Abstract
Misfolded secretory proteins are transported across the endoplasmic reticulum (ER) membrane into the cytosol for degradation by proteasomes. A large fraction of proteasomes in a cell is associated with the ER membrane. We show here that binding of proteasomes to ER membranes is salt sensitive, ATP dependent, and mediated by the 19S regulatory particle. The base of the 19S particle, which contains six AAA-ATPases, binds to microsomal membranes with high affinity, whereas the 19S lid complex binds weakly. We demonstrate that ribosomes and proteasomes compete for binding to the ER membrane and have similar affinities for their receptor. Ribosomes bind to the protein conducting channel formed by the Sec61 complex in the ER membrane. We co-precipitated subunits of the Sec61 complex with ER-associated proteasome 19S particles, and found that proteoliposomes containing only the Sec61 complex retained proteasome binding activity. Collectively, our data suggest that the Sec61 channel is a principal proteasome receptor in the ER membrane.
Keywords: endoplasmic reticulum, ER-associated degradation, proteasome, protein translocation, Sec61 channel
Introduction
The endoplasmic reticulum (ER) is the site of secretory protein biogenesis: nascent secretory proteins are targeted to and translocated into the ER where they fold, acquire covalent modifications, and oligomerize (Johnson and van Waes, 1999). A significant fraction of secretory proteins (10–75%, depending on the protein) fails to fold in the ER and is transported back to the cytosol where misfolded proteins are degraded by proteasomes (Ellgaard and Helenius, 2003; Sitia and Braakman, 2003). Up to 80% of proteasomes in a eukaryotic cell are associated with the nuclear envelope and the ER (Enenkel et al, 1998; Rivett, 1998). These proteasomes may be involved in misfolded secretory protein turnover, but evidence for this is lacking so far, nor has a proteasome receptor in the ER been identified.
Secretory protein transport across the ER membrane in both directions is mediated by proteinaceous channels whose principal component is the evolutionarily conserved Sec61 protein (Johnson and van Waes, 1999). Secretory protein import into the ER is primarily cotranslational in mammals, and both co- and post-translational in yeast (Johnson and van Waes, 1999). During cotranslational import, the Sec61 complex consisting of Sec61α, β, and γ in mammals (Sec61p, Sbh1p, and Sss1p, respectively, in yeast) binds ribosomes to the ER, which gives the ER its typical ‘rough' appearance (Kalies et al, 1994). Ribosome binding to the Sec61 channel is salt sensitive, but independent of the presence of a nascent chain, and high affinity (reported KD values range from 4 to 21 nM) (Kalies et al, 1994; Raden et al, 2000). In yeast, about 30% of the Sec61p in the ER membrane is associated with ribosomes (Pilon et al, 1998). Saturation of Sec61 channels in the ER membrane with translating ribosomes inhibits protein export from the ER lumen to the cytosol, suggesting that channels engaged in export cannot be bound to ribosomes at the same time (Schmitz et al, 2000).
Proteasomes are responsible for the majority of cytosolic protein degradation (Voges et al, 1999; Goldberg, 2003). Proteasome substrates can be wild-type proteins, a significant fraction of which misfolds early in biogenesis, wild-type proteins that have been damaged, for example by oxidation, proteins that are subject to regulated proteolysis, and misfolded mutant proteins (Goldberg, 2003). Many, but not all, proteasome substrates are covalently modified on lysine residues with ubiquitin (Voges et al, 1999; Goldberg, 2003). Proteasomes consist of two subparticles: the 20S core complex, which contains the proteolytically active subunits, tethered by the Ecm29 protein to the 19S regulatory particle (Leggett et al, 2002). The 19S particle can be subdivided into an eight-subunit base and an eight-subunit lid stabilized by a hinge protein, Rpn10p (Glickman et al, 1998; Leggett et al, 2002). The lid can be dissociated from the base by high salt (Glickman et al, 1998). The lid is required for deubiquitination of substrates prior to degradation, and is homologous to the COP9/signalosome complex and eIF3, both of which can bind to other large protein complexes (Glickman et al, 1998). The base consists of six nonequivalent ATPases (Rpt1p–6p) and two non-ATPase subunits, Rpn1p and Rpn2p (Glickman et al, 1998). Both Rpn1p and Rpn2p contain leucine-rich repeat domains, which typically mediate protein–protein interactions (Elsasser et al, 2002). Two binding partners for Rpn1p have been identified: the deubiquitinating enzyme Ubp6p and Rad23p, which is involved in DNA repair (Verma et al, 2000; Elsasser et al, 2002; Leggett et al, 2002). Interaction with Rpn1p is mediated by the ubiquitin-like domains of Ubp6p and Rad23p (Elsasser et al, 2002). The ATPases of the base of the 19S particle are involved in unfolding of degradation substrates, recognition of ubiquitin chains (Rpt5p), and opening of the proteolytic channel in the 20S core particle (Rpt2p) (Rubin et al, 1998; Kohler et al, 2001; Navon and Goldberg, 2001; Lam et al, 2002).
Proteins that have been recognized by the quality control machinery in the ER as dysfunctional are exported to the cytosol and degraded by proteasomes, a process known as ER-associated degradation (ERAD) (McCracken and Brodsky, 1996). Many ERAD substrates are ubiquitinated prior to degradation (Ellgaard and Helenius, 2003). Drugs that specifically inhibit proteolysis by proteasomes such as lactacystin can cause accumulation of misfolded secretory proteins in the cytoplasm or, in some instances, in the ER lumen (Wiertz et al, 1996b; Huppa and Ploegh, 1997; Yang et al, 1998). Mutations in proteolytic subunits of the yeast proteasome core particle (pre1 pre2) have a similar range of effects as proteasome inhibitors, suggesting that depending on the substrate protein, export and degradation may be coupled (Hiller et al, 1996; Loayza et al, 1998; Plemper et al, 1998). Mutations in subunits of the 19S particle (Rpt1p, Rpt6p, Rpn2p) cause ERAD defects (Mayer et al, 1998; Lee et al, 2004). A mutant in the 19S regulatory particle subunit Rpn1p (Hrd2p) was identified in a screen for mutants defective in degradation of ER-resident HMG-CoA reductase (Hampton et al, 1996). Collectively, these observations indicate that both the 19S and the 20S proteasome subparticles are involved in ERAD.
ERAD can be reconstituted in a cell-free system containing yeast microsomes loaded with an ERAD substrate, ATP, and cytosol (McCracken and Brodsky, 1996). In this system, cytosol can be replaced by purified proteasomes (Lee et al, 2004). Export and degradation can also be achieved separately: incubation of microsomes with ATP and 19S particles results in export of a misfolded secretory protein; subsequently added 20S particles can degrade the protein after its export has been completed (Lee et al, 2004).
Efficient removal of misfolded proteins from the ER is essential: accumulation of misfolded proteins in the ER triggers a signalling pathway from the ER to the nucleus, the unfolded protein response (UPR), which upregulates components of the protein translocation machinery and ER chaperones, but also proteins involved in vesicular secretory protein transport (Spear and Ng, 2001). Overexpression of misfolded proteins in yeast cells in which the UPR signalling pathway has been inactivated compromises essential cellular functions and is ultimately lethal (Spear and Ng, 2001). Mutant secretory proteins that polymerize in the ER and therefore cannot be transported to the cytosol for degradation lead to cell death even in the presence of a functional UPR (Carrell and Lomas, 1997). Formation of cytoplasmic, degradation-resistant protein aggregates either by mutant cytoplasmic proteins or by overexpressed misfolded secretory proteins exported from the ER can also be cytotoxic (Selkoe, 2003).
These observations suggest that efficient export of misfolded proteins from the ER must be concomitant with efficient degradation in the cytoplasm. The simplest way to achieve this is to mechanistically couple protein export through the protein translocation channel to proteasomal degradation in the cytosol. During cotranslational protein import into the ER, biosynthesis and translocation are synchronous, and the ribosome is docked to the protein translocation channel (Kalies et al, 1994). Here, we demonstrate that proteasomes can also bind directly to the protein translocation channel in the ER membrane. We analyze proteasome binding to purified ER membranes and show that proteasomes and ribosomes compete with each other for binding to the ER. We examine ER binding of the proteasome 20S core particle, the19S regulatory particle, and the 19S subparticles (base and lid) individually, and find that ER binding of proteasomes is ATP dependent and primarily mediated by the 19S regulatory particle base, which contains six AAA-ATPases. Using reconstituted proteoliposomes containing fractionated ER proteins, we define the Sec61 complex as a principal proteasome receptor in the ER membrane and propose a mechanism for misfolded protein export from the ER.
Results
Proteasomes bind to a site conserved in yeast and mammalian ER membranes
In order to characterize the binding of proteasomes to the ER membrane, we affinity-purified yeast proteasomes from a strain in which the Pre1p subunit of the 20S core particle was FLAG tagged (Verma et al, 2000). For the initial binding experiments, we isolated active 26S proteasomes in the presence of ATP from liquid nitrogen-lysed cells (Verma et al, 2000) (Figure 1A). Wild-type yeast microsomes were stripped of associated ribosomes by incubation with puromycin/high salt (PK-RM), and 10 eq of membranes was incubated in the presence of ATP with 2.5 pmol of 26S proteasomes for 20 min on ice followed by 10 min at room temperature. The samples were layered under a discontinuous sucrose gradient and after centrifugation fractions were analyzed by SDS–PAGE. The proteasomes in each fraction were detected by immunoblotting with a monoclonal anti-FLAG antibody. As shown in Figure 1B, proteasomes bound to and floated with yeast PK-RM (yeast PK-RM/26S, lanes 1 and 2). In the absence of membranes, proteasomes remained at the bottom of the gradient (26S, lanes 7–10). Proteasome binding to untreated yeast microsomes was two- to three-fold lower (not shown) than binding to puromycin/high-salt-washed membranes (Figure 1B).
Figure 1.
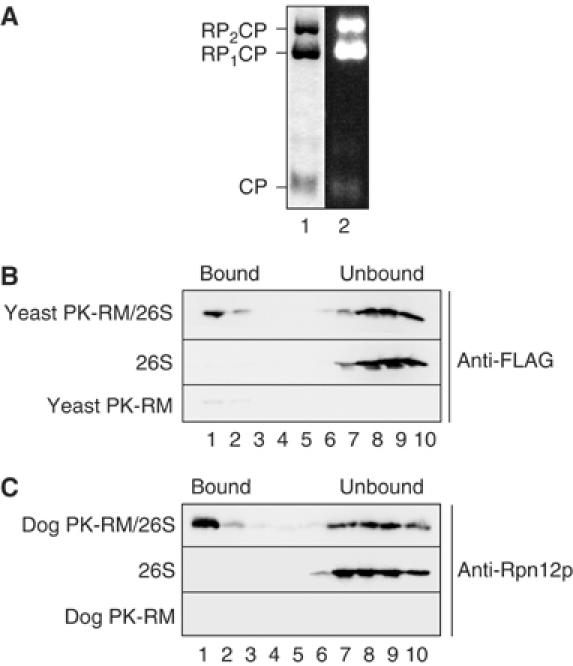
Proteasomes bind to a site conserved in yeast and mammalian ER membranes. (A) Purified yeast 26S proteasomes were separated by native gel electrophoresis and stained with Coomassie blue (lane 1), or the gel incubated with the fluorogenic proteasome substrate Suc-LLVY-AMC, and active proteasomes visualized on a UV transilluminator. CP, 20S core particle; RP1CP, 20S CP with one bound regulatory particle; RP2CP, 20S CP with two bound regulatory particles. (B) Yeast 26S proteasomes (2.5 pmol) were incubated with 10 eq of puromycin/high-salt-treated yeast microsomes (PK-RM) as described in Materials and methods. 1 eq equals 1 μl of microsomes of A280=50. The membranes were floated through 1.8 M sucrose cushions and the gradients fractionated from the top. Proteasomes in individual fractions were detected by SDS–PAGE and immunoblotting employing anti-FLAG antibody. Enhanced chemiluminescence and a CCD camera system (Raytest, Germany) were used as a detection system. The positions of proteasomes bound to membranes and unbound proteasomes are indicated. (C) Yeast 26S proteasomes (2.5 pmol) were bound to puromycin/high-salt-treated dog pancreas microsomes (10 eq). After binding, the samples were treated as in (B) and analyzed by SDS–PAGE and immunoblotting using anti-Rpn12p antibodies.
Proteasomes are also found associated with mammalian ER membranes, and proteasome structure is conserved between yeast and mammals (Rivett, 1998; Voges et al, 1999). We therefore asked if yeast proteasomes could also bind to mammalian microsomes. Puromycin/high-salt-treated dog pancreas microsomes (dog PK-RM) were incubated with purified 26S yeast proteasomes as above, and loaded under a 1.8 M sucrose cushion. After equilibrium centrifugation, proteins in each fraction were resolved by SDS–PAGE, and proteasomes detected with anti-Rpn12p antibody. As shown in Figure 1C, yeast 26S proteasomes bound to and floated with dog PK-RM, suggesting that the binding site for proteasomes at the ER is conserved between yeast and mammals.
Ribosomes and proteasomes compete for binding to the ER membrane
The ribosome binding site is one feature that is conserved between mammalian and yeast ER (Prinz et al, 2000). Therefore, we next asked whether proteasomes and ribosomes could compete for binding to the ER membrane. A constant amount of 26S proteasomes (2.5 pmol) was incubated with a constant amount of ER membranes (10 eq) in the absence or presence of increasing amounts of ribosomes. Membranes with bound proteasomes were sedimented through a sucrose cushion, and pellet (P), sucrose cushion (M), and supernatant fractions (S) were analyzed by SDS–PAGE and immunoblotting for the FLAG-tagged Pre1p subunit of the 20S core particle and the Rpt1p subunit of the 19S regulatory particle. As shown in Figure 2A, ribosomes efficiently competed with proteasomes for binding to the ER membrane, suggesting that ribosomes and proteasomes bind to the same receptor. We measured the affinity of proteasomes for their receptor in the ER membrane, and found a KD between 19 nM (detecting the 20S core particle with the anti-FLAG antibody) and 30 nM (detecting the 19S regulatory particle with the anti-Rpt1p antibody) (Figure 2B); these values are comparable to the KD of ribosome binding to the Sec61 channel (4–21 nM) (Kalies et al, 1994; Raden et al, 2000).
Figure 2.
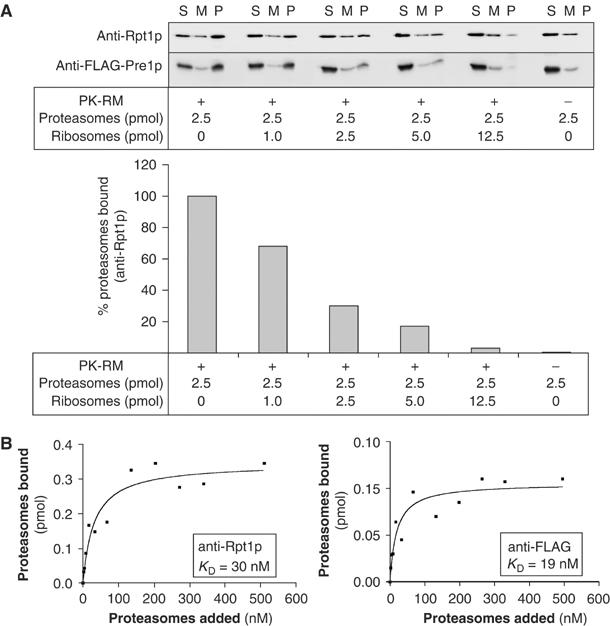
Ribosomes and proteasomes compete for binding to the ER membrane. (A) Yeast 26S proteasomes in which the Pre1p subunit of the 20S core particle was FLAG tagged were incubated with 10 eq of puromycin/high-salt-treated dog pancreas microsomes in the absence or presence of increasing amounts of ribosomes as indicated. The membranes were sedimented through a 0.75 M sucrose cushion, and free proteasomes in the supernatant (S), in the sucrose cushion (M), and proteasomes bound to membranes that had sedimented (P) were detected with anti-FLAG antibody (20S core particle) and anti-Rpt1p antibody (19S regulatory particle). Membrane-bound proteasomes in the pellets were quantified using the anti-Rpt1p signal and a CCD camera system (LAS1000 plus, Raytest, Germany). (B) Dog PK-RM (1 eq) were used in the proteasome binding assay at physiological salt concentrations with varying amounts of yeast 26S proteasomes. The samples were treated as in (A) and the amount of proteasomes bound to the microsomes was determined by SDS–PAGE and quantitative immunoblotting.
Proteasomes are associated with Sec61p in the ER membrane of Saccharomyces cerevisiae
Proteasome binding to evolutionarily conserved sites in yeast and mammalian ER (Figure 1), and competition of ribosomes with proteasomes for binding to the ER membrane (Figure 2) suggested that ribosomes and proteasomes bound to the same receptor, the Sec61 complex. In yeast, 80% of the proteasomes in a cell are associated with the nuclear envelope and the ER (Enenkel et al, 1998). We therefore asked if we could find an association between yeast proteasomes and the yeast Sec61 channel in microsomes isolated from S. cerevisiae. We prepared microsomes from wild-type yeast and from a strain in which the Rpn11p subunit of the proteasome 19S regulatory particle had been tagged with Protein A (Leggett et al, 2002). Membranes were solubilized with deoxy-BigCHAP and proteasomes and associated proteins precipitated with IgG-Sepharose under native conditions overnight at 4°C. The beads were washed, and proteins associated with the Protein A-tagged Rpn11p were eluted, resolved by SDS—PAGE, and detected by immunoblotting. As shown in Figure 3, Protein A-Rpn11p was efficiently precipitated under these conditions and another proteasome subunit, Rpt5p, was co-precipitated with IgG-Sepharose from a microsomal lysate of the strain containing Protein A-tagged Rpn11p (lanes 4 and 5, top panel), but not from a lysate of microsomes from an RPN11 wild-type strain (lanes 4 and 5, middle panel), suggesting that Protein A-tagged Rpn11p was incorporated into proteasome 19S regulatory particles, and that the co-precipitation of Rpt5p was specific. Sec61p specifically co-precipitated with Protein A-Rpn11p (Figure 3, lanes 4 and 5, top versus middle panel). In addition, the β and the γ subunits of the Sec61 complex, Sbh1/2p and Sss1p, co-precipitated with Protein A-Rpn11p (Figure 3, bottom panel, and not shown). Subunits of the Sec63 complex required for post-translational protein import into the yeast ER, Sec62p and Sec72p, were not detectably associated with Protein A-Rpn11p (Figure 3, bottom panel, and not shown). Collectively, our data suggest that the Sec61 complex mediates the interaction of both ribosomes and proteasomes with the ER membrane in intact yeast cells.
Figure 3.

Proteasomes interact with the Sec61 channel in the ER membrane. Yeast microsomes were isolated from a wild-type strain (RPN11) and a strain in which the Rpn11p subunit of the proteasome was Protein A tagged (Protein A-RPN11). Membranes were solubilized in DeoxyBigCHAP, and Protein A-tagged Rpn11p and associated proteins isolated by batch absorption to IgG-Sepharose. Protein A-Rpn11p and associated proteins were eluted from washed beads with 0.5 M acetic acid in two steps (HAc-eluate). Aliquots of microsomes, lysate, material not bound to IgG-Sepharose (supernatant), and HAc-eluates were separated by SDS–PAGE and individual proteasome subunits and associated proteins detected by immunoblotting with polyclonal antibodies. Proteins are indicated on the left and strain genotypes on the right. The asterisk marks a band nonspecifically labelled by the anti-Rpt5p antibody.
Proteasomes bind directly to the Sec61 complex
In order to determine whether proteasomes bound directly to the Sec61 complex, we performed proteasome binding experiments with intact dog pancreas ER membranes, proteoliposomes containing total ER proteins, and proteoliposomes reconstituted with purified Sec61 complex only (Kalies et al, 1994) (Supplementary Figure 1). Binding was quantified by flotation of the membranes as described above and immunoblotting with anti-FLAG antibodies for FLAG-Pre1p. We found that proteasomes bound to reconstituted proteoliposomes containing total ER protein with 80% efficiency compared to intact PK-RM, suggesting that membrane solubilization and reconstitution into proteoliposomes did not inactivate the proteasome receptor (Figure 4A). Proteoliposomes containing only the purified Sec61 complex retained proteasome binding activity, suggesting that the Sec61 complex is necessary and sufficient to bind proteasomes to the ER (Figure 4A). The reduced efficiency of proteasome binding to Sec61 proteoliposomes compared to total proteoliposomes may be due to either partial inactivation of the Sec61 complex during purification or absence of accessory factors that may promote proteasome binding to the channel in total proteoliposomes.
Figure 4.
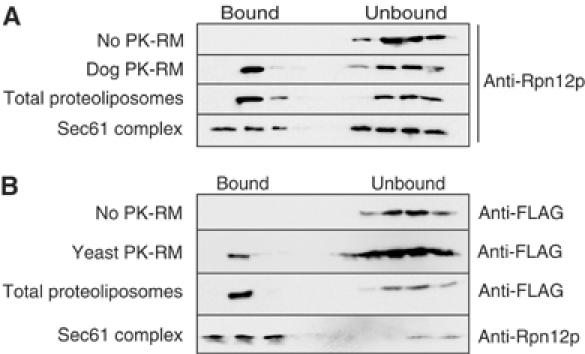
Proteasomes bind directly to the Sec61 complex. (A) Yeast 26S proteasomes (2.5 pmol) were bound to puromycin/high-salt-treated dog pancreas microsomes (10 eq), proteoliposomes reconstituted from total dog microsomal protein, and proteoliposomes containing only the mammalian Sec61 complex, as indicated. Bound and unbound fractions were analyzed by flotation through sucrose cushions, as in Figure 1B, and immunoblotting with anti-Rpn12p antibodies. (B) Yeast 26S proteasomes (2.5 pmol) were bound to puromycin/high-salt-treated yeast microsomes (10 eq), proteoliposomes reconstituted from total yeast microsomal protein, and proteoliposomes containing only the heterotrimeric yeast Sec61 complex (Sec61p, Sbh1p, Sss1p) as indicated. Bound and unbound fractions were analyzed by flotation through sucrose cushions, as in Figure 1B, and immunoblotting with anti-FLAG or anti-Rpn12p antibodies.
In mammalian and yeast ER membranes, ribosomes are bound to Sec61 channels formed by one or more heterotrimeric Sec61 complexes (Kalies et al, 1994; Beckmann et al, 2001; van den Berg et al, 2004). In yeast, the Sec61 complex can also associate with the Sec63 complex to form the heptameric Sec complex, which is required for post-translational protein import into the ER (Deshaies et al, 1991; Panzner et al, 1995). Ribosomes do not bind to the heptameric Sec complex, and the Sec63 complex is not involved in ERAD (Pilon et al, 1997; Plemper et al, 1997; Prinz et al, 2000). We asked whether in yeast the Sec61 complex was also sufficient for proteasome binding to the ER membrane. Wild-type yeast microsomal membranes were solubilized and total ER protein or purified Sec61 complex reconstituted into proteoliposomes. Binding of proteasomes to the proteoliposomes was assayed by flotation. As shown in Figure 4B, proteasomes bound to proteoliposomes reconstituted with purified yeast Sec61 complex, suggesting that the Sec63 complex was not required for binding of proteasomes to the yeast ER. The data shown in Figures 3 and 4 demonstrate that proteasomes like ribosomes bind to the Sec61 complex in the ER membrane of both yeast and mammalian cells. Our data do not exclude the existence of additional proteasome-interacting proteins in the ER membrane, but suggest that the Sec61 channel is a principal proteasome receptor in the ER.
Proteasome binding to the ER membrane is mediated by the 19S particle
We next characterized the interaction of proteasomes with the Sec61 channel in the ER membrane. Physiological binding should be affected by the salt concentration in the binding buffer. Using the flotation assay described above, we measured the binding of affinity-purified 26S proteasomes to ER membranes in buffers with increasing salt concentration. 26S proteasomes consist of 20S core particles and 19S regulatory particles, which behaved differently in this experiment: we found that at 150 mM potassium acetate, which is close to physiological salt concentration, binding of proteasome 19S regulatory particles to ER membranes was half-maximal (Figure 5A, black squares, Rpt1p), whereas binding of FLAG-tagged 20S core particles was only 20% (Figure 5A, open squares, FLAG). Our 26S proteasome preparation contained a fraction of dissociated 20S core particles (see Figure 1A), and some salt-induced dissociation of 26S particles may have occurred during the experiment, which would explain the differential binding of 19S and 20S particles to the ER membranes in this experiment. High concentrations of salt (500 mM) abolished binding of both proteasome subparticles and ribosomes to the ER membrane (Figure 5A; Kalies et al, 1994).
Figure 5.
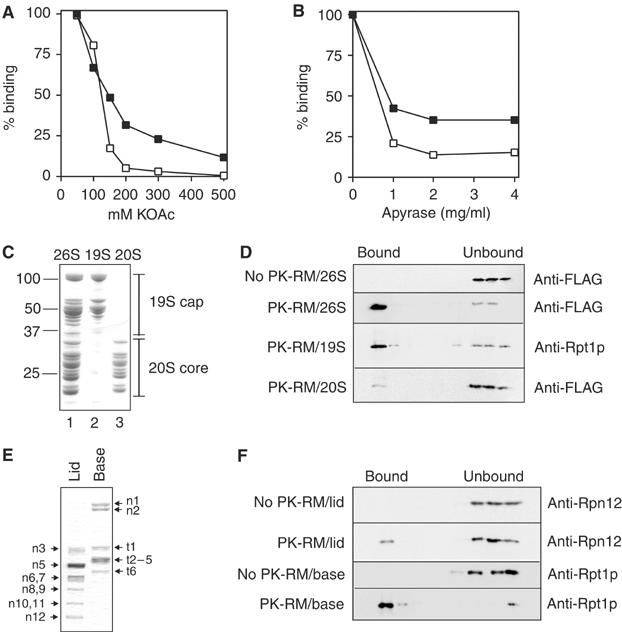
Proteasome binding to the ER membrane is mediated by the base of the 19S particle. (A) Yeast 26S proteasomes were bound to puromycin/high-salt-treated dog pancreas microsomes in the presence of 5 mM ATP and the indicated potassium acetate concentrations. Binding was quantified by flotation of membrane-bound proteasomes in sucrose gradients and immunoblotting for the indicated proteasome subunits (Pre1p-FLAG: open squares; Rpt1p: black squares). Binding was quantified using a CCD camera and binding at 100 mM potassium acetate was set to 100%. (B) Yeast 26S proteasomes were bound to puromycin/high-salt-treated dog pancreas microsomes in the presence of 120 mM potassium acetate and either 5 mM ATP (100%) or no ATP and in the presence of the indicated concentrations of apyrase. Samples were analyzed and binding quantified as in (A). (C) Yeast 26S proteasomes, 20S core particles, and 19S regulatory particles were purified from a strain in which the Pre1p core particle subunit was FLAG tagged. (D) Binding of yeast 26S, 20S, and 19S proteasome particles (2.5 pmol) to ER membranes (10 eq) was assessed by flotation as above. (E) 19S particles purified from a strain in which Rpt1p was FLAG tagged were dissociated into lid and base subparticles by incubation in high salt. 19S non-ATPase subunits Rpn1p, Rpn2p, etc. are indicated by n1, n2, etc.; ATPase subunits Rpt1p, Rpt2p, etc., are labelled t1, t2, etc. (F) Binding of 19S lid and base subparticles (2.5 pmol) to ER membranes (10 eq) was assessed by flotation as above.
The experiment shown in Figure 5A suggests that binding of proteasomes to the ER membrane may be mediated by the 19S regulatory particle, which in contrast to the 20S core particle contains six AAA-ATPase subunits. We therefore asked whether 26S proteasome binding to the ER membrane was ATP dependent. Binding experiments were performed in 120 mM potassium acetate and in the presence of 5 mM ATP, or in the absence of ATP and increasing amounts of apyrase. Binding of 26S proteasomes to ER membranes was nucleotide dependent (Figure 5B), but nucleotide depletion affected binding of the 20S core particle (open squares) more strongly than binding of the19S regulatory particle (black squares), presumably because ATP depletion also promotes dissociation of proteasomes into 19S and 20S subparticles (Verma et al, 2000).
Next, we prepared individual proteasome subparticles for binding experiments. 20S core particles were purified from the PRE1-FLAG strain in the absence of ATP (Figure 5C, lane 3; Verma et al, 2000). 19S regulatory particles were prepared by dissociating 26S proteasomes from the PRE1-FLAG strain after purification, or by directly purifying 19S particles from a strain in which Rpt1p was FLAG tagged (Figure 5C, lane 2; Verma et al, 2000) (Supplementary Figure 3). These 19S regulatory particle preparations were indistinguishable with respect to ER binding (not shown). Binding experiments were performed using the flotation method described above and equal molar amounts of either 20S proteasome core particles or 19S regulatory particles in buffer with 120 mM potassium acetate and in the presence of 5 mM ATP. As shown in Figure 5D, the 20S core particle had no significant affinity for the ER, whereas the 19S regulatory particle on its own efficiently bound to the ER and floated with the membranes (see also Supplementary Figure 2).
The 19S regulatory particle can be dissociated into two subparticles, the lid and the base (Glickman et al, 1998). The lid belongs to a family of large protein complexes, which have the capacity to interact with additional high-molecular-weight complexes, whereas the base contains the six proteasome AAA-ATPases involved in substrate unfolding and opening the central channel in the 20S core particle (Glickman et al, 1998; Rubin et al, 1998; Kohler et al, 2001; Navon and Goldberg, 2001; Benaroudj et al, 2003). We prepared 19S particles from the FLAG-RPT1 strain and dissociated lid and base by incubation in buffer containing 1 M NaCl and 1 mM EDTA (Figure 5E; Leggett et al, 2002). The lid preparation was dialyzed to remove the salt, and equimolar amounts of base and lid were used for binding to ER membranes. As expected from the ATP dependence of the interaction, the 19S base containing the AAA-ATPases bound almost quantitatively to ER membranes (Figure 5F). The lid preparation had significantly lower affinity for ER membranes, but a fraction bound reproducibly, suggesting that the lid contributes to the interaction of the 19S particle with the ER. We conclude that the 19S particle in the ATP-bound conformation mediates proteasome binding to the Sec61 channel in the ER membrane.
Discussion
We have shown here that the 19S regulatory particle of the proteasome, in addition to its role in substrate recognition and unfolding, and in opening the axial channel of the proteasome 20S core particle, mediates binding of proteasomes directly to the protein translocation channel in the ER membrane (Figures 4 and 5D) (Glickman et al, 1998; Kohler et al, 2001; Navon and Goldberg, 2001; Benaroudj et al, 2003). Proteasomes bind to the Sec61 channel with high affinity at physiological salt concentration (Figures 2B and 5A) and ribosomes compete with proteasomes for binding to the ER membrane (Figure 2A), suggesting that proteasomes and ribosomes may bind to the same domain of the protein conducting channel. In contrast to ribosome binding, proteasome binding to the ER is nucleotide dependent (Figure 5B), indicating that a specific conformation of AAA-ATPases of the base of the 19S regulatory particle is required for the interaction with the Sec61 channel. When we dissociated the 19S regulatory particle into subparticles, the base and the lid, we found that the AAA-ATPase containing base bound strongly to ER membranes whereas the lid on its own had limited affinity for the ER (Figure 5F).
AAA-ATPase complexes can extract degradation substrates from other membranes (Escherichia coli plasma membrane, mitochondria, chloroplasts) and proteasome-mediated extraction of membrane protein ERAD substrates has been proposed previously (Mayer et al, 1998; Langer, 2000). The AAA-ATPases of the 19S base are capable of translocating a substrate into the proteolytic core of the proteasome (Voges et al, 1999; Kohler et al, 2001; Navon and Goldberg, 2001); using the same mechanics, the 19S base may be able to extract misfolded proteins from the ER through the Sec61 channel. If the Sec61 channel and the 20S proteolytic core particle of the proteasome bound to opposite faces of the AAA-ATPase ring of the 19S base (as in Figure 6B), this configuration would be ideal to coordinate extraction of misfolded proteins from the ER with their degradation. This may be the optimal pathway for disposing of proteins that are prone to aggregation. In vitro proteasomes consisting of 20S core particle and 19S base can only degrade nonubiquitinated substrates; thus, ERAD mediated by direct coupling of the 20S core complex via the 19S base to the Sec61 channel may be independent of ubiquitylation (Glickman et al, 1998; Leggett et al, 2002). The degradation of mutant alpha-factor precursor, which is exported from the ER in vitro in a 19S regulatory particle-dependent fashion, is indeed independent of ubiquitin conjugation to the substrate (Werner et al, 1996; Lee et al, 2004).
Figure 6.
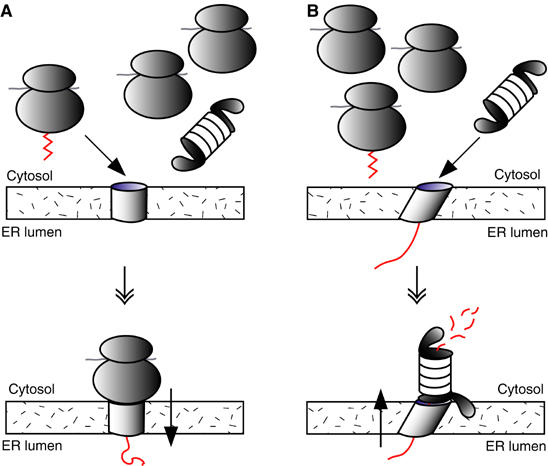
A model for proteasome-mediated export of proteins from the ER to the cytosol. (A) Cotranslational protein import into the ER. Yeast contain approximately 10 × more ribosomes than proteasomes. Ribosomes translating signal peptide containing proteins (zigzag, left) are targeted to the ER membrane by signal recognition particle (SRP; not shown); the signal peptide is released from SRP upon contact with the SRP receptor in the ER membrane and inserted into the Sec61 channel (bottom). This coordinated series of interactions may give ribosomes translating secretory proteins (left) an advantage over nontranslating ribosomes and proteasomes (right) for binding to inactive Sec61 channels in the ER membrane. (B) ERAD. Docking or insertion of a misfolded protein (red) into the Sec61 channel from the lumenal side may trigger a physical change in the Sec61 channel, which can be sensed at the cytoplasmic face of the ER membrane (top). This could either be a conformational change or an association of the channel with one or more accessory proteins required for ERAD; this physical change may increase the affinity of the channel for the 19S base, lead to proteasome docking, and export and degradation of the misfolded secretory protein (bottom).
The available data suggest that degradation itself is unlikely to be the driving force for misfolded protein export from the ER, since for many substrates, mutations in or inhibition of the proteases of the 20S core particle lead to accumulation of the export substrate in the cytosol or on the cytosolic face of the ER membrane (Hiller et al, 1996; Wiertz et al, 1996a; Huppa and Ploegh, 1997; Yang et al, 1998). Furthermore, Lee et al (2004) demonstrated recently that in the presence of ATP, the 19S regulatory particle can promote export of mutant alpha-factor precursor from the ER in a cell-free system in the absence of the 20S core particle; after export, the substrate was found associated with 19S particles. Mutations in genes encoding the 19S base AAA-ATPases (RPT1-6) and in its non-ATPase subunits (RPN1/2) affect ERAD: rpt6 (cim3) has a strong effect on the degradation of mutant alpha-factor precursor, whereas rpn2 (sen3) cells display a weaker defect (Lee et al, 2004; K Römisch, unpublished); rpt1 is defective in degradation of a Sec62p chimera with a proteasome degradation signal (Mayer et al, 1998); an rpn1 mutant (hrd2) was identified in a screen for mutants defective in the regulated degradation of HMG-CoA reductase (Hampton et al, 1996). Whether the degradation substrates accumulate in the cytosol or remain in the ER lumen in the 19S base mutants remains to be investigated. So far, mutations in genes encoding 19S lid subunits have not been identified in screens for ERAD mutants, and lid mutants have not been tested directly for effects on ERAD.
Complexes formed by the AAA-ATPase Cdc48p (p97 in mammals) are also associated with the ER, have been shown to be involved in ERAD of many substrates both by mutant analysis in yeast and by reconstitution experiments in a mammalian system, and have been proposed to promote protein export from the ER (Enenkel et al, 1998; Rivett, 1998; Ye et al, 2001; Braun et al, 2002; Jarosch et al, 2002). Given their structural homology, the proteasome base and the Cdc48p complex may both be able to interact with the Sec61 channel and may have analogous functions in driving protein export from the ER, but perhaps different substrate specificities. In the absence of ATP, p97 binds to two proteins in the mammalian ER membrane, VIMP and Derlin1; Derlin1 also interacts with an export intermediate of MHC class I heavy chain in the ER (Lilley and Ploegh, 2004; Ye et al, 2004). The gene encoding the yeast Derlin1 homolog, DER1, was first identified in a screen for ERAD mutants (Knop et al, 1996). In the presence of ATP and an ERAD substrate, the p97/Derlin1/VIMP complex may associate with Sec61p and promote the export of specific substrates (Jarosch et al, 2002; Ye et al, 2003). Alternatively, export may be mediated by the 19S base for all ERAD substrates, but only facultatively coupled to degradation by the 20S core particle; in this scenario, Cdc48p may act after export to maintain substrates degradation competent for the proteasome by preventing their aggregation, similar to the role of Cdc48p in the release of the cleaved transcription factor Spt23p from the cytoplasmic face of the ER membrane, or Cdc48p may serve as a polyubiquitin receptor and deliver substrates to the proteasome (Rape et al, 2001; Hartmann-Petersen et al, 2004; Verma et al, 2004).
A yeast cell contains approximately 2 × 105 ribosomes and 2 × 104 proteasomes, with little excess of 19S particles over 20S particles (Li et al, 2000; Kohler et al, 2001). Ribosomes translating signal peptide containing proteins are targeted to the ER membrane by SRP, which binds the signal peptide of the nascent protein (Johnson and van Waes, 1999). The signal peptide is released from SRP upon contact with the SRP receptor in the ER membrane and inserted into the Sec61 channel (Johnson and van Waes, 1999). This coordinated series of interactions may give ribosomes translating secretory proteins an advantage over nontranslating ribosomes and proteasomes for binding to inactive Sec61 channels in the ER membrane (Figure 6A). Docking or insertion of a misfolded protein into the Sec61 channel from the lumenal side, on the other hand, may trigger a physical change in the Sec61 channel, which can be sensed at the cytoplasmic face of the ER membrane (Figure 6B). This could either be a conformational change or an association of the channel with one or more accessory proteins required for ERAD, such as the Derlin1/VIMP complex or the ubiquitination machinery, or both; this physical change may increase the affinity of the channel for the 19S base, lead to proteasome docking, and export and degradation of the misfolded secretory protein (Figure 6B).
Materials and methods
Yeast strains
RJD1144 (MATa his3Δ200 leu2-3,112 lys2-801 trp1Δ63 ura3-52 PRE1FH∷Ylplac211(URA3)) and RJD1171 (MATa his3Δ200 leu2-3,112 lys2-801 trp1Δ63 ura3-52 RPT1FH∷Ylplac211(URA3)) (gifts from Ray Deshaies) were used for proteasome preparations as described by Verma et al (2000). SDL73 (MATα lys2-801 leu2-3,112 ura3-52 his3-Δ200 trp1-1 rpn11∷RPN11-TEVProA(HIS3)) was used for co-immunoprecipitation experiments (Leggett et al, 2002). YTX69 (MATa/αhis3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 can1-100/can1-100) was used as a source of yeast microsomes for binding and reconstitution experiments (Finke et al, 1996).
Yeast and dog pancreas microsomes, and proteoliposome preparation
Yeast microsomes were prepared as described by Pilon et al (1997). Dog pancreas microsomes and microsomes stripped of ribosomes by puromycin/high-salt treatment (PK-RM) were prepared as described by Gorlich et al (1992). Proteoliposomes were prepared essentially as described by Kalies et al (1994) with either total microsomal proteins or Sec61 complex only (Panzner et al, 1995).
Native co-precipitations
Microsomes from SDL73 (Protein A-RPN11; 500 μg protein) in 100 μl 50 mM Hepes–KOH, pH 7.4, 400 mM potassium acetate, 5 mM magnesium acetate, 10% glycerol, 5 mM ATP, 0.05% (vol/vol) β-mercaptoethanol, 5 μg/ml leupeptin, 0.5 μg/ml pepstatin, 1 mM aminobenzamidine, 2.5 μg/ml chymostatin, and 0.1 mM PMSF (solubilization buffer) (Pilon et al, 1998) were solubilized by adding 400 μl of 3% DeoxyBigCHAP (Calbiochem) in the same buffer and incubation on ice for 40 min. After centrifugation at 35 000 g for 30 min at 4°C, the lysate was diluted 1:4 in solubilization buffer, and 19S regulatory particles and associated proteins were precipitated with 200 μl of 50% IgG-Sepharose in the same buffer overnight at 4°C. The sample was transferred to a column, the flowthrough collected, and the Sepharose washed with 10 ml solubilization buffer containing 0.7% DeoxyBigCHAP. Bound proteins were eluted from the washed beads with 2 × 1 ml 0.5 M acetic acid, pH 3.4; eluates were lyophilized, heated for 10 min at 65°C in 2 × SDS sample buffer and analyzed by SDS–PAGE on 12% Nu-PAGE gels (Invitrogen) and immunoblotting with polyclonal antibodies against the N-terminus of Sec61p (1:5000), Rpt5p (1:25 000; Affiniti, UK), Sss1p, Sec62p, Sec63p (all 1:1000), Sec72p (1:750) (all from Randy Schekman), and Sbh1/2p (1:2000; from Jussi Jäntti; this antibody recognizes both Sbh1p and Sbh2p).
Purification of proteasomes and proteasome subparticles
26S proteasomes were purified from liquid nitrogen-lysed RJD1144 (PRE1-FLAG) in the presence of ATP as described (Verma et al, 2000). 19S regulatory particles were purified from RJD1171 (RPT1-FLAG) in the absence of ATP as described by Verma et al (2000) with the following modification: the washed anti-Flag M2 agarose beads with bound 19S particles were incubated in 50 mM Tris (pH 7.4), 1 mM MgCl2, 10% glycerol, 1 mM dithiothreitol (DTT), 500 mM NaCl, and 1 U/ml apyrase for 15 min at 30°C to remove residual bound 20S particles, prior to elution of the 19S particles with FLAG peptide. Base and lid were generated by incubating M2 agarose with bound 19S particles in 50 mM Tris (pH 7.5), 1 mM EDTA, and 1 M NaCl for 1 h at 23°C. The supernatant containing the lid was dialyzed and concentrated, and the base was eluted with FLAG peptide as above.
Proteasome activity assays
Proteasome samples were resolved by nondenaturing PAGE on 4% polyacrylamide gels, the gel incubated with the fluorescent peptidase substrate Suc-LLVY-AMC (0.1 mM for 10 min at 30°C), and proteasomes visualized on a UV transilluminator (Glickman et al, 1998).
Binding assays, flotation, and competition with ribosomes
Proteasome binding assays were carried out like the ribosome binding assays, essentially as described by Prinz et al (2000). Briefly, ribosome-stripped membranes (PK-RM) or proteoliposomes containing the indicated protein complexes were mixed with the indicated concentrations of 26S proteasomes (19S regulatory particles, 20S core particles, base particles, or lid particles, without or with competing ribosomes) in a final volume of 30 μl of 20 mM Hepes–KOH, pH 7.2, 120 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT, 5 mM ATP, and 250 mM sucrose. Samples were incubated for 20 min at 0°C followed by 10 min at room temperature, and mixed with 270 μl of the same buffer containing 2.3 M sucrose and ammonium acetate instead of potassium acetate. Samples were layered under an 800 μl 1.8 M sucrose cushion in the same buffer and overlaid with 200 μl binding buffer. After centrifugation in a TLS-55 rotor for 1 h at 4°C at 55 000 r.p.m., 130 μl fractions were collected from the top and analyzed by SDS–PAGE and immunoblotting. Alternatively, after binding, samples were centrifuged for 2 min through 0.1 ml of a 0.75 M sucrose cushion in a TLA100 rotor at 100 000 r.p.m. at 4°C and a supernatant fraction (50 μl from the top) and a cushion fraction were collected. The pellets were resuspended in 50 μl water and all fractions were analyzed by SDS–PAGE and immunoblotting.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Acknowledgments
We are grateful to Carol Harty for a batch of purified proteasomes, to Carl Mann and Randy Schekman for antibodies, to Ray Deshaies for strains, and to Enno Hartmann, Elena Miranda, and Candy Ng for helpful comments on the manuscript. We particularly thank Dan Finley for generously sharing numerous reagents and for advice regarding separation of 19S lid and base subparticles. K Römisch was funded by a Senior Fellowship from The Wellcome Trust (042216). K-U Kalies was funded by the DFG (Ka1444/1-3).
References
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL (2003) ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell 11: 69–78 [DOI] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S (2002) Role of the ubiquitin-selective CDC48 chaperone in ERAD of OLE1 and other substrates. EMBO J 21: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell RW, Lomas D (1997) Conformational disease. Lancet 350: 134–138 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R (1991) Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into the membrane-boumd multisubunit complex. Nature 349: 806–808 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GAG, Finley D (2002) Proteasome subunit Rpn1 binds ubiquitin-like domains. Nat Cell Biol 4: 725–730 [DOI] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM (1998) Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope–ER network in yeast. EMBO J 17: 6144–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke K, Plath K, Panzner S, Prehn S, Rapoport TA, Hartmann E, Sommer T (1996) A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J 15: 1482–1494 [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Croux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquiting-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Hartmann E, Kalies K-U, Rapoport TA (1992) A mammalian homlog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 71: 489–503 [DOI] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J (1996) Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 7: 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Petersen R, Wallace M, Hofmann K, Koch G, Johnson AH, Hendil KB, Gordon C (2004) The Ubx2 and Ubx3 cofactors direct Cdc48 activity to proteolytic and non-proteolytic ubiquitin-dependent processes. Curr Biol 14: 824–828 [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin–proteasome pathway. Science 273: 1725–1728 [DOI] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL (1997) The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity 7: 113–122 [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4: 134–139 [DOI] [PubMed] [Google Scholar]
- Johnson AE, van Waes MA (1999) The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol 15: 799–842 [DOI] [PubMed] [Google Scholar]
- Kalies K-U, Gorlich D, Rapoport TA (1994) Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J Cell Biol 126: 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH (1996) Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J 15: 753–763 [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D (2001) The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell 7: 1143–1152 [DOI] [PubMed] [Google Scholar]
- Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416: 763–767 [DOI] [PubMed] [Google Scholar]
- Langer T (2000) AAA proteases: cellular machines for degrading membrane proteins. Trends Biochem Sci 25: 247–251 [DOI] [PubMed] [Google Scholar]
- Lee RJ, Liu C, Harty C, McCracken AA, Romisch K, DeMartino GN, Thomas PJ, Brodsky JL (2004) The 19S cap of the 26S proteasome is sufficient to retro-translocate and deliver a soluble polypeptide for ER-associated degradation. EMBO J 23: 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D (2002) Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507 [DOI] [PubMed] [Google Scholar]
- Li Y, Moir RD, Sethy-Corachi IK, Warner JR, Willis IA (2000) Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol Cell Biol 20: 3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL (2004) A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429: 834–840 [DOI] [PubMed] [Google Scholar]
- Loayza D, Tam A, Schmidt WK, Michaelis S (1998) Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol Biol Cell 9: 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Braun T, Jentsch S (1998) Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J 17: 3251–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL (1996) Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol 132: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon A, Goldberg AL (2001) Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell 8: 1339–1349 [DOI] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81: 561–570 [DOI] [PubMed] [Google Scholar]
- Pilon M, Romisch K, Quach D, Schekman R (1998) Sec61p serves multiple roles in secretory precursor binding and translocation into the endoplasmic reticulum membrane. Mol Biol Cell 9: 3455–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K (1997) Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J 16: 4540–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388: 891–895 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Egner R, Kuchler K, Wolf DH (1998) Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem 273: 32848–32856 [DOI] [PubMed] [Google Scholar]
- Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU (2000) Evolutionarily conserved binding of ribosomes to the translocatin channel via the large ribosomal RNA. EMBO J 19: 1900–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raden D, Song W, Gilmore R (2000) Role of the cytoplasmic segments of Sec61alpha in the ribosome-binding and translocation-promoting activity of the Sec61 complex. J Cell Biol 150: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48 (UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107: 667–677 [DOI] [PubMed] [Google Scholar]
- Rivett AJ (1998) Intracellular distribution of proteasomes. Curr Opin Immunol 10: 110–114 [DOI] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakamar S, Finley D (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J 17: 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A, Herrgen H, Winkeler A, Herzog V (2000) Cholera toxin is exported from microsomes by the Sec61p complex. J Cell Biol 148: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D (2003) Folding proteins in fatal ways. Nature 426: 900–904 [DOI] [PubMed] [Google Scholar]
- Sitia R, Braakman I (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426: 891–894 [DOI] [PubMed] [Google Scholar]
- Spear E, Ng DTW (2001) The unfolded protein response: no longer just a special teams player. Traffic 2: 515–523 [DOI] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM, van Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X-ray structure of a protein-conducting channel. Nature 427: 36–44 [DOI] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometic analysi of affinity-purified proteasomes. Mol Biol Cell 11: 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Werner ED, Brodsky JL, McCracken AA (1996) Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA 93: 13797–13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL (1996a) The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84: 769–779 [DOI] [PubMed] [Google Scholar]
- Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL (1996b) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction [see comments]. Nature 384: 432–438 [DOI] [PubMed] [Google Scholar]
- Yang M, Omura S, Bonifacino JS, Weissman AM (1998) Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum (ER) in T cells: importance of oligosaccharide processing, ubiquitination, and proteasome-dependent removal from ER membranes. J Exp Med 187: 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER to the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2003) Function of the p97–Ufd1–Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of non-ubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA (2004) A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429: 841–847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3


