Abstract
The current model to explain the organization of the mammalian nervous system is based on studies of anatomy, embryology, and evolution. To further investigate the molecular organization of the adult mammalian brain, we have built a gene expression-based brain map. We measured gene expression patterns for 24 neural tissues covering the mouse central nervous system and found, surprisingly, that the adult brain bears a transcriptional “imprint” consistent with both embryological origins and classic evolutionary relationships. Embryonic cellular position along the anterior–posterior axis of the neural tube was shown to be closely associated with, and possibly a determinant of, the gene expression patterns in adult structures. We also observed a significant number of embryonic patterning and homeobox genes with region-specific expression in the adult nervous system. The relationships between global expression patterns for different anatomical regions and the nature of the observed region-specific genes suggest that the adult brain retains a degree of overall gene expression established during embryogenesis that is important for regional specificity and the functional relationships between regions in the adult. The complete collection of extensively annotated gene expression data along with data mining and visualization tools have been made available on a publicly accessible web site (www.barlow-lockhart-brainmapnimhgrant.org).
Keywords: database, development, evolution, gene expression profiling, inbred strains of mice
The adult nervous system achieves its mature form as the result of neuroectodermal cells committing to a specific fate and then segregating into distinct regional collectives of neurons that become fully functional through establishment of connections to other neurons. Our current understanding of brain architecture and organization is based on studies of embryology, anatomy, and evolution in which direct observation of anatomic structures was the foundation for postulated models of brain structure (1). Recent models of brain development and maturation consider relationships between different regions based on the expression of specific genes in assigning developmental origins of adult structures (2, 3). Here, we have constructed a regional gene expression atlas of the adult mouse brain and analyzed the quantitative results by using molecular classification algorithms.
Genome-wide gene expression profiling is a powerful technique for deriving information about specific brain regions (4, 5). This approach has been used to measure gene expression patterns in particular regions, subregions, or cell populations in the brain (6–11). Two previous studies have analyzed gene expression differences across multiple regions of the mammalian brain by using multiple strains or species (12, 13). However, the current study is the most extensive to date in terms of the number of genes and the coverage of different neural tissues. Our goal was to create a publicly accessible gene-based brain map with data sets, metadata, databases, and analysis tools available for use by the scientific community (5). As part of this work, we measured gene expression patterns for 24 neural tissues covering the adult mouse central nervous system plus 10 body regions. The gene expression data, along with data mining and visualization tools, are available on a publicly accessible web site (www.barlow-lockhartbrainmapnimhgrant.org). A large-scale, systematic, quantitative mouse brain gene expression database, called TeraGenomics, was built to house and provide access to all of the quantitative, region-specific gene expression data, along with quality control measures, anatomical information, strain information, dissection protocols, sample preparation information, and array hybridization parameters in accordance with MIAME (Minimal Information About a Microarray Experiment) (14).
Our goal in this study is to understand how regional gene expression patterns in the brain are related to brain architecture and organization. We sought to identify relationships between brain regions based on both shared and restricted gene expression patterns. The gene expression data were analyzed by using molecular classification algorithms, without prespecified anatomical information, to define relationships between brain structures. To our surprise, we found that the gene expression patterns of the adult brain have a transcriptional “imprint” that is consistent with embryological origins and classic evolutionary relationships between subregions of the cortex.
Materials and Methods
Tissue Collection. All animal procedures were performed according to protocols approved by The Salk Institute for Biological Studies and BrainCells Animal Care and Use Committees. Male A/J, C57BL/6J (B6), C3H/HeJ, and DBA/2J (DBA) mice were purchased from The Jackson Laboratory; male 129S6/SvEvTac (129) mice were purchased from Taconic Farms. All mice were purchased at an age of 7 weeks and housed individually for 1 week before being killed. Dissections were done between 1100 and 1700 h. Mice were killed by either cervical dislocation or decapitation, and dissected tissue, collected within 15 min of death, was directly frozen on dry ice and stored at -80°C. The following brain regions were collected: amygdala (Amg), bed nucleus of the stria terminalis (Bnst), CA1 region of the hippocampus, CA3 region of the hippocampus, cerebellum (Cb), choroid plexus from the fourth ventricle (cp4v), cortex (Cx), dentate gyrus (DG), diencephalon and midbrain excluding hypothalamus (Hy) (DiE-MD), entorhinal cortex (EntCx), hippocampal formation (HiF), Hy, inferior colliculus (IC), medulla oblongata (MO), motor cortex (MtrCx), olfactory bulbs (Olf), periaqueductal gray (Pag), perirhinal cortex (PrhCx), pituitary (Pit), pons, retina, spinal cord (SpCrd), striatum, and superior colliculus (SC). The following body regions were collected: adrenal glands, brown adipose tissue (retroperitoneal and interscapular), heart, kidney, liver, skeletal muscle (femoral), spleen, testes, thymus, and white adipose tissue (epididymal). (For a description of the samples, see Table 1, which is published as supporting information on the PNAS web site.)
To ensure that highly reproducible dissections were conducted for each region, bregma coordinates and anatomic boundaries defining each region were established based on the Paxinos and Franklin mouse brain atlas (15). A reference document was created that consisted of photographs and atlas bregma coordinates to illustrate the exact methods used to dissect each region, including step-by-step instructions (for an example, see Appendix 1, which is published as supporting information on the PNAS web site). The dissection reference documents accompany the processed microarray data as part of the MIAME (14)-compliant metadata housed in the publicly accessible relational database (www.barlow-lockhartbrainmapnimhgrant.org). The metadata contain 75 different fields of sample annotation, which include dissection protocols and anatomical information defining the bregma coordinates. (All dissection protocols and metadata are available at www.barlow-lockhartbrainmapnimhgrant.org; examples are available in Appendices 1–5 and Metadata 1–5, which are published as supporting information on the PNAS web site.) In addition, the anatomical hierarchy of the Neuro Names taxonomy (16) has been included as a user-friendly query tool within the database.
RNA Preparation. Total RNA was isolated according to the methods of Sandberg et al. (12). Tissues were placed into TRIzol (GIBCO/BRL) (added to the frozen tissues, ≈1 ml/100 mg of tissue) and homogenized (Polytron, Kinematica, Lucerne, Switzerland) at maximum speed for 90–120 s. Subsequent steps were performed according to the manufacturer's instructions for all tissues with the exception of spleen and white adipose, for which the Qiagen (Valencia, CA) RNeasy Mini Kit was used to clean up the total RNA after the TRIzol protocol. White adipose RNA was prepared by using a protocol kindly provided by Eric Muise and Yarek Hrywna of Merck. Tissues were added to 4 ml of TRIzol and homogenized for 90 s. After a 10-min incubation at room temperature, samples were spun for 10 min at 3,200 × g, and the top fat layer that resulted was removed. After the addition of chloroform, the samples were spun for 20 min at 3,200 × g. The rest of the protocol was performed according to the TRIzol instructions, and the Qiagen RNeasy Mini Kit was used to clean up the total RNA. Labeling of all samples, hybridization, and scanning were performed by using a modification of the protocol developed by Wodicka et al. (17) using the Affymetrix GeneChip MG_U74Av2 microarray (Affymetrix, Santa Clara, CA) that contains 12,422 probe sets corresponding to ≈12,000 genes and expressed sequence tags.
Database and Analysis Tools. After scanning the arrays with the Affymetrix GeneArray Scanner, the .cel files were uploaded, housed, and analyzed in the Teradata analytical relational database (Teradata, a division of NCR, Dayton, OH) with algorithms developed by our laboratory with the TeraGenomics software tool (Information Management Consultants, Reston, VA) (18). Additional analysis was performed with the freeware program bullfrog 10.2 (see ref. 19 for the original version; the current version is available at www.barlow-lockhartbrainmapnimhgrant.org), genespring 6 (Silicon Genetics, Redwood City, CA), the gene ontology tree machine (gotm) [http://genereg.ornl.gov/gotm (20)], and excel (Microsoft). The signals for each array were scaled to an overall target intensity of 200 (17), and arrays were normalized separately to the same average intensity based on the probe sets corresponding to the 60th to 90th percentile of hybridization signals.
Analysis Algorithms and Criteria. The algorithms and criteria used to analyze the gene expression data in TeraGenomics are described in Supporting Methods, which is published as supporting information on the PNAS web site.
Microarray Quality, Experimental Reproducibility, and False Positives. Given the large amount of data in the atlas, the quality of the samples were assessed at several steps, including total RNA quality (minimum yield was 10 μg with a 260/280 ratio in Tris-EDTA between 2.0 and 2.2), cRNA yield and quality (minimum cRNA yield was 0.66 μg/μl), and array hybridization quality control metrics (for all arrays, background was <200, raw Q < 5, scaling factor < 6, outliers < 500, percent present or marginal ≥ 45%, actin 3′/5′ < 2, and GAPDH 3′/5′ < 2), and by assessing the performance of replicates (Pearson correlation coefficient between replicates was required to be ≥0.97, and the number of genes scored as different between replicates was required to be <1% of the total number of probe sets on the arrays) (see Table 2, which is published as supporting information on the PNAS web site). To determine experimental reproducibility and false-positive rates, we compared independent samples from different animals from the same region and same strain. The set of criteria used to establish experimental reproducibility between replicates was a fold change of 1.5 or greater, a difference call of increase, marginal increase, decrease, or marginal decrease, and a signal change (scaled) of >30. Because these comparisons were between replicate groups, by definition, any genes returned as significantly different would be considered false positives (see Table 3, which is published as supporting information on the PNAS web site).
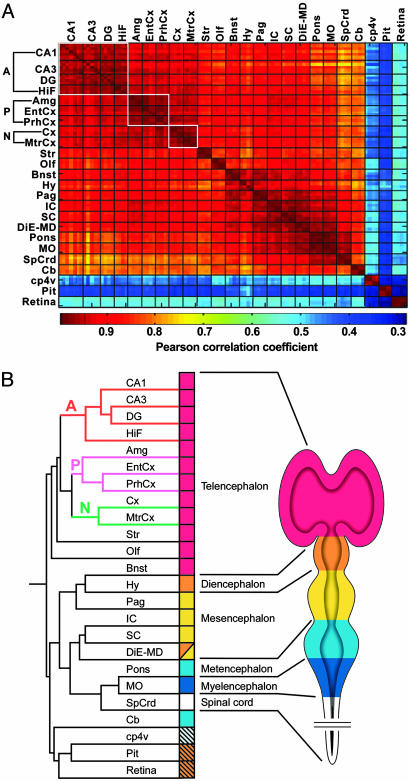
“Heat Map” and Cluster Analysis. A correlation matrix of brain region relatedness was generated for all 100 pairwise comparisons. From this analysis, we observed that the average intrastrain replicate R value (all regions) was 0.988 for B6, 0.987 for 129, and 0.978 for DBA. The average interstrain replicate R value was 0.974 for B6 versus 129, 0.961 for B6 versus DBA, and 0.963 for 129 versus DBA. Given the strong similarity between the intrastrain and interstrain comparisons within a particular brain region, for the purposes of this study, we averaged the data for each brain region independent of strain. The bullfrog software was used to identify the 7,852 probe sets that were called “present” and with a scaled signal of 35 or greater in at least one of the 24 neural tissues. The “most variable” genes from this subset were then identified by using an algorithm that normalized signal values across the 24 regions for each gene and ranked the genes from highest to lowest using the standard deviation of the normalized signals. The probe sets with a normalized standard deviation >0.15 were identified, yielding a total of 4,894 genes. matlab student 7.0 was used to generate a heat map, and genespring 6 was used to generate the clustering relationship based on the Pearson correlation for all pairwise comparisons of absolute signal intensity (Fig. 1; see also Table 4, which is published as supporting information on the PNAS web site).
Fig. 1.
The adult brain bears a gene expression imprint based on embryologic origin and classic evolutionary complexity. (A) Pearson correlation heat map matrix of all brain samples. The white boxes outline the classic evolutionarily related regions of the archicortex (A) (HiF, CA1, CA3, and DG), paleocortex (P) (Amg, EntCx, and PrhCx), and neocortex (N) (Cx and MtrCx). Samples with very similar gene expression profiles corresponding to a higher correlation coefficient are denoted by dark red, and map positions corresponding to brain regions with dissimilar gene expression profiles appear dark blue. (B) Unsupervised hierarchical cluster dendrogram. (Left) The dendrogram relating structures to one another. (Right) A schematic of the developing mouse brain with the five vesicle regions color-coded. The color chart shows the derivatives of these embryonic brain vesicles in the context of the dendrogram. The hatched boxes indicate brain structures formed by inductive events. A, archicortex; P, paleocortex; N, neocortex.
Identification of Region-Restricted or Region-Enriched Gene Expression Patterns. To identify genes with region-restricted or regionenriched expression patterns, probe sets that were called present and with a signal of 35 or greater in at least one sample were used in the analysis (8,156 probe sets). Data were analyzed for 22 mouse brain regions (excluding cp4v and Pit) to identify genes that are clearly expressed at detectable levels in only one to two distinct brain regions. Data from two different inbred mouse strains (two replicates per strain) were analyzed for each of the brain regions except retina. For retina, four samples from one strain were used in this analysis. Data files were exported from TeraGenomics, and a combination of filtering and “Venn” functions were used in bullfrog to identify region-restricted genes. Probe sets that were consistently detected as present in both strains for only one to two specific brain regions and consistently not detected in most other brain regions were identified. For genes to be categorized as region-specific or region-restricted, probe sets corresponding to those genes were required to meet a set of empirically derived selection criteria that were based on the “present” and “difference” calls (see the selection criteria described in Supporting Methods).
To allow searches for user-defined gene expression patterns, we developed algorithms in bullfrog to identify genes enriched in specific regions. For this purpose, the normalized signal intensities of the replicate samples were averaged. The “shape vector” analysis tool in bullfrog was used to identify probe sets with expression “vectors” [normalized signals across the 23 brain regions (excluding cp4v)] that were most highly correlated with an entered “ideal” pattern. For example, to find genes specifically enriched in the Amg, the “ideal shape” vector used was (1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0) for the regions Amg, Bnst, CA1, CA3, Cb, Cx, DG, EntCx, HiF, Hy, IC, DiE-MD, MO, MtrCx, Olf, Pag, PrhCx, Pit, pons, retina, SpCrd, striatum, and SC. The data were then sorted based on the correlation coefficient (R) between the observed and the ideal pattern to yield the list of genes with expression patterns that match the input pattern most closely. The enriched genes are shown in Table 5, which is published as supporting information on the PNAS web site.
Gene Ontology (GO) Analysis. The web-based program gotm [http://genereg.ornl.gov/gotm (20)] was used for the GO analysis. Excluding the genes specific for retina, Pit, and cp4v, 192 region-restricted and -enriched genes were analyzed by using gotm (see Table 6, which is published as supporting information on the PNAS web site, for a list of the genes). gotm was used to identify GO categories with representations significantly different from those expected by chance (P < 0.01). This analysis was carried out in October 2004.
Digital Brain Atlas. The digital atlas was generated from a 90-day-old C57BL/6N mouse brain that was prepared as follows. C57BL/6N mice (Charles River Breeding Laboratories) were anesthetized with Avertin (0.5 mg/g of body weight, i.p.) followed by transcardial perfusion with a light (10%) sucrose solution. The brains were removed, immediately frozen in isopentane at -30°C, and stored at -80°C until use. The brain chosen for the atlas was cryostat-cut (30 μm thick) in the coronal plane of section over a 2-day period, resulting in a total of 462 sections mounted onto 110 microscope slides. The sections, which span from the tip of the Olf to the end of the Cb, were Nissl-stained with a combination of cresyl violet and thionin, providing enhanced differentiation between neurons and glia. Brain sections for the atlas were then digitally acquired into a database by using in-house, java-based software, neurozoom (Neurome, La Jolla, CA), at a very high resolution of 1.3 μm/pixel. The individual image tiles were then stitched together.
In order for the atlas to be visualized in arbitrary planes of section and in a 3D virtual display (Fig. 2C), the digitized brain sections were aligned and are in register with a magnetic resonance microscopy (MRM) data file collected at 11.7 T from a formaldehyde-fixed C57BL/6N brain within the skull. The atlas sections were synchronized by surface alignments to the MRM file using center-alignment algorithms to register the atlas section contours to those from the MRM file. This MRM file is 1 of 10 similar MRM data sets for 90-day-old C57BL/6N male mice that we have captured and is a true representation of a C57BL/6N brain with minimal interindividual variance (21). The alignment allows the atlas to be viewed not only in the coronal plane of section in which it was generated, but also in the extrapolated sagittal and horizontal planes, which are dynamically constructed from slices of the coronal sections as well as orthogonal views with rotation.
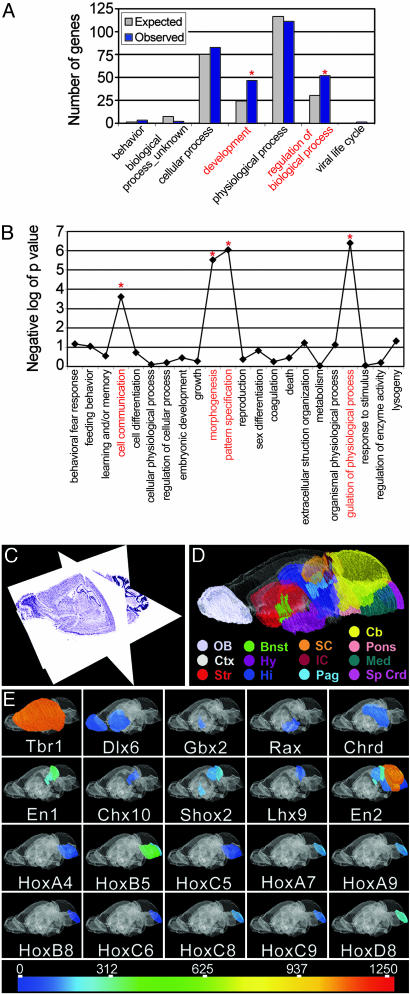
Fig. 2.
Genes with region-specific expression patterns function in development, pattern specification, and morphogenesis. (A) The abscissa indicates the functional categories from the gotm program. Within the GO biological process category, only “development” and “regulation of biological process” showed significant overrepresentation (*, P < 0.01). The ordinate indicates the number of genes observed in each category compared with the number of genes expected by chance. The significantly overrepresented categories are noted by an asterisk. (B) The GO subcategories in “development” from A that are significantly overrepresented in the set of genes with region-specific expression patterns. The GO categories are noted along the abscissa; the negative logarithm (base 10) of the P value is given along the ordinate. Functional categories significantly overrepresented are noted by an asterisk. (C) Reference brain atlas displayed in the three orthogonal planes. This Nissl-stained C57BL/6N mouse brain atlas comprises 462 coronal sections at 30-μm thickness, digitized at a resolution of 1.3 μm/pixel. The sagittal and horizontal planes are “virtual” sections dynamically constructed from the coronal sections. (D) 3D atlas of brain regions. Specific brain regions along the rostrocaudal neuraxis are color-coded. (E) The expression levels of the homeobox and other embryonic patterning genes expressed in the adult mouse brain are shown for each region. A complete list of these embryonic patterning genes is available upon request.
Graphical delineations of brain regions generated with neurozoom software are closed polygons that are overlaid on top of the coronal sections. For this study, 21 major and minor regions throughout the brain that match the regions dissected to obtain the mRNA samples were used for display, including Amg, Bnst, HiF, Cb, Cx, striatum, SC and IC, MO, Olf, Pag, and SpCrd (Fig. 2D). The 2D annotations of these regions were three-dimensionally reconstructed by using a surface triangulation algorithm. Data containing signal intensity values from gene expression microarray analyses were imported by the software and converted to a linear color scale with a high-to-low gradient range of red, orange, yellow, green, cyan, and blue (Fig. 2E). The 2D and 3D contours were filled with either a user-specified color or with a color corresponding to a value from the color scale representing the signal intensity. In some cases, the transparency of the color-filled contours was adjusted by using a scale from 0–100% transparent.
Results
Molecular Architecture of the Adult Brain. We have profiled gene expression patterns of 24 neural tissues and 10 body regions. In total, 150 array hybridization measurements were included in our data set. For a summary of the microarray data included in the atlas, the average quality control metrics by sample type, and the experimental reproducibility, see Tables 2 and 3.
A heat map was generated to look for similarities and differences in regional gene expression patterns based on the Pearson correlation coefficients calculated between all individual samples (Fig. 1 A). Replicate samples from a given brain region showed the most similar gene expression profiles of all of the sample groups, as demonstrated by the dark red diagonal line in Fig. 1 A, indicating the high level of reproducibility between independent replicate measurements.
Within the cortical subregions, three groups showed very similar gene expression patterns. Expression patterns for the brain regions that comprise the archicortex (“A” in Fig. 1) (CA1, CA3, and DG), paleocortex (“P” in Fig. 1) (Amg, EntCx, and PrhCx), and neocortex (“N” in Fig. 1) (Cx and MtrCx) were the most similar within their respective groups. Two other groups that showed very similar gene expression patterns include the Hy, Pag, IC, SC, and DiE-MD and the pons, MO, and SpCrd. Gene expression profiles of hindbrain regions (pons, MO, SpCrd, and Cb) were somewhat dissimilar to the profiles for structures of the forebrain and midbrain. We also noted that the patterns for three structures that develop as outpouchings of the brain (retina, Pit, and choroid plexus) were remarkably different not only from other brain structures, but also from each other. We found more similar gene expression patterns for regions that collectively shared a developmental origin [for example, DiE-MD brain structures (DiE-MD, Hy, Pag, IC, SC)]. These results demonstrate that position along the anterior–posterior axis of the neural tube is closely associated with, and may be a determinant of, the gene expression patterns in the adult structures.
To further explore the relationships between brain regions based on their gene expression profiles, unsupervised hierarchical clustering was performed (Fig. 1B). We hypothesized that clustering analysis might reveal brain region relatedness based on anatomy, embryology, or evolutionary relationships. The resulting dendrogram consisted of two main branches, with the telencephalic brain regions on one main branch, and the remaining regions clustered together on the second main branch. The first observation was that regions with shared cytoarchitectural features did not cluster together. The laminated structures (Olf, HiF, Cb, EntCx, PrhCx, MtrCx, Cx, and retina) were found on all branches of the dendrogram and were not more similar to each other than to other regions. We next compared the branch pattern of the dendrogram to the structures of a five-vesicle embryo. The majority of regions clustered together based on the embryologic region from which they were derived (Fig. 1B), demonstrating an overall region relatedness consistent with a classically defined, morphology-based embryological origin. We also noted a general preservation of the rostral–caudal axis suggested by the pattern of the heat map and the subdivisions of the dendrogram, where, for example, the neocortex is more related to the paleocortex than to the archicortex. It is important to emphasize that the observed relationships between brain regions based on expression patterns were robust and were not significantly influenced by the particular choice of genes or the strain of mouse used for the analysis.
Region-Restricted Gene Expression in the Adult Brain. To further investigate the embryological basis for the observed region-related gene expression patterns in the adult mouse brain, we focused on defining patterns that uniquely mark a particular region or set of structures. We used a set of analyses to identify genes with highly restricted expression patterns (12). In one analysis, we identified 93 genes that showed expression restricted to a region or specific subregions (see Table 7, which is published as supporting information on the PNAS web site), and in a separate analysis, we identified 129 genes that showed clear regional enrichment (see Table 5), yielding 192 unique genes in total. We hypothesized that these genes may perform functions related to regional specialization. Using gotm (20), we queried the set of region-specific and -enriched genes (omitting the genes restricted to the retina, Pit, and cp4v; for the list, see Table 6) to identify GO categories that were significantly overrepresented (P < 0.01). In the biological process category, genes were overrepresented for both “development” and “regulation of biological process” (Fig. 2 A). Within these two categories, we found an overrepresentation of genes involved in morphogenesis (P < 2.3 × 10-6), pattern specification (P < 1.43 × 10-6), and cell communication (P < 0.00025) (Fig. 2B). Thus, consistent with the embryonic imprint observed in the dendrogram, the GO categories for development, morphogenesis, and pattern specification were overrepresented in the list of region-specific genes. In particular, we observed a significant number of embryonic patterning and homeobox genes (e.g., Dlx6, Gbx2, Chrd, HoxA4, and HoxB5) with region-specific expression in the adult nervous system. Twenty-one of the 192 region-specific genes were embryonic patterning genes.
In studies of this type, with very large amounts of data, it is helpful to be able to visualize the data in a meaningful way. We and others have discussed the importance of methods to view data in three dimensions in the context of anatomy and/or brain circuitry (5). As a step toward this goal, we have taken the observations of embryonic patterning and homeobox gene expression in the adult brain and combined the quantitative expression data with a high-resolution, coordinate-based brain atlas that allowed us to visualize the gene expression relationships in the context of the whole brain rather than simply as a list of genes. These gene expression data were imported and visualized onto a 3D brain atlas representation (Fig. 2C) by using neurozoom software to provide a virtual in situ hybridization in which gene expression levels for specific brain regions are color-coded. Using this display technology, we viewed highly specific expression profiles for these embryonic patterning and homeobox genes throughout the neuraxis (Fig. 2 D and E). Other groups have proposed different methods to create 3D brain gene expression maps, such as voxelation, where RNA is extracted from spatially registered cubes within the brain (22). Our method differs from voxelation in that we used both standard stereotaxic coordinates and strict dissection protocols that respect natural boundaries, such that the gene expression patterns represent whole subregions rather than a mixture of multiple regions. Therefore, it is not necessary to use statistical methods such as singular value decomposition to delineate anatomical regions. With the 3D brain atlas presented here, the gene expression patterns of specific brain regions may be viewed alone, as a network with other specific regions, or in the context of the whole brain.
Discussion
Previous studies have shown that commitment to a specific lineage, specified in large part by anatomical position within the developing neural tube, involves the imprinting of a genetic program (23). Our expression data suggest that the imprinted genetic program is still evident in the mature brain. The concept of an imprinted genetic pattern has been strengthened by the identification of genes that mark morphogenetic fields during brain development (24, 25). The pattern of gene expression for a small set of genes for a particular brain region and its relatedness to patterns seen in other regions has been used extensively in developmental biology to help understand the embryologic origins and functional relationships between brain regions (2, 26). Analysis of the relationship between morphologically defined boundaries in brain development and domains defined by gene expression patterns has led to the identification of three major regions: the anterior region, midbrain–hindbrain boundary, and the rhombospinal region (1). The anterior region corresponds to the telencephalon, diencephalon, and anterior mesencephalon; the midbrain–hindbrain boundary is the origin of the Cb; and the rhombospinal region corresponds to the posterior mesencephalon, metencephalon, myelencephalon, and SpCrd (1).
Although the gene expression dendrogram observed in Fig. 1B did not have three main branches corresponding to these divisions, we observed two features of the dendrogram that were consistent with the three-region model. First, we observed that the Cb is on a branch distinct from the regions derived from the anterior or rhombospinal regions. Previously, the Cb was believed to be derived from the hindbrain structures along with the pons and medulla. However, it has recently been shown that the Cb is derived from the cells that meet at the midbrain–hindbrain junction (27). The clustering results are consistent with the findings that the Cb is not derived from the hindbrain. Nevertheless, the Cb is still more closely related to brainstem structures than nonbrainstem structures. It is also possible that the adult gene expression patterns of the Cb are so highly modified that they obscure the structure's developmental origins. Second, the brain structures comprising the rhombospinal region (pons, MO, and SpCrd) clustered together based on a high degree of expression pattern similarity. Notably, in a previous regional analysis of the Amg, gene expression patterns in specific amygdaloid nuclei were found to respect the ontogenetic origins of the subnuclei, which derive embryologically from both pallial and subpallial structures (7). Like the Amg, the Bnst is known to be a heterogenous structure, and in the embryo, the posterior Bnst occupies a wedge between the basal ganglia and the diencephalon (28). The neuroepithelium from which the posterior Bnst is derived lies lateral to where the anterior thalamus fuses with the hypothalamic portion of the third ventricle (29). This embryonic relationship between the Bnst and the diencephalon, specifically the Hy, appears to be observed in the gene expression patterns of the adult as demonstrated by the dendrogram (see Fig. 1B). These results suggest that although the expression pattern for many genes may change dramatically during development, the brain retains a degree of gene expression patterning established during embryogenesis that is important for maintaining regional specificity and functional relationships between brain regions in the adult.
The embryonic patterning and homeobox genes were found to be expressed in the adult brain with patterns that respected the domains and boundaries defined by the embryologic, gene expression, and classic evolutionary models of brain development and maturation; however, the evolutionary models remain controversial (Fig. 2 D and E) (30). Several studies of the developing brain have demonstrated that similar sets of genes are used to establish a particular anatomical region and to maintain the cell–cell relationships of the differentiated region (31). Thus, it may be that the roles of these genes in adulthood are similar to their roles during development. These roles include maintaining established phenotypes and connectivity of neuronal populations or preserving barriers to the inappropriate migration of neurons from one region to another. We speculate that these genes continue to play an important role in the regional specification of functional units in the adult brain.
The expression results and the analytical and visualization tools described here add to the expanding neurobiology tool chest and complement efforts to measure qualitative patterns of gene expression based on in situ hybridization (Mouse Brain Gene Expression Database project), reporter lines (32), and proteomics methods (Human Brain Proteome Project).
Supplementary Material
Acknowledgments
We thank Information Management Consultants for donation of the Teradata data warehouse and design and programming of the TeraGenomics database; Teradata/NCR for early support of the project; Neurome for donation of time and resources; Todd Carter for technical support; Selena Ellis-Vizcarra and Jamie Simon for technical assistance; Larry Swanson, Roland Stoughton, Todd Preuss, and David Anderson for helpful discussions; and Jim Velier for insights. This work was supported by National Institute of Neurological Disorders and Stroke Grant NS039601-04 (to C.B.) and National Institute of Mental Health Grant MH062344-03 (to C.B. and David J. Lockhart).
Author contributions: David J. Lockhart and C.B. designed research; I.H., J.A.E., L.W., J.A.D.R., R.T., W.T., R.H., B.S.S., and C.W. performed research; M.A.Z., R.S.B., Daniel J. Lockhart, J.F.R., W.G.Y., F.E.B., David J. Lockhart, and C.B. contributed new reagents/analytic tools; M.A.Z., I.H., J.A.E., L.W., R.S.B., J.F.R., W.G.Y., F.E.B., David J. Lockhart, and C.B. analyzed data; and M.A.Z., I.H., J.A.E., F.E.B., David J. Lockhart, and C.B. wrote the paper.
Abbreviations: Amg, amygdala; Bnst, bed nucleus of the stria terminalis; Cb, cerebellum; cp4v, choroid plexus from the fourth ventricle; Cx, cortex; DG, dentate gyrus; DiE-MD, diencephalon and midbrain excluding hypothalamus; EntCx, entorhinal cortex; GO, gene ontology; HiF, hippocampal formation; Hy, hypothalamus; IC, inferior colliculus; MO, medulla oblongata; MRM, magnetic resonance microscopy; MtrCx, motor cortex; Olf, olfactory bulbs; Pag, periaqueductal gray; Pit, pituitary; PrhCx, perirhinal cortex; SpCrd, spinal cord; SC, superior colliculus.
Data deposition: The data reported in this paper have been deposited in the database at the publicly accessible web site www.barlow-lockhartbrainmapnimhgrant.org.
References
- 1.Swanson, L. W. (2000) Trends Neurosci. 23, 519-527. [DOI] [PubMed] [Google Scholar]
- 2.Holland, L. Z. & Holland, N. D. (1999) Curr. Opin. Neurobiol. 9, 596-602. [DOI] [PubMed] [Google Scholar]
- 3.Puelles, L. & Rubenstein, J. L. (2003) Trends Neurosci. 26, 469-476. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart, D. J. & Barlow, C. (2001) Nat. Rev. Neurosci. 2, 63-68. [DOI] [PubMed] [Google Scholar]
- 5.Barlow, C. & Lockhart, D. J. (2002) Curr. Opin. Neurobiol. 12, 554-561. [DOI] [PubMed] [Google Scholar]
- 6.Zhao, X., Lein, E. S., He, A., Smith, S. C., Aston, C. & Gage, F. H. (2001) J. Comp. Neurol. 441, 187-196. [DOI] [PubMed] [Google Scholar]
- 7.Zirlinger, M., Kreiman, G. & Anderson, D. J. (2001) Proc. Natl. Acad. Sci. USA 98, 5270-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonaventure, P., Guo, H., Tian, B., Liu, X., Bittner, A., Roland, B., Salunga, R., Ma, X. J., Kamme, F., Meurers, B., et al. (2002) Brain Res. 943, 38-47. [DOI] [PubMed] [Google Scholar]
- 9.Kamme, F., Salunga, R., Yu, J., Tran, D. T., Zhu, J., Luo, L., Bittner, A., Guo, H. Q., Miller, N., Wan, J., et al. (2003) J. Neurosci. 23, 3607-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zirlinger, M. & Anderson, D. J. (2003) Genes Brain Behav. 2, 282-294. [DOI] [PubMed] [Google Scholar]
- 11.Lein, E. S., Zhao, X. & Gage F. H. (2004) J. Neurosci. 24, 3879-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandberg, R., Yasuda, R., Pankratz, D. G., Carter, T. A., Del Rio, J. A., Wodicka, L., Mayford, M., Lockhart, D. J. & Barlow, C. (2000) Proc. Natl. Acad. Sci. USA 97, 11038-11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khaitovich, P., Muetzel, B., She, X., Lachmann, M., Hellmann, I., Dietzsch, J., Steigele, S., Do, H. H., Weiss, G., Enard, W., et al. (2004) Genome Res. 14, 1462-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C. A., Causton, H. C., et al. (2001) Nat. Genet. 29, 365-371. [DOI] [PubMed] [Google Scholar]
- 15.Paxinos, G. & Franklin, K. B. J. (2001) The Mouse Brain in Stereotaxic Coordinates (Academic, San Diego).
- 16.Bowden, D. M. & Martin, R. F. (1995) Neuroimage 2, 63-83. [DOI] [PubMed] [Google Scholar]
- 17.Wodicka, L., Dong, H., Mittmann, M., Ho, M. H. & Lockhart, D. J. (1997) Nat. Biotechnol. 15, 1359-1367. [DOI] [PubMed] [Google Scholar]
- 18.Caceres, M., Lachuer, J., Zapala, M. A., Redmond, J. C., Kudo, L., Geschwind, D. H., Lockhart, D. J., Preuss, T. M. & Barlow, C. (2003) Proc. Natl. Acad. Sci. USA 100, 13030-13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zapala, M. A., Lockhart, D. J., Pankratz, D. G., Garcia, A. J., Barlow, C. & Lockhart, D. J. (2002) Genome Biol. 3, software0001.1-0001.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, B., Schmoyer, D., Kirov, S. & Snoddy, J. (2004) BMC Bioinformatics 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redwine, J. M., Kosofsky, B., Jacobs, R. E., Games, D., Reilly, J. F., Morrison, J. H., Young, W. G. & Bloom, F. E. (2003) Proc. Natl. Acad. Sci. USA 100, 1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown, V. M., Ossadtchi, A., Khan, A. H., Yee, S., Lacan, G., Melega, W. P., Cherry, S. R., Leahy, R. M. & Smith, D. J. (2002) Genome Res. 12, 868-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lumsden, A. & Krumlauf, R. (1996) Science 274, 1109-1115. [DOI] [PubMed] [Google Scholar]
- 24.Spemann, H. (1918) Zool. Jahr. Supp. 15, 1-48. [Google Scholar]
- 25.Spemann, H. & Mangold, H. (1924) Roux' Arch. f. Entw. Mech. 100, 599-638. [Google Scholar]
- 26.Shimamura, K., Hartigan, D. J., Martinez, S., Puelles, L. & Rubenstein, J. L. (1995) Development (Cambridge, U.K.) 121, 3923-3933. [DOI] [PubMed] [Google Scholar]
- 27.Wingate, R. J. (2001) Curr. Opin. Neurobiol. 11, 82-88. [DOI] [PubMed] [Google Scholar]
- 28.Bayer, S. A. (1987) J. Comp. Neurol. 265, 47-64. [DOI] [PubMed] [Google Scholar]
- 29.Altman, J. & Bayer, S. A. (1986) Adv. Anat. Embryol. Cell Biol. 100, 1-178. [PubMed] [Google Scholar]
- 30.Jarvis, E. D., Gunturkun, O., Bruce, L., Csillag, A., Karten, H., Kuenzel, W., Medina, L., Paxinos, G., Perkel, D. J., Shimizu, T., et al. (2005) Nat. Rev. Neurosci. 6, 151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasini, A. & Wilkinson, D. G. (2002) BioEssays 24, 427-438. [DOI] [PubMed] [Google Scholar]
- 32.Gong, S., Zheng, C., Doughty, M. L., Losos, K., Didkovsky, N., Schambra, U. B., Nowak, N. J., Joyner, A., Leblanc, G., Hatten, M. E., et al. (2003) Nature 425, 917-925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.