Abstract
The adapter SLP-76 is essential for thymocyte development. SLP-76−/− mice were reconstituted with SLP-76 deletion mutant transgenes to examine the role of SLP-76 domains in T cell development and function. The N-terminal domain deletion mutant completely failed to restore thymocyte development. Mice reconstituted with Gads-binding site and SH2 domain deletion mutants had decreased thymic cellularity, impaired transition from double to single positive thymocytes, and decreased numbers of mature T cells in the spleen. Calcium mobilization and extracellular signal-regulated protein kinase activation were decreased in the Gads-binding site mutant but almost normal in the SH2 domain mutant. T cells from both mutants failed to proliferate following T cell antigen receptor ligation. Nevertheless, both mutants mounted partial cutaneous hypersensitivity responses and normal T cell dependent IgG1 antibody responses. These results indicate differential roles for SLP-76 domains in T cell development, proliferation and effector functions.
SLP-76 is an adapter protein predominantly expressed in hematopoietic cells (1). SLP-76 has three distinct domains: an NH2-terminal domain (amino acids 1–155), a central proline rich domain (amino acids 156–421) that includes a Gads binding site (amino acids 224–244), and a C-terminal SH2 domain (amino acids 422–533). Phosphorylated tyrosine residues in the NH2-terminal domain bind SH2-domain containing proteins that include Vav, Nck, and the Tec kinase Itk (2–4). The SLP-76 central proline rich domain associates with SH3-containing proteins that include Gads and PLC-γ1. Upon T cell antigen receptor (TCR) stimulation, Gads recruits SLP-76 to linker of activated T cells (LAT). This translocates SLP-76 to glycolipid enriched microdomains (GEM) (5, 6). LAT through Grb2 interacts with Sos, a guanine nucleotide exchange factor for Ras GTPases, and may link SLP-76 to the Ras/mitogen-activated protein kinase (MAPK)/extracellular signal-regulated protein kinase (ERK) pathway. Finally, the SLP-76 SH2-domain interacts with phosphoproteins such as Fyb/SLAP130 and a 62-kDa phosphoprotein (7).
SLP-76−/− mice have a complete block in thymocyte development at the CD4−CD8−, double-negative stage and lack peripheral T cells (8, 9). SLP-76 plays an important role in T cell receptor signal transduction and T cell activation. SLP-76-deficient Jurkat cells exhibit severely impaired TCR/CD3 signaling with deficient PLC-γ1 activation, calcium mobilization, ERK phosphorylation, and IL-2 production (10). Overexpression of SLP-76 in a human T cell line (Jurkat) results in marked augmentation of TCR mediated activation of nuclear factor of activated T cells (NFAT) and IL-2 production (2). Single mutation of the three N-terminal domain tyrosine residues (amino acids 113, 128, and 145), that are phosphorylated after TCR stimulation, to phenylalanine has no effect on the ability of SLP-76 to augment the NFAT response. In contrast, double or triple mutants of these tyrosine residues and deletion mutants of the NH2-terminal region, the proline-rich Gads binding site, or the SH2-domain were inactive (11).
The role played by various domains of SLP-76 in T cell development and function in vivo remains unknown. In this study, we addressed this issue by reconstituting SLP-76−/− mice with wild-type (WT) SLP-76 and with SLP-76 functional domains deletion mutants.
Materials and Methods
SLP-76 Transgenic Mice.
The SLP-76 cDNA mutants Δ2–156, Δ224–244, and Δ421–533 (ΔSH2) were generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). WT and mutant SLP-76 cDNAs were subcloned in the BamHI site of the p1017 vector carrying the Lck proximal promoter (12). NotI linearized DNA was microinjected in fertilized C57BL/6 × SJL/J (B6SJL) mouse oocytes. Injected eggs were implanted into the oviducts of pseudopregnant foster mothers. Transgenic founder mice, identified by PCR using transgene-specific primers and confirmed by Southern blotting, were bred into the SLP-76 deficient (8) background by two rounds of back-crossing with SLP-76+/− mice. Transgenes and the endogenous SLP-76 locus were determined by PCR. All animal experiments were performed in compliance with the National Institutes of Health and institutional guidelines approved by Children's Hospital internal animal care and use committee.
Antibodies and Flow Cytometry Analysis.
Streptavidin-FITC, streptavidin-phycoerythrin (PE), and streptavidin-CyChrome and mAbs (unlabeled or labeled to FITC or PE) to mouse antigens were purchased from PharMingen. Cells were stained and analyzed on a FACSCalibur flow cytometer (Becton Dickinson) as described (8). Data on 5–20 × 105 viable, nonerythroid cells, as determined by forward versus side scatter, were collected. FACS analysis was performed on cells from at least three mice aged 6–8 weeks.
Rabbit Anti-SLP-76 Antibodies and Intracellular FACS Analysis.
The IgG fraction of rabbit antiserum against GST-SLP-76 fusion protein was enriched over a Protein G column and precleared by GST Sepharose. Thymocytes and splenic T cells were permeabilized and stained intracellularly, by using CytoPerm/CytoFix Kit (PharMingen), with FITC-conjugated anti SLP-76 antibody.
Western Blot Analysis.
Thymocytes (1 × 106) were lysed in sample buffer containing SDS and β-mercaptoethanol and proteins were separated on a 10% gel by PAGE, transferred to nitrocellulose membrane, and probed with rabbit anti-SLP-76 antibody, followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (Amersham Pharmacia).
Proliferation of Splenic T Cells.
Splenocytes were suspended in RPMI medium 1640 supplemented with L-glutamine, penicillin/streptomycin, and 10% FBS and cultured in triplicates with various stimuli. After 72 h, proliferation was assessed by [3H]thymidine uptake assay.
Measurement of [Ca2+]i.
Purified T cells (2 × 106/ml) were incubated with 4 μg/ml Fluo-4-AM (Molecular Probes) and 10 μg/ml Fura Red (Molecular Probes) in 0.5 ml Tyrode's buffer with 0.02% Pluronic F-127 (Sigma) and 1 mM Probenecid (Sigma). After 45 min at 37°C, cells were preloaded with 2 μg/ml anti-CD3ɛ antibody (Clone KT3, Serotec) on ice for 15 min, kept at room temperature for 30 min to allow cleavage of the AM esters, washed twice with Tyrode's buffer, then stimulated with 5 μg/ml goat F(ab′)2 anti-rat IgG (ICN/Cappel) and analyzed by flow cytometry. Intracellular Ca2+ concentration is indicated by Fluo-4AM/Fura Red fluorescence intensity ratio.
PLC-γ1 and ERK Phosphorylation.
Purified T cells were left unstimulated or were stimulated with 10 μg/ml anti-CD3ɛ (clone KT3, Serotec) crosslinked with F(ab′)2 goat anti-rat IgG for 5 min. For PLC-γ1, lysates were prepared in 1% Nonidet P-40 lysis buffer supplemented with protease and phosphatase inhibitors (13), and immunoprecipitated with anti-PLC-γ1 mAb (Santa Cruz Biotechnology). Precipitated proteins were resolved by SDS/10% PAGE, transferred on nitrocellulose membrane, and probed with indicated antibodies. For ERK assay, lysates were prepared in sample buffer containing SDS and β-mercaptoethanol and resolved as above. Activated ERK was detected using a phospho ERK1/2-specific mAb followed by reprobing the membrane with anti-ERK1 mAb (Santa Cruz Biotechnology).
Hapten-Induced Contact Hypersensitivity.
Mice were sensitized by the application of 100 μl of 2% oxazolone (Sigma) in ethanol to previously shaven abdominal skin. Five days later, 10 μl of 1% oxazolone were applied to the dorsal and ventral surfaces of the right ear. Ethanol was applied to the left ear. Ear thickness was measured after 24, 48, and 72 h, using a modified spring-loaded micrometer (Mitutoyo).
Antibody Responses.
Ten- to twelve-week-old mice were immunized intraperitoneally with 100 μg of ovalbumin (OVA) precipitated with alum and bled at day 21. Anti-OVA antibodies were detected by ELISA as described (14).
Results
Generation of Transgenic Mice.
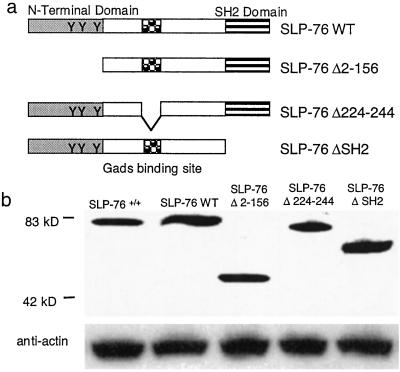
The cDNA constructs used to generate transgenic mice on SLP-76−/− background are shown in Fig. 1a. Non-transgene-bearing littermates were designated SLP-76+/+ and SLP-76−/−. Transgenic lines that expressed SLP-76 protein in thymocytes in amounts comparable to those expressed in SLP-76+/+ mice, as assessed by Western blotting (Fig. 1b) and FACS (see Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org), were expanded for study. Similar results were obtained when independent lines of SLP-76 WT (n = 2), SLP-76 Δ2–156 (n = 2), SLP-76 Δ224–244 (n=3), and SLP-76 ΔSH2 (n=4) mice were examined.
Figure 1.
SLP-76 mutants and their expression in thymocytes. (a) WT and SLP-76 deletion mutants used to reconstitute SLP-76−/− mice. (b) Expression of SLP-76 protein in 1 × 106 thymocytes assessed by Western blotting using rabbit anti-SLP-76 antibody. Actin was probed for loading control.
Thymocyte Development.
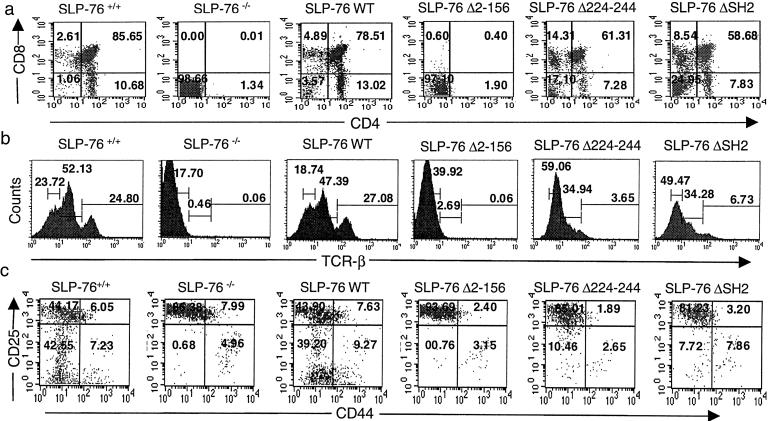
Thymic cellularity in SLP-76−/− mice was ≈1% that of SLP-76+/+ mice (1.6 × 106, n = 6). SLP-76−/− mice had no detectable CD4+CD8+ DP, CD4+CD8− single-positive (SP) or CD4−CD8+ SP thymocytes (Fig. 2a) and no detectable TCRβ+ cells (Fig. 2b). The SLP-76 WT transgene restored thymic cellularity (112 × 106, n = 4) and thymocyte development. The percentages of DN, DP, and SP cells and of TCRβ+ cells in SLP-76 WT mice were similar to SLP-76+/+ littermates (Fig. 2 a and b). SLP-76 Δ2–156 completely failed to rescue thymocyte cellularity (1.9 × 106, n = 4; Fig. 2 a and b). SLP-76 Δ224–244 and SLP-76 ΔSH2 restored thymic cellularity to approximately one third (35 × 106, n = 7) and one half normal (55 × 106, n = 8), respectively. Progression from DN to DP cells was partially impaired in SLP-76 Δ224–244 and SLP-76 ΔSH2 mice, as evidenced by an increase in the percentage of DN cells and a decrease in the percentage of DP cells (Fig. 2a). There was also a relative increase in the percentage of SP CD8+ cells. The percentage of TCRβ+ cells and the density of TCRβ on thymocytes were decreased compared with SLP-76+/+ and SLP-76 WT mice (Fig. 2b). In particular, percentages of TCRhi and TCRintermediate thymocytes were decreased in these mice.
Figure 2.
FACS analysis of thymocytes. Surface expression of (a) CD4 vs. CD8 on total thymocytes. The percentage of cells found in each quadrant is indicated. (b) TCRβ on total thymocytes. (c) CD44 and CD25 on DN thymocytes. Cells were triple stained with anti-CD44-FITC, anti-CD25-phycoerythrin, and a mixture of biotin-conjugated mAbs to CD3, CD4, CD8, B220, Mac1, and Gr-1, followed by streptavidin-CyChrome. Analysis was performed on gated CyChrome negative cells. Results are representative of three experiments.
The sequence of development of DN thymocytes is CD25−CD44+ → CD25+CD44+ → CD25+CD44− → CD25−CD44− cells (15). SLP-76−/− mice show a relative increase of the CD25+CD44− population, whereas CD25−CD44− cells, the most mature among DN T cell progenitors, are almost undetectable (8). This block was completely overcome by reconstitution with WT SLP-76 transgene (Fig. 2c). DN cells from SLP-76 Δ2–156 mice exhibited a block identical to that of SLP-76−/− mice (Fig. 2c). Reconstitution with SLP-76 Δ224–244 and SLP-76 ΔSH2 partially overcame the block in the transition from the CD25+CD44− to the CD25−CD44− stage.
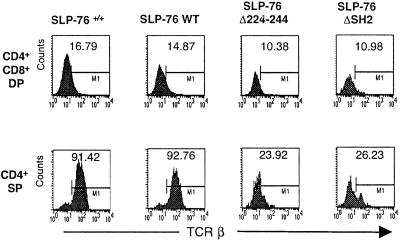
Expression of TCRβ and the CD69 Activation Markers on DP and SP Thymocytes.
Positive selection of thymocytes into SP cells is accompanied by up-regulation of TCR/CD3 and CD69 expression (16, 17). In both SLP-76+/+ and SLP-76 WT mice, expression of TCRβ and CD69 was up-regulated in SP cells (Fig. 3 and Fig. 9, which is published as supporting information on the PNAS web site). Expression of TCRβ and CD69 was also up-regulated on SP cells of SLP-76 Δ224–244 and SLP-76 ΔSH2 mice, but the percentages of TCRβ+ and CD69+ SP cells and density (MFI) of TCRβ and CD69 were lower than in SLP-76+/+ and SLP-76 WT controls.
Figure 3.
Expression of TCRβ on DP and SP thymocytes. Gated DP and SP cells were analyzed for expression of TCRβ after staining with anti-CD4-phycoerythrin, anti-CD8-CyChrome, and anti-TCRβ-FITC. Results are representative of three experiments.
Peripheral T Cells.
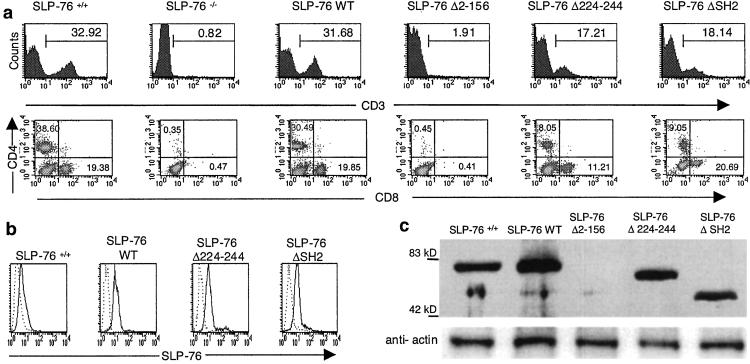
Spleens of SLP-76−/− contain no detectable CD3+, CD4+, or CD8+ cells (Fig. 4a). Introduction of the SLP-76 WT transgene restored splenic CD3+, CD4+, and CD8+ cells to normal. The SLP-76 Δ2–156 transgene completely failed to repopulate spleens with T cells. Both SLP-76 Δ224–244 and SLP-76 ΔSH2 restored splenic CD3+ T cells to ≈50% of normal. In both cases, the density of CD3 expression was about half of that of SLP-76+/+ T cells, and there was a relative increase in CD8+ cells (Fig. 4a). Similar phenotypes were obtained with blood mononuclear cells (data not shown).
Figure 4.
FACS analysis and SLP-76 expression in spleen cells. (a) Surface expression of CD3 and of CD4 and CD8 on spleen cells. Results are representative of three experiments. (b) Expression of SLP-76 protein in purified T cells by intracellular staining with FITC-conjugated rabbit anti-SLP-76 antibody (c) Expression of SLP-76 protein in 1 ×106 splenocytes assessed by Western blotting.
Intracellular FACS staining revealed that T cells from spleens of mice reconstituted with SLP-76 transgenes expressed SLP-76 proteins at comparable intensities (Fig. 4b). Western blotting revealed that the expressed SLP-76 proteins in splenocytes were of the expected sizes (Fig. 4c). These results indicate that the lck proximal promoter is driving sustained SLP-76 expression in mature T-cells.
T Cell Proliferation and IL-2 Receptor Expression.
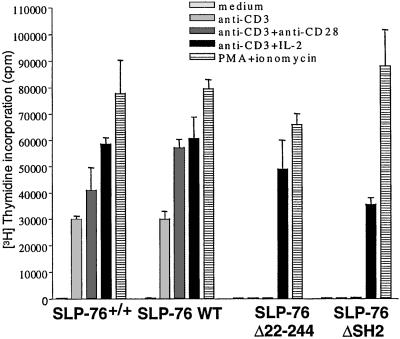
Splenocytes from SLP-76 WT mice proliferated normally to anti-CD3. In contrast, splenocytes from both SLP-76 Δ224–244 and SLP-76 ΔSH2 mice failed to proliferate to anti-CD3, over a wide range of concentrations (0.01 to 10 μg/ml; see Fig. 10, which is published as supporting information on the PNAS web site). The same T cells proliferated normally to PMA + ionomycin. Interactions between IL-2 and IL-2R play an important role in T cell proliferation and optimal production of IL-2 by T cells, and require co-stimulation via CD28 (18). IL-2, but not anti-CD28 mAb, corrected the failure of T cells from SLP-76 Δ224–244 and SLP-76 ΔSH2 mice to proliferate in response to immobilized anti-CD3 (Fig. 5). T cells from SLP-76 Δ224–244 and SLP-76 ΔSH2 mice poorly up-regulated IL-2Rα chain (CD25) expression following anti-CD3 stimulation (35% and 30% CD25+ cells, respectively) compared with T cells from SLP-76+/+ mice and SLP-76 WT mice (93% and 83% CD25+ cells, respectively). CD25 expression on these cells was enhanced by anti-CD28 (54% CD25+ cells for SLP-76 Δ224–244 and 43% CD25+ cells for SLP-76 ΔSH2) and IL-2 (59% CD25+ cells for SLP-76 Δ224–244 and 52% CD25+ cells for SLP-76 ΔSH2) (see Fig. 11, which is published as supporting information on the PNAS web site). Similar results were obtained for CD69 expression (data not shown).
Figure 5.
Anti CD3 induced proliferation of splenic T cells. Proliferation of purified T cells to anti-CD3 (coated at 5 μg/ml) in the presence or absence of anti-CD28 (coated at 5 μg/ml) and mouse rIL-2 (20 ng/ml), and to PMA (50 ng/ml) + ionomycin (500 μM). Results are representative of three experiments.
Intracellular Calcium Mobilization, PLC-γ1, and ERK Phosphorylation.
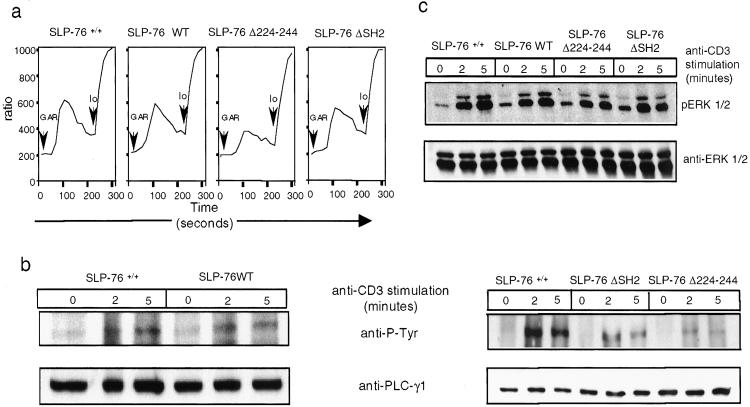
SLP-76 WT mice mobilized calcium normally following anti-CD3 ligation. Calcium mobilization was reduced in T cells from SLP-76 Δ224–244 mice, and minimally affected in T cells from SLP-76 ΔSH2 mice (Fig. 6a).
Figure 6.
Calcium flux, phosphorylation of PLC-γ1, and ERK following TCR/CD3 ligation in splenic T cells. (a) Ca2+ mobilization: T cells preloaded with Fluo-4-AM and Fura Red were preincubated with anti-CD3 (2 μg/ml), washed, then stimulated with 5 μg/ml goat F(ab′)2 anti-rat IgG (GAR) and analyzed by flow cytometry. Intracellular Ca2+ concentration is indicated by the Fluo-4AM/Fura Red fluorescence intensity ratio. Io, ionomycin. (b) PLC-γ1 tyrosine phosphorylation: PLC-γ1 immunoprecipitates from purified T cells stimulated for 0–5 min with crosslinked anti-CD3 were Western blotted with antiphosphoptyrosine mAb 4G10 (Upper), and reprobed with anti- PLC-γ1 (Lower). (c) ERK phosphorylation. Lysates from purified T cells stimulated as above were probed with phospho-ERK-specific antibody (Upper) and reprobed with ERK-specific antibody (Lower). Results shown are representative of three experiments.
Tyrosine phosphorylation of PLC-γ1 following TCR/CD3 ligation was normal in T cells from WT SLP-76 mice (Fig. 6b), but was decreased in T cells from SLP-76 Δ224–244 and SLP-76 ΔSH2 mice (Fig. 6b). This decrease was specific, because tyrosine phosphorylation of ZAP-70, which is upstream of SLP-76, was normal in these cells (see Fig. 12, which is published as supporting information on the PNAS web site).
ERK activation is impaired in SLP-76 deficient Jurkat cells (10). Phosphorylation of ERK1/2 following TCR/CD3 stimulation in T cells from SLP-76 WT mice was comparable to SLP- 76+/+ mice. Baseline ERK1/2 phosphorylation was increased in T cells from mice reconstituted with mutant transgenes. Induced ERK1/2 phosphorylation was ≈4-fold decreased in T cells from SLP-76 Δ224–244 mice (n = 3) and was less sustained (≈1.5-fold decreased) in T cells from SLP-76 ΔSH2 (n = 3; Fig. 6c).
In Vivo T Cell Effector Responses.
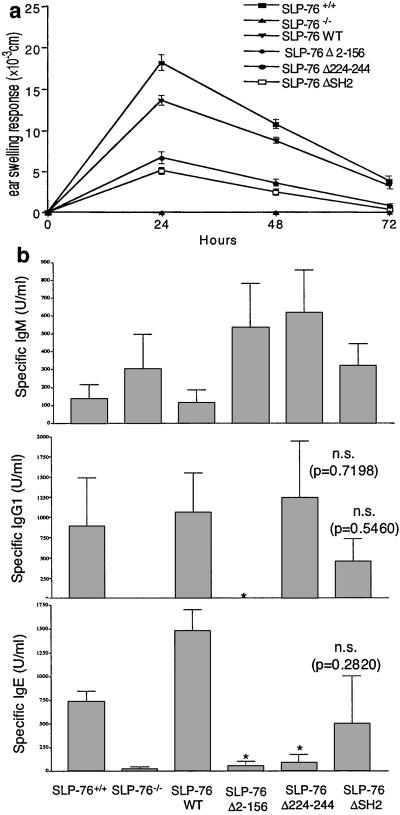
Oxazolone sensitized SLP-76−/− mice completely failed to exhibit ear swelling up to 72 h after hapten challenge. Ear swelling in SLP-76 WT mice was similar to that of SLP 76+/+ controls. There was partial ear swelling in both SLP-76 Δ224–244 and SLP-76 ΔSH2 mice (Fig. 7a).
Figure 7.
Hapten Induced Contact Hypersensitivity and antibody response to OVA. (a) Groups of four mice were sensitized with oxazolone and challenged with hapten (right ear) or ethanol (left ear). Results represent the difference in thickness between hapten- and vehicle-challenged ears. (b) Anti-ova IgM, IgG1, and IgE antibodies in the sera of mice (three mice for SLP-76−/− and SLP-76 Δ2–156, four mice for the others) immunized intraperitoneally with 100 μg OVA. Numbers indicates the P values in comparison to SLP-76+/+ mice (* indicates P < 0.01 by paired t test with Welch's correction; n.s. = not significant).
To determine the T helper responses, we measured the antibody response to OVA. All mice made an IgM antibody response comparable to that of SLP-76+/+ mice littermates (Fig. 7b). SLP-76−/− and SLP-76 Δ2–156 mice made no IgG1 antibody or IgE antibody responses to OVA. SLP-76 WT mice made IgG1 and IgE responses comparable to SLP-76+/+ controls. SLP-76 Δ224–244 made a normal IgG1 response, but had a significantly decreased IgE response to OVA. SLP-76 ΔSH2 mice made IgG1 and IgE responses to OVA comparable to SLP-76+/+ controls (Fig. 7b).
Discussion
The data presented demonstrate that SLP-76 domains play differential roles in mediating thymopoiesis and activation of mature T cells in vivo. The failure of the SLP-76 N-terminal domain mutant to restore thymocyte development in SLP-76−/− mice suggests that this domain is critical for signaling by the preTCR, which is required for the maturation of DN cells and their transition to DP cells. The block in SLP-76 Δ2–156 mice could be due to the inability of the SLP-76 mutant to recruit members of the Vav, Itk and/or Nck families, and/or additional proteins that may interact with the SLP-76 N-terminal domain (2–4). Neither disruption of individual or dual combinations of the Vav proteins Vav1 and Vav 2 (19–22), nor of the Tec kinases Itk, Tec, and Rlk/Txk (23, 24) blocks T cell development at the DN stage.
SLP-76 Δ224–244, and SLP-76 ΔSH2 behaved similarly in their ability to reconstitute T cell development and function in SLP-76−/− mice. Both mutants restored DN to DP and DP to SP transition in the thymus. This restoration was incomplete because thymic cellularity, CD25−CD44− mature DN cells, and DP cells were all decreased (Fig. 2). Furthermore, thymic selection into SP cells was affected, because these cells had decreased expression of TCRβ and of CD69, which are normally up-regulated upon maturation of DP into SP cells (Fig. 3). These data suggest that both Gads binding and SH2 domain interactions are necessary for optimal preTCR and TCR signaling in thymocytes. The skewing of SP thymocytes in these mice toward CD8+ cells is consistent with defective TCR signaling, because the strength of TCR signaling influences lineage commitment to CD4 and CD8 with former favored by TCR activation of ERK kinase (25). Although ERK activation was not measured in thymocytes, it was decreased in the splenic T cells of these mice.
Mature T cells were present in the blood, spleen, and lymph nodes of SLP-76 Δ224–244 and SLP-76 ΔSH2 mice and expressed the SLP-76 transgenes. This finding suggests that the lck promoter driving the transgene was active in mature T cells, as has been reported in other studies (26, 27). However, their percentage was decreased, and they had decreased expression of TCRβ and increased proportion of CD8+ cells (Fig. 4). T cells from SLP-76 Δ224–244 and SLP-76 ΔSH2 mice had decreased CD25 and CD69 expression following ligation of TCR/CD3 and completely failed to proliferate to anti-CD3 (Fig. 5). Up-regulation of CD25 expression and correction of the proliferative defect by IL-2 suggest that residual signaling via TCR/CD3 occurs in these cells.
Calcium mobilization was decreased in T cells from SLP-76 Δ224–244 mice, but minimally affected in SLP-76 ΔSH2 mice (Fig. 6a). This finding is consistent with findings in SLP-76 deficient Jurkat T cells transfected with SLP-76 mutants (13). The decreased response of SLP-76 Δ224–244 mice cannot simply be explained by lower level of expression of TCR/CD3, because SLP-76 ΔSH2 mice had a comparable low expression of TCR/CD3, yet fluxed calcium almost normally.
PLC-γ1 phosphorylation was decreased in T cells from SLP-76 Δ224–244 and SLP-76 ΔSH2 mice (Fig. 6b). Residual PLC-γ1 phosphorylation in SLP-76 Δ224–244 T cells suggests that phosphorylation of PLC-γ1 may occur in the absence of Gads mediated recruitment of SLP-76 to LAT. This phosphorylation may involve the recently demonstrated direct interaction of SLP-76 with PLC-γ1 (13). Decreased PLC-γ1 phosphorylation with almost normal calcium fluxes in SLP-76 ΔSH2 mice suggests that the remaining phosphorylation is sufficient for PLC-γ1 activation leading to normal calcium flux. A similar finding has been reported in SLP-76 deficient Jurkat cells reconstituted with a SLP-76 SH2 domain R448K point mutant (13).
Activation of the MAP kinase ERK was decreased in SLP-76 Δ224–244 mice (Fig. 6c). Decreased generation of DAG subsequent to decreased activation of PLC-γ may have resulted in impaired activation of the Ras exchange factor RasGRP, and impaired activation of the Ras/Raf/ERK pathway (28, 29). Erk activation was less affected in T cells from SLP-76 ΔSH2 mice, which mobilized calcium normally. Baseline phosphorylation of ERK was increased in mutant T cells, particularly in SLP-76 ΔSH2 T cells. It should be noted that deletion of the SH2 domain, in contrast to the R448K mutation that inactivates this domain, may not only prevent attachment of SLP-76 via SH2 to target proteins, but may also activate the adapter for signaling elements which are inhibited by the structure of the intact protein.
T cells from SLP-76 Δ224–244 and SLP-76 ΔSH2 mice mounted partial responses to specific antigen. These responses included partial CHS response to oxazolone, and normal IgG1 antibody response to the TD antigen OVA, with normal IgE response to OVA in SLP-76 ΔSH2 mice (Fig. 7). CHS involves the cytokines IFN-γ, IL-2, IL-4, and IL-10 (30). IgG1 and IgE responses to TD antigens requires IL-4 and CD40L:CD40 interactions between T and B cells (31). Our findings suggest that the Gads binding site and the SH2 domain of SLP-76 may be dispensable, at least partially, for the production of T cell cytokines involved in these T cell effector responses.
The phenotype of SLP-76 Δ224–244 mice is similar to that of Gads−/− mice, supporting the importance of the SLP-76-Gads-LAT complex in preTCR and TCR signaling. The defect in SLP-76 Δ224–244 mice is less severe than that observed in LAT−/− mice (32). This finding suggests that TCR signaling may proceed, albeit suboptimally, in the absence of SLP-76 recruitment to rafts. This may occur via other LAT-associated proteins, as suggested by the observation that a LAT mutant that fails to recruit SLP-76, partially restores T cell signaling in LAT deficient Jurkat cells (33).
T cells from SLP-76 ΔSH2 mice failed to proliferate following TCR ligation. This finding was unexpected given the ability of SLP-76 ΔSH2 to restore TCR signaling in SLP-76 deficient Jurkat T cells. This result suggests that proliferation of primary T cells has more complex requirements than activation of biochemical pathways and reporter gene expression in T cell lines. The SH2 domain of SLP-76 couples the TCR to FYB/SLAP130. FYB/SLAP130 has been implicated in actin cytoskeletal reorganization (34), which is critical for IL-2 production and T cell proliferation (35). Molecules other than FYB/SLAP130 that interact with the SH2 domain of SLP-76 may also be important for TCR signaling.
The differential role of SLP-76 domains in T cell development and function suggests that thymocyte maturation, and activation of mature T cells is regulated differently. Furthermore, the results obtained suggest different approaches to selectively interfere with T cell activation in immune-mediated diseases.
Supplementary Material
Acknowledgments
We thank Sheryl Freeman for technical assistance. This work was supported by U.S. Public Health Service Grant AI-35714. V.P. is a Charles A. King Trust fellow. M.A.F. is supported by a Spanish Ministry of Science and Technology postdoctoral grant (EX99 12368513).
Abbreviations
- LAT
linker of activated T cells
- TCR
T cell antigen receptor
- SP
single-positive
- ERK
extracellular signal-regulated protein kinase
- WT
wild type
- OVA
ovalbumin
References
- 1.Jackman J K, Motto D G, Sun Q, Tanemoto M, Turck C W, Peltz G A, Koretzky G A, Findell P R. J Biol Chem. 1995;270:7029–7032. doi: 10.1074/jbc.270.13.7029. [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Motto D G, Koretzky G A, Weiss A. Immunity. 1996;4:593–602. doi: 10.1016/s1074-7613(00)80485-9. [DOI] [PubMed] [Google Scholar]
- 3.Wunderlich L, Farago A, Downward J, Buday L. Eur J Immunol. 1999;29:1068–1075. doi: 10.1002/(SICI)1521-4141(199904)29:04<1068::AID-IMMU1068>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Bunnell S C, Diehn M, Yaffe M B, Findell P R, Cantley L C, Berg L J. J Biol Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 5.Liu S K, Fang N, Koretzky G A, McGlade C J. Curr Biol. 1999;9:67–75. doi: 10.1016/s0960-9822(99)80017-7. [DOI] [PubMed] [Google Scholar]
- 6.Ishiai M, Kurosaki M, Inabe K, Chan A C, Sugamura K, Kurosaki T. J Exp Med. 2000;192:847–856. doi: 10.1084/jem.192.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motto D G, Ross S E, Wu J, Hendricks-Taylor L R, Koretzky G A. J Exp Med. 1996;183:1937–1943. doi: 10.1084/jem.183.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pivniouk V, Tsitsikov E, Swinton P, Rathbun G, Alt F W, Geha R S. Cell. 1998;94:229–238. doi: 10.1016/s0092-8674(00)81422-1. [DOI] [PubMed] [Google Scholar]
- 9.Clements J L, Yang B, Ross-Barta S E, Eliason S L, Hrstka R F, Williamson R A, Koretzky G A. Science. 1998;281:416–419. doi: 10.1126/science.281.5375.416. [DOI] [PubMed] [Google Scholar]
- 10.Yablonski D, Kuhne M R, Kadlecek T, Weiss A. Science. 1998;281:413–416. doi: 10.1126/science.281.5375.413. [DOI] [PubMed] [Google Scholar]
- 11.Fang N, Motto D G, Ross S E, Koretzky G A. J Immunol. 1996;157:3769–3773. [PubMed] [Google Scholar]
- 12.Chaffin K E, Beals C R, Wilkie T M, Forbush K A, Simon M I, Perlmutter R M. EMBO J. 1990;9:3821–3829. doi: 10.1002/j.1460-2075.1990.tb07600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yablonski D, Kadlecek T, Weiss A. Mol Cell Biol. 2001;21:4208–4218. doi: 10.1128/MCB.21.13.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spergel J, Mizoguchi E, Brewer J, Martin T, Bhan A, Geha R. J Clin Invest. 1998;101:1614–1622. doi: 10.1172/JCI1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godfrey D I, Kennedy J, Suda T, Zlotnik A. J Immunol. 1993;150:4244–4253. [PubMed] [Google Scholar]
- 16.Swat W, Dessing M, von Boehmer H, Kisielow P. Eur J Immunol. 1993;23:739–746. doi: 10.1002/eji.1830230326. [DOI] [PubMed] [Google Scholar]
- 17.Bendelac A, Matzinger P, Seder R A, Paul W E, Schwartz R H. J Exp Med. 1992;175:731–742. doi: 10.1084/jem.175.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudd C E. Immunity. 1996;4:527–534. doi: 10.1016/s1074-7613(00)80479-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang R, Alt F W, Davidson L, Orkin S H, Swat W. Nature (London) 1995;374:470–473. doi: 10.1038/374470a0. [DOI] [PubMed] [Google Scholar]
- 20.Tarakhovsky A, Turner M, Schaal S, Mee P J, Duddy L P, Rajewsky K, Tybulewicz V L J. Nature (London) 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 21.Tedford K, Nitschke L, Girkontaite I, Charlesworth A, Chan G, Sakk V, Barbacid M, Fischer K D. Nat Immunol. 2001;2:548–555. doi: 10.1038/88756. [DOI] [PubMed] [Google Scholar]
- 22.Doody G M, Bell S E, Vigorito E, Clayton E, McAdam S, Tooze R, Fernandez C, Lee I J, Turner M. Nat Immunol. 2001;2:542–547. doi: 10.1038/88748. [DOI] [PubMed] [Google Scholar]
- 23.Liao X C, Littman D R, Weiss A. J Exp Med. 1997;186:2069–2073. doi: 10.1084/jem.186.12.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopal K, Sommers C L, Decker D C, Mitchell E O, Korthauer U, Sperling A I, Kozak C A, Love P E, Bluestone J A. J Exp Med. 1999;190:1657–1668. doi: 10.1084/jem.190.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bommhardt U, Basson M A, Krummrei U, Zamoyska R. J Immunol. 1999;163:715–722. [PubMed] [Google Scholar]
- 26.Fukuyama H, Adachi M, Suematsu S, Miwa K, Suda T, Yoshida N, Nagata S. J Immunol. 1998;160:3805–3811. [PubMed] [Google Scholar]
- 27.Wayne J, Suh H, Misulovin Z, Sokol K A, Inaba K, Nussenzweig M C. Immunity. 1994;1:95–107. doi: 10.1016/1074-7613(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 28.Ebinu J O, Stang S L, Teixeira C, Bottorff D A, Hooton J, Blumberg P M, Barry M, Bleakley R C, Ostergaard H L, Stone J C. Blood. 2000;95:3199–3203. [PubMed] [Google Scholar]
- 29.Dower N A, Stang S L, Bottorff D A, Ebinu J O, Dickie P, Ostergaard H L, Stone J C. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 30.Kondo S, Sauder D N. J Am Acad Dermatol. 1995;33:786–800. doi: 10.1016/0190-9622(95)91817-5. [DOI] [PubMed] [Google Scholar]
- 31.Castigli E, Alt F, Davidson L, Bottaro A, Mizoguchi E, Bhan A K, Geha R S. Proc Natl Acad Sci USA. 1994;91:12135–12139. doi: 10.1073/pnas.91.25.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Sommers C L, Burshtyn D N, Stebbins C C, DeJarnette J B, Trible R P, Grinberg A, Tsay H C, Jacobs H M, Kessler C M, et al. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Trible R P, Zhu M, Liu S K, McGlade C J, Samelson L E. J Biol Chem. 2000;275:23355–23361. doi: 10.1074/jbc.M000404200. [DOI] [PubMed] [Google Scholar]
- 34.Krause M, Sechi A S, Konradt M, Monner D, Gertler F B, Wehland J. J Cell Biol. 2000;149:181–194. doi: 10.1083/jcb.149.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valitutti S, Dessing M, Aktories K, Gallati H, Lanzavecchia A. J Exp Med. 1995;181:577–584. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.