Abstract
Actin cables, bundles of actin filaments that align along the long axis of budding yeast, are crucial for establishment of cell polarity. We fused green fluorescent protein (GFP) to actin binding protein 140 (Abp140p) and visualized actin cable dynamics in living yeast. We detected two populations of actin cables: (i) bud-associated cables, which extend from the bud along the mother-bud axis, and (ii) randomly oriented cables, which are relatively short. Time-lapse imaging of Abp140p–GFP revealed an apparent increase in the length of bud-associated actin cables. Analysis of movement of Abp140p–GFP fiduciary marks on bud-associated cables and fluorescence loss in photobleaching experiments revealed that this apparent elongation occurs by assembly of new material at the end of the cable within the bud and movement of the opposite end of the cable toward the tip of the mother cell distal to the bud. The rate of extension of the tip of an elongating actin cable is 0.29 ± 0.08 μm/s. Latrunculin A (Lat-A) treatment completely blocked this process. We also observed movement of randomly oriented cables around the cortex of cells at a rate of 0.59 ± 0.14 μm/s. Mild treatment with Lat-A did not affect the velocity of movement of randomly oriented cables. However, Lat-A treatment did increase the number of randomly oriented, motile cables per cell. Our observations suggest that establishment of bud-associated actin cables during the cell cycle is accomplished not by realignment of existing cables but by assembly of new cables within the bud or bud neck, followed by elongation.
Cross-linked bundles of actin filaments exist in many cell types, including the bristle and follicular nurse cell struts of Drosophila, the brush border of intestinal epithelial cells, the stereocilium of hair cells in the vertebrate ear, and the fertilization cone of the sea urchin egg. In all of these cases, the barbed end of the polarized actin bundle is embedded in a dense substance, and the bundle is assembled from this end (1). A similar structure may exist in budding yeast. Actin cables are bundles of filamentous actin (F-actin) that localize to the cell cortex (2, 3). Because myosin V proteins, barbed-end directed motor proteins (4), use actin cables as tracks for polarized particle movement, it is likely that actin cables consist of parallel filaments (5–8). Here, we describe studies showing dynamics of cross-linked bundles of F-actin in living yeast.
Actin cables are randomly distributed in nondividing yeast, but are oriented along the mother-bud axis during polarized growth from late G1 to M phase (2, 3). Studies with tropomyosin mutants, which show temperature-dependent loss of actin cables, implicate actin cables in polarized growth and polarized localization of actin patches, secretory vesicles, and the mitotic spindle (7–9). Furthermore, actin cables may function as tracks for movement of components from mother cell to bud, including ASH1 mRNA, vacuoles, mitochondria, and elements required for spindle alignment (5, 6, 8, 10).
Despite the critical roles of actin cables in cell polarity, the mechanism underlying cable formation and localization in vivo remains elusive. Latrunculin-A (Lat-A), an actin monomer-sequestering agent, disrupts actin cables within several minutes (11). This finding suggests that continuous polymerization and depolymerization of F-actin occurs in actin cables (1). In addition, previous studies revealed F-actin bundling proteins including tropomyosin (Tpm1p and Tpm2p) (12, 13) and fimbrin (Sac6p) (14) within actin cables. Fimbrin is important for organization and stabilization of cables, and tropomyosins are important for stabilizing cables (15).
Recently Asakura et al. (16) discovered that actin binding protein 140 (Abp140p) is an F-actin binding protein in budding yeast. Abp140p does not show homology to any known actin binding protein and is expressed in cells by fusion of two ORFs (YOR239W and YOR240W) by means of a +1 translational frameshift. Abp140p crosslinks F-actin in vitro and colocalizes with actin patches and cables. Cells carrying a deletion in ABP140 showed no abnormal phenotype.
To understand the mechanism underlying assembly and rearrangement of cables, we visualized actin cables in living yeast by using Abp140p fused to GFP. Here, we show the utility of GFP-tagged Abp140p for probing actin cables and report several aspects of actin cable dynamics in budding yeast.
Materials and Methods
DNA Manipulations for Green Fluorescent Protein (GFP) Tagging of Abp140p.
The C terminus of Abp140p was tagged with GFP (S65T) by using PCR-based insertion into the chromosomal copy of ABP140 (19). A PCR fragment containing regions homologous to the 3′ end of ABP140, and coding regions for GFP and the kanamycin resistance marker (KanMX6), was amplified from plasmid pFA6a-GFP (S65T)-kanMX6 with forward primer, 5′-GTACCGCTGCTGGGTACAAGCTGTGTTTGACGTTCCTCAAGCTGCTGCTGCTGCTCGGATCCCCGGGTTAATTAA-3′, and reverse primer, 5′-ATGATGAGAGAGGAGGTGGTACTTGTCTCAGAACTTCCTAGAATTCGAGCTCGTTTAAAC-3′. HA10–1b, ABY971, and ABY973 cells were transformed with the PCR product by using the lithium acetate method (18), and transformants that had integrated the PCR product were selected by growth on yeast extract/peptone/dextrose containing 200 μg/ml geneticin. The GFP-tagged yeast strains were characterized for correct integration of the tagging cassette at the ABP140 locus by using PCR and Western blot analysis. Fluorescence imaging of Abp140p–GFP was carried out as described (19).
Fluorescence Loss in Photobleaching (FLIP).
Samples were illuminated for image acquisition and photobleaching with a 25-mW argon laser on a Zeiss LSM510 laser scanning microscope. The laser was used at 0.1% and 6.2% of maximum power for image acquisition and photobleaching, respectively. The optical section thickness was 1 μm. An area of 1 μm2 was photobleached at a rate of 8 iterations per s at 5-s intervals. Images were collected during the intervals between each round of photobleaching. The time for each image acquisition was 1 s.
Quantitation of FLIP was performed by using LSM510 software (Zeiss). The relative fluorescence intensity (RFI) of the region of interest (ROI) is (Ft − Fb)/(F0 − Fb) × 100. F0 and Ft are the average fluorescence intensities of the ROI before photobleaching and at the various time points during the experiment. Fb is the background fluorescence intensity in areas adjacent to the ROI at each time point of image acquisition. The time needed to reach 50% of RFI (t1/2) was calculated by using a polynomial regression equation fitted to each data set. The average coefficient of determination in regression was 0.995.
Other.
All other methods including yeast strains used for this study, cell fixation and staining, and Lat-A treatment are published as supporting information on the PNAS web site, www.pnas.org.
Results
Abp140p–GFP Labels Actin Cables in Living Yeast.
We expressed Abp140p–GFP by tagging chromosomal ABP140 at its C terminus with GFP. Because ABP140–GFP was expressed in lieu of endogenous ABP140 and under control of the endogenous ABP140 promoter, it is likely that the tagged protein was expressed at wild-type levels. Expression of this Abp140p–GFP fusion protein had no obvious effect on cell size, shape, or growth rate (data not shown). Moreover, actin organization and structure were not affected in Abp140p–GFP-expressing cells (see below). Therefore, insertion of GFP into the C terminus of Abp140p does not appear to have a deleterious effect on cells.
To evaluate association of Abp140p–GFP with actin cytoskeletal structures in yeast, we compared fluorescence of Abp140p–GFP with F-actin stained by rhodamine phalloidin. Optical sections through the entire cell were acquired and each z-series was deconvolved, reconstructed, and projected onto a single image plane (Fig. 1A). Rhodamine-phalloidin staining of F-actin in fixed yeast revealed brightly stained actin patches and weakly stained actin cables. Although Abp140p–GFP localized to the same structures, Abp140p–GFP is a better marker for actin cables than for actin patches. Fluorescence from Abp140p–GFP in actin patches was significantly lower than that in actin cables.
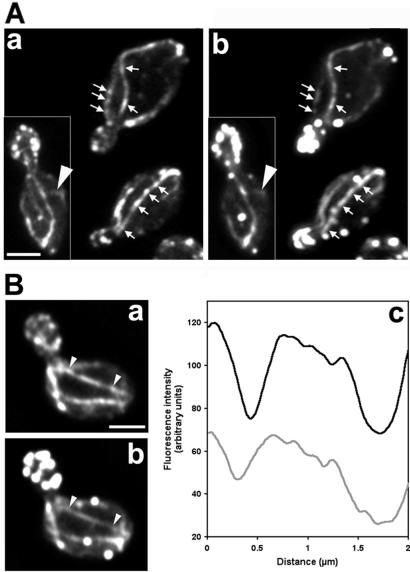
Figure 1.
(A) Colocalization of Abp140p–GFP (a) and the actin cytoskeleton (b). Cells (YCY006) expressing Abp140p–GFP were grown to midlog phase at room temperature (22–24°C) in lactate medium, fixed, and stained with rhodamine phalloidin to visualize F-actin structures. Fluorescence images of GFP and rhodamine were obtained at 0.3-μm z-intervals through the entire cell, deconvolved, and reconstructed. A two-dimensional projection of the three-dimensional reconstruction is shown. Arrows indicate regions along bud-associated actin cables that are brightly labeled with rhodamine phalloidin and Abp140p–GFP. Arrowhead shows a randomly oriented actin cable. Punctate structures in the mother cell and bud that show strong staining with rhodamine phalloidin and weak fluorescence with Abp140p–GFP are actin patches. (B) Fluorescence intensity of Abp140p–GFP and rhodamine phalloidin along an actin cable. The fluorescence images were obtained as in A, and fluorescence intensity was measured along the cable of Abp140p–GFP (a) and F-actin (b). The black and gray lines in c represent intensity of Abp140p–GFP and rhodamine phalloidin, respectively. The size of the area defining the actin cable for the measurement is 0.28 × 2.0 μm. (Bar, 1.5 μm.)
Further analysis of actin cables revealed two populations of fluorescent cables. Most cables were polarized in their orientation and associated with buds: they aligned along the mother-bud axis. In contrast to these bud-associated cables, cables without apparent polarity in their orientation were also detected. These randomly oriented cables were less abundant and shorter (≤1.5 μm in haploid yeast) than bud-associated cables and did not align along the mother-bud axis (arrowhead Fig. 1A).
Analysis of actin cables revealed that the intensity of fluorescence from rhodamine phalloidin was not uniform along the cables. Regions of strong and weak staining along actin cables were of variable length and periodicity and may reflect nonuniform amounts of F-actin along the cables, which was previously observed by Karpova et al. (20). As shown in Fig. 1A, Abp140p–GFP fluorescence variation along actin cables was very similar to that observed with rhodamine phalloidin. The position of bright spots of Abp140p–GFP on the cables always coincided with increased intensity of rhodamine phalloidin-stained F-actin. Quantitation showing parallel changes in fluorescence of Abp140p–GFP and rhodamine phalloidin along an actin cable (Fig. 1B) supports the notion that the amount of Abp140p–GFP is proportional to the amount of F-actin in cables.
Dependence of Abp140p–GFP Localization on Actin Cables.
F-actin structures are sensitive to Lat-A, an agent that produces loss of actin cables and patches within 2 min of exposure (11). We found that treatment of Abp140p–GFP-expressing cells with 200 μM Lat-A resulted in complete loss of fluorescent cable and patch structures and dispersion of Abp140p–GFP throughout the cytoplasm (Fig. 2A). Thus, localization of Abp140p–GFP fluorescence in bud-associated and randomly oriented cables depends on F-actin. Moreover, titration studies indicate that the Lat-A sensitivity of actin cables is similar in cells expressing tagged and untagged Abp140p (data not shown). Therefore, Abp140p–GFP does not have any obvious effect on actin cable dynamics.
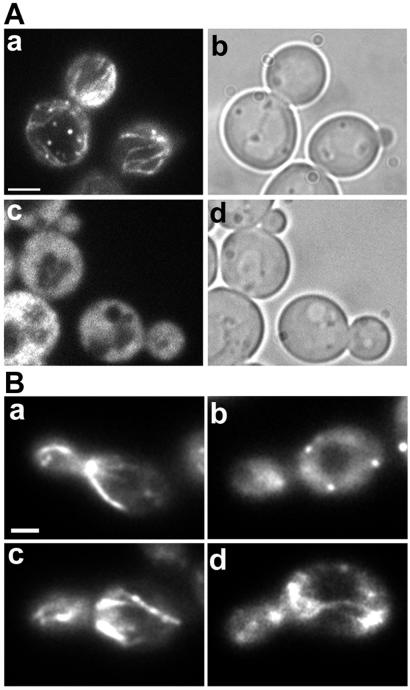
Figure 2.
(A) Abp140p–GFP localization to cable-like structures in living yeast is sensitive to Lat-A. Cells were grown as in Fig. 1 and incubated with 200 μM Lat-A dissolved in DMSO (c and d), or an equivalent amount (0.5%) of DMSO (a and b) for 2 min. Abp140p–GFP fluorescence (a and c) and phase-contrast images (b and d) are shown. (Bar, 1.5 μm.) (B) Destabilization of actin cables correlates with loss of Abp140p–GFP in cable-like structures in living yeast. Abp140p–GFP fluorescence in the actin cable mutant (tpm1–2 tpmΔ2) and control strain (TPM1 tpmΔ2) at 25°C (a and c, respectively) and 1 min after a shift from 25°C to 34.5°C (b and d). (Bar, 1 μm.)
To confirm localization of Abp140p to actin cables, we also tested the effect of specific loss of actin cables on Abp140p–GFP distribution in yeast. To do so, we took advantage of previous studies on the two tropomyosin proteins (Tpm1p and Tpm2p) of yeast (7). Those studies revealed that a cell carrying a mutation in the TPM1 gene and a deletion in the TPM2 gene (tpm1–2/tpm1–2 tpm2Δ/tpm2Δ) undergoes rapid loss of actin cables after shift to from permissive to restrictive temperatures. We will refer to this strain as the actin cable mutant. Because deletion of TPM2 alone has no obvious effect on actin structure or function, the isogenic strain carrying the TPM2 deletion (TPM1/TPM1 tpm2Δ/tpm2Δ) was used as the control for these studies.
In contrast to the control strain, which retained most cables after the temperature shift, the actin cable mutant underwent rapid, nearly complete loss of cables after shift to the restrictive elevated temperature (Fig. 2). Quantitation of the loss of Abp140p–GFP-labeled cables was performed after fixation of cells. At 25°C, 62.3% (n = 130) and 93.2% (n = 118) of cells showed GFP-labeled cables in actin cable mutant and control strains, respectively. When the cells were shifted to 34.5°C for 1 min, 100% of actin cable mutant cells (n = 155) completely lost GFP cables whereas the cables were still visible in 72.6% of the control strain (n = 105). The rapid disappearance of Abp140p–GFP cables in the actin cable mutant indicates that localization of Abp140p–GFP depends on structural integrity of actin cables. From these results, we concluded that Abp140p–GFP could be used as a probe for actin cables in living yeast.
Extension of Bud-Associated Actin Cables.
We used time-lapse imaging of Abp140p–GFP to study the dynamics of bud-associated actin cables in living yeast. Abp140p–GFP fluorescence was faint and photobleached easily. Therefore, it was difficult to acquire large numbers of images necessary for four-dimensional imaging (i.e., time-lapse imaging combined with three-dimensional reconstruction). Nonetheless, because actin cables are restricted to the cell cortex, it was possible to image entire actin cables within a single optical plane.
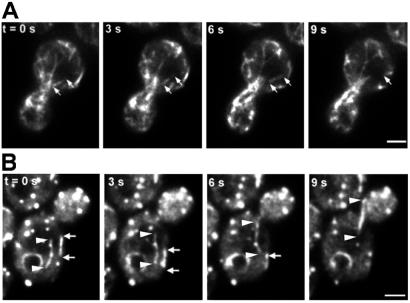
First, we observed an apparent increase in the length of bud-associated actin cables. The cable end nearer the bud neck did not appear to change position. However, we detected apparent extension of the other end of the actin cable toward the tip of the mother cell distal to the bud (Fig. 3A; Movie 1, which is published as supporting information on the PNAS web site). The rate of this extension was 0.29 ± 0.08 μm/s (n = 25).
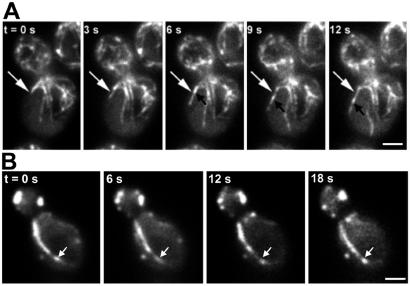
Figure 3.
Dynamics of bud-associated actin cables. (A) Extension of an Abp140p–GFP cable. Cells were grown as in Fig. 1. Fluorescence images of Abp140p–GFP were collected at a single plane of focus at 1.5-s intervals. White arrows point to the position of the tip of the elongating cable at 0 s. Black arrows indicate a fiduciary mark on the elongating cable that moves toward the end of the mother cell distal to the bud. (Bar, 1 μm.) (B) Immobilization of bud-associated actin cables in Lat-A-treated cells. Lat-A was added to a final concentration of 35 μM to a midlog phase liquid culture (OD600 = 1.0), and an aliquot of the cell suspension was placed on a microscope slide. Time-lapse images of Abp140p–GFP-labeled actin cables in a budding cell were acquired as in A. The total time from addition of Lat-A to acquisition of the first image was 2 min. Arrows indicate the position of a fluorescence fiduciary mark at 0 s. (Bar, 1 μm.)
Fluorescence speckle analysis has been used to study microtubule synthesis in vivo (21). We observed differences in fluorescence intensity of Abp140p–GFP along the length of actin cables. The position of bright spots of Abp140p–GFP on the cables always coincided with increased intensity of staining with rhodamine phalloidin (Fig. 1 A and B). Thus, although Abp140p–GFP is not the core subunit of the cable, it directly reflects increased actin density in the cable. Therefore, we used these bright spots of Abp140p–GFP as fiduciary marks to analyze longitudinal movement of cables.
Abp140p–GFP fiduciary marks always moved away from the bud, as the cable became longer (Fig. 3A). The average velocity of movement of these marks (0.29 ± 0.11 μm/s, n = 55) was similar to that of extension of the tip of the elongating actin cable (see Fig. 6). Movement of two fiduciary marks on a cable at the same rate and in the same direction was also observed in some cells (Fig. 7, which is published as supporting information on the PNAS web site).
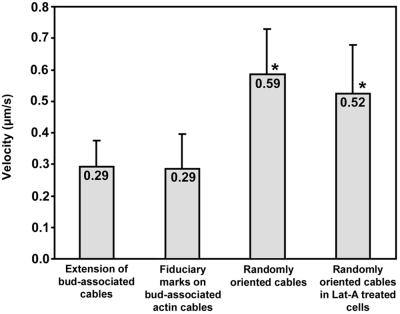
Figure 6.
Velocities of Abp140p–GFP cable movement. Cells were grown as in Fig. 1. Extension of cables was measured as the change in position of the tips of bud-associated cables away from the bud as a function of time. For movement of fiduciary marks, we monitored the time-dependent change in position of the center of bright fluorescent regions along Abp140p–GFP-labeled actin cables. All movements associated with extension of bud-associated actin cables and movement of fiduciary marks on bud-associated cables were unidirectional and directed toward the tip of the mother cell distal to the bud. To measure the velocity of the movement of randomly oriented actin cables either in untreated cells or in cells treated with Lat-A (35 μM, 2 min), we followed movement of the leading tips of moving actin cables. All velocities are averages of 10–20 measurements. Cables that displayed weak fluorescence or loss of fluorescence caused by photobleaching were excluded from these measurements. Error bars represent SD. * indicate velocities that were significantly different from the velocity of extension of bud-associated actin cables (P < 0.01, t test).
These findings suggest that the extension of the actin cable occurs by assembly at the end of the cable within the bud, followed by displacement of the cable from its point of origin toward the distal tip of the mother cell.
To test whether the apparent extension of the bud-associated cables depends on F-actin polymerization, we treated cells with a low concentration of Lat-A that resulted in partial inhibition of actin polymerization and partial disruption of actin cables. We detected short actin cables that extend from the bud and align along the mother-bud axis. Among more than 100 cells tested, we observed some undulation of cables; however, we did not detect any extension of these short bud-associated cables. Under these conditions, we also did not detect any movement of Abp140p–GFP fiduciary marks along the long axis of short, bud-associated actin cables (Fig. 3B). Thus, it appears that inhibition of actin polymerization blocks actin cable extension. This observation is consistent with the interpretation that this extension process is actin cable elongation.
The studies described above focus on the extension and movement of actin cables in the yeast mother cell during the S and G2 phases of the cell division cycle. We also detected extension of bud-associated actin cables in other stages of the cell cycle where polarized cell surface growth occurs (Fig. 8 B and D, which is published as supporting information on the PNAS web site). In unbudded cells that had completed bud site selection, Abp140p–GFP was concentrated at the ends of the actin cables closer to the incipient bud site, near the ring of actin patches. Actin cables were assembled within the incipient bud site and radiated toward the opposite end of the cell (Fig. 8B). In large-budded cells, we detected elongation of bud-associated actin cables in both mother cell and bud (Fig. 8D). At this stage, actin cables were assembled at the bud neck and radiated toward either the distal end of the mother cell or the bud tip. Schematic diagrams of these cable dynamics are shown in Fig. 8E.
FLIP of Abp140p–GFP.
The rapid rate of actin cable movement and slow rate of image capture on the imaging system used for photobleaching precluded analysis of the movement of photobleached zones on elongating actin cables. Instead, we used FLIP to study the dynamics of actin in Abp140p–GFP-labeled cells. We photobleached a 1-μm2 area in either the bud neck or tip of the mother cell distal to the bud site, and monitored loss of fluorescence in the middle third of mother cells (Fig. 4). If actin cables assemble in the bud and elongate into the mother cell, then we should detect actin-dependent movement of photobleached Abp140p–GFP from the bud to the mother cell.
Figure 4.
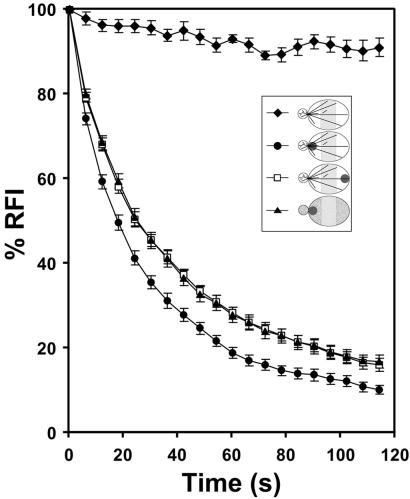
FLIP of Abp140p–GFP. Cells were prepared as in Fig. 3A. For Lat-A treatment, cells were exposed to 400 μM Lat-A for 5 min. Lat-A was solubilized in DMSO and added such that the final concentration of DMSO was 0.8%. The control cells were incubated with 0.8% DMSO for 5 min. The bleached area is shown as dark circles (Inset). Fluorescence of a region in the mother cell (shaded zone, Inset) was quantitated to obtain relative fluorescence intensity (defined in Materials and Methods). Fifteen cells with small buds were selected for each result. ⧫, unbleached; ●, bleached near bud neck; □, bleached near distal tip of mother cell; ▴, bleached near bud neck after Lat-A treatment. Error bars represent standard error.
To monitor the rate of actin-independent movement of Abp140–GFP from the bud to the mother cell, we carried out FLIP experiments in Lat-A-treated yeast. Control studies revealed that our image acquisition conditions did not produce significant loss of signal from Abp140–GFP. In contrast, sustained photobleaching in the bud neck of Lat-A-treated cells resulted in a decrease in the RFI of the region of interest in the mother cell. The average t1/2 of fluorescence loss with sustained photobleaching in the bud neck for 15 samples treated with Lat-A (400 μM) for 5 min was 26.1 s with a SD of 6.6 s. In contrast, the t1/2 of fluorescence loss with sustained photobleaching in the bud neck of control cells was 17.5 ± 5.1 s. Statistical analysis indicates that the FLIP rates were significantly different from each other in Lat-A-treated and untreated cells (P < 0.01). Thus, we observe that loss of F-actin results in a decrease in the rate of movement of Abp140p–GFP from the bud into the mother cell. Finally, to determine whether the rapid movement of Abp140–GFP in untreated cells was region-specific, we evaluated loss of fluorescence in control cells with repeated photobleaching in the tip of the mother cell distal to the bud site. Interestingly, the observed FLIP half-time (24.8 ± 5.5 s) was not significantly different from that observed upon photobleaching the bud neck in Lat-A-treated cells.
The rate of Abp140p–GFP movement from the bud to mother cell is significantly reduced upon destabilization of F-actin. This finding indicates that (i) movement of Abp140p–GFP from the bud neck to the mother cell is actin-dependent, not mediated simply by diffusion, and (ii) Abp140p–GFP does not rapidly equilibrate between actin cable-bound and unbound states. Both findings provide additional support for the notion that Abp140p–GFP fiduciary marks can be used to study actin cable dynamics. In addition, because the rate of FLIP upon photobleaching the distal tip of the mother cell was indistinguishable from that observed upon photobleaching the bud in Lat-A-treated cells, it is clear that the actin-dependent movement of Abp140p–GFP is predominantly unidirectional from the bud to the mother cell. Thus, our FLIP studies indicate that Abp140p–GFP (i) binds tightly to actin cables and (ii) shows actin-dependent movement from the bud to the mother cell and not from the mother toward the bud. These findings provide independent evidence that actin cables assemble in the bud and elongate from the bud into the mother cell.
Movement of Randomly Oriented Actin Cables.
Time-lapse imaging of randomly oriented, short actin cables revealed dynamics distinct from those of bud-associated actin cables (Fig. 5A). In contrast to bud-associated actin cables, which assemble at the assembly site and extend into the mother cell during elongation, randomly oriented actin cables move rapidly without any obvious change in cable length. This movement was unidirectional, parallel to the long axis of the motile cable and restricted to the cell cortex. Because these short cables do not align along the mother-bud axis, they moved in all directions throughout the cell cortex. The velocity of movement of the randomly oriented cables ranged from 0.27 to 0.83 μm/s with an average velocity of 0.59 ± 0.14 μm/s (n = 16) (Fig. 6). This velocity is significantly higher than the rate of extension of long actin cables from the bud site.
Figure 5.
Dynamics of randomly oriented actin cables. (A) Movement of randomly oriented actin cables in an untreated cell. Cells were grown as in Fig. 1. Time-lapse images of Abp140p–GFP-labeled actin cables in a budding cell were acquired as in Fig. 3A. The beginning and end of a motile cable are marked with white arrows. (B) Randomly oriented actin cable movement in Lat-A -treated cells. Lat-A treatment and image acquisitions were performed as in Fig. 3B. The ends of a cable moving toward the bud are marked with arrows and arrowheads. The ends of a cable moving away from the bud are marked with white arrows. (Bars, 1 μm.)
Extension of bud-associated actin cables was inhibited by mild treatment with Lat-A. Under the same conditions, we detected an increase in the number of motile and randomly oriented actin cables (Fig. 5B; Movie 2, which is published as supporting information on the PNAS web site). The randomly oriented cables detected in Lat-A-treated cells displayed movements similar to that of randomly oriented cables in untreated cells, that is, unidirectional, parallel to the long axis of the cable, and in all directions relative to the mother-bud axis. The velocity of this cable movement, 0.52 ± 0.15 μm/s, was also similar to that of randomly oriented cable movement in untreated cells (Fig. 6). Finally, randomly oriented actin cables are not abundant in untreated yeast, but increase in numbers under conditions that block actin cable elongation and dampen actin dynamics. Therefore, Lat-A treatment increases the pool of randomly oriented cables by destabilizing or perturbing bud-associated, elongating cables.
Discussion
In this study, actin cables were visualized in living yeast cells by using GFP fused to Abp140p. When expressed from the ABP140 chromosomal locus, Abp140p–GFP localizes to actin cables and has no obvious effect on cell growth or on organization, polarization, or dynamics of the actin cytoskeleton. We detected two populations of actin cables in budding yeast in live cells by using Abp140p–GFP and in fixed cells by using fluorochrome-coupled phalloidin. Bud-associated actin cables, which are more abundant, emerge from the bud and align along the mother-bud axis. Some actin cables are randomly distributed within the yeast cell cortex. Here, we describe the dynamics of bud-associated and randomly oriented actin cables in living yeast.
Elongation of Bud-Associated Actin Cables.
We detected apparent elongation of bud-associated actin cables. During this process, the end of the cable within the bud did not appear to change position. Instead, we observed apparent extension of the other end of the cable away from the bud. Moreover, we found that fiduciary marks of Abp140p–GFP on actin cables moved at the same velocity and in the same direction as the tip of the elongating cable. The possibility of polarized transport of fiduciary marks along the cables seems unlikely, because this process is blocked by Lat-A, an agent that binds to actin monomers and inhibits actin polymerization. The simplest interpretation of these observations is that actin cable assembly occurs at specific points within the cell, which we will call the assembly site. Elongation of these bud-associated actin cables occurs by addition of new material to the end of the cable at the assembly site. As a result, actin cables grow from the assembly site toward the end of the mother cell distal to the bud. Time-lapse imaging of elongating cables in yeast at different stages in the cell division cycle revealed that actin cable assembly sites are located in (i) the presumptive bud site in unbudded cells, (ii) small- and medium-sized buds, and (iii) the bud neck in cells bearing large buds.
This interpretation is supported by FLIP experiments. Abp140p–GFP that was photobleached in the bud displayed actin-dependent movement into the mother cell. Interestingly, Abp140p–GFP that was photobleached in the tip of the mother cell distal to the bud did not display rapid, actin-dependent movement toward the bud.
Yet to be determined is the detailed mechanism underlying actin cable assembly. It is possible that assembly occurs by polymerization of F-actin on the end of the cable within the assembly site. Localized polymerization of F-actin occurs in a variety of cells, including at the leading edge of motile cells and filopodia of neurons (22, 23). If actin cable elongation is driven by actin polymerization in yeast, then the fast-growing (barbed) end of actin filaments within cables is probably directed toward the bud. This interpretation is consistent with previous findings that myosin V proteins transport cargo specifically toward the bud by using actin cables as tracks (5, 7, 8).
Alternatively, it is possible that actin cable assembly in yeast occurs by a mechanism similar to that of actin bundle assembly in the bristle and follicular nurse cell struts of Drosophila: incorporation of pre-existing F-actin onto one end of actin cables (1). In this case, the location of barbed and pointed ends of actin filaments within actin cables could be determined by proteins that mediate F-actin bundling within the nascent cable (e.g., fimbrin and tropomyosin). In any case, because the majority of actin cable assembly occurs at assembly sites in the presumptive bud, the bud or the bud neck, there is a polarity of assembly, although not necessarily an intrinsic polarity, in cables.
Retrograde flow of newly assembled actin cables in filopodia is driven by myosin (24–26). Therefore, while assembly contributes to movement of elongating actin cables from the bud to the mother cell in yeast, it is possible that other factors including motor molecules contribute to this motility event.
Finally, because actin cables are disrupted within seconds after treatment with Lat-A, an agent that binds to G-actin and prevents its polymerization, it is clear that actin cables are highly dynamic structures that undergo assembly and disassembly. Although the mechanism of actin cable disassembly is not well understood, our evidence indicates that disassembly of actin cables does not occur at the tips as they move toward the distal end of mother cells. First, we do not detect any apparent shrinkage of cables. Second, the tip of actin cables and fiduciary marks along the cables move toward the distal end of the mother cell at the same average velocities.
Movement of Randomly Oriented Actin Cables.
We found that randomly oriented actin cables move around the cell cortex (Fig. 5). There are striking differences between this movement and that of bud-associated actin cables. First, the velocity of randomly oriented cable movement is significantly faster than that of extension of the end of an elongating, bud-associated actin cable. Second, in contrast to elongation of actin cables, which is blocked by Lat-A treatment, we find that randomly oriented actin cable movement is not significantly affected by treatment of yeast with Lat-A, until the point when the cables themselves disassemble.
These observations support the model that these randomly oriented cables are not undergoing detectable elongation. Moreover, because dampening actin dynamics with Lat-A has no effect on this type of actin cable movement, it is likely that this movement is accomplished by the migration of entire cables, not by actin treadmilling through continuous actin polymerization at one end and depolymerization at the other end. Although there is no direct evidence for a role of motor molecules in this process, movement of randomly oriented actin cables resembles sliding of F-actin on myosin molecules: it is unidirectional and parallel to the long axis of the cable (27).
What is the relationship between these two populations of actin cables? Because Lat-A treatment blocks elongation of bud-associated actin cables, and produces an increase in the number of motile, randomly oriented actin cables, it is possible that randomly oriented cables are produced by destabilization or fragmentation of bud-associated cables. This interpretation is supported by the observation that actin cables fragment upon instantaneous dampening of actin dynamics (20). Thus, it appears that some actin cables, even those that are produced by destabilization of bud-associated actin cables, can move, and that other cables, namely bud-associated actin cables, do not move. We propose that (i) elongating, bud-associated cables are immobilized at the assembly site, and (ii) this immobilization is required for actin cable assembly and alignment of these cables along the mother-bud axis.
Supplementary Material
Acknowledgments
We thank members of the Pon laboratory for critical evaluation of the manuscript; Dr. P. Crews for Lat-A; Drs. M. Longtine and J. Pringle for tagging cassette DNA; and Dr. A. Bretscher for tropomyosin mutants. This work was supported by research grants to L.A.P. from the National Institutes of Health (GM45735) and the American Cancer Society (RPG-97–163-04-CSM). Some imaging experiments were carried out in the Optical Imaging Facility of the Herbert Irving Comprehensive Cancer Center at Columbia University. The facility was established with instruments financed by National Institutes of Health Shared Instrumentation Grants S10RR10506 and S10RR13701 and the Lieber Foundation. The operation of the facility is supported in part by National Institutes of Health Grant P30CA13696.
Abbreviations
- Abp140p
actin binding protein 140
- F-actin
filamentous actin
- GFP
green fluorescent protein
- Lat-A
Latrunculin-A
- FLIP
fluorescence loss in photobleaching
- RFI
relative fluorescence intensity
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.DeRosier D J, Tilney L G. J Cell Biol. 2000;148:1–6. [PMC free article] [PubMed] [Google Scholar]
- 2.Adams A E M, Pringle J R. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amberg D C. Mol Biol Cell. 1998;9:3259–3262. doi: 10.1091/mbc.9.12.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheney R E, O'Shea M K, Heuser J E, Coelho M V, Wolenski J S, Espreafico E M, Forscher P, Larson R E, Mooseker M S. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 5.Hill K L, Catlett N L, Weisman L S. J Cell Biol. 1996;135:1535–1549. doi: 10.1083/jcb.135.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takizawa P A, Sil A, Swedlow J R, Herskowitz I, Vale R D. Nature (London) 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 7.Pruyne D W, Schott D H, Bretscher A. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, Pruyne D, Huffaker T C, Bretscher A. Nature (London) 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- 9.Karpova T S, Reck-Peterson S L, Elkind N B, Mooseker M S, Novick P J, Cooper J A. Mol Biol Cell. 2000;11:1727–1737. doi: 10.1091/mbc.11.5.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon V R, Karmon S L, Pon L A. Cell Motil Cytoskeleton. 1997;37:199–210. doi: 10.1002/(SICI)1097-0169(1997)37:3<199::AID-CM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Bretscher A. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- 13.Drees B, Brown C, Barrell B G, Bretscher A. J Cell Biol. 1995;128:383–392. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drubin D G, Miller K G, Botstein D. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belmont L D, Drubin D G. J Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asakura T, Sakaki T, Nagano F, Satoh A, Obaishi H, Nishioka H, Imamura H, Hotta K, Tanaka K, Nakanishi H, Takai Y. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- 17.Longtine M S, McKenzie M, 3rd, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Ito H, Jukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith M G, Swamy S R, Pon L A. J Cell Sci. 2001;114:1505–1513. doi: 10.1242/jcs.114.8.1505. [DOI] [PubMed] [Google Scholar]
- 20.Karpova T S, McNally J G, Moltz S L, Cooper J A. J Cell Biol. 1998;142:1501–1517. doi: 10.1083/jcb.142.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterman-Storer C M, Salmon E D. Biophys J. 1998;75:2059–2069. doi: 10.1016/S0006-3495(98)77648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchison T J, Cramer L P. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- 23.Katoh K, Hammar K, Smith P J S, Oldenbourg R. Mol Biol Cell. 1999;10:197–210. doi: 10.1091/mbc.10.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallavarapu A, Mitchison T. J Cell Biol. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C H, Espreafico E M, Mooseker M S, Forscher P. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 26.Welch M D, Mallavarapu A, Rosenblatt J, Mitchison T J. Curr Opin Cell Biol. 1997;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- 27.Kron S J, Spudich J A. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.