Summary
Engineered ascorbate peroxidase, APEX2, is widely applied for the identification of intracellular molecule-molecule interaction analyses. Here, we present a protocol for identifying interactors of RNA-binding proteins (RBPs) in living HeLa cells using the APEX2 fusion construct. We describe steps for generation of RBP-APEX2, proximity biotin labeling, and preparation of labeled molecules for mass spectrometry analysis. This protocol may be applicable to other cell cultures and RBPs of interest.
For complete details on the use and execution of this protocol, please refer to Uozumi et al.1
Subject areas: Bioinformatics, Cell Biology, Proteomics, Protein expression and purification
Graphical abstract

Highlights
-
•
Instructions for generating an RNA-binding protein (RBP)-APEX2 fusion construct
-
•
Steps for performing proximity biotinylation via RBP-APEX2 in mammalian cells
-
•
Protocol for avidin-based purification of biotinylated RBP interactors
-
•
Guidance for identifying RBP interactors by mass spectrometry
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Engineered ascorbate peroxidase, APEX2, is widely applied for the identification of intracellular molecule-molecule interaction analyses. Here, we present a protocol for identifying interactors of RNA-binding proteins (RBPs) in living HeLa cells using the APEX2 fusion construct. We describe steps for generation of RBP-APEX2, proximity biotin labeling, and preparation of labeled molecules for mass spectrometry analysis. This protocol may be applicable to other cell cultures and RBPs of interest.
Before you begin
A plant-derived ascorbate peroxidase APEX was originally generated as a tag to gain high-resolution images of electron microscopy analysis.2 The mutated peroxidase APEX2 was developed to improve the sensitivity of proximity labeling.3 In the presence of hydrogen peroxide (H2O2), APEX2 catalyzes the radicalization of biotin-phenol (BP) and allows radical BP to bond to endogenous molecules.4 Since radical BP is short-lived, APEX2 restricts the biotin labeling of endogenous molecules to occur within the proximal radius (<20–25 nm) of APEX2 fusion protein of interest; therefore, APEX2 analysis allows the characterization of spatial omics mapping using living organisms.3,4,5,6,7 To date, APEX2 proximity labeling assay has been used for proteome mapping of mitochondria,4,5 chromatins,8 lysosomes,9 and stress granules,10 or application to transcriptome analysis namely APEX-seq.11,12,13
Heterogeneous nuclear ribonucleoproteins (hnRNPs) compose a family of RNA-binding proteins (RBPs) having various roles including RNA processing, splicing, and nuclear-cytoplasmic transports.14 hnRNPs have been reported to bind and interact with endogenous RNAs and RBPs through their RNA binding and low complexity (LC) domains with relatively weak interactions, which can promote liquid-liquid phase separation (LLPS).15,16
Although immunoprecipitation assay is a widely used method for capturing interacting molecules, its targets are limited to high-affinity molecules. In contrast, in APEX2 proximity labeling assay, the expression of APEX2-fused protein allows the comprehensive identification of molecules within the proximal area of the peroxidase. Our protocol outlines specific steps for APEX2 proximity labeling using hnRNPA3-APEX2 construct in cultured HeLa cells; however, it can also be applied to other RBPs of interest, such as TDP-43,17 and other mammalian cell species.
Part A (steps 1–20) described below is required to ensure that the experimental system is functioning properly in your laboratory before proceeding with the biotin-labeling procedure for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (Part B; steps 21–33).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-hnRNPA3 (WB: 1:1,000, IF: 1:200) | Abcam | Cat# ab78300; RRID: AB_2041662 |
| Mouse anti-V5 tag (WB: 1:1,000, IF: 1:1500) | Abcam | Cat# ab27671; RRID: AB_471093 |
| Rabbit anti-β-actin (WB: 1:1,000) | Cell Signaling Technology | Cat# 4970; RRID: AB_2223172 |
| Goat anti-rabbit IgG (H + L) HRP conjugate | Promega | Cat# W401B |
| Goat anti-mouse IgG (H + L) HRP conjugate | Promega | Cat# W402B |
| Bacterial and virus strains | ||
| NEB stable competent E. coli (high efficiency) | New England Biolabs | Cat# C3040 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM, high glucose, GlutaMAX supplement | Gibco | Cat# 10566-016 |
| Fetal bovine serum | Sigma-Aldrich | Cat# 172012 |
| Lipofectamine LTX reagent with PLUS reagent | Invitrogen | Cat# 15338100 |
| Biotin-phenol | AdipoGen | Cat# CDX-B0270 |
| Hydrogen peroxide solution (30%) | Nacalai Tesque | Cat# 18411-25 |
| Sodium ascorbate | Nacalai Tesque | Cat# 11692-52 |
| Sodium azide | FUJIFILM Wako | Cat# 199-11095 |
| Trolox | Sigma-Aldrich | Cat# 238813 |
| Deoxycholic acid | Sigma-Aldrich | Cat# D2510 |
| NP-40 | Nacalai Tesque | Cat# 1855124 |
| Dithiothreitol | Nacalai Tesque | Cat# 14128-04 |
| cOmplete, EDTA-free protease inhibitor cocktail | Roche | Cat# 11873580001 |
| Ammonium bicarbonate | Sigma-Aldrich | Cat# 09830 |
| I-Block protein-based blocking reagent | Invitrogen | Cat# T2015 |
| Biotin | Sigma-Aldrich | Cat# B4501 |
| Streptavidin-horseradish peroxidase (HRP) conjugate | Invitrogen | Cat# SA10001 |
| Alexa Fluor 488 streptavidin conjugate (1:4,000) | Invitrogen | N/A |
| 30% acrylamide/bis solution, 37.5:1 | Bio-Rad | Cat# 1610158 |
| 4%-paraformaldehyde phosphate-buffered solution | Nacalai Tesque | Cat# 09154 |
| Critical commercial assays | ||
| DC protein assay kit II | Bio-Rad | Cat# 5000112 |
| Silver stain MS kit | FUJIFILM Wako | Cat# 299-58901 |
| Experimental models: Cell lines | ||
| HeLa cell (human) | ATCC | Cat# CCL-2 |
| Recombinant DNA | ||
| pcDNA5/FRT/TO | Invitrogen | Cat# V652020 |
| pcDNA5/FRT/TO-hnRNPA3-V5-APEX2 | Uozumi et al.1 | N/A |
| Software and algorithms | ||
| Scaffold software | https://www.proteomesoftware.com/ | N/A |
| Other | ||
| Countess 3 automated cell counter | Invitrogen | N/A |
| Bioruptor II | Sonic Bio | N/A |
| Immobilon-P PVDF membrane | Merck Millipore | Cat# IPVH00010 |
| Mini trans-blot cell | Bio-Rad | Cat# 1703930JA |
| SnakeSkin dialysis tube (10-kDa molecular weight cutoff) | Thermo Fisher Scientific | Cat# 68100 |
| SnakeSkin dialysis tubing clip | Thermo Fisher Scientific | Cat# 68011 |
| NanoLINK streptavidin magnetic bead (1.0 μm) (10 mg/mL) | Vector Laboratories | Cat# M-1002 |
| DynaMag-2 magnet | Invitrogen | Cat# 12321D |
| Novex WedgeWell 10%, Tris-glycine gel | Thermo Fisher Scientific | Cat# XP00105BOX |
| Amersham Imager 680 | GE Healthcare | N/A |
| Confocal laser scanning microscope LSM710 or LSM980 with Airyscan2 Multiplex | Carl Zeiss | N/A |
WB, western blot; IF, immunofluorescence.
Materials and equipment
100 mM BP
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| Biotin-phenol | 100 mM | 36.35 mg | – |
| DMSO | N/A | 1 mL | – |
| Total | N/A | 1 mL | Sonicate to fully dissolve. |
Divide into aliquots and store at −20°C for several months.
12 mM H2O2 Solution
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| 9.8 M (30%) H2O2 | 12 mM | 12.24 μL | H2O2 is hazardous to human health. |
| 10× PBS | 1× | 1 mL | – |
| Ultra-pure water | N/A | Up to 10 mL | – |
| Total | N/A | 10 mL | – |
Prepare fresh.
10× RIPA Buffer
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| Triton X-100 | 10% | 25 mL | – |
| Deoxycholic acid | 5% | 12.5 g | – |
| NaCl | 1.5 M | 21.9 g | – |
| 20% Sodium dodecyl sulfate | 1% | 12.5 mL | Sodium dodecyl sulfate is hazardous to human health. |
| 1 M Tris-HCl (pH 8.0) | 250 mM | 62.5 mL | – |
| Ultra-pure water | N/A | Up to 250 mL | – |
| Total | N/A | 250 mL | – |
Dilute in ultra-pure water and use.
Divide into 50 mL of aliquots and store at −20°C for up to six months (10×) or 4°C for several months (1×).
Quenching Buffer
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| 500 mM TROLOX | 5 mM | 600 μL | Prepare in DMSO and sonicate to dissolve. |
| 1 M Sodium ascorbate | 10 mM | 600 μL | – |
| 1 M Sodium azide | 10 mM | 600 μL | Sodium azide is hazardous to human health. |
| 10× PBS | 1× | 6 mL | – |
| Ultra-pure water | N/A | 52.2 mL | – |
| Total | N/A | 60 mL | – |
Prepare fresh.
10× TBST
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 7.4) | 250 mM | 500 mL |
| NaCl | 1.37 M | 160 g |
| KCl | 26.8 mM | 4 g |
| Ultra-pure water | N/A | up to 2,000 mL |
| Total | N/A | 2,000 mL |
Before use, dilute in ultra-pure water and add 20% Tween-20 into 1×TBST to a final concentration of 0.1%.
Store for several months at room temperature (20°C–25°C).
Lysis Buffer
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| 500 mM TROLOX | 5 mM | 80 μL | Prepare in DMSO and sonicate to dissolve. |
| 1 M Sodium ascorbate | 10 mM | 80 μL | – |
| 1 M Sodium azide | 10 mM | 80 μL | Sodium azide is hazardous to human health. |
| 1 M Tris-HCl (pH 7.4) | 50 mM | 400 μL | – |
| 0.4% Sodium dodecyl sulfate | 0.2% | 4 mL | Sodium dodecyl sulfate is hazardous to human health. |
| 5 M NaCl | 500 mM | 800 μL | – |
| 1 M Dithiothreitol | 1 mM | 8 μL | – |
| 25× cOmplete protease inhibitor EDTA free | 1× | 320 μL | – |
| Ultra-pure water | N/A | 2.23 mL | – |
| Total | N/A | 8 mL | – |
Fresh preparation is recommended, but it can be stored at −20°C for several months.
Dialysis Buffer
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| 1 M Tris-HCl (pH 7.4) | 50 mM | 150 mL | – |
| 5 M NaCl | 250 mM | 150 mL | – |
| 10% Sodium dodecyl sulfate | 0.1% | 30 mL | Sodium dodecyl sulfate is hazardous to human health. |
| Triton X-100 | 1% | 3 mL | – |
| Ultra-pure water | N/A | up to 3,000 mL | – |
| Total | N/A | 3,000 mL | – |
Prepare fresh.
Wash Buffer 1
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| 10% Sodium dodecyl sulfate | 2% | 6 mL | Sodium dodecyl sulfate is hazardous to human health. |
| Ultra-pure water | N/A | 24 mL | – |
| Total | N/A | 30 mL | – |
Store at room temperature (20°C–25°C) for several months.
Wash Buffer 2
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 7.4) | 50 mM | 1.5 mL |
| 1 M NaCl | 100 mM | 3 mL |
| 1% Deoxycholic acid | 0.1% | 3 mL |
| 10% Triton X-100 | 1% | 3 mL |
| 500 mM EDTA | 1 mM | 60 μL |
| Ultra-pure water | N/A | 19.44 mL |
| Total | N/A | 30 mL |
Prepare fresh.
Wash Buffer 3
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 7.4) | 10 mM | 300 μL |
| 1 M NaCl | 250 mM | 7.5 mL |
| 1% Deoxycholic acid | 0.5% | 15 mL |
| NP-40 | 0.5% | 150 μL |
| 500 mM EDTA | 1 mM | 60 μL |
| Ultra-pure water | N/A | 6.99 mL |
| Total | N/A | 30 mL |
Prepare fresh.
Wash Buffer 4
| Reagent | Final concentration | Amount |
|---|---|---|
| Ammonium bicarbonate | 50 mM | 118.59 mg |
| Ultra-pure water | N/A | Up to 30 mL |
| Total | N/A | 30 mL |
Fresh preparation is recommended, but it can be stored at room temperature (20°C–25°C) for up to 1 month.
5× Protein Loading Buffer
| Reagent | Final concentration | Amount | Comment |
|---|---|---|---|
| 1 M Tris-HCl (pH 7.4) | 250 mM | 6.25 mL | – |
| Sodium dodecyl sulfate | 5% | 1.25 g | Sodium dodecyl sulfate is hazardous to human health. |
| Glycerol | 50% | 12.5 mL | – |
| Dithiothreitol | 450 mM | 1.73 g | – |
| Bromophenol blue | 0.08% | 20.0 mg | – |
| Ultra-pure water | N/A | Up to 25 mL | – |
| Total | N/A | 25 mL | – |
Divide into aliquots and store at −20°C for up to 3 months.
CRITICAL: Wear experimental groves and clothing through experiments.
CRITICAL: For mass spectrometry analysis, use MS-grade ultra-pure water in the whole experiment.
Step-by-step method details
Part A: Setting up and validating experimental conditions
Designing RBP-APEX2 construct (steps 1 and 2)
Timing: >2 weeks
This section describes the steps for designing RBP-APEX2 fusion construct.
-
1.Generate RBP-APEX2 fusion constructs.
-
a.Clone APEX2 sequence3 to the 5′- or 3′-end of RBP of interest DNA sequence (Figure S1). Some linker sequences can be added if needed.
 CRITICAL: The position of APEX2 fusion in the RBP of interest should be carefully considered depending on the distinctive structure in the protein of interest, such as low-complexity domains or RNA recognition motifs.
CRITICAL: The position of APEX2 fusion in the RBP of interest should be carefully considered depending on the distinctive structure in the protein of interest, such as low-complexity domains or RNA recognition motifs. CRITICAL: In addition to APEX2, an epitope tag sequence should be fused to the RBP of interest. This will facilitate the detection of APEX2-fused RBP expression with anti-epitope tag antibodies. In our study, the V5-tag sequence (amino acid sequence: GKPIPNPLLGLDST) was fused to the 3′-end of hnRNPA3 (Figure S1).
CRITICAL: In addition to APEX2, an epitope tag sequence should be fused to the RBP of interest. This will facilitate the detection of APEX2-fused RBP expression with anti-epitope tag antibodies. In our study, the V5-tag sequence (amino acid sequence: GKPIPNPLLGLDST) was fused to the 3′-end of hnRNPA3 (Figure S1). -
b.Insert RBP-APEX2 into an arbitrary mammalian expression plasmid by restriction enzymes.Note: In our setup, we inserted hnRNPA3-V5-APEX2 fragment at HindIII/BamHI restriction enzyme sites upon pcDNA5/FRT/TO (Invitrogen). See also Uozumi et al. (2024).1
-
a.
-
2.
Confirm plasmid sequences by Sanger sequencing.
Confirmation of RBP-APEX2 fusion protein expression (steps 3–8)
Timing: <1 week
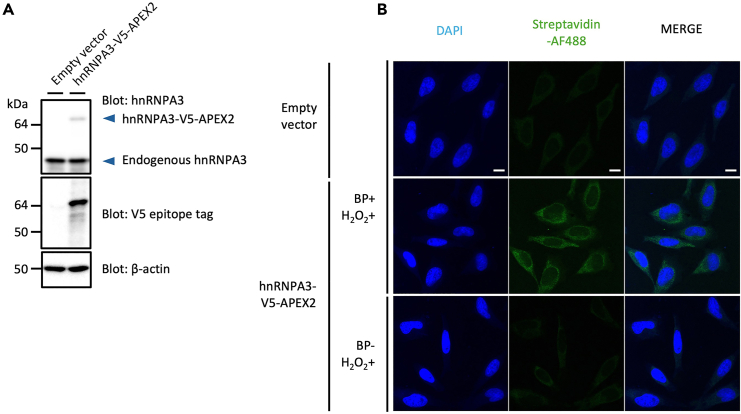
This section describes the steps for plasmid transfection and confirmation of its expression by western blot (WB) analysis (Figure 1).
Figure 1.
hnRNPA3-V5-APEX2 expression and biotin-labeling enzymatic activities in HeLa cells
(A) Detection of hnRNPA3-V5-APEX2 expression by western blotting using anti-hnRNPA3, anti-V5 tag, and anti-β-actin antibodies.
(B) Spatial characterization images of biotin labeling with the streptavidin-Alexa Fluor 488 (AF488) conjugate. Scale bar = 10 μm. BP+/−, cells with (+) or without (−) biotin-phenol treatment; H2O2+, cells with hydrogen peroxide treatment (1 min).
Day 1.
-
3.Seed the cells and incubate them.
-
a.Adjust HeLa cell concentration to 1.30 × 105 cells/mL with growth medium (DMEM supplemented with 10% FBS).Note: Warm the medium to 37°C before use.
-
b.Seed 500 μL of them into each well of a 24-well plate.
-
c.Incubate the cells for 12–20 h at 37°C in an incubator maintained with 5% CO2.
-
a.
Day 2.
-
4.Transfect the plasmid into the cells.
-
a.Replace 500 μL of growth medium with fresh (DMEM supplemented with 10% FBS).Note: Warm the medium to 37°C before use.
-
b.Introduce RBP-APEX2 plasmid into HeLa cells (70–85% confluency) using Lipofectamine LTX (Invitrogen) according to manufacturing instructions.Note: In our setup, 400–600 ng of plasmid per each well is suitable. The amount of the plasmid is adjustable depending on the levels of desired protein expression. See also troubleshooting.
 CRITICAL: Prepare and introduce mock plasmid as a negative control.
CRITICAL: Prepare and introduce mock plasmid as a negative control. -
c.Incubate the cells for 6 h at 37°C in an incubator maintained with 5% CO2.
-
d.Replace 500 μL of growth medium with fresh (DMEM supplemented with 10% FBS).Note: Warm the medium to 37°C before use.
-
e.Incubate the cells for 12–20 h at 37°C in an incubator maintained with 5% CO2.
-
a.
Day 3.
-
5.
Aspirate growth medium.
-
6.
Wash each well with 500 μL of 1× phosphate-buffered saline (PBS).
-
7.
Aspirate 1× PBS.
Pause Point: The plate can be stored at −80°C for several weeks.
-
8.Perform WB assay.
-
a.Lyse cells with 100 μL of 1× RIPA Buffer per well.
-
b.Use the cell lysate for WB analysis blotting with antibodies against the RBP of interest and the epitope tag.
-
c.Obtain images using Amersham Imager 680 (GE Healthcare).
-
a.
Characterization of APEX2 time-dependent biotinylation efficacy (steps 9–15)
Timing: <1 week
This section describes the steps for confirmation of the time-dependent biotinylation efficacy of APEX2-fused RBP (Figure 2).
Figure 2.
hnRNPA3-V5-APEX2 fusion protein promotes biotin labeling of endogenous molecules in BP/H2O2 treatment HeLa cells
(A) APEX2 biotinylation process was promoted in hnRNPA3-V5-APEX2-expressing HeLa cells stimulated with H2O2 for 1 min or longer. Staining of whole lysate proteins (Ponceau) and β-actin as a housekeeping protein was performed.
(B) A low concentration of streptavidin-HRP (1:10000) failed to detect biotin-labeled molecules even in the APEX2 fusion-expressing cells. BP, biotin-phenol; H2O2, hydrogen peroxide. See also troubleshooting.
Days 1 and 2.
-
9.
Prepare the transfected cells as described in steps 3 and 4.
Day 3.
-
10.Perform APEX2 biotin-labeling assay.
-
a.Aspirate growth medium.
-
b.Add 500 μL of 250 μM BP-containing medium (DMEM supplemented with 10% FBS) into each well.Note: Warm the medium to 37°C before use.
-
c.Incubate the cells for 30 min at 37°C in an incubator maintained with 5% CO2.
-
d.Add H2O2 (final concentration 2 mM) to each well and tap the plate briefly to mix.
-
e.Incubate the cells for 1–30 min at room temperature (20°C–25°C).
 CRITICAL: Always prepare cells lacking BP or H2O2 treatments as negative controls (Figure 2).
CRITICAL: Always prepare cells lacking BP or H2O2 treatments as negative controls (Figure 2). CRITICAL: Prepare H2O2 solution immediately before use.Note: The concentrations of BP/H2O2 are adjustable when the signals are too high or low. See also troubleshooting.Note: We usually perform 1-minute treatments, but the incubation time should be optimized depending on the experimental setup. Longer incubation may increase nonspecific biotinylation signals, but shorter incubation may result in insufficient biotinylation signals.
CRITICAL: Prepare H2O2 solution immediately before use.Note: The concentrations of BP/H2O2 are adjustable when the signals are too high or low. See also troubleshooting.Note: We usually perform 1-minute treatments, but the incubation time should be optimized depending on the experimental setup. Longer incubation may increase nonspecific biotinylation signals, but shorter incubation may result in insufficient biotinylation signals. -
f.Quench the biotinylation activity of APEX2.
-
i.Aspirate growth medium.
-
ii.Treat the cells with 500 μL of ice-cold Quencher Buffer on ice for 1 min.
-
iii.Repeat this step two more times.
 CRITICAL: Prepare Quencher Buffer immediately before use.
CRITICAL: Prepare Quencher Buffer immediately before use.
-
i.
-
a.
-
11.
Aspirate Quencher Buffer.
-
12.
Wash each well with 500 μL of 1× PBS.
-
13.
Aspirate 1× PBS.
Pause Point: The plate can be stored at −80°C for several weeks.
-
14.Lyse the cells and perform SDS-PAGE.
-
a.Treat the cells with 100 μL of 1× RIPA Buffer per well for 15 min on ice.
-
b.Scrape the cells and collect cell lysates into 1.5-mL tubes.
-
c.Sonicate the cell lysate for 10 min (ON 30 s/OFF 30 s, 10 cycles; Power High) (BIORUPTOR II) (Sonic Bio). Maintain the sample cold during the sonication on an ice bath.
 Pause Point: The lysates can be stored at −80°C for several weeks.
Pause Point: The lysates can be stored at −80°C for several weeks. -
d.Unfold and denature proteins in the lysate.
-
i.Add 25 μL of 5× Protein Loading Buffer to each sample.
-
ii.Vortex samples for 5–10 s.
-
iii.Boil the samples for 10 min at 100°C.
-
iv.Place the sample tubes on ice immediately and cool for 3 min.
-
i.
-
e.Centrifuge samples for 10 s (microcentrifuges, 3500 × g) at room temperature (20°C–25°C).
-
f.Carefully load 15–20 μL of samples onto a 10% acrylamide Tris-Glycine gel.Note: Vortex sample for 5–10 sec before load.Note: The amount of loading proteins should be optimized depending on the experimental setup.
-
g.Run and separate proteins in the gel for 90 min at a constant 120 V.Alternatives: The running time can be extended until protein separation is complete.Note: The voltage and running time are adjustable depending on the experimental setup.
-
a.
-
15.Perform Streptavidin-HRP blotting assay.
-
a.Wet-transfer WB assay.
-
i.Retrieve the gel and place it on a polyvinylidene fluoride (PVDF) membrane.
-
ii.Transfer proteins onto the PVDF membrane for 60 min at a constant current of 0.4 A in a transfer tank filled with transfer buffer. Maintain the apparatus cold with ice during the transfer.
 CRITICAL: To normalize and quantify the streptavidin-HRP signal intensity, prepare PVDF membranes for detecting housekeeping protein (e.g. β-actin or GAPDH) or Ponceau staining; here, we describe a protocol for housekeeping protein detection.
CRITICAL: To normalize and quantify the streptavidin-HRP signal intensity, prepare PVDF membranes for detecting housekeeping protein (e.g. β-actin or GAPDH) or Ponceau staining; here, we describe a protocol for housekeeping protein detection.
-
i.
-
b.Membrane blocking.
-
i.After transfer, place the membrane into I-Block (Invitrogen) blocking solution (supplemented with 0.02% Triton X-100).
-
ii.For streptavidin-HRP detection, gently shake the membrane for 12–20 h at 4°C
-
iii.For housekeeping protein detection, gently shake the membrane for 1 h at room temperature (20°C–25°C).
 CRITICAL: Confirm that the blocking solution is biotin-free.
CRITICAL: Confirm that the blocking solution is biotin-free.
-
i.
-
c.(Steps only for housekeeping protein detection) Incubation in the primary antibody solution.
-
i.After 1 h of blocking, discard the I-Block solution.
-
ii.Place the membrane in primary antibody (diluted to desired concentration within blocking solution supplemented with 0.02% Triton X-100) solution.
-
iii.Gently shake the membrane for 12–20 h at 4°C
-
i.
-
a.
Day 4.
-
d.Discard the blocking solution for streptavidin-HRP detection (continued from step 15.b.ii) and the primary antibody solution for housekeeping protein detection (continued from step 15.c).
-
e.Wash the membrane.
-
i.Place the membranes into 1× TBST buffer.
-
ii.Shake gently at room temperature (20°C–25°C) for 5 min.
-
iii.Discard 1× TBST buffer.
-
iv.Repeat this step two more times.
-
i.
-
f.Incubation in the HRP-conjugate solution.
-
i.Place the membrane in streptavidin-HRP (diluted 1:5000 within blocking solution supplemented with 0.02% Triton X-100) (Invitrogen) solution.
-
ii.Gently shake it for 1 h at room temperature (20°C–25°C).
-
iii.For housekeeping protein detection, place it in HRP-conjugated secondary antibody solution (diluted to desired concentration within blocking solution supplemented with 0.02% Triton X-100) and gently shake it for 1 h at room temperature (20°C–25°C).
-
i.
-
g.Discard both streptavidin-HRP and secondary antibody solution.
-
h.Wash the membrane.
-
i.Place the membranes into 1× TBST buffer.
-
ii.Shake gently at room temperature (20°C–25°C) for 10 min.
-
iii.Discard 1× TBST buffer.
-
iv.Repeat this step two more times.
-
i.
-
i.Detect HRP signals with ECL Western Blotting Detection Reagents (Cytiva). Obtain electrochemiluminescence images using Amersham Imager 680 (GE Healthcare).
-
d.
Confirm intracellular localization of RBP-APEX2 fusion protein (steps 16 and 17)
Timing: <1 week
This section describes the steps for plasmid transfection and confirmation of RBP-APEX2 localization in HeLa cells by immunofluorescence analysis (Figure 3).
Figure 3.
Fusion of APEX2 does not alter the localization of hnRNPA3
Representative immunofluorescence images show the nuclear-dominant localization of hnRNPA3 in fusion construct-expressing HeLa cells. Scale bar = 20 μm.
Days 1 and 2.
-
16.
Prepare the transfected cells as described in steps 3–4.
Note: For immunofluorescence analysis, the cells should be seeded onto a round coverslip in each well.
Day 3.
-
17.Perform immunofluorescence assay.
-
a.Wash the cells with 1× PBS as described in steps 5–7.
-
b.Fix the cell with 250 μL of 4% (w/v) PFA for 15 min at room temperature (20°C–25°C).
 CRITICAL: PFA is hazardous to human health.
CRITICAL: PFA is hazardous to human health. -
c.Aspirate PFA.
-
d.Wash each well with 500 μL of 1× PBS.
 Pause Point: The plate can be stored at 4°C for several weeks. Avoid drying while pausing.Note: Dispose of PFA in accordance with local and national regulations.
Pause Point: The plate can be stored at 4°C for several weeks. Avoid drying while pausing.Note: Dispose of PFA in accordance with local and national regulations. -
e.Use the cells for immunofluorescence analysis with antibodies against the RBP of interest and/or an epitope tag.
-
f.Obtain immunofluorescence images using confocal laser scanning microscopes.
-
a.
Characterize spatial enzymatic activities of APEX2 (steps 18–20)
Timing: <1 week
This section describes the steps for confirmation of APEX2 spatial biotinylation activities by immunofluorescence (Figure 1).
Days 1 and 2.
-
18.
Prepare the transfected cells as described in steps 3–4.
Note: For immunofluorescence analysis, the cells should be seeded onto a round glass coverslip in each well.
Day 3.
-
19.
Perform APEX2 biotin labeling as described in step 10.
-
20.Perform immunofluorescence assay.
-
a.Wash and fix the cells as described in steps 17.a-d.
 Pause Point: The plate can be stored at 4°C for several weeks. Avoid drying while pausing.
Pause Point: The plate can be stored at 4°C for several weeks. Avoid drying while pausing. -
b.Use the cells for immunofluorescence analysis with Alexa Fluor streptavidin conjugate (Invitrogen).
-
c.Obtain immunofluorescence images using confocal laser scanning microscopes.
-
a.
Part B: Detailed steps of proximity biotin labeling for identification of endogenous interactors of RNA-binding proteins in mammalian cells
Cell seeding and expression of RBP-APEX2 fusion protein (steps 21 and 22)
Timing: 2 days
The purpose of this section is to introduce the APEX2 fusion construct into cultured HeLa cells by lipofection.
Day 1.
-
21.Seed the cells and incubate them.
-
a.Adjust HeLa cell concentration to 1.30 × 105 cells/mL with growth medium (DMEM supplemented with 10% FBS).Note: Warm the medium to 37°C before use.
-
b.Seed 10 mL of them into a 10-cm culture dish.
-
c.Incubate the cells for 12–20 h at 37°C in an incubator maintained with 5% CO2.
-
a.
Day 2.
-
22.Transfect the plasmid into the cells.
-
a.Replace 10 mL of growth medium with fresh (DMEM supplemented with 10% FBS).Note: Warm the medium to 37°C before use.
-
b.Introduce RBP-APEX2 plasmid into HeLa cells (70–85% confluency) using Lipofectamine LTX (Invitrogen) according to manufacturing instructions.Note: In our setup, 10–15 μg of plasmid per 10-cm dish is suitable. See also troubleshooting.
 CRITICAL: Prepare cells lacking BP treatment as a negative control.
CRITICAL: Prepare cells lacking BP treatment as a negative control. -
c.Incubate the cells for 6 h at 37°C in an incubator maintained with 5% CO2.
-
d.After 6 h, replace 10 mL of growth medium with fresh (DMEM supplemented with 10% FBS).Note: Warm the medium to 37°C before use.
-
e.Incubate the cells for 12–20 h at 37°C in an incubator maintained with 5% CO2.
-
a.
Incubation of cells with BP/H2O2 and proximity biotin labeling (steps 23–26)
Timing: 1.5 h
The purpose of this section is to label RBP-APEX2 proximity proteins with BP in cultured mammalian cells.
Day 3.
-
23.Perform APEX2 biotin-labeling treatment.
-
a.Aspirate growth medium.
-
b.Replace the growth medium with 10 mL of 250 μM BP-containing medium (DMEM supplemented with 10% FBS).Note: Warm the medium to 37°C before use.
-
c.Incubate the cells for 30 min at 37°C in an incubator maintained with 5% CO2.
-
d.Add H2O2 (final concentration 2 mM) to the medium and tap the culture dish briefly to mix.
-
e.Incubate the cells for 1 min at room temperature (20°C–25°C).
 CRITICAL: Prepare H2O2 solution immediately before use.Note: The concentrations of BP/H2O2 are adjustable when the signals are too high or low. See also troubleshooting.Note: We usually perform 1-minute treatments, but the incubation time should be optimized depending on the experimental setup. Longer incubation may increase nonspecific biotinylation signals, but shorter incubation may result in insufficient biotinylation signals.
CRITICAL: Prepare H2O2 solution immediately before use.Note: The concentrations of BP/H2O2 are adjustable when the signals are too high or low. See also troubleshooting.Note: We usually perform 1-minute treatments, but the incubation time should be optimized depending on the experimental setup. Longer incubation may increase nonspecific biotinylation signals, but shorter incubation may result in insufficient biotinylation signals. -
f.Quench the biotinylation activity of APEX2.
-
i.Aspirate growth medium.
-
ii.Treat the cells with 5 mL of ice-cold Quencher Buffer on ice for 1 min.
-
iii.Repeat this step two more times.
 CRITICAL: Prepare Quencher Buffer immediately before use.
CRITICAL: Prepare Quencher Buffer immediately before use.
-
i.
-
a.
-
24.
Aspirate Quencher Buffer.
-
25.
Wash each dish with 10 mL of 1× PBS.
CRITICAL: Wash the cells gently so that they do not detach from the dish.
-
26.
Aspirate 1× PBS.
Preparation of cell lysate and dialysis to remove excess BP (steps 27 and 28)
Timing: 1.5 days
The purpose of this section is to remove unreacted BP from the cell lysate.
-
27.Prepare the cell lysate.
-
a.Lyse cells with 1.5 mL of Lysis Buffer for 15 min on ice.
-
b.Scrape the cells and collect cell lysate into a 2-mL tube.
-
c.Sonicate the cell lysate (ON 60 s/OFF 30 s, 21 cycles; Power High) (BIORUPTOR II) (Sonic Bio). Maintain the sample cold during the sonication on an ice bath.
-
d.Centrifuge cell lysate at 21500 × g for 12 min at 4°C
-
a.
-
28.Dialyze the cell lysate.
-
a.Collect 1 mL of the supernatant into a new 2-mL tube.
-
b.Add the same volume of 50 mM Tris-HCl (pH 7.4).
 CRITICAL: Avoid collecting the supernatant that is close to the pellet.
CRITICAL: Avoid collecting the supernatant that is close to the pellet. -
c.Load sample into SnakeSkin dialysis tubes (10-kDa MWCO) (Thermo Fisher Scientific).
-
d.Place the dialysis tubes within 700 mL of Dialysis Buffer and stir for 1 h at 4°C
-
e.Replace the Dialysis Buffer with 700 mL of fresh buffer and spin with a magnetic bar for 1 h at 4°C. After 1 h of dialysis, repeat this step once more.
-
f.Replace the Dialysis Buffer with fresh buffer again and stir for 12–20 h at 4°C
-
a.
Pull-down of labeled molecules and electrophoresis (steps 29–33)
Timing: 2 days
The purpose of this section is to pull-down biotinylated proteins using streptavidin magnetic beads and separate them by SDS-PAGE. Proteins separated in the gels are visualized with silver staining and used for mass spectrometry analysis.
Day 1.
-
29.Perform pull-down assay using streptavidin magnetic beads.
-
a.After dialysis, collect and put the dialyzed samples into new 1.5-mL tubes.
-
b.Quantify the protein concentration in the sample.
-
i.Determine the protein concentration in each sample using a detergent-compatible protein assay (based on the Lowry method) according to manufacturing protocols.
 CRITICAL: Bicinchoninic Acid (BCA) assay for protein quantification cannot be used for this step due to the presence of a high concentration of ascorbate in the samples. If you must use BCA assay in this step, see Hung et al. (2016).5
CRITICAL: Bicinchoninic Acid (BCA) assay for protein quantification cannot be used for this step due to the presence of a high concentration of ascorbate in the samples. If you must use BCA assay in this step, see Hung et al. (2016).5
-
ii.Adjust the protein concentration to 360 μg/mL with 50 mM Tris-HCl (pH 7.4).
-
i.
-
c.Put 1 mL of adjusted samples into 1.5-mL tubes.
-
d.Add 30 μL of streptavidin magnetic beads (10 mg/mL) per sample.Note: Vortex the streptavidin beads for 5–10 sec before use.
-
e.Incubate the samples for 12–20 h at 4°C with gentle agitation. As a negative control, add 30 μL of magnetic beads (but not dialyzed lysate) into 1 mL of 50 mM Tris-HCl (pH 7.4) and incubate under the same condition.
 CRITICAL: Prepare an arbitrary amount of input controls.
CRITICAL: Prepare an arbitrary amount of input controls.
-
a.
Day 2.
Note: Perform the same treatment in steps 30–33 for all negative control samples.
-
30.Wash the beads solution.
-
a.Wash Buffer treatment.
-
i.Pellet the beads using a magnetic rack.
-
ii.Remove the supernatant.
-
iii.Resuspend the beads with ice-cold Wash Buffer 1.
-
iv.Agitate the beads for 5 min at 4°C.
-
i.
-
b.Repeat this step three more times. Use ice-cold Wash Buffer 2 through 4 sequentially.
-
a.
-
31.Perform SDS-PAGE.
-
a.Elute the biotinylated proteins from the streptavidin magnetic beads.
-
i.Add Protein Elution Buffer (containing 5.0 μL of 5× Protein Loading Buffer, 2.5 μL of 200 mM Dithiothreitol, 8.3 μL of 6 mM Biotin, and 9.2 μL of ultra-pure water) to each sample.
-
ii.Boil the samples for 10 min at 100°C.
-
iii.Place the sample tubes on ice immediately and cool for 3 min.
-
i.
-
b.Centrifuge samples for 10 s (microcentrifuges, 3500 × g) at room temperature (20°C–25°C).
-
c.Carefully load 15–20 μL of samples onto a 10% acrylamide Tris-Glycine gel.Note: Vortex sample for 5–10 sec before load.Note: The amount of loading proteins should be optimized depending on the experimental setup.
 CRITICAL: For MS analysis, the use of pre-cast gels and Protein Elution buffer with no contaminating proteins is very critical to minimize contamination of unintended proteins from the environment. Note that the buffer used for PAGE depends on the composition of the gels.
CRITICAL: For MS analysis, the use of pre-cast gels and Protein Elution buffer with no contaminating proteins is very critical to minimize contamination of unintended proteins from the environment. Note that the buffer used for PAGE depends on the composition of the gels. CRITICAL: Load input samples to the gel to evaluate pull-down efficacy.
CRITICAL: Load input samples to the gel to evaluate pull-down efficacy. CRITICAL: Load BP untreated samples to the gel as negative controls.
CRITICAL: Load BP untreated samples to the gel as negative controls. CRITICAL: Load samples eluted from streptavidin magnetic beads without cell lysates as a negative control.
CRITICAL: Load samples eluted from streptavidin magnetic beads without cell lysates as a negative control. -
d.Run and separate proteins in the gel for 90 min at a constant 120 V.Alternatives: The running time can be extended until protein separation is complete.Note: The voltage and running time are adjustable depending on the experimental setup.
-
a.
-
32.
After electrophoresis, retrieve the gel and use it for MS-compatible silver staining according to the manufacturing protocol.
CRITICAL: For silver staining, make sure that using MS-grade ultra-pure water.
Note: When the protein separation is unclear on silver staining, SDS denaturation and/or the amount of loading sample should be optimized.
Note: Dispose of the silver staining solution in accordance with local and national regulations and manufacturing instructions.
Pause Point: The gel can be stored at 4°C for up to one week in ultra-pure water away from drying and light.
-
33.
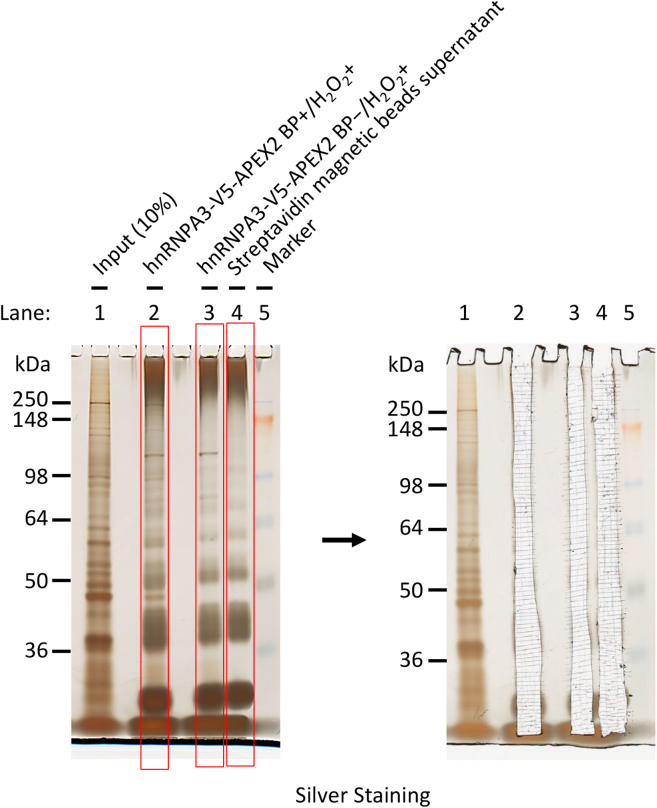
Use the silver-stained gel for LC-MS/MS analysis (Figure 4) (see quantification and statistical analysis).
Figure 4.
Representative image of the silver-stained gel used for LC-MS/MS analysis
After dialysis and incubation with magnetic beads, the HeLa cell lysate samples were separated in the acrylamide Tris-Glycine gel. (Left image) BP-treated hnRNPA3-V5-APEX2-expressing sample lane (lane 2) showed multiple specific band patterns compared to BP treatment-lacking (lane 3) and magnetic beads supernatant (lane 4). Boxed lanes are subjected to the following LC-MS/MS. (Right image) Each lane (2, 3, and 4) was cut out and used for LC-MS/MS analysis. Input, cell lysate without incubation with magnetic beads solution; BP+/−, cells with (+) or without (−) biotin-phenol treatment; H2O2+, cells with hydrogen peroxide treatment (1 min).
Expected outcomes
The expression levels and intracellular localization of endogenous RBP of interest should not be altered by the APEX2 fusion; WB analysis using anti-RBP antibodies would show two band patterns of endogenous and introduced RBPs with no degradation pattern. As APEX2 is approximately 27 kDa, the band of RBP-APEX2 fusion is expected to be detected at a position 27 kDa larger than the intrinsic RBP by that size (Figure 1A). Immunofluorescence analysis should confirm that the localization of RBP-APEX2 has not altered compared to the endogenous RBP. Using hnRNPA3-APEX2 construct, we confirmed that the APEX2 fusion does not alter the nuclear dominant intracellular distribution of hnRNPA3 (Figure 3).
The time-course assay characterizes that the cells treated with H2O2 for 1 min are sufficient for biotin labeling, and the labeling does not occur when either the APEX2 construct, BP, or H2O2 is omitted (Figure 2A) (see troubleshooting).1,5,7 When quantifying the signal intensity in regions blotted with streptavidin-HRP, it should be normalized with housekeeping proteins (e.g., β-actin or GAPDH) or total protein levels by Ponceau staining (Figure 2A). For detailed quantification methods, see Uozumi et al. (2024).1
Immunofluorescence imaging with AlexaFluor streptavidin conjugate is expected to show high signals in the cytoplasm and faint signals in the nucleus in cells (Figure 1B).1 This is because of the poor nuclear membrane permeability of BP due to its charge of the functional group4,18; for alternative biotin-labeling methods instead of using BP, see Tan et al. (2020)7 or Padrón et al. (2019).11
Silver staining shows specific band patterns in BP-treated pull-down sample lanes. Nonspecific multiple bands are also detected in the sample lanes eluted from streptavidin magnetic beads only, which may be due to endogenous biotin-binding proteins (e.g., carboxylases) or proteins contained in the magnetic bead solution. These negative control lanes should also be cut out and analyzed by mass spectrometry to accurately identify the labeled RBP-interacting proteins (Figure 4) (see quantification and statistical analysis).
Quantification and statistical analysis
The quantification methods we have demonstrated are described below, but procedures such as LC-MS and data processing can be replaced by other suitable protocols or outsourced if LC-MS is not available.
Liquid chromatography and mass spectrometry
LC-MS/MS analysis was performed on a Q-Exactive mass spectrometer (Thermo Fisher Scientific) and UltiMate 3000 Nano LC system (Thermo Fisher Scientific). The samples were injected into an ESI-column (0.075 × 150 mm). The experimental analyses were conducted with 300 nL/min mobile phase flow rate, water containing 0.1% formic acid (Solvent A), and acetonitrile containing 0.1% formic acid (Solvent B). The following gradient profile was used for the LS-MS/MS method: (min:B%) 0:5; 5:5; 100:30. Data were acquired in data-dependent analysis mode. The ionization was performed on a nano-ESI source, positive mode, and 1.8 kV capillary voltage; the collision energy was set to 32.
Data processing
Protein Identification was processed using Mascot Distiller (v2.5) and Mascot Server (v2.5) (Matrix Science). Data searching was performed on UniProt (AA) (February 5, 2017) and UniProt-GOA (ver. 159) databases. Digestion enzymes were trypsin and lysyl endopeptidase; parameters were set to a maximum of one missed cleavage. Fixed modifications were carbamidomethylation (Cys) and variable modifications were oxidation (Met) and deamidation (Asn, Gln). Precursor and fragment mass tolerance were 10 ppm and 0.01 Da. The peptide spectrum was validated using a false discovery rate (FDR) threshold of <0.1%.
With Scaffold software, quantitative values (QV) for each detected protein were quantified by spectrum counting. For each technical replicate, we obtained QV of the BP-treated samples and the negative controls and calculated their averages of QV (AQV). The baseline-corrected AQV was obtained by subtraction of the negative control average from the BP average. For detailed methods, see Uozumi et al. (2024).1
Limitations
The difficulty in biotin labeling in the nucleus is due to the low membrane permeability of BP.4,7 Additionally, the presence of a nuclear export signal within APEX2 may be responsible for the cytoplasmic dominance of the labeling. The further engineered mutant of APEX2, APEX2-L242A (namely APEX3), may be useful to facilitate labeling reactions in the nucleus.19
It has been reported that the concentrations of BP and H₂O₂ affect the proximity range of APEX2 biotin-labeling activity7; however, we have not evaluated which proximity range is maintained in our system.
Troubleshooting
Problem 1
APEX2 fusion construct is not expressed in the cells (steps 8 and 17).
Potential solution
If protein expression is not confirmed by blotting or staining with antibodies against the epitope tag included in the construct, refer to the information below (related to steps 1 and 2).
-
•
Make sure the direction and translation codon frame of the insert is correct.
-
•
Make sure that the translation initiation codon and Kozak sequence are located upstream of the construct.
-
•
The sequence of the fusion construct should be validated to ensure that no stop codons are located in the middle.
Problem 2
The expression level of APEX2 fusion construct is too low or high (steps 8 and 17).
Potential solution
-
•
In case the expression level is too low, the expression level can be increased by changing the amount of plasmid and methods of transfection (steps 4 and 22).
-
•
The concentration of seeded cells can be optimized depending on the type and condition of the cells. In addition, cell lines should be regularly checked to ensure they are not infected with Mycoplasma spp (steps 3 and 21).
-
•
The quality of the purified plasmid solution should be checked for too excess contamination. With measuring plasmid concentration using a spectrophotometer, it is necessary to verify that there is little or no protein or phenol carry-over by checking the A260/A280 absorbance ratio.
-
•
In case the expression level is too high, the amount of plasmid should be reduced to approximately 200 ng or less per well (24-well plate) (step 4).
-
•
The expression levels of proteins can also be affected by promoters in the plasmid backbone (step 1).
Problem 3
The amount of biotinylated protein detected is too low in the APEX2-fusion-expressing cells, or a significant amount of biotinylated protein is detected in cells without H2O2 treatment (steps 10 and 23).
Potential solution
In the below cases, the BP/H2O2 incubation assay is incorrectly performed.
-
•
Make sure that BP is fully dissolved within the solvent and medium, and H2O2 solution is prepared immediately before use (steps 10.d and 23.d).
-
•
If the assay still fails, adjust the BP/H2O2 concentrations and/or BP incubation time (steps 10.b-e and 23.b-e).
-
•
In case the blotting patterns are not clear, adjust the streptavidin-HRP concentration (steps 15.f). In our setup, a low concentration of streptavidin-HRP (<1:10000) failed to detect biotin-labeled molecules by blotting (Figure 2B).
-
•
Methanol fixation is not suitable for APEX2.5 If the biotin-labeling activity cannot be confirmed by the AlexaFluor conjugate under methanol fixation, paraformaldehyde fixation can be performed (step 17).
-
•
The biotin-labeling process may still occur in H2O2 untreated cells if quenching is inadequate. Quenching steps are crucial for removing unbonded BP and H2O2 left inside the culture wells or dishes and completely stopping biotinylation activities. Do not omit these steps and use an ice-cold fresh quenching solution. If the quenching is still insufficient, increase the number of wash steps (steps 10.f and 23.f).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kohji Mori (kmori@psy.med.osaka-u.ac.jp).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be answered by the technical contact, Kohji Mori (kmori@psy.med.osaka-u.ac.jp).
Materials availability
Plasmids generated in this study will be available with a completed material transfer agreement.
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report the original code.
Acknowledgments
We thank Shinji Tagami, Yuya Kawabe, Shiho Gotoh, Tesshin Miyamoto, Koujin Miura, Yuki Aoki, Shizuko Kondo, and Tomoko Yamashita for their constructive discussion and technical help. We also thank Umihito Nakagawa and Ayako Nishioka (The Center of Medical Innovation and Translational Research, Osaka University) for performing LC-MS/MS. The graphical abstract was created with BioRender (https://www.biorender.com).
K.M. was supported by the JSPS KAKENHI grant numbers JP20H03602, JP20H05927, JP22K19492, and JP24K02382; JST FOREST program grant number JPMJFR200Z; AMED grant number 23dk0207066h0001; SENSHIN Medical Research Foundation; and Takeda Science Foundation. M.I. was supported by AMED grant numbers JP21ek0109510h0001 and 22bm0804034h0001.
Author contributions
R.U., conceptualization, methodology, validation, investigation, and writing – original draft; K.M., conceptualization, methodology, writing – review and editing, and funding acquisition; S.A., methodology and writing – review and editing; M.I., supervision, writing – review and editing, and funding acquisition.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2024.103368.
Supplemental information
References
- 1.Uozumi R., Mori K., Gotoh S., Miyamoto T., Kondo S., Yamashita T., Kawabe Y., Tagami S., Akamine S., Ikeda M. PABPC1 mediates degradation of C9orf72-FTLD/ALS GGGGCC repeat RNA. iScience. 2024;27 doi: 10.1016/j.isci.2024.109303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., Ellisman M.H., Ting A.Y. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., Ting A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee H.W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung V., Udeshi N.D., Lam S.S., Loh K.H., Cox K.J., Pedram K., Carr S.A., Ting A.Y. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 2016;11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trinkle-Mulcahy L. Recent advances in proximity-based labeling methods for interactome mapping. F1000Res. 2019;8 doi: 10.12688/f1000research.16903.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan B., Peng S., Yatim S.M.J.M., Gunaratne J., Hunziker W., Ludwig A. An Optimized Protocol for Proximity Biotinylation in Confluent Epithelial Cell Cultures Using the Peroxidase APEX2. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X.D., Tu L.-C., Mir A., Rodriguez T., Ding Y., Leszyk J., Dekker J., Shaffer S.A., Zhu L.J., Wolfe S.A., Sontheimer E.J. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9–APEX2. Nat. Methods. 2018;15:433–436. doi: 10.1038/s41592-018-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao Y.C., Fernandopulle M.S., Wang G., Choi H., Hao L., Drerup C.M., Patel R., Qamar S., Nixon-Abell J., Shen Y., et al. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell. 2019;179:147–164.e20. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.-C., Krach F., Yang D., Sen A., et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padrón A., Iwasaki S., Ingolia N.T. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol. Cell. 2019;75:875–887.e5. doi: 10.1016/j.molcel.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fazal F.M., Han S., Parker K.R., Kaewsapsak P., Xu J., Boettiger A.N., Chang H.Y., Ting A.Y. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell. 2019;178:473–490.e26. doi: 10.1016/j.cell.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R., Zou Z., Wang W., Zou P. Metabolic incorporation of electron-rich ribonucleosides enhances APEX-seq for profiling spatially restricted nascent transcriptome. Cell Chem. Biol. 2022;29:1218–1231.e8. doi: 10.1016/j.chembiol.2022.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Bampton A., Gittings L.M., Fratta P., Lashley T., Gatt A. The role of hnRNPs in frontotemporal dementia and amyotrophic lateral sclerosis. Acta Neuropathol. 2020;140:599–623. doi: 10.1007/s00401-020-02203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y., Protter D.S.W., Rosen M.K., Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calabretta S., Richard S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem. Sci. 2015;40:662–672. doi: 10.1016/j.tibs.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Schreiber K.J., Kadijk E., Youn J.-Y. Exploring Options for Proximity-Dependent Biotinylation Experiments: Comparative Analysis of Labeling Enzymes and Affinity Purification Resins. J. Proteome Res. 2024;23:1531–1543. doi: 10.1021/acs.jproteome.3c00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannix K.M., Starble R.M., Kaufman R.S., Cooley L. Proximity labeling reveals novel interactomes in live Drosophila tissue. Development. 2019;146 doi: 10.1242/dev.176644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker J.T., Auerbach A.A., Harris R.S. APEX3 – An Optimized Tool for Rapid and Unbiased Proximity Labeling. J. Mol. Biol. 2023;435 doi: 10.1016/j.jmb.2023.168145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report the original code.