Abstract
Moth species have evolved integral membrane desaturases that exhibit a wide diversity in substrate specificity, as well as in regiospecificity and stereospecificity of the unsaturated products. We report here the cloning and expression of a single desaturase from the sex pheromone gland of the light brown apple moth, Epiphyas postvittana, that makes E11 isomers of monounsaturated (E11-16 and E11-14) fatty acids and a diunsaturated (E9,E11-14) fatty acid. In the pheromone gland, the monoene precursor is made available by β oxidation of E11-16 acid with a subsequent two-carbon loss to E9-14 acid. A functional assay using a baculovirus expression system required addition of myristic acid and E9-14 acid precursors to demonstrate the unusual regiospecificity and stereospecificity of this desaturase. The amino acid sequence of this desaturase has ≈61% identity to that of Z11-desaturases from two other insect species, and only ≈48% identity to the metabolic Z9-desaturases in those species. A pheromone-gland Z9-desaturase gene also was found with the light brown apple moth that differed in its deduced amino acid sequence (66% identity) with the metabolic Z9-desaturase from fat body in this species.
The most common desaturases in nature are those that produce palmitoleic acid (Z9-16:Acid) and oleic acid (Z9-18:Acid). Although soluble desaturases of plants and integral membrane desaturases of yeast, fungi, and animals produce these same unsaturated acids, they belong to different multifunctional classes and have different consensus motifs (1). Most progress on the structure of desaturases has been achieved with soluble desaturases, as structure–function analyses using molecular, biochemical, and crystallographic approaches have provided the basis for the engineering of plant enzymes with altered substrate specificities (2, 3) and reaction outcomes for improved oil crops (4). The more abundant integral membrane desaturases, however, have proved to be recalcitrant to characterization. The lack of a crystal structure has shifted the focus to the characterization of membrane desaturases with diverse substrate specificities, regiospecificities and stereospecificities.
The evolution of diverse membrane desaturases in female moth abdominal glands for production of specific sex pheromone components has provided the opportunity to generate sequence information on many diverse desaturases. Several unusual Z11-desaturases have already been characterized for the cabbage looper moth, Trichoplusia ni (5), and the corn earworm moth, Helicoverpa zea (6), and in this article we describe the characterization and function of an unusual E-desaturase from the sex pheromone gland of the light brown apple moth (LBAM), Epiphyas postvittana. Not only does LBAM have unusual regiospecificities and stereospecificities in its production of (E)-11 isomers, but it also utilizes both saturated and monounsaturated 14-carbon precursor fatty acids to produce E11-14:Acid and E9,E11-14:Acid. In addition, two Z9-desaturase genes were characterized from this species—one from fat body tissue and one from pheromone gland tissue.
Materials and Methods
Insect Tissue Collection and Poly(A)+ RNA Isolation.
Female LBAM moths were obtained from pupae shipped from New Zealand. Fat bodies and pheromone glands were carefully dissected from 2- to 3-day-old female moths and stored at −80°C. Poly(A)+ RNA (mRNA) was isolated and purified from fat bodies and pheromone glands by using an mRNA Isolation kit (Ambion) according to the procedures recommended by the manufacturer.
Cloning of Desaturase cDNA from Fat Body.
One microliter of mRNA isolated from fat bodies was denatured and transcribed to single-stranded cDNA (ss-cDNA) by reverse transcriptase with a SMART RACE (rapid amplification of cDNA ends) cDNA Amplification kit (CLONTECH). The ss-cDNA was diluted (1:10; final volume, 200 μl) and used as template for PCR to obtain the central region of a desaturase gene. Two degenerate primers, PR1 and PR2 (Table 1), which were designed based on highly conserved regions of desaturase genes, were used. A 50-μl standard reaction contained 1× Advantage 2 reaction buffer, 1× Advantage 2 DNA polymerase mix (CLONTECH), 0.2 mM each dNTP, 0.5 μM PR1, 0.5 μM PR2, and 5 μl of diluted ss-cDNA. The PCR was performed under the following conditions: 94°C denature, 1 min; 33 cycles of 94°C, 30 s; 56°C, 30 s; and 72°C, 3 min with a final extra 10-min extension at 72°C. The PCR product was ligated directly to a linearized TOPO TA PCR 2.1 vector (Invitrogen) and the ligated vector was transformed into Top 10 competent cells (Invitrogen). Colony PCR was used to screen positive colonies with the insert, and plasmids in selected positive colonies were amplified and sequenced. The central region (LBAM-FBZ9-CR) was obtained and compared with other central regions of desaturase genes to confirm its possibility of being a desaturase gene fragment.
Table 1.
Primers used for RACE PCR and ORF
| Degenerate primers |
| PR1: 5′-ATYACHGCCGGKKMYCAYMG-3′ |
| PR2: 5′-GGRAABDYGTGRTGGWAGTT-3′ |
| Primers for LBAM-FBZ9 |
| LBAM-FB-1: 5′-CTTCAACACTACTGCCTTCCAGGA-3′ |
| LBAM-FB-2: 5′-TGCTGACCCCCACAACGC-3′ |
| LBAM-FB-3: 5′-GCGTTGTGGGGGTCAGCA-3′ |
| LBAM-FB-4: 5′-TCCTGGAAGGCAGTAGTGTTGAAG-3′ |
| LBAM-FB-5: 5′-CCCGTCGACATGGCACCTAACGTAACT-3′ |
| LBAM-FB-6: 5′-CCTCTAGATTAATCATCTTTAGGGTTA-3′ |
| Primers for LBAM-PGZ9 |
| LBAM-PG1-1: 3′-AAGTAGGCGTTCCAAAGCGATTCTC-5′ |
| LBAM-PG1-2: 3′-CAGCACGGGGTCAGAGCAGAG-5′ |
| LBAM-PG1-3: 3′-CAAGGCCAAGGGACACACCATC-5′ |
| LBAM-PG1-4: 3′-CCCGTCGAAACCAAGCCAGTATC-5′ |
| LBAM-PG1-5: 3′-ACTGGATCCATGCCTCCTCAAGGGCAGCCTC-5′ |
| LBAM-PG1-6: 3′-GTGGAGCTCTCATTCAGTCTTTTCGGGGTTG-5′ |
| Primers for LBAM-PGE11 |
| LBAM-PG2-1: 3′-GGCGGCGTTGTGAGGGTCGGCGTCC-5′ |
| LBAM-PG2-2: 3′-TTAATGGCACTGTTTTGGAATGAC-5′ |
| LBAM-PG2-3: 3′-CAGAGACCACCGATCGCACCACAAGTAT-5′ |
| LBAM-PG2-4: 3′-AACGCTTGGCACATCACG-5′ |
| LBAM-PG2-5: 3′-AGAGAATTCATTATGGCTCCAAACGTAGAAGAAA-5′ |
| LBAM-PG2-6: 3′-GCCTCTAGATTACTCATCTTGCGTAGATATA-5′ |
Sequence information of LBAM-FB-CR was used to design specific primers for RACE PCR. First-round 3′ RACE PCR was performed with gene-specific primer LBAM-FB-1 (Table 1) and the Universal Primer Mix (UPM-Long: 5′-CTAATACGACTCATCATAGGGCAAGCAGTGGTAACAACGCAGAGT-3′ and UPM-Short: 5′-CTAATACGACTCATCATAGGGC-3′) from the Smart RACE cDNA Amplification kit. The second-round 3′ RACE PCR was performed by using the first-round PCR products as templates with the Nested Universal Primer (NUP: 5′-AAGCAGTGGTAACAACGCAGAGT-3′) from the kit and the nested gene-specific primers LBAM-FB-2 (Table 1). With a GeneRacer kit (Invitrogen), another ss-cDNA library was constructed, which was used as template to run first-round 5′ RACE PCR by using a gene-specific primer LBAM-FB-3 (Table 1) along with 5′ primer (5′-CGACTGGAGCACGAGGACACTGA-3′) from the kit. The first-round PCR product was used as template to run the second-round PCR with gene-specific primer LBAM-FB-4 (Table 1) and 5′ nested primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) that is also from the kit. The second-round PCR products were ligated directly to TOPO TA PCR 2.1 vector for sequencing.
Cloning of Desaturase cDNAs from Pheromone-Gland Tissue.
One microliter of mRNA isolated from pheromone gland was used to construct the ss-cDNA library as described above. The ss-cDNA library and degenerate primers PR1 and PR2 were used to amplify the central regions of desaturase sequences with the same conditions described above. The PCR products were cloned directly to the linearized TOPO TA PCR 2.1 vector and sequenced. Two central regions (LBAM-PGZ9-CR and LBAM-PGE11-CR) were obtained and used to design gene-specific primers for 3′ and 5′ RACE PCR. The first-round 5′ RACE PCR was performed with UPM and LBAM-PG1-1 (Table 1) for LBAM-PGZ9, and with universal primer mix (UPM) and LBAM-PG2-1 (Table 1) for LBAM-PGE11. The first-round PCR products were used as templates for the second-round PCR with NUP and LBAM-PG1-2 (Table 1) for LBAM-PGZ9, and with nested universal primer (NUP) and LBAM-PG2-2 (Table 1) for LBAM-PGE11. The first- and second-round 3′ RACE PCR were performed similarly with LBAM-PG1-3 and LBAM-PG1-4 (Table 1) for LBAM-PGZ9 and with LBAM-PG2-3 and LBAM-PG2-4 (Table 1) for LBAM-PGE11. The second-round RACE PCR products were cloned into the TOPO TA PCR 2.1 vector for sequencing.
Functional Assay of Δ9-Desaturases in YEpOLEX Expression System.
Two gene-specific primers, LBAM-FB-5 and LBAM-FB-6 (Table 1), were designed to amplify the ORF of LBAM-FBZ9 from the fat body cDNA library. Two other gene-specific primers, LBAM-PG1-5 and LBAM-PG1-6 (Table 1), were designed to amplify the ORF of LBAM-PGZ9 from the pheromone gland cDNA library. A single clear band for each of the ORFs was obtained. The PCR products were digested and ligated to linearized YEpOLEX plasmids and transformed into Top 10 competent cells for positive colony screening. After colony PCR, the recombinant plasmids in positive colonies were amplified, purified, and sequenced, and consensus clones for YEpOLEX-LBAM-FBZ9-ORF and YEpOLEX-LBAM-PGZ9-ORF were generated for functional expression.
The competent cells of mutant yeast strain L8-14C (7) were cultured as described (8) and transformed with YEpOLEX-LBAM-FBZ9-ORF and YEpOLEX-LBAM-PGZ9-ORF, respectively, by the standard method (9). The transformed cells were able to grow on yeast extract/peptone/dextrose (YPD) plate without complementation by additional unsaturated fatty acids (UFAs). These cells were inoculated into 30 ml of YPD medium and grown at 30°C overnight with shaking (300 rpm). The induced cells were transferred to a 50-ml sterile centrifuge tube and spun at 1,500 × g for 5 min. The cell pellet was washed two times with 0.2% BSA, transferred to a 1.5-ml tube, and spun briefly to remove as much liquid as possible. The washed yeast cells were lysed with 1 ml of Y-PER Yeast Protein Extraction Reagent (Pierce) with brief vortex and 10-min agitation at room temperature. The cell debris was collected by centrifugation at 13,000 × g for 10 min and extracted with 1 ml of chloroform/methanol (2:1) at room temperature for 1 h. The solvent was decanted from the debris and evaporated under nitrogen. The oily residue was extracted twice with 0.5 ml of 10% boron trichloride/methanol, and the combined extracts were heated at 100°C for 30 min. The resulting fatty acid methyl esters were extracted with 1 ml of hexane, and the solution was concentrated under nitrogen for analyses by GC/MS. The double bond position in the products was confirmed by MS analysis of the dimethyl disulfide (DMDS) adducts (10).
Functional Assay of E11-Desaturase in a Bac-to-Bac Baculovirus Expression System.
Two gene-specific primers, LBAM-PG2-5 and LBAM-PG2-6 (Table 1), were designed to obtain the ORF of LBAM-PGE11. This ORF was cloned to pFastBac1 vector (GIBCO/BRL) and sequenced. The transfer of the expression cassette in the recombinant plasmid (pFastBac1-LBAM-PGE11-ORF) to the baculovirus genome (bacmid DNA) was carried out in DH10Bac competent cells (GIBCO/BRL) by using Tn7 site-specific transposition. Transfection of the recombinant baculovirus to Sf21 insect cells (GIBCO/BRL) was performed according to the procedures described by the supplier. The amplified recombinant baculoviruses were harvested from cell culture medium (5 ml) in a 25-mm2 cell culture flask (Corning) at 72 h after transfection. The original virus was amplified in Sf21 cells. In a 75-mm2 cell culture flask (Corning), a mixture containing 1 ml of the virus medium and 2 ml Sf-900 II SFM (GIBCO/BRL) was used to infect Sf21 cells, which were 3–4 days old in midlogarithmic phase with a viability of >97%. The flask was incubated at 28°C with slow agitation (three or four times per min) for 1 h, then 12 ml of Sf-900 II SFM was added to the flask, which was then incubated at 28°C for 2 days.
Myristic acid methyl ester (MAME), E9-14:Acid, and E11-14:Acid were added (0.5 mM each) in separate experiments as possible precursors in the desaturase reaction. After 2 days incubation at 28°C, the cells were collected by spinning in a 50-ml tube at 500 × g for 5 min. The cell pellet was washed two times with 0.9% NaCl containing 0.2% BSA, transferred to a 1.5-ml tube, and extracted with 1 ml of chloroform/methanol (2:1) at room temperature for 1 h. Procedures described above were used for base methanolysis and DMDS reaction of fatty acids. The resulting fatty acid methyl esters were analyzed by GC/MS and double-bond positions were confirmed by MS analysis of the DMDS adduct. Positions of the conjugated double bonds in the diene were confirmed by GC/MS analysis of their 4-methyl-1,2,4-triazoline-3,5-dione (MTAD) derivatives (11, 12).
Results
Isolation and Sequencing of LBAM-FBZ9, LBAM-PGZ9, and LABM-PGE11.
Reverse transcription (RT)-PCR was used to amplify a 572-bp fragment (LBAM-FBZ9-CR) by two degenerate primers from the fat-body cDNA library. Comparison to the central regions of other Z9-desaturase genes showed that this fragment has high identity to other insect Z9-desaturases, such as that of T. ni (72%) (8) and H. zea (77%) (6). Another two 569-bp fragments (LBAM-PGZ9-CR and LBAM-PGE11-CR) were obtained from the pheromone-gland cDNA library by using the same degenerate primers. LBAM-PGZ9-CR has high identity to the central regions of other insect Z9-desaturase genes (74% and 71% to T. ni and H. zea, respectively). The other central region, LBAM-PGE11-CR, has a lower identify to the Z9-desaturases of T .ni (63%) and H. zea (61%), but a higher identity to the central regions of Z11-desaturases in T. ni (68%) and H. zea (69%) (5, 6). These results showed that these three fragments could be from desaturase genes.
A full-length cDNA sequence (LBAM-FBZ9) was obtained by combining the central region (LBAM-FB-CR) along with 3′ and 5′ RACE fragments that were amplified by RACE PCR. This cDNA spans 1,510 nt and contains an ORF of 1,059 nt, which encodes a protein with 352 aa. The deduced amino acid sequence from LBAM-FBZ9 has high identity to other known insect Δ9-desaturases (66% to T. ni Z9 and 86% to H. zea Z9) (Fig. 1). Two full-length cDNAs (LBAM-PGZ9 and LBAM-PGE11) were obtained from the pheromone-gland cDNA library. LBAM-PGZ9 spans 1,705 nt with an ORF of 1,062 nt, which encodes a protein with 353 aa. This deduced amino acid sequence has higher identity to other insect Δ9-desaturases (65% to T. ni Z9, 63% to H. zea Z9, and 66% to LBAM-FBZ9) than to the Δ11-desaturases (53% to T. ni Z11 and 54% to H. zea Z11) (Fig. 1). LBAM-PGE11 spans 1,665 nt with an ORF of 999 nt, which encodes a protein with 332 aa. The deduced amino acid sequence of LBAM-PGE11 has higher identity to the Δ11-desaturases (61% to T. ni Z11 and 62% to H. zea Z11) and lower identity to the Δ9-desaturases (48% to T. ni Z9 and 47% to H. zea Z9) (Fig. 1). The deduced amino acid sequence of LBAM-PGE11 has 51% identity to that of LBAM-FBZ9 and 46% identity to that of LBAM-PGZ9 (Fig. 1).
Figure 1.
Comparison of deduced amino acid sequences of LBAM (LBAM-FBZ9, LBAM-PGZ9, and LBAM-PGE11) to those of Z9- and Z11-desaturases from H. zea (H. zea Z9 and H. zea Z11) and T. ni (T. ni Z9 and T. ni Z11). The consensus residues are shaded with solid black. The three histidine domains are boxed.
Functional Assay of Z9-Desaturases.
The mutant ole1 yeast cells used for the functional assay are desaturase-deficient and cannot grow if UFAs are not complemented in the medium. In this study, yeast cells transformed with YEpOLEX-LBAM-FBZ9-ORF or YEpOLEX-LBAM-PGZ9-ORF plasmids grew well in YPD medium without addition of UFAs. GC/MS analysis of the UFA methyl esters generated by transmethylation of acyl compounds extracted from the yeast cells showed that LBAM-FBZ9 desaturase produced Z9-16:Acid and Z9-18:Acid in a 10:7 ratio, and that LBAM-PGZ9 desaturase produced these acids in a 1:4 ratio. Trace amounts of DMDS adducts for Z9-14:Me, Z9-15:Me, and Z9-17:Me methyl esters were also found as products from both desaturases in single ion monitoring scans for the characteristic Δ9 fragment at m/z 217.
Functional Assay of the E11-Desaturase in a Bac-to-Bac Baculovirus Expression System.
GC/MS analysis of the UFA methyl esters generated by transmethylation of acyl compounds extracted from Sf21 insect cells infected with bacmid viruses, which were transposed with pFastBac1-LBAM-PGE11-ORF, showed that this desaturase produced both E11-14:Acid and E11-16:Acid. When MAME was added to the Sf21 cell medium as precursor, the amount of E11-14:Acid increased and E11-16:Acid decreased (Fig. 2). Regiospecificity for Δ11 desaturation was shown by MS analysis of DMDS adducts, and stereospecificity for the E configuration was shown by comparison of GC retention times to synthetic standards.
Figure 2.
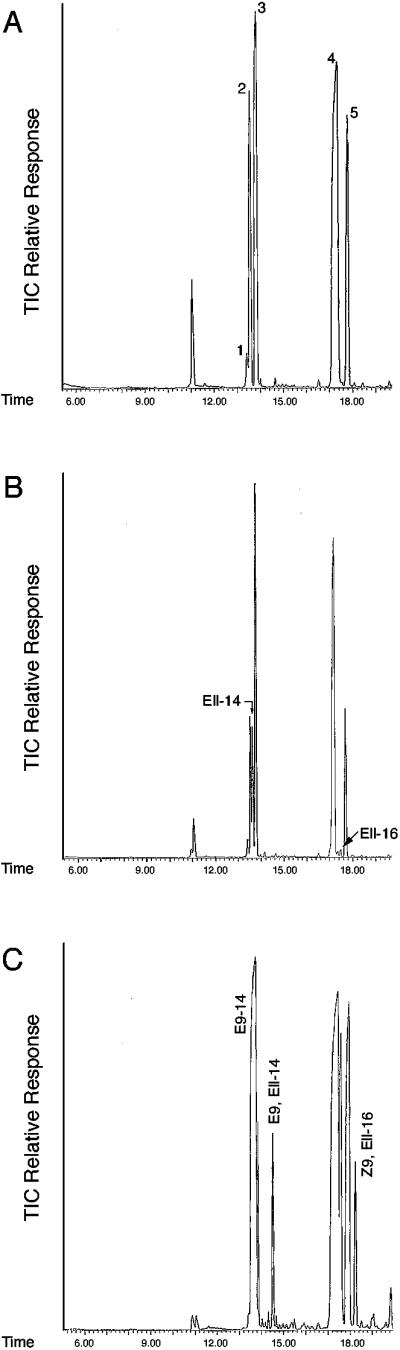
GC/MS analysis of methyl esters. TIC, total ion current; retention time is in minutes. (A) Control Sf21 cells infected with bacmid viruses, which were transposed with pFastBac1, produced acids: 1, Z7-14; 2, Z9-14; 3, C14; 4, Z9-16; 5, C16. Sf21 cells infected with bacmid viruses, which were transposed with pFastBac1-LBAM-PGE11-ORF, exhibited additional peaks in B for E11-14 and E11-16 acids, and in C for E9,E11-14 acids when E9-14:Acid was added as a precursor, in addition to peaks for E9-14:Acid and its chain-elongation product, E11-16:Acid, which is desaturated by the Z9-desaturase of the Sf21 cells to produce Z9,E11-16:Acid.
When E9-14:Acid was added to the medium, a 14-carbon acid conjugated diene was produced (Fig. 2). GC/MS analysis of the MTAD derivatives of the products showed that the product was E9,E11-14:Acid (Fig. 3). The derivative exhibited a molecular ion at 351 and major ions at 196 and 322 m/z, which are all indicative of a 9,11-conjugated system of a 14-carbon methyl ester. The GC retention time was the same as that of the E,E isomer when compared with the four synthetic diene isomers. Chain elongation reactions in the control insect cells produced Z11-16:Acid from the natural Z9-14:Acid, and E11-16:Acid from the added E9-14:Acid. The Δ9-desaturase of the insect cells was able to use E11-16:Acid as a precursor and produce Z9,E11-16:Acid (Fig. 3). This same phenomenon also was found in a yeast expression system (pYES2) used for other desaturases (13). Apparently, precursors containing an E double bond are structurally similar to saturated precursor acids with these Δ9-desaturases. Chain-shortening reactions were present in the insect cells, but not in yeast cells, as evidenced by production of Z7-14:Acid from Z9-16:Acid in the insect cells.
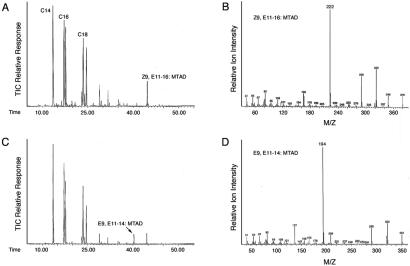
Figure 3.
Confirmation of the conjugated diene compounds by formation of MTAD adducts. (A and B) Control Sf21 cells with addition of E9-14:Acid produced a diene with retention time (A) and diagnostic ions (B) for Z9,E11-16. (C and D) Sf21 cells infected with bacmid viruses, which were transposed with pFastBac1-LBAM-PGE11-ORF, produced an additional diene with retention time (C) and diagnostic ions (D) for E9,E11-14:Acid.
Addition of E11-14:Acid as a precursor also yielded a 14-carbon conjugated diene, but the same product was produced by control insect cells. GC/MS analysis of the MTAD derivative showed that the product was Z9,E11-14:Acid produced by the Z9-desaturase present in the insect cells (similarly produced by the Δ9-desaturase in Saccharomyces cerevisiae cells with the pYES2 expression system with addition of E11-14:Acid). The Z9,E11-14:Acid was chain elongated to Z11,E13-16:Acid and the E11-14:Acid was chain elongated to E13-16:Acid. The E11-desaturase did not produce any E,E-isomer product with the E11-14:Acid precursor.
Discussion
Evolution of membrane desaturases in moth sex pheromone glands for production of species-specific pheromone components has generated many diverse enzymes with different regiospecificities and stereospecificities. Knowledge of these desaturase structures would be invaluable in defining active-site amino acids involved in the different specificities. Two Z11-desaturases have already been characterized by means of their corresponding genes (5, 6), and herein we report on the characterization of a gene for a Δ11-desaturase that has stereospecificity for producing fatty acids containing E isomers. Of additional interest for structure/activity studies is the specificity of this desaturase for saturated and monounsaturated 14-carbon acid precursors. E fatty acids are not as common in nature as those containing a Z isomer, and the only gene characterized for an E-desaturase has been for a soluble desaturase that produces a Z/E mix of Δ8 and Δ9 isomers of phytosphingenines (14).
Research on pheromone biosynthetic pathways in various moth species has provided information on the existence of desaturases with different specificities. With the LBAM, a tortricid species found in Australia and New Zealand, several biosynthetic pathways can be postulated for production of its pheromone components, E11-14:OAc and E9,E11-14:OAc (fatty alcohol acetate esters). However, a study (15) involving deuterium-labeled intermediates showed that E11-14:Acid was produced directly from myristic acid by E11 desaturation, and that the diene was produced by the following pathway: E11-16:Acid, which is produced by E11 desaturation of palmitic acid, is chain-shortened to E9-14:Acid, and then E11 desaturation of this intermediate produces the diene, E9,E11-14:Acid. Although there are three different E11 products involved in the biosynthesis of these pheromone components, the present findings show that a single desaturase is responsible for all three unsaturated acid intermediates.
Production of E11-16:Acid and E11-14:Acid by a single desaturase is consistent with the range of precursors used by other desaturases, which usually involves mainly C16 and C18 precursors, but also can use C17, C15, and C14 precursors, as evidenced by the products of the LBAM-FBZ9 desaturase. However, it was surprising to find that the same desaturase could also use an E9-14:Acid precursor and produce the diene, E9,E11-14:Acid. Apparently, the E9 acid is sufficiently similar to the saturated acid to be accepted by the E11-desaturase. This lack of specificity for a precursor acid containing an E double bond also was found with the metabolic Z9-desaturase in insect and yeast cells. Although the Z9-desaturase normally produces Z9-14:Acid, Z9-16:Acid, and Z9-18:Acid, it can also produce the dienes, Z9,E11-14:Acid and Z9,E11-16:Acid, if provided with E11-14:Acid or E11-16:Acid as precursors. A comparison of Δ9- and Δ11-desaturase structures (Fig. 1) shows that there are a number of amino acids that are consistently different between the two groups. The E11-desaturase with specificity mainly for C14 precursors should be invaluable in assessing amino acids located on the cytoplasmic face for targets in studies, such as site-directed mutagenesis, domain switching, or combinatorial saturation mutagenesis (3), to determine specific sites that affect different specificities.
Acknowledgments
We thank Anne Barrington (Hort Research, Auckland, New Zealand) for shipping LBAM pupae, and Ping Wang for his technical assistance with the baculovirus expression system. This work was supported by National Science Foundation Grant IBN-9870669.
Abbreviations
- LBAM
light brown apple moth
- E11-14:Acid
(E)-11-tetradecenoic acid
- E9,E11-14:Acid
(E,E)-9,11-tetradecadienoic acid (fatty acids of other configurations, chain lengths, and positions of unsaturation are named similarly)
- RACE
rapid amplification of cDNA ends
- UFA
unsaturated fatty acid
- ss-cDNA
single stranded cDNA
- DMDS
dimethyl disulfide
- MTAD
4-methyl-1,2,4-triazoline-3,5-dione
- MAME
myristic acid methyl ester
- RT
reverse transcription
- YPD
yeast extract/peptone/dextrose
Footnotes
References
- 1.Shanklin J, Cahoon E B. Annu Rev Plant Physiol Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 2.Cahoon E B, Lindqvist Y, Schneider G, Shanklin J. Proc Natl Acad Sci USA. 1997;94:4872–4877. doi: 10.1073/pnas.94.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whittle E, Shanklin J. J Biol Chem. 2001;276:21500–21505. doi: 10.1074/jbc.M102129200. [DOI] [PubMed] [Google Scholar]
- 4.Somerville C R, Browse J A. Trends Cell Biol. 1996;6:148–153. doi: 10.1016/0962-8924(96)10002-7. [DOI] [PubMed] [Google Scholar]
- 5.Knipple D C, Rosenfield C L, Miller S J, Liu W, Tang J, Ma P W K, Roelofs W L. Proc Natl Acad Sci USA. 1998;95:15287–15292. doi: 10.1073/pnas.95.26.15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfield C-L, You K-M, Marsella-Herrick P, Roelofs W L, Knipple D C. Insect Biochem Mol Biol. 2001;31:949–964. doi: 10.1016/s0965-1748(01)00043-1. [DOI] [PubMed] [Google Scholar]
- 7.Stukey J E, McDonough V M, Martin C E. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- 8.Liu W, Ma P W K, Marsella-Herrick P, Rosenfield C L, Knipple D C, Roelofs W L. Insect Biochem Mol Biol. 1999;29:435–443. doi: 10.1016/s0965-1748(99)00020-x. [DOI] [PubMed] [Google Scholar]
- 9.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buser H R, Arn H, Guerin P, Rauscher S. Anal Chem. 1983;55:818–822. [Google Scholar]
- 11.Young D C, Vouros P, Holick M F. J Chromatogr. 1990;522:295–302. doi: 10.1016/0021-9673(90)85199-6. [DOI] [PubMed] [Google Scholar]
- 12.McElfresh J S, Millar J G. J Chem Ecol. 1999;25:687–709. [Google Scholar]
- 13.Hao, G., Liu, W., O'Connor, M. & Roelofs, W. L. (2002) Insect Biochem. Mol. Biol.32, in press. [DOI] [PubMed]
- 14.Sperling P, Zähringer U, Heinz E. J Biol Chem. 1998;273:28590–28596. doi: 10.1074/jbc.273.44.28590. [DOI] [PubMed] [Google Scholar]
- 15.Foster S P, Roelofs W L. Experientia. 1990;46:269–273. [Google Scholar]