might be more important for cellular functionality than their absolute serum concentration. 15 While sIgE quantification lacks such qualitative information it also omits the detection of potentially protective serum factors including allergen‐specific IgG. If not carefully interpreted serological assays can thus lead to a false positive diagnosis, causing unnecessary food avoidance strategies for patients.
Functional cell‐based allergy tests, which evaluate the activation of allergic effector cells including basophils or mast cells, have been emerging as promising alternatives to clinical in vivo and serological in vitro approaches. Among these, the basophil activation test (BAT) has gained recognition as a practical tool for diagnosing food allergies, including peanut, 16 milk, 17 and egg 18 allergy, in the clinical setting. 19 , 20 Our recently published results from the markers of nut allergy study (MONAS) have demonstrated that the BAT shows high accuracy in the diagnosis of peanut and tree nut allergies. 21 The BAT has even been reported to be superior to other diagnostic tests in discriminating between peanut allergy or tolerance and to reduce the need for OFCs. 22 Despite all these promising features including the possibility to obtain objective and quantifiable results, broad implementation of the BAT has been hampered by several limitations. These mainly include the variability between donor reactivity among which the issue of so‐called nonresponders (i.e., lack of basophil activation upon FcεRI‐dependent stimulation) is most problematic 23 as well as logistical challenges that arise from the necessity for fresh blood samples to be processed within maximally 24 h. 24 Mast cell activation tests (MAT) have more recently gained a lot of traction as potential diagnostic tool to overcome such limitations. 25 , 26 , 27 , 28 , 29 Most importantly, the MAT is based on patient serum, which can be used to passively sensitize in vitro cultured mast cells, eliminating the need for fresh blood samples, allowing prospective as well as retrospective analysis of samples, and simultaneously reducing donor variability. The introduction of a novel MAT technology, using genetically engineered mouse mast cell progenitors that stably express the human high‐affinity IgE receptor, FcεRIα, and that are conditionally immortalized with the homeobox B8 gene (Hoxb8 MCs), has further facilitated the implementation of the MAT as diagnostic tool. 30 Preliminary studies have demonstrated that the new Hoxb8 MAT is a promising approach to provide standardized and robust diagnostic results for various allergens including those found in foods. 30
In this study, we aimed to assess the clinical utility of the Hoxb8 MAT in diagnosing peanut allergies, using serum samples from clinically confirmed allergic and nonallergic children and adolescents. For this purpose, we took advantage of the existing Toronto site MONA study cohort which allowed us to assess the diagnostic performance of the Hoxb8 MAT assay in a blinded fashion compared to SPT, sIgE measurements, BAT, and OFC outcomes. This approach enabled us to comprehensively assess the prospective diagnostic accuracy of the Hoxb8 MAT in a real‐world setting.
2. METHODS
2.1. Study population
Sera from 112 patients, who were prospectively enrolled in the MONA study and had clear clinical data on the peanut allergic status (80 peanut allergic children and adolescents and 32 nonallergic controls) were used to assess reactivity to peanut extract with the Hoxb8 MAT in a blinded setting. Allergic patients were classified based on positive OFC, or a pretest probability of >90% allergic reaction (i.e., SPT ≥8 mm and/or sIgE ≥15kUA/L). OFC was performed in an open‐label fashion as previously described 21 and according to internally standardized clinical protocols. Nonallergic controls were defined as patients who either passed an oral food challenge due to a suspicion of peanut allergy or consumed peanut regularly and were enrolled due to suspicion or proven tree nut allergy in the MONAS cohort as reported. 21 Amongst the 112 samples, BAT results were available for a total of 96 individuals including 69 allergic samples and 27 nonallergic control samples (Table S1). The 27 nonallergic control samples contained sera from 12 sensitized (≥0.35 kU/L sIgE against peanut) but tolerant (clinically nonreactive) individuals (Figure S1 and Table S2). Samples used in the BAT and Hoxb8 MAT were collected at the same timepoint. Research ethics board (REB) approval was obtained (REB 1000053791). Patient provided informed consent and studies were performed in accordance with the Helsinki Declaration. Serum samples were received at University of Bern completely blinded under an MTA.
2.2. Skin prick test (SPT), allergen‐specific IgE measurements (sIgE), and disease severity assessment
Previously acquired SPT and sIgE results from MONAS 21 have been used in this study for direct comparison to Hoxb8 MAT results. From the 96 samples, for which BAT data were available, SPT data for peanut extract (ALK‐Abelló) was previously collected for 47 individuals. Furthermore, sIgE measurements were performed using ImmunoCAP (Thermo Fisher) allergen testing as part of the clinical workup in MONAS. ImmunoCAP data from a total of 67 individuals was available (Figure 1A). Specific IgE measurements for the basophil nonresponder serum samples was determined using the ALEX platform from MADx. Disease severity based on the Astier classification (grade 1–5) 31 was available for 56 individuals (29 PA and 27 NA).
FIGURE 1.
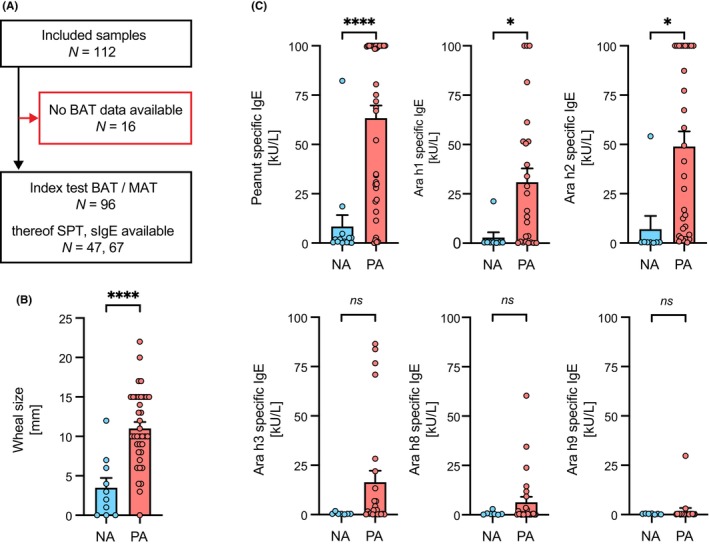
Study population with comparison of skin prick test and allergen‐specific IgE measurement results between clinically confirmed nonallergic controls and peanut allergic patients. (A) Blinded MONA study serum samples (n = 112) from the Toronto site with available clinical status were transferred to the University of Bern. Serum samples for which BAT data was recorded in MONAS (n = 96) were included in the clinical performance analysis of the Hoxb8 MAT and in direct comparison to BAT. Amongst those SPT information for 47 samples and sIgE measurements for 67 samples has been available. (B) Wheal size measurement results from skin prick test compared between nonallergic controls (NA: N = 10; blue) and peanut allergic patients (PA: N = 37; red). (C) Comparison of allergen‐specific IgE measurements by ImmunoCAP for peanut extract, Ara h1, Ara h2, Ara h3, Ara h8, and Ara h9 between nonallergic controls (NA: N = 16; blue) and peanut allergic patients (PA: N = 51; red). Data are shown as individual data points and means ± SEMs. *p < .05, ****p < .001, ns, not significant.
2.3. BAT
Previously acquired BAT results from the MONA study have been used and bioinformatically reanalyzed in this study for direct comparison to Hoxb8 MAT results. BAT has been performed as previously described. 21 Briefly, heparinized whole blood was drawn and stored at room temperature until processed within <6 h. Whole blood was stimulated with up to 7 serial 10‐fold dilutions (0.001–1000 ng/mL) with peanut extract (ALK‐Abelló). The BAT was performed according to the manufacturer's instructions (BÜHLMANN Laboratories AG, Basel, Switzerland). Flow cytometric analysis was performed with a CytoFLEX (Beckman Coulter, USA). Additionally, sera from BAT nonresponders (four positive responders in the negative control; three nonresponders to the positive control), who were previously identified in MONAS, were transferred to the University of Bern for this study.
2.4. Hoxb8 MAT
Hoxb8 MC progenitors that were cultured in RPMIc/IL3 medium containing 4‐hydroxytamoxifen (4‐OHT) were washed in PBS, resuspended in RPMIc/IL‐3 medium without 4‐OHT and reseeded at 7.5 × 104 cells/ml in a culture flask for 5 days to differentiate mature Hoxb8 MCs. For the Hoxb8 MAT, 5 × 104 Hoxb8 MCs per condition/well were seeded in a 96‐well round bottom plate. Cells were centrifuged at 600 × g for 5 min and resuspended in preprocessed serum samples for overnight passive sensitization. Preprocessing included a buffer exchange into activation medium using 2 mL ultrafiltration spin columns with an MWCO of 100 kDa (VivaSpin, Sartorius, Germany). The columns were used according to the manufacturer's instructions. Briefly, 300 μL of serum was topped up with activation medium to yield 1.5 mL of solution. The mixture was centrifuged at 4000 × g until a >5‐fold reduction in volume was achieved. This step was repeated two more times. In the end the solution was filled up with activation medium to the 300 μL of initial starting volume of the serum sample. Cells were sensitized with preprocessed serum samples overnight. Subsequently, without wash step the cells were stimulated with 6 serial 10‐fold dilutions (0.001–1000 ng/mL) with peanut extract (provided by ALK‐Abelló) plus a staining antibody for anti‐CD107a (clone: 1D4B, BioLegend). Stimulation was performed for 25 min at 37°C in the presence of 5% CO2. After washing the cells with FACS buffer, they were resuspended in 200 μL FACS buffer and acquired on a CytoFLEX S 4L 13C (B2‐R3‐V4‐Y4) plus 96 DW plate loader (Beckman Coulter Life Sciences, CA, USA), and results were evaluated with FlowJo Version 10.1 (FlowJo, OR, USA). To ensure robustness and reproducibility of results generated with mature Hoxb8 MCs from different progenitor line thawings at various differentiation timepoints, we performed quality control experiments, in which cells were sensitized with a titration of NIP‐specific humanized JW8‐IgE (0.001–5 μg/mL) and stimulated with 100 ng/mL NIP22‐BSA (Figure S2). Furthermore, to address whether preprocessing of the serum samples affected their IgE content, we performed an IgE sandwich ELISA. For this purpose, a dilution series of recombinant monoclonal Sus11‐IgE (0.5–100 μg/mL) was measured on solid phase immobilized anti‐IgE Le27 (5.4 μg/mL). Biotinylated anti‐human IgE (MabTech, clones 107, 182, 101) followed by streptavidin poly‐HRP (ThermoFisher) was used for detection (Figure S3A). Once the assay was established, IgE levels were measured in unprocessed versus processed serum samples for two concentrations of spiked Sus11‐IgE (100 ng/mL and 1000 ng/mL) in a nonallergic serum and for three peanut allergic sera (Figure S3B). Absorption values were determined by optical density measurement at 450 nm wavelength.
2.5. Proteins, antibodies, and media
Antibody for detection of Hoxb8 MC activation: monoclonal rat anti‐mouse CD107a APC and PE (clone 1D4B, BioLegend, CA, USA). Activation medium: RPMI‐1640 w/stable glutamine, 2.0 g/L NaHCO3 (Seraglob, Bioswisstec AG, Schaffhausen, Switzerland) complemented with 10% Hyclone FCS (Fisher Scientific, Hampton, NH), 100 U/mL penicillin, 100 mg/mL streptomycin (1003 penicillin/streptomycin, Gibco), 10 mM HEPES buffer solution (stock‐solution 1 mol, Gibco), 1 mM sodium pyruvate (stock‐solution 100 mM, 1003, Gibco), 4 mM L‐glutamine (stock‐solution 200 mM, 1003, Gibco), 13 nonessential amino acids (stock‐solution 1003, Gibco), 30 ng/ mL mouse recombinant IL‐3 (Peprotech), and 50 mM 2‐mercaptoethanol (stock‐solution 14.3 mol, Merck, Darmstadt, Germany). RPMIc: RPMI1640 medium AQmedia (Sigma‐Aldrich, St Louis, Mo) complemented with 10% FCS Sera Pro (Pan Biotech, Aidenbach, Germany), 100 U/mL penicillin, 100 mg/mL streptomycin (1003 penicillin/streptomycin, Gibco by Sigma‐Aldrich), and 50 mM 2‐mercaptoethanol (stock‐solution 14.3 mol, Merck, Darmstadt, Germany). RPMIc/IL3: RPMIc plus 10% WEHI‐3b conditioned medium.
2.6. Statistical analyses
SPT and sIgE measurements results of PA and NA groups were compared using Student's unpaired t‐test. The simple linear regression model was used to correlate SPT, sIgE measurements, disease severity, and maximal BAT results with maximal Hoxb8 MAT results. Two‐way ANOVA with Sidak's multiple comparison was used to compare BAT and Hoxb8 MAT results at individual allergen concentrations between PA and NA groups. Performance of SPT, sIgE measurements, BAT and Hoxb8 MAT was examined against the clinically validated allergic status of individual samples using receiver‐operating characteristic (ROC) curves via either the R package cutpointr (version 1.1.2) to find the optimal cut point or by application of a logistic regression models in connection with the R package ROCR (version 1.0–11). GraphPad Prism (Version 10; Pad Software, La Jolla, CA, USA) and R (Version 4; R Core Team 2023) were used for statistical testing.
3. RESULTS
3.1. Clinical characterization of study population
This investigation included patient samples from the Toronto site of MONAS 21 with confirmed allergy status regarding peanut allergy and available serum samples. In the total of 96 serum samples 69 were classified as peanut allergic (PA) and 27 as nonallergic controls (NA) based on positive OFC, or a pretest probability of >90% allergic reaction (Figure 1A and Table S1). Twelve out of the 27 nonallergic control samples contained sIgE against peanut ≥0.35 kU/L and can thus be considered as sensitized but tolerant (Figure S1 and Table S2). Amongst the 96 serum samples, SPT as well as sIgE measurements were readily available from 47 and 67 patients, respectively (Figure 1A,B and Table S1). PA and NA differed significantly regarding SPT wheal size (Figure 1B) and sIgE serum levels against whole peanut extract or the seed storage proteins Ara h 1 and Ara h 2 (Figure 1C). Additionally, the performance of SPT and sIgE measurements to correctly identify the allergic status was assessed using receiver‐operating characteristic (ROC) curves. Based on the calculated optimal cut offs for each allergen, the diagnostic accuracy was highest for IgE against Ara h2 with an area under the ROC curve (AUROC) of 0.91, which outperformed both sIgE measurement against whole peanut extract and SPT with peanut extract (both AUROC of 0.89) (Table S3).
3.2. Hoxb8 MAT discriminates between peanut allergic and nonallergic children and adolescents
All samples had been stored at −80°C for more than 12 months before they were analyzed in the previously described Hoxb8 MAT 30 in a blinded manner, and all experiments were analyzed by a biostatistician outside of the lab of the two principal investigators. The primary goal of this study was to assess the diagnostic performance of the serum based Hoxb8 MAT in comparison to the whole blood based BAT using 96 samples (69 PA; 27 NA controls). Mature Hoxb8 MCs were passively sensitized with preprocessed sera and stimulated with 6 serial 10‐fold dilutions (0.01–1000 ng/mL) of peanut extract. The percentage of activated mast cells was quantified by the detection of CD107a using flow cytometry. Mature Hoxb8 MCs sensitized with samples from peanut allergic patients showed a significantly higher percentage of activated mast cells upon stimulation with 10–1000 ng/mL peanut extract as compared to those with samples of the nonallergic controls (Figure 2A). All PA patient sera, except five, followed an allergen dose‐dependent activation curve (Figure 2B), while all samples in the NA population, except one, showed no activation. Interestingly, the only false positive sample featured remarkably high sIgE against peanut extract (82.3 kU/L), Ara h1 (21.2 kU/L), and Ara h2 (54.1 kU/L), consumes peanut regularly at relevant amounts without being on an OIT and displays, mild, intermittent skin reactivity. The very patient also displayed BAT reactivity. In addition to this individual, the NA group contained 11 additional samples from sensitized but tolerant participants, which all remained negative in the Hoxb8 MAT (Figure S1 and Table S2). Sera from peanut allergic individuals with as little as 0.49 kU/L or 0.27 kU/L specific IgE to peanut extract or Ara h2, respectively, showed activation above the optimal cut off and were correctly classified as allergic.
FIGURE 2.
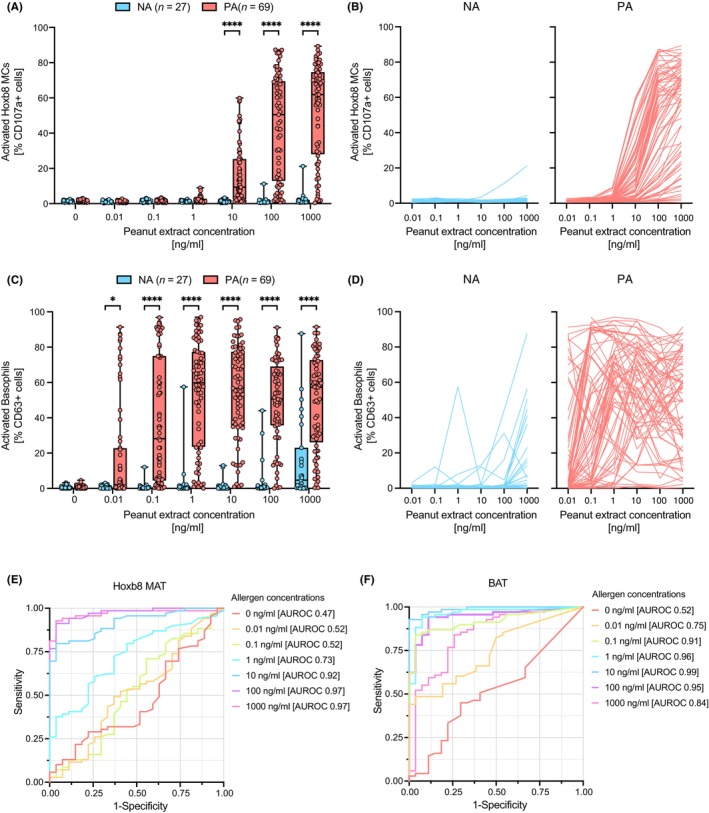
Performance analysis of Hoxb8 MAT and BAT to differentiate between clinically confirmed nonallergic controls and peanut allergic patients. Passively sensitized Hoxb8 MCs (A and B) or whole blood (C and D) were stimulated with peanut allergen extract at various doses (0–1000 ng/mL). Comparison of activated Hoxb8 MCs or blood basophils in the nonallergic (NA) and peanut allergic (PA) group on a population basis is shown (A and C). Dose–response curves of activated Hoxb8 MCs and blood basophils for individual patient samples are represented in two separate subpanels (B and D). Nonallergic controls are depicted in blue, while peanut allergic patients are represented in red. (E and F) ROC curve analyses for Hoxb8 MAT and BAT results across different allergen concentrations (color coded) are depicted and AUROC values are indicated. Data are shown as individual data points and means ± SEMs. *p < .05, ****p < .001.
Similarly, the percentage of CD63 (i.e., LAMP‐3) positive basophils in whole blood of PA patients were significantly higher compared to NA patient samples upon allergen challenge at concentrations ranging from 0.1 to 1000 ng/mL (Figure 2C). Overall, the allergen dose response curves on the individual sample level showed more variability in the BAT (Figure 2D), with several samples in the NA group becoming activated at allergen‐doses >100 ng/mL and various samples in the PA group showing inverse dose‐dependency or the so‐called “hook effect”.
To assess and re‐evaluate the diagnostic performance of the Hoxb8 MAT and the BAT in the identification of peanut allergic patients in our cohort, we performed ROC curve analyses for both assays including the data from all 96 tested samples at each individual allergen concentration (0–1000 ng/mL). For the Hoxb8 MAT the AUROC curves demonstrated a consistent allergen dose‐dependent increase of diagnostic accuracy which hit a plateau at the 100 ng/mL peanut allergen dose (AUROC of 0.97) and stayed at this level for 1000 ng/mL peanut allergen (Figure 2E and Table 1). The diagnostic accuracy of the BAT was high at low peanut allergen concentration (e.g., 0.1 and 1 ng/mL), reached its maximum at 10 ng/mL (AUROC of 0.99) and then declined at 100 and 1000 ng/mL of peanut allergen (Figure 2F and Table 1). A direct comparison between BAT and Hoxb8 MAT results at allergen concentrations with the highest diagnostic accuracy (10 ng/mL for BAT and 1000 ng/mL for MAT) shows moderate but highly significant linear correlation (Figure S4).
TABLE 1.
Diagnostic accuracy of BAT and Hoxb8 MAT.
| Diagnostic test | Optimal cut‐off | AUROC | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| BAT 0.01 ng/ml (% CD63) | 2.00 | 0.75 | 0.49 | 0.96 | 0.97 | 0.42 |
| BAT 0.1 ng/ml (% CD63) | 2.61 | 0.92 | 0.84 | 0.96 | 0.98 | 0.70 |
| BAT 1 ng/ml (% CD63) | 2.50 | 0.96 | 0.94 | 0.93 | 0.97 | 0.86 |
| BAT 10 ng/ml (% CD63) | 13.06 | 0.99 | 0.93 | 1.00 | 1.00 | 0.84 |
| BAT 100 ng/ml (% CD63) | 5.59 | 0.95 | 0.94 | 0.89 | 0.96 | 0.86 |
| BAT 1000 ng/ml (% CD63) | 17.10 | 0.84 | 0.84 | 0.74 | 0.89 | 0.65 |
| Hoxb8 MAT 0.01 ng/ml (% CD107a) | 1.02 | 0.52 | 0.49 | 0.63 | 0.77 | 0.33 |
| Hoxb8 MAT 0.1 ng/ml (% CD107a) | 1.13 | 0.52 | 0.71 | 0.44 | 0.77 | 0.38 |
| Hoxb8 MAT 1 ng/ml (% CD107a) | 1.24 | 0.73 | 0.72 | 0.63 | 0.83 | 0.47 |
| Hoxb8 MAT 10 ng/ml (% CD107a) | 1.98 | 0.92 | 0.80 | 0.96 | 0.98 | 0.65 |
| Hoxb8 MAT 100 ng/ml (% CD107a) | 2.58 | 0.97 | 0.91 | 0.96 | 0.98 | 0.81 |
| Hoxb8 MAT 1000 ng/ml (%CD107a) | 5.20 | 0.97 | 0.93 | 0.96 | 0.98 | 0.84 |
3.3. Hoxb8 MC activation correlates with SPT, sIgE, and disease severity
To better understand, if the data we generated using Hoxb8 MC passively sensitized with serum samples correlate with results from allergy diagnosis tests routinely performed in clinics, we conducted linear regression analyses between the maximal activation of Hoxb8 MCs achieved upon allergen stimulation and either SPT, sIgE measurements, or disease severity as graded by Astier's classification. 31 Only a weak (r = 0.3), yet statistically significant, relationship between the Hoxb8 MC activation and SPT results was found (Figure 3A). Furthermore, there was a strong but imperfect correlation with sIgE measurements against peanut extract (r = 0.91) and Ara h2 (r = 0.81) (Figure 3B–D). While the correlation between disease severity and allergen stimulation in the Hoxb8 MAT got linearly stronger with increasing allergen concentrations used, this correlation showed a hook effect with optimal allergen concentration of 10 ng/mL peanut extract for the BAT (Figure S5 and Figure 3E). Importantly, results from both cellular assays showed significant correlations with disease severity (Table S4).
FIGURE 3.
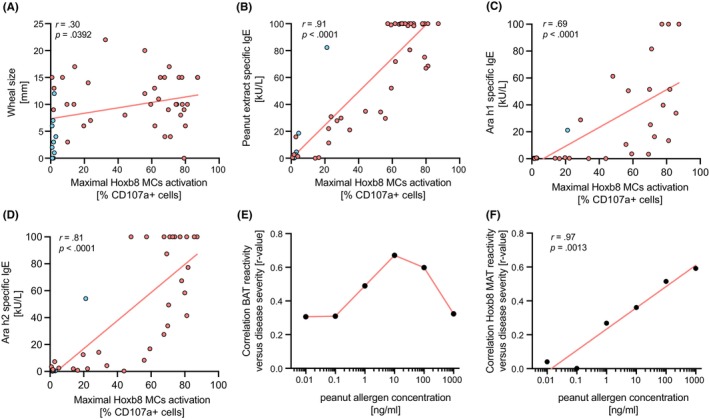
Correlation analysis between skin prick test or allergen‐specific IgE measurements and maximal activation signal in the Hoxb8 MAT. Wheal size results from skin prick test (A) or allergen specific IgE measurement for peanut extract (B), Ara h1 (C), and Ara h2 (D) from nonallergic controls (red dots) and allergic patients (blue dots) were plotted against the maximal activation signal from the Hoxb8 MAT and a linear correlation analysis (red line) was performed. The correlation coefficient (r) as well as the significance of the correlation (p) are indicated for each subpanel. (E) The strength of the linear relationship (r‐value) between the maximal Hoxb8 MAT activation and the disease severity according to Astier classification was plotted against the different allergen concentration used (0.01–1000 ng/mL) and a linear correlation analysis (red line) was performed.
3.4. The Hoxb8 MAT accurately captures BAT nonresponders
To investigate whether the Hoxb8 MAT could potentially overcome the diagnostic limitation of basophils that do not respond to FcεRI‐dependent activation in the BAT, we tested sera from seven patients (Table S5) that were previously classified as not meeting the quality controls in MONAS. 21 Four out of these seven samples stemmed from clinically confirmed peanut allergic patients (i.e., NR1, NR3, NR4, and NR5). Three samples were from nonallergic individuals (i.e., NR2, NR6, and NR7). Importantly, all four samples from the PA group showed dose‐dependent activation in the Hoxb8 MAT, whereas no activation upon allergen challenge was observed in the three samples from the nonallergic individuals (Figure 4). These results highlight that nonresponder basophil samples have been accurately classified based on the Hoxb8 MAT outcome.
FIGURE 4.
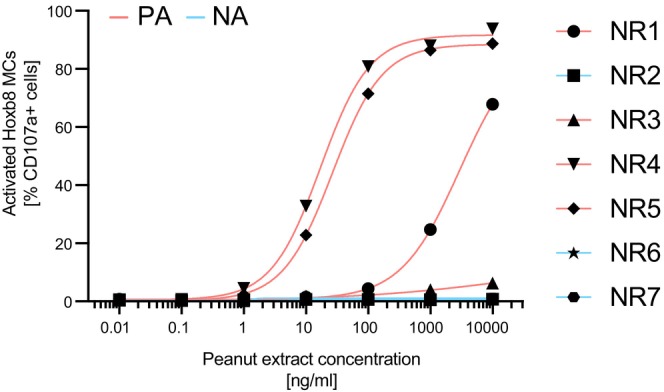
Analysis of seven BAT nonresponder samples with the Hoxb8 MAT. Seven sera from previously identified BAT nonresponders (NR1‐7) were measured on passively sensitized Hoxb8 MCs. The clinically confirmed allergy status of the individual patient samples is color coded (peanut allergic, PA: red; nonallergic, NA: blue; no information).
3.5. The Hoxb8 MAT performs similar as the BAT in clinical utility assessment
Further, we directly compared the diagnostic performance of the Hoxb8 MAT to the BAT, SPT, and sIgE measurement all using peanut extract as an allergen source. While a mixed effect model integrating 10 and 100 ng/mL of peanut allergen stimulus has previously been reported to be most accurate for the BAT (AUROC 0.975), 21 we here used a logistic regression model taking into consideration all peanut allergen concentration (0.01–1000 ng/mL) as variables for the Hoxb8 MAT as well as for the BAT. Different sample numbers had to be used to calculate the diagnostic accuracy of each test due to limitations in serum sample volume and data availability. However, the Hoxb8 MAT and the BAT outperformed SPT and sIgE measurements in this analysis (Figure 5 and Table S3). Both cellular assays (i.e., Hoxb8 MAT and BAT) were highly accurate with similar diagnostic performance (AUROC 0.95 and 0.96, respectively).
FIGURE 5.
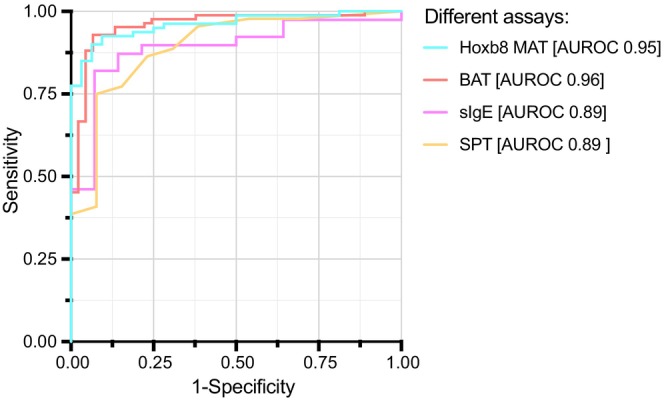
Direct comparison of performance analyses between different diagnostic tests. Direct ROC curve analysis for SPT and sIgE against peanut extract compared with ROC curve analysis of logistic regression models with Hoxb8 MAT and BAT results including all peanut allergen concentrations are depicted (color coded) and AUROC values are indicated.
4. DISCUSSION
Peanut allergy is one of the most frequent causes of severe food‐associated anaphylaxis. To prevent or minimize such systemic, potentially life‐threatening allergic reactions, accurate and reliable diagnostic tools that are easy to implement and safe are urgently required. Moreover, diagnostic accuracy and the reliability of negative values in sensitized individuals is needed to avoid unnecessary diagnosis with all the consequences that come with the diagnosis of peanut allergy. In this study, we sought to investigate the clinical utility of the recently developed in vitro Hoxb8 MAT 30 for the diagnosis of peanut allergies and to compare it to established and validated clinical diagnostic tests including SPT, sIgE measurements, and BAT. For this purpose, we analyzed serum samples that have been prospectively collected in the context of MONAS and were subsequently stored long term in a frozen state (−80°C) in a blinded manner. Importantly, the BAT has previously shown high accuracy in the diagnosis of peanut allergy in multiple studies. 9 , 11 , 16 , 21 , 22 , 32 The results shown here strongly suggest that the Hoxb8 MAT performs as well as the BAT in terms of diagnostic accuracy in a mixed cohort of severe and clear peanut allergic patients and those with an equivocal history including sensitized but tolerant individuals. While both tests accurately classified 26 out of 27 NA controls, the Hoxb8 MAT correctly identified 64 and the BAT 65 out of 69 PA individuals at their respective optimal cutoff values. At the same time, the cellular assays were more accurate as commonly performed tests like the SPT and sIgE measurement.
While the idea to perform MAT for diagnostic purposes is not new, 29 , 33 its broader implementation has been hampered by scalability limitations. In one study, Bahri et al. generated mature mast cells from isolated human primary blood stem cells. 33 Their results concerning diagnostic accuracy were similar to those we present here using Hoxb8 MCs. However, their assay relied on a sophisticated mast cell differentiation protocol, which required 8–10 weeks to generate a limited number of cells available for the diagnostic assay. Another challenge of this approach is the variability between different blood stem cell donors, which might affect assay robustness and diagnostic reproducibility, unless cells of various donors are pooled to minimize variability each time. These issues have been resolved with the Hoxb8 MC system, as mature mast cells can be repetitively and robustly differentiated from the same immortalized mast cell progenitor stock at unlimited numbers within only 5 days. In another study, Santos et al. have used the human LAD2 34 mast cell line for the diagnosis of peanut allergies. 34 While the result also looked encouraging, LAD2 cells generally feature slow growth characteristics with a doubling time of 2 weeks. 35 Hoxb8 MC progenitors double roughly once every 24 h. 30
Hoxb8 MCs showed optimal cut‐off values ≤5.2%, which is comparable to the cut offs reported for BAT, 9 , 11 , 32 as well as maximal activation of >89% in our diagnostic performance study. While other common cell lines such as rat basophilic leukemia cells (e.g., RBL‐2H3 or RBL‐SX38) can be used in degranulation experiments by quantifying released enzymatic mediators in the culture supernatant with an indirect colorimetric assay (i.e., β‐hexosaminidase activity quantification), these cells grow in an adherent manner and can thus not be used in flow cytometric analysis. Importantly, their maximal degranulation capacity lies around 40%–60%. 36 The findings presented here suggest that the Hoxb8 MAT might overcome prior limitations of other MAT approaches or allergic effector cell lines, while maintaining excellent diagnostic accuracy.
From a methodological standpoint the BAT and the MAT both represent similar cell‐based functional in vitro tests, which rely on flow cytometric identification of activated allergic effector cells upon allergen stimulation. However, there are major differences between these two assays. 28 The BAT is performed with patient's whole blood, which must be processed and measured within maximally 24 h to ensure valid results. 24 The MAT, on the contrary, relies on serum samples that can be stored in the freezer over months (even years) and analyzed in large batches. This is particularly important for large studies or multicenter clinical trials as the analysis of serum samples can be done retrospectively in a centralized manner. The serum samples included in this study were analyzed in just 2 days. Thus, the Hoxb8 MAT clearly has the potential to overcome logistical challenges that have hampered a broader implementation of the BAT in clinics so far.
Direct comparison of activation results between the BAT and the Hoxb8 MAT revealed that the BAT requires roughly 100‐times lower allergen concentrations to induce comparable percentage of cell activation. This suggests that the releasability threshold of endogenous basophils is lower compared to passively sensitized Hoxb8 MCs. One potential explanation for this observation could be that primary human basophils express higher FcɛRI levels. Indeed, an average level of 250′000 receptors per cell (rpc) has been reported on basophils of atopic individuals. 37 Thus, there are roughly 2.7 times more FcɛRI on the cell surface of basophils than passively sensitized Hoxb8 MCs (i.e., ~90′000 rpc). 30 At the same time, the BAT features significantly higher interindividual variability and nonspecific activation in nonallergic individuals at high allergen concentrations, which is likely due to the lower releasability threshold or higher cellular sensitivity. Moreover, the BAT showed a bell‐shaped activation curve (hook effect) in which high allergen concentrations led to lower activation, which has not been observed in the Hoxb8 MAT at the tested allergen concentrations. In general, it is our understanding that the cellular sensitivity is irrelevant when it comes to the comparison of diagnostic accuracies between the BAT and MAT. In other words, the absolute concentration of allergen needed to activate the cells is secondary to the optimal discrimination of activated versus nonactivated cells at a given allergen concentration in samples from allergic and nonallergic individuals.
Despite losing intrinsic cellular information (i.e., patient's own versus lab grown allergic effector cells), the Hoxb8 MAT showed comparable diagnostic accuracy as the BAT in this study, further supporting the notion that humoral components contained in the serum are sufficiently representative of a patient's allergy status. Furthermore, in approximately 10%–15% of cases the BAT results in false negative or false positive outcomes with basophils that do not show any activation upon FcεRI‐mediated stimulation or with unspecific activation of basophils in the negative control. 21 , 38 , 39 It has been demonstrated that basophils of so‐called nonresponders feature an impaired signaling cascade with low tyrosine kinase Syk levels. 40 , 41 These individuals pose an important diagnostic challenge in the daily clinical allergy workup. It is also important to note, that many diagnostic performance studies using BAT exclude these nonresponder samples and thereby do not entirely represent the real‐life situation. 16 , 18 , 21 , 22 As the initial dataset we used in this study did not contain nonresponder samples, the diagnostic accuracy of the BAT in our ROC curve analysis might be overestimated as in many other studies. Importantly, we provided strong evidence in a separate experiment showing that the Hoxb8 MAT is able of accurately capturing these nonresponder samples and might thereby overcomes this current diagnostic issue. 29
Compared to the well‐established sIgE measurements (e.g., ImmunoCAP) the Hoxb8 MAT requires living cells and is thus more laborious to perform. At the same time, it is a functional assay that is more representative of the in vivo situation than an immunoassay that assesses the level but not functionality of particular IgE or IgG antibodies. The Hoxb8 MAT integrates multiple quantitative and qualitative variables that play an important role in the allergic response like IgE clonality, affinity and epitope specificity and the same for potentially protective IgG present in serum. While it will take several days to get a Hoxb8 MAT result, the physician receives test results of an SPT within 1 h. However, the discomfort associated with performing SPT including extended supervision of the patient in the clinic will be more extensive compared to an ex vivo test based on patient serum.
Overall, this study has demonstrated that the Hoxb8 MAT features excellent diagnostic accuracy and outperformed both SPT and sIgE measurements in the diagnosis of peanut allergy. While it represents a valuable alternative to established allergy diagnosis tests further studies including other allergens and allergen components will be conducted in near future.
AUTHOR CONTRIBUTIONS
NZ, RvB, NR, and MS performed Hoxb8 MAT and did data entry. AC analyzed data and critically reviewed the manuscript. JAH, XY, and CMD performed BAT and did data entry. LD cross checked clinical accuracy and contributed to clinical data entry. JEMU cross checked clinical accuracy and contributed to clinical data entry, contributed to recruitment, and contributed to manuscript revision. TK contributed to Hoxb8 MC generation, provided advise on study design and critically reviewed the manuscript. AE and TE codesigned the study, analyzed data, cocontributed to generation of the manuscript, and cosupervised the project and collaboration. All authors reviewed the results and commented on the manuscript.
FUNDING INFORMATION
TK and AE received grant support from the Swiss Innovation Agency Innosuisse, Switzerland (grant no. 52202.1 IP‐LS). AE received grant support from the Swiss National Science Foundation (grant no. 310030_219361). TE and JEM received grant support from the Food Allergy and Anaphylaxis Program at the Hospital for Sick Children.
CONFLICT OF INTEREST STATEMENT
NBZ is a cofounder, shareholder, and employee of ATANIS Biotech AG. AC has nothing to declare. RvB is an employee of ATANIS Biotech AG. NR, MS, JAH, XY, CMD, and LD have nothing to disclose. JEMU reports grants and personal fees from ALK‐Abelló A/S, personal fees from Bausch Health, personal fees from Kaleo and Pharming Group N.V, grants from DBV Technologies, grants from Regeneron and Sanofi, and grants from Food Allergy Anaphylaxis Programme (SickKids), outside the submitted work; and Section Chair of Food Allergy and Anaphylaxis, Canadian Society of Allergy and Clinical Immunology; Healthcare Advisory Board, Food Allergy Canada. TK is a cofounder, shareholder, and board member of ATANIS Biotech AG. AE is a cofounder, consultant, and shareholder of ATANIS Biotech AG and Excellergy, Inc. AE received grants from ATANIS Biotech AG, Nestlé, Novartis AG, and BÜHLMANN Laboratories AG. TE reports grants from CIHR, FWF, Food Allergy and Anaphylaxis Program Sickkids, ALK. He is site PI of company sponsored trials by DBV, Novartis and Stallergenes Greer, EFSA and FARE. Personal fees from Danone/Nutricia/Milupa, ThermoFisher, Aimmune, Stallergenes Greer, ALK, MADx and Nonfinancial support from Novartis and MADx, all outside the submitted work. He is an associate editor of Allergy.
Supporting information
Figures S1–S5.
Tables S1–S5.
ACKNOWLEDGEMENTS
We thank all the people from the Kaufmann, Eggel and Eiwegger laboratories. We also acknowledge members of ATANIS Biotech AG, Prof. Dr. med. Jean‐Pierre Kinet, Charlène Niogret, Marianne Zwicker and especially Antonia Ferreira and Sophie Buchser for valuable scientific discussions and support with experimental procedures and protocols. Open access funding provided by Universitat Bern.
1.
MONAS working group: Heimo Breiteneder, Chiara Palladino, Klara Schmidthaler, Delvin So, Marta Ponce, Jean‐Soo Lee, Lisa Hung, Adelle R. Atkinson, Vy H.D. Kim, Alireza Berenjy, Maria Asper, David Hummel, Samantha Wong, Mara Alexanian‐Farr, Ahuva Magder, Zsolt Szépfalusi.
Bachmeier‐Zbären N, Celik A, van Brummelen R, et al. Clinical utility analysis of the Hoxb8 mast cell activation test for the diagnosis of peanut allergy. Allergy. 2025;80:215‐226. doi: 10.1111/all.16341
See Appendix for MONAS working group.
Noemi Bachmeier‐Zbären and Alper Celik contributed equally to this work.
Alexander Eggel and Thomas Eiwegger jointly supervised this work.
Contributor Information
Alexander Eggel, Email: alexander.eggel@unibe.ch.
Thomas Eiwegger, Email: thomas.eiwegger@kl.ac.at.
MONAS working group:
Heimo Breiteneder, Chiara Palladino, Klara Schmidthaler, Delvin So, Marta Ponce, Jean‐Soo Lee, Lisa Hung, Adelle R. Atkinson, Vy H.D. Kim, Alireza Berenjy, Maria Asper, David Hummel, Samantha Wong, Mara Alexanian‐Farr, Ahuva Magder, and Zsolt Szépfalusi
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Loh W, Tang MLK. The epidemiology of food allergy in the global context. Int J Environ Res Public Health. 2018;15:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lieberman JA, Gupta RS, Knibb RC, et al. The global burden of illness of peanut allergy: a comprehensive literature review. Allergy. 2021;76:1367‐1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prescott SL, Pawankar R, Allen KJ, et al. A global survey of changing patterns of food allergy burden in children. World Allergy Organ J. 2013;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hourihane JO, Kilburn SA, Dean P, Warner JO. Clinical characteristics of peanut allergy. Clin Exp Allergy. 1997;27:634‐639. [PubMed] [Google Scholar]
- 5. Cummings AJ, Knibb RC, King RM, Lucas JS. The psychosocial impact of food allergy and food hypersensitivity in children, adolescents and their families: a review. Allergy. 2010;65:933‐945. [DOI] [PubMed] [Google Scholar]
- 6. Blok BMJD, Vlieg‐Boerstra BJ, Elberink JNGO, et al. A framework for measuring the social impact of food allergy across Europe: a EuroPrevall state of the art paper: social impact of food allergy across Europe. Allergy. 2007;62:733‐737. [DOI] [PubMed] [Google Scholar]
- 7. Foong R‐X, Dantzer JA, Wood RA, Santos AF. Improving diagnostic accuracy in food allergy. J Allergy Clin Immunol Pract. 2021;9:71‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sindher SB, Long A, Chin AR, et al. Food allergy, mechanisms, diagnosis and treatment: innovation through a multi‐targeted approach. Allergy. 2022;77:2937‐2948. [DOI] [PubMed] [Google Scholar]
- 9. Santos AF, Riggioni C, Agache I, et al. EAACI guidelines on the diagnosis of IgE‐mediated food allergy. Allergy. 2023;78:3057‐3076. doi: 10.1111/all.15902 [DOI] [PubMed] [Google Scholar]
- 10. Boyce JA, Assa'ad A, Burks AW, et al. Guidelines for the diagnosis and Management of Food Allergy in the United States: summary of the NIAID‐sponsored expert panel report. J Allergy Clin Immunol. 2010;126:1105‐1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muraro A, Werfel T, Hoffmann‐Sommergruber K, et al. EAACI Food Allergy and Anaphylaxis Guidelines: diagnosis and management of food allergy. Allergy. 2014;69:1008‐1025. doi: 10.1111/all.12429 [DOI] [PubMed] [Google Scholar]
- 12. Kattan JD, Sicherer SH. Optimizing the diagnosis of food allergy. Immunol Allergy Clin. 2015;35:61‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santos AF, Brough HA. Making the most of in vitro tests to diagnose food allergy. J Allergy Clin Immunol: Pr. 2017;5:237‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. NIAID‐Sponsored Expert Panel , Boyce JA, Assa'ad A, et al. Guidelines for the diagnosis and Management of Food Allergy in the United States: report of the NIAID‐sponsored expert panel. J Allergy Clin Immunol. 2010;126:S1‐S58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hemmings O, Niazi U, Kwok M, James LK, Lack G, Santos AF. Peanut diversity and specific activity are the dominant IgE characteristics for effector cell activation in children. J Allergy Clin Immunol. 2021;148:495‐505.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santos AF, Bergmann M, Brough HA, et al. Basophil activation test reduces oral food challenges to nuts and sesame. J Allergy Clin Immunol Pract. 2021;9:2016‐2027.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruinemans‐Koerts J, Schmidt‐Hieltjes Y, Jansen A, Savelkoul HFJ, Plaisier A, Setten P. The basophil activation test reduces the need for a food challenge test in children suspected of IgE‐mediated cow's milk allergy. Clin Exp Allergy. 2019;49:350‐356. [DOI] [PubMed] [Google Scholar]
- 18. Krawiec M, Radulovic S, Foong R, et al. Diagnostic utility of allergy tests to predict baked egg and lightly cooked egg allergies compared to double‐blind placebo‐controlled food challenges. Allergy. 2023;78:2510‐2522. doi: 10.1111/all.15797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoffmann HJ, Santos AF, Mayorga C, et al. The clinical utility of basophil activation testing in diagnosis and monitoring of allergic disease. Allergy. 2015;70:1393‐1405. [DOI] [PubMed] [Google Scholar]
- 20. Santos AF, Kulis MD, Sampson HA. Bringing the next generation of food allergy diagnostics into the clinic. J Allergy Clin Immunol Pract. 2022;10:1‐9. [DOI] [PubMed] [Google Scholar]
- 21. Duan L, Celik A, Hoang JA, et al. Basophil activation test shows high accuracy in the diagnosis of peanut and tree nut allergy: the markers of nut allergy study. Allergy. 2021;76:1800‐1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Santos AF, Douiri A, Bécares N, et al. Basophil activation test discriminates between allergy and tolerance in peanut‐sensitized children. J Allergy Clin Immunol. 2014;134:645‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kleine‐Tebbe J, Erdmann S, Knol EF Jr, Macglashan DW Jr, Poulsen LK, Gibbs BF. Diagnostic tests based on human basophils: potentials, pitfalls and perspectives. Int Arch Allergy Immunol. 2006;141:79‐90. [DOI] [PubMed] [Google Scholar]
- 24. Mukai K, Gaudenzio N, Gupta S, et al. Assessing basophil activation by using flow cytometry and mass cytometry in blood stored 24 hours before analysis. J Allergy Clin Immunol. 2017;139:889‐899.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bachmeier‐Zbären N, Kinet J‐P, Kaufmann T, Eggel A. Mast cell activation test. Reference Module in Food Science; 2022. doi: 10.1016/b978-0-323-96018-2.00032-8 [DOI] [Google Scholar]
- 26. Bahri R, Bulfone‐Paus S. Mast cell activation test (MAT). Methods Mol Biol. 2020;2163:227‐238. [DOI] [PubMed] [Google Scholar]
- 27. Elst J, van der Poorten M‐LM, Gasse ALV, et al. Mast cell activation tests by flow cytometry: a new diagnostic asset? Clin Exp Allergy. 2021;51:1482‐1500. [DOI] [PubMed] [Google Scholar]
- 28. Ebo DG, Bahri R, Tontini C, et al. Mast cell versus basophil activation test in allergy: current status. Clin Exp Allergy. 2024;54:378‐387. doi: 10.1111/cea.14487 [DOI] [PubMed] [Google Scholar]
- 29. Santos AF, Couto‐Francisco N, Bécares N, Kwok M, Bahnson HT, Lack G. A novel human mast cell activation test for peanut allergy. J Allergy Clin Immunol. 2018;142:689‐691.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zbären N, Brigger D, Bachmann D, et al. A novel functional mast cell assay for the detection of allergies. J Allergy Clin Immunol. 2021;149:1018‐1030.e11. [DOI] [PubMed] [Google Scholar]
- 31. Astier C, Morisset M, Roitel O, et al. Predictive value of skin prick tests using recombinant allergens for diagnosis of peanut allergy. J Allergy Clin Immunol. 2006;118:250‐256. [DOI] [PubMed] [Google Scholar]
- 32. Riggioni C, Ricci C, Moya B, et al. Systematic review and meta‐analyses on the accuracy of diagnostic tests for IgE‐mediated food allergy. Allergy. 2023;79:324‐352. doi: 10.1111/all.15939 [DOI] [PubMed] [Google Scholar]
- 33. Bahri R, Custovic A, Korosec P, et al. Mast cell activation test in the diagnosis of allergic disease and anaphylaxis. J Allergy Clin Immunol. 2018;142:485‐496.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kirshenbaum AS, Akin C, Wu Y, et al. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FcepsilonRI or FcgammaRI. Leuk Res. 2003;27:677‐682. [DOI] [PubMed] [Google Scholar]
- 35. Kirshenbaum AS, Petrik A, Walsh R, et al. A ten‐year retrospective analysis of the distribution, use and phenotypic characteristics of the LAD2 human mast cell line. Int Arch Allergy Immunol. 2014;164:265‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rujitharanawong C, Yoodee S, Sueksakit K, et al. Systematic comparisons of various markers for mast cell activation in RBL‐2H3 cells. Cell Tissue Res. 2022;390:413‐428. [DOI] [PubMed] [Google Scholar]
- 37. MacGlashan D. FceRI density and spontaneous secretion from human basophils. PLoS One. 2017;12:e0179734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Santos AF, Alpan O, Hoffmann H. Basophil activation test: mechanisms and considerations for use in clinical trials and clinical practice. Allergy. 2021;76:2420‐2432. [DOI] [PubMed] [Google Scholar]
- 39. Hemmings O, Kwok M, McKendry R, Santos AF. Basophil activation test: old and new applications in allergy. Curr Allergy Asthma Rep. 2018;18:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kepley CL, Youssef L, Andrews RP, Wilson BS, Oliver JM. Syk deficiency in nonreleaser basophils. J Allergy Clin Immunol. 1999;104:279‐284. [DOI] [PubMed] [Google Scholar]
- 41. Kepley CL, Youssef L, Andrews RP, Wilson BS, Oliver JM. Multiple defects in fc epsilon RI signaling in Syk‐deficient nonreleaser basophils and IL‐3‐induced recovery of Syk expression and secretion. J Immunol. 2000;165:5913‐5920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S5.
Tables S1–S5.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


