Abstract
Huntington's disease is caused by the expansion of CAG repeats coding for a polyglutamine tract in the huntingtin protein. The major pathological feature found in Huntington's disease neurons is the presence of detergent-insoluble ubiquitinated inclusion bodies composed of the huntingtin protein. However, the mechanisms that underlie inclusion body formation, and the precise relationship between inclusion bodies and events that initiate toxicity, remain unclear. Here, we analyzed the effects of drugs or genetic mutations that disrupt the microtubule cytoskeleton in a Saccharomyces cerevisiae model of the aggregation of an amino-terminal polyglutamine-containing fragment of huntingtin exon 1 (HtEx1). Treatment of yeast with drugs that disrupt microtubules resulted in less than 2% of the detergent-insoluble HtEx1 observed in mock-treated cells and prevented the formation of large juxtanuclear inclusion bodies. Disruption of microtubules also unmasked a potent glutamine length-dependent toxicity of HtEx1 under conditions where HtEx1 exists in an entirely detergent-soluble nonaggregated form. Results from the yeast model paralleled those from neuronal pheochromocytoma cells, where disruption of microtubules eliminated the formation of juxtanuclear and intranuclear inclusion bodies by HtEx1. Our results suggest that active transport along microtubules may be required for inclusion body formation by HtEx1 and that inclusion body formation may have evolved as a cellular mechanism to promote the sequestration or clearance of soluble species of HtEx1 that are otherwise toxic to cells.
Huntington's disease (HD) is caused by dominant mutation of a gene encoding the protein huntingtin, a 350-kDa protein of unknown but essential function. The genetic mutation that underlies HD, and at least eight other inherited neurodegenerative diseases, is the expansion of CAG triplet repeats coding for polyglutamine stretches in the affected proteins (reviewed in ref. 1). Expanded polyglutamine repeats are thought to result in conformational changes in the proteins that lead to misfolding, aggregation, inclusion body formation, and eventual neuronal cell death. Inclusion bodies composed of proteins with expanded polyglutamine repeats are observed in the brains of affected individuals and in cell and animal models of polyglutamine expansion diseases (reviewed in ref. 1). Despite evidence supporting a causal link among aggregation, inclusion body formation, and disease pathogenesis, it has not been directly established that protein aggregation and inclusion body formation are pathogenic cellular events.
Several lines of evidence suggest that protein aggregation and inclusion body formation in and of themselves are insufficient for initiation of disease pathogenesis. In a cellular model of huntingtin aggregation, treatment of cells with antiapoptotic compounds prevented neuronal cell death without suppressing inclusion body formation (2). In the same model, overexpression of a dominant negative mutant of the ubiquitin-conjugating enzyme Cdc34 significantly decreased inclusion body formation but increased cell death (2). In a mouse model of spinocerebellar ataxia type 2 (SCA-2), mice developed pathology in the absence of detectable inclusion bodies (3). Similarly, inclusion bodies were not observed in postmortem brain tissue from patients with SCA-2 (3). The most compelling evidence against a toxic role for inclusion bodies comes from Drosophila models of polyglutamine-induced neurodegeneration in which overexpression of chaperones prevented disease pathology in the absence of a visible effect on inclusion body formation (4, 5).
The subcellular localization of huntingtin with microtubules and synaptic vesicles has led to the proposal that huntingtin may function in vesicle trafficking (6, 7). Huntingtin cofractionates with microtubules in vitro through two rounds of assembly and disassembly (8). Huntingtin has also been linked indirectly to the microtubule cytoskeleton through its interactions with the huntingtin-associated protein HAP1, a neuronal protein that binds to huntingtin in a polyglutamine length-dependent manner (9). Biochemical and yeast two-hybrid data showed that HAP1 interacts with dynactin p150Glued, a component of the cytoplasmic dynein motor responsible for microtubule-dependent retrograde transport (10, 11). Collectively, these results implicate the microtubule cytoskeleton and microtubule-based transport as important cellular targets for modulating huntingtin localization, aggregation, and inclusion body formation in cells.
We reasoned that aggregation and inclusion body formation of huntingtin with an expanded polyglutamine repeat may be an active cellular process that requires an intact microtubule cytoskeleton. By using drugs or mutations in yeast that disrupt the microtubule cytoskeleton, we demonstrate here that assembled microtubules are indeed required for the formation of SDS-insoluble, juxtanuclear, and intranuclear inclusion bodies in yeast and neuronal models of huntingtin exon 1 (HtEx1) aggregation.
Materials and Methods
Yeast Strains, Plasmids, Media, and Reagents.
Unless otherwise noted, all chemical reagents were acquired from Sigma. Unless otherwise noted, all experiments were performed in the yeast strain DS10 (MATa GAL2 his3-11,15 leu2-3,112 lys1 lys2 Δtrp1 ura3-52; labeled TUB2 throughout the manuscript) (12). The genotype of the benomyl-resistant strain DBY2303 is MATα ura3-52 ade2-101 tub2-402 (13). The genotype of the tub4–1 strain is MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 tub4-1 (also known as ESM208) (14). The genotype of the tub4–32 strain is MATα his3Δ200 leu2-3, 112 lys2-801 tub4–32 ura3-52 (also known as TSY 498) (15). Cells were grown in synthetic complete media at 30°C. Yeast were transformed with YEp105-HD constructs (16). These constructs express exon 1 of the human huntingtin gene with 20, 39, or 53 glutamines under the control of the CUP1 promoter/CYC1 terminator, with a 15-aa linker at the N terminus that includes a c-myc epitope.
Experimental Protocol for Transient Disruption of Cytoskeletal Structures with Drugs in Yeast.
Cells transformed with the YEp105-HD constructs were grown to mid-log phase (OD600 0.5) in synthetic complete media and then treated with 400 μM CuSO4 and either 20 μg/ml benomyl, 100 μg/ml nocodazole, 100 μg/ml thiabendazole, 100 μM latrunculin A (Calbiochem), or 20 μg/ml cytochalasin B. After 3 h at 30°C, cells were harvested by centrifugation (for 5 min at 1,500 × g), washed with sterile water, and resuspended in fresh media lacking drugs but containing 400 μM CuSO4. Six hours after recovery from drug treatment, cells were harvested by centrifugation (for 5 min at 1,500 × g). Preparation of yeast cell lysates with glass beads and filter-trap assays were performed essentially as described (16). In brief, 10 μg of total yeast lysate was boiled in 2% SDS and loaded onto a cellulose acetate membrane with a 0.2 μm pore size that had been prewashed with 0.1% SDS. After 3 washes with 0.1% SDS, aggregates were detected by using the anti-c-myc Ab and the enhanced chemiluminescence reagent (Amersham Pharmacia Biotech). Double-label immunofluorescence was performed as described (16), except that monoclonal mouse anti-c-myc (clone 9E10, Santa Cruz Biotechnology), rabbit anti-Tub2 antisera (kind gift of Frank Solomon, Massachusetts Institute of Technology, Cambridge, MA), goat anti-rabbit IgG-FITC conjugate (Santa Cruz Biotechnology), and goat anti-mouse IgG-AlexaFluor594 conjugate (Molecular Probes) were used as primary and secondary Abs.
Expression of Green Fluorescent Protein (GFP)-HtEx1-104Q in Pheochromocytoma (PC12) Cells.
The mammalian expression plasmid pCDNA3-1 encoding GFP-tagged HtEx1 with 104 glutamine repeats (GFP-HtEx1-104Q), was a generous gift from Alexsey Kazantsev and David Housman (18). PC12 cells were transfected with pCDNA3-1-GFP-HtEx1-104Q by using lipofectamine transfection reagent (GIBCO/BRL) according to the manufacturer's protocol. Transfected cells were suspended in 2 ml of growth medium, seeded on glass coverslips, and analyzed for GFP fluorescence at 24 h after transfection.
Fluorescence Imaging of GFP-HtEx1-104Q in PC12 Cells.
Cells on coverslips were fixed and permeabilized as described (19). Cells were treated with 1 μM nocodazole or 50 ng/ml of cytochalasin D 18 h before fixation to disrupt the microtubule and the actin cytoskeleton, respectively. Cells were stained with rhodamine phalloidin (Molecular Probes) to visualize actin filament distribution. Cells were visualized with a Nikon fluorescence microscope and a Bio-Rad confocal scanning imaging system.
In Situ SDS Treatment of PC12 Cells That Express GFP-HtEx1-104Q.
The in situ SDS treatment of PC12 cells was performed essentially as described (18). Briefly, PC12 cells on coverslips that had been treated with DMSO, nocodazole, or cytochalasin D were fixed with 2% paraformaldehyde for 1 h at room temperature and then washed with PBS ± 0.1% SDS for 20 min at room temperature. Cells were then processed for fluorescence microscopy. For the quantification of inclusion bodies formed by PC12 cells that express GFP-HtEx1-104Q, at least 150 cells were counted (double-blind) for the presence of juxtanuclear or intranuclear inclusion bodies.
Results
An Intact Microtubule Cytoskeleton Is Required for the Aggregation of HtEx1-53Q in Yeast.
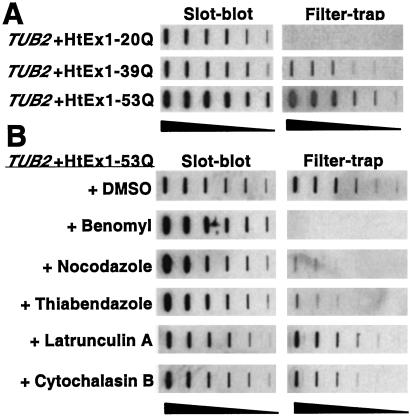
We first asked whether an intact microtubule or actin cytoskeleton is required for the aggregation of HtEx1 with an expanded polyglutamine repeat in a yeast model of HtEx1 aggregation. In this model, yeast cells express HtEx1 with a polyglutamine repeat in the normal (20Q) or disease-causing (39Q, 53Q) range (16). A membrane filter-trap assay was used to detect SDS-insoluble aggregates in yeast cells (17). Consistent with previous studies (16, 20), expression of HtEx1 in yeast resulted in a glutamine length-dependent aggregation only in lysates that express HtEx1 with a polyglutamine repeat in the expanded disease-causing range (≥ 39Q) (Fig. 1A).
Figure 1.
Requirement of an intact microtubule cytoskeleton for the formation of SDS-insoluble HtEx1-53Q aggregates in yeast. (A) Glutamine length-dependent aggregation of HtEx1 in yeast. Yeast cells (TUB2) were grown and induced for HtEx1 expression with CuSO4 as described in Materials and Methods. After 6 h of induction at 30°C, protein extracts were made and analyzed for HtEx1 expression in slot-blots and for SDS-insoluble aggregates in filter-trap assays. (B) Treatment of yeast with drugs that disrupt microtubules decreases the amount of SDS-insoluble HtEx1-53Q aggregates detected in filter-trap assays. Cells transformed with HtEx1-53Q were grown and treated transiently with microtubule (benomyl, nocodazole, thiabendazole) or actin (latrunculin A, cytochalasin B) drugs as described in Materials and Methods. After 6 h of recovery in media lacking drugs, protein lysates were made and analyzed by slot-blot and filter-trap assays as in A. Shown are two-fold serial dilutions starting with 10 μg of total lysate.
Cells that express HtEx1-53Q were next treated transiently with the microtubule-depolymerizing drugs benomyl, nocodazole, or thiabendazole, or with the actin filament-disrupting drugs latrunculin A or cytochalasin B. The effects of these drugs on HtEx1-53Q aggregation were evaluated by using the filter-trap assay. Treatment of cells with microtubule drugs caused a dramatic decrease (>98%) in the level of SDS-insoluble HtEx1-53Q aggregates detected by the filter-trap assay, although the expression level of HtEx1-53Q in microtubule drug-treated cells as monitored by slot-blot analyses was slightly higher than that observed in mock-treated cells (Fig. 1B). In contrast, treatment of cells with actin drugs caused a minimal effect on HtEx1-53Q aggregation and expression levels (Fig. 1B). Fluorescence microscopy of tubulin and actin staining in cells that expressed HtEx1-53Q showed that all drugs tested were effective at the doses used in these studies (data not shown).
An Intact Microtubule Cytoskeleton Is Required for the Juxtanuclear Localization of HtEx1-53Q Inclusion Bodies in Yeast.
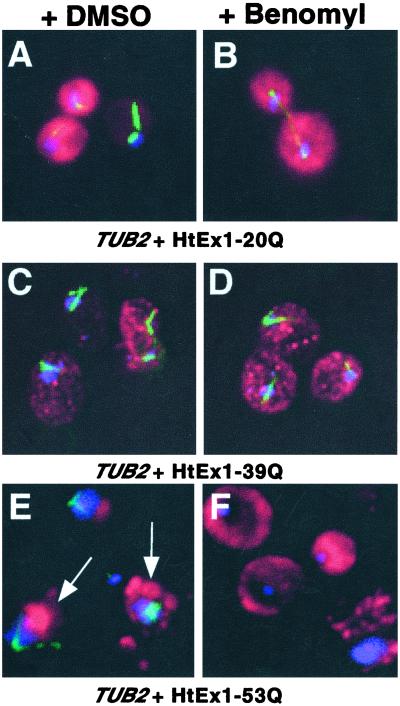
Immunofluorescence microscopy was used to visualize the staining pattern for HtEx1 in yeast in DMSO- (mock) and benomyl-treated cells (Fig. 2). In cells that express HtEx1 with a nonexpanded repeat (20Q), a diffuse cytoplasmic staining for HtEx1 was observed after recovery from treatment with DMSO or benomyl (Fig. 2 A and B). In DMSO- and benomyl-treated cells that express HtEx1-39Q, a glutamine repeat length that is on the border of causing disease, numerous small fluorescent foci dispersed throughout the cytoplasm were observed (Fig. 2 C and D). In contrast, in DMSO-treated cells that express HtEx1-53Q, large, punctate, and often juxtanuclear fluorescent structures were observed (Fig. 2E, see arrows), a pattern that is consistent with previous studies of HtEx1-53Q localization in yeast (16). Benomyl-treated cells that express HtEx1-53Q displayed two different fluorescence patterns: a diffuse cytoplasmic staining similar to cells that express HtEx1 with nonexpanded polyglutamine repeats (compare Fig. 2 F and B), and the staining of numerous smaller foci distributed throughout the cytoplasm, which resembled the fluorescence patterns observed in cells that express HtEx1-39Q (compare Fig. 2 D and F).
Figure 2.
An intact microtubule cytoskeleton is required for the juxtanuclear localization of HtEx1-53Q inclusion bodies in yeast. Cells treated transiently with DMSO (A, C, and E) or benomyl (B, D, and F) were harvested, fixed, and reacted with anti-c-myc and antitubulin Abs to visualize HtEx1 (red) and microtubules (green), respectively, and 4,6-diamino-2-phenylindole (DAPI) to show DNA (blue). (A and B) TUB2 + HtEx1-20Q, (C and D) TUB2 + HtEx1-39Q, (E and F) TUB2 + HtEx1-53Q. The arrows shown in E denote the juxtanuclear localization of HtEx1-53Q.
Disruption of Microtubules with Drugs in Yeast Cells That Express HtEx1 Unmasks a Glutamine Length-Dependent Toxicity.
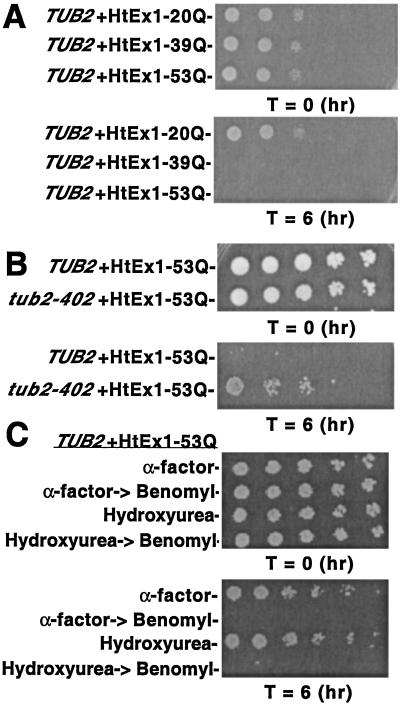
Under standard growth conditions, expression of HtEx1 with normal or expanded polyglutamine repeats has no effect on cell viability in yeast (16, 20). However, we observed that transient treatment with benomyl resulted in a glutamine length-dependent toxicity that was observed only for HtEx1 that contains expanded polyglutamine repeats in the disease-causing range (Fig. 3A). Treatment of cells with drugs that disrupt the actin cytoskeleton had no effect on viability (data not shown).
Figure 3.
Transient depolymerization of microtubules with benomyl results in a glutamine length-dependent cell cycle-independent toxicity of HtEx1 in yeast. (A and B) Yeast cells (TUB2 or tub2-402) were treated transiently with 20 μg/ml of benomyl to depolymerize microtubules as described in Materials and Methods. (C) Yeast cells (TUB2) were grown and treated with 5 μM α-factor or 0.1 M hydroxyurea to arrest cells at G1 and S phases of the cell cycle, respectively. After 3 h the cells were harvested, washed with sterile water, and resuspended in fresh media with and without 20 μg/ml of benomyl. Cells without benomyl treatment were allowed to recover for 6 h and then were tested for viability in spotting assays. After 3 h, benomyl-treated cells were harvested, washed in sterile water, resuspended in fresh media, and allowed to recover for 6 h. (A–C) Shown are 5-fold serial dilutions starting with equal numbers of cells before (T = 0) and after (T = 6) drug treatment/CuSO4 induction. Cells were spotted on plates containing synthetic complete media ± 400 μM CuSO4 and were incubated at 30°C for 3 days.
A benomyl-resistant yeast strain (tub2-402) that expresses HtEx1-53Q displayed normal viability after benomyl treatment, demonstrating that the glutamine-length dependent toxicity observed in yeast required the specific action of benomyl to depolymerize microtubules (Fig. 3B). The tub2-402 strain contains a single point mutation in the TUB2 gene, rendering it resistant to the effects of benomyl at the concentrations used in these studies (13). The level of SDS-insoluble aggregates observed by filter-trap assays in tub2-402 cells that express HtEx1-53Q after transient benomyl treatment was similar to that observed in its mock-treated parental control strain (data not shown).
Benomyl treatment is known to activate the mitotic checkpoint in budding yeast and arrest cells at M phase in the cell cycle (21). Quantification of cell morphology by phase-contrast microscopy demonstrated that the majority of benomyl-treated cells that express HtEx1-53Q remained arrested as large budded cells at the mitotic checkpoint 24 h after recovery from treatment (64% vs. 31% for cells that express HtEx1-20Q and were also treated transiently with benomyl) (data not shown). These results suggest that the observed loss of cell viability is not likely to be caused by a weakening of the mitotic checkpoint machinery.
In an effort to understand the mechanism of toxicity in our yeast model, we next tested whether accumulation of HtEx1-53Q was toxic to yeast that were arrested at different stages during the cell cycle. Cells that express HtEx1-53Q were treated with the yeast mating pheromone α-factor or DNA replication inhibitor hydroxyurea, which cause arrests at G1 and S phases in the cell cycle, respectively. Cell morphology determined by phase contrast microscopy indicated that α-factor and hydroxyurea were effective at inducing their respective cell cycle arrests (data not shown). Expression of HtEx1-53Q in cells arrested at G1 or S phases by α-factor or hydroxyurea, respectively, had only a modest effect on cell viability (Fig. 3C). Nevertheless, when cells that express HtEx1-53Q were first treated with α-factor or hydroxyurea, then released to media containing benomyl, and finally allowed to recover in media lacking drugs for 6 h, a potent decrease in cell viability was observed (Fig. 3C). Collectively, these results suggest that toxicity of HtEx1-53Q in our yeast model of HtEx1 aggregation does not depend on the cell cycle, or cell cycle arrests, but does depend on the depolymerization of microtubules.
Yeast with Temperature-Sensitive (Ts) Mutations of TUB4 (γ-tubulin) Accumulate Lower Levels of SDS-insoluble HtEx1-53Q.
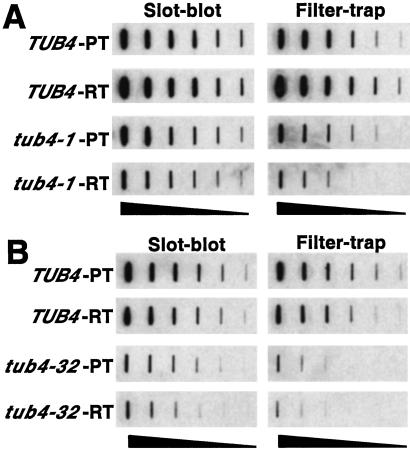
The gene encoding γ-tubulin in Saccharomyces cerevisiae, TUB4, is essential and Tub4 is a major component of the spindle pole body (SPB), the cellular structure required for organizing and nucleating cytoplasmic and nuclear microtubules in budding yeast. Several Ts mutants of TUB4 that have been described have defects in microtubule organization and nucleation from the SPB (14, 15). HtEx1-53Q was expressed in wild-type (TUB4) and mutant (tub4-1, tub4-32) yeast strains for 6 h at the permissive (23°C) and restrictive (37°C) temperatures, after which filter-trap assays were performed on cell lysates to quantify SDS-insoluble aggregates (Fig. 4). Although TUB4 and tub4-1 cells accumulated approximately equivalent levels of SDS-insoluble HtEx1-53Q at the permissive temperature, at least an 88% decrease (3 2-fold dilutions) in the level of SDS-insoluble HtEx1-53Q was observed in tub4-1 cells shifted to the restrictive temperature in comparison to TUB4 cells (Fig. 4A). A more severe effect was observed in tub4-32 cells, where approximately a 97% decrease in the level of SDS-insoluble HtEx1-53Q was observed in comparison to TUB4 cells at the restrictive temperature (Fig. 4B). The expression levels of HtEx1-53Q in TUB4 and tub4-1 cells were approximately equivalent at the permissive temperature as judged by slot-blot analysis of total cell lysates (Fig. 4A). Although the expression levels of HtEx1-53Q in tub4-1 and tub4-32 cells were slightly lower than their TUB4 counterparts at the restrictive temperatures, the levels were still sufficient to form detectable SDS-insoluble aggregates in these strains (Fig. 4 A and B).
Figure 4.
Temperature-sensitive yeast strains with mutations of TUB4 (encoding γ-tubulin) accumulate lower levels of SDS-insoluble HtEx1-53Q. Yeast cells (TUB4, tub4-401, or tub4-32) that express HtEx1-53Q were grown in synthetic complete media at 23°C to mid-log phase (OD600 0.5) at which point 400 μM CuSO4 was added to the media to induce HtEx1-53Q expression. After 6 h of incubation at 23°C (permissive temperature, PT) or 37°C (restrictive temperature, RT), protein extracts were prepared by glass bead lysis and analyzed for HtEx1-53Q expression in slot-blots and for SDS-insoluble aggregates in filter-trap assays as in Fig. 1A. Shown are 2-fold serial dilutions starting with 10 μg of total lysate for tub4-1 and parental control strain TUB4 (A) and tub4-32 and parental control strain TUB4 (B).
An Intact Microtubule Cytoskeleton Is Required for Juxtanuclear and Intranuclear Inclusion Body Formation by GFP-HtEx1-104Q in Neuronal PC12 Cells.
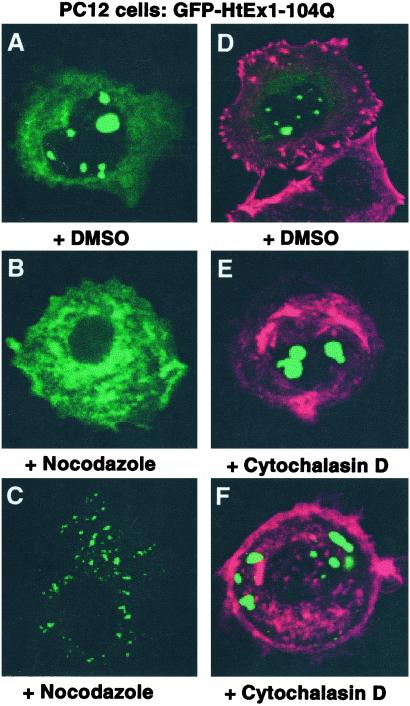
The effects of drugs that disrupt the microtubule and actin cytoskeleton were also evaluated in a PC12 model of HtEx1 aggregation (Fig. 5). Cells were transfected with a construct (pCDNA3-1-GFP-HtEx1-104Q) that expresses HtEx1 with 104 glutamines fused to GFP under the control of a cytomegalovirus-based promoter (18). Six hours after transfection, cells were treated with DMSO, nocodazole, or cytochalasin D. After 18 h of incubation at 37°C, cells were fixed and processed to visualize GFP (labeled green) and actin (labeled red) fluorescence (Fig. 5). In cells treated with DMSO, GFP-HtEx1-104Q fluorescence appears in large, punctate, juxtanuclear, and intranuclear structures (Fig. 5 A and D). A low level of background cytoplasmic fluorescence is also observed in these cells. In contrast, nocodazole-treated cells that express GFP-HtEx1-104Q displayed two different fluorescence patterns: a diffuse cytoplasmic staining (Fig. 5B) similar to cells that express HtEx1 with nonexpanded polyglutamine repeats (data not shown), and the staining of numerous smaller foci distributed throughout the cytoplasm but not in the nucleus (Fig. 5C). Similar results were observed when Chinese hamster ovary cells that express GFP-HtEx1-104Q were treated with nocodazole (data not shown). At the concentration used in these experiments, nocodazole was effective at depolymerizing microtubules (data not shown). PC12 cells that express GFP-HtEx1-104Q were also treated with cytochalasin D, which targets actin, in a manner identical to that described for nocodazole. Although cytochalasin D was fully active at disrupting actin filament organization, it had no observable effect on the localization or formation of inclusion bodies by GFP-HtEx1-104Q (Fig. 5 D–F). The expression level of GFP-HtEx1-104Q in PC12 cells treated with DMSO, nocodazole, or cytochalasin D was approximately equivalent as determined by Western immunoblots (data not shown).
Figure 5.
An intact microtubule cytoskeleton is required for the juxtanuclear and intranuclear localization of GFP-HtEx1-104Q inclusion bodies in PC12 cells. PC12 cells were transfected with pCDNA3-1-GFP-HtEx1-104Q that expresses HtEx1 with 104 glutamine repeats fused to GFP under the control of a cytomegalovirus-based promoter (18). Six hours after transfection, cells were treated with DMSO (A and D), nocodazole (B and C), or cytochalasin D (E and F). After 18 h of incubation at 37°C, cells were fixed and analyzed for GFP (green) or actin (red) fluorescence. B and C and E and F represent two independent examples of PC12 cells that express GFP-HtEx1-104Q and were treated with nocodazole or cytochalasin D, respectively.
We next evaluated the effects of SDS on the in situ fluorescence pattern of PC12 cells that expressed GFP-HtEx1-104Q and were treated with DMSO or nocodazole. In a previous study with the identical expression vector, a striking resistance was reported to in situ solubilization with SDS for inclusion bodies formed by GFP-HtEx1-104Q in mammalian cells (18). Cell counts were performed to quantify the effects of nocodazole and SDS treatments on juxtanuclear and intranuclear inclusion body formation by GFP-HtEx1-104Q in PC12 cells (Table 1). Nocodazole treatment resulted in a statistically significant decrease in the number of inclusion bodies observed in PC12 cells in comparison to cells treated with DMSO alone (Table 1), whereas treatment with the actin drug cytochalasin D was without effect (data not shown). A similar effect was also observed if nocodazole-treated cells were washed in situ with SDS (Table 1).
Table 1.
Quantification of GFP-HtEx1-104Q fluorescence in PC12 cells
| Treatment
|
Inclusion bodies
|
||
|---|---|---|---|
| Nocodazole | SDS | Juxtanuclear | Intranuclear |
| – | – | 32.6 ± 2.3 | 4.8 ± 0.6 |
| – | + | 31.5 ± 0.9 | 4.7 ± 0.6 |
| + | – | 9.7 ± 0.5 | 0.5 ± 0.1 |
| + | + | 4.9 ± 1.8 | 0.1 ± 0.1 |
Shown is the percentage of cells that exhibited fluorescence staining for large, juxtanuclear, and intranuclear aggregates. The results shown are the means and SDs from three independent experiments where ≥150 cells were counted (double-blind) for each experimental condition.
Discussion
The precise relationship between inclusion bodies and neuronal pathology in HD remains controversial. Inclusion bodies have been suggested to be intimately involved in disease pathology, to be benign side-products of the cellular machinery that neither help nor hinder disease pathogenesis, or even to serve a protective function (1, 22). Although a specific subset of striatal neurons is most affected in HD, huntingtin inclusion bodies are also observed in nonaffected regions of the brain and even in nonneuronal cells (23–25). These results suggest that the cellular machinery and mechanisms involved in huntingtin aggregation and inclusion body formation are not specific to the neurons that are most vulnerable in HD. Despite numerous studies of huntingtin aggregation in diverse cell types, it has yet to be established whether aggregation and inclusion body formation of the huntingtin protein is an active process regulated by cellular machinery or occurs purely by a diffusion-limited process. Here, we provide evidence that an intact microtubule cytoskeleton is critically required for the formation of SDS-insoluble aggregates and inclusion bodies in yeast and neuronal models of HtEx1 aggregation. Disassembly of microtubules by drugs or genetic mutations in yeast unmasks a glutamine length-dependent toxicity that is attributable to an entirely soluble nonaggregated form of HtEx1. These results suggest that inclusion body formation may have evolved as an active cellular defense mechanism to protect against the toxic effects of the soluble mutant HtEx1.
Microtubules are required for the formation of perinuclear inclusion bodies by the cystic fibrosis transmembrane conductance regulator (CFTR), the protein affected in this disease, and by mutant forms of superoxide dismutase (SOD) that cause familial amyotrophic lateral sclerosis (26–28). Johnston et al. (26, 27) named the inclusion bodies formed by CFTR and mutant SOD “aggresomes.” The structures formed by HtEx1 with expanded polyglutamine repeats in our cell models of HtEx1 aggregation are similar to aggresomes, and our results support the hypothesis that aggresome formation may represent a general cellular response to the presence of an excess of misfolded proteins (26). Our studies also suggest somewhat surprisingly that an intact microtubule cytoskeleton may also be required for the formation of intranuclear inclusion bodies composed of expanded HtEx1 in neuronal cells. Although the mechanisms underlying nuclear transport and inclusion body formation of HtEx1 remain uncharacterized, it is possible that disassembly of microtubules prevents the nuclear accumulation of HtEx1 to a concentration sufficient to initiate inclusion body formation, or perhaps microtubules may transport other factors required for the intranuclear aggregation of HtEx1.
If inclusion body formation in HD indeed represents an active cellular process, it still cannot be excluded that inclusion bodies are the toxic species that initiate disease pathogenesis. In a cell-based model, aggresome formation by a cytosolic protein chimera (GFP-250) led to disruption of the normal architecture of the Golgi apparatus and disorganization of microtubule arrays (29), suggesting that microtubule-based trafficking may be impaired in the presence of an aggresome. It has also recently been demonstrated that juxtanuclear inclusion bodies formed by HtEx1 completely inhibit proteasome function in mammalian cells (30). Furthermore, although our results suggest that cells may normally attempt to remove toxic misfolded proteins by forming inclusion bodies, this process may inadvertently sequester other proteins such as chaperones or transcription factors that are critical for normal cellular functions.
Despite these results, it is important to note that in several animal models of neurodegenerative disease caused by polyglutamine expansion, cell dysfunction is known to precede the appearance of inclusion bodies and pathology, suggesting that inclusion body formation may occur only after the toxic insult has already taken place (31–33). Consistent with these observations, we showed that a completely SDS-soluble and apparently nonaggregated form of HtEx1-53Q confers cellular toxicity in the absence of assembled microtubules in yeast. Disassembly of microtubules in our PC12 model of HtEx1 aggregation also resulted in a small but reproducible increase in polyglutamine length-dependent toxicity (data not shown). Although other investigators have reported varying degrees of polyglutamine length-dependent toxicity in the presence of assembled microtubules in animal cells, the mechanisms underlying toxicity remain poorly understood. Future experiments will be required to determine whether the toxicity we observe in the absence of assembled microtubules shares similarities to that observed in other cellular models of huntingtin aggregation. Collectively, our results are consistent with the hypothesis that the formation of early misfolding intermediates of huntingtin, which are detergent-soluble, is linked to initiation of disease pathogenesis.
Acknowledgments
We thank Tim Stearns, Elmar Schiebel, and Tim Huffaker for kindly providing yeast strains; Frank Solomon for the β-tubulin antisera; and Michael DeVit for useful comments and suggestions. P.J.M. and C.D.S. are supported by the Hereditary Disease Foundation under the auspices of the “Cure HD” Initiative. S.F. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- HD
Huntington's disease
- HtEx1
huntingtin exon 1
- GFP
green fluorescent protein
- PC12
pheochromocytoma
References
- 1.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 3.Huynh D P, Figueroa K, Hoang N, Pulst S M. Nat Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- 4.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 5.Kazemi-Esfarjani P, Benzer S. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 6.Gutekunst C A, Levey A I, Heilman C J, Whaley W L, Yi H, Nash N R, Rees H D, Madden J J, Hersch S M. Proc Natl Acad Sci USA. 1995;92:8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velier J, Kim M, Schwarz C, Kim T W, Sapp E, Chase K, Aronin N, DiFiglia M. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- 8.Tukamoto T, Nukina N, Ide K, Kanazawa I. Brain Res Mol Brain Res. 1997;51:8–14. doi: 10.1016/s0169-328x(97)00205-2. [DOI] [PubMed] [Google Scholar]
- 9.Li X J, Li S H, Sharp A H, Nucifora F C, Jr, Schilling G, Lanahan A, Worley P, Snyder S H, Ross C A. Nature (London) 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 10.Engelender S, Sharp A H, Colomer V, Tokito M K, Lanahan A, Worley P, Holzbaur E L, Ross C A. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 11.Li S H, Gutekunst C A, Hersch S M, Li X J. J Neurosci. 1998;18:1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 13.Huffaker T C, Thomas J H, Botstein D. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spang A, Geissler S, Grein K, Schiebel E. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marschall L G, Jeng R L, Mulholland J, Stearns T. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muchowski P J, Schaffar G, Sittler A, Wanker E E, Hayer-Hartl M K, Hartl F U. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. . (First Published June 20, 2000; 10.1073/pnas.140202897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanker E E, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 18.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Souza-Schorey C, Li G, Colombo M I, Stahl P D. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 20.Krobitsch S, Lindquist S. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straight A F, Murray A W. Methods Enzymol. 1997;283:425–440. doi: 10.1016/s0076-6879(97)83035-2. [DOI] [PubMed] [Google Scholar]
- 22.Tobin A J, Signer E R. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- 23.Kuemmerle S, Gutekunst C A, Klein A M, Li X J, Li S H, Beal M F, Hersch S M, Ferrante R J. Ann Neurol. 1999;46:842–849. [PubMed] [Google Scholar]
- 24.Gutekunst C A, Li S H, Yi H, Mulroy J S, Kuemmerle S, Jones R, Rye D, Ferrante R J, Hersch S M, Li X J. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sathasivam K, Hobbs C, Turmaine M, Mangiarini L, Mahal A, Bertaux F, Wanker E E, Doherty P, Davies S W, Bates G P. Hum Mol Genet. 1999;8:813–822. doi: 10.1093/hmg/8.5.813. [DOI] [PubMed] [Google Scholar]
- 26.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston J A, Dalton M J, Gurney M E, Kopito R R. Proc Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. . (First Published October 24, 2000; 10.1073/pnas.220417997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wigley W C, Fabunmi R P, Lee M G, Marino C R, Muallem S, DeMartino G N, Thomas P J. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Mata R, Bebok Z, Sorscher E J, Sztul E S. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bence N F, Sampat R M, Kopito R R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 31.Reddy P H, Williams M, Charles V, Garrett L, Pike-Buchanan L, Whetsell W O, Jr, Miller G, Tagle D A. Nat Genet. 1998;20:198–202. doi: 10.1038/2510. [DOI] [PubMed] [Google Scholar]
- 32.Hodgson J G, Agopyan N, Gutekunst C A, Leavitt B R, LePiane F, Singaraja R, Smith D J, Bissada N, McCutcheon K, Nasir J, et al. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 33.Guidetti P, Reddy P H, Tagle D A, Schwarcz R. Neurosci Lett. 2000;283:233–235. doi: 10.1016/s0304-3940(00)00956-3. [DOI] [PubMed] [Google Scholar]