Abstract
Gene identification, also known as gene finding or gene recognition, is among the important problems of molecular biology that have been receiving increasing attention with the advent of large scale sequencing projects. Previous strategies for solving this problem can be categorized into essentially two schools of thought: one school employs sequence composition statistics, whereas the other relies on database similarity searches. In this paper, we propose a new gene identification scheme that combines the best characteristics from each of these two schools. In particular, our method determines gene candidates among the ORFs that can be identified in a given DNA strand through the use of the Bio-Dictionary, a database of patterns that covers essentially all of the currently available sample of the natural protein sequence space. Our approach relies entirely on the use of redundant patterns as the agents on which the presence or absence of genes is predicated and does not employ any additional evidence, e.g. ribosome-binding site signals. The Bio-Dictionary Gene Finder (BDGF), the algorithm’s implementation, is a single computational engine able to handle the gene identification task across distinct archaeal and bacterial genomes. The engine exhibits performance that is characterized by simultaneous very high values of sensitivity and specificity, and a high percentage of correctly predicted start sites. Using a collection of patterns derived from an old (June 2000) release of the Swiss-Prot/TrEMBL database that contained 451 602 proteins and fragments, we demonstrate our method’s generality and capabilities through an extensive analysis of 17 complete archaeal and bacterial genomes. Examples of previously unreported genes are also shown and discussed in detail.
INTRODUCTION
As a testimony to the accelerated pace of genome sequencing projects, almost 80 complete genomes have been deposited in the public databases to date, whereas many more genomes are currently at various stages of sequencing. Consequently, the automated identification of the protein coding regions in a newly sequenced genome is attracting increasing attention.
Accurate gene prediction is of relevance to many biological applications. For example, the predicted coding regions can be used to generate probes for a DNA microarray, they can form the basis for knockout experiments, the candidate proteins corresponding to these predicted genes might be used as new drug targets, etc. In this paper, we focus on the prokaryotic gene identification problem. Gene identification is also known as ‘gene discovery’, ‘gene recognition’ or ‘gene finding’—the latter is the term we use in this discussion.
With the exception of a handful of reported instances in archaeal organisms, splicing does not generally occur in prokaryotes. Thus, the problem of gene identification in these organisms is generally considered to be simpler than its eukaryotic counterpart. The schemes which have been proposed over the years have permitted great advances in the in silico prediction of genes in prokaryotic genomes but, arguably, have shortcomings. As such, the demand for increasingly accurate prediction schemes continues.
Over the years, a large number of methods have been proposed that address the gene finding problem. These methods can be largely divided into two categories. The methods in the first category make use of the statistics of DNA sequences to determine the location of genes. In fact, it was observed very early that the statistical properties of nucleotide usage differ inside DNA regions which code for genes and outside those regions: the concept of the CpG island (1) is a demonstration of such a difference in statistical behavior. Among the gene identification methods that make use of this observation, those which are based on Markov models are the most popular to date (2–5).
The second category comprises methods that are based on similarity search in large databases of genomic information (6–11). These methods carry out database searches in an effort to determine DNA regions (respectively amino acid sequences) that share similarities with the DNA regions (respectively amino acid translations) of ORFs from the genome under consideration. For details on such techniques, the reader is referred to one of the many review papers on the topic: several of these papers also address the gene identification problem in eukaryotic organisms (12–17).
Despite the notable success of the methods that have been developed over the years, each of these two basic strategies has its own shortcomings. Statistical methods such as those based on Markov models can identify coding regions whose statistical behavior is similar to that of the used training set. If no appropriate training set is available, one resorts to using sets derived from database searches, or simply assumes that very long ORFs do code for genes. The statistics of coding regions often differ from organism to organism and, ideally, if one wishes to achieve high prediction ratios using such approaches then one ought to employ models with organism-dependent parameters. Essentially, a different Markov model must be built for each targeted genome. Moreover, short genes (e.g. <60–80 amino acids) cannot be predicted reliably using statistical methods. Finally, genes that are statistically distinct from other genes of the same organism, e.g. genes that are the result of horizontal transfer (18,19), typically represent challenges for methods based on statistical schemes.
Unlike statistical methods, similarity-based approaches are more effective in finding short genes or genes that are statistically distinct from the majority of the genes in the organism being studied. The implicit assumption here is that similar genes or similar proteins are already present in the databases that are searched. Clearly, there is no dependence on training sets since no such sets are needed. Problems can arise if the shared similarity between a candidate gene and its database counterpart is very low and not detectable. In general, similarity-based methods have an improved ability to determine the correct location of genes over statistical methods, a very desirable property for gene finding tools.
Given that the best characteristics of these two categories complement one another, genome sequencing projects typically use representative methods from both categories to generate results (20).
In what follows, we present a new method which borrows the best attributes from similarity searches, while at the same time relying on implicit sequence statistics as in the case of Markov models. Our method builds on a new paradigm that describes amino acid sequences with the help of patterns that are present in these sequences. The patterns are derived by processing very large public databases of amino acid sequences with the help of an unsupervised discovery algorithm. It is worth noting that our method does not make any use of additional evidence, e.g. ribosome-binding sites, in order to decide the presence, absence and location of genes. Clearly, incorporating such information provides additional constraints that can further increase the quality and accuracy of the results generated by a gene finding algorithm. Nonetheless, in this first installment of our approach, we have decided against incorporating such modules. We made this decision because of our desire to present a clear assessment of our method’s potential as a generic, alternative scheme to gene finding. We plan to incorporate such additional modules in follow-up work.
In Methods and Algorithms we describe our approach in detail. The Experimental Details and Results contains details on the experimental setup as well as a presentation and analysis of the results obtained from applying Bio-Dictionary Gene Finder (BDGF), our algorithm’s implementation, to 17 archaeal and bacterial genomes. The presentation concludes with a discussion.
METHODS AND ALGORITHMS
In this section, we present methodological details and give information on the algorithms that we have employed.
Notation and definitions
Let Σ denote the alphabet of all 20 amino acids. When processing an input dataset containing a collection of strings from Σ+ with the Teiresias algorithm (21,22), we can succinctly capture the patterns that can be discovered with the regular expression L(L{‘.∏})*L where L = (ΣU[ΣΣ*Σ]), and ‘.’ is a ‘don’t care’ character which stands for any character in Σ. In other words, the generated patterns can either be a single alphabet symbol or strings that begin and end with a symbol or a bracket with two or more characters, and contain an arbitrary combination of zero or more residues, brackets with at least two alphabet characters, and don’t care characters. A bracket denotes a ‘one of’ choice, i.e. [CPM] denotes exactly one of C, P or M. Also, a bracket can have a minimum of two alphabet characters but obviously not more than │Σ│ –1.
A pattern t is called an <L,W> pattern with (L ≤ W) if every substring of t of length W which begins and ends with a literal comprises L or more positions that are occupied by literals. The smallest length of an <L,W> pattern is obviously equal to L whereas its maximum length is unbounded. Any given choice for the parameters L and W has direct bearing on the degree of remaining similarity among the instances of the sequence fragments that the pattern captures: the smaller the value of the ratio L/W, the lower the degree of local similarity. Also associated with each pattern t is its support which is equal to the number of t’s instances in the processed input database. Finally, K denotes the minimum required support and represents the minimum number of instances that a pattern t must have before it can be reported.
The Bio-Dictionary
In previous work, we introduced and discussed in detail the concept of the Bio-Dictionary (23–25). The Bio-Dictionary is a collection of patterns that we refer to as seqlets (for ‘small sequences’) and which completely describes and accounts for the sequence space of natural proteins at the amino acid level. The seqlets are derived by processing a large public database of proteins and fragments using the Teiresias algorithm (21,22) and for an appropriate choice of L, W and K. In Rigoutsos et al. (23), we described our computation of the Bio-Dictionary from the GenPept release from February 10, 1999 using L = 6, W = 15 and K = 2. The processed input contained ∼387 000 sequences amounting to a grand total of ∼120 000 000 amino acids. The computation resulted in a Bio-Dictionary that at the time comprised ~26 000 000 seqlets and accounted for (i.e. covered) 98.12% of the amino acid positions in the processed input. The reader is referred to that publication for details regarding the computation, example seqlets with discussion, and an extensive description of possible applications. Indeed, the availability of such a complete collection of seqlets permits one to effectively and successfully tackle a number of tasks that include similarity searching (26), functional annotation (25), phylogenetic domain analysis (24), gene identification and others. Below, we describe gene identification in detail.
The key idea behind dictionary-driven gene finding
As mentioned already, the Bio-Dictionary concept seeks to substitute a given database of proteins and fragments such as GenPept or SwissProt/TrEMBL (27) by an equivalent collection of regular expressions (= seqlets) each of which represents combinations of amino acids that appear two or more times in the processed input. To the extent that the input sequences in such a public database correspond to a representative sampling of the sequence space of natural proteins, the seqlets of the Bio-Dictionary represent an exhaustive collection of intra- and inter-family signals that are discovered in an unsupervised and exhaustive manner. Our computation of intra- and inter-family signals becomes possible due to the fact that during processing we consider all publicly available sequences without any of the filtering that one encounters in databases such as PROSITE (28), Pfam (29) or PRINTS (30) and which is based on the sequences’ known functional behavior. The properties of the Teiresias algorithm guarantee that all <L,W> patterns will be discovered and that they have the maximum possible extent and specificity.
Two requirements need to be fulfilled for the Bio-Dictionary approach to be successful in tackling the kinds of problems in which we are interested. First and foremost, the input to be processed should be a large and diverse collection of proteins and fragments [see Rigoutsos et al. (24) for details], a condition that can be satisfied given the large number of completed and ongoing genome sequencing projects which contribute to the public databases. Second, the pattern discovery process should be able to generate patterns that are specific enough, not accidental, and account for as much of the processed input as possible. As we have demonstrated previously (23) this is indeed possible, thus the second requirement is satisfied as well.
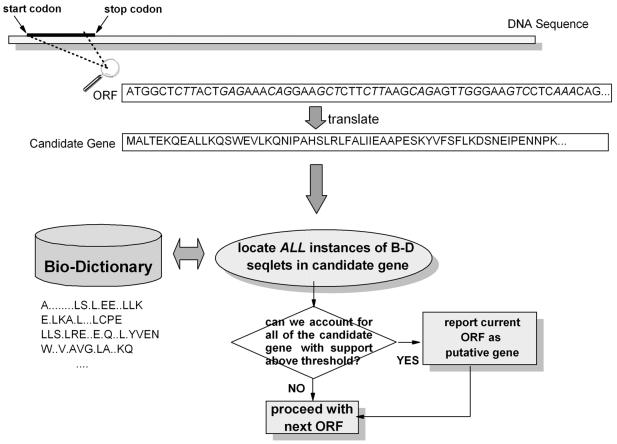
Given this description, our strategy for gene finding should now be evident. Figure 1 graphically depicts the basic idea. First, we compute all possible ORFs in each of the three reading frames and for both the forward and reverse strands of the given DNA sequence. Clearly, the number of true coding regions will be a proper subset of this collection of ORFs. Then, for each ORF we generate its amino acid translation: if the ORF under consideration is indeed a coding one, then we should be able to locate instances of many of the Bio-Dictionary’s seqlets across the span of the ORF’s translation, and vice versa. If the number of seqlets that we can locate exceeds a predetermined threshold, we report the ORF as a putative gene. We discuss the details of thresholding below; as a rule of thumb, the higher the number of Bio-Dictionary seqlets that can be found in a given ORF, the more likely it is that the ORF is coding for a gene.
Figure 1.
The basic idea behind our dictionary-driven gene finding method.
Incorporating a weighting scheme
The basic strategy for gene identification which we just described is straightforward. It is easy to see that in addition to the number of seqlets that can be located within the translation of an ORF, the very composition of these seqlets can have an impact when deciding whether the ORF codes for a gene. In general, any two Bio-Dictionary seqlets that match an amino acid translation affect this final decision differently. One can think of each seqlet being associated with a specific score: by summing up the scores of the individual seqlets that match an ORF, we can compute a quality measure that will allow us to determine whether to report the ORF as a putative gene or to discard it. Next, we examine how to appropriately weigh each of the seqlets.
Let T = {t1,t2,...,tn} be the complete collection of seqlets in a given Bio-Dictionary. Let us consider the amino acid translation s of an ORF from a given DNA sequence and let l be the length of s. We say that a seqlet matches at position j of the amino acid sequence s if an instance of the seqlet can be found beginning at the jth location of s. For example, G..G.GK[ST]TL matches the sequence MTHVLIKGAGGSGKSTLAFW beginning at position 8 of the sequence. Let ![]() denote the set of those seqlets that match beginning at position j of s, and let
denote the set of those seqlets that match beginning at position j of s, and let ![]() be the set T\
be the set T\![]() . Also let Ts = {
. Also let Ts = {![]() } denote the concatenated list of
} denote the concatenated list of ![]() values for all j(1 ≤ j ≤ l). Similarly, let T ′s = {
values for all j(1 ≤ j ≤ l). Similarly, let T ′s = {![]() } denote the concatenated list of
} denote the concatenated list of ![]() values for all j(1 ≤ j ≤ l). Note that
values for all j(1 ≤ j ≤ l). Note that ![]() values for different js can contain the same seqlet, thus Ts is in general a multiset. Similarly, T ′s can also be a multiset.
values for different js can contain the same seqlet, thus Ts is in general a multiset. Similarly, T ′s can also be a multiset.
Let pi be the probability that seqlet ti matches a database protein or fragment at a fixed location, and qi be the probability that ti matches the amino acid translation of a non-coding ORF at a fixed location. If all the seqlets in the Bio-Dictionary are assumed to be statistically independent then the probability that a given ORF corresponds to an actual gene will be equal to rPs /[rPs + (1 – r)Qs] where Ps = ![]() , Qs =
, Qs = ![]() , and r is the ratio of ORFs within the given ORF set that correspond to actual genes.
, and r is the ratio of ORFs within the given ORF set that correspond to actual genes.
Let us examine this independence assumption a little further by considering two specific seqlets that contain don’t care characters and have several instances in the database from which they were derived. It is easy to see that because of the don’t care characters the seqlets can be overlapping and matching the sequence under consideration (the ‘query’) even though they are derived from two distinct groups of unrelated sequences—in such a case, the seqlets cannot really be considered dependent. Of course, it can also happen that the seqlets are overlapping and matching the query and they are also derived from related groups of sequences in the processed database—in this case, the two seqlets would be dependent. Frequently though, two seqlets would be dependent only as a result of a small subset of sequences that is shared by the two groups of input sequences that gave rise to the two seqlets in the first place. It is conceivable that one could keep track of such situations for all groups of seqlets whose instances overlap within some sequences of the database. Something like this would of course require exceptionally demanding book- keeping (in terms of required space and time) and would make the application of our approach prohibitive for problems of practical size. It should be clear that the combinations of seqlets that ought to be considered dependent due to overlaps of some of their database instances are substantially fewer than the combinations of seqlets that are independent. Moreover, a given amino acid position of the query is typically ‘covered’ by 5–10 seqlets, a small number compared with the total number of seqlets that will match somewhere within it. Thus, any seqlet dependence that manifests itself in the outlined gene finding process will involve only a small number of seqlets each time and only because of a small fraction of the total number of their instances in the database from which they were originally derived. Consequently, the assumption of independence is a reasonable simplifying approximation that also allows for speedy computations and, as evidenced by the results we present in the next section, does not have any noticeable adverse impact on our results.
We can use the ratio Rs = Ps /Qs to determine the likelihood that a candidate ORF corresponds to a gene. Note that the probability will be >0.5 if Rs > 1 and r = 0.5. Let N = ![]() (1 – pi) and N′ =
(1 – pi) and N′ = ![]() (1 – qi). N denotes the probability that no seqlet matches at a fixed position of an actual protein, and N′ the probability that no seqlet matches at a fixed position of the amino acid translation derived from a non-coding ORF. Recall that the Bio-Dictionary is derived from a database of proteins and fragments, thus N is smaller than N′ in most cases. Considering that the number of seqlets that match at a given position is much smaller than the number of seqlets not matching at the same position, and that the pis and qis are very small numbers, we can approximate the terms
(1 – qi). N denotes the probability that no seqlet matches at a fixed position of an actual protein, and N′ the probability that no seqlet matches at a fixed position of the amino acid translation derived from a non-coding ORF. Recall that the Bio-Dictionary is derived from a database of proteins and fragments, thus N is smaller than N′ in most cases. Considering that the number of seqlets that match at a given position is much smaller than the number of seqlets not matching at the same position, and that the pis and qis are very small numbers, we can approximate the terms ![]() and
and ![]() by Nl and N′l respectively. Thus we can use Rs′ = (Ps′/Qs′)(N/N′)l instead of Rs, where Ps′ =
by Nl and N′l respectively. Thus we can use Rs′ = (Ps′/Qs′)(N/N′)l instead of Rs, where Ps′ = ![]() and Qs′ =
and Qs′ =![]() . Note that if N ∼ N′, we can use
. Note that if N ∼ N′, we can use ![]() = P′s/Q′s instead of
= P′s/Q′s instead of ![]() .
.
By definition, ORFs do not contain any stop codons internally, and consequently long ORF-like stretches are not likely to be random. Let L denote the probability that a stop codon is observed at a fixed position of a random DNA sequence. Since there exist three stop codons among the 64 possible codons and assuming that all four possible nucleotides appear with equal probability in the random sequence, then L is equal to 3/64. With this in mind, we can use Rs′′′ = R′/(1 – L)l = (Ps′/Qs′)Ml where M = N/N′(1 – L) instead of Rs′. If M ∼ 1, we can use Rs′′ = Ps′/Qs′ instead of Rs″′.
Let wi = log pi – log qi be the weight associated with seqlet ti. Let us also consider the sum of weights of the seqlets matching anywhere in the translation s of an ORF as the measure Ws that is characteristic of the coding quality of the ORF under consideration. It is easy to see that we can write the following equation for the coding quality measure of an ORF: ![]() = log Rs′′. If M cannot be ignored, we can instead use the following expression: W′s = Ws + l × log M = log Rs′′′ to define the coding quality of an ORF. In actuality, the value of l × log M is far smaller than the value of Ws, and we can safely ignore the term during the actual computations.
= log Rs′′. If M cannot be ignored, we can instead use the following expression: W′s = Ws + l × log M = log Rs′′′ to define the coding quality of an ORF. In actuality, the value of l × log M is far smaller than the value of Ws, and we can safely ignore the term during the actual computations.
At times, we are faced with a situation where we have multiple start codons matching the same stop codon and must decide which start/stop pair to report. Our solution amounts to picking the start codon that will result in the highest value for the coding quality measure. Due to the fact that seqlets can also have negative associated weights, and even if we ignore the log M term, it should be evident that selecting the start codon in such a way will not necessarily result in the reporting of the longest ORF as coding.
On a related note, ATG is the most frequently used start codon but it is not the only one. Consequently, it is inappropriate to treat the different start codons in a uniform manner. Let {c1,c2,...,ck} denote the set of possible start codons. Let fi be the probability that ci is the start codon of a randomly chosen coding region, f ′i be the probability that ci is observed in non-coding regions, and gi be log fi – log f ′i. We can then use Ws + gi instead of Ws as the measure of coding quality for the amino acid translation s of an ORF that is initiated by the start codon ci.
In order to compute the coding quality measure, we need the values for pis and qis. The most natural way to obtain these values is to compute them with the help of actual genes and non-coding ORFs. We can calculate the actual seqlet occurrences in the regions annotated as coding in a given training set and derive the needed pi values; we can then compute each seqlet’s occurrences in ORFs that are not designated as coding in a training set and derive the qi values. The values for the fis and f ′is can be computed in a similar manner.
But how can these values be obtained in the absence of a training set? For the pis we can use the probabilities computed with the help of the protein database from which the Bio-Dictionary is derived. Alternatively, we can compute them using very long ORFs instead of actual coding regions. For the qis we can use non-ORF regions, or we can estimate the probability of random occurrence based on an appropriately chosen amino acid bias.
Once we have attached a corresponding coding-quality measure to each ORF we can decide which ORFs correspond to putative genes by appropriately setting a threshold value. The higher the value of the measure we associate with a given ORF the more likely it is that the ORF is a coding one.
Removing encapsulated genes coded in different frames
Sometimes one encounters ORFs whose span completely encapsulates other ORFs in one or more of the remaining five reading frames. Occasionally, our method will give comparable, high scores to a pair of ORFs where one of the ORFs completely includes the other. It is believed that not both members of such pairs of coding regions can correspond to actual genes. In these situations, we use the ORF score to sub-select, and report the one with the higher score. Note that most of the time, the ORFs selected and reported in this manner correspond to the longer member of the pair. A similar approach is also employed by Glimmer (2).
Matching seqlets to a sequence of amino acids
The Bio-Dictionary we use in our experiments contains ∼30 000 000 seqlets (see the following section for more details). All of these seqlets need to be checked against a large number of amino acid translations from all ORFs on both strands of a given complete genome. It is thus important that this operation be carried out as efficiently as possible so as to reduce the overall computational requirements of our method. There exist many efficient linear-time algorithms for searching exact strings (31), but since seqlets include don’t care characters these algorithms are not applicable here. In this section, we present and discuss two algorithms for quickly determining whether a seqlet matches a given amino acid sequence. We can address this problem in one of two generic ways: we can either preprocess the amino acid sequence, or we can preprocess the Bio-Dictionary seqlets with which we will be searching.
Most of the work that has appeared in the literature revolved around the preprocessing of the sequence which is assumed to be fixed (31). Moreover, the number of patterns whose instances were sought in the sequence was substantially smaller than the collection of patterns we are interested in.
Note that in our work, the set of seqlets is fixed, known in advance and very large. On the other hand, the amino acid sequences that we need to examine are numerous and not known in advance. Moreover, the average length of such a query sequence is just several hundred amino acids. It thus appears more effective to preprocess the contents of the Bio-Dictionary. One approach would be to build hash indices out of seqlets so as to reduce the number of seqlets that need to be examined at each position of the amino acid sequence; however, it is not immediately clear how to build such indices for patterns that have variable numbers of don’t care characters as is the case of the seqlets.
Similarly to what is done in traditional dictionary searches without don’t care characters, we can build the index with the help of the first few characters of a seqlet. For example, the seqlet G..G.GK[ST]TL could be indexed through G..G if we consider indices that are built using the first four characters of the seqlet. If one of the positions used to build the index is occupied by a bracket expression like [ST] it is much simpler to replace the contents of that position by a don’t care character. This method is rather effective and can in fact be improved further. In the case of G..G.GK[ST]TL, it would seem more appropriate to use one of GK.T or K.TL as the seqlet’s four-character-induced index instead of the first characters G..G. Since GK.T and K.TL contain fewer don’t care characters, they should generally appear fewer times than G..G in an arbitrary protein.
Keeping these observations in mind, we next propose a novel effective scheme for generating indices out of seqlets. Let us define an (l,w) subpattern of a seqlet t as follows: first, we replace all brackets in the seqlet by don’t care characters and let t′ denote the modified seqlet. The (l,w) subpattern is a minimal substring of t′ such that its length is less than w and it contains l characters that are not don’t care characters. Notice that we cannot always find such (l,w) subpatterns; if one such subpattern exists, we select it and form the index for t. If such a subpattern does not exist we find the largest value l′ such that an (l′,w) subpattern exists in t′ and use this subpattern to form the seqlet’s index: for example, GK.T is a (3,4) subpattern of G..G.GK[ST]TL.
If a seqlet t is indexed by an (l,w) subpattern, we have to check this seqlet approximately n/│Σ│l times when we consider a random protein of length n that has uniform amino acid bias. We can of course reduce searching time by setting l to a large value. But, at each position, we must examine patterns that are indexed by any of ![]() possible (l′,w) subpatterns for all l′ ≤ l. The total number of patterns to be checked at any one position is thus O(2w); if we set w and l to large values, the search will be slow. In the section entitled ‘Performance of the seqlet search scheme’ we will describe how to choose appropriate values for l and w. Note that if there exist seqlets without any fixed character (e.g. [LK]..[ST][GKT]..[AERST][ELK]..[AVL]) we must check them at each position in addition to the seqlets hashed by subpatterns as above.
possible (l′,w) subpatterns for all l′ ≤ l. The total number of patterns to be checked at any one position is thus O(2w); if we set w and l to large values, the search will be slow. In the section entitled ‘Performance of the seqlet search scheme’ we will describe how to choose appropriate values for l and w. Note that if there exist seqlets without any fixed character (e.g. [LK]..[ST][GKT]..[AERST][ELK]..[AVL]) we must check them at each position in addition to the seqlets hashed by subpatterns as above.
If we know the frequency of appearance of individual amino acids in query sequences, we can use it to estimate the probability of appearance for each subpattern. In our case, we can use it to further improve the performance. For example, in the case of G..G.GK[ST]TL, if we know that the amino acid G appears less frequently than the amino acid L, we can assume that GK.T is also rarer than K.TL, and thus we should hash the seqlet with the help of GK.T. There can be cases where some (l′,w) subpattern is estimated to be less frequent than an (l,w) subpattern although l′ < l. In such cases, we should use the former, more rare subpattern. We can easily search for such a subpattern in time that is linear to the size of the seqlet. In the experiments below, and for given values of l and w, we use as a hash index the least frequent (l′,w) subpattern, with l′ ≤ l, as can be estimated from a given known amino acid bias.
EXPERIMENTAL DETAILS AND RESULTS
In this section, we describe and report on the results we obtained with BDGF, the implementation of our gene-finding algorithm. BDGF was applied to several complete archaeal and bacterial genomes. We begin by describing the building of the Bio-Dictionary that we use and the parameter choices for our search scheme that determines possible matches of a given seqlet in the amino acid translation of an ORF.
Generation of the Bio-Dictionary
With the help of the Teiresias algorithm (21,22), we computed an instance of the Bio-Dictionary for the June 12, 2000 release of the SwissProt/TrEMBL (27) database. The processing was carried in the manner that is outlined in Rigoutsos et al. (23). As a matter of fact, we used Teiresias with a setting of L = 6, W = 15 and K = 2. The justification for this choice of values for L and W is the result of earlier extensive analysis and was described previously (26). The Bio-Dictionary that resulted from this processing contained 29 397 880 seqlets. The instances of these seqlets accounted for 98.10% of the amino acid positions in the processed database. It is this collection that we used in our experiments.
It should be noted that the SwissProt/TrEMBL release that we used to build our Bio-Dictionary is >1.5 years old and chronologically preceded the releases of some of the test genomes that we processed for our experiments. This was an intentional choice on our part and was meant to demonstrate our method’s generality and ability to extrapolate.
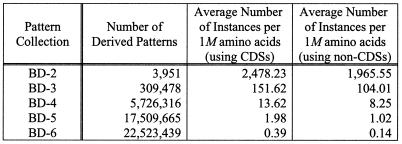
Some of the seqlets in the employed Bio-Dictionary have rather long spans. These are typically important patterns, generated by putative proteins that are coded by genes in distinct genomes from the same phylogenetic domain. Because these patterns appear infrequently, it is difficult to compute their weights in the absence of an extraordinarily large training set. For our experiments, we handle this situation as follows: we replace seqlets that have k fixed characters by k – s + 1 seqlets each of which contained s fixed characters, for some choice of the value s (see below). Let S{i..j} denote a subpattern of a seqlet S that starts at the ith fixed character and ends at the jth fixed character. We replace S by subpatterns S{1..s}, S{2..s + 1},...,S{k – s + 1..k}. Any duplicate seqlets that appear in the resulting collection are removed before further processing. Heretofore, and for simplicity purposes, we will use the shorthand notation BD-i to refer to the derived pattern collections that are constructed as above by setting s to i. The collections that we used in our experiments were BD-4 and BD-6.
Table 1 shows statistics for collections BD-i with i assuming values between 2 and 6 inclusive. In particular, for each BD-i the table lists the following items: (i) the number of seqlet-derived patterns contained in BD-i—as expected, and because identical patterns are removed after splitting, the number is small for small values of i; and (ii) the average numbers of instances of a derived pattern per 1 000 000 amino acids, computed from experiments against coding sequences (column 3) and non-coding sequences (column 4) from the 17 genomes we used for our experiments—as expected, the number of instances is much higher in coding sequences than in non-coding ones.
Table 1. Statistics for the seqlet-derived pattern collections used in our experiments.
See text for additional discussion.
The total lengths for the coding and non-coding sequence sets for the 17 genomes that we used in our experiments are ∼30 and ∼94 Mb respectively (note that the seemingly large sizes of theses datasets are due to the existence of six reading frames). If we make use of all 17 organisms for training, the corresponding coding and non-coding sequence sets have sufficiently large sizes to permit the computation of representative weights, even for the patterns in the BD-6 collection. On the other hand, if only one or a handful of genomes are available, the corresponding sizes for these two sets are not substantial to permit the generation of representative weights for the BD-5 and BD-6 collections. Thus, a given choice for the training set indirectly dictates which collections BD-i can be used to carry out any planned gene finding experiments.
Performance of the seqlet search scheme
As we have already mentioned, it is important that we be able to quickly determine which of the seqlets of the Bio-Dictionary are contained in the amino acid translation of a given ORF. To this end, we present experimental results on the performance of the search scheme that we described in the previous section, and for different combinations of the parameters l and w.
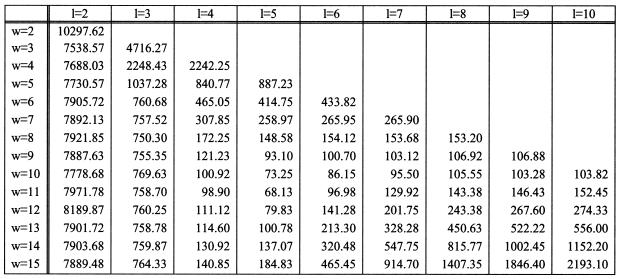
For the purposes of benchmarking the method’s performance, we used the (i) original unmodified Bio-Dictionary that contained 29 397 880 patterns and (ii) 100 proteins from Escherichia coli as queries. The average span of the seqlets in this Bio-Dictionary is 13.15 positions whereas the average number of fixed characters in these seqlets is 7.41. The average length of the 100 query proteins is 340.61 with the shortest and longest sequences having lengths of 21 and 1073 amino acids respectively. The goal of the experiment is to find all the Bio-Dictionary seqlets that have instances in the 100 query proteins. As it turns out, a total of 302 349 Bio-Dictionary seqlets appear in the cumulative collection of 100 proteins, with each amino acid position participating in the instances of 8.88 distinct seqlets, on average. Table 2 shows the computation time required to determine all these matches and for the parameters l and w assuming values in the range 2 ≤ l ≤ 10, 2 ≤w ≤ 15 (l ≤ w). All of the experiments are carried out on a single IBM RS64III processor with a clock speed of 450 MHz. For these experiments, we constructed a hash index assuming an amino acid bias that results from a uniformly distributed random sequence of nucleotides. The best performance is obtained when l = 5 and w = 11: with these settings, we can process a 500 amino acid query and determine the subset of the Bio-Dictionary’s ∼30 000 000 seqlets that have instances in the query in ∼1 s.
Table 2. Sample search times (in s) using (l,w)-subpattern-based indexing and for various choices of l and w.
See text for additional details.
Table S1 (Supplementary Material) shows the average number of seqlets that are examined for instances in the query sequence. Note that only a subset of the Bio-Dictionary’s seqlets will actually match somewhere in the query. As anticipated, the number of seqlets that need to be examined decreases as l and w grow larger, an expected result. Also, the search time does not decrease when l >5 or w >11. The explanation can be found in Table S2 (see Supplementary Material) where we show the maximum number of subpatterns that need to be checked at each position of a given protein: this number increases as l and w increase. As a matter of fact, the actual search times are roughly proportional to 1.6 × x + y, where x is the number of the checked subpatterns at each position and y is the average number of the actually checked seqlets at each position. Note that x is sometimes larger than y because subpatterns exist that must be checked but for which no seqlet has been hashed. We can easily compute the expected number of checked seqlets against random sequences with a given amino acid bias, and thus can estimate appropriate values for l and w in this manner.
For completeness purposes, we also carried out experiments where we searched using hash keys derived from the prefixes of seqlets. The results are shown in Table S3 (see Supplementary Material), for various values of the prefix length. This scheme does not perform as well as our (l,w)-based hashing scheme and the best result of 237 s that is obtained for a prefix length of 11 is far too slow to be useful; thus, we abandoned this prefix scheme idea.
Gene finding results on archaeal and bacterial genomes
In this section, we outline and discuss the capabilities of our gene-finding method by reporting the results we obtain from processing 17 complete genomes with BDGF.
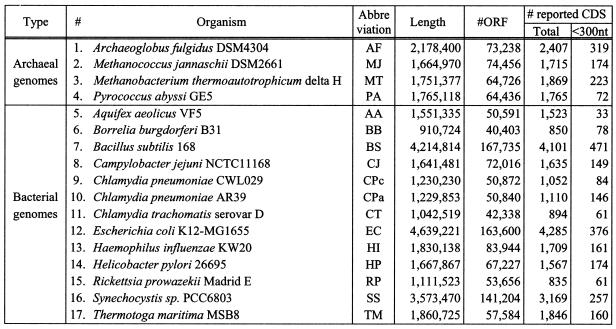
Genome identities. Of the 17 genomes we used in our experiments, four were archaeal (Archaeoglobus fulgidus, Methanococcus jannaschii, Methanobacterium thermoautotrophicum and Pyrococcus abyssi) whereas the remaining 13 were bacterial. Table 3 shows relevant information for these genomes including the genome length in nucleotides, the number of all identifiable ORFs that are >18 nt (i.e. six amino acids), and the number of annotated coding regions that have been reported in the public databases for each genome. We should mention at this point that each of these genomes may contain additional actual coding regions that to date have remained unreported. Also, one should not lose sight of the fact that the annotated (= reported) coding regions are for the most part putative and have typically been reported without verification via wet laboratory experiments.
Table 3. Details of the 17 genomes used in our experiments.
Quantifying the quality of our predictions. There exist several ways in which one can evaluate the performance of a gene finding algorithm. But the algorithm’s sensitivity and specificity remain the most important measures. Sensitivity, often referred to as the prediction rate, is defined as the ratio of the number of genes predicted by the algorithm over the number of genes that have been reported in the public databases. Specificity is defined as the ratio of the number of predicted genes that are also reported in the public databases over the number of all genes that the algorithm has predicted.
Clearly, one can generate very appealing, large values for the sensitivity of the algorithm simply by lowering the employed decision thresholds. But this is typically done at the expense of introducing false positives in the output which will in turn lead to decreased values for specificity. The opposite situation is also possible: one can choose thresholds in a way that will result in high specificity values at the expense of sensitivity; i.e. many actual genes will not be reported. Sensitivity and specificity are competing goals and, ideally, any proposed algorithm must aim at achieving simultaneous high values for both of these measures.
In addition to an algorithm’s specificity and sensitivity, also of interest is the cardinality of the collection of genes that have been predicted by the algorithm and satisfy the following two conditions: (i) the predicted genes are not among the genes that have been reported in the public databases; and (ii) the predicted genes have substantial similarity to one or more protein/cDNA sequences contained in the public repositories. Naturally, this collection forms a subset of the results that would otherwise be characterized as ‘false positives’.
The existence of genes in several distinct genomes that also exhibit similarity to a gene predicted by a given algorithm adds support to the hypothesis that this gene is indeed correctly predicted. In what follows we use the term ‘hits’ to refer to the members of the special subset of predicted genes that also satisfy conditions (i) and (ii) above. In our experiments, we determined whether a predicted gene satisfied condition (ii) by using both the FASTA (32) and the BLAST (33) algorithms: with default threshold and matrix settings we carried our similarity searches against the release of SwissProt/TrEMBL (27) from September 21, 2001. A query was considered to generate a hit in the searched database if one or more of the reported results had associated E(.) values that were ≤1.0e–4 for FASTA and 1.0e–3 for BLASTP (these specific threshold values were recommended by one of the reviewers). In addition to running FASTA and BLAST, we carried out a CD search for conserved domains using rpsblast and the Conserved Domain Database from February 28, 2002 (34): the E(.) value threshold we used here was equal to 1.0e–4.
In all cases that are described below, we quantified the performance of our approach by simultaneously reporting the values of the following three measures: ‘sensitivity,’ ‘specificity’ and ‘hits’.
How we built and used our training and test sets. As we explained previously, an appropriate training set is needed in order to compute weights for all the seqlets in the Bio-Dictionary. The experiments that we carried out and whose results are presented in detail in the section entitled ‘Prediction results on the various genomes’ were meant to mimic the very wide spectrum of situations that a researcher may encounter in a real-world setting.
First, we divided each genome into two equal-length parts: we used the second half of the genome as a training set and the first half as a test set (case 1). In spirit, this is a test similar to what has been previously reported in the literature (2,5). However, it should be stressed that what we use in this case to derive weights for our seqlets is a mere 50% of a genome whereas we test our prediction capability on the remaining 50% of it. The size of the training set we use in this set of experiments is much smaller than what has been typically employed to train previously reported methods. This was an intentional decision meant to showcase our system’s capabilities.
We also carried out experiments without using any a priori knowledge for the training sets. This is discussed in detail in case 2 below. The purpose of these experiments was to determine how well our algorithm works in the absence of such information.
The next group of experiments (cases 3a and 3b) was designed to examine the performance of our method in the case where the seqlets’ weights were not genome specific. For each test genome, we derived weights for the seqlets by training with a collection of several complete genomes that did not include the genome under consideration; we then applied our method on this test genome. These jack-knifing experiments are typically too severe for statistical methods such as those based on Markov models. It is for this reason that many web implementations of previously reported, statistics-based methods often provide several parameter settings derived from training on various genomes: users are asked to select the appropriate settings to be used by the algorithm. The experiments for cases 3a and 3b were carried out using BD-4, i.e. the derived pattern collection was constructed by setting the parameter s to 4 (see section ‘Generation of the Bio-Dictionary’).
In order to determine the impact of the different pattern collections on the results, we repeated the experiments of cases 3a and 3b using BD-6, i.e. the collection constructed by setting the parameter s to 6. Cases 4a and 4b correspond to this set of experiments and are the counterparts of cases 3a and 3b respectively.
Finally, for our last group of experiments (case 5) we used all 17 genomes to assign weights to the seqlets, then tested the resulting non-genome-specific system by processing each of the genomes in turn.
As we noted in the Introduction, in this first incarnation of our system we do not make use of any additional information (e.g. promoter information that can either be computed or retrieved from the public databases) to constrain the discovery of genes and start sites. Nonetheless, as evidenced by the results that we report in cases 4 and 5 (see section ‘Prediction results on the various genomes’), our start site prediction rates are comparable in quality, and at times superior to those that have been previously reported in the literature (5,35–37). What is more, our results are achieved using a system that is generic and not genome specific. We will revisit the topic of start-site prediction at the end of this section.
Format of the reported results. In the tables that follow, and for each processed genome, we report the total number of genes predicted by our algorithm in column 2; we also indicate how many of these genes are <300 nt in column 3. In our studies, we threshold at that score value for which the number of predicted genes is equal to the number of annotated genes in the public databases—clearly this threshold value is different for each genome. Note that in this case, the exhibited sensitivity and specificity are equal; this common value is shown in column 4. For a subset of the genes that are predicted by our algorithm there is no corresponding database entry characterizing them as such. Column 5 shows how many of the predicted genes fall into this category, whereas column 6 indicates how many of these genes are <300 nt. With the help of FASTA, BLAST and CD-search we report how many of these genes are in fact hits (recall the definition from section ‘Quantifying the quality of our predictions’) in columns 7, 9 and 11 respectively; the number of hits which correspond to gene predictions that are <300 nt is listed in columns 8, 10 and 12 respectively. Finally, in each of the result tables we also report on our ability to correctly predict the start sites for the reported genes through comparison with the existing database annotations. In column 13, we show the number of genes whose start site is correctly predicted; and in column 14 we list the same figure as a percentage of reported genes.
Prediction results on the various genomes. We now report on the results of our method in five experimental settings. The experiments were designed so as to mimic the type of situations that a real-world researcher is likely to encounter, and showcase the performance of our approach across a wide spectrum of settings.
Case 1. BD-4 and weights derived from the second half of each genome only. BD-4 was used in this case. For each genome, the seqlets’ weights were obtained by training on the second half of the genome. Gene prediction was carried out on the first half. Table S4 (see Supplementary Material) shows the results for this experiment: in all 17 cases, the sensitivity/specificity value ranged between 90.1 and 95.1%. Also, ∼33% of the additional putative genes reported by our method corresponded to hits, i.e. we could find statistically significant similarities with proteins in the September 21, 2001 installment of the SwissProt/TrEMBL database. The implication of this is that the actual gene prediction rate is likely to be even higher than what we report here. With respect to the start site prediction rate, the rates of correctly predicted start sites in this case range from 55.6 to 84.2%. Recall that we currently make no use of any promoter information.
Case 2. BD-4 and weights derived from using long ORFs only. The previous experiment assumes the availability of annotations for at least some of the actual coding genes of a target genome. But what if such information is not available? In such a situation the only recourse is to derive the seqlet weights by restricting ourselves to the very long ORFs that can be identified in the genome which is being processed; the implicit assumption here is that long ORFs are more likely than short ORFs to be coding for genes and can thus be used as training sets. Similar heuristics have been employed by other groups as well (2,38).
In this case, we used as a training set for the coding regions all the ORFs that were >600 nt and which were not included within other longer ORFs. As a training set for the non-coding regions we used all the ORFs that were <200 nt and which occasionally (and incorrectly) include bona fide coding ORFs. We again used BD-4 as the collection of patterns for which to derive weights. As will become evident after we have described our complete set of experiments, it is not necessary to carry out this kind of training when dealing with a new genome—we have simply included case 2 for the purpose of completeness of description.
Table S5 (see Supplementary Material) shows the results of this experiment. In this case, the sensitivity and specificity value ranged from 89.9 to 95.6%. Similarly to case 1, ∼32% of the additional predicted genes have significant similarities with other database entries. It is notable that despite the fact that the amount of information we used for training purposes was substantially less than that we used in case 1, the performance levels remained essentially unchanged.
Case 3a. BD-4 and weights derived from fixed 4 of 17 genomes (leave-many-out). Another realistic situation is the one where users will carry out gene prediction on newly sequenced genomes for which little or no information is yet available. The situation can be facilitated if a phylogenetically similar genome already exists in the public databases but this is not always going to be the case. This particular experiment is meant to simulate the situation where the genomes that are already available in the public databases are few and rather distant from the ones that are being examined. To this end, we used the union of the full CDS lists that were reported for Bacillus subtilis, Campylobacter jejuni, Helicobacter pylori and Rickettsia prowazekii to derive the weights for BD-4’s patterns and subsequently tested our method by predicting genes for the remaining 13 genomes of our genome collection.
Case 3b. BD-4 and weights derived from 16 of 17 genomes (jack-knifing or leave-one-out). Here we study the jack-knife variation of case 3a: prior to carrying out gene prediction for each of the 17 genomes of the collection, we used the union of the full CDS lists from the remaining 16 genomes to derive the weights for BD-4’s patterns.
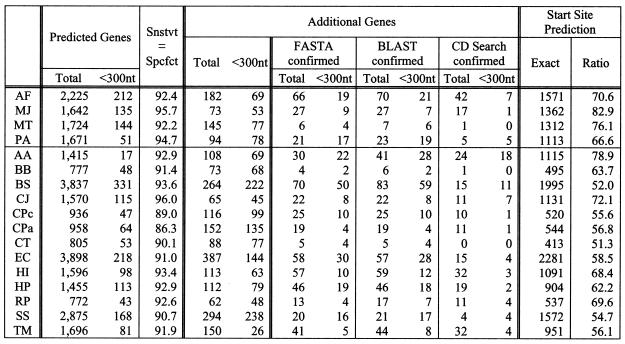
Tables S6 (see Supplementary Material) and 4 show the results for cases 3a and 3b, respectively. As one would intuitively expect, the weights that are derived from 16 genomes are more representative (case 3b). Consequently, the prediction rates reported in Table 4 are superior to those reported in Table S6. The sensitivity/specificity value was in the range of 86.3–94.7%, exceeding the 90% mark for the majority of the genomes. Approximately 23% of the additional putative genes reported by our algorithm correspond to hits.
Table 4. Gene prediction results for case 3b.
BD-4 was used and the weights were derived from training using the remaining 16 genomes (‘jack-knifing’ or ‘leave-one-out’ scenario).
This set of experiments (i.e. cases 3a and 3b) is particularly interesting in light of observations made previously in the literature according to which the performance of statistical methods typically deteriorates if the method is used with parameters derived from a phylogenetically distant genome. This deterioration reflects merely the overall difference in the statistical properties of the respective genomes.
Unlike statistical approaches, our method depends largely on the statistical properties of verified and putative proteins and exhibits resilience to statistical variability. Thus, the conclusion of this set of experiments is that if the weights of the seqlets have been appropriately derived we expect to be able to reach very high prediction levels in a manner that will be relatively independent of the studied organism. This is indeed corroborated by the results shown in Tables S6 and 4.
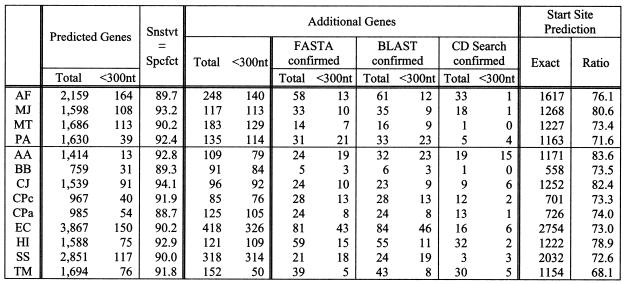
Case 4a. BD-6 and weights derived from fixed 4 of 17 genomes (leave-many-out). In this experiment, we repeat the experiment of case 3a but now using the BD-6 collection. In general, with smaller values for s one expects that the derived collection of patterns will be more sensitive but will capture less ‘structure’. On the other hand, larger s values will give rise to a situation where potentially fewer seqlets match the putative amino acid translations and there is an associated increased difficulty to appropriately compute the seqlets’ weights. Table 5 shows the results of this experiment.
Table 5. Gene prediction results for case 4a.
Unlike case 3a, we used BD-6 and the weights were derived from training using only four of the 17 genomes (‘leave-many-out’ scenario). The four genomes that were used for training were: Bacillus subtilis, Campylobacter jejuni, Helicobacter pylori and Rickettsia prowazekii.
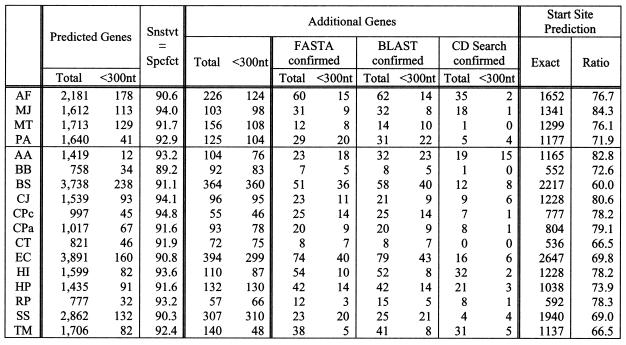
Case 4b. BD-6 and weights derived from 16 of 17 genomes (jack-knifing). Here we repeat the experiment of case 3b but this time using the seqlets from the BD-6 collection: results are shown in Table 6.
Table 6. Gene prediction results for case 4b.
Unlike case 3b, we used BD-6 and the weights were derived from training using the remaining 16 genomes (‘jack-knifing’ or ‘leave-one-out’ scenario).
When we compare the results from cases 3a/3b with those from cases 4a/4b we conclude that for most of the genomes in our collection, and for the training carried out as described above, the BD-4 collection will result in better performance; a notable exception is represented by the Chlamydophila and Chlamydia species for which BD-6 gives better results.
An additional observation is that the number of genomes for which BD-6 performs better than BD-4 increases as the size of the training set increases. The very important ramification of this is that, if we have access to a rather large training set, our method will exhibit better prediction performance when used in conjunction with BD-s sets corresponding to larger s values.
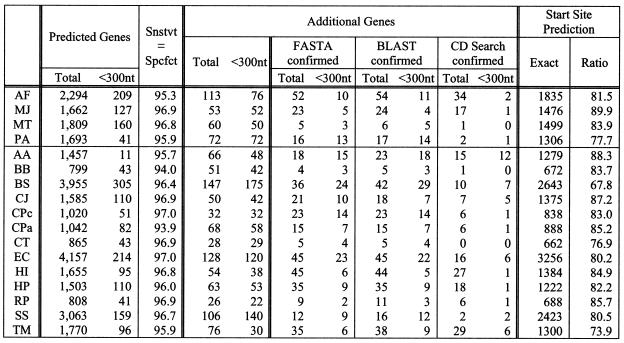
Case 5. BD-6 and weights derived from all 17 genomes. The four experimental cases above provided sufficient information that permitted us to generate optimal results with our method. In particular, (i) we should use the BD-6 collection since it is bound to perform better than BD-4 as more and more information is deposited in the public databases; and (ii) during training, the weights should be derived from all of the previously published coding/non-coding information for all available genomes. With these two observations at hand, we carry out this last set of experiments noting that it is indicative of the levels of prediction quality we can expect when we use a good training set, i.e. one derived from many complete genomes.
The results for this experiment are shown in Table 7. The achieved sensitivity/specificity value is in the 93.9–97.0% range with most of the genomes exceeding the 95% mark. Invariably, and similarly to all of the above experiments, a substantial percentage of the additional putative genes that our method reports correspond to hits, i.e. they can be corroborated with the help of sequence similarities to entries in the public databases.
Table 7. Gene prediction results for case 5.
This is the optimal setting where we used BD-6 and the weights were derived from training using all 17 genomes.
Another very notable result that is reported in Table 7 has to do with our ability to correctly predict the start sites of genes. As can be seen, the ratio of correctly predicted start sites ranges between 80 and 90% across almost all studied genomes. More importantly, this ratio is achieved simultaneously with very high specificity and sensitivity values and without making use of any promoter information.
We conclude by stressing a very important point: our method derives and uses a single set of weights for the seqlets in the Bio-Dictionary and these weights are not genome specific.
On the prediction of start sites. As we mentioned already, the accurate prediction of start codon sites is a notoriously difficult problem. To derive the various ratios for our experiments, we made use of the gene starts that are reported in the annotated database entries for each processed genome. Although this kind of information is generally correct, errors are known to exist. Thus, we also carried out a verification step of our start site prediction rates using experimentally validated genes. In particular, we focused on the 1248 experimentally validated genes from B.subtilis (36) and computed the ratio of start sites that were correctly predicted by our algorithm when using the pattern collections and weights described in cases 4b and 5 above [in other words, we used the BD-6 collection of patterns with weights derived from (i) jack-knifing and (ii) optimal computations from all 17 genomes]. Of the 1248 genes, we correctly determined the start sites for 797 (case 4b) and 898 genes (case 5) respectively, or 63.9 and 72.0% of the processed set respectively. These numbers closely match the corresponding entries for the entire B.subtilis genome in Tables 6 (case 4b) and 7 (case 5) respectively; in fact, they are slightly higher than what is listed in these tables. This verification lends more support to the correctness of our start site predictions.
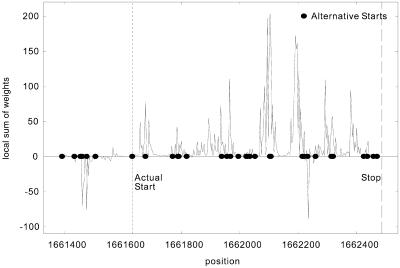
We conclude the discussion on start sites by noting that our gene prediction algorithm can also be used for start site localization. In Figure 2 we show a graph that depicts for each position i in the neighborhood of an E.coli coding sequence the local sum of the weights for the seqlets that match starting at position i. To obtain the cumulative score corresponding to position i, the local sums from i through the stop codon position must be added together. The seqlet weights used here are the ones from case 5 above and correspond to what we consider to be an optimal setting. The true start site of the coding region as well as alternative start sites are shown in this plot. Note how the local sum of the seqlets’ weights is very low just prior to the true start codon then ‘jumps’ abruptly to a much higher value immediately after it. These score jumps can be exploited to predict the start sites of predicted genes. And as the results for case 5 have shown, we can achieve high ratios of correct start site prediction simultaneously with high levels of specificity and sensitivity. We are in the process of incorporating information from promoter regions to our system and expect that the overall prediction capability of our method will improve further.
Figure 2.
Example of start site prediction using a coding sequence from E.coli—see text for more details.
Some notes on the quality of our ‘additional gene’ predictions. In order to demonstrate that our approach does make a tangible advance to the gene finding problem, we discuss next two specific examples.
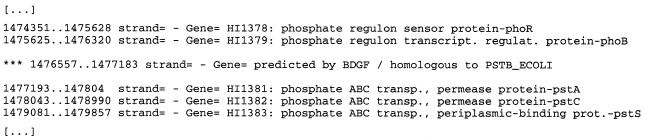
The first example comes from the processing of the Haemophilus influenzae genome with BDGF. One of the ORFs that we predict is found on the reverse strand of the genome in the region 1476557..1477183; the alleged corresponding gene product is 206 amino acids long. Quick analysis shows that this sequence is essentially identical to PSTB_ECOLI, a phosphate transport, atp-binding protein from E.coli. The following MUSCA-derived (39) alignment shows the similarity—conserved amino acids have been colored based on their hydropathy (Scheme 1).
Scheme 1.
The ability of our method to correctly identify and report this particular gene is very important for the following reason: until the late part of 2001 the list of genes and their positions in the genome that accompanied the GenBank entry for H.influenzae included no gene prediction for this region (recall that the input database out of which we build our pattern collection dates back to June 12, 2000) (Scheme 2).
Scheme 2.
The proteins pstS, pstC, pstA and pstB constitute an intra-membrane complex that acts as a high-affinity transport system for inorganic phosphates. This system permits the transport of the phosphates through the inner membrane and into the cell. The four proteins pstS, pstC, pstA and pstB form an operon; similarly, phoR and phoB form a second, regulatory operon. The expression of the genes in these two operons is controlled by the level of inorganic phosphates in the cell.
Note that the gene that we predicted is coded by the same strand (= reverse) as the remaining components of the two operons. Moreover, the localization and hypothesized identity for our predicted gene are further supported by the fact that all the members of these two operons are present, appear in the same order, and have been reported in the genome of Pastereulla multocida; in fact, the relevant segment of the list of the predicted genes for P.multocida reads as in Scheme 3. As can be seen, in the P.multocida instance of the two operons, the gene that codes for pstB is located between the genes coding for phoB and pstA, similarly to the H.influenzae genome.
Scheme 3.
The entry for pstB was added to the list of H.influenzae genes prior to the October 19, 2001 update of the Entrez entry for this genome. The value of this example lies in showcasing our ability to make a correct gene prediction for H.influenzae as early as June 12, 2000, i.e. the date on which we compiled the knowledge base that was used to derive our patterns. With time, the database from which we build our pattern collection will increase in size and become more rich further improving our gene prediction ability.
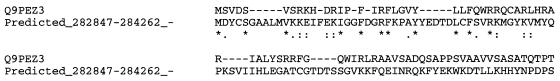
Our second example comes from the genome of A.fulgidus. One of our gene predictions involves an ORF that appears on the reverse strand in region 282847..284262; based on the list of genes that were reported with the original GenBank submission of A.fulgidus, this particular region is gene-empty (Scheme 4).
Scheme 4.
A straightforward analysis reveals the existence of a hypothetical protein, Q9PEZ3, in Xylella fastidiosa which shares extensive similarities with our predicted gene. A ClustalW (40) alignment between our predicted gene and Q9PEZ3 is shown in Scheme 5.
Scheme 5.
The very high degree of similarity and the fact that the corresponding involved organisms belong to distinct phylogenetic domains (archaea and bacteria respectively) provides strong support for the correctness of this gene prediction.
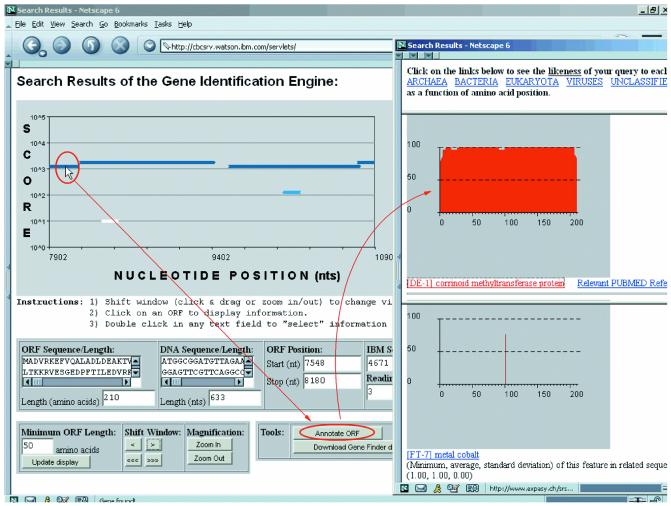
Web server for this gene finding algorithm. BDGF, the implementation of our gene finding algorithm, has been made available via the World Wide Web. The implementation uses the patterns of the BD-6 collection and optimal weight settings derived from 17 genomes. The server can be accessed by visiting the URL http://cbcsrv.watson.ibm.com/Tgi.html and is operational around the clock; the server runs on a single IBM RS64III processor with a 450 MHz clock, and can process 250 000 nt in all six reading frames in a little over 60 s. Upon completion of the computation, the results are presented to the user via a graphical user interface an instance of which is shown in Figure 3. The predicted genes are color-coded depending on the score they have been assigned. The interface allows the user to navigate around the processed DNA sequence and to zoom in/out of the regions of interest. Once an ORF has been selected with the help of the mouse, its location on the processed sequence is reported together with its nucleotide composition, length and amino acid translation. The amino acid translation of a selected ORF can be annotated interactively. Also, the minimum length of an ORF that will be reported is controlled by the user: its default value is 50 amino acids (i.e. 150 nt). The complete list of ORFs that have been predicted by BDGF together with their position, length and associated score can also be downloaded from the same page.
Figure 3.
Result page of the web-based graphical user interface. The reported ORFs are color coded according to their scores. Selecting an ORF with the cursor permits the user to see its starting and ending positions, the score that the ORF has received, length and nucleotide composition as well as its amino acid translation. ORFs can be annotated interactively and regions of the genome can be further explored by zooming in or out using the corresponding buttons (http://cbcsrv.watson.ibm.com/Tgi.html).
DISCUSSION AND CONCLUSION
In this paper, we described a new method for solving the gene identification problem. Our method begins with the Bio-Dictionary, a collection of patterns that is generated from processing very large public databases with the help of the Teiresias algorithm. The collection accounts completely for the processed input, and discovers genes by making use of this set of patterns alone. The method is augmented by associating each of the used patterns with automatically derived weights. These weights are genome independent and thus remain fixed across genomes.
Through a series of carefully designed experiments we extensively explored various settings that mimicked real-world situations, and determined the optimal settings for our gene finding approach. As evidenced by reported experimental results from 17 archaeal and bacterial genomes, our method can predict genes very accurately. The method achieves sensitivity and specificity values that are simultaneously very high while at the same time achieving a high rate of correctly predicted start sites. Notably, no promoter or other information is brought to bear during our determination of the genes and/or start sites.
We demonstrated the capabilities of our method to extrapolate by intentionally relying upon a Bio-Dictionary that was built from the June 12, 2000 release of SwissProt/TrEMBL, i.e. a public collection of sequences that is by now >1.5 years old. We nonetheless applied the resulting system to genomes whose ORF translations were included in SwissProt/TrEMBL either only in part or not at all, with exceptional results.
We should note that in addition to correctly discovering and reporting those ORFs that have already been listed in the public databases as putative genes, our method determines additional candidate genes in essentially all of the genomes that were used in the experiments: for a substantial fraction of these previously unreported genes, and with the help of FASTA, BLAST and CD-search, we determined similarities with amino acid sequences contained in a very recent release of SwissProt/TrEMBL. Such similarities further support the hypothesis that these ORFs ought to have been reported as putative genes in the first place.
We are currently in the process of pursuing several related topics that include the determination of the performance impact using Bio-Dictionaries built from selected collections of proteins and not from full-size datasets; the determination of a compact collection of seqlets for the express purpose of efficient gene identification; extending our strategy to the case of eukaryotic genomes, etc.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank the anonymous reviewers for their insightful comments and feedback on the manuscript. The authors would also like to thank Stephen Chin-Bow and Tien Huynh for designing and implementing the graphical user interface of the Web server for BDGF.
REFERENCES
- 1.Bird A. (1987) CpG islands as gene markers in the vertebrate nucleus. Trends Genet., 3, 342–347. [Google Scholar]
- 2.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borodovsky M. and McIninch J. (1993) GeneMark: parallel gene recognition for both DNA strands. Comput. Chem., 17, 123–133. [Google Scholar]
- 5.Lukashin A.V. and Borodovsky,M. (1998) GeneMark.hmm: new solutions for gene identification. Nucleic Acids Res., 26, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frishman D., Mironov,A., Mewes,H.-W. and Gelfand,M. (1998) Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res., 26, 2941–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badger J.H. and Olsen,G.J. (1999) CIRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol., 16, 512–524. [DOI] [PubMed] [Google Scholar]
- 8.Bafna V. and Huson,D.H. (2000) The conserved exon method for gene finding. Proc. Int. Conf. Intell. Syst. Mol. Biol., 8, 3–12 [PubMed] [Google Scholar]
- 9.Gelfand M.S., Mironov,A.A. and Pevzner,P. (1996) Gene recognition via spliced alignment. Proc. Natl Acad. Sci. USA, 93, 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gish W. and States,D.J. (1993) Identification of protein coding regions by database similarity search. Nature Genet., 3, 266–272. [DOI] [PubMed] [Google Scholar]
- 11.Robinson K., Gilbert.W. and Church.G. (1994) Large-scale bacterial gene discovery by similarity search. Nature Genet., 7, 205–214. [DOI] [PubMed] [Google Scholar]
- 12.Burge C. and Karlin,S. (1998) Finding the genes in genomic DNA. Curr. Opin. Struct. Biol., 8, 346–354. [DOI] [PubMed] [Google Scholar]
- 13.Burset M. and Guigo,R. (1996) Evaluation of gene structure prediction programs. Genomics, 34, 353–367. [DOI] [PubMed] [Google Scholar]
- 14.Claverie J.M. (1997) Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet., 6, 1735–1744. [DOI] [PubMed] [Google Scholar]
- 15.Claverie J.M. (1998) Computational methods for exon detection. Mol. Biotechnol., 10, 27–48. [DOI] [PubMed] [Google Scholar]
- 16.Fickett J.W. (1996) The gene identification problem: an overview for developers. Comput. Chem., 20, 103–118. [DOI] [PubMed] [Google Scholar]
- 17.Fickett J.W. and Hatzigeorgiou,A.G. (1997) Eukaryotic promoter recognition. Genome Res., 7, 861–878. [DOI] [PubMed] [Google Scholar]
- 18.Kehoe M.A., Kapur,V., Whatmore,A.M. and Musser,J.M. (1996) Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol., 4, 436–443. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen K.M., Bones,A.M., Smalla,K. and van Elsas,J.D. (1998) Horizontal gene transfer from transgenic plants to terrestrial bacteria —a rare event? FEMS Microbiol. Rev., 22, 79–103. [DOI] [PubMed] [Google Scholar]
- 20.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J.F., Dougherty,B.A., Merrick,J.M. et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae. Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 21.Rigoutsos I. and Floratos,A. (1998) Combinatorial pattern discovery in biological sequences: the Teiresias algorithm. Bioinformatics, 14, 55–67. [DOI] [PubMed] [Google Scholar]
- 22.Rigoutsos I. and Floratos,A. (1998) Motif discovery without alignment or enumeration. Proceedings of the 2nd ACM International Conference on Computational Molecular Biology (RECOMB’98). ACM Press, New York, NY, 221–227.
- 23.Rigoutsos I., Floratos,A., Ouzounis,C., Gao,Y. and Parida,L. (1999) Dictionary building via unsupervised hierarchical motif discovery. Proteins, 37, 264–277. [DOI] [PubMed] [Google Scholar]
- 24.Rigoutsos I., Floratos,A., Parida,L., Gao,Y. and Platt,D. (2000) The emergence of pattern discovery techniques in computational biology. Metab. Eng., 2, 159–177. [DOI] [PubMed] [Google Scholar]
- 25.Rigoutsos I., Gao,Y., Floratos,A. and Parida,L. (1999) Building dictionaries of 1D and 3D motifs by mining the unaligned 1D sequences of 17 archaeal and bacterial genomes. Proc. Int. Conf. Intell. Syst. Mol. Biol., 223–233. [PubMed] [Google Scholar]
- 26.Floratos A., Rigoutsos,I., Parida,L. and Gao,Y. (1999) Sequence homology detection through large scale pattern discovery. Proceedings of the 3rd ACM International Conference on Computational Molecular Biology (RECOMB’99). ACM Press, New York, NY, pp.164–169.
- 27.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attwood T.K., Flower,D.R., Lewis,A.P., Mabey,J.E., Morgan,S.R., Scordis,P., Selley.J.N. and Wright,W. (1999) PRINTS prepares for the new millennium. Nucleic Acids Res., 27, 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gusfield D. (1997) Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. Cambridge University Press, Cambridge, UK.
- 32.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchler-Bauer A., Panchenko,A.R., Shoemaker,B.A., Thiessen,P.A., Geer,L.Y. and Bryant,S.H. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frishman D., Mironov,A. and Gelfand,M. (1999) Starts of bacterial genes: estimating the reliability of computer predictions, Gene, 234, 257–265. [DOI] [PubMed] [Google Scholar]
- 36.Hannenhalli S.S., Hayes,W.S., Hatzigeorgiou,A.G. and Fickett,J.W. (1999) Bacterial start site prediction. Nucleic Acids Res., 27, 3577–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shmatkov A.M., Melikyan,A.A., Chernousko,F.L. and Borodovsky,M. (1999) Finding prokaryotic genes by the ‘frame-by-frame’ algorithm: targeting gene starts and overlapping genes. Bioinformatics, 15, 874–886. [DOI] [PubMed] [Google Scholar]
- 38.Audic S. and Claverie,J.-M. (1998) Self-identification of protein-coding regions in microbial genomes. Proc. Natl Acad. Sci. USA, 95, 10026–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parida L., Floratos,A. and Rigoutsos,I. (1999) An approximation algorithm for alignment of multiple sequences using motif discovery. Journal of Combinatorial Optimization, 3, 247–275. [Google Scholar]
- 40.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.