Abstract
Conformationally compromised oncogenic mutants of the tumor suppressor protein p53 can, in principle, be rescued by small molecules that bind the native, but not the denatured state. We describe a strategy for the rational search for such molecules. A nine-residue peptide, CDB3, which was derived from a p53 binding protein, binds to p53 core domain and stabilizes it in vitro. NMR studies showed that CDB3 bound to p53 at the edge of the DNA binding site, partly overlapping it. The fluorescein-labeled peptide, FL-CDB3, binds wild-type p53 core domain with a dissociation constant of 0.5 μM, and raises the apparent melting temperatures of wild-type and a representative oncogenic mutant, R249S core domain. gadd45 DNA competes with CDB3 and displaces it from its binding site. But this competition does not preclude CDB3 from being a lead compound. CDB3 may act as a “chaperone” that maintains existing or newly synthesized destabilized p53 mutants in a native conformation and then allows transfer to specific DNA, which binds more tightly. Indeed, CDB3 restored specific DNA binding activity to a highly destabilized mutant I195T to close to that of wild-type level.
More than 50% of human cancers have missense mutations in the gene coding for the tumor suppressor p53 that result in its inactivation (1). Nearly all such mutations are in the DNA-binding core domain (p53C) (1). The six most frequent cancer-associated mutations are the “hot-spots” R175H, G245S, R248Q, R249S, R273H, and R282W. These mutations can be divided into two categories: (i) DNA-contact mutations (R248 and R273) that result in loss of DNA-binding residues, and (ii) “structural mutations” that result in structural changes in p53C (2). An assessment of the mutation database (3, 4), based on thermodynamic stability and DNA binding properties of the mutants, classifies three broad phenotypes: (i) DNA-contact mutations that have little effect on folding or stability (e.g., R273H); (ii) mutations that cause a local distortion, mainly in proximity to the DNA-binding site (e.g., R249S), which are usually destabilized by <2 kcal/mol; and (iii) mutations that cause global unfolding (e.g., mutations in the β sandwich) that are destabilized by >3 kcal/mol.
Owing to the large frequency of p53 mutants in cancers, a promising strategy in cancer therapy is rescue of p53 mutants and restoration of the tumor suppression activity (reviewed in refs. 4 and 5). Different classes of mutants require different rescue strategies. DNA contact mutants need the introduction of functional groups that will establish new contacts with the DNA, compensating for the missing contacts. Globally unfolded mutants could be rescued by stabilizing agents that will lead to refolding of the mutant. In principle, any molecule that will bind the native, but not the denatured state, will cause the equilibrium to be shifted toward the native state. A small molecule, CP-31398, has been found by random screening to bind and stabilize the core domain and has been reported to be an effective anticancer agent (6).
Here, we test the feasibility of stabilizing p53C by small peptides that bind its native state and which may act as leads for drugs. Such peptides may be found rationally by examining known complexes of p53 with its binding proteins, and so avoid random screening. This strategy has been neglected because of the likelihood of the inhibition of the natural activity of p53 by the small molecule. Nevertheless, we have found a small peptide that did stabilize p53 by binding at the edge of its DNA-binding site, and could restore sequence-specific DNA-binding activity to the highly destabilized mutant I195T to near wild-type levels.
Materials and Methods
Peptide Synthesis.
The peptides were synthesized using a 432A Synergy peptide synthesizer (Applied Biosystems), using standard Fmoc chemistry, and were purified using reverse-phase HPLC [Waters 600 equipped with a 996 PDA detector and a preparative reverse-phase C8 column (Vydac, Hesperia, CA)]. The gradient was 100%A to 100%B in 35 min (A = 0.1% TFA in water, B = 95% acetonitrile/5% water/0.1%TFA). The purified peptides were characterized by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) MS and had the expected Mr. The fluorescein-labeled core domain binding peptide FL-CDB3 was purchased from Graham Bloomberg (University of Bristol, U.K.).
Protein Expression and Purification.
Human p53C wild type and mutants (residues 94–312) and human tetrameric p53 (residues 94–360) were cloned, expressed, and purified as described (7). 15N-labeled human p53C was produced as described (8).
NMR Spectroscopy.
Samples for NMR experiments contained 15N-labeled p53C at a concentration of 225 μM and the corresponding CDB peptide at a final concentration of 2–2.5 mM in 150 mM KCl, 5 mM DTT, and 5% D2O in 25 mM sodium phosphate buffer, pH 7.2. All 1H–15N heteronuclear sequential quantum correlation (HSQC) spectra were acquired as described (8).
Binding Studies by Surface Plasmon Resonance.
A BIACORE 2000 equipped with a sensor chip SA (BIAcore, Uppsala, Sweden) was used to screen the peptides for p53 core domain binding, and to quantify the binding of p53 core domain to peptide CDB3. Biotinylated CDB peptides were immobilized and the binding of p53 was studied. Immobilization and binding measurements were performed at 10°C (unless otherwise stated) with 50 mM Hepes (pH 7.2)/5 mM DTT as running buffer. The streptavidin surface of the chip was activated as described by the manufacturer. Biotinylated peptides (1.5–4.0 mM peptide in buffer containing 0.13 M NaCl) were immobilized at 5 μl/min until the level of saturation. Flow cell 1 was used as a background for the change in bulk refractive index.
To screen for binding to immobilized peptides, we measured the association of p53 (0.36–18 μM) for 15 min at 10 μl/min. Bound protein was dissociated by a regeneration cycle of 1–3 min with 1 M NaCl.
The binding affinity of p53 for immobilized CDB3 was estimated from the half saturation concentration of binding isotherm with varying concentrations of p53 core domain (0.019–0.19 μM). The binding association was measured for 5 min at 30 μl/min and 20°C. A two-state equation was fitted to the relative responses versus the logarithm of the p53 concentrations (KALEIDAGRAPH, Abelbeck Software, Reading, PA).
The binding affinity of soluble, unlabeled CDB3 was determined by competition experiments (9). The samples contained 0.20 μM p53 with various concentrations of CDB3 (0.030–120 μM). Association data were collected after 1 h of incubation at 20°C (5 min, 30 μl/min). Control samples containing p53 only were run as references. The initial association rate (over the first 150 s) was estimated by fitting a linear equation to data (BIAEVALUATION 3.1, BIAcore AB, Uppsala, Sweden). The data were analyzed according to a 1:1 binding model (9), using KALEIDAGRAPH. Control experiment showed that association rate of binding is proportional to the concentration of p53 (0.19–1.9 μM).
Fluorescence Anisotropy Measurements.
All anisotropy measurements were performed with fluorescein-labeled CDB3 (FL-CDB3, sequence FL-REDEDEIEW-NH2) at 10°C, using a Perkin–Elmer LS-50b luminescence spectrofluorimeter equipped with a Hamilton microlab M dispenser controlled by laboratory software. The peptide (≈5 μM, 900 μl) was dissolved in 50 mM Hepes buffer (pH 7.2)/5 mM DTT. Fluorescence anisotropy was measured on excitation at 480 nm (bandwidth 8 nm) and emission at 525 nm (bandwidth 2.5 nm).
To determine the dissociation constant for FL-CDB3 complexed with various p53C constructs FL-CDB3 (900 μl, ≈5 μM) was placed in the cuvette and the appropriate p53C construct (240 μl, ≈50 μM) was placed in the dispenser. Additions of 3 μl of protein were titrated into the peptide solution about every 1 min, the solution was stirred for 30 s, and the anisotropy was measured. Dissociation constants for the FL-CDB3-p53C complex were calculated by fitting the anisotropy and fluorescence titration curves (corrected for dilution) to a simple 1:1 equilibrium model (unpublished data).
DNA-binding experiments were performed similarly with 140 mM NaCl added to the buffer. Fluorescein-labeled gadd45 30-mer DNA (15 nM) and fluorescein-labeled random DNA (7 nM) were used.
Anisotropy was measured in competition experiments to study (indirectly) how CDB3 derivatives or gadd45 30-mer DNA compete with the fluorescein-labeled CDB3 for the binding site of p53 core domain (wavelengths as above, slit widths of excitation10 nm, and emission 8 nm). A stock solution of unlabeled CDB3 was titrated into a cuvette containing 900 μl 2.0 μM p53 and 0.50 μM FL-CDB3. For DNA, stock solutions of 5 and 25 μM were used.
The concentrations of p53-FL-CDB3 complex and free FL-CDB3 before addition of competitor were calculated using the Kd of 0.53 μM. The change in concentration of free FL-CDB3 after addition of competitor peptide aliquot was derived from the change in the anisotropy. The concentration of CDB3-p53C complex was calculated from the total concentration of p53C. When unlabeled CDB3 was in large excess over p53C, the fraction of free peptide could be approximated to its total concentration and Kd was derived from the equilibrium equation.
Differential Scanning Calorimetry (DSC).
DSC experiments were performed using a Microcal VP-DSC microcalorimeter (Microcal, Amherst, MA). Temperatures from 5–95°C were scanned at a rate of 60°/h, using a Hepes buffer (pH 7.2)/1 mM DTT, which also served for baseline measurements. Samples of wild-type and mutant p53C (6–15 μM) in the presence or absence of FL-CDB3 (15–80 μM) in the above buffer were prepared and then degassed for 15 min before each experiment. A pressure of 25 psi (1.56 atm) was applied to the cell. The data were analyzed using ORIGIN software (Microcal).
Results
Design of Potential p53C Binding Peptides.
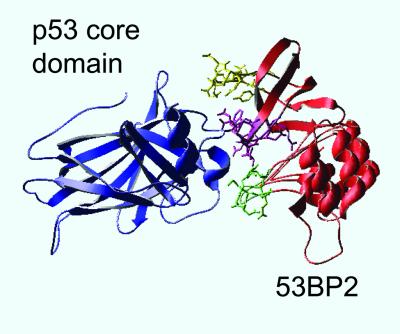
A prime source of peptides that could bind the native state of p53 is from p53-binding proteins. A rare example of a complex of a protein-p53 that has been solved at high resolution is the p53C-53BP2 complex (10). 53BP2 is a p53 binding protein (11) that enhances p53-mediated transactivation, and specifically stimulates the apoptotic function of p53 (12–14). 53BP2 binds p53C in its DNA binding site, with three loops making the contacts with p53 (ref. 10; Fig. 1). Three peptides corresponding to these three loops were synthesized and tested (CDB1–CDB3; see Table 1).
Figure 1.
Crystal structure of the p53C (blue)–53BP2 (red) complex (coordinates taken from ref. 10) with the three 53BP2-derived peptides synthesized for this study highlighted: CDB1 (residues 422–428), green; CDB2 (residues 469–477), yellow; CDB3 (residues 490–498), purple. The picture was generated using SWISSPDB VIEWER (23).
Table 1.
Peptides tested for binding p53 core domain
| Peptide | Derived from | Sequence |
|---|---|---|
| Peptides derived from 53BP2 | ||
| CDB1 | 53BP2 422–428 | MTYSDMQ-NH2 |
| CDB2 | 53BP2 469–477 | YEPQNDDEL-NH2 |
| CDB3 | 53BP2 490–498 | REDEDEIEW-NH2 |
| Peptides derived from p53 | ||
| CDB4 | p53 81–100 | TPAAPAPAPSWPLSSSVPSQ-NH2 |
| CDB5 | phospho-Ser-378 p53 369–383 | LKSKKGQSTpSRHKKL-NH2 |
| CDB6 | phospho-Ser-378 p53 361–383 | GSRAHSSHLKSKKGQSTpSRHKKL-NH2 |
| CDB7 | p53 369–383 | LKSKKGQSTSRHKKL-NH2 |
| CDB8 | p53 361–383 | GSRAHSSHLKSKKGQSTSRHKKL-NH2 |
| CDB9 | p53 81–94 | TPAAPAPAPSWPLS-NH2 |
| CDB10 | p53 76–94 | APAAPTPAAPAPAPSWPLS-NH2 |
| CDB11 | fusion p53 82–94 and 369–383 with a GG linker | PAAPAPAPSWPLSGGLKSKKGQSTSRHKKL-NH2 |
A second potential source for core domain binding peptides are sequences within p53 itself that bind the core domain and regulate its activity. Two such regions within p53 are the C-terminal domain (amino acids 363–393; ref. 15) and the proline-rich domain (amino acids 54–94; ref. 16). Several overlapping peptides corresponding to both regions were synthesized (CDB4 and CDB7–CDB10 in Table 1). Because Ser-378 within the C-terminal domain is known to undergo phosphorylation (17), phosphopeptides derived from this region were also synthesized (CDB5 and CDB6 in Table 1). It was suggested that the C-terminal and the proline-rich domains can bind the core domain only in presence of each other (18), and thus a fusion peptide between these domains was also designed (CDB11 in Table 1).
Screening of the CDB Peptides for Binding p53C.
We used heteronuclear NMR spectroscopy for initial screening for binding of the peptides to p53C to monitor any changes in the backbone 1H and 15N resonances of 15N-labeled p53C (8). Chemical shift changes were observed only with CDB2 and CDB3, implicating binding of only these peptides to p53C. We used surface plasmon resonance for initial estimates of the affinity for p53C. Peptides CDB1, -2, -3, -9, and -11 were resynthesized with a biotin label attached to their N terminus, and immobilized onto a streptavidin (SA) sensor chip. p53C had the tightest binding to CDB3, in good agreement with the NMR data. The concentration of p53C for 50% binding was estimated to be 200 nM. There was no significant binding to CDB1 or CDB9. CDB3 was chosen as a lead peptide for further experiments.
Characterization of CDB3-p53C Binding Site.
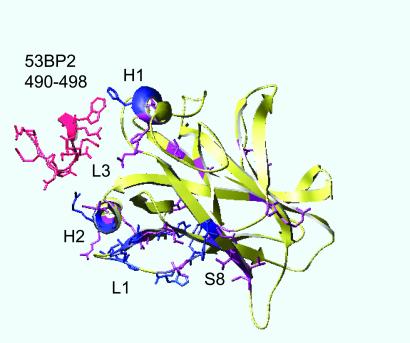
Chemical shift differences between the spectra of the bound and unbound p53C were used to identify the site in p53C where CDB3 bound. Changes of backbone 1H–15N resonances for each residue between the bound and the unbound states were found mainly in loop 1, helix 2, and strand 8 (Fig. 2), which are located at one edge of the DNA-binding site. The residues that had a significant chemical shift (above five times the standard deviation: Δ∂ > 0.25 ppm for 15N and Δ∂ > 0.05 ppm for 1H, color-coded blue) were: F113, H115, G117, T118, V122, and T123 (from loop 1), H233 (from strand 8), G279 and R280 (both from helix 2), Y126, H178, H296, and E298 (from the rest of the protein). Residues that revealed moderate chemical shifts (Δ∂ differences between 2.5 times and 5 times the standard deviation: 0.125 < Δ∂ < 0.25 ppm for 15N, and 0.025 < Δ∂ < 0.05 ppm for 1H, color-coded purple) were: L114 and C124 (from loop 1), C229, T230, and T231 (all from strand 8), R282 and R283 (from helix 2), Y103, N131, T140, V143, C176, H179, I195, G199, R202, N239, I251, R267, C277, K291, and L299 (from the rest of the protein). CDB3, as a free peptide (color-coded red in Fig. 2), bound p53C in a different location from that of the original loop within 53BP2, which binds in the middle of the DNA-binding site (between loop 3 and the other side of helix 2; ref. 10).
Figure 2.
Chemical shift changes (Δ∂) in p53C on binding to CDB3. Deviations above five times the standard deviation (Δ∂ > 0.25 ppm for 15N and Δ∂ > 0.05 ppm for 1H) were considered significant (color-coded blue). Δ∂ differences between 2.5 times and 5 times the standard deviation (0.125 < Δ∂ < 0.25 ppm for 15N, 0.025 < Δ∂ < 0.05 ppm for 1H) were considered as minor (color-coded purple), and Δ∂ differences below 2.5 times the standard deviation (Δ∂ < 0.125 ppm for 15N and Δ∂ < 0.025 ppm for 1H) were considered insignificant (color-coded yellow). See text for residue number details. CDB3 in its original position in the 53BP2-p53 complex is shown in red (coordinates taken from ref. 10). The picture was generated using SWISSPDB VIEWER.
Fluorescence anisotropy titrations were used to determine accurately the dissociation constant for the p53C–CDB3 complex at 10°C. p53C (residues 94–312) was titrated into fluorescein-labeled CDB3 (FL-CDB3) and changes in anisotropy of the labeled peptide (Fig. 3) as well as the total fluorescence at 525 nm were monitored. The initial anisotropy value for the labeled peptide was 0.04, and the limiting value for the FL-CDB3-p53C complex was 0.20. The binding curve was fitted to a 1:1 simple equilibrium model, and Kd was found to be 0.53 ± 0.09 μM (Table 2). To confirm that FL-CDB3 binds tetrameric p53, and not only isolated core domain, Kd for the binding to the tetrameric p53 construct (residues 94–360) was determined in the same way and was found to be 0.77 ± 0.09 μM (data not shown).
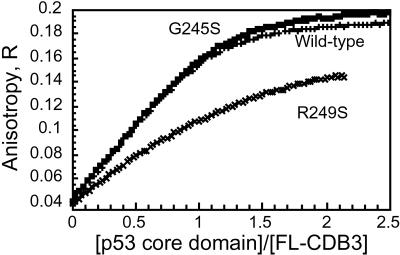
Figure 3.
CDB3 binding to p53C analyzed by fluorescence anisotropy. Wild-type and mutant p53C were titrated into a fluorescein-labeled CDB3 (4.6 μM; see Materials and Methods for details).
Table 2.
Kd values for FL-CDB3 binding to wild-type and mutant p53
| Protein | Conditions | Kd, μM |
|---|---|---|
| Wild-type core (94–312) | 0.53 ± 0.09 | |
| Wild-type core + tet (94–363) | 0.77 ± 0.09 | |
| Wild-type core | 4 M urea | 61 ± 10 |
| Wild-type core | 2 M Gdm Cl | >1,000 |
| G245S (94–312) | 0.57 ± 0.09 | |
| G245S (94–312) | 4 M urea | 39 ± 4 |
| G245S (94–312) | 2 M Gdm Cl | >1,000 |
| R249S (94–312) | 3.3 ± 0.5 |
Kd values were determined from the anisotropy and fluorescence at 525 nm following titration of p53 into fluorescein-labelled CDB3. The temperature was 10°C and the buffer was 50 mM Hepes (pH 7.2)/5 mM DTT.
The binding of FL-CDB3 to two p53C mutants was measured: to G245S, which is 95% folded at 37°C and is destabilized by 1.21 kcal/mol at 10°C; and to R249S, which is 85% folded at 37°C and is distorted and destabilized by 1.92 kcal/mol at 10°C (3, 7). At 10°C both mutants are expected to be in a native-like conformation. Kd values, from fluorescence anisotropy (see Fig. 3), were 0.57 ± 0.09 μM for G245S and 3.3 ± 0.5 μM for R249S.
Tighter Binding of Fluorescein-Labeled CDB3.
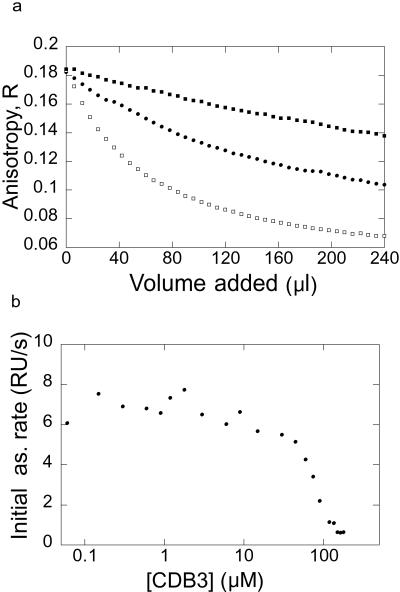
To determine whether attaching the different labels (fluorescein and biotin) to CDB3 N terminus alters Kd, we measured the dissociation constants for the unlabeled peptide by competition BIAcore (Fig. 4b) and competition with FL-CDB3 analyzed by anisotropy (Fig. 4a). The unlabeled peptide had a Kd of 37 μM (Fig. 4a). Biotinylation of the N terminus improved the affinity (compared with the unlabeled peptide) 3-fold for solution measurements (Kd = 12 μM; see Fig. 4a) and even more for the immobilized sample − BIAcore assays (apparent Kd = 0.2 μM; data not shown). Perhaps the unlabeled peptide was bound more weakly because of the positive charge on its N-terminal α-amino group, which might cause an electrostatic repulsion form the positively charged protein surface. Attaching a label on the N terminus of the peptide eliminates this charge and might add hydrophobic interactions that improve binding.
Figure 4.
Binding competition experiments. (a) Competition experiment where unlabeled or biotinylated CDB3 were titrated into 0.50 μM fluorescein-labeled CDB3 and 2.0 μM p53C wild-type (■ and □, 0.26 mM and 2.6 mM unlabeled CDB3, respectively, and ●, 0.24 mM biotinylated CDB3). (b) Titration of CDB3 binding to p53C by Competition BIAcore. The concentration of free p53C (reflected by association rate in binding to immobilized CDB3) was analyzed by BIAcore after incubation of 0.2 μM p53C and various concentrations of free CDB3.
FL-CDB3 Stabilized p53C and Raised Its Apparent Tm.
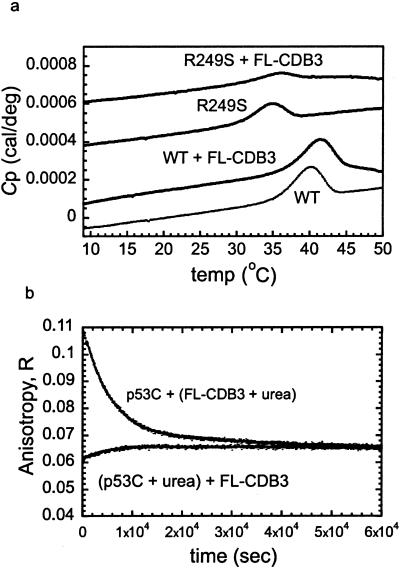
Differential scanning calorimetry (DSC) was used to detect stabilization of p53C by FL-CDB3. The thermal denaturation of p53C is irreversible, and thus only an apparent melting temperature (Tm) can be determined (7), but increase in stability can be correlated with increase in the apparent Tm. The apparent Tm of wild-type p53C was 40.1°C, and increased by 1.5° in the presence of the peptide FL-CDB3 (Fig. 5a). The apparent Tm of the mutant R249S was raised from 34.9°C to 35.9°C by the peptide.
Figure 5.
Stabilization of p53C by FL-CDB3. (a) Differential scanning calorimetry (irreversible denaturation). The apparent Tm of wild-type and R249S core domain in the presence or absence of FL-CDB3 was determined as described in Materials and Methods. For the wild-type core domain, Tm = 40.1°C in the absence of the peptide and 41.6°C in its presence. For R249S, Tm = 34.9°C in the absence of the peptide and 35.9°C in its presence. Raw data are shown and are offset for clarity. (b) Time-dependent anisotropy studies of the binding of CDB3 to wild-type p53C in presence of 3 M urea (reversible denaturation). Wild-type p53C (5 μM) was preincubated overnight with 3 M urea, then mixed with FL-CDB3 (5 μM), and the anisotropy change over time was monitored. As a control, the same protein (5 μM) was mixed with 3 M urea and with FL-CDB3 (5 μM) without preincubation and anisotropy changes over time were monitored.
FL-CDB3 Induced Refolding of Equilibrium-Denatured p53C.
The ability of FL-CDB3 to refold p53C under equilibrium denaturation conditions was monitored using fluorescence anisotropy. p53C was incubated overnight at 10°C in 3 M urea, under which conditions it is predominantly denatured. Then it was mixed with FL-CDB3 in 3 M urea, and the changes in anisotropy over time were monitored (Fig. 5b). The initial anisotropy value for the labeled peptide was 0.04. Upon mixing the peptide with p53C a rapid binding event took place, leading to the formation of a FL-CDB3-p53C complex. The anisotropy values for the complex following preincubation overnight with 3 M urea were 0.06–0.07, far below the limiting anisotropy value for the bound complex at these concentrations, which was 0.17 (estimated from Fig. 3), because under these conditions most of the protein was denatured and did not bind the peptide. There was an increase in the anisotropy over time, as the peptide induced protein refolding by mass action (Fig. 5b). On mixing p53C and FL-CDB3 (5 μM each) with 3 M urea without preincubation overnight, unfolding took place reaching the same endpoint. Overall, FL-CDB3 induced refolding of p53C and in its presence the equilibrium shifted toward the native state. Binding of FL-CDB3 to p53C became progressively weaker with increasing [urea], consistent with both a lack of binding to the denatured state and weaker binding to the native state (data not shown). Thus, the stabilizing effects in 3 M urea were not as pronounced as they would be in water alone.
DNA Competed with FL-CDB3 for p53C Binding.
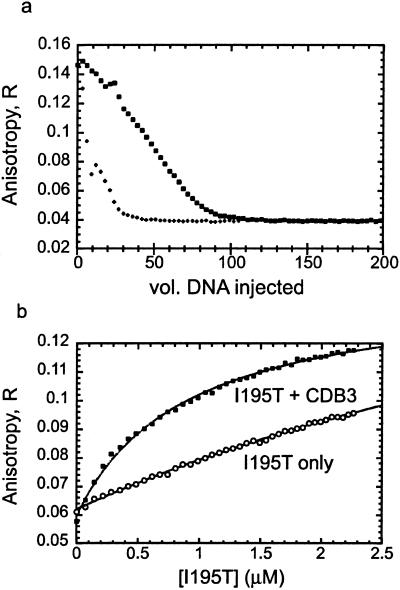
From the NMR data (Fig. 2) it seems that CDB3 bound p53C at the edge of the DNA binding site, suggesting at least a partial overlap between the two binding sites. We measured the competition between the binding of FL-CDB3 and gadd45 DNA to p53C by using competition fluorescence anisotropy. DNA (5 and 25 μM) displaced the peptide completely from the binding site, indicating overlap between the DNA and peptide binding sites (Fig. 6a).
Figure 6.
Effect of CDB3 on DNA binding of p53C. (a) DNA competes with FL-CDB3 on p53C binding. 30-mer gadd45 DNA (+, 25 μM; □, 5 μM) was titrated into a mixture of p53C-FL-CDB3 as described in Materials and Methods. (b) CDB3 restores DNA binding to the I195T mutant. I195T (10 μM) was preincubated for 1 h in the presence (■) and the absence (○) of 100 μM CDB3 and titrated into 15 nM fluorescein-labeled 30-mer gadd45 DNA. Dissociation constants were calculated from a fit to a 1:1 binding model.
CDB3 Restored Sequence-Specific DNA Binding to the Highly Destabilized p53 Mutant I195T.
We tested whether CDB3 can restore sequence-specific DNA binding activity to p53C mutants by observing its effect on the β-sandwich mutant I195T, which is highly destabilized by 4.1 kcal/mol (3) and has poor DNA-binding affinity. I195T (10 μM) was incubated for 1 h at 10°C in presence of CDB3 (100 μM) (or its absence) and titrated into fluorescein-labeled gadd45 DNA in presence of the same peptide concentration. In the absence of peptide, I195T bound gadd45 DNA with Kd = 6 μM (Fig. 6b). After incubation with CDB3, the binding improved 6-fold, and Kd was 1 μM, which is close to the value of 0.8 μM for the wild type. As expected, CDB3 did not affect DNA binding of the completely native wild-type p53C (data not shown).
To confirm that the restoration of DNA binding is sequence-specific, we repeated the experiments with the random double-stranded DNA sequence fluorescein-AATATGGTTTGAATAAAGAGTAAAGATTTG. Binding of I195T to this sequence was very weak, and was not improved, but rather inhibited, by the peptide (data not shown).
Discussion
We report a 9-residue peptide, CDB3, and its biotin and fluorescein-labeled derivatives that bind and stabilize p53C. CDB3 is derived from residues 490–498 of 53BP2, which constitute one of its binding loops for p53. The most striking properties of CDB3 and its derivatives are their abilities to: stabilize wild-type and mutant p53C, as shown by raising their apparent melting temperatures; induce refolding of reversibly denatured p53C; and restore sequence-specific DNA binding to a highly destabilized p53C mutant. Thus, a small peptide can stabilize p53C and restore its sequence-specific DNA binding simply by binding its native state but not the denatured state and shifting the equilibrium toward the native form.
Defining a General Target Site Within p53C for Stabilizing Peptides.
The search strategy used in this study was successful for discovering a peptide that binds p53C in a defined site. Being derived from a protein that binds p53C in its DNA-binding site, CDB3 itself binds also in the DNA-binding site, although in a slightly different location. The CDB3-binding site within p53C, as mapped by NMR chemical shift analysis, is situated at the edge of the DNA-binding site and consists of three structural elements (loop 1, helix 2, and the edge of strand 8), which are remote sequentially but close spatially. This site might serve as a specific target for core-domain-stabilizing molecules. Its advantage as such is its location in proximity to the DNA-binding site, enabling a local stabilizing effect in that site. Indeed, the NMR data shows that CDB3 binding generates a strong localized effect on the edge of the DNA-binding site within p53C, whereas DNA binding results in shifts that are spread throughout the whole protein structure (data not shown).
CDB3, as a free peptide, did not bind p53C exactly in the same location as the parent loop in the 53BP2 protein (10). The original 53BP2 loop binds the core domain between helix 2 and loop 3 (10), whereas CDB3 binds at the other side of helix 2, close to loop1. A possible explanation is that the CDB3-binding site might also be an alternative binding site for 53BP2, and the two binding sites might have a regulatory role. Alternatively, owing to its high negative charge, CDB3 as a free peptide might act partly as a “DNA-mimic” that binds the positively charged surface of the DNA-binding site.
Implications for Rescue of p53C Mutants.
CDB3 was found to bind two p53C hot-spot mutants: G245S, which is weakly destabilized (3), and R249S, which is distorted in the DNA-binding region (3, 8). The affinity of FL-CDB3 to G245S was the same as for the wild type. Binding to R249S was weaker, but still in the low micromolar range.
CDB3-like compounds could be used in principle for the rescue of mutants that are weakly destabilized (e.g., G245S) and mutants that are at body temperature globally unfolded (e.g., I195T) and are unable to bind DNA (see above). Indeed, we demonstrated that CDB3 can restore sequence-specific DNA binding to the highly destabilized I195T mutant. Peptides such as CDB3 cannot be used to rescue DNA contact mutants. Other strategies, which involve introduction of residues or small molecules that contribute the missing interactions, should be used for rescue of these mutants.
The mode of action of CDB3 is different from that of the previously reported C-terminal peptides (19–22). CDB3 stabilizes p53 by binding its native but not its denatured state, whereas the C-terminal peptides specifically regulate the activity and the DNA binding of p53C. CDB3 and its labeled derivative FL-CDB3 are lead compounds, and they can be used as a basis for the future design of peptides and small molecules that have a larger stabilizing effect on p53C. We have applied the key biophysical measurements that successfully detected the binding and stabilization of p53C by CDB3 to the drug CP-31398 (6), but all of the results were negative (T.M.R, V. J. N. Bykov, S.M.V.F., G. Selivanova, K. G. Wiman, and A.R.F., unpublished data).
Chaperone Strategy.
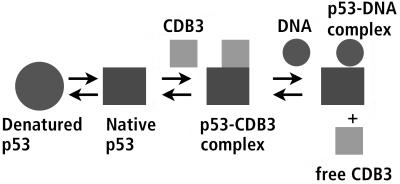
The inherent drawback of using a natural binding site for a drug is that it competes with the natural ligand. Thus, it might be thought that the competition between DNA and CDB3 peptide would preclude it from being of use as a lead. But this need not be so. Because the binding of DNA itself stabilizes p53C, and it binds very tightly, stabilization by a peptide such as CDB3 is needed only for mutants where DNA binding is impaired because mutant p53 is in denatured conformation. Once the protein has bound DNA, the peptide is not needed any more. The ability of CDB3 to induce refolding of p53C, together with the observation that DNA can displace it from p53, led us to propose a “chaperone” mechanism for rescuing a denatured mutant (Fig. 7). CDB3 binds an unfolded or distorted mutant that is unable to bind DNA, either immediately on biosynthesis or later for reversibly denatured mutants, and shifts the equilibrium toward the native state. Then DNA can bind the protein, displacing the peptide, which is free again to bind another protein molecule. We have demonstrated this mechanism for the highly destabilized mutant I195T. Binding of this mutant to the gadd45 DNA was improved 6-fold in presence of CDB3, to the level of wild-type p53C (Fig. 6b). Random DNA, on the other hand, remained unaffected. The competition between DNA and peptide should favor DNA even better in vivo because the peptide binds independently to each site in a tetramer of p53, but DNA binds far more tightly to the native tetramer because of cooperativity, thus allowing DNA to displace the drug more easily. CDB3 may well be a lead compound for designing drugs to stabilize p53.
Figure 7.
“Chaperone” strategy for rescue of p53. A schematic model of the proposed mechanism of action for CDB3. See text for details.
Acknowledgments
We thank Dr Mark Bycroft for useful discussions. A.F. is supported by Long-Term Fellowship LT00056/2000-M from the Human Frontier Science Program. L.O.H. is supported by Wenner-Gren Foundations, Stockholm. T.M.R. is the recipient of Herchel Smith and Medical Research Council (UK) scholarships. This work was also partly supported by the Cancer Research Campaign (London). D.B.V. was supported by Postdoctoral Fellowship LT0318/1998-M from the Human Frontier Science Program.
Abbreviations
- CDB
core domain binding
- FL
fluorescein-labeled
- p53C
p53 core domain
References
- 1.Hainaut P, Hollstein M. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 2.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 3.Bullock A N, Henckel J, Fersht A R. Oncogene. 2000;19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 4.Bullock A N, Fersht A R. Nat Cancer Rev. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 5.Hupp T R, Lane D P, Ball K L. Biochem J. 2000;352:1–17. [PMC free article] [PubMed] [Google Scholar]
- 6.Foster B A, Coffey H A, Morin M J, Rastinejad F. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 7.Bullock A N, Henckel J, DeDecker B S, Johnson C M, Nikolova P V, Proctor M R, Lane D P, Fersht A R. Proc Natl Acad Sci USA. 1997;94:14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong K B, DeDecker B S, Freund S M, Proctor M R, Bycroft M, Fersht A R. Proc Natl Acad Sci USA. 1999;96:8438–8442. doi: 10.1073/pnas.96.15.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nieba L, Krebber A, Pluckthun A. Anal Biochem. 1996;234:155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- 10.Gorina S, Pavletich N P. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 11.Iwabuchi K, Bartel P L, Li B, Marraccino R, Fields S. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuels-Lev Y, O'Connor D J, Bergamaschi D, Trigiante G, Hsieh J K, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 13.Iwabuchi K, Li B, Massa H F, Trask B J, Date T, Fields S. J Biol Chem. 1998;273:26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 14.Lopez C D, Ao Y, Rohde L H, Perez T D, O'Connor D J, Lu X, Ford J M, Naumovski L. Mol Cell Biol. 2000;20:8018–8025. doi: 10.1128/mcb.20.21.8018-8025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayle J H, Elenbaas B, Levine A J. Proc Natl Acad Sci USA. 1995;92:5729–5733. doi: 10.1073/pnas.92.12.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller-Tiemann B F, Halazonetis T D, Elting J J. Proc Natl Acad Sci USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takenaka I, Morin F, Seizinger B R, Kley N. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- 18.Kim A L, Raffo A J, Brandt-Rauf P W, Pincus M R, Monaco R, Abarzua P, Fine R L. J Biol Chem. 1999;274:34924–34931. doi: 10.1074/jbc.274.49.34924. [DOI] [PubMed] [Google Scholar]
- 19.Abarzua P, LoSardo J E, Gubler M L, Spathis R, Lu Y A, Felix A, Neri A. Oncogene. 1996;13:2477–2482. [PubMed] [Google Scholar]
- 20.Selivanova G, Iotsova V, Okan I, Fritsche M, Strom M, Groner B, Grafstrom R C, Wiman K G. Nat Med. 1997;3:632–638. doi: 10.1038/nm0697-632. [DOI] [PubMed] [Google Scholar]
- 21.Selivanova G, Ryabchenko L, Jansson E, Iotsova V, Wiman K G. Mol Cell Biol. 1999;19:3395–3402. doi: 10.1128/mcb.19.5.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hupp T R, Sparks A, Lane D P. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 23.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]